- Division of Nematology, ICAR-Indian Agricultural Research Institute, New Delhi, India
Root-knot nematodes (Meloidogyne incognita) cause substantial yield losses in vegetables worldwide, and are difficult to manage. Continuous withdrawal of environmentally-harmful nematicides from the global market warrants the need for novel nematode management strategies. Utility of host-delivered RNAi has been demonstrated in several plants (Arabidopsis, tobacco, and soybean) that exhibited resistance against root-knot and cyst nematodes. Herein, a M. incognita-specific protease gene, cathepsin L cysteine proteinase (Mi-cpl-1), was targeted to generate tomato transgenic lines to evaluate the genetically modified nematode resistance. In vitro knockdown of Mi-cpl-1 gene led to the reduced attraction and penetration of M. incognita in tomato, suggesting the involvement of Mi-cpl-1 in nematode parasitism. Transgenic expression of the RNAi construct of Mi-cpl-1 gene resulted in 60–80% reduction in infection and multiplication of M. incognita in tomato. Evidence for in vitro and in vivo silencing of Mi-cpl-1 was confirmed by expression analysis using quantitative PCR. Our study demonstrates that Mi-cpl-1 plays crucial role during plant-nematode interaction and plant-mediated downregulation of this gene elicits detrimental effect on M. incognita development, reinforcing the potential of RNAi technology for management of phytonematodes in crop plants.
Introduction
Root-knot nematodes (Meloidogyne spp.) are one of the potential constraints for cultivation of vegetables particularly in the tropical and subtropical countries. In India, Meloidogyne incognita race 1 is most prevalent causing considerable damage especially in solanaceous and cucurbit crops (Jain et al., 2007). The infective second stage juvenile (J2) penetrates at the root tip zone and migrates intercellularly until it reaches the differentiating vascular cylinder. During invasion, nematodes inject a cascade of effector proteins of esophageal gland origin into plant cells via its stylet (Hassan et al., 2010). These effectors are thought to be involved in host pathogen interaction starting from the host recognition process to degradation of plant cell walls in order to facilitate the migration of nematode, culminating in the establishment of hypermetabolic, multinucleate feeding cell (giant cell, GC) which serves as the permanent food source for nematode development and reproduction (Davis et al., 2008). Cortical cells surrounding the GC are proliferated through hyperplasia to form the gall. Due to the formation of knots or galls around the feeding site in vascular tissue, upward translocation of water and nutrient in the root is affected, resulting in the reduction of crop yield (Moens et al., 2009).
Existing management practices such as the use of nematicides are posing a threat on the environment and are costly. Therefore, resorting to the environmentally benign and cost-effective nematode management strategies is the preferred alternative. In the recent years, RNA interference (RNAi) has emerged as a potential tool to manage the crop pathogens through host-induced gene silencing (HIGS) approach (Koch and Kogel, 2014). Plethora of nematode genes was knocked down using HIGS approach, causing reduction in parasitic success of root-knot and cyst nematodes in different crop plants (Dutta et al., 2015). Due to its precise selectivity for the target organism with least side effects, RNAi can be utilized as a remarkable tool to develop nematode resistant transgenic plants.
Proteinases are ubiquitous proteolytic enzymes that cleave the internal peptide bonds within proteins and peptides, found in a wide range of organisms such as bacteria, plants, invertebrates and vertebrates. In case of parasitic helminths, papain superfamily of cysteine proteinases (i.e., cathepsins) has drawn the most attention (Tort et al., 1999). Based on the presence and absence of a distinctive set of amino acids within the polypeptide, phylogenetic analysis identified more than 10 subdivisions within the cathepsin superfamily including cathepsin B, C, L, and Z, among which cathepsin L and Z-like proteases are exclusively present in many parasitic nematodes, and have potential roles in invasion and feeding on host tissues, molting, and evasion of innate host defenses (Santamaria et al., 1998; Koiwa et al., 2000; Shompole and Jasmer, 2001).
Among the plant-parasitic species, cysteine proteinase activity was detected in potato cyst nematode (Globodera pallida), while cathepsin like cysteine proteinase activity was identified in the intestine of feeding females of soybean cyst nematode, Heterodera glycines (Lilley et al., 1996). Neveu et al. (2003) characterized a cathepsin L protease full length cDNA (Mi-cpl-1) from infective J2 of M. incognita, expression of which was shown to occur exclusively in the developmental stages (i.e., J2 and females), indicating the possible role of Mi-cpl-1 in nematode development. Transcripts of the Mi-cpl-1 accumulated specifically in the intestinal cells of nematodes, suggesting their involvement in the digestive function of nematodes.
Induction of RNAi upon ingestion of double-stranded RNA (dsRNA) was proved to be effective in the free-living nematode, Caenorhabditis elegans, during in vitro experiments (Fire et al., 1998). Subsequently, cathepsin L-like cysteine proteinases were used as the target gene in a number of in vitro RNAi studies. According to Hashmi et al. (2002), RNAi of Ce-cpl-1, a C. elegans cathepsin L protease, resulted in embryonic lethality and delayed the growth of larvae to egg producing adults, indicating the activity of Ce-cpl-1 is correlated with the embryogenesis and post-embryonic development process of C. elegans. RNAi of cysteine protease genes (falcipain-1 and -2) caused severe morphological abnormalities in the malaria parasite, Plasmodium falciparum (Malhotra et al., 2002). In case of human filarial parasite (Onchocerca volvulus), RNAi of two specific genes, i.e., cathepsin L (Ov-cpl) and cathepsin Z (Ov-cpz) inhibited the molting of third stage (L3) to fourth stage larvae (L4) (Lustigman et al., 2004).
For the functional validation of cysteine proteinase genes in plant nematodes, dsRNA delivery was accomplished by soaking the nematodes with dsRNA solution mixed with the neurotransmitters like resorcinol, octopamine, serotonin etc. Silencing of the cysteine proteinases of H. glycines (hgcp-I) and G. pallida (gpcp-I) led to the altered sexual fate and less recovery of egg laying females (Urwin et al., 2002). A reduction in the transcript abundance along with the reduced infectivity was observed while in vitro RNAi was used to investigate the function of Mi-cpl-1 gene in M. incognita. Downregulation of Mi-cpl-1 gene led to the reduced parasitic success of M. incognita (Shingles et al., 2007).
Despite the reports of in vitro studies, cysteine proteinase genes have not yet been extensively targeted for the HIGS approach. In an isolated report, tobacco transgenic lines expressing dsRNA for Mi-cpl-1 gene imparted partial resistance to M. incognita race 3 (Antonino de Souza Júnior et al., 2013). In the current investigation, Mi-cpl-1 gene was knocked down using in vitro as well as in planta RNAi approach to analyse the function of this important protease in plant-nematode interaction. Tomato transgenic lines were generated which exhibited resistance to M. incognita race 1. In addition to that, single copy transgenic events were generated and target gene small RNAs were detected in the transgenic roots, through Southern and northern analysis, respectively. Reduction in the transcript level of Mi-cpl-1 was detected in the females that developed in the transgenic plants.
Materials and Methods
Culturing of Nematodes
A pure culture of M. incognita (Kofoid & White) Chitwood race 1 was maintained on tomato (Solanum lycopersicum L. cv. Pusa Ruby) in a glasshouse. Egg masses were collected from the roots of 2 months old plant using sterilized forceps and were kept for hatching in a double-layered paper tissue supported on a molded sieve of wire gauze in a Petri dish containing distilled water (Hooper, 1986). Freshly hatched J2s were used for all the experiments.
Selection of Target Region of Mi-cpl-1 Gene
A pairwise alignment of full length sequence of Mi-cpl-1 showed 66, 68, 63, and 59% identity with the other homologous gene sequences including Gp-cpl-1 of G. pallida, Hg-cpl-1 of H. glycines, Rr-cpl-1 of Rotylenchulus reniformis and Ce-cpl-1 of C. elegans, respectively, at the nucleotide level using ClustalW programme (Larkin et al., 2007). The probability of silencing is reduced if the two related genes are <90% homologous (Sharp, 2001) and even a single nucleotide mismatch between a siRNA molecule and its target mRNA may prevent the RNAi process (Elbashir et al., 2001). Therefore, silencing of Mi-cpl-1 may not induce any off-target effects on non-target organisms. A 366 bp sequence (spanning between 787 and 1152 bp of Mi-cpl-1 gene) conserved across the similar gene family of other nematode species, was identified as the target sequence for RNAi construct development (Figure 1).
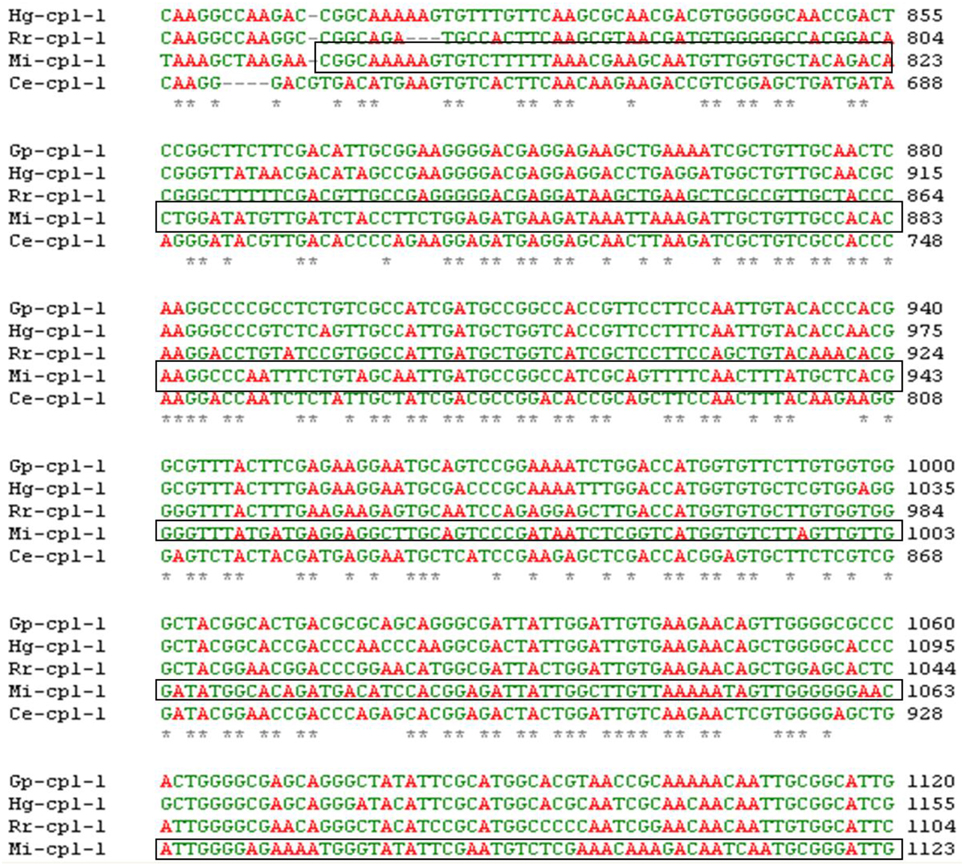
Figure 1. Multiple sequence alignment of Mi-Cpl-1 gene with the homologous gene sequences of different nematode species. *Shows the conserved region across the nematode species. Boxed portion indicates the target region of Mi-cpl-1 for RNAi studies. GenBank ID for Mi-Cpl-1 (1152 bp), Gp-Cpl-1 (1305 bp), Hg-Cpl-1 (1356 bp), Rr-Cpl-1 (1264 bp) and Ce-Cpl-1 (1192 bp) are AJ557572.1, AY999065.1, AY554271.1, AY999066.1, and NM_074798, respectively.
Cloning and Sequencing of Mi-cpl-1 Gene
Total RNA was isolated from J2s using Trizol reagent (Invitrogen) and digested with DNaseI (New England Biolabs) to remove the contaminating DNA molecules. The quantity and quality of the extracted RNA molecule were assessed using Nanodrop ND-1000 spectrophotometer (Thermo Scientific). Approximately 400 ng of purified RNA was reverse transcribed to cDNA using cDNA synthesis Kit (Superscript VILO, Invitrogen). A 366 bp region of Mi-cpl-1 (GenBank ID: AJ557572) was amplified from the cDNA using the primers as given in Table 1. The fresh PCR product was cloned into pGEM-T Easy (Promega, USA) using standard protocol and transformed into Escherichia coli DH5α competent cells (New England Biolabs). The plasmid was recovered from positive colonies (QIAGEN Plasmid Miniprep kit) and sequenced (ABI SOLiD sequencing platform) to ascertain the identity of the insert.
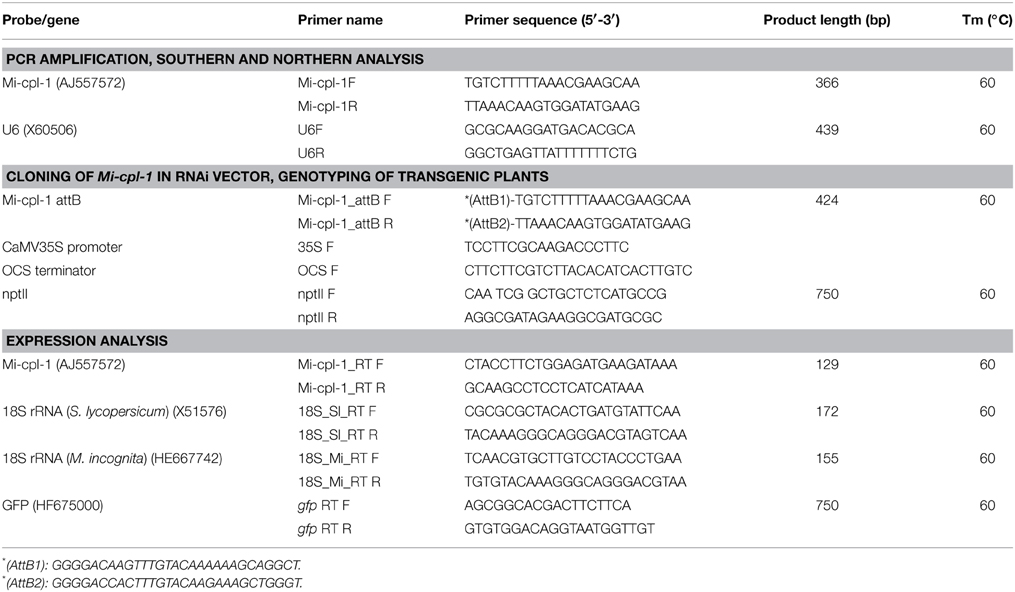
Table 1. List of primers and probes used for cloning, PCR amplification, Southern and northern hybridization.
Comparative Expression of Mi-cpl-1 in Different Life Stages of M. incognita
To quantify the expression of Mi-cpl-1 throughout the life cycle of M. incognita, quantitative real-time PCR (qRT-PCR) was performed with cDNA from different developmental stages (eggs, J2s and mature females). Total RNA was isolated and synthesized to cDNA as explained above. qRT-PCR was performed in a realplex2 thermal cycler (Eppendorf) using SYBR Green Supermix kit (Eurogentec). Reaction mixture for each sample contained a final volume of 10 μl, containing 5 μl of SYBR Green PCR Master mix (Eurogentec), 750 nM of each primer and 1.5 ng of cDNA. To normalize the gene expression level 18S rRNA, a constitutively expressed gene was used as the reference. The amplification reactions were run using the following programme: a hot start of 95°C for 5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. After 40 cycles a melt curve analysis or dissociation programme (95°C for 15 s, 60°C for 15 s, followed by a slow ramp from 60°C to 95°C) was acquired to ensure the specificity of amplification. At least two biological and three technical replicates were used for each of the samples. After obtaining the Ct values the 2−ΔΔCT method was used to quantify the relative fold change in gene expression (Livak and Schmittgen, 2001), and student's t-test was performed. Primer details are given in Table 1.
Preparation of dsRNA for In Vitro RNAi
Mi-cpl-1 gene was PCR amplified from the pGEM-T clone using M13 primers. Purified PCR product (2 μg) was used as the template to synthesize the sense and antisense strands of Mi-cpl-1 using T7 and Sp6 transcription kits (Ambion). Synthesis of dsRNA was done by mixing of two ssRNAs followed by incubation at 65°C for 10 min and 37°C for 30 min. Confirmation of dsRNA synthesis was done by running a 2 μl aliquot on 1% agarose gel. Subsequently, dsRNA of an unrelated gene, gfp (green fluorescent protein), cloned in pGEM-T vector was generated to be used as the negative control.
In Vitro RNAi of Mi-cpl-1
To trace the efficiency of dsRNA uptake by nematodes, FITC (Fluorescein isothiocyanate) was used. Freshly hatched M. incognita J2s were soaked in 1 mg ml−1 Mi-cpl-1 dsRNA in a soaking buffer containing FITC (0.1 mg ml−1) and 50 mM octopamine, incubated for 6 h in dark on a rotator at room temperature. J2s incubated in soaking buffer alone and in soaking buffer with gfp dsRNA were used as the controls to demonstrate the target-specific silencing. After incubation, J2s were rinsed five times with sterile water by centrifugation. Treated nematodes were observed using a Zeiss Axiocam MRm fluorescence microscope to monitor the uptake efficiency.
To examine the Mi-cpl-1 silencing effect on nematodes, attraction and migration assay was conducted on pluronic gel (Wang et al., 2009). Three ml of 23% pluronic gel F-127 (Sigma) evenly mixed with approximately 1000 dsRNA fed J2s was pipetted onto 35 × 10 mm Petri dish. Tomato (cv. Pusa Ruby) seedling with 1 cm long root tip was placed at the center of the dish and incubated at 27°C. Attraction of J2 toward root tip was monitored at 4 h and 8 h. After 24 h roots were stained with acid fuchsin to record the number of J2s that had invaded the root (Byrd et al., 1983). Three replicates were taken for each treatment and repeated at least twice. Data were subjected to One-Way ANOVA and CRD test followed by Duncan's multiple-comparison test with significance level at P < 0.05 using SAS software (version 9.3).
To analyse the echamber were inoculated with xpression of Mi-cpl-1 gene in the dsRNA fed nematodes, total RNA was isolated from nematodes treated with dsRNA of Mi-cpl-1, dsRNA of gfp and soaking buffer alone. qRT-PCR of the synthesized cDNA was performed as described above. At least two biological and three technical replicates were used for each of the samples. Expression was calculated using 2−ΔΔCT, and student's t-test was performed.
Preparation of RNAi Construct of Mi-cpl-1 for HIGS
The pHELLSGATE12 vector (RNAi Gateway ready) was obtained from Commonwealth Scientific and Industrial Research Organization (CSIRO), Australia (http://www.csiro.au/Organisation-Structure/Divisions/Plant-Industry/RNAi/Plant vectors.aspx) (Helliwell and Waterhouse, 2003). Partial sequence of Mi-cpl-1 (366 bp) was initially amplified from pGEM-T clone with attB1 and attB2 sites at the upstream and downstream of the gene sequence, respectively, through PCR. Primer details are given in Table 1. AttB flanked PCR product of Mi-cpl-1 was sub-cloned into the entry vector pDONR221, using BP clonase enzyme. Mi-cpl-1 fragment was subsequently cloned into the binary vector pHELLSGATE12 in sense and antisense orientation separated by an intron, using recombination-based Gateway cloning technique, mediated by the LR clonase enzyme (Invitrogen). The recombinant clone was transformed to E. coli DH5α cells. To confirm the target gene orientation, colony PCR was carried out with four different sets of primers (gene-specific forward and reverse; CaMV 35S promoter forward and attB2 reverse; CaMV 35S terminator forward and attB2 reverse; nptII forward and reverse primers). To ensure the orientation of the insert, PCR products were sequenced.
Binary plasmid (pHELLSGATE12) containing the RNAi construct was mobilized to the competent cells of Agrobacterium tumefaciens strain GV3101 using freeze and thaw method (Jyothishwaran et al., 2007). Positive clones were selected through colony PCR and maintained in Yeast-Peptone-Agar medium with selection against the antibiotics rifampicin (25 μg ml−1), gentamicin (50 μg ml−1) and spectinomycin (50 μg ml−1).
Co-Transformation and Selection of Tomato Explants
Tomato (cv. Pusa Ruby) seeds were surface sterilized with 70% ethanol for 5 min and 1% NaOCl for 2 h, followed by four times washing with sterile water. Seeds were germinated on MS agar (pH 5.8) medium. Leaf explants of 1 cm2 cut from the fortnight old tomato seedling were used for Agrobacterium-mediated transformation. Explants were plated on pre-cultivation medium (MS + 0.5 mg L−1 IAA + 0.5 mg L−1 Zeatin). After 3 days, pre-cultivated explants were infected with A. tumefaciens (GV3101) cells harboring the RNAi construct for 5 min. Agroinfected leaves were dried by soaking in sterile tissue paper and plated on co-cultivation media (MS + 0.5 mg L−1 IAA + 0.5 mg L−1 Zeatin). After 2 days, co-cultured leaf discs were transferred to the selection medium (MS + 0.5 mg L−1 IAA + 0.5 mg L−1 Zeatin + 100 mg L−1 kanamycin + 250 mg L−1 cefotaxime). Selection plates were incubated at 25°C with a 16/8 h light/dark cycle. After 25 days, shoots that emerged from the explants were excised and sub-cultured in the fresh selection medium at 15 days interval for further shoot elongation. For root initiation, elongated shoots were kept in the rooting media (1/2 MS + 0.5 mg L−1 NAA + 100 mg L−1 kanamycin + 250 mg L−1 cefotaxime). The established plants with hardened roots were transferred to the transgenic glasshouse facility of ICAR-IARI, New Delhi for further growth and T0 seeds were produced.
DNA Isolation and PCR Confirmation of Primary Transgenic Plants
Genomic DNA was isolated from the fresh leaves of all the T0 events using NucleoSpin Plant II DNA extraction kit (Macherey-Nagel) following the manufacturer's protocol. PCR confirmation of the transgene was done with different sets of primers (gene-specific forward and reverse; CaMV 35S promoter forward and attB2 reverse; CaMV 35S terminator forward and attB2 reverse; nptII forward and reverse primers) (Table 1).
Southern Blot for Transgenic Tomato Lines
To ensure the T-DNA integration, 15 μg DNA of each PCR-positive tomato line was digested with 50 U BamHI (New England Biolabs) for 16 h at 37°C. Digested DNA was resolved on 0.8% agarose gel and transferred by capillary action onto a nitrocellulose membrane (BioRad Zeta probe) in 10X saline sodium citrate (SSC) buffer [1.5 M NaCl, 0.15 M sodium citrate (pH 7.0)]. Fragment of Mi-cpl-1 gene (366 bp) labeled with [α-32P]-dCTP using mega prime DNA labeling kit (Amersham Pharmacia Biotech), was used as the probe. Probe was purified with Sephadex G-50 column (GE Healthcare Life Sciences) following manufacturer's instructions. Membrane was UV-crosslinked and hybridized overnight with radioactive probe in the hybridization buffer [2 M Na2HPO4 (pH 7.2), 10% sodium dodecyl sulfate (SDS), 0.5 M EDTA (pH 7.0)] at 65°C. Later, membrane was gradually washed three times in 3X SSC and 0.1% SDS; 0.5X SSC and 0.1% SDS followed by 0.1X SSC and 0.1% SDS, for 30 min each at 65°C (Southern, 1975), exposed to Fujifilm (Kodak) for 1–5 days at −80°C and developed. All experiments were conducted twice and showed similar results.
Expression Analysis of Mi-cpl-1 in T1 Transgenic Plants
T0 seeds of tomato plant were germinated on MS media supplemented with 100 mg L−1 kanamycin and the seedlings were transferred to the medium containing ½MS, 0.25 mg L−1 GA3 and 100 mg L−1 kanamycin. Matured plants were subsequently transferred to 300 ml pots containing autoclaved soil and soil rite in 3:1 ratio. T1 plants were kept in a growth chamber at 28°C, 70% RH and 16: 8 h light: dark photoperiod. DNA was isolated from the fresh leaves of T1 plants as stated above and PCR confirmation of the transgene was done using different sets of primers (Table 1).
To analyse the transcript abundance of Mi-cpl-1 gene in T1 plants, total RNA was isolated from fresh leaves of PCR positive events and reverse transcribed to cDNA. qRT-PCR analysis was performed as mentioned above. Average ΔCT values were obtained by calculating the difference between the Ct mean of Mi-cpl-1 and 18S rRNA gene. At least two biological and three technical replicates were taken for each of the samples. Primer details are given in Table 1.
Northern Blot for Transgenic Tomato Lines
Total RNA and small RNA were isolated from the fresh leaves of PCR-positive events using Nucleospin miRNA isolation Kit (Macherey-Nagel). 10 μg total RNA of each sample was denatured by heating at 68°C for 10 min, separated on 2% high resolution MetaPhor agarose gel, and transferred to a nitrocellulose membrane as described above. Similarly, denatured small RNA was separated on 15% denaturing polyacrylamide gel and transferred to a membrane. Probes Mi-cpl-1 and U6 (small nuclear RNA as positive control) were synthesized by PCR using primers Mi-cpl-1F, Mi-cpl-1R, U6F, and U6R, respectively (Table 1), and radiolabeled with [α-32P]-dCTP by mega prime DNA labeling kit (Amersham Pharmacia Biotech). Membranes were UV-crosslinked and hybridized with the purified probes overnight at 42°C in hybridization buffer. Membranes were washed and X-ray films were developed as described above. All experiments were conducted twice and showed similar results.
Bioefficacy Studies of T1 Plants Expressing dsRNA of Mi-cpl-1 against M. incognita
A fortnight old T1 plants kept in growth chamber were inoculated with approximately 500 freshly hatched J2s in the vicinity of the root zone, and incubated at 28°C, 70% relative humidity and 16: 8 h light: dark photoperiod. Plants were harvested 35 days post-inoculation (DPI), and roots were washed free of soil. Observations were taken on total number of galls, females, egg masses, and eggs per egg mass for each plant. In addition, to determine the effect of HIGS on M. incognita reproductive potential, nematode multiplication factor [(number of eggmasses × number of eggs per egg mass) ÷ nematode inoculum level] was calculated for each transgenic event. All the data were compared with the wild type plants inoculated with similar number of M. incognita J2s, grown under similar conditions. Infection assay was set up as randomized complete block design with six replicates per treatment. Data were subjected to One-Way ANOVA and CRD test followed by Duncan's multiple-comparison test with significance level at P < 0.05 using SAS software (version 9.3). Experiment was conducted twice and showed similar results.
Expression Analysis of Mi-cpl-1 in Nematodes Extracted from Transgenic Plants
Mature females feeding on the roots of wild type and transgenic RNAi plants were isolated at 35 days post-infection stage by macerating the infected galled root tissue under the microscope using forceps. Extracted nematodes were frozen immediately in liquid N2 and stored at −80°C. Total RNA was isolated and reverse transcribed to cDNA as described above. Transcript accumulation of Mi-cpl-1 was analyzed by qRT- PCR. Primer details are given in Table 1. Two biological and three technical replicates were used. Expression was calculated using 2−ΔΔCT, and student t-test was performed.
Results
Differential Expression of Mi-cpl-1 Transcript in Different Stages of M. incognita
Sequencing and BLAST analysis of the cloned fragment of Mi-cpl-1 (366 bp) gene amplified from the cDNA of M. incognita, revealed 100% similarity to the reported coding sequence of Mi-cpl-1. To determine the transcription pattern of Mi-cpl-1 throughout the nematode development process, stage-specific expression of Mi-cpl-1 was investigated in eggs, J2s and mature females of M. incognita, using qRT-PCR. Using the expression level of Mi-cpl-1 in eggs as reference, Mi-cpl-1 was upregulated 2.2-fold in infective J2 in terms of fold change values, while no significant (P < 0.05) difference in expression between the eggs and females could be observed (Figure 2). Higher transcript accumulation of Mi-cpl-1 in infective J2 indicates that, function of Mi-cpl-1 is crucial at the early stage of nematode development, during which nematodes find the suitable host, penetrate the root and establish a compatible relationship with the host tissue, leading to formation of GC.
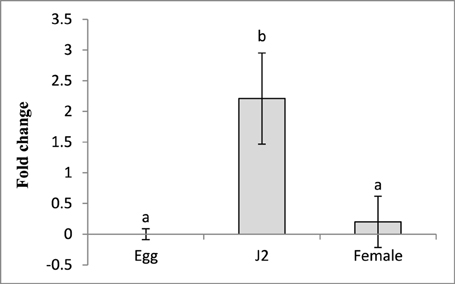
Figure 2. Relative abundance of the transcript of Mi-cpl-1 gene in different developmental stages of M. incognita. Using 2-ΔΔCT method relative expression level was quantified. Using the transcript level of Mi-cpl-1 in eggs as reference, expression of Mi-cpl-1 was found to be significantly higher at J2 stage. Each bar represents the mean ±SE of n = 3, and bars with different letters denote a significant difference at P < 0.05, student's t-test.
In Vitro RNAi of Mi-cpl-1 in M. incognita J2
To study the silencing of Mi-cpl-1 in vitro, M. incognita J2s were soaked in a buffer containing dsRNA of Mi-cpl-1 gene. Fluorescence microscopic observations revealed that the J2s had efficiently ingested the FITC marker and likely also the dsRNA molecules (Figure 3A). According to qRT-PCR results, approximately 76% reduction in mRNA abundance was observed in Mi-cpl-1 dsRNA fed J2s compared to J2s treated with soaking buffer alone or gfp dsRNA. There was no significant (P < 0.05) difference between the expression level of J2s treated with soaking buffer alone and gfp dsRNA (Figure 3B).
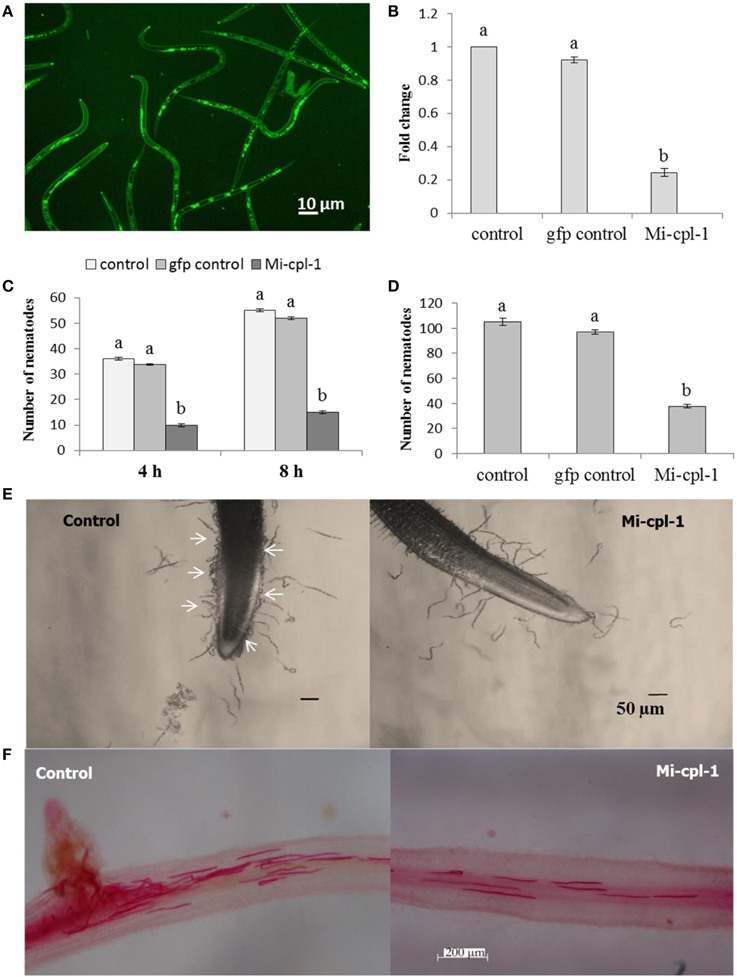
Figure 3. Effect of in vitro RNAi of Mi-cpl-1 in M. incognita J2. (A) Fluorescence microscopy showing the ingestion of FITC in M. incognita J2 after 6 h treatment with octopamine (50 mM) in soaking buffer. (B) Real-time PCR analysis of Mi-cpl-1 transcript abundance. Expression was quantified as fold change values calculated by 2-ΔΔCT method. (C) Attraction of J2s to tomato root tip at different time points (4 h and 8 h) in pluronic gel. (D) Nematode penetration of tomato root after 24 h. (E) Attraction assay in pluronic gel. Arrows indicate control J2s were attracted in greater number toward tomato root tip compared to dsRNA fed J2s. (F) Invasion of M. incognita J2 inside tomato roots after 24 h. Tomato roots supported less number of dsRNA treated J2s compared to control (Scale bar = 200 μm). Nematodes were stained with acid fuchsin (Byrd et al., 1983). Control-treatment with soaking buffer alone; gfp control-treatment with soaking buffer containing 1 mg ml−1 gfp dsRNA; Mi-cpl-1-treatment with soaking buffer containing 1 mg ml−1 Mi-cpl-1 dsRNA. Each bar represents the mean ±SE of n = 3, and bars with different letters denote a significant difference at P < 0.05.
To further evaluate the effect of Mi-cpl-1 RNAi, chemotactic behavior of J2s was assessed using attraction assay of dsRNA treated nematodes toward tomato root tip in pluronic gel (Wang et al., 2009). Silencing of Mi-cpl-1 had negative impact on nematode chemotaxis as J2s treated with Mi-cpl-1 dsRNA were attracted to tomato root tip in significantly (P < 0.05) lesser number than the control worms at 4 h and 8 h (Figures 3C,E). After 24 h penetration study was carried out, the results of which corroborated with the attraction experiment (Figures 3D,F).
These findings suggest that Mi-cpl-1 RNAi effects are significant at the transcript level as soaking buffer alone or soaking buffer with gfp dsRNA did not affect the dsRNA phenotypes and Mi-cpl-1 displayed no genetic redundancy.
Molecular Analysis of Primary Transgenic Events
Recombinant pHELLSGATE12 containing dsRNA construct of Mi-cpl-1 (Figure S1) was transformed into the tomato plants (Figure S2), and T0 plants were generated. To ensure the complete integration of T-DNA into the genome of S. lycopersicum, initial genotyping of T0 plants were carried out using PCR analysis. Presence of gene specific, sense, antisense and selectable marker fragment was detected in all the events (Figure S3).
To determine the T-DNA integration pattern in transgenic plants, genomic Southern analysis was performed with six PCR-confirmed events. Lines M4 and M10 had multiple insertions of the RNAi transgene, while M2, M5, M8, and M12 showed single copy integration pattern. No hybridization signal was detected in the wild type and empty vector control plants (Figure 4).
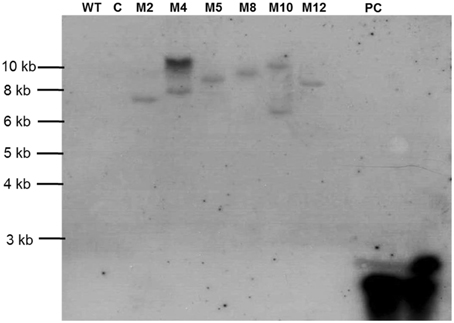
Figure 4. Southern blot for transgenic tomato harboring dsRNA of Mi-cpl-1. Lines used include Tomato “Pusa Ruby” wild type (WT), transgenic empty pHELLSGATE12 vector control (C) and transgenic pHELLSGATE12-Mi-cpl-1 (M2, M4, M5, M8, M10, M12). Probe Mi-cpl-1 (PC) was used for the hybridization.
Based on the outcome of integration studies, four events carrying the hairpin construct of Mi-cpl-1 (M2, M5, M8, and M12) were subjected to expression and bioefficacy analysis in the successive generation. T1progeny plants were generated in the growth chamber by germinating the seeds of T0plants in the presence of antibiotic kanamycin. T1plants were genotyped by PCR and expected fragments were detected, which indicates the stable integration and inheritance of RNAi transgene in the progeny plants.
Expression Analysis of Mi-cpl-1 in T1 Transgenic Events
In order to validate the expression of dsRNA transcript and its abundance, qRT-PCR analysis of the selected events (M2, M5, M8, and M12) was carried out. An increased transcript abundance of Mi-cpl-1 was recorded in all the events, and among them event M2 had the highest expression level in terms of average ΔCT values (Figure 5).
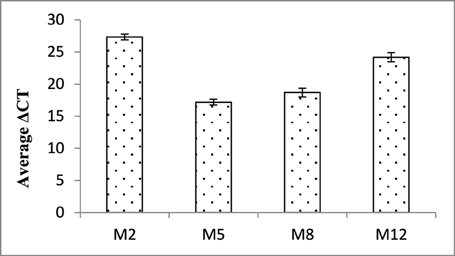
Figure 5. Expression analysis of Mi-cpl-1 gene in T1 plants. ΔCT values were calculated using the difference in the Ct mean of Mi-cpl-1 and 18S rRNA gene. Each bar represents the mean ±SE of n = 3.
As a key component of HIGS, expression of siRNA of Mi-cpl-1 was demonstrated by northern analysis in four transgenic events (M2, M5, M8, and M12). As expected, presence of Mi-cpl-1 siRNA could not be detected in the wild type and empty vector control plants (Figure 6).
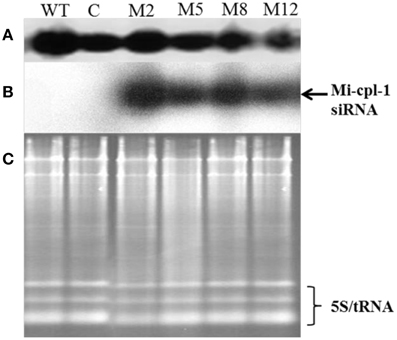
Figure 6. Production of Mi-cpl-1-specific small RNAs in transgenic tomato plants.(A) Northern blot for U6 small nuclear RNA as control, (B) Northern blot showing Mi-cpl-1 siRNAs hybridizing with probe Mi-cpl-1, and (C) Agarose gel showing total RNA loading control in Tomato “Pusa Ruby” wild type (WT), transgenic empty pHELLSGATE12 vector control (C), and transgenic pHELLSGATE12-Mi-cpl-1 (M2, M5, M8, M12) plants.
Evaluation of T1 Plants for Resistance Against M. incognita
To assess whether in planta expression of Mi-cpl-1 dsRNA exhibits resistance to M. incognita, four T1 tomato lines (M2, M5, M8, and M12) harboring the RNAi construct were inoculated with 500 M. incognita J2s per plant. In comparison to wild type plants no apparent phenotypical variation was observed in transgenic plants. At 35 DPI, the average number of galls per plant was reduced significantly (P < 0.05) by 45.6–65.2% in RNAi lines M8, M5, M12, and M2 compared to the wild type (Figure 7, Table 2). Accordingly, there was a reduction in number of females and the percentage reduction ranged between 40.2–61.8% in RNAi lines compared to the wild type (Figure 7, Table 2). Post-parasitic juveniles of M. incognita become exposed to the target dsRNA while they ingest the plant cell contents during GC formation. Dynamic maintenance of GC entails the development of the juvenile into an egg-laying female and no establishment of GC may lead to nematode death or to development as the non-feeding male. Therefore, plant-mediated silencing of Mi-cpl-1 gene resulted in reduced root galling due to impeded developmental progression of M. incognita and eventually, lesser number of developed females was observed in the transgenic plants compared to the wild type plants (Figures 8A,B). However, mature females extracted from both transgenic and wild type plant were similar in shape and size (Figure 8C). Comparable results were obtained when the fecundity of nematodes was measured in RNAi lines as number of egg masses and number of eggs per egg mass, with significant reductions of 41.3–64% and 31.9–46.8% (P < 0.05) compared to the wild type, respectively (Figure 7, Table 2). Last but not least, the nematode multiplication factor (MF) which determines the reproductive fitness and parasitic success of a given nematode was significantly (P < 0.05) reduced by 60–80.8% in RNAi lines compared to the wild type plants (Figure 7, Table 2). In essence, host-delivered RNAi of Mi-cpl-1 gene confers resistance to M. incognita by inducing negative effect on nematode infection, development and reproduction.
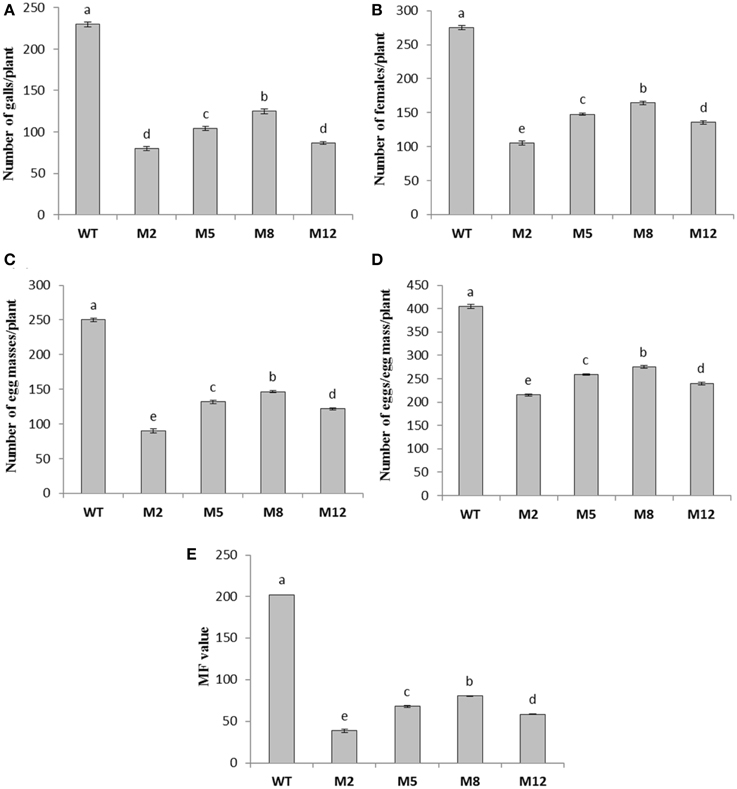
Figure 7. Effect of host induced gene silencing of Mi-cpl-1 on development and reproduction of M. incognita. Relative number of galls (A), females (B), egg masses (C), eggs/egg mass (D) and the respective multiplication factor (MF) of M. incognita (E) in different transgenic events (M2, M5, M8, and M12) and wild type plants (WT) at 35 DPI. Each bar represents the mean ±SE of n = 4, and bars with different letters denote a significant difference at P < 0.05, CRD test.
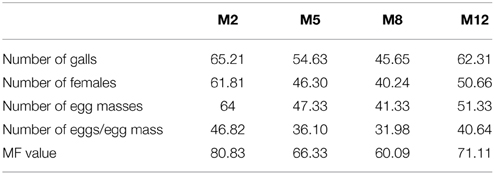
Table 2. Percentage reduction in different parameters of nematode development and reproduction on transgenics (M2, M5, M8, and M12) compared to the wild type plants.
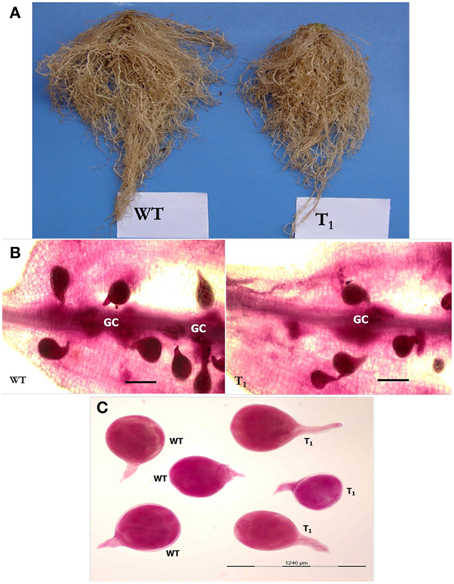
Figure 8. Comparison of M. incognita infection in transgenic (T1) and wild type (WT) tomato plants at 35 DPI. (A) Intensity of galling was comparatively higher in control plants than the transgenic plants. (B) Stained females of M. incognita inside the tomato root. Transgenic plants had supported less number of females compared to wild type plants. GC—giant cell (Scale bar = 500 μm). (C) Size and shape of mature females of M. incognita isolated from tomato roots. Nematodes were stained with acid fuchsin (Byrd et al., 1983).
To investigate the effect of HIGS on the target gene abundance in nematodes, qRT-PCR was performed to analyse the possible change in transcript level of Mi-cpl-1 in M. incognita females harvested from T1transgenic lines (M2 and M12) at 35 DPI. Using 18S ribosomal DNA as reference, it was found that the expression level of Mi-cpl-1 in M. incognita females that developed on RNAi lines M2 and M12 was reduced significantly (P < 0.05) by 59 and 51%, respectively, compared to females extracted from wild type plants (Figure 9).
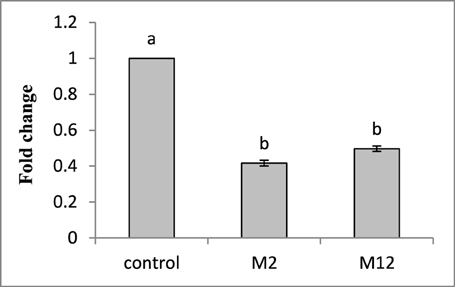
Figure 9. Transcript levels of Mi-cpl-1 gene in M. incognita females that developed in the dsRNA expressing transgenic tomato lines. Using 18S rRNA gene as reference, expression was quantified as fold change values calculated by 2-ΔΔCT method. Each bar represents the mean ±SE of n = 3, and bars with different letters denote a significant difference at P < 0.05, student's t-test.
Discussion
The parasitic success of M. incognita largely depends on the exploitation of its arsenal of parasitism genes to circumvent the plant immune responses and to induce and maintain the GC in the host vascular tissue. The current study was devised to downregulate an essential gene of M. incognita using HIGS approach and to assess the knock-down effect on nematode biology. According to the available data, stable transformation of agronomically important crops to enhance the RNAi-mediated resistance against nematodes has been accomplished in tobacco (Nicotiana tabacum L.), soybean [Glycine max (L.) Merr.] and potato (S. tuberosum L.), with varying degree of success (Steeves et al., 2006; Yadav et al., 2006; Fairbairn et al., 2007; Klink et al., 2009; Ibrahim et al., 2011; Papolu et al., 2013; Dinh et al., 2014). The targeted nematode genes in some of these reports are conserved in other animals, and due to their potential off-target effects the engineered crops may prove to be ineligible for field trial. Cathepsin L-like cysteine proteinases are an attractive group of candidate genes for RNAi-induced downregulation, not only because they are crucial for parasitic aspects of plant-nematode interaction, but they lack significant homology to genes in other organisms. In an earlier report, tobacco transgenic lines containing the dsRNA (201 bp) and fusion (600 bp) construct of Mi-cpl-1 gene had been generated, although nematodes feeding on those transgenics did not affect the gall formation or egg-mass production by females (Antonino de Souza Júnior et al., 2013). As reviewed by Sharp (2001), long dsRNA sequences may lead to greater RNAi effect compared to short dsRNAs. Conversely, longer dsRNA molecule (>400 bp) causes less effective silencing as they may not readily be assimilated in the nematode body (Dalzell et al., 2009). Therefore, in the present study, stable tomato transgenic lines overexpressing a 366 bp dsRNA construct of Mi-cpl-1 gene have been developed to elucidate the role of this gene in plant-nematode interaction. To the best of our knowledge, this is the first detailed investigation to address the function of Mi-cpl-1 gene in M. incognita-S. lycopersicum interaction using both in vitro and in vivo RNAi strategy.
Initially, to investigate the differential expression of Mi-cpl-1 transcripts throughout the developmental stage of M. incognita, qRT-PCR analysis was used. The transcript level of Mi-cpl-1 was relatively higher in pre-parasitic J2 than females and eggs. This finding is in agreement with the RT-PCR data reported by Neveu et al. (2003). Shingles et al. (2007) demonstrated the similar expression of Mi-cpl-1 in pre-parasitic J2, young and mature female of M. incognita, and slightly lower expression in older females, based on RT-PCR data. According to Antonino de Souza Júnior et al. (2013), transcript abundance of Mi-cpl-1 was comparatively higher in eggs and parasitic juveniles than females and pre-parasitic J2 of M. incognita, as observed by qRT-PCR. These discrepancies might have resulted from the sequence polymorphisms between populations of M. incognita. Using in situ hybridization assay, activity of this enzyme was detected in the intestinal cells of pre-parasitic J2 (Neveu et al., 2003) and females (Shingles et al., 2007). Combining all these stage-specific expression data, it can be speculated that M. incognita development (J2 to J3 to J4 to female) is largely dependent on the smooth functioning of this digestive enzyme. Developmental stages of M. incognita occur in close interaction with the host root tissues, indicating Mi-cpl-1 is indeed related to the parasitic phase of plant-nematode interaction.
In vitro RNAi can be used for functional validation of specific nematode genes and to assess their suitability for HIGS approach. Using this strategy, efficient suppression of the target genes with corresponding aberrant phenotypes have been successfully documented in M. incognita (Rosso et al., 2005; Huang et al., 2006; Dubreuil et al., 2007; Shingles et al., 2007; Papolu et al., 2013; Dong et al., 2014), M. javanica (Adam et al., 2008; Gleason et al., 2008), G. pallida (Urwin et al., 2002; Kimber et al., 2007), G. rostochiensis (Chen et al., 2005), H. glycines (Urwin et al., 2002; Lilley et al., 2005; Bakhetia et al., 2007), H. schachtii (Vanholme et al., 2007), Pratylenchus spp. (Joseph et al., 2012; Tan et al., 2013), Bursaphelenchus xylophilus (Park et al., 2008; Cheng et al., 2010) etc. Therefore, in the present investigation, Mi-cpl-1 gene was silenced in vitro to analyse its function in nematode behavior. qRT-PCR results revealed that silencing of Mi-cpl-1 gene leads to 76% transcript knock-down in the dsRNA treated worms compared to the control J2s. Target-specific downregulation of Mi-cpl-1 gene was also confirmed due to the absence of RNAi effect in the gfp dsRNA treated worms. To determine the phenotypic changes in nematodes induced by in vitro RNAi, chemotactic behavior of J2s toward tomato root tip was studied. In compliance with the expression data, dsRNA treated M. incognita J2s were attracted to and invaded the tomato root in lesser number compared to control worms. Hence, reduction in infection ability of J2 could be attributed to the nutritional deficiency experienced by the nematodes due to depletion in Mi-cpl-1 enzyme.
Furthermore, tomato plants were transformed to express the Mi-cpl-1 dsRNA and challenge inoculated with M. incognita J2 to delineate the function of Mi-cpl-1 in nematodes through HIGS. Substantial reduction in infection, development and reproduction of M. incognita was observed in the transgenic lines compared to the wild type plants. Nematode multiplication factor which determines its successful establishment in the host plants was reduced significantly in the transgenic lines. No apparent morphological variation was observed in tomato lines compared to the wild type plants, indicating the focused RNAi effect. In addition, qRT-PCR analysis revealed the higher expression of transgenes in RNAi lines which commensurate with the bioefficacy data. Finally, the detection of target gene siRNA in the tomato plants using northern analysis provided the ultimate evidence for plant-mediated RNAi of Mi-cpl-1 gene that minimized the nematode virulence and reproduction. Therefore, in planta reduction of Mi-cpl-1 transcripts diminished the parasitic success of M. incognita in tomato. Silencing of a nematode effector gene (16D10) has been reported by Huang et al. (2006) to lower the number of galls and egg masses by 63–90% and 69–93%, respectively, in Arabidopsis plants infected with M. incognita, M. javanica, M. arenaria or M. hapla. This classical report has been regarded as the commencement of HIGS strategy targeting plant-parasitic nematodes. Since then, several nematode genes have been targeted to generate the hairpin expressing transgenic plants with variable levels of resistance against phytoparasitic nematodes (Dutta et al., 2015), and the level and range of resistance conferred by tomato lines to M. incognita in the current study is comparable with those reports.
In order to evaluate the long-term effects of RNAi on nematode development, females of M. incognita that developed on the plants expressing Mi-cpl-1 dsRNA were extracted and subjected to qRT-PCR analysis. A substantial reduction in Mi-cpl-1 expression was recorded in females extracted from RNAi plants and consequently, the nematodes that fed on the transgenic plants were less successful in further development. It can be hypothesized that the silencing effect of Mi-cpl-1 in M. incognita is systemic and propagates upon the uptake of host-derived dsRNAs/siRNAs through stylet to the entire body. RNAi effect is transmitted as the nematode subsequently molts to the female stage and ultimately the offspring having defunct Mi-cpl-1 gene maybe produced. The heritable nature of RNAi was previously described in C. elegans (Grishok et al., 2000), and may occur in plant nematodes in similar fashion (Fairbairn et al., 2007; Sindhu et al., 2009; Papolu et al., 2013; Dinh et al., 2014). It remains inconclusive whether M. incognita ingested the host-derived dsRNAs and processed them into siRNAs using Dicer enzyme or (if) they directly took up the plant-processed siRNAs in the current study. Either or both may be possible as root-knot nematodes are known to efficiently ingest relatively large biomolecules (Urwin et al., 1997; Li et al., 2007; Zhang et al., 2012).
Although the transgenic delivery of dsRNA/siRNA of Mi-cpl-1 gene via plant cells to M. incognita J2 elicited developmental retardation in the feeding nematodes, none of the tomato lines exhibited complete resistance in the current study. The reason could be explained by the fact that nematode genes may have additive, combinatorial or redundant function in the plant-nematode interface. Therefore, to achieve complete and durable nematode resistance more than one nematode gene should be targeted for simultaneous interference. In this direction, effectiveness of combinatorial RNAi was investigated in H. glycines (Bakhetia et al., 2008) and M. incognita (Antonino de Souza Júnior et al., 2013). In either case, however, desired results could not be obtained perhaps because high level of dsRNA delivery saturates the dsRNA processing ability of nematodes. Nevertheless, crossing of different RNAi lines resulting in the co-expression of compound dsRNA constructs in the same transgenic plant enhances the efficacy of RNAi (Huang et al., 2006; Charlton et al., 2010). This strategy ensures the continuous delivery of bioactive RNA species to the feeding nematodes.
Classical breeding approaches are time-consuming and additional impediments including linkage drag, compatibility barrier, and gene loss during introgression are associated with it (Hermsen, 1994). Biosafety-related issues associated with the transgenic crops expressing nematicidal proteins, such as cystatins or cry proteins have not been addressed yet. In view of this, RNAi-based transgenics have been a viable option due to the lack of impact of RNAi on non-target species and no transgenic protein is produced in RNAi plants (Atkinson et al., 2009). The RNAi-based genetic engineering strategy presented here could provide an attractive alternative to breed root-knot nematode-resistant tomatoes. The biosafety level of this novel strategy can be improved by using tissue-specific promoters that can efficiently deliver the nematode dsRNAs into the active feeding cells. To date, no crop varieties with resistance to Meloidogyne spp. have yet been bioengineered for field trial owing to the lack of awareness of the damage potential of phytonematodes among the plant breeders (Atkinson et al., 2012). Given that how destructive nematodes can be (Jones et al., 2013), it is imperative that pyramiding of nematode genes with the other pathogen genes in the same crop plant may provide unprecedented broad-spectrum resistance to various pests and pathogens. To ensure the global food security, future studies should focus on the evaluation of RNAi technology in staple crops such as cereals and pulses to manage the plant nematodes and to explore the suitability of this technology at the field level.
Author Contributions
Conceived and designed the experiments: TD, UR, AS. Performed the experiments: TD, PP, PB, DC. Analyzed the data: TD. Wrote the manuscript: TD.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank ICAR-IARI, New Delhi, India for funding and providing the infrastructural facilities. We extend our gratitude to CSIRO, Australia for providing the RNAi binary vector, pHELLSGATE12. Special thanks to Nagavara Prasad Gantasala, Sagar Banerjee, Rakesh Kumar, Ramanna Koulagi and Yogesh Thorat for their help in the execution of the experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2015.00260/abstract
References
Adam, M. A. M., Phillips, M. S., Jones, J. T., and Blok, V. C. (2008). Characterisation of the cellulose-binding protein Mj-cbp-1 of the root knot nematode, Meloidogyne javanica. Physiol. Mol. Plant Pathol. 72, 21–28. doi: 10.1016/j.pmpp.2008.05.002
Antonino de Souza Júnior, J. D., Coelho, R. R., Lourenço, I. T., da Rocha Fragoso, R., Viana, A. A. B., de Macedo, L. L. P., et al. (2013). Knocking-down Meloidogyne incognita proteases by plant-delivered dsRNA has negative pleiotropic effect on nematode vigor. PLoS ONE 8:e85364. doi: 10.1371/journal.pone.0085364
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Atkinson, H. J., Lilley, C. J., and Urwin, P. E. (2012). Strategies for transgenic nematode control in developed and developing world crops. Curr. Opin. Biotechnol. 23, 251–256. doi: 10.1016/j.copbio.2011.09.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Atkinson, H. J., Urwin, P. E., and Hussey, R. S. (2009). “Plant biotechnology and control,” in Root-knot nematodes, eds R. N. Perry, M. Moens, and J. L. Starr (Wallingford;Oxfordshire: CAB International), 338–362.
Bakhetia, M., Urwin, P. E., and Atkinson, H. J. (2007). qPCR analysis and RNAi define pharyngeal gland cell-expressed genes of Heterodera glycines required for initial interactions with the host. Mol. Plant Microbe Interact. 20, 306–312. doi: 10.1094/MPMI-20-3-0306
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bakhetia, M., Urwin, P. E., and Atkinson, H. J. (2008). Characterisation by RNAi of pioneer genes expressed in the dorsal pharyngeal gland cell of Heterodera glycines and the effects of combinatorial RNAi. Int. J. Parasitol. 38, 1589–1597. doi: 10.1016/j.ijpara.2008.05.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Byrd, D. W., Kirkpatrick, T., and Barker, K. R. (1983). An improved technique for clearing and staining plant tissues for detection of nematodes. J. Nematol. 15, 142–143.
Charlton, W. L., Harel, H. Y. M., Bakhetia, M., Hibbard, J. K., Atkinson, H. J., and McPherson, M. J. (2010). Additive effects of plant expressed double-stranded RNAs on root-knot nematode development. Int. J. Parasitol. 40, 855–864. doi: 10.1016/j.ijpara.2010.01.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, Q., Rehman, S., Smant, G., and Jones, J. T. (2005). Functional analysis of pathogenicity proteins of the potato cyst nematode Globodera rostochiensis using RNAi. Mol. Plant Microbe Interact 18, 621–625. doi: 10.1094/MPMI-18-0621
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cheng, X. Y., Dai, S. M., Xiao, L., and Xie, B. Y. (2010). Influence of cellulase gene knockdown by dsRNA interference on the development and reproduction of the pine wood nematode, Bursaphelenchus xylophilus. Nematology 12, 225–233. doi: 10.1163/138855409X12469541205044
Dalzell, J. J., McMaster, S., Johnstone, M. J., Kerr, R., Fleming, C. C., and Maule, A. G. (2009). Non-nematode-derived double-stranded RNAs induce profound phenotypic changes in Meloidogyne incognita and Globodera pallida infective juveniles. Int. J. Parasitol. 39, 1503–1516. doi: 10.1016/j.ijpara.2009.05.006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Davis, E. L., Hussey, R. S., and Baum, T. J. (2008). Parasitism proteins in nematode-plant interactions. Curr. Opin. Plant Biol. 11, 360–366. doi: 10.1016/j.pbi.2008.04.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dinh, P. T. Y., Zhang, L., Brown, C. R., and Elling, A. A. (2014). Plant-mediated RNA interference of effector gene Mc16D10L confers resistance against Meloidogyne chitwoodi in diverse genetic backgrounds of potato and reduces pathogenicity of nematode offspring. Nematology 16, 669–682. doi: 10.1163/15685411-00002796
Dong, L., Li, X., Huang, L., Gao, Y., Zhong, L., Zheng, Y., et al. (2014). Lauric acid in crown daisy root exudate potently regulates root-knot nematode chemotaxis and disrupts Mi-flp-18 expression to block infection. J. Exp. Bot. 65, 131–141. doi: 10.1093/jxb/ert356
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dubreuil, G., Magliano, M., Deleury, E., Abad, P., and Rosso, M. N. (2007). Transcriptome analysis of root-knot nematode functions induced in the early stages of parasitism. New Phytol. 176, 426–436. doi: 10.1111/j.1469-8137.2007.02181.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dutta, T. K., Banakar, P., and Rao, U. (2015). The status of RNAi-based transgenics in plant nematology. Front. Microbiol. 5:760. doi: 10.3389/fmicb.2014.00760
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Elbashir, S. M., Martinez, J., Patkaniowska, A., Lendeckel, W., and Tuschl, T. (2001). Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20, 6877–6888. doi: 10.1093/emboj/20.23.6877
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fairbairn, D. J., Cavallaro, A. S., Bernard, M., Mahalinga-Iyer, J., Graham, M. W., and Botella, J. R. (2007). Host-delivered RNAi: an effective strategy to silence genes in plant parasitic nematodes. Planta 226, 1525–1533. doi: 10.1007/s00425-007-0588-x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi: 10.1038/35888
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gleason, C. A., Liu, Q. L., and Williamson, V. M. (2008). Silencing a candidate nematode effector gene corresponding to the tomato resistance gene Mi-1 leads to acquisition of virulence. Mol. Plant Microbe Interact. 21, 576–585. doi: 10.1094/MPMI-21-5-0576
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Grishok, A., Tabara, H., and Mello, C. C. (2000). Genetic requirements for inheritance of RNAi in C. elegans. Science 287, 2494–2497. doi: 10.1126/science.287.5462.2494
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hashmi, S., Britton, C., Liu, J., Guiliano, D. B., Oksov, Y., and Lustigman, S. (2002). Cathepsin L is essential for embryogenesis and development of Caenorhabditis elegans. J. Biol. Chem. 277, 3477–3486. doi: 10.1074/jbc.M106117200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hassan, S., Behm, C. A., and Mathesius, U. (2010). Effectors of plant parasitic nematodes that reprogram root cell development. Funct. Plant Biol. 37, 933–942. doi: 10.1071/FP10112
Helliwell, C., and Waterhouse, P. (2003). Constructs and methods for high throughput gene silencing in plants. Methods 30, 289–295. doi: 10.1016/S1046-2023(03)00036-7
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hermsen, J. G. T. (1994). “Introgression of genes from wild species, including molecular and cellular approaches,” in, Potato Genetics, eds J. E. Bradshaw and G. R. Mackay (Wallingford;Oxfordshire: CAB International), 515–538.
Hooper, D. J. (1986). “Extraction of free-living stages from soil,” in Laboratory Methods for Work with Plant and Soil Nematodes, eds J. F. Southey (London: Ministry of Agriculture, Fisheries and Food), 5–30.
Huang, G., Allen, R., Davis, E. L., Baum, T. J., and Hussey, R. S. (2006). Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci. U.S.A. 103, 14302–14306. doi: 10.1073/pnas.0604698103
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ibrahim, H. M., Alkharouf, N. W., Meyer, S. L., Aly, M. A., and Gamal, E. A. K. (2011). Post-transcriptional gene silencing of root-knot nematode in transformed soybean roots. Exp. Parasitol. 127, 90–99. doi: 10.1016/j.exppara.2010.06.037
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jain, R. K., Mathur, K. N., and Singh, R. V. (2007). Estimation of losses due to plant parasitic nematodes on different crops in India. Ind. J. Nematol. 37, 219–220.
Jones, J. T., Haegeman, A., Danchin, E. G. J., Gaur, H. S., Helder, J., Jones, M. G. K., et al. (2013). Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 14, 946–961. doi: 10.1111/mpp.12057
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Joseph, S., Gheysen, G., and Subramaniam, K. (2012). RNA interference in Pratylenchus coffeae: knock down of Pc-pat-10 and Pc-unc-87 impedes migration. Mol. Biochem. Parasitol. 186, 51–59. doi: 10.1016/j.molbiopara.2012.09.009
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jyothishwaran, G., Kotresha, D., Selvaraj, T., Srideshikan, S. M., Rajvanshi, P. K., and Jayabaskaran, C. (2007). A modified freeze-thaw method for efficient transformation of Agrobacterium tumefaciens. Cur. Sci. 6, 770–772.
Kimber, M. J., McKinney, S., McMaster, S., Day, T. A., Fleming, C. C., and Maule, A. G. (2007). flp gene disruption in a parasitic nematode reveals motor dysfunction and unusual neuronal sensitivity to RNA interference. FASEB J. 21, 1233–1243. doi: 10.1096/fj.06-7343com
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Klink, V. P., Kim, K. H., Martins, V., Macdonald, M. H., Beard, H. S., Alkharouf, N. W., et al. (2009). A correlation between host-mediated expression of parasite genes as tandem inverted repeats and abrogation of development of female Heterodera glycines cyst formation during infection of Glycine max. Planta 230, 53–71. doi: 10.1007/s00425-009-0926-2
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Koch, A., and Kogel, K. H. (2014). New wind in the sails: improving the agronomic value of crop plants through RNAi-mediated gene silencing. Plant Biotechnol. J. 12, 821–831. doi: 10.1111/pbi.12226
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Koiwa, H., Shade, R. E., Zhu-Salzman, K., D'Urzo, M. P., Murdock, L. L., Bressan, R. A., et al. (2000). A plant defensive cystatin (soyacystatin) targets cathepsin L like digestive cysteine proteinases in the larval midgut of western corn rootworm (Diabrotica virgifera virgifera). FEBS Lett. 471, 67–70. doi: 10.1016/S0014-5793(00)01368-5
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). ClustalW and ClustalX version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Li, X. Q., Wei, J. Z., Tan, A., and Aroian, R. V. (2007). Resistance to root-knot nematode in tomato roots expressing a nematicidal Bacillus thuringiensis crystal protein. Plant Biotechnol. J. 5, 455–464. doi: 10.1111/j.1467-7652.2007.00257.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lilley, C. J., Goodchild, S. A., Atkinson, H. J., and Urwin, P. E. (2005). Cloning and characterisation of a Heterodera glycines aminopeptidase cDNA. Int. J. Parasitol. 35, 1577–1585. doi: 10.1016/j.ijpara.2005.07.017
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lilley, C. J., Urwin, P. E., McPherson, M. J., and Atkinson, H. J. (1996).Characterization of intestinally active proteinases of cyst-nematodes. Parasitology 113, 415–424. doi: 10.1017/S0031182000066555
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lustigman, S., Zhang, J., Liu, J., Oksov, Y., and Hashmi, S. (2004). RNA interference targeting cathepsin L and Z-like cysteine proteases of Onchocerca volvulus confirmed their essential function during L3 molting. Mol. Biochem. Parasitol. 138, 165–170. doi: 10.1016/j.molbiopara.2004.08.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Malhotra, P., Dasaradhi, P. V. N., Kumar, A., Mohmmed, A., Agrawal, N., Bhatnagar, R. K., et al. (2002). Double-stranded RNA-mediated gene silencing of cysteine proteases (falcipain-1 and -2) of Plasmodium falciparum. Mol. Microbiol. 45, 1245–1254. doi: 10.1046/j.1365-2958.2002.03105.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Moens, M., Perry, R. N., and Starr, J. L. (2009). “Meloidogyne spp.-a diverse group of novel and important plant parasite,” in Root-knot nematodes, eds R. N. Perry, M. Moens, and J. L. Starr, (Wallingford;Oxfordshire: CAB International), 1–17.
Neveu, C., Abad, P., and Castagnone-Sereno, P. (2003). Molecular cloning and characterization of an intestinal cathepsin L protease from plant-parasitic nematode Meloidogyne incognita. Physiol. Mol. Plant Pathol. 63, 159–165. doi: 10.1016/j.pmpp.2003.10.005
Papolu, P. K., Gantasala, N. P., Kamaraju, D., Banakar, P., Sreevathsa, R., and Rao, U. (2013). Utility of host delivered RNAi of two FMRF amide like peptides, flp-14 and flp-18, for the management of root-knot nematode, Meloidogyne incognita. PLoS ONE 8:e80603. doi: 10.1371/journal.pone.0080603
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Park, J. E., Lee, K. Y., Lee, S. J., Oh, W. S., Jeong, P. Y., Woo, T., et al. (2008). The efficiency of RNA interference in Bursaphelenchus xylophilus. Mol. Cells 26, 81–86.
Rosso, M. N., Dubrana, M. P., Cimbolini, N., Jaubert, S., and Abad, P. (2005). Application of RNA interference to root-knot nematode genes encoding esophageal gland proteins. Mol. Plant Microbe Interact. 18, 615–620. doi: 10.1094/MPMI-18-0615
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Santamaria, I., Velasco, G., Pendas, A. M., Fueyo, A., and Lopez-Otin, C. (1998). Cathepsin Z, a novel human cysteine proteinase with a short propeptide domain and a unique chromosomal location. J. Biol. Chem. 273, 16816–16823. doi: 10.1074/jbc.273.27.16816
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sharp, P. A. (2001). RNA interference-2001. Genes Dev. 15, 485–490. doi: 10.1101/gad.880001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shingles, J., Lilley, C. J., Atkinson, H. J., and Urwin, P. E. (2007). Meloidogyne incognita: molecular and biochemical characterisation of a cathepsin L cysteine proteinase and the effect on parasitism following RNAi. Exp. Parasitol. 115, 114–120. doi: 10.1016/j.exppara.2006.07.008
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shompole, P. S., and Jasmer, D. P. (2001). Cathepsin B-like cysteine proteases confer intestinal cysteine protease activity in Haemonchus contortus. J. Biol. Chem. 276, 2928–2934. doi: 10.1074/jbc.M007321200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sindhu, A. S., Maier, T. R., Mitchum, M. G., Hussey, R. S., and Davis, E. L. (2009). Effective and specific in planta RNAi in cyst nematodes: expression interference of four parasitism genes reduces parasitic success. J. Exp. Bot. 60, 315–324. doi: 10.1093/jxb/ern289
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Southern, E. M. (1975). Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98, 503–517. doi: 10.1016/S0022-2836(75)80083-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Steeves, R. M., Todd, T. C., Essig, J. S., and Trick, H. N. (2006). Transgenic soybeans expressing siRNAs specific to a major sperm protein gene suppress Heterodera glycines reproduction. Funct. Plant Biol. 33, 991–999. doi: 10.1071/FP06130
Tan, C. H. J., Jones, M. G. K., and Fosu, N. (2013). Gene silencing in root lesion nematodes (Pratylenchus spp.) significantly reduces reproduction in a plant host. Exp. Parasitol. 133, 166–178. doi: 10.1016/j.exppara.2012.11.011
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tort, J., Brindley, P. J., Knox, D., Wolfe, K. H., and Dalton, J. P. (1999). Proteinases and associated genes of parasitic helminths. Adv. Parasitol. 43, 161–266. doi: 10.1016/S0065-308X(08)60243-2
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Urwin, P. E., Lilley, C. J., and Atkinson, H. J. (2002). Ingestion of double-stranded RNA by pre-parasitic juvenile cyst nematodes leads to RNA interference. Mol. Plant Microbe Interact. 15, 747–752. doi: 10.1094/MPMI.2002.15.8.747
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Urwin, P. E., Moller, S. G., Lilley, C. J., McPherson, M. J., and Atkinson, H. J. (1997). Continual green fluorescent protein monitoring of cauliflower mosaic virus 35S promoter activity in nematode-induced feeding cells in Arabidopsis thaliana. Mol. Plant Microbe Interact. 10, 394–400. doi: 10.1094/MPMI.1997.10.3.394
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vanholme, B., Van Thuyne, W., Vanhouteghem, K., De Meutter, J., Cannoot, B., and Gheysen, G. (2007). Molecular characterization and functional importance of pectate lyase secreted by the cyst nematode Heterodera schachtii. Mol. Plant Pathol. 8, 267–278. doi: 10.1111/j.1364-3703.2007.00392.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, C. L., Lower, S., and Williamson, V. M. (2009). Application of Pluronic gel to the study of root-knot nematode behaviour. Nematology 11, 453–464. doi: 10.1163/156854109X447024
Yadav, B. C., Veluthambi, K., and Subramaniam, K. (2006). Host-generated double stranded RNA induces RNAi in plant-parasitic nematodes and protects the host from infection. Mol. Biochem. Parasitol. 148, 219–222. doi: 10.1016/j.molbiopara.2006.03.013
Zhang, F., Peng, D., Ye, X., Yu, Z., Hu, Z., Ruan, L., et al. (2012). In vitro uptake of 140 kDa Bacillus thuringiensis nematicidal crystal proteins by the second stage juvenile of Meloidogyne hapla. PLoS ONE 7:e38534. doi: 10.1371/journal.pone.0038534
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: root-knot nematode, Mi-cpl-1, RNAi, transgenics, resistance
Citation: Dutta TK, Papolu PK, Banakar P, Choudhary D, Sirohi A and Rao U (2015) Tomato transgenic plants expressing hairpin construct of a nematode protease gene conferred enhanced resistance to root-knot nematodes. Front. Microbiol. 6:260. doi: 10.3389/fmicb.2015.00260
Received: 17 February 2015; Accepted: 16 March 2015;
Published: 01 April 2015.
Edited by:
Essaid Ait Barka, Reims University, FranceReviewed by:
Helene Sanfacon, Pacific Agri-Food Research Centre, CanadaLaura Ellen Rose, Heinrich-Heine University, Germany
Copyright © 2015 Dutta, Papolu, Banakar, Choudhary, Sirohi and Rao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tushar K. Dutta, Division of Nematology, ICAR-Indian Agricultural Research Institute, Room No-332, New Delhi-110012, IndiabmVtYWlhcmlAZ21haWwuY29t
 Tushar K. Dutta
Tushar K. Dutta Pradeep K. Papolu
Pradeep K. Papolu