- 1Moreau Lab, School of Earth Sciences, Faculty of Science, University of Melbourne, Melbourne, VIC, Australia
- 2Department of Microbiology and Immunology, Peter Doherty Institute for Infection and Immunity, University of Melbourne, Melbourne, VIC, Australia
Our primary research paper (Mu et al., 2014) demonstrated selective changes to a deep subsurface prokaryotic community as a result of CO2 stress. Analyzing geochemical and microbial 16S rRNA gene profiles, we evaluated how in situ prokaryotic communities responded to increased CO2 and the presence of trace organic compounds, and related temporal shifts in phylogeny to changes in metabolic potential. In this focused review, we extend upon our previous discussion to present analysis of taxonomic unit co-occurrence profiles from the same field experiment, to attempt to describe dynamic community behavior within the deep subsurface. Understanding the physiology of the subsurface microbial biosphere, including how key functional groups integrate into the community, will be critical to determining the fate of injected CO2. For example, community-wide network analyses may provide insights to whether microbes cooperatively produce biofilm biomass, and/or biomineralize the CO2, and hence, induce changes to formation porosity or changes in electron flow. Furthermore, we discuss potential impacts to the feasibility of subsurface CO2 storage of selectively enriching for particular metabolic functions (e.g., methanogenesis) as a result of CO2 injection.
Introduction
The carbon capture and storage (CCS) technology known as “geosequestration,” or injection of large volumes of supercritical CO2 (scCO2) into deep aquifers, has the potential to impact subsurface microbial community dynamics. Such impacts include changes to microbially-mediated terminal electron accepting processes that may, in turn, affect the geochemistry and mineralogy of the CO2 storage aquifer. Subsequent mineral precipitation or dissolution reactions, for example, could result in a decrease or increase in porosity that leads to redistribution of injected scCO2 (Gadd, 2010) or undesirable changes in groundwater chemistry. Other impacts may include methanogenesis as a result of locally increased dissolved CO2 levels under lowered pH conditions (Sato et al., 2013), or the inhibition of enzymatic carbon monoxide (CO) oxidation with broad consequences for microbial community carbon utilization and electron flow (Ragsdale, 2004; Techtmann et al., 2009). Accumulation of excess CO may increase the activity of acetotrophic methanogens, as CO is a metabolic intermediate and its concentration is known to be inversely related to that of methane in anaerobic digesters (Krzycki and Zeikus, 1984; Hickey and Switzenbaum, 1990). In fact, uncertainty currently exists around whether deep subsurface microbial responses to scCO2 injection will result in positive or negative effects on geosequestration. As an example, biofilm formation has been observed under scCO2 in laboratory bioreactors (e.g., Mitchell et al., 2009), and some researchers have proposed that this process may be stimulated in CO2 storage aquifers to help “lock in” scCO2 and prevent its migration as a buoyant plume. In contrast, other studies have observed that scCO2 injection led to the growth of methanogens (Morozova et al., 2011) and therefore raise the question of whether geosequestration could result in enhanced methane generation and potential leakage. Clearly, understanding the changes in microbial community composition and dynamics throughout geosequestration experiments, especially those conducted in situ under quasi-realistic conditions or in real CO2 storage aquifers, will provide useful insights for understanding the fate of scCO2.
KEY CONCEPT 1. Biofilm
Microbial biofilms are a community of mixed or single-isolate microbes encapsulated in a self-produced extracellular biopolymer matrix. Biofilms exhibit greater tolerance to CO2 stress, and have the potential to alter biogeochemical processes and reduce reservoir porosity.
Previous efforts to study microbial responses to scCO2 exposure have focused on characterizing the subsurface microbial community at the individual taxonomic level (Morozova et al., 2011; Bordenave et al., 2012; Lavalleur and Colwell, 2013). However, the resolution of these studies make apparent the need for metagenomic and systems biology -based analyses, such as functional gene and co-occurrence profiling, with a focus on elucidating potential syntrophic associations of importance to CO2 storage. We need to understand the microbial biosphere beyond simple biodiversity characterizations because unrelated lineages have been shown to converge in functional similarity or complementarity where lineage-environment associations are stable (Chaffron et al., 2010; Gadd, 2010). For example, different average genome sizes suggesting the presence of multiple taxa have been observed in redox selective environments (Raes et al., 2007; Angly et al., 2009), and dissimilatory Fe(III) reduction might be a common metabolic feature in deep subsurface petroleum reservoirs that are host to a range of anaerobic thermophiles and hyperthermophiles (Slobodkin et al., 1999). Another example might include observations of close spatial association between sulfate-reducing and sulfide-oxidizing bacteria or archaea in anoxic or acidic environments (Bond et al., 2000; Loy et al., 2004; Moreau et al., 2010). Furthermore, increasing consideration should be placed on incorporating the potential impacts derived from operational-dependent engineering, such as the emplacement of injection and sampling wells, into the analysis pipeline, as these can influence subsurface microbial community composition and function (Morozova et al., 2011; Bordenave et al., 2012; Lavalleur and Colwell, 2013; Mu et al., 2014). The same principles of systems biology apply to the study of many other subsurface environments, such as hydrocarbon rich reservoirs (Dojka et al., 1998; Golby et al., 2011; Joshi et al., 2014) and radioactive waste storage sites (Nazina et al., 2004; Chi Fru and Athar, 2008).
KEY CONCEPT 2. Metagenomic
Metagenomics involves the bioinformatic analysis of high-throughput sequencing data of whole community genomic material. Determining the functional gene profile will provide insights to potential biogeochemical processes that underpin microbial community dynamics.
KEY CONCEPT 3. Systems biology
Systems biology (systeomics) takes a holistic view of the biological system and assesses the community at a higher-order level. Analyses typically include computationally intensive co-occurrence profiling, and microbe-metabolism network associations.
KEY CONCEPT 4. Syntrophy
Also known as microbial cross-feeding, syntrophy describes the process by which one species feeds off the metabolic products of another species, thus potentially forming dependent relationships between phylogenetically distantly related microorganisms.
KEY CONCEPT 5. Operational-dependent engineering
The engineering required for CO2 geosequestration includes, for example, injection and sampling wells, which play an integral part of the project and may influence the geochemical milieu. Residual organic compounds from the drilling fluid used during emplacement of injection wells can influence microbial community composition and function.
Current literature is largely based on in vitro experiments that utilize representative geologic material to simulate CO2 geosequestration, and results indicate that single-isolate microbial biofilms exhibit greater tolerance to scCO2 stress compared to the planktonic phase of growth (Mitchell et al., 2008). Analysis of the in vitro community structure also showed the subsequent predominance of different taxonomic groups following exposure to CO2 stress (Mitchell et al., 2009). While it is essential to conduct in vitro studies, it is also important to conduct in situ field-scale experiments to (1) validate results obtained from lab-scale studies and (2) provide biological relevance in the context of real geology. To this extent, a field-based scCO2 sequestration experiment was conducted in the 1.4 km-deep Paaratte Formation of the Otway Basin, Australia (Paterson et al., 2013). One hundred and fifty tons of mixed scCO2 and groundwater was injected into the sandstone Paaratte aquifer over 4 days. Our primary research (Mu et al., 2014) detected changes in microbial community structure prior to scCO2 injection, which revealed a general shift from Firmicutes to Proteobacteria concurrent with the disappearance of polyethylene glycols (PEGs) that were interpreted as residual from drilling fluid used during the emplacement of the CO2 injection well. Furthermore, the persistence of Carboxydocella, Comamonadaceae, and Sphingomonadaceae after scCO2 injection suggested that these groups could adapt to the changes in groundwater chemistry resulting from the CO2 geosequestration experiment, including decreases in pH and temperature of 2.6 log units and 5.8°C, respectively, and an increase in CO2 concentration from 148 parts per million (ppm) to 1410 ppm. However, issues of activity (mRNA), metabolic function (transcriptome, proteome) and syntrophic interactions (network associations) of the subsurface biosphere still remain. Considering many environmental microbes are uncultivable in the laboratory (Rappé and Giovannoni, 2003; Schloss and Handelsman, 2004), scientists will need to employ multiple omics-based measurements (e.g., metagenomics, transcriptomics, etc.) and bioinformatic analyses (e.g., taxonomic co-occurrence profiling) coupled to geochemical measurements, to understand microbial physiology and the dynamic cycling of nutrients. Furthermore, to broaden our analysis, inductively coupled plasma mass spectrometry (ICP-MS) was performed to determine potential toxicity effects of CO2-induced mobilization of trace metals from the sediments into the water phase. With the exception of Fe, there were no discernable changes to the concentration of dissolved trace metals (including Ca, K, Mg, Al, Ba, Cu, Mn, Sr, and Zn) in Paaratte groundwater within the respective pre- and post-CO2 injection phases. Therefore, the observed changes to microbial community structure are unlikely to be attributable to potential metal toxicity. However, it is acknowledged that our results may not be universal for the reasons of different redox states and sediment types at different CO2-receiving reservoirs (Ardelan et al., 2009) where lowered pH conditions may facilitate the mobilization of trace elements. Therefore, this review aims to address some of the key issues, and to highlight the key gaps, in our understanding of environmental microbial responses to CO2 stress, and also present new analysis of microbial networks during a field-based scCO2 sequestration experiment.
Materials and Methods
CO2CRC Otway Stage 2B
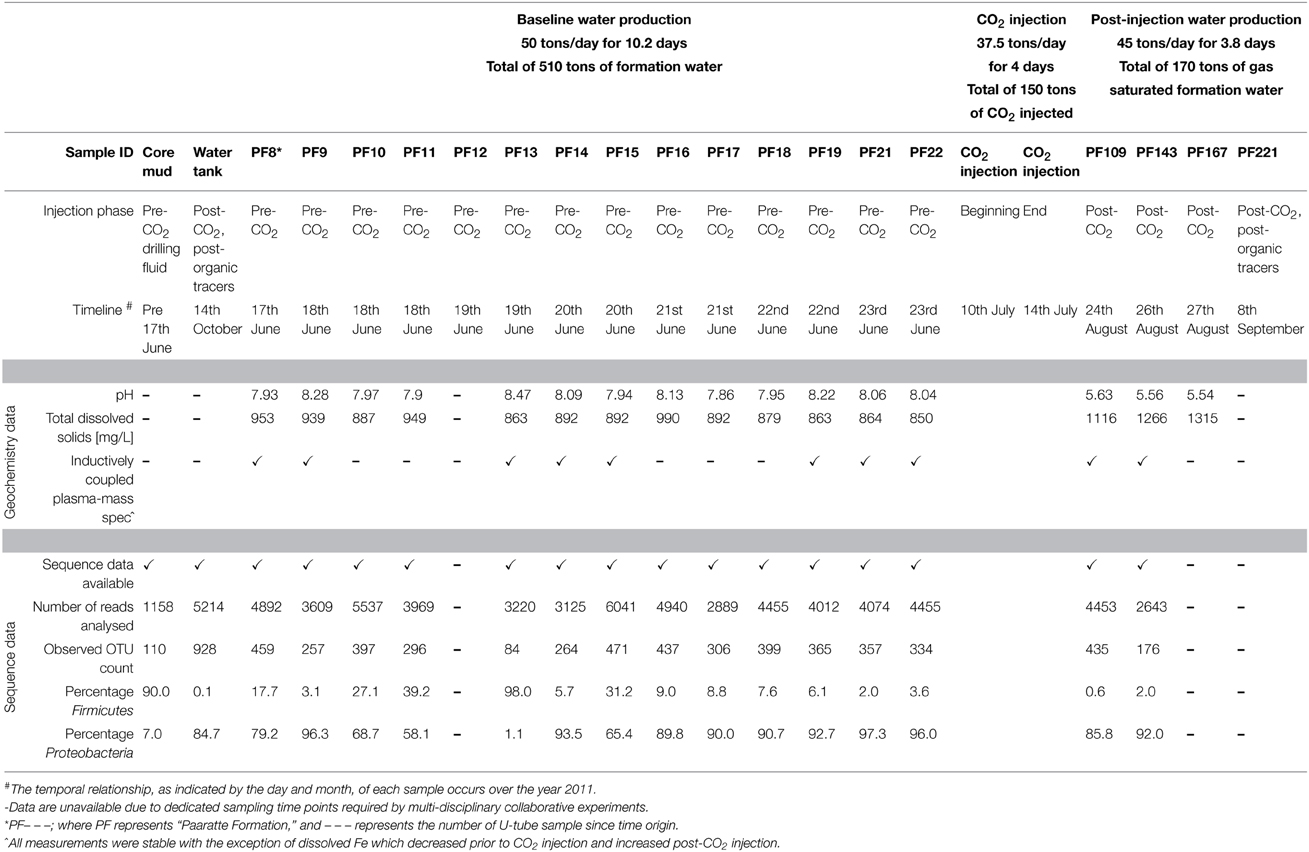
The Cooperative Research Centre for Greenhouse Gas Technologies (CO2CRC) Otway Stage 2B field experiment was conducted to ascertain the residual CO2 storage capacity of a sandstone aquifer, 1400 m true vertical depth sub-sea (TVDSS) in the Paaratte Formation (Otway Basin, Southeastern Australia), to assess geosequestration as a means to mitigate atmospheric CO2 pollution (Paterson et al., 2013). A summary of the scCO2 injection experiment is provided in Table 1.
Pristine water samples held under in situ conditions were obtained via a novel U-tube sampling system (see Section In Situ Sampling of the Deep Subsurface Biosphere for further information on the system) over the course of the scCO2 injection (150 tons) event for baseline geochemical analyses and genomic DNA extraction. A total of 79 U-tube water samples were collected. Whole community 16S rRNA gene profile analysis is described in detail in Mu et al. (2014). Briefly, biomass was concentrated, as 50 ml aliquots, on to 0.22 μm nylon filter membranes (Merk Millipore) using vacuum filtration, and processed for gDNA extraction using the MoBio Powersoil DNA extraction kit. Whole community gDNA was extracted onsite within 12 h of sampling. However, given the logistics of the sampling schedule post-scCO2 injection, filter membranes with concentrated biomass for nucleic acid extraction were stored on site in RNAprotect Bacteria Reagent (QIAGEN) at −20°C until processing could be performed in the laboratory. Whole community gDNA were amplified with native universal small subunit 803 forward and universal SSU1392w reverse (5′- ACG GGC GGT GWG TRC -3′) primers using High-fidelity OneTaq DNA polymerase mastermix (New England Biolabs). SSU803F primer is a combination of 803Fa 5′- TTA GAT ACC CTG GTA GTC -3′; 803Fb 5′- TTA GAT ACC CSG GTA GTC -3′; 803Fc 5′- TTA GAT ACC CYH GTA GTC -3′; 803Fd 5′- TTA GAG ACC CYG GTA GTC-3′; in a ratio of 2:1:1:1 for 803Fa:b:c:d. Amplicon sequencing was performed by the Australian Centre for Ecogenomics (ACE; University of Queensland, Australia) using barcoded 454 pyrosequencing (Roche). The QIIME bioinformatics pipeline was employed to analyse sequence data, assign taxonomy, and to determine phylogenetic distributions of each microbial community (Caporaso, 2010). Sequence data were clustered into operational taxonomic units at 97% pairwise identity using the UCLUST (Edgar, 2010) seed-based algorithm. A representative sequence from each OTU was aligned using the PyNAST tool (Caporaso et al., 2010) and queried against the Ribosomal Database Project (Wang et al., 2007) for taxonomy assignment. Results from the taxonomic classification using the Ribosomal Database Project classifier (Mu et al., 2014) through the QIIME tool (Caporaso, 2010) was analyzed to compute statistical dependence of each of the microbial orders, pre- and post-CO2 injection. The otu.association function from the Mothur software (version 1.27; Schloss et al., 2009) was used to calculate Spearman's rank correlation values of the taxonomic units based on their relative abundance percentages across all samples. Visualization of the corresponding networks with a coefficient cutoff value of 0.5, and one standard deviation away from the mean (i.e., 0.84), were aided by Cytoscape version 2.8.3 (Cline et al., 2007; Saito et al., 2012). The nodes represent microbial orders, while an edge indicates associations between connecting nodes. The degree sorted circle layout was imposed on the network to indicate a decreasing degree of associations between nodes (i.e., the number of nodes one OTU associates with) proceeding in a counter clockwise direction with the point of origin at the 180° position. Subnetworks within a community are denoted with alphabetical characters. The correlation test used in this study is the same as that found in the Phoenix 2 software for 16S rRNA gene analyses (Soh et al., 2013).
KEY CONCEPT 6. U-tube
A hydraulically sealed sampling system used to collect deep subsurface water and gas samples under in situ conditions. In situ water samples are collected and stored in high-pressure stainless steel cylinders (Swagelok) for downstream microbial and geochemical analyses.
Results
Community Network Analyses
The Paaratte Formation microbial community separated into five sub-networks (Spearman's rank correlation ≥ 0.5; p < 0.025) for samples obtained during the pre-CO2 injection phase (Figure 1). Sub-networks A, B, C, and D consisted of two taxonomical orders each, while, Sub-network E contained 52 orders. Pseudomonadales/Pseudomonas (Order/Genus), the predominant Proteobacteria, associated with an uncharacterized taxon in Sub-network A. Similarly, Clostridiales/Carboxydocella (Order/Genus), an anaerobic thermophile, associated with an uncharacterized taxon in Sub-network B. Sphingomonadales/Sphingomonadaceae (Order/Family) and Burkholderiales/Comamonadaceae (Order/Family), which were shown to proliferate in relative abundance during the post-CO2 injection phase (Mu et al., 2014), are illustrated in Sub-network E to have an underlying association with 87% of the microbial community at the order level of classification. However, the degree of association for Sphingomonadaceae and Burkholderiales was relatively weak as the orders were positioned further away from point origin. Furthermore, sulfur cycling-associated taxa, i.e., Acidithiobacillales and Desulfitobacterales (Table 2) were integrated into Sub-network E (Figure 1). The microbial community as visualized at a Spearman's rank correlation ≥ 0.84 (p < 0.005) illustrated a strong interaction between Methanomicrobiales and Myxococcales (Table 2, and Figure 2 sub-network G). The most abundant OTUs, Pseudomonadales/Pseudomonas and Clostridiales/Carboxydocella, were absent from the entire network at a Spearman's rank correlation cutoff ≥ 0.84 (p < 0.005; Figure 2).
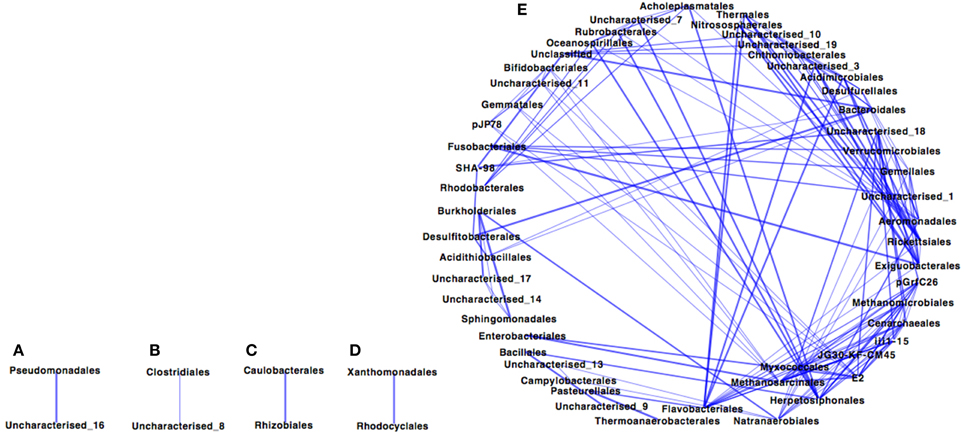
Figure 1. Co-occurrence profile of the pre-CO2 injection Paaratte Formation microbial community with a Spearman's rank correlation of ≥0.5 (p < 0.025). Results from the taxonomic classification using the Ribosomal Database Project classifier through the QIIME tool was analyzed to compute statistical dependence of each of the microbial orders pre-CO2 injection. Spearman's Rank correlation values were calculated based on the relative abundance percentages of all taxonomic units across all samples using the otu.association function from the Mothur software (version 1.27). Networks were visualized using Cytoscape version 2.8.3. The nodes represent microbial orders, while an edge indicates an association between connecting nodes. The degree sorted circle layout was imposed on the network to indicate a decreasing degree of association between nodes proceeding in a counter clockwise direction (point of origin at the 180° position). That is to say the operational taxonomic unit at the 180° position associates with more OTUs than the others. Sub-networks are denoted with alphabetic characters.
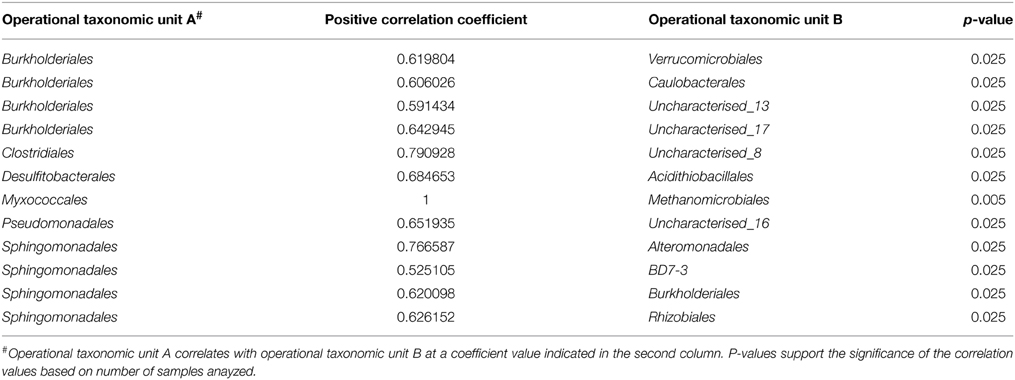
Table 2. Spearman's rank correlation coefficient and corresponding p-values for associating operational taxonomic units.
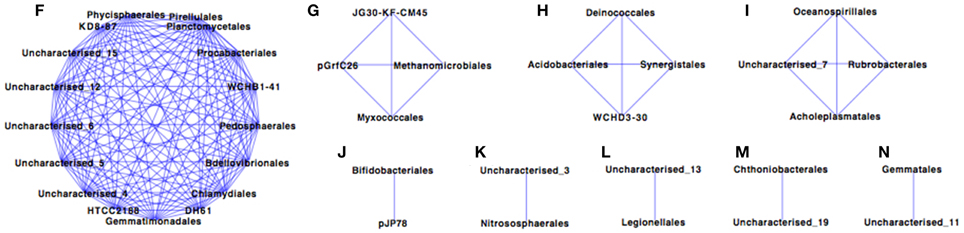
Figure 2. Co-occurrence profiling of the pre- and post-CO2 injection Paaratte Formation microbial community with a Spearman's rank correlation of ≥0.84. Results from the taxonomic classification using the Ribosomal Database Project classifier through the QIIME tool was analyzed to compute statistical dependence of each of the microbial orders pre- and post-CO2 injection. Spearman's Rank correlation values were calculated based on the relative abundance percentages of all taxonomic units across all samples using the otu.association function from the Mothur software (version 1.27). Networks were visualized using Cytoscape version 2.8.3. The nodes represent microbial orders, while an edge indicates an association between connecting nodes. The degree sorted circle layout was imposed on the network to indicate a decreasing degree of association between nodes proceeding in a counter clockwise direction (point of origin at the 180° position).
Discussion
Logistics of a Carbon Geosequestration Project
Ongoing investigations are aimed at understanding the feasibility of capturing and sequestering industrial volumes of CO2 within the Earth's subsurface, in an attempt to reduce the impacts of anthropogenic climate change. Carbon capture and storage is steadily proving to be a viable solution in cutting global CO2 emissions (Benson and Surles, 2006; Gibbins and Chalmers, 2008). However, a number of factors need to be considered before injecting many kilotons of CO2 into the subsurface, including, for example, site selection (Bachu, 2000, 2002; Ennis-King et al., 2011), geochemical and biological monitoring & verification (Stalker et al., 2009; Réveillère and Rohmer, 2011; Schacht et al., 2011; Jenkins et al., 2012; Noble et al., 2012), engineering and sampling representative material for analysis (Freifeld et al., 2005; Kharaka et al., 2006; Freifeld, 2009), and the mechanism by which CO2 is sequestered (Li et al., 2006; Saadatpoor et al., 2009). The Australian CO2CRC Otway project provides insights into factors associated with field-scale experimental design (Sharma et al., 2009; Boreham et al., 2011; Underschultz et al., 2011; Haese et al., 2013) that may have implications for understanding subsurface biosphere responses to CO2 geosequestration. More details about each of these factors can be found in the original references, and several are discussed below.
Engineering Derived Influences on Biodiversity
In the context of geosequestration, “engineering” includes controlling the flow rate of water sampling, the injection rate of gas-saturated water, and the use of drilling fluid during the emplacement of injection and sampling wells. Operational-dependent engineering can have indirect impacts on the characterization of the microbial community. For example, a high flow rate of water sampling may force attached bacterial cells (sessile population) to detach into pore waters for analysis (MacDonald et al., 1999). Whereas, low flow rates may only be sampling planktonic community members for analysis due to reduced shearing forces (MacDonald et al., 1999), and thereby under-represent the complete microbial biosphere. Surface-associated physiochemical properties could also influence the affinity of cell attachment as a function of hydrophobicity (Donlan, 2002; Mitchell et al., 2009). An understanding of site-specific hydrogeology, and collection of both fluid phase and core samples (Kolbel-Boelke et al., 1988; Godsy et al., 1992; Bekins et al., 1999), will help to evaluate microbial community responses to increased CO2 levels.
In Situ Sampling of the Deep Subsurface Biosphere
Understanding the microbial biosphere of deep subsurface environments requires precise and “sterile” sampling of representative geologic material, which is often difficult to achieve because very few techniques allow for subsurface samples to be retained under in situ pressure and temperature conditions (e.g., Schlumberger's MDT syringe-like tool; and evacuated Kuster samplers, Kuster Company, Long Beach, Ca, USA; Freifeld et al., 2005; Kharaka et al., 2006). Therefore, to obtain geochemically pristine water samples held under in situ conditions, while not compromising the stability of injected CO2, a novel, hydraulically sealed “U-tube” sampling system (Freifeld et al., 2005; Freifeld, 2009) was employed in our primary research paper (Mu et al., 2014). The U-tube system produces formation water to the surface using directive flow of high-pressure nitrogen gas into specialized pressure cylinders independent of the injection line. Furthermore, engineering of the U-tube allows for the isolation, and storage of formation water under conditions that are characteristic of subsurface environments. This sampling mechanism is particularly important for deep, anoxic subsurface environments where even slight exposure to oxygen can be harmful to the sampled microorganisms (Fredrickson and Onstott, 1996). Therefore, by using the U-tube system the microbial community may be analyzed as close to its in situ physiological state, whether it is through culture-dependent and/or -independent assays.
Residual Sequestration of CO2
Residual sequestration is characterized by the formation of plumes when free-phase CO2 migrates and becomes trapped by capillary pressure from the water in the pore spaces between the rocks. The mechanism thus prevents CO2 from flowing, and effectively sequesters the injected CO2 (Nghiem et al., 2009; Zhang et al., 2011; Shamshiri and Jafarpour, 2012). Residual trapping was the method of sequestration used in our primary research paper. The mechanics of residual trapping alludes to the possibility of creating microenvironments that may select for, and/or provide the conditions in which, different syntrophic microbial networks co-occur within the reservoir. Indeed, regions with trapped CO2 can affect the immediate environments geochemistry (e.g., pH, solubility), and in turn alter microbe-surface interactions. The concept of microenvironments is discussed further in the following sections.
It is probable that over geological time scales the buoyant CO2 plume may escape residual sequestration, migrate upwards and subsequently affect microbial community structure and dynamics. Therefore, recent studies (Oppermann et al., 2010; Morales and Holben, 2014) have investigated the impacts of CO2 leakage on surface soil communities. Similar to our original research (Mu et al., 2014), Morales and Holben (2014) reported a community structure predominated by Proteobacteria and low abundances of Firmicutes after exposure to CO2. Furthermore, they measured a statistically significant decrease in species richness, albeit only 3%. Reduced species richness has implications in ecosystem functional potential whereby a loss of diversity can translate to a loss in ecological functions (Tilman, 1999; Loreau et al., 2001; Petchey and Gaston, 2006), thus, affecting the cycling of carbon and other energy sources.
In Vitro Studies
It is difficult to ascertain whether subsurface microorganisms biomineralize and/or convert the injected CO2 into biomass. Therefore, in vitro bioreactor studies are conducted to elucidate the mechanisms behind biogeochemical processes that play key roles in determining the fate of injected CO2. Of particular interest is how microbial biofilms might impact community dynamics and the sequestration of CO2; it is known that microbial biofilms have the potential to tolerate short exposures to scCO2 stress, and reduce reservoir porosity (Mitchell et al., 2008). For example, Mitchell and colleagues hypothesized that the protective effects of extracellular polymeric substances—through offering mass transport resistance, and large surface areas—resulted in the resilience of Bacillus mojavensis biofilms after exposure to scCO2 stress (Mitchell et al., 2008). Using a high-pressure flow reactor (1290 psi, 32°C) and electron microscopy, they also showed that sandstone cores, initially inoculated with biofilm forming Shewanella frigidimarina, were later populated with viable, sandstone-native, biofilm-forming cells of B. mojavensis and Citrobacter sp., after scCO2 and nutrient starvation stress. Observation that the microbial community changes after CO2 stress indicates the importance in understanding which taxa are being selected for, and their physiology, in order to predict their responses to increased CO2 levels. However, many in vitro analyses have so far been restricted to phenotypic assessment, which falls short of the resolution required to predict the behavior of biofilms and planktonic populations. It is therefore crucial that we understand the molecular mechanisms (i.e., genomic, metagenomic, transcriptomic, metabolomic, and proteomic) behind biofilm formation, biofilm-induced calcium carbonate precipitation (Phillips et al., 2013), and general microbial tolerance to CO2 stress, in order to control and utilize microorganisms for remediation purposes, such as long-term sealing of fractures in subsurface storage reservoirs. A recent study analyzing the transcriptome of a model sulfate-reducing microorganism, Desulfovibrio vulgaris, identified that cells up-regulated the transcription of certain amino-acids related to osmotic stress responses, and genes associated with chemotaxis (e.g., flagella subunits), in response to elevated CO2 pressures (Wilkins et al., 2014). However, scientists are routinely tasked with collecting adequate amounts of quality nucleic acid, in the form of biomass, from the deep subsurface to conduct the aforementioned omics measurements; modifying the U-tube system to collect significantly larger volumes of subsurface waters may be one option. Nevertheless, the real value in resolving such technical issues associated with omics-based measurements will be reflected upon when the physiological roles (i.e., transcriptome, proteome) that microorganisms play in the dynamic cycling of biogeochemically relevant elements, particularly the physiology of prokaryotes that are uncultivable in vitro, are determined (Handelsman, 2004; Lasken, 2012).
Field-Scale Studies
There are a number of factors that might contribute to apparent disconnects between data generated from laboratory-based experiments and those obtained from field-scale projects, such as, hydrogeology (e.g., flow rates), rates of CO2-induced mobilization of contaminants (including trace metals), CO2-induced pH changes, unknown native reservoir gas composition, and intrusion of groundwater from nearby aquifers (refer to Harvey et al., 2013 for a detailed review on the geochemical implications of geosequestration). Such parameters have implications on evaluating the feasibility and potential of certain geological formations as CO2 storage sites for the reason that they can significantly influence the physiological state of the microbial community, and in turn alter the cycling of biogeochemically-relevant elements (Chapelle, 2000; Allen et al., 2007). For example, certain net redox reactions may be strongly acid consuming and thus highly sensitive to pH shifts (e.g., manganese and iron reduction), while others (e.g., sulfate reduction and methanogenesis) are less sensitive to CO2 induced pH changes (Bethke et al., 2011). Therefore, to better account for the inherent difficulties in detailing the intricate dynamics of the subsurface microbial biosphere, advances in sampling systems, such as the U-tube system (Freifeld et al., 2005; Freifeld, 2009) which provides fluid and gas samples retained under in situ conditions, can perhaps yield the necessary insights to design “geomicrobially representative” synthetic communities for in vitro culturing assays. Furthermore, there needs to be an ongoing process whereby results from laboratory experiments are analyzed and interpreted alongside data collected from in situ field-scale projects. More specifically, the experimental design of laboratory-based studies should incorporate site-specific geology and microbiology.
Valid concerns have been raised about whether studied subsurface communities are representative, especially when aerobic and facultative anaerobic taxa are detected in bioinformatic analyses of what should presumably be anaerobic environments (Balkwill, 1989; Sinclair and Ghiorse, 1989; Chandler et al., 1997; Biddle et al., 2008; An et al., 2013; Kimes et al., 2013). An et al. (2013) showed that hydrocarbon resource environments (HRE) commonly harbor an unexpected population of aerobic taxa and genes. The authors reasoned that some HREs might have available oxygen through infiltrating precipitation-derived waters (Andriashek and Atkinson, 2007; An et al., 2013). Furthermore, oxygenated groundwaters could be more prevalent in the subsurface than what is presumed and hence create oxic microenvironments that could allow aerobic respiration to persist in predominantly reducing environments (Winograd and Robertson, 1982). Alternatively, we speculate that operational–dependent engineering, such as sampling wells, may seed aerobic communities (or facultative anaerobes) into the subsurface. Because these communities, including eukaryotes (Orsi et al., 2013; Rédou et al., 2014), may play a significant role in the cycling of trace elements, they need to be assessed as apart of the ecosystem (e.g., CO2 geosequestration, oil recovery etc.). That is to say, for example, CO2 geosequestration will not be complete without the emplacement, and utilization of injection and sampling wells. Therefore, we must include potential engineering-derived communities and site-specific metadata (e.g., geochemistry [dissolved oxygen], lithology, hydrogeology/groundwater recharge, spatial and temporal heterogeneities) in the analysis process.
In order to predict what might happen to injected CO2, and understand the microbial influences on geosequestration at a field-scale level, Bordenave et al. (2012) characterized the baseline microbial community of a salt cavern (Baker Hughes, Alberta, Canada), while Lavalleur and Colwell (2013) focused on a basalt formation (Wallula pilot Eastern Washington State, USA). Pyrosequencing analysis showed that the anaerobic salt cavern was predominated by a halophilic, and thermophilic community. Further analysis by Bordenave et al. (2012), indicated that homoacetogenic activity was present at high and low salt concentrations, while methanogenesis was only present at low salt concentrations in the presence of H2. Similarly, Lavalleur and Colwell (2013) characterized the pre-CO2 injection biosphere of a basalt formation and demonstrated, through pyrosequencing, that Proteobacteria, Firmicutes and Actinobacteria were the predominant taxa. Furthermore, the closest known relatives as determined by 16S rRNA gene sequence similarity suggested the presence of H2-oxidizers, methylotrophs, sulfate reducers, methanotrophs, and methanogens (Lavalleur and Colwell, 2013), and thus, implied that hydrogen and single carbon-compounds might play significant roles in sustaining the deep biosphere (Stevens and McKinley, 1995). A study by Morozova et al. (2011) characterizing the microbial community of a siltstone and sandstone environment using fluorescent in situ hybridization (bacteria, archaea, and SRB 16S rRNA probes) and 16S rRNA gene fingerprinting (single-strand conformation polymorphism, and denaturing gradient gel electrophoresis) showed that fermentative halophilic bacteria (Haloanaerobium sp., Halobacteroidaceae) and SRB (Desulfohalobium sp., Desulfotomaculum sp.) were the dominant members of the community. This study represented one of the first efforts to characterize the biosphere throughout a CO2 geosequestration project, but lacked the sensitivity of high throughput DNA sequencing. Combining high throughput sequencing and organic geochemistry analyses, our principle research paper evaluated how in situ bacterial populations responded to increased CO2 levels and the presence of residual organic compounds, and related the temporal shift in taxonomic grouping (i.e., Firmicutes to Proteobacteria) to a switch in metabolic potential (Fermentation of residual organic compounds to respiration; Mu et al., 2014). Furthermore, our results suggested the potential for enhanced scCO2 tolerance, including changes in response to the associated variables (e.g., pH, temperature, and salinity), by Comamonadaceae and Sphingomonadaceae.
High variability exists in microbial community structure for all sites analyzed to date for geosequestration and other subsurface environments. The observed diversity of functional groups within geochemically defined environments might obscure syntrophic behaviors that are not readily discernable through relative abundance analyses of taxonomy data alone. This observation supports the need to incorporate systems biology approaches to develop an understanding of the links that underpin the networks and interactions of subsurface microbial communities in response to anthropogenic change. A community-wide approach that incorporates the analysis of microbial co-occurrences might reveal characteristics such as microbial cross-feeding that links phylogenetically distant microbes in a syntrophic relationship. For example, the degradation of natural organic matter in the subsurface by fermenting microbes generates bicarbonate (HCO−3), hydrogen (H2), and simple organic compounds (e.g., CH3COO−), which in turn may be utilized by hydrogenotrophic and acetotrophic methanogens to produce methane (Demirel and Scherer, 2008), or by sulfate and iron reducers as substrates for anaerobic respiration (Nealson and Saffarini, 1994). Another example might include competitive exclusion, a phenomenon that gives functional groups of microbes with a higher potential on the thermodynamic redox ladder an advantage over groups lower on the ladder that compete for the same electron donor (Lovley and Phillips, 1987; Chapelle and Lovley, 1992; Hoehler et al., 1998; Heimann et al., 2010; Bethke et al., 2011). We need to understand community functional dynamics, rather than solely taxonomic diversity, in order to delineate community functions and understand how the microbial biosphere responds to perturbation events such as CO2 injection and/or trace metal contaminants.
Co-occurrence Profiling Reveal Insights into Microbial Community Functions
Preliminary results from co-occurrence analyses indicate that the Paaratte Formation microbial community may have underlying similarities in biogeochemical pathways and function as an interactive consortium. An association of the majority of the community (Figure 1 Network E) suggests a high degree of interdependency between the taxonomic units. However, separation of the community into five networks during the pre-CO2 injection phase (Figure 1) alludes to the presence of different co-occurring system behaviors, including narrow and syntrophic associations. The following subsections provide a brief overview of two main metabolic functions frequently characterized at geosequestration sites.
Methanogenesis
A pertinent question in carbon geosequestration is, “Will the injected CO2 stimulate a community that is overrepresented by microorganisms that can convert CO2 to CH4?”. Previous studies have highlighted the presence of methanogens pre-perturbation in subsurface biospheres that are targeted for CO2 geosequestration (Bordenave et al., 2012; Lavalleur and Colwell, 2013). Furthermore, methane is shown to be in high concentrations in basaltic formations (1,270 m depth; Stevens et al., 1993; Stevens and McKinley, 1995), and any leakage from CO2 injection-induced events will also trigger the leakage of native reservoir gases (Harvey et al., 2013). Methane is a primary contributor to climate change and is more potent as a greenhouse gas when compared to CO2 (Lashof and Ahuja, 1990; Shindell et al., 2009). Therefore, the concern for CCS becomes a question of, “Are we solving one environmental problem (atmospheric CO2 pollution) at the expense of creating another (e.g., methane pollution)?”
Microbes are important terminal oxidizers during the anaerobic mineralization of organic matter to CO2 and CH4 in low sulfate environments. Methanogenesis occurs slowly in the presence of sulfate-reducing bacteria because sulfate reducers have a higher affinity for hydrogen and acetate, and there is a higher energy yield from sulfate reduction (Schonheit et al., 1982; Lovley and Klug, 1983). Therefore, SRB tend to outcompete methanogens. However, Morozova et al. (2011) demonstrated the temporary dominance of methanogenic archaea over SRB during CO2 storage in a saline aquifer (Ketzin, Germany). The observation by Morozova and colleagues highlights the question of potential stimulated methane production and subsequently, the potential for CO2 and methane co-leakage from a storage aquifer. Preliminary co-occurrence analyses of microbial taxa from the Paaratte Formation (Figure 3), however, demonstrate the association (>84% co-occurrence) between Methanomicrobiales and Myxococcales (Table 2; Figure 2). Methanomicrobiales is a CO2-reducing methanogen that uses H2 (or formate) as the reducing agent (Sakai et al., 2012). On the other-hand, Myxococcales is an organism that feeds solely on insoluble organic substances (Dawid, 2000; Zhou et al., 2014), and has been proposed to associate with methanogens through dependent predation on methanotrophs (Osaka et al., 2008). Therefore, co-occurrence of the two taxa suggests that some methane cycling may have occurred with a subsequent contribution to (recycled) biomass.
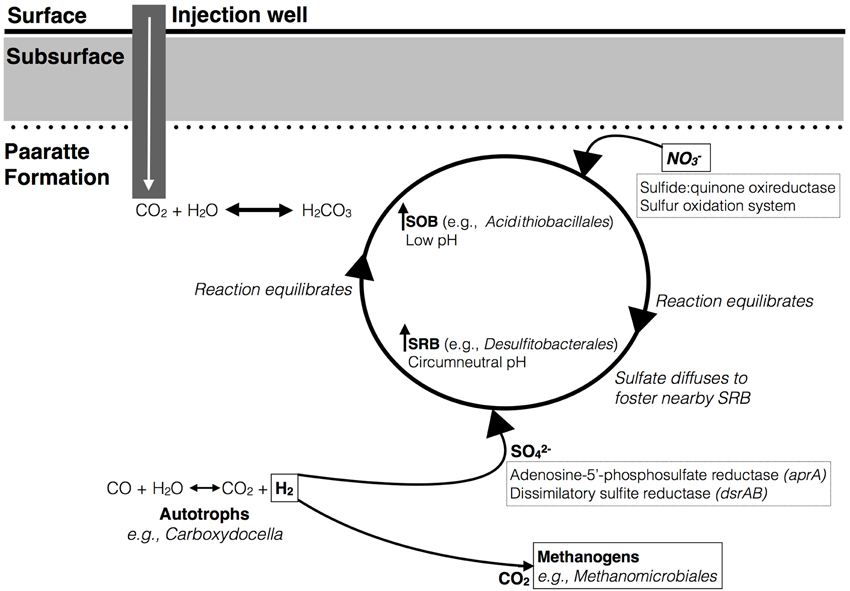
Figure 3. A working model to explain the co-occurrence of sulfur-oxidizing and -reducing bacteria in network analyses. A decrease in groundwater pH as a result of CO2 injection selects for growth of sulfide-oxidizing bacteria (SOB). The oxidation of sulfur may be coupled to nitrate as a terminal electron acceptor to proceed anaerobically. This reaction produces sulfate that can diffuse to allow for the activity of nearby sulfate-reducing bacteria (SRB) under circumneutral pH conditions. Autotrophs (e.g., carboxydotrophs) are a source of H2 for sulfate-reducers to metabolize the available SO2−4. The genes that encode for the potential enzymes responsible for oxidizing and reducing sulfur are respectively highlighted (in dashed boxes). Furthermore, it is illustrated that carboxydotrophs can also supply methanogens with CO2 and H2 required for methanogenesis.
The results provide a basis for testing the hypothesis that a dependent relationship can form around methanogenesis during a CO2 geosequestration project. Further culture-based experiments should therefore include targeted analysis of autotrophic consumption of CO2 and methane production.
KEY CONCEPT 7. Autotrophy
Autotrophy describes the process by which microorganisms produce complex organic compounds from simple inorganic carbon molecules.
Cyclic Metabolism of Sulfur
Sulfur exists in multiple oxidation states with an eight-electron difference between the two end members, from sulfide to sulfate. Multiple microbial members are required to carry out the series of reaction pathways of sulfur oxidation intermediates (Vairavamurthy et al., 1993; Detmers et al., 2001; Friedrich et al., 2005). Preliminary analysis of the Paaratte Formation has suggested a co-occurrence of sulfide-oxidizing bacteria (SOB) Acidithiobacillales, and sulfur-reducing bacteria, Desulfitobacterales (Table 2). Associations between SOB and SRB in the environment have been described in the past (Elshahed et al., 2003). A working model is presented in Figure 3, which hypothesizes an association between the SRB and SOB within microenvironments created by the residually trapped CO2. We speculate that a geochemical link establishes between the sulfur cyclers within the reservoir whereby the decrease in pH as a result of CO2 injection selects for SOB (Harrison, 1984). This reaction may produce sulfate that can diffuse to foster the activity of nearby SRB under circumneutral pH conditions. Autotrophs (e.g., Carboxydocella) are potential sources of H2 for sulfate-reducers to metabolize the available SO2−4 (Techtmann et al., 2009). However, results from our original research suggest the potential inhibition of CO-oxidation by the thermophilic autotroph, Carboxydocella, as a result of the CO2 injection project. The inhibition of enzymatic CO-oxidation could therefore have broad implications for the cycling of carbon, including methanogenesis, and sulfur within the Paaratte Formation (Figure 3). Furthermore, the microbial oxidation of sulfide, to recycle sulfur in its most reduced state to more oxidized forms, is preferentially facilitated by nitrate as the electron acceptor under anoxic conditions (Canfield and Thamdrup, 1994; Thamdrup et al., 1994; Habicht and Canfield, 1997; Bruchert and Pratt, 1999). Therefore, metagenomic analysis of sulfide:quinone oxireductase and the sulfur oxidation system (Poser et al., 2014), along with adenosine-5′-phosphosulfate reductase (aprA) (Meyer and Kuever, 2007) and dissimilatory sulfite reductase (dsrAB) genes (Geets et al., 2006) may provide further insights (Figure 3).
Concluding Remarks
Many subsurface environments targeted for CO2 storage have shown differences in microbial community composition. Thus, by using an analytical approach that considers potential syntrophic behaviors, this focused review on the geomicrobiology of geosequestration provides insights into more specialized biogeochemical processes—processes and relationships that would otherwise be omitted from solely taxonomical analyses—to reveal the complexity of what appears to be an extreme but simple environment (i.e., relatively low biomass, reduced conditions, high pressure and salinity). Co-occurrence profiling of the Paaratte Formation suggests the lack of an interdependency of the predominant OTUs with the majority of the biosphere, which implies a high degree of functional redundancy such that their removal may not affect the overall dynamics of the community. The review also highlights the need to consider anthropogenic activities in the subsurface as an engineered system in order to comprehend microbial community function and dynamics. Recent studies (e.g., Mitchell et al., 2008, 2009; Bordenave et al., 2012; Lavalleur and Colwell, 2013; Lau et al., 2014; Wilkins et al., 2014) are laying a foundation upon which we can start to build our understanding of the complexities of the subsurface biosphere. Indeed, future studies need to incorporate multi-omic approaches in order to unravel the interactions (the interactome; Baker, 2013) that connect subsurface biogeochemical processes. More importantly, the co-occurrence profiling approach described in this Focused Review can be a powerful in silico technique through which hypotheses are generated and specific subsurface populations tested in vitro, so as to start to address the gaps in our understanding of the complete geomicrobiological response to CO2 geosequestration.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Prof. Richard A. Strugnell and Assoc. Prof. Timothy P. Stinear for laboratory resources and helpful advice on our manuscript. Special thanks to all reviewers of the manuscript as their comments and suggestions added to the quality of the current paper. Furthermore, we acknowledge funding from the Commonwealth of Australia and industry sponsors through the CO2CRC program.
Author Biography

References
Allen, J. P., Atekwana, E. A., Atekwana, E. A., Duris, J. W., Werkema, D. D., and Rossbach, S. (2007). The microbial community structure in petroleum-contaminated sediments corresponds to geophysical signatures. Appl. Environ. Microbiol. 73, 2860–2870. doi: 10.1128/AEM.01752-06
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
An, D., Caffrey, S. M., Soh, J., Agrawal, A., Brown, D., Budwill, K., et al. (2013). Metagenomics of hydrocarbon resource environments indicates aerobic taxa and genes to be unexpectedly common. Environ. Sci. Technol. 47, 10708–10717. doi: 10.1021/es4020184
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Andriashek, L. D., and Atkinson, N. (2007). “Buried channels and glacial-drift aquifers in the Fort McMurray region, northeast Alberta,” in Earth Sciences Report, Alberta Geological Survey, Alberta Energy and Utilities Board (Edmonton, AB), 1–170.
Angly, F. E., Willner, D., Prieto-Davó, A., Edwards, R. A., Schmieder, R., Vega-Thurber, R., et al. (2009). The GAAS metagenomic tool and its estimations of viral and microbial average genome size in four major biomes. PLoS Comput. Biol. 5:e1000593. doi: 10.1371/journal.pcbi.1000593
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ardelan, M. V., Steinnes, E., Lierhagen, S., and Linde, S. O. (2009). Effects of experimental CO2 leakage on solubility and transport of seven trace metals in seawater and sediment. Sci. Total Environ. 407, 6255–6266. doi: 10.1016/j.scitotenv.2009.09.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bachu, S. (2000). Sequestration of CO2 in geological media: criteria and approach for site selection in response to climate change. Energy Convers. Manage. 41, 953–970. doi: 10.1016/S0196-8904(99)00149-1
Bachu, S. (2002). Sequestration of CO2 in geological media in response to climate change: road map for site selection using the transform of the geological space into the CO2 phase space. Energy Convers. Manage. 43, 87–102. doi: 10.1016/S0196-8904(01)00009-7
Baker, M. (2013). The 'omes puzzle. Nature 494, 416–419. doi: 10.1038/494416a
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Balkwill, D. L. (1989). Numbers, diversity, and morphological characteristics of aerobic, chemoheterotrophic bacteria in deep subsurface sediments from a site in South Carolina. Geomicrobiol. J. 7, 33–52. doi: 10.1080/01490458909377848
Bekins, B. A., Godsy, E. M., and Warren, E. (1999). Distribution of microbial physiologic types in an aquifer contaminated by crude oil. Microb. Ecol. 37, 263–275. doi: 10.1007/s002489900149
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Benson, S. M., and Surles, T. (2006). Carbon dioxide capture and storage: an overview with emphasis on capture and storage in deep geological formations. Proc. IEEE 94, 1795–1805. doi: 10.1109/JPROC.2006.883718
Bethke, C. M., Sanford, R. A., Kirk, M. F., Jin, Q., and Flynn, T. M. (2011). The thermodynamic ladder in geomicrobiology. Am. J. Sci. 311, 183–210. doi: 10.2475/03.2011.01
Biddle, J. F., Fitz-Gibbon, S., Schuster, S. C., Brenchley, J. E., and House, C. H. (2008). Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc. Natl. Acad. Sci. U.S.A. 105, 10583–10588. doi: 10.1073/pnas.0709942105
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bond, P. L., Smriga, S. P., and Banfield, J. F. (2000). Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl. Environ. Microbiol. 66, 3842–3849. doi: 10.1128/AEM.66.9.3842-3849.2000
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bordenave, S., Chatterjee, I., and Voordouw, G. (2012). Microbial community structure and microbial activities related to CO2 storage capacities of a salt cavern. Intergovern. Panel Clim. Change 81, 82–87. doi: 10.1016/j.ibiod.2012.08.001
Boreham, C., Underschultz, J., Stalker, L., Kirste, D., Freifeld, B., Jenkins, C., et al. (2011). Monitoring of CO2 storage in a depleted natural gas reservoir: gas geochemistry from the CO2CRC Otway Project, Australia. Int. J. Greenhouse Gas Control 5, 1039–1054. doi: 10.1016/j.ijggc.2011.03.011
Bruchert, V., and Pratt, L. M. (1999). Stable sulfur isotopic evidence for historical changes of sulfur cycling in estuarine sediments from Northern Florida. Aquat. Geochem. 5, 249–268. doi: 10.1023/A:1009661812641
Canfield, D., and Thamdrup, B. (1994). The production of 34S-depleted sulfide during bacterial disproportionation of elemental sulfur. Science 266, 1973–1975. doi: 10.1126/science.11540246
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Caporaso, J. G. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Caporaso, J. G., Bittinger, K., Bushman, F. D., DeSantis, T. Z., Andersen, G. L., and Knight, R. (2010). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267. doi: 10.1093/bioinformatics/btp636
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chaffron, S., Rehrauer, H., Pernthaler, J., and von Mering, C. (2010). A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res. 20, 947–959. doi: 10.1101/gr.6104521.109
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chandler, D. P., Li, S.-M., Spadoni, C. M., Drake, G. R., Balkwill, D. L., Fredrickson, J. K., et al. (1997). A molecular comparison of culturable aerobic heterotrophic bacteria and 16S rDNA clones derived from a deep subsurface sediment. FEMS Microbiol. Ecol. 23, 131–144. doi: 10.1111/j.1574-6941.1997.tb00397.x
Chapelle, F. H. (2000). The significance of microbial processes in hydrogeology and geochemistry. Hydrogeol. J. 8, 41–46. doi: 10.1007/PL00010973
Chapelle, F. H., and Lovley, D. R. (1992). Competitive exclusion of sulfate reduction by Fe(III)-reducing bacteria: a mechanism for producing discrete zones of high-iron ground water. Ground Water 30, 1–8. doi: 10.1111/j.1745-6584.1992.tb00808.x
Chi Fru, E., and Athar, R. (2008). In situ bacterial colonization of compacted bentonite under deep geological high-level radioactive waste repository conditions. Appl. Microbiol. Biotechnol. 79, 499–510. doi: 10.1007/s00253-008-1436-z
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cline, M. S., Smoot, M., Cerami, E., Kuchinsky, A., Landys, N., Workman, C., et al. (2007). Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382. doi: 10.1038/nprot.2007.324
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dawid, W. (2000). Biology and global distribution of myxobacteria in soils. FEMS Microbiol. Rev. 24, 403–427. doi: 10.1111/j.1574-6976.2000.tb00548.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Demirel, B., and Scherer, P. (2008). The roles of acetotrophic and hydrogenotrophic methanogens during anaerobic conversion of biomass to methane: a review. Rev. Environ. Sci. Biotechnol. 7, 173–190. doi: 10.1007/s11157-008-9131-1
Detmers, J., Bruchert, V., Habicht, K. S., and Kuever, J. (2001). Diversity of sulfur isotope fractionations by sulfate-reducing prokaryotes. Appl. Environ. Microbiol. 67, 888–894. doi: 10.1128/AEM.67.2.888-894.2001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dojka, M. A., Hugenholtz, P., Haack, S. K., and Pace, N. R. (1998). Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64, 3869–3877.
Donlan, R. M. (2002). Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8, 881–890. doi: 10.3201/eid0809.020063
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Elshahed, M. S., Senko, J. M., Najar, F. Z., Kenton, S. M., Roe, B. A., Dewers, T. A., et al. (2003). Bacterial diversity and sulfur cycling in a mesophilic sulfide-rich spring. Appl. Environ. Microbiol. 69, 5609–5621. doi: 10.1128/AEM.69.9.5609-5621.2003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ennis-King, J., Dance, T., Xu, J., Boreham, C., Freifeld, B., Jenkins, C., et al. (2011). The role of heterogeneity in CO2 storage in a depleted gas field: history matching of simulation models to field data for the CO2CRC Otway Project, Australia. Energy Procedia 4, 3494–3501. doi: 10.1016/j.egypro.2011.02.276
Fredrickson, J. K., and Onstott, T. C. (1996). Microbes deep inside the Earth. Sci. Am. 275, 68–73. doi: 10.1038/scientificamerican1096-68
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Freifeld, B. (2009). The U-tube: a new paradigm for borehole fluid sampling. Sci. Drill. 8, 41–45. doi: 10.2204/iodp.sd.8.07.2009
Freifeld, B. M., Trautz, R. C., Kharaka, Y. K., Phelps, T. J., Myer, L. R., Hovorka, S. D., et al. (2005). The U-tube: a novel system for acquiring borehole fluid samples from a deep geologic CO2 sequestration experiment. J. Geophys. Res. 110:B10203. doi: 10.1029/2005JB003735
Friedrich, C. G., Bardischewsky, F., Rother, D., Quentmeier, A., and Fischer, J. (2005). Prokaryotic sulfur oxidation. Curr. Opin. Microbiol. 8, 253–259. doi: 10.1016/j.mib.2005.04.005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gadd, G. M. (2010). Metals, minerals and microbes: geomicrobiology and bioremediation. Microbiology 156, 609–643. doi: 10.1099/mic.0.037143-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Geets, J., Borremans, B., Diels, L., Springael, D., Vangronsveld, J., van der Lelie, D., et al. (2006). DsrB gene-based DGGE for community and diversity surveys of sulfate-reducing bacteria. J. Microbiol. Methods 66, 194–205. doi: 10.1016/j.mimet.2005.11.002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gibbins, J., and Chalmers, H. (2008). Carbon capture and storage. Energy Policy 36, 4317–4322. doi: 10.1016/j.enpol.2008.09.058
Godsy, E. M., Goerlitz, D. F., and Grbic-Galic, D. (1992). Methanogenic biodegradation of creosote contaminants in natural and simulated ground-water ecosystems. Ground Water 30, 232–242. doi: 10.1111/j.1745-6584.1992.tb01795.x
Golby, S., Ceri, H., Gieg, L. M., Chatterjee, I., Marques, L. L. R., and Turner, R. J. (2011). Evaluation of microbial biofilm communities from an Alberta oil sands tailings pond. FEMS Microbiol. Ecol. 79, 240–250. doi: 10.1111/j.1574-6941.2011.01212.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Habicht, K. S., and Canfield, D. (1997). Sulfur isotope fractionation during bacterial sulfate reduction in organic-rich sediments. Geochim. Cosmochim. Acta 61, 5351–5361. doi: 10.1016/S0016-7037(97)00311-6
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Haese, R. R., LaForce, T., Boreham, C., Ennis-King, J., Freifeld, B. M., Paterson, L., et al. (2013). Determining residual CO2 saturation through a dissolution test—results from the CO2CRC Otway Project. Energy Procedia 37, 5379–5386. doi: 10.1016/j.egypro.2013.06.456
Handelsman, J. (2004). Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68, 669–685. doi: 10.1128/MMBR.68.4.669-685.2004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Harrison, A. P. J. (1984). The acidophilic thiobacilli and other acidophilic bacteria that share their habitat. Annu. Rev. Microbiol. 38, 265–292. doi: 10.1146/annurev.mi.38.100184.001405
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Harvey, O. R., Qafoku, N. P., Cantrell, K. J., Lee, G., Amonette, J. E., and Brown, C. F. (2013). Geochemical implications of gas leakage associated with geologic CO2 storage—a qualitative review. Environ. Sci. Technol. 47, 23–36. doi: 10.1021/es3029457
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Heimann, A., Jakobsen, R., and Blodau, C. (2010). Energetic constraints on H2-dependent terminal electron accepting processes in anoxic environments: a review of observations and model approaches. Environ. Sci. Technol. 44, 24–33. doi: 10.1021/es9018207
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hickey, R. F., and Switzenbaum, M. S. (1990). Behavior of carbon monoxide as a trace component of anaerobic digester gases and methanogenesis from acetate. Environ. Sci. Technol. 24, 1642–1648. doi: 10.1021/es00081a003
Hoehler, T. M., Alperin, M. J., Albert, D. B., and Martens, C. S. (1998). Thermodynamic control of hydrogen concentrations in anoxic sediments. Geochim. Cosmochim. Acta 62, 1745–1756. doi: 10.1016/S0016-7037(98)00106-9
Jenkins, C. R., Cook, P. J., Ennis-King, J., Underschultz, J., Boreham, C., Dance, T., et al. (2012). Safe storage and effective monitoring of CO2 in depleted gas fields. Proc. Natl. Acad. Sci. U.S.A. 109, E35–E41. doi: 10.1073/pnas.1107255108
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Joshi, M. N., Dhebar, S. V., Dhebar, S. V., Bhargava, P., Pandit, A., Patel, R. P., et al. (2014). Metagenomics of petroleum muck: revealing microbial diversity and depicting microbial syntrophy. Arch. Microbiol. 196, 531–544. doi: 10.1007/s00203-014-0992-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kharaka, Y. K., Cole, D. R., Hovorka, S. D., Gunter, W., Knauss, K. G., and Freifeld, B. M. (2006). Gas-water-rock interactions in frio formation following CO2 injection: implications for the storage of greenhouse gases in sedimentary basins. Geology 34, 577–580. doi: 10.1130/G22357.1
Kimes, N. E., Callaghan, A. V., Aktas, D. F., Smith, W. L., Sunner, J., Golding, B. T., et al. (2013). Metagenomic analysis and metabolite profiling of deep-sea sediments from the Gulf of Mexico following the Deepwater Horizon oil spill. Front. Microbiol. 4:50. doi: 10.3389/fmicb.2013.00050
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kolbel-Boelke, J., Anders, E. M., and Nehkorn, A. (1988). Microbial communities in the saturated groundwater environment II: diversity of bacterial communities in a pleistocene sand aquifer and their in vitro activities. Microb. Ecol. 16, 31–48. doi: 10.1007/BF02097403
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Krzycki, J. A., and Zeikus, J. G. (1984). Acetate catabolism by Methanosarcina barkeri: hydrogen-dependent methane production from acetate by a soluble cell protein fraction. FEMS Microbiol. Lett. 25, 27–32. doi: 10.1111/j.1574-6968.1984.tb01369.x
Lashof, D. A., and Ahuja, D. R. (1990). Relative contributions of greenhouse gas emissions to global warming. Nature 344, 529–531. doi: 10.1038/344529a0
Lasken, R. S. (2012). Genomic sequencing of uncultured microorganisms from single cells. Nat. Rev. Microbiol. 10, 631–640. doi: 10.1038/nrmicro2857
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lau, M. C. Y., Cameron, C., Magnabosco, C., Brown, C. T., Schilkey, F., Grim, S., et al. (2014). Phylogeny and phylogeography of functional genes shared among seven terrestrial subsurface metagenomes reveal N-cycling and microbial evolutionary relationships. Front. Microbiol. 5:531. doi: 10.3389/fmicb.2014.00531
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lavalleur, H. J., and Colwell, F. S. (2013). Microbial characterization of basalt formation waters targeted for geological carbon sequestration. FEMS Microbiol. Ecol. 85, 62–73. doi: 10.1111/1574-6941.12098
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Li, Z., Dong, M., Li, S., and Huang, S. (2006). CO2 sequestration in depleted oil and gas reservoirs—caprock characterization and storage capacity. Energy Convers. Manage. 47, 1372–1382. doi: 10.1016/j.enconman.2005.08.023
Loreau, M., Naeem, S., Inchausti, P., Bengtsson, J., Grime, J. P., Hector, A., et al. (2001). Biodiversity and ecosystem functioning: current knowledge and future challenges. Science 294, 804–808. doi: 10.1126/science.1064088
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lovley, D. R., and Klug, M. J. (1983). Sulfate reducers can outcompete methanogens at freshwater sulfate concentrations. Appl. Environ. Microbiol. 45, 187–192.
Lovley, D. R., and Phillips, E. J. P. (1987). Competitive mechanisms for inhibition of sulfate reduction and methane production in the zone of ferric iron reduction in sediments. Appl. Environ. Microbiol. 53, 2636–2641.
Loy, A., Kusel, K., Lehner, A., Drake, H. L., and Wagner, M. (2004). Microarray and functional gene analyses of sulfate-reducing prokaryotes in low-sulfate, acidic fens reveal cooccurrence of recognized genera and novel lineages. Appl. Environ. Microbiol. 70, 6998–7009. doi: 10.1128/AEM.70.12.6998-7009.2004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
MacDonald, T. R., Kitanidis, P. K., McCarty, P. L., and Roberts, P. V. (1999). Effects of shear detachment on biomass growth and in situ bioremediation. Ground Water 4, 555–563. doi: 10.1111/j.1745-6584.1999.tb01142.x
Meyer, B., and Kuever, J. (2007). Molecular analysis of the diversity of sulfate-reducing and sulfur-oxidizing prokaryotes in the environment, using aprA as functional marker gene. Appl. Environ. Microbiol. 73, 7664–7679. doi: 10.1128/AEM.01272-07
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mitchell, A. C., Phillips, A. J., Hiebert, R., Gerlach, R., Spangler, L. H., and Cunningham, A. B. (2009). Biofilm enhanced geologic sequestration of supercritical CO2. Int. J. Greenhouse Gas Control 3, 90–99. doi: 10.1016/j.ijggc.2008.05.002
Mitchell, A. C., Phillips, A. J., Hamilton, M. A., Gerlach, R., Hollis, W. K., Kaszuba, J. P., et al. (2008). Resilience of planktonic and biofilm cultures to supercritical CO2. J. Supercrit. Fluids 47, 318–325. doi: 10.1016/j.supflu.2008.07.005
Morales, S. E., and Holben, W. E. (2014). Simulated geologic carbon storage leak reduces bacterial richness and alters bacterial community composition in surface soil. Soil Biol. Biochem. 76, 286–296. doi: 10.1016/j.soilbio.2014.05.018
Moreau, J. W., Zierenberg, R. A., and Banfield, J. F. (2010). Diversity of dissimilatory sulfite reductase genes (dsrAB) in a salt marsh impacted by long-term acid mine drainage. Appl. Environ. Microbiol. 76, 4819–4828. doi: 10.1128/AEM.03006-09
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Morozova, D., Zettlitzer, M., Let, D., and Würdemann, H. (2011). Monitoring of the microbial community composition in deep subsurface saline aquifers during CO2 storage in Ketzin, Germany. Energy Procedia 4, 4362–4370. doi: 10.1016/j.egypro.2011.02.388
Mu, A., Boreham, C., Leong, H. X., Haese, R. R., and Moreau, J. W. (2014). Changes in the deep subsurface microbial biosphere resulting from a field-scale CO2 geosequestration experiment. Front. Microbiol. 5:209. doi: 10.3389/fmicb.2014.00209
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nazina, T. N., Kosareva, I. M., Petrunyaka, V. V., Savushkina, M. K., Kudriavtsev, E. G., Lebedev, V. A., et al. (2004). Microbiology of formation waters from the deep repository of liquid radioactive wastes Severnyi. FEMS Microbiol. Ecol. 49, 97–107. doi: 10.1016/j.femsec.2004.02.017
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nealson, K. H., and Saffarini, D. (1994). Iron and manganese in anaerobic respiration: environmental significance physiology and regulation. Annu. Rev. Microbiol. 48, 311–343. doi: 10.1146/annurev.mi.48.100194.001523
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nghiem, L., Yang, C., Shrivastava, V., Kohse, B., Hassam, M., and Card, C. (2009). Risk mitigation through the optimization of residual gas and solubility trapping for CO2 storage in saline aquifers. Energy Procedia 1, 3015–3022. doi: 10.1016/j.egypro.2009.02.079
Noble, R. R. P., Stalker, L., Wakelin, S. A., Pejcic, B., Leybourne, M. I., Hortle, A. L., et al. (2012). Biological monitoring for carbon capture and storage—a review and potential future developments. Int. J. Greenhouse Gas Control 10, 520–535. doi: 10.1016/j.ijggc.2012.07.022
Oppermann, B. I., Michaelis, W., Blumenberg, M., Frerichs, J., Schulz, H. M., Schippers, A., et al. (2010). Soil microbial community changes as a result of long-term exposure to a natural CO2 vent. Geochim. Cosmochim. Acta 74, 2697–2716. doi: 10.1016/j.gca.2010.02.006
Orsi, W., Biddle, J. F., and Edgcomb, V. (2013). Deep sequencing of subseafloor eukaryotic rRNA reveals active fungi across marine subsurface provinces. PLoS ONE 8:e56335. doi: 10.1371/journal.pone.0056335
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Osaka, T., Ebie, Y., Tsuneda, S., and Inamori, Y. (2008). Identification of the bacterial community involved in methane-dependent denitrification in activated sludge using DNA stable-isotope probing. FEMS Microbiol. Ecol. 64, 494–506. doi: 10.1111/j.1574-6941.2008.00473.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Paterson, L., Boreham, C., Bunch, M., Dance, T., Ennis-King, J., Freifeld, B., et al. (2013). Overview of the CO2CRC Otway residual saturation and dissolution test. Energy Procedia 37, 6140–6148. doi: 10.1016/j.egypro.2013.06.543
Petchey, O. L., and Gaston, K. J. (2006). Functional diversity: back to basics and looking forward. Ecol. Lett. 9, 741–758. doi: 10.1111/j.1461-0248.2006.00924.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Phillips, A. J., Lauchnor, E., Eldring, J. J., Esposito, R., Mitchell, A. C., Gerlach, R., et al. (2013). Potential CO2 leakage reduction through biofilm-induced calcium carbonate precipitation. Environ. Sci. Technol. 47, 142–149. doi: 10.1021/es301294q
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Poser, A., Vogt, C., Knöller, K., Ahlheim, J., Weiss, H., Kleinsteuber, S., et al. (2014). Stable sulfur and oxygen isotope fractionation of anoxic sulfide oxidation by two different enzymatic pathways. Environ. Sci. Technol. 48, 9094–9102. doi: 10.1021/es404808r
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Raes, J., Korbel, J. O., Lercher, M. J., von Mering, C., and Bork, P. (2007). Prediction of effective genome size in metagenomic samples. Genome Biol. 8:R10. doi: 10.1186/gb-2007-8-1-r10
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ragsdale, S. W. (2004). Life with carbon monoxide. Crit. Rev. Biochem. Mol. Biol. 39, 165–195. doi: 10.1080/10409230490496577
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rappé, M. S., and Giovannoni, S. J. (2003). The uncultured microbial majority. Annu. Rev. Microbiol. 57, 369–394. doi: 10.1146/annurev.micro.57.030502.090759
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rédou, V., Ciobanu, M. C., Pachiadaki, M. G., Edgcomb, V., Alain, K., Barbier, G., et al. (2014). In-depth analyses of deep subsurface sediments using 454-pyrosequencing reveals a reservoir of buried fungal communities at record-breaking depths. FEMS Microbiol. Ecol. 90, 908–921. doi: 10.1111/1574-6941.12447
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Réveillère, A., and Rohmer, J. (2011). Managing the risk of CO2 leakage from deep saline aquifer reservoirs through the creation of a hydraulic barrier. Energy Procedia 4, 3187–3194. doi: 10.1016/j.egypro.2011.02.234
Saadatpoor, E., Bryant, S. L., and Sepehrnoori, K. (2009). New trapping mechanism in carbon sequestration. Transp. Porous Med. 82, 3–17. doi: 10.1007/s11242-009-9446-6
Saito, R., Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P.-L., Lotia, S., et al. (2012). A travel guide to Cytoscape plugins. Nat. Methods 9, 1069–1076. doi: 10.1038/nmeth.2212
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sakai, S., Ehara, M., Tseng, I. C., Yamaguchi, T., Brauer, S. L., Cadillo-Quiroz, H., et al. (2012). Methanolinea mesophila sp. nov., a hydrogenotrophic methanogen isolated from rice field soil, and proposal of the archaeal family Methanoregulaceae fam. nov. within the order Methanomicrobiales. Int. J. Syst. Evol. Microbiol. 62, 1389–1395. doi: 10.1099/ijs.0.035048-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sato, K., Kawaguchi, H., and Kobayashi, H. (2013). Bio-electrochemical conversion of carbon dioxide to methane in geological storage reservoirs. Energy Convers. Manage. 66, 343–350. doi: 10.1016/j.enconman.2012.12.008
Schacht, U., Regan, M., Boreham, C., and Sharma, S. (2011). CO2CRC Otway project–soil gas baseline and assurance monitoring 2007–2010. Energy Procedia 4, 3346–3353. doi: 10.1016/j.egypro.2011.02.256
Schloss, P. D., and Handelsman, J. (2004). Status of the microbial census. Microbiol. Mol. Biol. Rev. 68, 686–691. doi: 10.1128/MMBR.68.4.686-691.2004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schonheit, P., Kristjansson, J. K., and Thauer, R. K. (1982). Kinetic mechanism for the ability of sulfate reducers to out-compete methanogens for acetate. Arch. Microbiol. 132, 285–288. doi: 10.1007/BF00407967
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shamshiri, H., and Jafarpour, B. (2012). Controlled CO2 injection into heterogeneous geologic formations for improved solubility and residual trapping. Water Resour. Res. 48:W02530. doi: 10.1029/2011WR010455
Sharma, S., Cook, P., Berly, T., and Lees, M. (2009). The CO2CRC Otway project: overcoming challenges from planning to execution of Australia's first CCS project. Energy Procedia 1, 1965–1972. doi: 10.1016/j.egypro.2009.01.256
Shindell, D. T., Faluvegi, G., Koch, D. M., Schmidt, G. A., Unger, N., and Bauer, S. E. (2009). Improved attribution of climate forcing to emissions. Science 326, 716–718. doi: 10.1126/science.1174760
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sinclair, J. L., and Ghiorse, W. C. (1989). Distribution of aerobic bacteria, protozoa, algae, and fungi in deep subsurface sediments. Geomicrobiol. J. 7, 15–31. doi: 10.1080/01490458909377847
Slobodkin, A. I., Jeanthon, C., Haridon, S. L., Nazina, T., Miroshnichenko, M., and Bonch-Osmolovskaya, E. (1999). Dissimilatory reduction of Fe(III) by thermophilic bacteria and archaea in deep subsurface petroleum reservoirs of Western Siberia. Curr. Microbiol. 39, 99–102. doi: 10.1007/s002849900426
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Soh, J., Dong, X., Caffrey, S. M., Voordouw, G., and Sensen, C. W. (2013). Phoenix 2: a locally installable large-scale 16S rRNA gene sequence analysis pipeline with web interface. J. Biotechnol. 167, 393–403. doi: 10.1016/j.jbiotec.2013.07.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Stalker, L., Boreham, C., Underschultz, J., Freifeld, B., Perkins, E., Schacht, U., et al. (2009). Geochemical monitoring at the CO2CRC Otway project: tracer injection and reservoir fluid acquisition. Energy Procedia 1, 2119–2125. doi: 10.1016/j.egypro.2009.01.276
Stevens, T. O., and McKinley, J. P. (1995). Lithoautotrophic microbial ecosystems in deep basalt aquifers. Science 270, 450–454. doi: 10.1126/science.270.5235.450
Stevens, T. O., McKinley, J. P., and Fredrickson, J. K. (1993). Bacteria assocaited with deep, alkaline, anaerobic groundwaters in Southeast Washington. Microb. Ecol. 25, 35–50. doi: 10.1007/BF00182128
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Techtmann, S. M., Colman, A. S., and Robb, F. T. (2009). “That which does not kill us only makes us stronger”: the role of carbon monoxide in thermophilic microbial consortia. Environ. Microbiol. 11, 1027–1037. doi: 10.1111/j.1462-2920.2009.01865.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Thamdrup, B., Fossing, H., and Jorgensen, B. B. (1994). Manganese, iron, and sulfur cycling in a coastal marine sediment, Aarhus Bay, Denmark. Geochim. Cosmochim. Acta 58, 5115–5129. doi: 10.1016/0016-7037(94)90298-4
Tilman, D. (1999). The ecological consequences of changes in biodiversity: a search for general principles. Ecology 80, 1455–1474. doi: 10.2307/176540
Underschultz, J., Boreham, C., Dance, T., Stalker, L., Freifeld, B., Kirste, D., et al. (2011). CO2 storage in a depleted gas field: an overview of the CO2CRC Otway project and initial results. Int. J. Greenhouse Gas Control 5, 922–932. doi: 10.1016/j.ijggc.2011.02.009
Vairavamurthy, A., Manowitz, B., Luther, G. W. III, and Jeon, Y. (1993). Oxidation state of sulfur in thiosulfate and implications for anaerobic energy metabolism. Geochim. Cosmochim. Acta 57, 1619–1623. doi: 10.1016/0016-7037(93)90020-W
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wilkins, M. J., Hoyt, D. W., Marshall, M. J., Alderson, P. A., Plymale, A. E., Markillie, L. M., et al. (2014). CO2 exposure at pressure impacts metabolism and stress responses in the model sulfate-reducing bacterium Desulfovibrio vulgaris strain Hildenborough. Front. Microbiol. 5:507. doi: 10.3389/fmicb.2014.00507
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Winograd, I. J., and Robertson, F. N. (1982). Deep oxygenated ground water: anomaly or common occurrence. Science 216, 1227–1230. doi: 10.1126/science.216.4551.1227
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, Y., Freifeld, B., Finsterle, S., Leahy, M., Ennis-King, J., Paterson, L., et al. (2011). Estimating CO2 residual trapping from a single-well test: experimental design calculations. Energy Procedia 4, 5044–5049. doi: 10.1016/j.egypro.2011.02.477
Zhou, X. W., Li, S. G., Li, W., Jiang, D. M., Han, K., Wu, Z. H., et al. (2014). Myxobacterial community is a predominant and highly diverse bacterial group in soil niches. Environ. Microbiol. Rep. 6, 45–56. doi: 10.1111/1758-2229.12107
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: CO2 geosequestration, deep subsurface, microbial response, CODH, systems biology, network analysis, methanogenesis, sulfur cycling
Citation: Mu A and Moreau JW (2015) The geomicrobiology of CO2 geosequestration: a focused review on prokaryotic community responses to field-scale CO2 injection. Front. Microbiol. 6:263. doi: 10.3389/fmicb.2015.00263
Received: 26 November 2014; Accepted: 16 March 2015;
Published: 09 April 2015.
Edited by:
Lisa Y. Stein, University of Alberta, CanadaReviewed by:
Virginia P. Edgcomb, Woods Hole Oceanographic Institution, USAPeter Croot, National University of Ireland, Galway, Ireland
Jim K. Fredrickson, Pacific Northwest National Laboratory, USA
Copyright © 2015 Mu and Moreau. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence:am1vcmVhdUB1bmltZWxiLmVkdS5hdQ==
 Andre Mu
Andre Mu John W. Moreau
John W. Moreau