- 1Computational and Quantitative Biology, Sorbonne Universités, UPMC Univ Paris 06, UMR 7238, Paris, France
- 2Centre National de la Recherche Scientifique, LBPA, UMR 8113, ENS Cachan, Cachan, France
- 3Centre National de la Recherche Scientifique, UMR 7238, Paris, France
Science has a close but very complex relationship with technology (Latour, 1987). A simple phenomenon is that technology enables science by offering tools that provide new data or new kinds of data. In other cases, aspects or views of the empirical world may remain invisible until technology builds something that unveils them to the eyes of the scientific community. On a deeper level, building something may be a form of understanding. For example “complex networks” became prominent in all sectors of science in the late 1990s, at the time that the Internet became a common tool for research and for society at large. Before then, networks had been restricted for decades to smaller niches. This change was accompanied by a thrust of high throughput technologies to collect new data, but arguably many of the “network” data had already been available for many years.
On a smaller scale, we want to suggest here that so called “smart polymers” (Galaev and Mattiasson, 1999; Kumar et al., 2007) could be a promising technological metaphor for the behavior of the bacterial nucleoid. We want to explore the analogy with the similarly “intelligent” behavior shaped into bacterial nucleoids by natural selection.
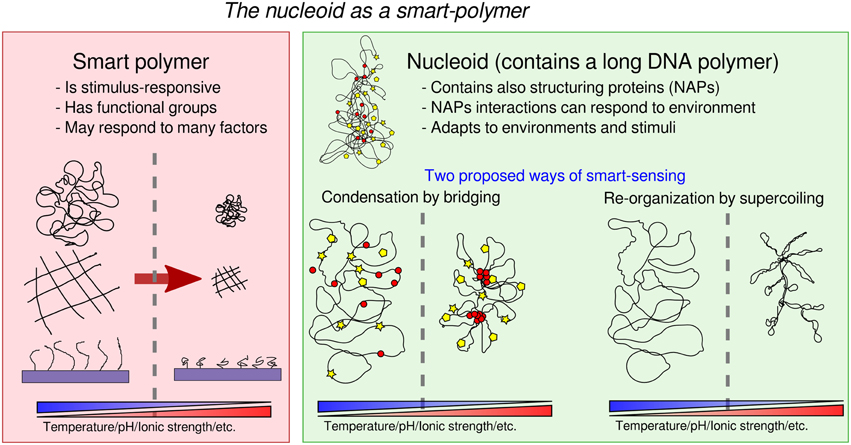
But first, what is a smart polymer, and what does it do? In soft-matter physics, “smart,” or “stimulus-responsive,” polymers are technological polymer systems designed to effect a variety of responsive behaviors to external stimuli (Figure 1). Smart polymers respond to the environment they are in. They are engineered to be sensitive to a number of factors, such as solvency, temperature, humidity, pH, light, electrical and magnetic field, and to effect mechanical and chemical changes (Galaev and Mattiasson, 1999; Kumar et al., 2007; Chen and Chang, 2014). They can be realized as linear free chains in solution, or as surface-grafted brushes or gels. Usually, response to stimuli is achieved through the addition of specific reactive functional groups and side chains, or by the use of graft-and-block copolymers (two different polymers grafted together) with different chemical properties (e.g., hydrophyly). Effective smart polymers typically undergo large changes (e.g., conformational transitions) in response to just small changes in the environment (e.g., pH, temperature, ionic strength). One way to achieve this behavior is through the introduction of “pre-programmed” phase transitions. For example, the polymer undergoes a reversible collapse after an external stimulus is applied. The reversibility of this change may also be an important property, allowing to detect changes in both directions. To fix the ideas, a prevalent use for smart polymers is targeted drug delivery. A smart-polymer system may control the release of drugs until the desired target is reached, and it is sensed by either a chemical or physical response triggering the release of the drug by “uncaging” it. For example, a polymer site-specific conjugation to specific amino acid sites may induce a trigger in the concentration of a targeted protein (Hoffman et al., 2000). It is then evident that the bacterial nucleoid can be seen as a smart polymer (Dillon and Dorman, 2010; Muskhelishvili et al., 2010; Benza et al., 2012; Kleckner et al., 2014). Its degree of compaction and conformation are modulated by the cell's growth conditions and in response to specific external cues (Figure 1). It is a complex system made of a long DNA polymer associated with RNA and proteins that may play at least two roles: adapt the shape of the nucleoid through both specific and non-specific DNA binding, and change the physical properties of DNA through dynamic changes in DNA topology.
First, the abundant nucleoid associated proteins (“NAPs,” e.g., focusing on E. coli, Dps, Fis, H-NS, IHF, HU, and the condensin MukBEF), can act as “functional groups” (Luijsterburg et al., 2006; Dillon and Dorman, 2010; Ohniwa et al., 2011) to plastically modify the genome conformation. Of particular interest are NAP-mediated bridging interactions (Wiggins et al., 2009) (e.g., from Fis, H-NS, and MukBEF in E. coli), which can thus act as “functional groups” in the nucleoid. In particular, H-NS is known respond to temperature, salt concentration and pH (La Teana et al., 1991; Atlung and Ingmer, 1997; Amit et al., 2003; Dorman, 2004; Ono et al., 2005; Stella et al., 2006). and Fis has been implicated in adaptation to favorable growth conditions and quorum sensing (Lenz and Bassler, 2007). Additionally, some NAPs may operate both as monomers and as oligomers, introducing the possibility of cooperativity in the formation of higher order complexes (Luijsterburg et al., 2006; Skoko et al., 2006; Lim et al., 2012). On theoretical grounds, looped domain formation offers the opportunity of producing a very rich phase behavior (Leibler, 1980; Borisov and Halperin, 1996, 1997; Kantor and Kardar, 1996; Camacho and Schanke, 1997), as exploited in recent models motivated by the study of the organization of chromatin (Junier et al., 2010; Barbieri et al., 2013; Brackley et al., 2013). Biologically, one can imagine that the collapse and swelling of selected genomic regions by bridging proteins may be tuned to be switch-like (Scolari and Cosentino Lagomarsino, 2015) in order to be differentially controlled by the cell. While the mechanisms has not yet been studied in detail, domain formation is well-documented in bacterial chromosomes (Espéli and Boccard, 2006; Espéli et al., 2008; Dame et al., 2011). Such mechanism may account for the observed correlation between the position of genetic loci along the chromosome and their position in the cell (Mercier et al., 2008; Wiggins et al., 2010), it may help the resolution of the identity of segregating sister chromosomes (Lesterlin et al., 2012; Junier et al., 2014), as well as play a role in explaining observed “abrupt” transitions in chromosome arrangements (Joshi et al., 2011; Fisher et al., 2013; Javer et al., 2014). Additionally, NAPs that do not bridge specifically such as Dps may also trigger switch-like collapse (Zimmerman, 2006), and NAPs that do not bridge but exhibit cooperative clustering may also affect the global nucleoid state by affecting key parameters such as effective stiffness (Luijsterburg et al., 2006).
Second, the action of specific DNA enzymes such as topoisomerases and gyrases changes the polymer's mechanical properties through changes in DNA topology. Nucleoids are composed of topologically unlinked dynamic domain structures, forming plectonemes and toroids (Trun and Marko, 1998). Torsional constraints can be generated by active processes, such as DNA replication and transcription (Le et al., 2013), and stabilized by bridging NAPs, such as Fis and H-NS (Schneider et al., 2001). Together, supercoiling and nucleoid organization can affect gene expression (Breier and Cozzarelli, 2004; Postow et al., 2004; Travers and Muskhelishvili, 2005; Blot et al., 2006; Dillon and Dorman, 2010) and, in turn, expression of specific regulators may affect the concentration or the activity of the genes setting nucleoid conformation resulting in feedback loops that can lead to more robust nucleoid conformations. Also, NAPs and supercoiling regulation by enzymes may interact in complex ways (Dorman, 2013a). Clearly, such an object has higher computational power than any current technological smart polymer, because it is also able to control the elements leading to its self-organization, which may inspire new technology. The coexistence of two parallel mechanisms of regulation through polymer organization (mainly supercoiling and growth) and through conventional protein binding may be an important feature of the nucleoid. A series of studies on E. coli investigated the interactions between these mechanisms arguing the presence of two different codes overlapped at different at levels on DNA, and possibly evolving at different time-scales, carrying, respectively, a “digital” and an “analog” information (Sobetzko et al., 2012; Dorman, 2013b; Muskhelishvili and Travers, 2013; Sobetzko et al., 2013). Finally this system is able to rapidly evolve in response to adaptation to recurring changes (Crozat et al., 2010), possibly improving the efficiency and the speed of the programmed conformational changes.
We propose that this technological parallel could also be useful in the reverse direction, to reframe the current biological knowledge in a physical perspective. Indeed, the smart polymer analogy does not by itself add new knowledge to the long list of biological information already acquired on the nucleoid. However, it may help us putting the same knowledge in a different perspective, and treat the same information in more precise and quantitative ways using the tools of soft-matter physics. This may lead to defining new questions, and ultimately to reaching new knowledge. For example, new biomimetic “constructive” approaches using purified DNA and NAPs may be defined to explore the resulting phase diagram in a controlled fashion (Maurer et al., 2009; Pelletier et al., 2012; Thacker et al., 2014), and to achieve a physical understanding of how robustness and response to changes are encoded in such structures.
Funding
This work was supported by the International Human Frontier Science Program Organization, grants RGY0069/2009-C and RGY0070/2014.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Amit, R., Oppenheim, A. B., and Stavans, J. (2003). Increased bending rigidity of single DNA molecules by h-ns, a temperature and osmolarity sensor. Biophys. J. 84, 2467–2473. doi: 10.1016/S0006-3495(03)75051-6
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Atlung, T., and Ingmer, H. (1997). H-ns: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24, 7–17. doi: 10.1046/j.1365-2958.1997.3151679.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Barbieri, M., Chotalia, M., Fraser, J., Lavitas, L.-M., Dostie, J., Pombo, A., et al. (2013). A model of the large-scale organization of chromatin. Biochem. Soc. Trans. 41, 508–512. doi: 10.1042/BST20120238
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Benza, V. G., Bassetti, B., Dorfman, K. D., Scolari, V. F., Bromek, K., Cicuta, P., et al. (2012). Physical descriptions of the bacterial nucleoid at large scales, and their biological implications. Rep. Prog. Phys. 75:076602. doi: 10.1088/0034-4885/75/7/076602
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Blot, N., Mavathur, R., Geertz, M., Travers, A., and Muskhelishvili, G. (2006). Homeostatic regulation of supercoiling sensitivity coordinates transcription of the bacterial genome. Embo. Rep. 7, 710–715. doi: 10.1038/sj.embor.7400729
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Borisov, O. V., and Halperin, A. (1996). Micelles of polysoaps: the role of bridging interactions. Macromolecules 29, 2612–2617. doi: 10.1021/ma951565w
Borisov, O. V., and Halperin, A. (1997). Polysoaps: the signatures of intrachain self assembly in solvents. Macromol. Symp. 117, 99–107. doi: 10.1002/masy.19971170114
Brackley, C. A., Taylor, S., Papantonis, A., Cook, P. R., and Marenduzzo, D. (2013). Nonspecific bridging-induced attraction drives clustering of DNA-binding proteins and genome organization. Proc. Natl. Acad. Sci. U.S.A. 110, E3605–E3611. doi: 10.1073/pnas.1302950110
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Breier, A. M., and Cozzarelli, N. R. (2004). Linear ordering and dynamic segregation of the bacterial chromosome. Proc. Natl. Acad. Sci. U.S.A. 101, 9175–9176. doi: 10.1073/pnas.0403722101
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Camacho, C. J., and Schanke, T. (1997). From collapse to freezing in random heteropolymers. Europhys. Lett. 37, 603. doi: 10.1209/epl/i1997-00197-2
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, J.-K., and Chang, C.-J. (2014). Fabrications and applications of stimulus-responsive polymer films and patterns on surfaces: a review. Materials 7, 805–875. doi: 10.3390/ma7020805
Crozat, E., Winkworth, C., Gaffé, J., Hallin, P. F., Riley, M. A., Lenski, R. E., et al. (2010). Parallel genetic and phenotypic evolution of DNA superhelicity in experimental populations of Escherichia coli. Mol. Biol. Evol. 27, 2113–2128. doi: 10.1093/molbev/msq099
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dame, R. T., Kalmykowa, O. J., and Grainger, D. C. (2011). Chromosomal macrodomains and associated proteins: implications for DNA organization and replication in gram negative bacteria. PLoS Genet. 7:e1002123. doi: 10.1371/journal.pgen.1002123
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dillon, S. C., and Dorman, C. J. (2010). Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195. doi: 10.1038/nrmicro2261
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dorman, C. J. (2004). H-ns: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2, 391–400. doi: 10.1038/nrmicro883
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dorman, C. J. (2013a). Co-operative roles for DNA supercoiling and nucleoid-associated proteins in the regulation of bacterial transcription. Biochem. Soc. Trans. 41, 542–547. doi: 10.1042/BST20120222
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dorman, C. J. (2013b). Genome architecture and global gene regulation in bacteria: making progress towards a unified model? Nat. Rev. Microbiol. 11, 349–355. doi: 10.1038/nrmicro3007
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Espéli, O., and Boccard, F. (2006). Organization of the Escherichia coli chromosome into macrodomains and its possible functional implications. J. Struct. Biol. 156, 304–310. doi: 10.1016/j.jsb.2006.07.010
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Espéli, O., Mercier, R., and Boccard, F. (2008). DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol. Microbiol. 68, 1418–1427. doi: 10.1111/j.1365-2958.2008.06239.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fisher, J. K., Bourniquel, A., Witz, G., Weiner, B., Prentiss, M., and Kleckner, N. (2013). Four-dimensional imaging of E. coli nucleoid organization and dynamics in living cells. Cell 153, 882–895. doi: 10.1016/j.cell.2013.04.006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Galaev, I. Y., and Mattiasson, B. (1999). ‘Smart’ polymers and what they could do in biotechnology and medicine. Trends Biotechnol. 17, 335–340. doi: 10.1016/S0167-7799(99)01345-1
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hoffman, A. S., Stayton, P. S., Bulmus, V., Chen, G., Chen, J., Cheung, C., et al. (2000). Really smart bioconjugates of smart polymers and receptor proteins. J. Biomed. Mater. Res. 52, 577–586. doi: 10.1002/1097-4636(20001215)52:4<577::AID-JBM1>3.0.CO;2-5
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Javer, A., Kuwada, N. J., Long, Z., Benza, V. G., Dorfman, K. D., Wiggins, P. A., et al. (2014). Persistent super-diffusive motion of Escherichia coli chromosomal loci. Nat. Commun. 5:3854. doi: 10.1038/ncomms4854
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Joshi, M. C., Bourniquel, A., Fisher, J., Ho, B. T., Magnan, D., Kleckner, N., et al. (2011). Escherichia coli sister chromosome separation includes an abrupt global transition with concomitant release of late-splitting intersister snaps. Proc. Natl. Acad. Sci. U.S.A. 108, 2765–2770. doi: 10.1073/pnas.1019593108
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Junier, I., Boccard, F., and Espli, O. (2014). Polymer modeling of the E. coli genome reveals the involvement of locus positioning and macrodomain structuring for the control of chromosome conformation and segregation. Nucleic Acids Res. 42, 1461–1473. doi: 10.1093/nar/gkt1005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Junier, I., Martin, O., and Képès, F. (2010). Spatial and topological organization of DNA chains induced by gene co-localization. PLoS Comput. Biol. 6:e1000678. doi: 10.1371/journal.pcbi.1000678
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kantor, Y., and Kardar, M. (1996). Collapse of randomly linked polymers. Phys. Rev. Lett. 77:4275. doi: 10.1103/PhysRevLett.77.4275
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kleckner, N., Fisher, J. K., Stouf, M., White, M. A., Bates, D., and Witz, G. (2014). The bacterial nucleoid: nature, dynamics and sister segregation. Curr. Opin. Microbiol. 22C, 127–137. doi: 10.1016/j.mib.2014.10.001
Kumar, A., Srivastava, A., Galaev, I. Y., and Mattiasson, B. (2007). Smart polymers: physical forms and bioengineering applications. Prog. Polym. Sci. 32, 1205–1237. doi: 10.1016/j.progpolymsci.2007.05.003
La Teana, A., Brandi, A., Falconi, M., Spurio, R., Pon, C. L., and Gualerzi, C. (1991). Identification of a cold shock transcriptional enhancer of the Escherichia coli gene encoding nucleoid protein h-ns. Proc. Natl. Acad. Sci. U.S.A. 88, 10907–10911. doi: 10.1073/pnas.88.23.10907
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Latour, B. (1987). Science in Action: How to Follow Scientists and Engineers through Society. Cambridge, MA: Harvard University Press.
Le, T. B. K., Imakaev, M. V., Mirny, L. A., and Laub, M. T. (2013). High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342, 731–734. doi: 10.1126/science.1242059
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Leibler, L. (1980). Theory of microphase separation in block copolymers. Macromolecules 13, 1602–1617. doi: 10.1021/ma60078a047
Lenz, D. H., and Bassler, B. L. (2007). The small nucleoid protein fis is involved in Vibrio cholerae quorum sensing. Mol. Microbiol. 63, 859–871. doi: 10.1111/j.1365-2958.2006.05545.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lesterlin, C., Gigant, E., Boccard, F., and Espeli, O. (2012). Sister chromatid interactions in bacteria revealed by a site-specific recombination assay. EMBO J. 31, 3468–3479. doi: 10.1038/emboj.2012.194
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lim, C. J., Whang, Y. R., Kenney, L. J., and Yan, J. (2012). Gene silencing h-ns paralogue stpa forms a rigid protein filament along DNA that blocks DNA accessibility. Nucleic Acids Res. 40, 3316–3328. doi: 10.1093/nar/gkr1247
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Luijsterburg, M. S., Noom, M. C., Wuite, G. J., and Dame, R. T. (2006). The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J. Struct. Biol. 156, 262–272. doi: 10.1016/j.jsb.2006.05.006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Maurer, S., Fritz, J., and Muskhelishvili, G. (2009). A systematic in vitro study of nucleoprotein complexes formed by bacterial nucleoid-associated proteins revealing novel types of DNA organization. J. Mol. Biol. 387, 1261–1276. doi: 10.1016/j.jmb.2009.02.050
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mercier, R., Petit, M.-A., Schbath, S., Robin, S., Karoui, M. E., Boccard, F., et al. (2008). The matp/mats site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135, 475–485. doi: 10.1016/j.cell.2008.08.031
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Muskhelishvili, G., Sobetzko, P., Geertz, M., and Berger, M. (2010). General organisational principles of the transcriptional regulation system: a tree or a circle? Mol. BioSyst. 6, 662–676. doi: 10.1039/b909192k
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Muskhelishvili, G., and Travers, A. (2013). Integration of syntactic and semantic properties of the DNA code reveals chromosomes as thermodynamic machines converting energy into information. Cell. Mol. Life Sci. 70, 4555–4567. doi: 10.1007/s00018-013-1394-1
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ohniwa, R. L., Ushijima, Y., Saito, S., and Morikawa, K. (2011). Proteomic analyses of nucleoid-associated proteins in Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis, and Staphylococcus aureus. PLoS ONE 6:e19172. doi: 10.1371/journal.pone.0019172
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ono, S., Goldberg, M. D., Olsson, T., Esposito, D., Hinton, J. C. D., and Ladbury, J. E. (2005). H-ns is a part of a thermally controlled mechanism for bacterial gene regulation. Biochem. J. 391(Pt 2), 203–213. doi: 10.1042/BJ20050453
Pelletier, J., Halvorsen, K., Ha, B.-Y., Paparcone, R., Sandler, S. J., Woldringh, C. L., et al. (2012). Physical manipulation of the Escherichia coli chromosome reveals its soft nature. Proc. Natl. Acad. Sci. U.S.A. 109, E2649–E2656. doi: 10.1073/pnas.1208689109
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Postow, L., Hardy, C., Arsuaga, J., and Cozzarelli, N. (2004). Topological domain structure of the Escherichia coli chromosome. Genes Dev. 18, 1766–1779. doi: 10.1101/gad.1207504
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schneider, R., Lurz, R., Lder, G., Tolksdorf, C., Travers, A., and Muskhelishvili, G. (2001). An architectural role of the Escherichia coli chromatin protein fis in organising DNA. Nucleic Acids Res. 29, 5107–5114. doi: 10.1093/nar/29.24.5107
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Scolari, V. F., and Cosentino Lagomarsino, M. (2015). Combined collapse by bridging and self-adhesion in a prototypical polymer model inspired by the bacterial nucleoid. Soft. Matter. 11, 1677–1687. doi: 10.1039/C4SM02434F
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Skoko, D., Yoo, D., Bai, H., Schnurr, B., Yan, J., McLeod, S. M., et al. (2006). Mechanism of chromosome compaction and looping by the Escherichia coli nucleoid protein fis. J. Mol. Biol. 364, 777–798. doi: 10.1016/j.jmb.2006.09.043
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sobetzko, P., Glinkowska, M., Travers, A., and Muskhelishvili, G. (2013). DNA thermodynamic stability and supercoil dynamics determine the gene expression program during the bacterial growth cycle. Mol. BioSyst. 9, 1643–1651. doi: 10.1039/c3mb25515h
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sobetzko, P., Travers, A., and Muskhelishvili, G. (2012). Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc. Natl. Acad. Sci. U.S.A. 109, E42–E50. doi: 10.1073/pnas.1108229109
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Stella, S., Falconi, M., Lammi, M., Gualerzi, C. O., and Pon, C. L. (2006). Environmental control of the in vivo oligomerization of nucleoid protein h-ns. J. Mol. Biol. 355, 169–174. doi: 10.1073/pnas.1108229109
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Thacker, V. V., Bromek, K., Meijer, B., Kotar, J., Sclavi, B., Cosentino Lagomarsino, M., et al. (2014). Bacterial nucleoid structure probed by active drag and resistive pulse sensing. Integr. Biol. 6, 184–191. doi: 10.1039/c3ib40147b
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Travers, A., and Muskhelishvili, G. (2005). DNA supercoiling - a global transcriptional regulator for enterobacterial growth? Nat. Rev. Microbiol. 3, 157–169. doi: 10.1038/nrmicro1088
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Trun, N. J., and Marko, J. F. (1998). Architecture of a bacterial chromosome (review). Am. Soc. Microbiol. News 64:276.
Wiggins, P. A., Cheveralls, K. C., Martin, J. S., Lintner, R., and Kondev, J. (2010). Strong intranucleoid interactions organize the Escherichia coli chromosome into a nucleoid filament. Proc. Natl. Acad. Sci. U.S.A. 107, 4991–4995. doi: 10.1073/pnas.0912062107
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wiggins, P. A., Dame, R. T., Noom, M. C., and Wuite, G. J. L. (2009). Protein-mediated molecular bridging: a key mechanism in biopolymer organization. Biophys. J. 97, 1997–2003. doi: 10.1016/j.bpj.2009.06.051
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zimmerman, S. B. (2006). Cooperative transitions of isolated Escherichia coli nucleoids: implications for the nucleoid as a cellular phase. J. Struct. Biol. 153, 160–175. doi: 10.1016/j.jsb.2005.10.011
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: genome organization, bacterial nucleoid, nucleoid-associated proteins, supercoiling, smart-polymers, polymer sensors
Citation: Scolari VF, Sclavi B and Cosentino Lagomarsino M (2015) The nucleoid as a smart polymer. Front. Microbiol. 6:424. doi: 10.3389/fmicb.2015.00424
Received: 27 February 2015; Accepted: 21 April 2015;
Published: 08 May 2015.
Edited by:
Conrad L. Woldringh, University of Amsterdam, NetherlandsReviewed by:
Remus T. Dame, Leiden University, NetherlandsGeorgi Muskhelishvili, Jacobs University, Germany
Copyright © 2015 Scolari, Sclavi and Cosentino Lagomarsino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marco Cosentino Lagomarsino,bWFyY28uY29zZW50aW5vLWxhZ29tYXJzaW5vQHVwbWMuZnI=
†Present Address: Vittore F. Scolari, Spatial Regulation of Genomes Group, Department of Genomes and Genetics, Institut Pasteur, Paris, France
 Vittore F. Scolari
Vittore F. Scolari Bianca Sclavi
Bianca Sclavi Marco Cosentino Lagomarsino
Marco Cosentino Lagomarsino