- 1Institute of Infection, Immunity and Inflammation, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow, UK
- 2Michael Smith Laboratories, University of British Columbia, Vancouver, BC, Canada
Enterohaemorrhagic Escherichia coli (EHEC) binds to host tissue and intimately attaches to intestinal cells using a dedicated type III secretion system (T3SS). This complex multi-protein organelle is encoded within a large pathogenicity island called the locus of enterocyte effacement (LEE), which is subject to extensive regulatory control. Over the past 15 years we have gained a wealth of knowledge concerning how the LEE is regulated transcriptionally by specific, global and phage encoded regulators. More recently, significant advances have been made in our understanding of how specific signals, including host or microbiota derived metabolic products and various nutrient sources, can affect how the LEE-encoded T3SS is regulated. In this review we discuss regulation of the LEE, focusing on how these physiologically relevant signals are sensed and how they affect the expression of this major virulence factor. The implications for understanding the disease process by specific regulatory mechanisms are also discussed.
Introduction—Enterohaemorrhagic E. coli O157:H7
Enterohaemorrhagic Escherichia coli is a subset of the Shiga-toxigenic E. coli (STEC) group of pathogens. STEC are capable of causing disease that ranges from mild watery diarrhea to hemorrhagic colitis (HC) and in extreme scenarios hemolytic uremic syndrome (HUS; Nataro and Kaper, 1998; Croxen et al., 2013). HUS occurs as a result of Shiga-like toxin (Stx) exposure, a virulence factor carried by members of the STEC that is capable of ribosomal inhibition in susceptible cells. Subsets of STEC vary in their serotype and virulence factor reservoir with E. coli O157:H7 being the most extensively studied EHEC, and STEC for that matter, responsible for regular outbreaks of foodborne illness worldwide. Classically, O157:H7 infections have been associated with contaminated meat products but cases also appear in relation to contaminated fresh produce (Riley et al., 1983). Ruminants, most notably cattle, are the primary host for EHEC O157:H7, which they colonize asymptomatically at the recto-anal junction using the LEE-encoded T3SS (Karmali et al., 1983a,b; Orskov et al., 1987; Naylor et al., 2003). Cattle lack Gb3 receptors (the Stx receptor found on human endothelial cells) and are therefore not susceptible to Stx-associated pathogenesis (Pruimboom-Brees et al., 2000). Cattle provide a natural reservoir for EHEC O157:H7 and a direct link to subsequent infection of the human host.
The repertoire of virulence factors in EHEC O157:H7 includes a T3SS and its set of associated effector proteins, the Stx toxin as mentioned above and the pO157 plasmid, which have been reviewed extensively elsewhere (Croxen and Finlay, 2010; Croxen et al., 2013). Evidence for horizontal acquisition of virulence factors was highlighted in a study characterizing the complete genome sequence of the E. coli O157:H7 strain EDL933 (Perna et al., 2001). Genome sequence analysis revealed that EHEC contains hundreds of introgressed segments or “O-islands,” many of which carry virulence factors such as the LEE pathogenicity island (PAI). The study also characterized genomic regions related to known bacteriophage, including BP-933W encoding stx2, and a selection of cryptic prophage. These elements were found to encode various virulence related proteins such as T3SS effector proteins (Perna et al., 2001). Acquisition of horizontally acquired virulence factors can transform a harmless commensal into a niche specific pathogen. However, acquired virulence traits are not enough to confer pathogenicity alone, they must be adapted to the regulatory network of the cell for efficient expression in a temporal manner (Tan et al., 2015). Once appropriately integrated into the bacterial regulatory circuits these virulence factors can provide a competitive advantage for the emergent pathogen over the resident microbiota to thrive in this specific niche.
The LEE Encoded Type III Secretion System
Escherichia coli O157:H7 forms attaching and effacing (A/E) lesions on host epithelial cells. This phenotype is characterized by intimate attachment to the host cell surface, subversion of the host actin cytoskeleton and raised “pedestal” formation (Moon et al., 1983; McDaniel et al., 1995), a hallmark of O157:H7 pathogenesis. Similarly, enteropathogenic E. coli (EPEC) and the mouse pathogen Citrobacter rodentium also confer the A/E phenotype on host cells (Moon et al., 1983; Schauer and Falkow, 1993; Croxen et al., 2013; Collins et al., 2014). The ability to form A/E lesions has been attributed to the carriage of the ∼35 kb LEE PAI, which contains all the necessary genes for the formation of a functional T3SS (McDaniel et al., 1995; McDaniel and Kaper, 1997; Wong et al., 2011). The LEE has a GC content of 38.3%, which is in contrast to the 50.8% that constitutes the E. coli chromosome, suggesting that the LEE was acquired by horizontal gene transfer (McDaniel et al., 1995; McDaniel and Kaper, 1997; Elliott et al., 1998). Indeed, whole contribution of the LEE to the A/E phenotype was confirmed when the entire LEE from the EPEC strain E2348/69 was introduced to the non-pathogenic K-12 strain resulting in the ability to form A/E lesions (McDaniel and Kaper, 1997). In contrast, the EHEC LEE is incapable of conferring virulence on K-12 suggesting that LEE adapts to different genetic backgrounds in unique ways (Elliott et al., 1999).
The LEE contains 41 open reading frames (ORFs) that include genes encoding the T3SS basal apparatus and secretion machinery, the translocon needle filament subunit EspA (Knutton et al., 1998), the translocon pore proteins EspD and EspB (Ide et al., 2001), the major adhesin intimin Eae and its cognate “translocated intimin receptor” Tir (Kenny et al., 1997b), a lytic transglycosylase EtgA to clear peptidoglycan localized around the forming T3SS (Burkinshaw et al., 2015), a selection of secreted effector proteins and cellular chaperones (reviewed, in Frankel et al., 1998 and Wong et al., 2011). As well as LEE encoded effector proteins this T3SS is also capable of translocating a large number of non-LEE encoded effectors, located throughout the chromosome on horizontally acquired genetic elements, that are important for the virulence process (Tobe et al., 2006; Deng et al., 2012). The 41 ORFs can be largely organized into five polycistronic operons, annotated LEE1 through LEE5 with LEE1, LEE2, and LEE3 encoding the major structural components of the T3SS, LEE4 encoding the translocon pore and LEE5 encoding Tir and Intimin (Elliott et al., 1998). ORF 1 on the LEE1 operon encodes the master regulator Ler (LEE encoded regulator) through which activation of operons LEE2 to LEE5 is mediated (Mellies et al., 1999; Sánchez-SanMartín et al., 2001; Haack et al., 2003). A second master regulatory system, GrlRA, is located between the LEE1 and LEE2 operons, is activated by Ler and forms a regulatory feedback loop on LEE1 expression (Deng et al., 2004). Thus, the LEE contains all the necessary equipment to form a functional T3SS, the structure of which has been recently reviewed in detail (Büttner, 2012), capable of mediating the A/E phenotype.
Expression of the LEE is tightly regulated in response to multiple stimuli. Host temperature of 37°C provides optimal LEE expression as well as the exponential growth phase (Rosenshine et al., 1996). Experimentally, the LEE can be induced by growth in tissue culture media (DMEM or MEM-HEPES) as these conditions mimic the physiological environment (Roe et al., 2003). A detailed study by Kenny et al. (1997a) attributed in vitro secretion to no single component but rather a combination of temperature, pH, osmolarity, calcium, iron and salt concentrations.
Transcriptional regulation of the LEE is extremely complex. It is controlled at the core level by two integral regulatory systems encoded within the LEE, Ler and GrlRA (Deng et al., 2004). Numerous other systems such as nucleoid regulators, stress response regulators and various environmental sensing systems make contributions to regulation of the LEE. Chromosomal and phage encoded genetic elements also feed into the LEE creating layers of specific control on the system. Mellies et al. (2007) have previously reviewed regulation of the LEE in great detail for EHEC and EPEC. In this review, we summarize the key points of LEE regulation in relation to EHEC primarily whilst also commenting on EPEC and C. rodentium where appropriate. We provide an up to date summary of the regulatory systems involved in LEE control and later focus in more detail on the specific responses of this PAI to various physiologically relevant stimuli that EHEC can encounter during colonization of the bovine or human hosts. It is important to emphasize that LEE regulation has both common elements and distinct differences between the pathotypes of EHEC, EPEC or C. rodentium that will be mentioned in this review, and this can be attributed to the different environments encountered within the host that they primarily colonize.
Master Regulation of the LEE
Transcription of the LEE is orchestrated largely by the ∼15 kDa protein Ler that activates transcription at each of the subsequent LEE operons as well as non-operonic members of the LEE such as map, espG, and escD (Mellies et al., 1999; Elliott et al., 2000; Sánchez-SanMartín et al., 2001; Haack et al., 2003). The importance of Ler for virulence was demonstrated by way of a deletion mutant in EHEC strain 86-24. This mutant was impaired in its ability to secrete effectors and form A/E lesions on host cells. Additionally, the ler mutant strain showed a decreased expression of non-LEE encoded virulence factors by western blot (Elliott et al., 2000). StcE, a non-T3SS secreted metalloprotease encoded on the pO157 plasmid, was demonstrated to be under the control of Ler, implying a global control of virulence gene regulation by Ler (Lathem et al., 2002). Additionally, a number of non-LEE encoded effector proteins that are translocated into the host cell via the T3SS are also under direct control by Ler (Roe et al., 2007; Holmes et al., 2012). A comprehensive study by Deng et al. (2004) systematically analyzed the role of each ORF in virulence by deletion within the LEE of C. rodentium. This work further highlighted the absolute importance of Ler for virulence by the T3SS in a mouse model of infection, thus expanding the knowledge of LEE regulation to other pathogens (Deng et al., 2004).
Transcriptional regulation of ler specifically is complex. The LEE1 operon contains an unusually long upstream leader sequence of ∼170 bases (Islam et al., 2012). In EHEC, EPEC and C. rodentium transcription is driven through a promoter designated P1 (distal) however, in EHEC transcription has been demonstrated to be driven from not one but two promoters, designated P1 and P2 (proximal), as well as a cryptic promoter (P1A) that overlaps P1 (Sperandio et al., 2002a; Islam et al., 2011b). The precise contributions of each promoter are unclear but investigations into the activity of the LEE1 promoter region has identified that P1 is likely to be the major promoter of the LEE1 operon in EHEC with the P2 promoter being active to a lesser extent (Islam et al., 2011a). Ler has also been demonstrated to elicit negative autoregulation on the LEE1 promoter as well as positively drive expression from operons LEE1 through LEE5, presumably as a mechanism of optimizing LEE expression levels to steady-state during the infection process (Berdichevsky et al., 2005).
The GrlRA Regulatory Feedback Loop
The analysis of systematic LEE mutations by Deng et al. (2004) revealed a second LEE-encoded regulatory system that was indispensable for the LEE function. The work identified a positive regulator of ler expression, named GrlA (global regulator of ler activation), and a negative repressor of ler, GrlR (global regulator of ler repression; Deng et al., 2004). GrlA forms a positive feedback loop with Ler to maintain steady activation of the LEE under positive conditions, whereas GrlR was postulated to inhibit GrlA mediated activation of ler in order to tightly control LEE expression (Barba et al., 2005). Recently the structure of GrlR and a GrlR/GrlA complex has been solved and a mechanism of Ler regulation by this system proposed. GrlR forms a dimeric structure in solution that binds GrlA, stabilizes it and inhibits transcriptional activation of the LEE1 operon (Padavannil et al., 2013). This complements other findings that suggest under certain conditions, such as those favoring LEE expression, GrlR is cleaved by the ClpXP protease, freeing GrlA and allowing ler transcriptional activation (Iyoda and Watanabe, 2005). Despite these findings, the precise mechanism governing GrlR antirepression is unknown. An overview of LEE master regulation is illustrated in Figure 1.
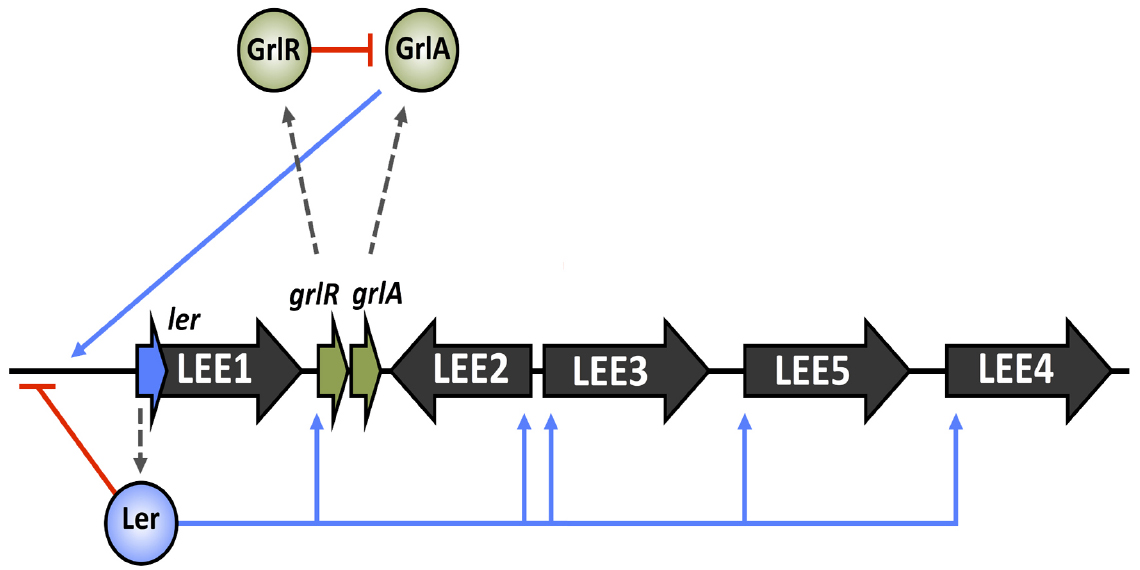
Figure 1. Core regulation of the LEE island. LEE operons 1–5 are indicated in black with the ler and grlRA ORFs highlighted in blue and green respectively. Broken gray arrows correspond to translated products from these genes. Blue lines represent input points of positive transcriptional regulation whereas red blunt arrows indicate negative regulation.
Regulation of the LEE by Bacterial Nucleoid Associated Proteins
Nucleoid-associated proteins (NAPs) are found across all lineages of life (Eukarya, Bacteria, and Archaea) and play global roles in nucleoid maintenance. The DNA-binding and shaping ability of NAPs allows them to affect more than just chromosome architecture and they have emerged as global regulators of transcription (Dillon and Dorman, 2010). Perhaps of most relevance to the LEE is the histone-like nucleoid structuring protein (H-NS). This regulator forms DNA-bridges over extended stretches of DNA in the vicinity of promoters under its control, thus blocking or trapping DNA polymerase and “silencing” the promoter (Dame et al., 2006; Grainger et al., 2006; Dorman, 2007). H-NS silencing is biased toward foreign AT rich segments of the DNA and is thought of as a global repressor of horizontally acquired genetic elements (Navarre et al., 2006; Oshima et al., 2006). Overcoming H-NS repression is often an adaptation of pre-existing global regulators but can occur via horizontally acquired elements also, as is the case for Ler (Abe et al., 2008). The LEE is repressed by H-NS under non-inducing conditions such as low temperature. Ler acts as an H-NS antagonist by displacement from the LEE thus relieving H-NS mediated silencing (Umanski et al., 2002).
As well as H-NS, other NAPs have been reported to contribute to LEE regulation. Integration host factor (Ihf) is a dimeric NAP composed of alpha and beta subunits. Ihf is capable of wrapping DNA and affecting transcription on a genome-wide scale. A global DNA binding study of Ihf in E. coli K-12 revealed that Ihf was capable of binding with sequence-specificity to ∼30% of all operons in E. coli K-12, however, not all of these operons were transcriptionally affected in an Ihf mutant background suggesting accessory regulation (Dillon and Dorman, 2010; Prieto et al., 2012). Ihf positively promotes ler expression by binding upstream of the LEE1 promoter, aiding H-NS displacement and promoting full LEE expression (Friedberg et al., 1999). In contrast, the NAP Hha represses ler transcription by a proposed DNA binding mimicry mechanism, concentrating H-NS repression on the LEE (Sharma and Zuerner, 2004; Madrid et al., 2007). The factor for inversion stimulation (Fis) is responsible for early exponential phase transcriptional regulation and has been demonstrated to affect ler and LEE4 expression in EPEC (Goldberg et al., 2001). Collectively, the cellular NAPs have been adapted by the cell maintain control over the LEE. NAP input to the LEE is illustrated in Figure 2.
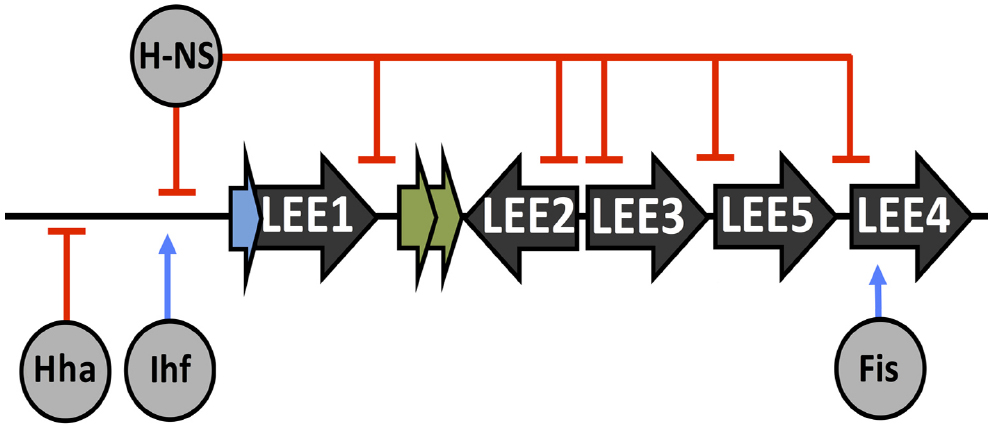
Figure 2. Regulation of the LEE by nucleoid associated proteins (NAPs). Points of transcriptional activation (blue arrow) or repression (red blunt arrow) by NAPs, which are labeled and indicated in gray.
Cross-talk between Genetic Regulatory Elements and the LEE
The LEE, like many horizontally acquired virulence factors, must be regulated appropriately to ensure efficient expression in a timely manner. There are a number of global regulators, both chromosomally encoded and horizontally acquired that contribute to LEE regulation (illustrated in Figure 3). In EPEC, the perABC family of AraC-like transcriptional regulators are encoded on the EAF virulence plasmid but this system is not found in EHEC (Mellies et al., 1999). Instead, three functional homologs of the direct EPEC Ler activator PerC, pchABC encoded on lambdoid prophages, were identified in O157:H7 and found to be required for full virulence (Iyoda and Watanabe, 2004). Expression of these Pch regulators was required for full LEE activation during growth in tissue culture medium (LEE-inducing) or interaction with host cells. Furthermore, Abe et al. (2008) comprehensively demonstrated the binding capacity of Pch to LEE associated ORFs, non-LEE associated effectors and a variety of other global sites including those involved in acid resistance and NAPs.
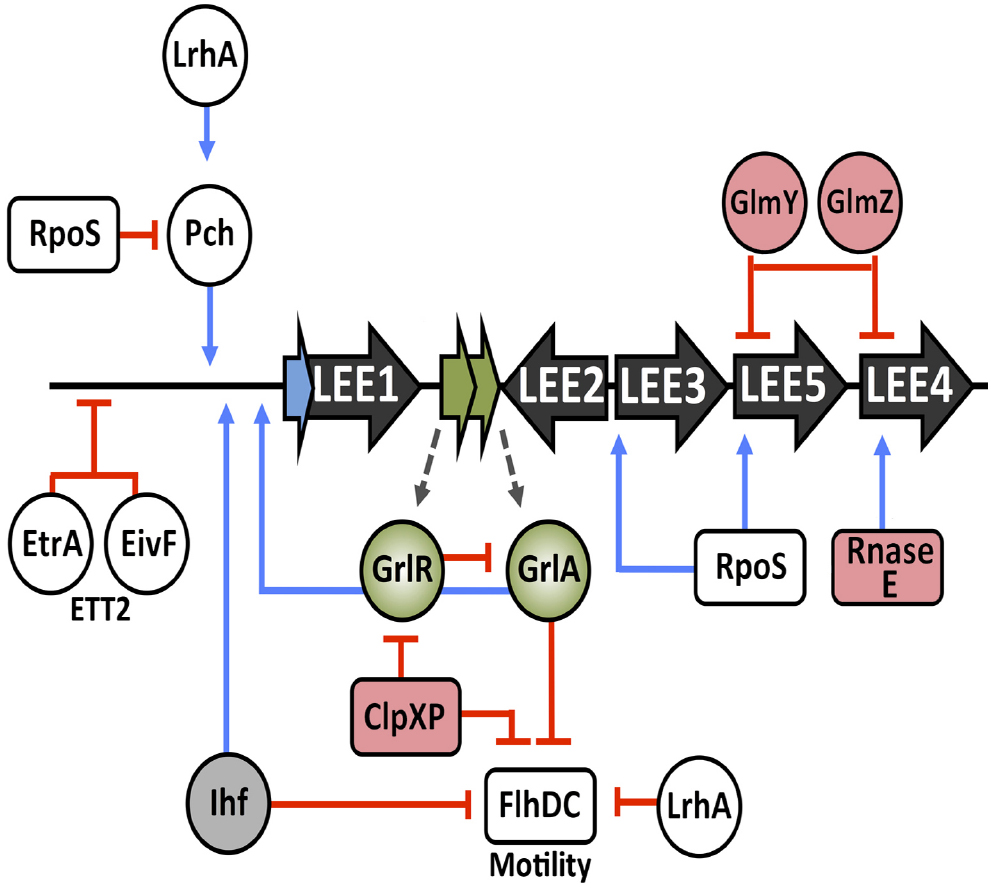
Figure 3. Transcriptional cross-talk of cellular regulators and post-transcriptional control of the LEE. Inputs from various transcriptional regulators, indicated in white, and the cross-regulation associated with them. Regulators of the ETT2 system and motility are labeled accordingly. Post-transcriptional regulators are shaded in pink, Ihf is shaded in gray and GrlRA are shaded in green. Blue lines represent positive regulation whereas red blunt arrows indicate negative regulation.
Cross-regulation between the LEE and flagella is an intriguing concept. Classically, EHEC are known to be non-motile under LEE-inducing conditions in vitro (Tobe et al., 2011). The H7 flagellum has adhesive properties to the bovine epithelium, mediating initial attachment to host cells in the early stages of infection followed by a regulatory switch in LEE activation and subsequent intimate attachment via the T3SS (Mahajan et al., 2009). As discussed above, GrlR repression of GrlA is removed by the ClpXP protease, thus promoting LEE expression by GrlA (Padavannil et al., 2013). Furthermore, ClpXP is capable of post-translationally inactivating the flagellar master regulator, FlhDC (Kitagawa et al., 2011). Additionally, GrlA transcriptionally represses flhDC resulting in two independent mechanisms of motility inhibition under LEE inducing conditions (Iyoda et al., 2006). Ihf, which is required for LEE1 expression has also been shown mediate flhDC repression (Friedberg et al., 1999; Yona-Nadler et al., 2003). LrhA is a LysR type transcriptional regulator known to repress motility in a K-12 background. LrhA is important for LEE1 activation by directly activating PchA and PchB (Lehnen et al., 2002; Honda et al., 2009).
Genome sequencing of O157:H7 revealed the presence of a second cryptic Type III secretion system designated ETT2 (Hayashi et al., 2001; Perna et al., 2001). Functionality and the role of the ETT2 system is controversial with the locus being subject to widespread mutational attrition (Ren et al., 2004). However, two regulators encoded by the system, EtrA and EivF, were found to be functional repressors of the LEE in EHEC. This study highlighted the cross-talk between two independently acquired PAI and was hypothesized to mediate avoiding the expression of two T3SSs simultaneously. The work also demonstrates the ability of remaining genes from degenerate systems to have residual effects on the cell and play roles in regulation (Zhang et al., 2004).
Hfq, the small RNA (sRNA) chaperone, is a global post-transcriptional regulator of gene expression. The LEE was found to be negatively regulated through Hfq-mediated modulation of the ler and grlRA mRNA transcripts (Hansen and Kaper, 2009; Shakhnovich et al., 2009). Conversely, in strain 86-24, Hfq was attributed to an indirect activation of the LEE in this genetic background (Kendall et al., 2011). Strain to strain variation in LEE expression by global regulators has also been documented in relation to the stationary phase sigma factor RpoS. In 86-24 LEE3 and tir were found to be dependent on RpoS (Sperandio et al., 1999). This is in contrast to studies using the Sakai O157:H7 strain that reported repression of the LEE by RpoS (Iyoda and Watanabe, 2005). Global gene expression profiling of an rpoS mutant in EDL933 revealed a dependency of ler expression on RpoS but many LEE encoded ORFs were unaffected, whereas analysis of an rpoS mutant in the mouse pathogen C. rodentium revealed a positive influence of RpoS on LEE-mediated infection in vivo (Dong and Schellhorn, 2009; Dong et al., 2009). Although, the role of RpoS in LEE regulation is influenced by growth conditions these studies collectively demonstrate strain-to-strain variation in virulence factor expression depending on the genetic background examined.
Post-transcriptional regulation of the LEE is a less explored but relevant topic. As mentioned, Hfq has been reported to regulate the LEE in a strain-specific manner. A number of studies have detailed the role of small regulatory RNAs (sRNA) in control of LEE expression. The RpoS stimulatory sRNA, DsrA, stimulates the expression of ler in an RpoS dependent manner (Laaberki et al., 2006). Recently, two sRNAs involved in activation of glucosamine biosynthesis (GlmY and GlmZ) were found to play global roles in gene expression that included post-transcriptional regulation of the LEE4 and LEE5 operons as well as the non-LEE encoded effectors NleA and EspFu providing further evidence of the complexity of LEE control (Gruber and Sperandio, 2014, 2015). Transcription from the LEE4 operon, encoding a combination of T3SS components and regulators has also been demonstrated to be under post-transcriptional modification by the cellular endonuclease Rnase E allowing specific regulation of protein production from these transcripts (Lodato and Kaper, 2009).
The Adaptable GAD Regulators and the Response to Nitric Oxide
Foodborne pathogens must survive ingestion and passage through the stomach to reach their intestinal niche. E. coli employs specific mechanisms to counteract the harsh acidic environment of the stomach, such as the glutamate-decarboxylase (GAD) acid stress response (De Biase et al., 1999). The GAD system is regulated globally by multiple factors including H-NS, PchA, PchB, and RpoS (Atlung and Ingmer, 1997; De Biase et al., 1999; Giangrossi et al., 2005). The system is a complex signaling cascade that ultimately senses environmental pH and responds accordingly by influencing transcription of the acid tolerance gene network (Masuda and Church, 2003).
Aside from regulating acid resistance, transcriptional regulators of the GAD system have shown adaptability to transcriptional control of the LEE. GadX has been demonstrated to directly repress perA in EPEC, thus inhibiting PerC activation of the LEE (Shin et al., 2001). In EHEC, a transposon mutant screen identified GadE and YhiF mutants that displayed increased activity at the LEE2 and LEE4 operons by a mechanism independent of ler. The mutants were increased in their ability to colonize host cells and infect mice (Tatsuno et al., 2003). As discussed above, both horizontally acquired and chromosomally encoded regulators can have direct and indirect effects on LEE expression. Tree et al. (2011) described the prophage-encoded secretion regulators (Psr) found on numerous O-islands in O157:H7. The mechanism of regulation involved GadE and YhiF mediated Psr repression of the LEE, largely through repression of the LEE2, LEE3, and LEE5 operons. Furthermore, the study demonstrated direct binding of purified GadE to the LEE1 and LEE2/3 promoters indicating both direct and indirect control over LEE expression. It was proposed that the Psr system has been adapted to promote the expression of prophage-encoded effectors that employ the T3SS for translocation into the host cell. This is achieved by repressing LEE-encoded effectors at later time points during colonization (Tree et al., 2011). Based on these reports it was hypothesized that the GAD regulators have adapted to control the LEE as well as the acid stress regulon. The reason for this adaptation is likely to inhibit the expression of colonization factors under conditions of stress, such as the acidic environment of the stomach. Once enteric pathogens have passed through the stomach and into the intestine, this more favorable environment allows lifting of GAD repression on the LEE. The latter study demonstrates not only the ability of the GAD system to regulate the LEE in response to the environment but also demonstrate further how horizontally acquired regulators such as Psr can override pre-existing regulatory networks to mediate regulation of virulence factors in efficient ways.
Nitric oxide (NO) is a key mediator of the host innate immune response (Foley and O’Farrell, 2003). Recently, GadX and GadE were implicated in NO mediated repression of the LEE through a complex regulatory interplay with the nitrite sensitive repressor NsrR. The work described how under LEE-inducing conditions GadE, and therefore GadX, is repressed and NsrR activates the LEE. In this context, NO exposure lifts NsrR repression of GadE and results in activation of LEE1 and LEE4/LEE5 by GadX and GadE respectively. In a manner similar to the GAD system, the LEE is therefore repressed in the stomach due to the abundance of NO found in gastric juices and the GAD regulators are recruited to enhance this response (Branchu et al., 2014). These studies highlight the adaptive power of the GAD regulators to mediate not only the response to acid stress but also temporal expression of the LEE within the host (Figure 4).
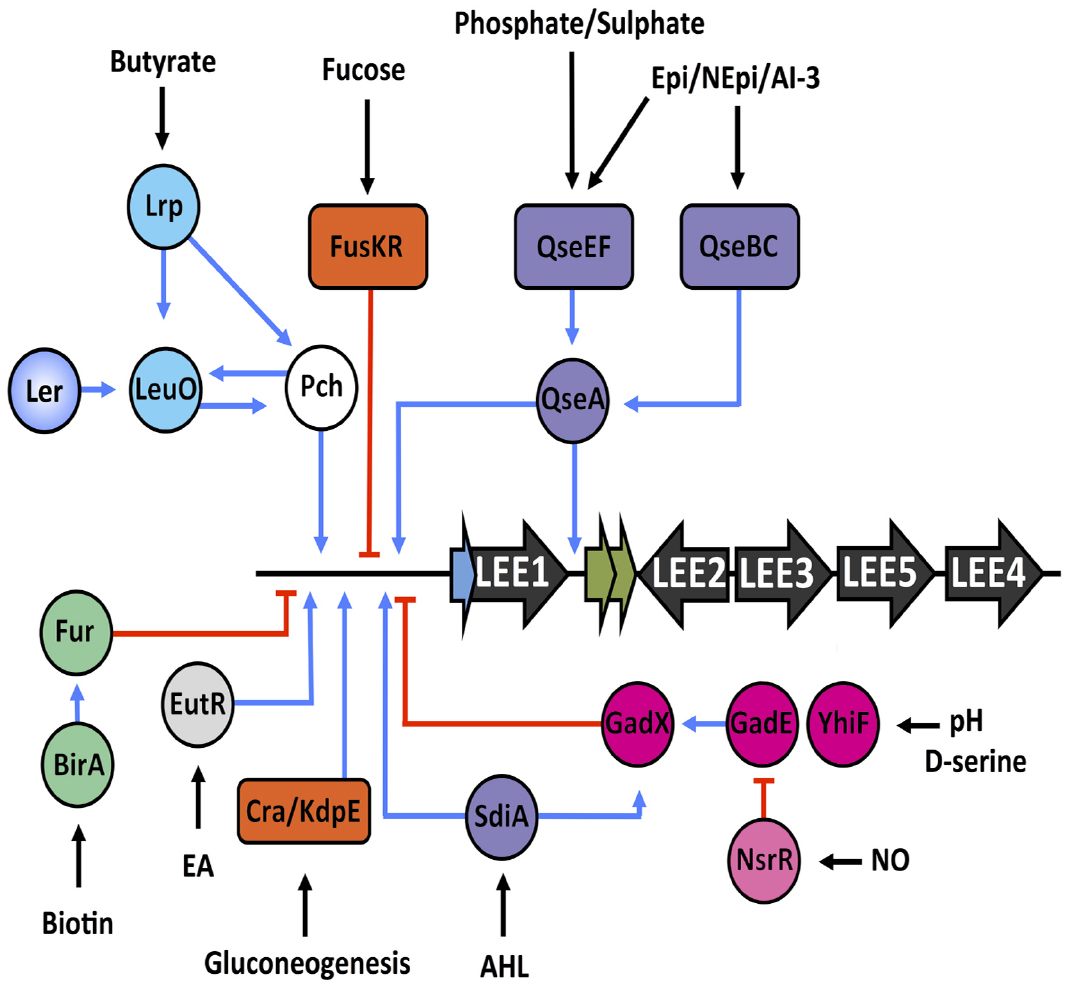
Figure 4. Regulatory map of various signals encountered by EHEC within the host. Black arrows indicate the signal being sensed and the regulatory systems are color coded as follows—quorum sensing of hormones and hormone-like signals (purple), nutrient sensing (orange), nitric oxide sensing (pink), pH and D-serine regulation (magenta), ethanolamine sensing (light gray), butyrate sensing (aqua), and biotin sensing (green). Blue lines represent input points of positive transcriptional regulation whereas red blunt arrows indicate negative regulation.
Chemical Signaling in a Complex Environment—LEE Regulation by Quorum Sensing
Interspecies communication by way of quorum sensing (QS) involves the production of hormone-like signals by bacterial populations that are subsequently sensed and perceived by two-component regulatory systems (TCS). These systems consists of a surface exposed Histidine-kinase sensor protein that autophosphorylates in response to certain stimuli. This phosphate is subsequently transferred to a cellular response regulator and the signals are transmitted to the genome for differential regulation of signal-sensitive genes (Hughes and Sperandio, 2008). QS however, has evolved to allow the sensing of not only bacterially derived signals, such as the auto-inducer 3 signaling molecule (AI-3), but also host-derived hormones, namely the stress hormones epinephrine (Epi) and norepinephrine (NEpi), in a synergistic manner (Walters and Sperandio, 2006). Sensing of these host-derived cues provides EHEC with important inputs into the physiology of the host and resident microbiota. Two TCSs (QseBC and QseEF) involved in QS have been described in detail and play key roles in regulation of virulence and motility in EHEC and signals are transmitted through two LysR type transcriptional regulators, QseA and QseD (Sperandio et al., 1999, 2002b; Russell et al., 2007; Sharp and Sperandio, 2007; Habdas et al., 2010). QseA can both directly and indirectly activate expression of the LEE1 operon by binding to both the P1 and P2 promoter regions. Additionally, QseA has been demonstrated to directly activate expression of O-islands encoding non-LEE effectors (Kendall et al., 2010). Epi, NEpi and AI-3 feed in via the sensor kinase QseC, thus phosphorylating the response regulator QseB (Clarke et al., 2006). QseB in turn positively regulates motility through direct interaction with the flhDC regulatory region (Clarke and Sperandio, 2005). QseEF can sense Epi, phosphate and sulfate then signal to QseA in a QseC dependent manner (Reading et al., 2007, 2009).
Enterohaemorrhagic Escherichia coli can also sense hormone-like signals produced by members of the microbiota. SdiA is a LuxR-like transcriptional regulator has the remarkable ability of responding to acyl-homoserine lactones produced by neighboring bacteria but not EHEC (Hughes et al., 2010; Nguyen et al., 2013). SdiA represses the LEE in response to these signals but also activates expression of GadX known to inhibit LEE1 expression as EHEC passage through the stomach. The rumen of cattle is very abundant in acyl-homoserine lactones thus limiting colonization of EHEC to more distal sites of the gastrointestinal tract, such as the recto-anal junction (Kanamaru et al., 2000; Naylor et al., 2003; Hughes et al., 2010).
Competition for Nutrients and the Expression of Virulence
The intestine is a complex environment where commensals have adapted to a specific niche with specific nutrients. Incoming pathogens are therefore unwanted visitors and are in direct competition for the limited nutrient supply of this environment. Chang et al. (2004) have described by systematic mutagenesis of metabolic pathways that only mutation in sugar utilization pathways affected competitive colonization of E. coli in a mouse model. Gluconate was found to be the primary carbon source for K-12 with glucuronate, mannose, fucose and ribose contributing to niche maintenance (Chang et al., 2004). Utilization of unique sugar sources therefore allows pathogenic E. coli to establish distinct niches within this competitive environment. The researchers have recently proposed “The Restaurant Hypothesis” which states that inhabitants of mixed biofilm populations can locally dictate which nutrients are available (Leatham-Jensen et al., 2012; Meador et al., 2014). In order to overcome competition for limited nutrients, EHEC utilizes both glycolytic and gluconeogenic carbon sources to temporally regulate initial colonization and long term establishment respectively (Miranda et al., 2004). Expression of ler is directly inhibited by glycolytic by-products and promoted by gluconeogenic conditions respectively by the catabolite/osmotic stress responsive Cra/KdpE system (Njoroge et al., 2012). The binding capacity of Cra and KdpE to the LEE1 regulatory region is dependent on signal transduction the Histidine kinase KdpD, with the phosphorylation state of KdpE being determined by the concentration of glucose in the environment.
In a study investigating the transcriptomic profiles of EHEC persistence in the bovine small intestine, the importance of gluconeogenic metabolism was assessed. The study found regulons involved in ethanolamine (EA), urea and amino acid utilization were all upregulated under these in vivo conditions (Bertin et al., 2014). This study agreed with an earlier observation that EA, a cell membrane component released into the gut lumen, provided EHEC with a competitive advantage over the resident microbiota, which normally do not metabolize EA (Bertin et al., 2011). More recently, EA was confirmed as a signal for EutR, the transcriptional regulator, which drives expression of the LEE1 operon and EA metabolic genes independently of one another (Kendall et al., 2012; Luzader et al., 2013). EA has also been attributed to activation of putative O157:H7 fimbrial-like loci, supporting the theory that EA plays a physiologically relevant role in EHEC colonization (Gonyar and Kendall, 2014).
Fucose is a sugar found in abundance (∼100 μM) in the human or bovine intestine and is one of the preferred carbon sources utilized by E. coli for maintenance of intestinal colonization (Chang et al., 2004). Fucose is derived from host glycans by the gut commensal Bacteroides thetaiotaomicron. EHEC utilizes the FusKR TCS to avoid competition with resident E. coli for this carbon source. This system has been recently acquired in the evolutionary timeline and is regulated in response to the presence of epithelial mucus. The system acts to directly repress the LEE1 operon and indirectly repress fucose utilization genes in response to high concentrations of fucose available in the intestinal mucus layer, thus limiting the possibility of unnecessary competition for this carbon source. Once in close contact with the intestinal epithelium, a niche devoid of commensal E. coli, QS systems activate LEE1 expression and lift fucose mediated repression of the LEE, therefore allowing establishment of A/E associated colonization of the host epithelium (Pacheco et al., 2012).
These studies provide elegant examples of how nutrient metabolism and virulence must be co-regulated to work together but not dependently on each other in order to carefully control niche-specific virulence in response to particular environmental nutritional content. A summary of environmental signals that feed into the regulation of the LEE is found in Figure 4.
A Link between Virulence and Metabolism—Regulation by Short Chain Fatty Acids and Biotin in the Host Intestinal Tract
As well as carbon and nitrogen sources, the intestine is abundant in short chain fatty acids (SCFAs) such as butyrate, acetate and propionate that can reach concentrations of up to 140 mM (Cummings and Macfarlane, 1991). SCFAs are end products of bacterial fermentation of dietary carbohydrates such as resistant starches or fiber and are thought to provide a nutrient source for maintenance of a healthy colon of both humans and animals (Wong et al., 2006). Studies have also shown that SCFAs play a distinct role in LEE regulation being adapted as niche-specific signals helping to trigger colonization of EHEC. Nakanishi et al. (2009) showed that butyrate could significantly increase LEE expression in a PchA dependent manner at concentrations that were not inhibitory to growth. This regulation required another regulator, the leucine-responsive protein (Lrp), which is a global transcription factor necessary for nitrogen metabolism in E. coli (Nakanishi et al., 2009). Lrp has been shown in K-12 to act at 138 different binding sites including that of lrhA (Cho et al., 2008). Thus, butyrate activation of the LEE may be via an Lrp-LrhA-PchA signaling cascade. Recently, Takao et al identified the global LysR type transcriptional regulator LeuO as a mediator in the Lrp dependent system. They showed that butyrate induced leuO and pchA via Lrp, subsequently activating ler via PchA. Ler and PchA were capable of directly activating leuO suggesting the system forms a positive feedback loop upon leuO induction (Takao et al., 2014).
Short chain fatty acids not only regulate virulence associated with the LEE but also motility. Addition of SCFAs to growth in DMEM, conditions promoting LEE expression that are normally repressive of motility, activated motility in EHEC. Butyrate was capable of simultaneously activating motility and LEE expression in an Lrp dependent manner. This occurred by direct activation of the flhDC flagellar master regulators. Acetate and propionate on the other hand could not induce LEE expression and induced motility further down the regulatory hierarchy. Due to the known adhesive properties of the H7 flagellum these findings further bolster the proposed role of flagella in initial attachment to the host epithelia (Tobe et al., 2011). In a study characterizing the protein targets of a class of type 3 secretion inhibitors, the salicylidene acylhydrazides (SA), a number of putative cellular targets were implicated in regulation of virulence (Wang et al., 2011). One of these was AdhE, an alcohol dehydrogenase. Deletion of adhE resulted in a pleiotropic phenotype—overproduction of extracellular acetate, expression of non-functional flagella and post-transcriptional inhibition of the T3SS. The findings highlight the fine balance required between central metabolism and regulation of virulence factors in order to appropriately function in the environment. It is known that cattle contain high concentrations of SCFAs particularly in the rumen. It is therefore conceivable that lower concentrations of acetate in the rectum, the major colonization site for EHEC in cattle, would promote T3SS mediated attachment more efficiently (Beckham et al., 2014).
Similarly to SCFAs, the distribution of the essential cofactor biotin within the intestine has recently been proposed to be an important determinant of EHEC niche specification. Yang et al. (2015) revealed that biotin concentrations in the small intestine were 14-fold higher than that of the large intestine, the preferred colonization site of EHEC, and that this correlated with a 31.5-fold higher EHEC bacterial count associated with the large intestine during infection of mice. The study proposed a mechanism of niche specification whereby the biotin protein ligase BirA indirectly repressed the LEE through a global regulator called Fur. This repression was characterized in vitro and was induced by biotin concentrations lower than that found in the small intestine but higher than that of the large intestine, consistent with their proposal. Furthermore, mice fed with a biotin rich diet during EHEC infection significantly reduced the amount of EHEC associated colonization when compared to mice maintained on a normal diet. This study revealed novel insights into LEE regulation by a physiologically relevant signal but also suggests the potential for modulating diet with a view to limiting EHEC infections. The effects were also found to be EHEC specific with EPEC not being responsive to biotin levels through this regulation (Yang et al., 2015). This further highlights the adaptive nature of LEE regulation in a pathotype specific manner.
D-serine as a Niche Determinant of Pathogenic E. coli
D-amino acids have classically been seen as integral members of the bacterial peptidoglycan layer, with other roles being obscure due to the nature of D-amino acids not being utilized by bacterial ribosomes. Recently, studies have revealed that non-canonical D-amino acids are actively produced by diverse bacterial species and are used to regulate cell wall composition during different stages of growth (Lam et al., 2009; Cava et al., 2011). D-serine is a niche specific metabolite that is abundant in extra-intestinal sites of the human body, such as the urinary tract and the brain (Anfora et al., 2007; Wolosker et al., 2008). Indeed, D-serine acts as a neurotransmitter in the human brain but the high concentrations in urine are correlated to endogenous and exogenous sources (Nagata et al., 2007). For instance, processed foods are known to be concentrated in the d-isomeric form of amino acids (Man and Bada, 1987).
Owing to its abundance in the urinary tract. D-serine acts as a positive fitness trait and regulator of virulence genes in urinary pathogenic bacteria such as uropathogenic E. coli and Staphylococcus saprophyticus (Roesch et al., 2003; Anfora et al., 2007; Haugen et al., 2007; Sakinç et al., 2009; Korte-Berwanger et al., 2013). In contrast, EHEC cannot use D-serine as a carbon source due to a genetic replacement of the D-serine tolerance locus (Moritz and Welch, 2006). This replacement is widespread among the E. coli phylogeny and is almost always correlated with LEE-positive pathogens even though normal gut commensals can retain this locus. Analysis of the genome sequence of 1591 E. coli isolates revealed that carriage of both the LEE and the D-serine tolerance locus is an extremely rare event occurring in only 1.6% of cases. In accordance with this it was found that D-serine downregulates the LEE through indirect modulation of pre-existing transcriptional regulators Ihf and YhiF. The effect on LEE expression was also found to be independent of the ability to metabolize D-serine, explaining the lack of necessity to retain the D-serine tolerance locus in EHEC and separating metabolism from virulence. Uropathogenic E. coli do not encode the LEE T3SS and it was therefore proposed that the inhibitory action of D-serine was used by EHEC as an environmental signal to limit these pathogens to the intestine, an environment lacking an abundance of D-serine, thus reducing competition with extraintestinal E. coli pathogens (Connolly et al., 2015). This study further highlighted the impact that host metabolism can have on niche specification of bacterial pathogens.
The Impact of the Microbiome on EHEC Infection
Enterohaemorrhagic Escherichia coli must actively compete with the resident microbiota in order to establish colonization of the host (Kamada et al., 2012). EHEC must penetrate the intestinal lumen in order to reach its preferred colonization site at the epithelial surface. This “zone of clearance” between the lumen and the cell surface is largely devoid of resident bacteria and the localized gluconeogenic conditions here promote LEE expression (Miranda et al., 2004; Vaishnava et al., 2011; Kamada et al., 2012). The microbiome is composed of trillions of bacteria from hundreds of species that have evolved to form a synergistic relationship with the host (Eckburg et al., 2005; Gill et al., 2006). In a sense, this relationship provides the first barrier of defense against invading pathogens. There is a wealth of recent research that highlights how diet can influence the composition of the microbiota and this in turn can have strong implications on the susceptibility of an individual to enteric infection given the essential roles of the microbiota in host metabolism and immunity (Turnbaugh and Gordon, 2009; Kau et al., 2011; Wu et al., 2011).
The majority of research on EHEC virulence regulation to date has involved detailed mapping of transcriptional regulatory networks in vitro complimented with in vivo infection models. However, some recent studies have revealed fascinating insights into the roles of the microbiota in the infection process. A study investigating the protective effect of species belonging to the bacterial genus Bifidobacteria revealed concentrations of acetate that were significantly higher in mice associated with these strains than non-protective Bifidobacteria. Species of Bifidobacteria were previously implicated in a protective manner by probiotic inhibition of EHEC. The mechanism of this protection involves the production of acetate in a glucose-independent manner associated with the distil part of the colon. To complement these findings the researchers also found that mice fed with a diet rich in acetylated starch had a significantly increased concentration of acetate in their feces and strongly suggested that targeted modulation of the diet should be considered as a therapeutic strategy to limit EHEC infections (Fukuda et al., 2011, 2012).
As discussed above the presence of favorable sugar sources can enhance disease progression of EHEC. Consistent with this, Curtis et al. (2014a) demonstrated that B. thetaiotaomicron, a major constituent of the intestinal microbiota, is involved in enriching the environment with simple monosaccharide sugars capable of being readily utilized by E. coli strains. The group had previously determined the mechanistic basis behind this, activation of the sugar sensitive transcription factor Cra, which leads to increased transcription of the LEE (Njoroge et al., 2012). In this study however, they identified a specific gluconeogenic substrate of Cra, succinate, which was responsible for increased disease severity in mice infected with C. rodentium. In contrast, antibiotic treated mice devoid of B. thetaiotaomicron were less susceptible to severe infection strongly suggesting that naturally produced sugars enhance the outcome of infection (Curtis et al., 2014a).
Exploiting Virulence Regulation in the Quest for Novel Therapeutic Strategies
Resistance of bacterial pathogens to antibiotics is no longer an emerging theme but is now considered a global health concern (Davies and Davies, 2010). In an effort to combat antibiotic resistance much emphasis has been placed on developing the concept of antivirulence therapy. This can be defined as the inhibition of non-essential virulence factors without being detrimental to normal cell growth (Rasko and Sperandio, 2010). Several classes of antivirulence compounds have been described thus far as and have been recently reviewed in detail (Beckham and Roe, 2014; Zambelloni et al., 2015). Targeting regulatory systems is a promising strategy to inhibit the activity of the virulence factors. For instance, an inhibitor compound of QseC, LED209, acts by allosterically interacting with this receptor thus blocking the QseC signaling cascade required for expression of multiple virulence factors. The compound also elicits promising broad spectrum activity against multiple pathogens that all utilize the QseC signaling system to regulate virulence (Rasko et al., 2008; Curtis et al., 2014b).
Specifically targeting the T3SS is well established in the field of antivirulence given the non-essential nature of this appendage. The SA class of inhibitor compounds have been extensively described and are efficient inhibitors of Type III secretion in EHEC (Tree et al., 2009; Wang et al., 2011). The mechanism of action of these inhibitors is unclear but a study outlining the transcriptional effects of EHEC treated with two different SA compounds under conditions that stimulate the LEE resulted in inhibition of type 3 secretion but an increase in motility. Given the known cross-regulation between these two systems one could postulate that the mechanism of inhibition is targeting the regulatory circuit of the LEE as opposed to the T3SS itself.
The LEE of C. rodentium confers the A/E phenotype upon this mouse-specific pathogen therefore allowing C. rodentium to be used as a model for EHEC infections. This is a unique pathogen however, and despite the core regulation of the LEE being under the control of similar mechanisms, such as Ler and GrlRA, C. rodentium specific regulation of the LEE has been extensively described (Deng et al., 2004). RegA is a C. rodentium specific AraC-like transcriptional regulator controlling gene expression on a global scale (Hart et al., 2008). Over 40 ORFS are under the control of RegA including GrlRA. The system is also highly responsive to bicarbonate ions, which are involved in intestinal homeostasis (Yang et al., 2008, 2009). Bicarbonate is secreted to neutralize stomach acid and this is in turn sensed by RegA to regulate gene expression appropriately (Kaunitz and Akiba, 2006). This control feeds into the LEE in a similar manner to the GAD system to correctly express the T3SS under favorable environmental conditions. As well as characterizing RegA transcriptional regulation in detail, researchers have also targeted RegA directly as a therapeutic strategy. The team characterized a compound, Regacin, that specifically inhibits DNA binding of RegA thus was capable of reducing LEE expression and protecting mice from C. rodentium infection (Yang et al., 2013). This study provides an excellent model for screening and elucidation of mechanisms involving antivirulence compounds while providing proof of concept that targeting regulation of the LEE directly is a viable option for new therapeutics.
Finally, exploitation of our knowledge of metabolism and how it regulates virulence provides yet another exciting route to explore. As discussed in this review, the LEE is responsive to numerous physiologically relevant signals including host or microbiota derived metabolites. This makes sense considering how important microbiome-associated homeostasis is to a healthy functioning gut. With this in mind it is important to emphasize the idea of dietary supplementation in combatting EHEC infection. As mentioned above, Yang et al. (2015) reduced EHEC infection in mice treated with a biotin enriched diet whereas Fukuda et al. (2011) successfully treated mice against EHEC infection using acetylated starch feed, mimicking the effects of protective members of the microbiota that naturally produce this metabolite. Supplementary zinc has also been used to limit diarrheal infection in children (Sazawal et al., 1995; Crane et al., 2007). Zinc was shown to indirectly reduce transcription of the LEE, and thus expression of the T3SS in EPEC, by a mechanism involving activation of envelope stress (Mellies et al., 2012). Zinc is also important for immune homeostasis in the host and has a very low toxicity suggesting that increased administration of supplementary zinc may be a useful strategy to limiting diarrheal infection associated with LEE-positive pathogens (Prasad, 2007; Bolick et al., 2014).
Conclusion
Nearly two decades of research on the LEE and how it is regulated has resulted in extensive knowledge of the system it encodes, the effects that A/E lesions and effector translocation have on the host, as well as how bacterial regulatory networks adapt to control this acquired genetic element appropriately. EHEC, EPEC, and C. rodentium have integrated the LEE island into their chromosomes and are capable of controlling its expression intuitively to respond to the environment as they travel through the host digestive system. There are many barriers to overcome along this journey such as survival in extreme pH and competition with the microbiota for limited nutrients, all the while responding to niche specific signals in order to regulate colonization appropriately. The detailed knowledge of how systems such as the QS network and microbiota associated metabolism affect regulation of the LEE has mediated the emergence of novel ways of targeting this virulence factor. These “bottom-up” approaches involving antivirulence compounds is an exciting field but much work remains to be done. What are the mechanisms of action of such strategies? How do they affect the pathogen and the host? Will this relationship lead to selective pressure for compound resistance? We are beginning to unravel these questions and advancing technologies are allowing more progressive ways of studying transcriptional regulation. For instance, the majority of work carried out on virulence factor regulation to date has been performed in vitro but in vivo regulation is much less explored. The application of next generation technologies such as RNA-sequencing (RNA-seq) to in vivo studies is allowing infection-relevant transcriptomic networks to be studied, as described for the bacterial pathogens Vibrio cholerae, Aggregatibacter actinomycetemcomitans, and more recently Yersinia pseudotuberculosis persistent infection (Mandlik et al., 2011; Jorth et al., 2013; Avican et al., 2015). Host and pathogen whole transcriptome profiling using RNA-sequencing (dual RNA-seq) is also emerging as the next step in infection-relevant gene expression studies and has been successfully applied to uropathogenic E. coli infected mouse macrophage samples to provide a comprehensive view of host-pathogen transcriptional responses to infection (Westermann et al., 2012; Mavromatis et al., 2015). The knowledge we have gained on LEE regulation is significant and continued work in this area in combination with next generation techniques will reveal more about pathogen adaptation to the host, strain specific regulatory factors and shed more light on the intricate relationship between virulence and metabolism, which has the potential to lead to new therapeutic strategies such as dietary supplementation to limit bacterial infections.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by an MRC funded studentship. We thank Professor Richard Cogdell for his support and supervision during the duration of this studentship.
References
Abe, H., Miyahara, A., Oshima, T., Tashiro, K., Ogura, Y., Kuhara, S., et al. (2008). Global regulation by horizontally transferred regulators establishes the pathogenicity of Escherichia coli. DNA Res. 15, 13–23. doi: 10.1093/dnares/dsm033
Anfora, A. T., Haugen, B. J., Roesch, P., Redford, P., and Welch, R. A. (2007). Roles of serine accumulation and catabolism in the colonization of the murine urinary tract by Escherichia coli CFT073. Infect. Immun. 75, 5298–5304. doi: 10.1128/IAI.00652-07
Atlung, T., and Ingmer, H. (1997). H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24, 7–17. doi: 10.1046/j.1365-2958.1997.3151679.x
Avican, K., Fahlgren, A., Huss, M., Heroven, A. K., Beckstette, M., Dersch, P., et al. (2015). Reprogramming of yersinia from virulent to persistent mode revealed by complex in vivo rna-seq analysis. PLoS Pathog. 11:e1004600. doi: 10.1371/journal.ppat.1004600
Barba, J., Bustamante, V. H., Flores-Valdez, M. A., Deng, W., Finlay, B. B., and Puente, J. L. (2005). A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA. J. Bacteriol. 187, 7918–7930. doi: 10.1128/JB.187.23.7918-7930.2005
Beckham, K. S. H., Connolly, J. P. R., Ritchie, J. M., Wang, D., Gawthorne, J. A., Tahoun, A., et al. (2014). The Metabolic enzyme AdhE controls the virulence of Escherichia coli O157:H7. Mol. Microbiol. 93, 199–211. doi: 10.1111/mmi.12651
Beckham, K. S. H., and Roe, A. J. (2014). From screen to target: insights and approaches for the development of anti-virulence compounds. Front. Cell. Infect. Microbiol. 4:139. doi: 10.3389/fcimb.2014.00139
Berdichevsky, T., Friedberg, D., Nadler, C., Rokney, A., Oppenheim, A., and Rosenshine, I. (2005). Ler is a negative autoregulator of the LEE1 operon in enteropathogenic Escherichia coli. J. Bacteriol. 187, 349–357. doi: 10.1128/JB.187.1.349-357.2005
Bertin, Y., Deval, C., de la Foye, A., Masson, L., Gannon, V., Harel, J., et al. (2014). The gluconeogenesis pathway is involved in maintenance of enterohaemorrhagic Escherichia coli O157:H7 in bovine intestinal content. PLoS ONE 9:e98367. doi: 10.1371/journal.pone.0098367
Bertin, Y., Girardeau, J. P., Chaucheyras-Durand, F., Lyan, B., Pujos-Guillot, E., Harel, J., et al. (2011). Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ. Microbiol. 13, 365–377. doi: 10.1111/j.1462-2920.2010.02334.x
Bolick, D. T., Kolling, G. L., Moore, J. H., de Oliveira, L. A., Tung, K., Philipson, C., et al. (2014). Zinc deficiency alters host response and pathogen virulence in a mouse model of enteroaggregative Escherichia coli-induced diarrhea. Gut Microbes 5, 618–627. doi: 10.4161/19490976.2014.969642
Branchu, P., Matrat, S., Vareille, M., Garrivier, A., Durand, A., Crépin, S., et al. (2014). NsrR, GadE, and GadX interplay in repressing expression of the Escherichia coli O157:H7 LEE pathogenicity island in response to nitric oxide. PLoS Pathog. 10:e1003874. doi: 10.1371/journal.ppat.1003874
Burkinshaw, B. J., Deng, W., Lameignère, E., Wasney, G. A., Zhu, H., Worrall, L. J., et al. (2015). Structural analysis of a specialized type III secretion system peptidoglycan-cleaving enzyme. J. Biol. Chem. 290, 10406–10417. doi: 10.1074/jbc.M115.639013
Büttner, D. (2012). Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol. Mol. Biol. Rev. 76, 262–310. doi: 10.1128/MMBR.05017-11
Cava, F., de Pedro, M. A., Lam, H., Davis, B. M., and Waldor, M. K. (2011). Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J. 30, 3442–3453. doi: 10.1038/emboj.2011.246
Chang, D.-E., Smalley, D. J., Tucker, D. L., Leatham, M. P., Norris, W. E., Stevenson, S. J., et al. (2004). Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. U.S.A. 101, 7427–7432. doi: 10.1073/pnas.0307888101
Cho, B.-K., Barrett, C. L., Knight, E. M., Park, Y. S., and Palsson, B. Ø. (2008). Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 105, 19462–19467. doi: 10.1073/pnas.0807227105
Clarke, M. B., Hughes, D. T., Zhu, C., Boedeker, E. C., and Sperandio, V. (2006). The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 10420–10425. doi: 10.1073/pnas.0604343103
Clarke, M. B., and Sperandio, V. (2005). Transcriptional regulation of flhDC by QseBC and sigma (FliA) in enterohaemorrhagic Escherichia coli. Mol. Microbiol. 57, 1734–1749. doi: 10.1111/j.1365-2958.2005.04792.x
Collins, J. W., Keeney, K. M., Crepin, V. F., Rathinam, V. A. K., Fitzgerald, K. A., Finlay, B. B., et al. (2014). Citrobacter rodentium: infection, inflammation and the microbiota. Nat. Rev. Microbiol. 12, 612–623. doi: 10.1038/nrmicro3315
Connolly, J. P., Goldstone, R. J., Burgess, K., Cogdell, R. J., Beatson, S. A., Vollmer, W., et al. (2015). The host metabolite D-serine contributes to bacterial niche specificity through gene selection. ISME J. 9, 1039–1051. doi: 10.1038/ismej.2014.242
Crane, J. K., Naeher, T. M., Shulgina, I., Zhu, C., and Boedeker, E. C. (2007). Effect of zinc in enteropathogenic Escherichia coli infection. Infect. Immun. 75, 5974–5984. doi: 10.1128/IAI.00750-07
Croxen, M. A., and Finlay, B. B. (2010). Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8, 26–38. doi: 10.1038/nrmicro2265
Croxen, M. A., Law, R. J., Scholz, R., Keeney, K. M., Wlodarska, M., and Finlay, B. B. (2013). Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 26, 822–880. doi: 10.1128/CMR.00022-13
Cummings, J. H., and Macfarlane, G. T. (1991). The control and consequences of bacterial fermentation in the human colon. J. Appl. Bacteriol. 70, 443–459. doi: 10.1111/j.1365-2672.1991.tb02739.x
Curtis, M. M., Hu, Z., Klimko, C., Narayanan, S., Deberardinis, R., and Sperandio, V. (2014a). The gut commensal bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe 16, 759–769. doi: 10.1016/j.chom.2014.11.005
Curtis, M. M., Russell, R., Moreira, C. G., Adebesin, A. M., Wang, C., Williams, N. S., et al. (2014b). QseC inhibitors as an antivirulence approach for Gram-negative pathogens. MBio 5, e02165. doi: 10.1128/mBio.02165-14
Dame, R. T., Noom, M. C., and Wuite, G. J. L. (2006). Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature 444, 387–390. doi: 10.1038/nature05283
Davies, J., and Davies, D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433. doi: 10.1128/MMBR.00016-10
De Biase, D., Tramonti, A., Bossa, F., and Visca, P. (1999). The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 32, 1198–1211.
Deng, W., Puente, L., Gruenheid, S., Li, Y., Vallance, B. A., Va, A., et al. (2004). Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. U.S.A. 101, 3597–3602. doi: 10.1073/pnas.0400326101
Deng, W., Yu, H. B., de Hoog, C. L., Stoynov, N., Li, Y., Foster, L. J., et al. (2012). Quantitative proteomic analysis of type III secretome of enteropathogenic Escherichia coli reveals an expanded effector repertoire for attaching/effacing bacterial pathogens. Mol. Cell. Proteomics 11, 692–709. doi: 10.1074/mcp.M111.013672
Dillon, S. C., and Dorman, C. J. (2010). Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195. doi: 10.1038/nrmicro2261
Dong, T., Coombes, B. K., and Schellhorn, H. E. (2009). Role of RpoS in the virulence of Citrobacter rodentium. Infect. Immun. 77, 501–507. doi: 10.1128/IAI.00850-08
Dong, T., and Schellhorn, H. E. (2009). Global effect of RpoS on gene expression in pathogenic Escherichia coli O157:H7 strain EDL933. BMC Genomics 10:349. doi: 10.1186/1471-2164-10-349
Dorman, C. J. (2007). H-NS, the genome sentinel. Nat. Rev. Microbiol. 5, 157–161. doi: 10.1038/nrmicro1598
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638. doi: 10.1126/science.1110591
Elliott, S. J., Sperandio, V., Girón, J. A., Mellies, J. L., Wainwright, L., Steven, W., et al. (2000). The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68, 6115–6126. doi: 10.1128/IAI.68.11.6115-6126.2000
Elliott, S. J., Wainwright, L. A., Timothy, K., Jarvis, K. G., Deng, Y., Lai, L., et al. (1998). The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia. Mol. Microbiol. 28, 1–4.
Elliott, S. J., Yu, J., and Kaper, J. B. (1999). The cloned locus of enterocyte effacement from enterohemorrhagic Escherichia coli O157:H7 is unable to confer the attaching and effacing phenotype upon E. coli K-12. Infect. Immun. 67, 4260–4263.
Foley, E., and O’Farrell, P. H. (2003). Nitric oxide contributes to induction of innate immune responses to gram-negative bacteria in Drosophila. Genes Dev. 17, 115–125. doi: 10.1101/gad.1018503
Frankel, G., Phillips, A. D., Rosenshine, I., Dougan, G., Kaper, J. B., and Knutton, S. (1998). Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30, 911–921.
Friedberg, D., Umanski, T., Fang, Y., and Rosenshine, I. (1999). Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 34, 941–952. doi: 10.1046/j.1365-2958.1999.01655.x
Fukuda, S., Toh, H., Hase, K., Oshima, K., Nakanishi, Y., Yoshimura, K., et al. (2011). Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547. doi: 10.1038/nature09646
Fukuda, S., Toh, H., Taylor, T. D., Ohno, H., and Hattori, M. (2012). Acetate-producing bifidobacteria protect the host from enteropathogenic infection via carbohydrate transporters. Gut Microbes 3, 449–454. doi: 10.4161/gmic.21214
Giangrossi, M., Zattoni, S., Tramonti, A., De Biase, D., and Falconi, M. (2005). Antagonistic role of H-NS and GadX in the regulation of the glutamate decarboxylase-dependent acid resistance system in Escherichia coli. J. Biol. Chem. 280, 21498–21505. doi: 10.1074/jbc.M413255200
Gill, S. R., Pop, M., Deboy, R. T., Eckburg, P. B., Turnbaugh, P. J., Samuel, B. S., et al. (2006). Metagenomic analysis of the human distal gut microbiome. Science 312, 1355–1359. doi: 10.1126/science.1124234
Goldberg, M. D., Johnson, M., Hinton, J. C. D., and Williams, P. H. (2001). Role of the nucleoid-associated protein Fis in the regulation of virulence properties of enteropathogenic Escherichia coli. Mol. Microbiol. 41, 549–559. doi: 10.1046/j.1365-2958.2001.02526.x
Gonyar, L. A., and Kendall, M. M. (2014). Ethanolamine and choline promote expression of putative and characterized fimbriae in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 82, 193–201. doi: 10.1128/IAI.00980-13
Grainger, D. C., Hurd, D., Goldberg, M. D., and Busby, S. J. W. (2006). Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 34, 4642–4652. doi: 10.1093/nar/gkl542
Gruber, C. C., and Sperandio, V. (2014). Posttranscriptional control of microbe-induced rearrangement of host cell actin. MBio 5, e01025–13. doi: 10.1128/mBio.01025-13
Gruber, C. C., and Sperandio, V. (2015). Global analysis of posttranscriptional regulation by GlmY and GlmZ in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 83, 1286–1295. doi: 10.1128/IAI.02918-14
Haack, K. R., Robinson, C. L., Miller, K. J., Fowlkes, J. W., and Mellies, J. L. (2003). Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect. Immun. 71, 384–392. doi: 10.1128/IAI.71.1.384-392.2003
Habdas, B. J., Smart, J., Kaper, J. B., and Sperandio, V. (2010). The LysR-type transcriptional regulator QseD alters type three secretion in enterohemorrhagic Escherichia coli and motility in K-12 Escherichia coli. J. Bacteriol. 192, 3699–3712. doi: 10.1128/JB.00382-10
Hansen, A.-M., and Kaper, J. B. (2009). Hfq affects the expression of the LEE pathogenicity island in enterohaemorrhagic Escherichia coli. Mol. Microbiol. 73, 446–465. doi: 10.1111/j.1365-2958.2009.06781.x
Hart, E., Yang, J., Tauschek, M., Kelly, M., Wakefield, M. J., Frankel, G., et al. (2008). RegA, an AraC-like protein, is a global transcriptional regulator that controls virulence gene expression in Citrobacter rodentium. Infect. Immun. 76, 5247–5256. doi: 10.1128/IAI.00770-08
Haugen, B. J., Pellett, S., Redford, P., Hamilton, H. L., Roesch, P. L., and Welch, R. A. (2007). In vivo gene expression analysis identifies genes required for enhanced colonization of the mouse urinary tract by uropathogenic Escherichia coli strain CFT073 dsdA. Infect. Immun. 75, 278–289. doi: 10.1128/IAI.01319-06
Hayashi, T., Makino, K., Ohnishi, M., Kurokawa, K., Ishii, K., Yokoyama, K., et al. (2001). Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8, 11–22. doi: 10.1093/dnares/8.1.11
Holmes, A., Lindestam Arlehamn, C. S., Wang, D., Mitchell, T. J., Evans, T. J., and Roe, A. J. (2012). Expression and regulation of the Escherichia coli O157:H7 effector proteins NleH1 and NleH2. PLoS ONE 7:e33408. doi: 10.1371/journal.pone.0033408
Honda, N., Iyoda, S., Yamamoto, S., Terajima, J., and Watanabe, H. (2009). LrhA positively controls the expression of the locus of enterocyte effacement genes in enterohemorrhagic Escherichia coli by differential regulation of their master regulators PchA and PchB. Mol. Microbiol. 74, 1393–1341. doi: 10.1111/j.1365-2958.2009.06937.x
Hughes, D. T., and Sperandio, V. (2008). Inter-kingdom signalling: communication between bacteria and their hosts. Nat. Rev. Microbiol. 6, 111–120. doi: 10.1038/nrmicro1836
Hughes, D. T., Terekhova, D. A., Liou, L., Hovde, C. J., Sahl, J. W., Patankar, A. V., et al. (2010). Chemical sensing in mammalian host-bacterial commensal associations. Proc. Natl. Acad. Sci. U.S.A. 107, 9831–9836. doi: 10.1073/pnas.1002551107
Ide, T., Laarmann, S., Greune, L., Schillers, H., Oberleithner, H., and Schmidt, M. A. (2001). Characterization of translocation pores inserted into plasma membranes by type III-secreted Esp proteins of enteropathogenic Escherichia coli. Cell. Microbiol. 3, 669–679. doi: 10.1046/j.1462-5822.2001.00146.x
Islam, M. S., Bingle, L. E. H., Pallen, M. J., and Busby, S. J. W. (2011a). Organization of the LEE1 operon regulatory region of enterohaemorrhagic Escherichia coli O157:H7 and activation by GrlA. Mol. Microbiol. 79, 468–483. doi: 10.1111/j.1365-2958.2010.07460.x
Islam, M. S., Pallen, M. J., and Busby, S. J. W. (2011b). A cryptic promoter in the LEE1 regulatory region of enterohaemorrhagic Escherichia coli: promoter specificity in AT-rich gene regulatory regions. Biochem. J. 436, 681–686. doi: 10.1042/BJ20110260
Islam, M. S., Shaw, R. K., Frankel, G., Pallen, M. J., and Busby, S. J. W. (2012). Translation of a minigene in the 5′ leader sequence of the enterohaemorrhagic Escherichia coli LEE1 transcription unit affects expression of the neighbouring downstream gene. Biochem. J. 441, 247–253. doi: 10.1042/BJ20110912
Iyoda, S., Koizumi, N., Satou, H., Lu, Y., Saitoh, T., Ohnishi, M., et al. (2006). The GrlR-GrlA regulatory system coordinately controls the expression of flagellar and LEE-encoded type III protein secretion systems in enterohemorrhagic Escherichia coli. J. Bacteriol. 188, 5682–5692. doi: 10.1128/JB.00352-06
Iyoda, S., and Watanabe, H. (2004). Positive effects of multiple pch genes on expression of the locus of enterocyte effacement genes and adherence of enterohaemorrhagic Escherichia coli O157: H7 to HEp-2 cells. Microbiology 150, 2357–2571. doi: 10.1099/mic.0.27100-0
Iyoda, S., and Watanabe, H. (2005). ClpXP protease controls expression of the type III protein secretion system through regulation of RpoS and GrlR levels in enterohemorrhagic Escherichia coli. J. Bacteriol. 187, 4086–4094. doi: 10.1128/JB.187.12.4086-4094.2005
Jorth, P., Trivedi, U., Rumbaugh, K., and Whiteley, M. (2013). Probing bacterial metabolism during infection using high-resolution transcriptomics. J. Bacteriol. 195, 4991–4998. doi: 10.1128/JB.00875-13
Kamada, N., Kim, Y.-G., Sham, H. P., Vallance, B. A., Puente, J. L., Martens, E. C., et al. (2012). Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336, 1325–1329. doi: 10.1126/science.1222195
Kanamaru, K., Tatsuno, I., Tobe, T., and Sasakawa, C. (2000). SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 38, 805–816. doi: 10.1046/j.1365-2958.2000.02171.x
Karmali, M. A., Petric, M., Lim, C., Fleming, P. C., and Steele, B. T. (1983a). Escherichia coli cytotoxin, haemolytic-uraemic syndrome, and haemorrhagic colitis. Lancet 2, 1299–1300.
Karmali, M. A., Steele, B. T., Petric, M., and Lim, C. (1983b). Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet 1, 619–620.
Kau, A. L., Ahern, P. P., Griffin, N. W., Goodman, A. L., and Gordon, J. I. (2011). Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336. doi: 10.1038/nature10213
Kaunitz, J. D., and Akiba, Y. (2006). Review article: duodenal bicarbonate—mucosal protection, luminal chemosensing and acid-base balance. Aliment. Pharmacol. Ther. 24(Suppl. 4), 169–176. doi: 10.1111/j.1365-2036.2006.00041.x
Kendall, M. M., Gruber, C. C., Parker, C. T., and Sperandio, V. (2012). Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. MBio 3, e00050–12. doi: 10.1128/mBio.00050-12
Kendall, M. M., Gruber, C. C., Rasko, D. A., Hughes, D. T., and Sperandio, V. (2011). Hfq virulence regulation in enterohemorrhagic Escherichia coli O157:H7 strain 86-24. J. Bacteriol. 193, 6843–6851. doi: 10.1128/JB.06141-11
Kendall, M. M., Rasko, D. A., and Sperandio, V. (2010). The LysR-type regulator QseA regulates both characterized and putative virulence genes in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 76, 1306–1321. doi: 10.1111/j.1365-2958.2010.07174.x
Kenny, B., Abe, A., Stein, M., and Finlay, B. B. (1997a). Enteropathogenic Escherichia coli protein secretion is induced in response to conditions similar to those in the gastrointestinal tract. Infect. Immun. 65, 2606–2612.
Kenny, B., DeVinney, R., Stein, M., Reinscheid, D. J., Frey, E. A., and Finlay, B. B. (1997b). Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91, 511–520.
Kitagawa, R., Takaya, A., and Yamamoto, T. (2011). Dual regulatory pathways of flagellar gene expression by ClpXP protease in enterohaemorrhagic Escherichia coli. Microbiology 157, 3094–3103. doi: 10.1099/mic.0.051151-0
Knutton, S., Rosenshine, I., Pallen, M. J., Nisan, I., Neves, B. C., Bain, C., et al. (1998). A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17, 2166–2176. doi: 10.1093/emboj/17.8.2166
Korte-Berwanger, M., Sakinc, T., Kline, K., Nielsen, H. V, Hultgren, S., and Gatermann, S. G. (2013). Significance of the D-serine-deaminase and D-serine metabolism of Staphylococcus saprophyticus for virulence. Infect. Immun. 81, 4525–4533. doi: 10.1128/IAI.00599-13
Laaberki, M.-H., Janabi, N., Oswald, E., and Repoila, F. (2006). Concert of regulators to switch on LEE expression in enterohemorrhagic Escherichia coli O157:H7: interplay between Ler, GrlA, HNS and RpoS. Int. J. Med. Microbiol. 296, 197–210. doi: 10.1016/j.ijmm.2006.02.017
Lam, H., Oh, D.-C., Cava, F., Takacs, C. N., Clardy, J., de Pedro, M. A., et al. (2009). D-amino acids govern stationary phase cell wall remodeling in bacteria. Science 325, 1552–1555. doi: 10.1126/science.1178123
Lathem, W. W., Grys, T. E., Witowski, S. E., Torres, A. G., Kaper, J. B., Tarr, P. I., et al. (2002). StcE, a metalloprotease secreted by Escherichia coli O157:H7, specifically cleaves C1 esterase inhibitor. Mol. Microbiol. 45, 277–288. doi: 10.1046/j.1365-2958.2002.02997.x
Leatham-Jensen, M. P., Frimodt-Møller, J., Adediran, J., Mokszycki, M. E., Banner, M. E., Caughron, J. E., et al. (2012). The streptomycin-treated mouse intestine selects Escherichia coli envZ missense mutants that interact with dense and diverse intestinal microbiota. Infect. Immun. 80, 1716–1727. doi: 10.1128/IAI.06193-11
Lehnen, D., Blumer, C., Polen, T., Wackwitz, B., Wendisch, V. F., and Unden, G. (2002). LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol. Microbiol. 45, 521–532. doi: 10.1046/j.1365-2958.2002.03032.x
Lodato, P. B., and Kaper, J. B. (2009). Post-transcriptional processing of the LEE4 operon in enterohaemorrhagic Escherichia coli. Mol. Microbiol. 71, 273–290. doi: 10.1111/j.1365-2958.2008.06530.x
Luzader, D. H., Clark, D. E., Gonyar, L. A., and Kendall, M. M. (2013). EutR is a direct regulator of genes that contribute to metabolism and virulence in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 195, 4947–4953. doi: 10.1128/JB.00937-13
Madrid, C., Balsalobre, C., García, J., and Juárez, A. (2007). The novel Hha/YmoA family of nucleoid-associated proteins: use of structural mimicry to modulate the activity of the H-NS family of proteins. Mol. Microbiol. 63, 7–14. doi: 10.1111/j.1365-2958.2006.05497.x
Mahajan, A., Currie, C. G., Mackie, S., Tree, J., McAteer, S., McKendrick, I., et al. (2009). An investigation of the expression and adhesin function of H7 flagella in the interaction of Escherichia coli O157:H7 with bovine intestinal epithelium. Cell. Microbiol. 11, 121–137. doi: 10.1111/j.1462-5822.2008.01244.x
Man, E. H., and Bada, J. L. (1987). Dietary D-amino acids. Annu. Rev. Nutr. 7, 209–225. doi: 10.1146/annurev.nu.07.070187.001233
Mandlik, A., Livny, J., Robins, W. P., Ritchie, J. M., Mekalanos, J. J., and Waldor, M. K. (2011). RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 10, 165–174. doi: 10.1016/j.chom.2011.07.007
Masuda, N., and Church, G. M. (2003). Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48, 699–712. doi: 10.1046/j.1365-2958.2003.03477.x
Mavromatis, C. H., Bokil, N. J., Totsika, M., Kakkanat, A., Schaale, K., Cannistraci, C. V., et al. (2015). The co-transcriptome of uropathogenic Escherichia coli-infected mouse macrophages reveals new insights into host-pathogen interactions. Cell. Microbiol. 17, 730–746. doi: 10.1111/cmi.12397
McDaniel, T. K., Jarvis, K. G., Donnenberg, M. S., and Kaper, J. B. (1995). A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 92, 1664–1668. doi: 10.1073/pnas.92.5.1664
McDaniel, T. K., and Kaper, J. B. (1997). A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23, 399–407.
Meador, J. P., Caldwell, M. E., Cohen, P. S., and Conway, T. (2014). Escherichia coli pathotypes occupy distinct niches in the mouse intestine. Infect. Immun. 82, 1931–1938. doi: 10.1128/IAI.01435-13
Mellies, J. L., Barron, A. M. S., and Carmona, A. M. (2007). Enteropathogenic and enterohemorrhagic Escherichia coli virulence gene regulation. Infect. Immun. 75, 4199–4210. doi: 10.1128/IAI.01927-06
Mellies, J. L., Elliott, S. J., Sperandio, V., Donnenberg, M. S., and Kaper, J. B. (1999). The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33, 296–306.
Mellies, J. L., Thomas, K., Turvey, M., Evans, N. R., Crane, J., Boedeker, E., et al. (2012). Zinc-induced envelope stress diminishes type III secretion in enteropathogenic Escherichia coli. BMC Microbiol. 12:123. doi: 10.1186/1471-2180-12-123
Miranda, R. L., Conway, T., Leatham, M. P., Chang, D. E., Norris, W. E., Allen, J. H., et al. (2004). Glycolytic and gluconeogenic growth of Escherichia coli O157:H7 (EDL933) and E. coli K-12 (MG1655) in the mouse intestine. Infect. Immun. 72, 1666–1676. doi: 10.1128/IAI.72.3.1666-1676.2004
Moon, H. W., Whipp, S. C., Argenzio, R. A., Levine, M. M., and Giannella, R. A. (1983). Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41, 1340–1351.
Moritz, R. L., and Welch, R. A. (2006). The Escherichia coli argW-dsdCXA genetic island is highly variable, and E. coli K1 strains commonly possess two copies of dsdCXA. J. Clin. Microbiol. 44, 4038–4048. doi: 10.1128/JCM.01172-06
Nagata, Y., Sato, T., Enomoto, N., Ishii, Y., Sasaki, K., and Yamada, T. (2007). High concentrations of D-amino acids in human gastric juice. Amino Acids 32, 137–140. doi: 10.1007/s00726-006-0262-9
Nakanishi, N., Tashiro, K., Kuhara, S., Hayashi, T., Sugimoto, N., and Tobe, T. (2009). Regulation of virulence by butyrate sensing in enterohaemorrhagic Escherichia coli. Microbiology 155, 521–530. doi: 10.1099/mic.0.023499-0
Nataro, J. P., and Kaper, J. B. (1998). Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11, 142–201.
Navarre, W. W., Porwollik, S., Wang, Y., McClelland, M., Rosen, H., Libby, S. J., et al. (2006). Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313, 236–238. doi: 10.1126/science.1128794
Naylor, S. W., Low, J. C., Besser, T. E., Mahajan, A., Gunn, G. J., Pearce, M. C., et al. (2003). Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect. Immun. 71, 1505–1512. doi: 10.1128/IAI.71.3.1505-1512.2003
Nguyen, Y. N., Sheng, H., Dakarapu, R., Falck, J. R., Hovde, C. J., and Sperandio, V. (2013). The acyl-homoserine lactone synthase YenI from Yersinia enterocolitica modulates virulence gene expression in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 81, 4192–4199. doi: 10.1128/IAI.00889-13
Njoroge, J. W., Nguyen, Y., Curtis, M. M., Moreira, C. G., and Sperandio, V. (2012). Virulence meets metabolism: Cra and KdpE gene regulation in enterohemorrhagic Escherichia coli. MBio 3, e00280–12. doi: 10.1128/mBio.00280-12
Orskov, F., Orskov, I., and Villar, J. A. (1987). Cattle as reservoir of verotoxin-producing Escherichia coli O157:H7. Lancet 2, 276.
Oshima, T., Ishikawa, S., Kurokawa, K., Aiba, H., and Ogasawara, N. (2006). Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 13, 141–153. doi: 10.1093/dnares/dsl009
Pacheco, A. R., Curtis, M. M., Ritchie, J. M., Munera, D., Waldor, M. K., Moreira, C. G., et al. (2012). Fucose sensing regulates bacterial intestinal colonization. Nature 492, 113–117. doi: 10.1038/nature11623
Padavannil, A., Jobichen, C., Mills, E., Velazquez-Campoy, A., Li, M., Leung, K. Y., et al. (2013). Structure of GrlR-GrlA complex that prevents GrlA activation of virulence genes. Nat. Commun. 4, 2546. doi: 10.1038/ncomms3546
Perna, N. T., Plunkett, G., Burland, V., Mau, B., Glasner, J. D., Rose, D. J., et al. (2001). Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409, 529–533. doi: 10.1038/35054089
Prieto, A. I., Kahramanoglou, C., Ali, R. M., Fraser, G. M., Seshasayee, A. S. N., and Luscombe, N. M. (2012). Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12. Nucleic Acids Res. 40, 3524–3537. doi: 10.1093/nar/gkr1236
Pruimboom-Brees, I. M., Morgan, T. W., Ackermann, M. R., Nystrom, E. D., Samuel, J. E., Cornick, N. A., et al. (2000). Cattle lack vascular receptors for Escherichia coli O157:H7 Shiga toxins. Proc. Natl. Acad. Sci. U.S.A. 97, 10325–10329. doi: 10.1073/pnas.190329997
Rasko, D. A., Moreira, C. G., Li, D. R., Reading, N. C., Ritchie, J. M., Waldor, M. K., et al. (2008). Targeting QseC signaling and virulence for antibiotic development. Science 321, 1078–1080. doi: 10.1126/science.1160354
Rasko, D. A., and Sperandio, V. (2010). Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 9, 117–128. doi: 10.1038/nrd3013
Reading, N. C., Rasko, D. A., Torres, A. G., and Sperandio, V. (2009). The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 106, 5889–5894. doi: 10.1073/pnas.0811409106
Reading, N. C., Torres, A. G., Kendall, M. M., Hughes, D. T., Yamamoto, K., and Sperandio, V. (2007). A novel two-component signaling system that activates transcription of an enterohemorrhagic Escherichia coli effector involved in remodeling of host actin. J. Bacteriol. 189, 2468–2476. doi: 10.1128/JB.01848-06
Ren, C.-P., Chaudhuri, R. R., Fivian, A., Bailey, C. M., Antonio, M., Barnes, W. M., et al. (2004). The ETT2 gene cluster, encoding a second type III secretion system from Escherichia coli, is present in the majority of strains but has undergone widespread mutational attrition. J. Bacteriol. 186, 3547–3560. doi: 10.1128/JB.186.11.3547-3560.2004
Riley, L. W., Remis, R. S., Helgerson, S. D., McGee, H. B., Wells, J. G., Davis, B. R., et al. (1983). Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308, 681–685. doi: 10.1056/NEJM198303243081203
Roe, A. J., Tysall, L., Dransfield, T., Wang, D., Fraser-Pitt, D., Mahajan, A., et al. (2007). Analysis of the expression, regulation and export of NleA-E in Escherichia coli O157:H7. Microbiology 153, 1350–1360. doi: 10.1099/mic.0.2006/003707-0
Roe, A. J., Yull, H., Naylor, S. W., Martin, J., Smith, D. G. E., Gally, D. L., et al. (2003). Heterogeneous surface expression of EspA translocon filaments by Escherichia coli O157:H7 is controlled at the posttranscriptional level. Infect. Immun. 71, 5900–5909. doi: 10.1128/IAI.71.10.5900-5909.2003
Roesch, P. L., Redford, P., Batchelet, S., Moritz, R. L., Pellett, S., Haugen, B. J., et al. (2003). Uropathogenic Escherichia coli use D-serine deaminase to modulate infection of the murine urinary tract. Mol. Microbiol. 49, 55–67. doi: 10.1046/j.1365-2958.2003.03543.x
Rosenshine, I., Ruschkowski, S., and Finlay, B. B. (1996). Expression of attaching/effacing activity by enteropathogenic Escherichia coli depends on growth phase, temperature, and protein synthesis upon contact with epithelial cells. Infect. Immun. 64, 966–973.
Russell, R. M., Sharp, F. C., Rasko, D. A., and Sperandio, V. (2007). QseA and GrlR/GrlA regulation of the locus of enterocyte effacement genes in enterohemorrhagic Escherichia coli. J. Bacteriol. 189, 5387–5392. doi: 10.1128/JB.00553-07
Sakinç, T., Michalski, N., Kleine, B., and Gatermann, S. G. (2009). The uropathogenic species Staphylococcus saprophyticus tolerates a high concentration of D-serine. FEMS Microbiol. Lett. 299, 60–64. doi: 10.1111/j.1574-6968.2009.01731.x
Sánchez-SanMartín, C., Bustamante, V. H., Calva, E., and Puente, J. L. (2001). Transcriptional regulation of the orf19 gene and the tir-cesT-eae operon of enteropathogenic Escherichia coli. J. Bacteriol. 183, 2823–2833. doi: 10.1128/JB.183.9.2823-2833.2001
Sazawal, S., Black, R. E., Bhan, M. K., Bhandari, N., Sinha, A., and Jalla, S. (1995). Zinc supplementation in young children with acute diarrhea in India. N. Engl. J. Med. 333, 839–844. doi: 10.1056/NEJM199509283331304
Schauer, D. B., and Falkow, S. (1993). Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infect. Immun. 61, 2486–2492.
Shakhnovich, E. A., Davis, B. M., and Waldor, M. K. (2009). Hfq negatively regulates type III secretion in EHEC and several other pathogens. Mol. Microbiol. 74, 347–363. doi: 10.1111/j.1365-2958.2009.06856.x
Sharma, V. K., and Zuerner, R. L. (2004). Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 186, 7290–7301. doi: 10.1128/JB.186.21.7290-7301.2004
Sharp, F. C., and Sperandio, V. (2007). QseA directly activates transcription of LEE1 in enterohemorrhagic Escherichia coli. Infect. Immun. 75, 2432–2440. doi: 10.1128/IAI.02003-06
Shin, S., Castanie-Cornet, M. P., Foster, J. W., Crawford, J. A., Brinkley, C., and Kaper, J. B. (2001). An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microbiol. 41, 1133–1150. doi: 10.1046/j.1365-2958.2001.02570.x
Sperandio, V., Li, C. C., and Kaper, J. B. (2002a). Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect. Immun. 70, 3085–3093. doi: 10.1128/IAI.70.6.3085-3093.2002
Sperandio, V., Torres, A. G., and Kaper, J. B. (2002b). Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43, 809–821. doi: 10.1046/j.1365-2958.2002.02803.x
Sperandio, V., Mellies, J. L., Nguyen, W., Shin, S., and Kaper, J. B. (1999). Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 96, 15196–15201.
Takao, M., Yen, H., and Tobe, T. (2014). LeuO enhances butyrate-induced virulence expression through a positive regulatory loop in enterohaemorrhagic Escherichia coli. Mol. Microbiol. 93, 1302–1313. doi: 10.1111/mmi.12737
Tatsuno, I., Nagano, K., Taguchi, K., Rong, L., Mori, H., and Sasakawa, C. (2003). Increased adherence to Caco-2 cells caused by disruption of the yhiE and yhiF genes in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 71, 2598–2606. doi: 10.1128/IAI.71.5.2598-2606.2003
Tan, A., Petty, N. K., Hocking, D., Bennett-Wood, V., Wakefield, M., Praszkier, J., et al. (2015). Evolutionary adaptation of an AraC-like regulatory protein in Citrobacter rodentium and Escherichia species. Infect. Immun. 83, 1384–1395. doi: 10.1128/IAI.02697-14
Tobe, T., Beatson, S. A., Taniguchi, H., Abe, H., Bailey, C. M., Fivian, A., et al. (2006). An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc. Natl. Acad. Sci. U.S.A. 103, 14941–14946. doi: 10.1073/pnas.0604891103
Tobe, T., Nakanishi, N., and Sugimoto, N. (2011). Activation of motility by sensing short-chain fatty acids via two steps in a flagellar gene regulatory cascade in enterohemorrhagic Escherichia coli. Infect. Immun. 79, 1016–1024. doi: 10.1128/IAI.00927-10
Tree, J. J., Roe, A. J., Flockhart, A., McAteer, S. P., Xu, X., Shaw, D., et al. (2011). Transcriptional regulators of the GAD acid stress island are carried by effector protein-encoding prophages and indirectly control type III secretion in enterohemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 80, 1349–1365. doi: 10.1111/j.1365-2958.2011.07650.x
Tree, J. J., Wang, D., McInally, C., Mahajan, A., Layton, A., Houghton, I., et al. (2009). Characterization of the effects of salicylidene acylhydrazide compounds on type III secretion in Escherichia coli O157:H7. Infect. Immun. 77, 4209–4220. doi: 10.1128/IAI.00562-09
Turnbaugh, P. J., and Gordon, J. I. (2009). The core gut microbiome, energy balance and obesity. J. Physiol. 587, 4153–4158. doi: 10.1113/jphysiol.2009.174136
Umanski, T., Rosenshine, I., and Friedberg, D. (2002). Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology 148, 2735–2744.
Vaishnava, S., Yamamoto, M., Severson, K. M., Ruhn, K. A., Yu, X., Koren, O., et al. (2011). The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258. doi: 10.1126/science.1209791
Walters, M., and Sperandio, V. (2006). Autoinducer 3 and epinephrine signaling in the kinetics of locus of enterocyte effacement gene expression in enterohemorrhagic Escherichia coli. Infect. Immun. 74, 5445–5455. doi: 10.1128/IAI.00099-06
Wang, D., Zetterström, C. E., Gabrielsen, M., Beckham, K. S. H., Tree, J. J., Macdonald, S. E., et al. (2011). Identification of bacterial target proteins for the salicylidene acylhydrazide class of virulence-blocking compounds. J. Biol. Chem. 286, 29922–29931. doi: 10.1074/jbc.M111.233858
Westermann, A. J., Gorski, S. A., and Vogel, J. (2012). Dual RNA-seq of pathogen and host. Nat. Rev. Microbiol. 10, 618–630. doi: 10.1038/nrmicro2852
Wolosker, H., Dumin, E., Balan, L., and Foltyn, V. N. (2008). D-amino acids in the brain: D-serine in neurotransmission and neurodegeneration. FEBS J. 275, 3514–3526. doi: 10.1111/j.1742-4658.2008.06515.x
Wong, A. R. C., Pearson, J. S., Bright, M. D., Munera, D., Robinson, K. S., Lee, S. F., et al. (2011). Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol. Microbiol. 80, 1420–1438. doi: 10.1111/j.1365-2958.2011.07661.x
Wong, J. M. W., de Souza, R., Kendall, C. W. C., Emam, A., and Jenkins, D. J. A. (2006). Colonic health: fermentation and short chain fatty acids. J. Clin. Gastroenterol. 40, 235–243. doi: 10.1097/00004836-200603000-00015
Wu, G. D., Chen, J., Hoffmann, C., Bittinger, K., Chen, Y.-Y., Keilbaugh, S. A., et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108. doi: 10.1126/science.1208344
Yang, B., Feng, L., Wang, F., and Wang, L. (2015). Enterohemorrhagic Escherichia coli senses low biotin status in the large intestine for colonization and infection. Nat. Commun. 6, 6592. doi: 10.1038/ncomms7592
Yang, J., Dogovski, C., Hocking, D., Tauschek, M., Perugini, M., and Robins-Browne, R. M. (2009). Bicarbonate-mediated stimulation of RegA, the global virulence regulator from Citrobacter rodentium. J. Mol. Biol. 394, 591–599. doi: 10.1016/j.jmb.2009.10.033
Yang, J., Hart, E., Tauschek, M., Price, G. D., Hartland, E. L., Strugnell, R. A., et al. (2008). Bicarbonate-mediated transcriptional activation of divergent operons by the virulence regulatory protein, RegA, from Citrobacter rodentium. Mol. Microbiol. 68, 314–327. doi: 10.1111/j.1365-2958.2008.06171.x
Yang, J., Hocking, D. M., Cheng, C., Dogovski, C., Perugini, M. A., Holien, J. K., et al. (2013). Disarming bacterial virulence through chemical inhibition of the DNA binding domain of an AraC-like transcriptional activator protein. J. Biol. Chem. 288, 31115–31126. doi: 10.1074/jbc.M113.503912
Yona-Nadler, C., Umanski, T., Aizawa, S.-I., Friedberg, D., and Rosenshine, I. (2003). Integration host factor (IHF) mediates repression of flagella in enteropathogenic and enterohaemorrhagic Escherichia coli. Microbiology 149, 877–884. doi: 10.1099/mic.0.25970-0
Zambelloni, R., Marquez, R., and Roe, A. J. (2015). Development of antivirulence compounds: a biochemical review. Chem. Biol. Drug Des. 85, 43–55. doi: 10.1111/cbdd.12430
Zhang, L., Chaudhuri, R. R., Constantinidou, C., Hobman, J. L., Patel, M. D., Jones, A. C., et al. (2004). Regulators encoded in the Escherichia coli type III secretion system 2 gene cluster influence expression of genes within the locus for enterocyte effacement in enterohemorrhagic E. coli O157:H7. Infect. Immun. 72, 7282–7293. doi: 10.1128/IAI.72.12.7282-7293.2004
Keywords: virulence, regulation, secretion, host, environmental, Escherichia coli
Citation: Connolly JPR, Finlay BB and Roe AJ (2015) From ingestion to colonization: the influence of the host environment on regulation of the LEE encoded type III secretion system in enterohaemorrhagic Escherichia coli. Front. Microbiol. 6:568. doi: 10.3389/fmicb.2015.00568
Received: 01 April 2015; Accepted: 24 May 2015;
Published: 05 June 2015.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Victoria Auerbuch, University of California, Santa Cruz, USAMatthew S. Francis, Umeå University, Sweden
Copyright © 2015 Connolly, Finlay and Roe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrew J. Roe, Institute of Infection, Immunity and Inflammation, College of Medical, Veterinary and Life Sciences, University of Glasgow, Room B214, Glasgow G12 8TA, UK,YW5kcmV3LnJvZUBnbGFzZ293LmFjLnVr
 James P. R. Connolly
James P. R. Connolly