- 1Key Laboratory of Quality and Safety Risk Assessment for Aquatic Products on Storage and Preservation (Shanghai), China Ministry of Agriculture, College of Food Science and Technology, Shanghai Ocean University, Shanghai, China
- 2Department of Food Science and Technology, The Ohio State University, Columbus, OH, USA
Vibrio parahaemolyticus is a causative agent of serious human seafood-borne gastroenteritis disease and even death. In this study, for the first time, we obtained the secretomic profiles of seven V. parahaemolyticus strains of clinical and food origins. The strains exhibited various toxic genotypes and phenotypes of antimicrobial susceptibility and heavy metal resistance, five of which were isolated from aquatic products in Shanghai, China. Fourteen common extracellular proteins were identified from the distinct secretomic profiles using the two-dimensional gel electrophoresis (2-DE) and liquid chromatography tandem mass spectrometry (LC-MS/MS) techniques. Of these, half were involved in protein synthesis and sugar transport of V. parahaemolyticus. Strikingly, six identified proteins were virulence-associated factors involved in the pathogenicity of some other pathogenic bacteria, including the translation elongation factor EF-Tu, pyridoxine 5′-phosphate synthase, σ54 modulation protein, dihydrolipoyl dehydrogenase, transaldolase and phosphoglycerate kinase. In addition, comparative secretomics also revealed several extracellular proteins that have not been described in any bacteria, such as the ribosome-recycling factor, translation elongation factor EF-Ts, phosphocarrier protein HPr and maltose-binding protein MalE. The results in this study will facilitate the better understanding of the pathogenesis of V. parahaemolyticus and provide data in support of novel vaccine candidates against the leading seafood-borne pathogen worldwide.
Introduction
Vibrio parahaemolyticus is a Gram-negative bacterium that thrives in marine, estuarine and aquaculture environments worldwide (Broberg et al., 2011; Letchumanan et al., 2014). The bacterium is a causative agent of serious human seafood-borne gastroenteritis disease and even death (Boyd et al., 2008; Ceccarelli et al., 2013). Previous research indicated that pathogenic bacteria establish infection, elicit diseases and survive in hostile environments via a large armamentarium of virulence mechanisms (Chen et al., 2012). Pathogenic V. parahaemolyticus strains have been reported to produce two major toxic proteins, thermostable direct haemolysin (TDH) and TDH-related haemolysin (TRH), in the human gastrointestinal tract where it elicits diarrhea disease (Boyd et al., 2008). Nevertheless, approximately 90–99% of V. parahaemolyticus isolates of environmental origins were detected negative for the two major toxic genes (e.g., Su and Liu, 2007; Martinez-Urtaza et al., 2008; Flores-Primo et al., 2014; Haley et al., 2014; Letchumanan et al., 2015). Environmental isolates lacking tdh and/or trh are also highly cytotoxic to human gastrointestinal cells, indicating other virulence factors exist (Raghunath, 2014). Due to the intricacy of the pathogenesis of V. parahaemolyticus, there exists a clear need for a complete understanding of the other virulence-associated factors of the bacterium.
Virulence factors are secreted by bacterial secretion systems (Wooldridge, 2009). Numerous previous studies have been conducted to characterize effector proteins secreted by toxic V. parahaemolyticus isolates, which possess virulence-associated type III and type VI secretion systems (T3SS and T6SS) (Makino et al., 2003). Recently, several T3SS-secreted proteins have been characterized, which manipulated host cell mitogen activated protein kinase for critical steps in the pathogenesis of V. parahaemolyticus (Ono et al., 2006; Matlawska-Wasowska et al., 2010). For example, VopQ encoded by the VP1680 gene of V. parahaemolyticus RIMD2210633 (GenBank accession no. NC_004603.1, NC_004605.1) induced PI3-kinase-independent autophagy and antagonized phagocytosis (Burdette et al., 2009), while VopS (VP1686) induced toll-like receptor-independent apoptosis in macrophage through NF-kappaB inhibition (Bhattacharjee et al., 2006; Yarbrough et al., 2009). They also suppressed inflammasome activation by interfering with host autophagy signaling (Higa et al., 2013). In addition, VopV (Vpa1357) was an F-actin-binding effector and required for V. parahaemolyticus-induced enterotoxicity (Hiyoshi et al., 2011), while VopZ (Vpa1336) mediated pathogenesis by independently enabling intestinal colonization and inhibiting the transforming growth factor β-activated kinase activation (Zhou et al., 2013). Recent research also revealed cell density- and quorum sensing-dependent expression of T6SS2 in V. parahaemolyticus (Wang et al., 2013).
To date, very few information is available in secreted proteins of non-toxic bacterial strains, including V. parahaemolyticus. Herein, for the first time, we investigated the secretomes of seven V. parahaemolyticus strains of clinical and food origins. The strains exhibited various toxic genotypes and resistant phenotypes. Fourteen common extracellular proteins were characterized using the two-dimensional gel electrophoresis (2-DE) and liquid chromatography tandem mass spectrometry (LC-MS/MS) techniques, most of which have not been reported previously in V. parahaemolyticus. The information in this study sheds light on the pathogenesis of the leading seafood-borne pathogen worldwide.
Materials and Methods
V. parahaemolyticus Isolation and Identification
V. parahaemolyticus isolation and identification were performed according to the methods described previously (Song et al., 2013). Aquatic products including shrimps and shellfish were sampled from fish markets in Shanghai, China in 2011–2014. DNA sequencing was carried out by Shanghai Sangon Biological Engineering Technology and Services Co., Ltd. (Shanghai, China). The 16S rDNA sequences of the strains identified in this study were deposited in the GenBank database under the accession numbers from KP696473 to KP696477.
Susceptibility to Antimicrobial Agents and Heavy Metals
V. parahaemolyticus strains were measured for in vitro susceptibility to 10 antimicrobial agents and heavy metals according to the methods described previously (Song et al., 2013). The heavy metals used in this study included: NiCl2, CrCl3, CdCl2, PbCl2, CuCl2, ZnCl2, MnCl2 and HgCl2 (Sinopharm Chemical Reagent Co., Ltd, Shanghai, China).
Pulsed-field Gel Electrophoresis (PFGE) Analysis
V. parahaemolyticus strains were individually cultured in Luria-Bertani (LB) broth (Beijing Land Bridge Technology Co., Ltd., Beijing, China) (pH 7.0, 1% NaCl) aerobically at 37°C with shaking at 180 rpm overnight. Genomic DNA was isolated using the CHEF Bacterial DNA Plug Kit (Bio-Rad Laboratories Inc., Hercules, CA, USA) according to the manufacturer's instructions. Briefly, each agarose plug was prepared by mixing an equal volume of the bacterial cell culture and melted 2% CleanCut agarose provided by the Kit, and then digested with NotI restriction endonulease (Japan TaKaRa BIO, Dalian Company, Dalian, China) at 37°C overnight according to the manufacturer's instructions. The resulting DNA fragments were resolved using the CHEF Mapper system (Bio-Rad Laboratories Inc.) with 0.5 × Tris-borate ethylene diamine tetraacetic acid (EDTA) buffer (Shanghai Sangon Biological Engineering Technology and Services Co., Ltd.). Electrophoresis was performed at 6 V/cm at 14°C with a field angle of 120° using 1% SeaKem® Gold Agarose gel (Lonza, Basel, Switzerland). The pulse time was ramped from 10 to 35.03 s over 19 h. Following electrophoresis, the gels were stained with 0.5 μg/ml ethidium bromide (Shanghai Sangon Biological Engineering Technology and Services Co., Ltd.). The PFGE patterns were visualized under short-wavelength UV light (260 nm) and imaged using the UVP EC3 Imaging system (UVP LLC, Upland, CA, USA).
Isolation of Extracellular Proteins of V. parahaemolyticus Strains
Extracellular proteins of V. parahaemolyticus strains were isolated according to the method described by Ono et al. with minor modifications (Ono et al., 2006). Briefly, V. parahaemolyticus strains were individually cultured in the LB broth (pH 8.5, 3% NaCl) to mid-logarithmic phase at 37°C without shaking. Growth curves were determined using the Synergy 2 Multi-Mode Microplate Reader (BioTek Instruments, Inc., Winooski, VT, USA) as described previously (Sun et al., 2014). Bacterial protease inhibitors complex (Shanghai Sangon Biological Engineering Technology and Services Co., Ltd.) was added to each bacterial cell culture, which was then centrifuged at 12,000 rpm for 5 min at 4°C. The supernatant was filtered through 0.22 μm pore-size membranes (Millipore, Bedford, MA, USA) to remove residual bacterial cells. The filtrate was precipitated by adding trichloracetic acid (TCA) to a final concentration of 10% (vol/vol), and then incubated on ice overnight. The extracellular proteins were collected by centrifugation at 12,000 rpm for 30 min at 4°C. The pellet was washed with ice-cold acetone for five times, air-dried and stored at −80°C. Protein concentration was measured using the BCA Protein Assay Kit (Shanghai Sangon Biological Engineering Technology and Services Co., Ltd.) with bovine serum albumin as the standard.
2-DE Analysis
The 2-DE analysis was performed according to the manufacturer's instructions for E. coli (Bio-Rad Laboratories Inc.) with slight modifications such as bacterial culture and collection conditions. Extracellular protein pellets were individually dissolved with 1 ml of rehydration solution containing 8 M urea, 4% (wt/vol) 3-[3-cholamidopropyl-dimethylammonio-1-propanesulfonate (CHAPS) (Bio-Rad Laboratories Inc.)], 2 mM tributyl phosphine (TBP) (Bio-Rad Laboratories Inc.), 0.2% (vol/vol) Bio-Lyte 3/10 ampholyte (Bio-Rad Laboratories Inc.) and 0.0002% (wt/vol) bromophenol blue, and then centrifuged at 12,000 rpm for 10 min at room temperature to remove undissolved residues. Isoelectric focusing (IEF) was performed as the first-dimension separation with the immobilized pH gradient gel (IPG) strip (pH 3-10/NL, 17 cm) (Bio-Rad Laboratories Inc.). Aliquots of each 300 μl of protein sample were individually applied to the strips (pH 3-10/NL, 17 cm) and positive rehydrated for 17 h at 17°C. After rehydration, IEF of the samples was run with a 8-step program: 50, 100, 500 and 1000 V for 1 h with slow ramping, 2000 and 4000 V for 1 h with linear ramping, 6000 V for 3 h with rapid ramping, and 10,000 V with rapid ramping until 70,000 V-hr was reached. Following the electrophoresis in the first dimension, the IPG gel strip was incubated in an equilibration buffer I [6 M urea, 0.05 M Tris, 2% SDS, 20% glycerol, 2% (wt/vol) dithiothreitol] for 15 min, and then washed for a further 15 min with an equilibration buffer II [6 M urea, 0.05 M Tris, 2% SDS, 20% glycerol, 2.5% (wt/vol) iodoacetamide (Sigma-Aldrich, MO, USA)].
The second-dimension separation was performed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 15% separation gel using a Mini-PROTEANW electrophoresis cell (Bio-Rad Laboratories Inc.) as described previously (Shi et al., 2013). After electrophoresis, the gel was stained using the Silver Stain Plus Kit (Bio-Rad Laboratories Inc.) according to the manufacturer's instructions. The stained gels were stored in 1% acetic acid at 4°C. The assays were performed in triplicates. The PDQuest Advanced-8.0.1 software (Bio-Rad Laboratories Inc.) was used for the detection and analysis of protein spots on the 2-DE profiles.
LC-MS/MS Analysis
The visible and discriminative protein spots were individually excised from the 2-DE gels and then digested by freshly prepared Sequencing Grade Modified Trypsin (Promega, Madison, WI, USA) at 37°C overnight. The resulting peptides were analyzed at Beijing Protein Innovation (Beijing, China) using liquid chromatography tandem mass spectrometry (LC-MS) with a serially coupled microcolumn in a Paradigm HPLC system (Shimadzu Inc., Kyoto, Japan) coupled with a microTOF-QII mass spectrometer (Bruker, Billerica, Germany) (LC-MS/MS). The data collection was performed using the Bruker Data Analysis 4.0 software (Bruker, Billerica, MA, Germany).
Data Analysis
Data analysis was carried out at Beijing Protein Innovation. The collected LC-MS/MS data files were converted to the Mascot generic format (mgf). The combined mgf-files derived from triplicate analyses were sent to the Mascot server (Version 2.3.01, Matrix Science, London, UK) for automated peptide identification using the UniProt database. The following Mascot settings were used: carbamidomethyl was specified as a fixed modification; methionine oxidation was specified with variable modifications; two missed cleavages were allowed. The precursor and fragment mass tolerance was set to 0.1 Da, and the significance threshold was p < 0.05.
Results
V. parahaemolyticus Isolation and Identification
Aquatic products were sampled from fish markets in Shanghai in 2011–2014. Pure cultures of V. parahaemolyticus isolates were identified according to the instructions of the China Government Standard (GB17378-2007) and the Standard of the Bacteriological Analytical Manual (BAM) of U.S. Food and Drug Administration (8th Edition, Revision A, 1998). A total of 334 V. parahaemolyticus strains were obtained and detected positive for the thermolabile hemolysin gene (tlh) by polymerase chain reaction (PCR). Toxin-related genes were also examined by PCR. In most cases, the V. parahaemolyticus strains were featured as not virulent, since amplification of toxic tdh gene was negative, whereas approximately 97.1% of the isolates yielded no product for the trh gene. Secretomes of five representative V. parahaemolyticus strains isolated from the most common shrimps sold in Shanghai, China were assessed in this study (Table 1). Of these, V. parahaemolyticus Chn201 and Chn204 strains were detected as the trh-positive genotype, while the other strains were negative for the toxic genes (Table 1).
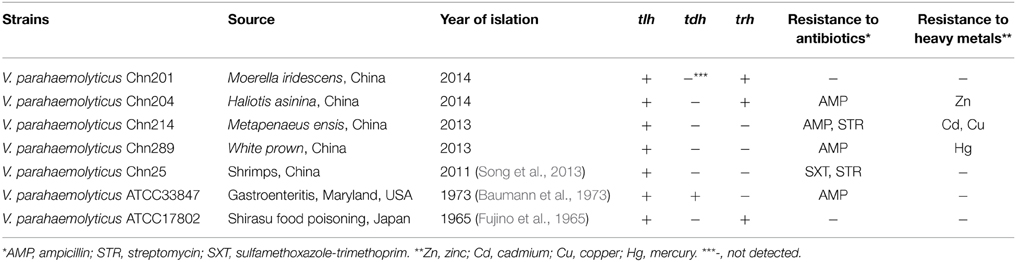
Table 1. Toxic genotypes and phenotypes of antimicrobial susceptibility and heavy metal resistance of the V. parahaemolyticus strains.
PFGE-based Genotypes of the V. parahaemolyticus Strains
In order to compare the secretomes of toxic and non-toxic V. parahaemolyticus strains, we used two standard toxic strains in this study, including the ATCC33847 (tdh+trh−) and ATCC17802 (tdh−trh+) isolated from clinical and food-poisoning cases, respectively (Fujino et al., 1965; Baumann et al., 1973). Genomic DNA of the seven V. parahaemolyticus strains was individually digested with the restriction endonuclease NotI, and the resulting DNA fragments were resolved by PFGE. This analysis revealed different genomic fingerprints of the strains tested (Figure 1). Clustering analysis of the genomic fingerprints revealed seven distinguishable NotI-PFGE types, demonstrating that the strains chosen for the secretomic analysis exhibited various genotypes.
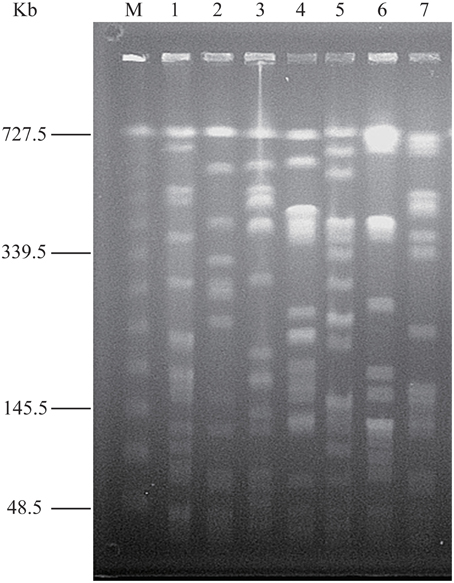
Figure 1. NotI-PFGE genotyping of the V. parahaemolyticus strains. Lane M: Lambda Ladder PFG Marker (48.5–727.5 kb, New England Biolabs, Beijing Company, Beijing, China); Lane 1-7: V. parahaemolyticus Chn25, ATCC17802, Chn201, Chn204, Chn214, Chn289 and ATCC33847 strains, respectively.
Antimicrobial Susceptibility and Heavy Metal Resistance of the V. parahaemolyticus Strains
Antimicrobial susceptibility of the V. parahaemolyticus strains were determined and the results were summarized in Table 1. All the strains were susceptible to three of the 10 antimicrobial agents tested, including chloramphenicol, gentamicin and tetracycline. Of these, Chn201 (tdh−trh+) and ATCC17802 (tdh−trh+) strains were susceptible to all the 10 drugs. In contrast, ampicillin resistance was the most predominant among the strains examined in this study.
Susceptibility of the strains to heavy metals including cadmium (Cd), chromium (Cr), copper (Cu), mercury (Hg), manganese (Mn), nickel (Ni), lead (Pb) and zinc (Zn) was also determined (Table 1). Chn214 strain displayed resistance phenotypes to Cd and Cu, while Chn204 and Chn289 strains resistance to Zn and Hg, respectively. In contrast, all the strains were susceptible to the four heavy metals tested, including Ni, Cr, Mn and Pb. Moreover, Chn25, Chn201, ATCC33847 and ATCC17802 strains showed no resistance to all the heavy metals tested.
Secretome Profiles of the V. parahaemolyticus Strains
To gain an insight into the possible growth dynamics of the V. parahaemolyticus strains with various genotypes and phenotypes, we determined growth curves of the bacteria cultured in LB broth (pH 8.5, 3% NaCl) at 37°C without shaking. As illustrated in Figure S1 (Supplementary Material), the growth of ATCC33847 stain of clinical origin was notably slower than the other strains. However, no obvious difference in growth was observed under the same condition among the other strains.
The supernatant of the cultures at mid-logarithmic growth phase with OD600nm values ranging from 0.5 to 0.6 were collected, and extracellular proteins were isolated, and analyzed by 2-DE. This analysis uncovered distinct secretome profiles (Figure 2), showing various numbers of visible protein spots. The patterns yielded from three independent 2-DE gels per biological sample were consistent (the data not shown). Based on the consensus patterns, one interesting observation was that toxic ATCC33847 and ATCC17802 strains appeared to secret more extracellular proteins (56–59) than the five strains derived from aquatic products (20–36), including Chn201 and Chn204 strains with the trh+ toxic genotype. In addition, 16 protein spots, designated vps1 to vps16, were observed at similar locations on all the 2-DE patterns derived from the strains tested (Figure 2, marked with the same numbers). These protein spots were excised from the 2-DE gels and digested with the trypsin. The resulting peptides were further identified by LC-MS/MS.
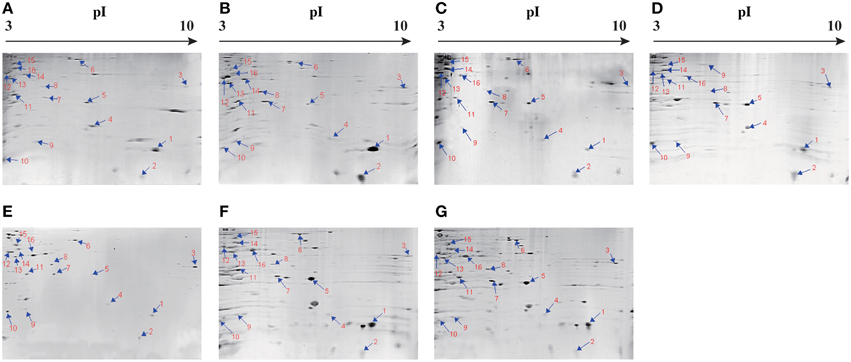
Figure 2. Isolation of extracellular proteins of the V. parahaemolyticus strains by 2-DE. (A–G): V. parahaemolyticus Chn201, Chn204, Chn214, Chn289, Chn25, ATCC33847 and ATCC17802, respectively. The protein spots marked with the same numbers in red were characterized by LC-MS/MS analysis. pI, isoelectric point.
Identification of Common Extracellular Proteins of the V. parahaemolyticus Strains
The results yielded from LC-MS/MS analysis were summarized in Table 2. Total 14 protein sequences were successfully obtained. The metabolism-related proteins constituted the largest proportion of the identified proteins shared among the V. parahaemolyticus strains tested. Of these, strikingly, approximately 43% were associated with protein synthesis of the bacterium. For example, the protein spots vps3 and vps4 were identified as the components S2 and S6 of the 30S subunit of bacterial ribosome, respectively, while the vps9 was the component L9 of the 50S subunit. In addition, the vps5 was a ribosome-recycling factor (RRF), moreover, two other translation-associated proteins, vps15 and vps16, were also found. The former was an elongation factor Tu (EF-Tu), while the latter matched an elongation factor Ts (EF-Ts), a guanosine nucleotide exchange factor for EF-Tu.
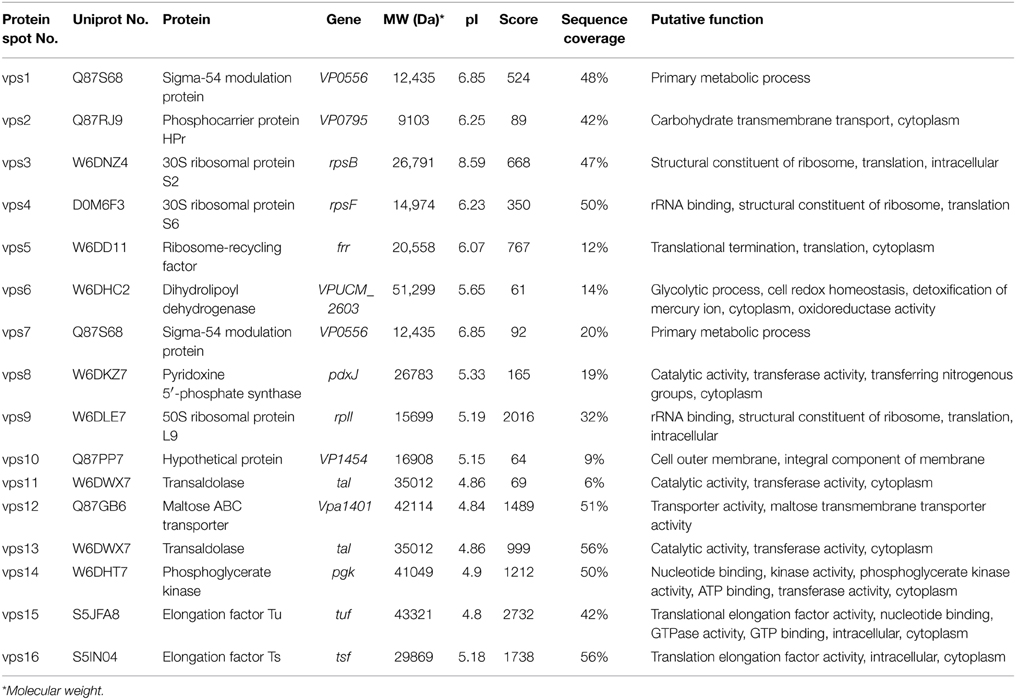
Table 2. Identification of the protein spots from vps1 to vps16 on the secretome profiles by LC-MS/MS analysis.
The protein spot vps8 was identified as a pyridoxine 5′-phosphate synthase (PdxJ) that catalyzes the final-step reaction of vitamin B6 de novo synthesis, a complicated ring closure reaction between the two acyclic compounds 1-deoxy-D-xylulose-5-phosphate and aminoacetone-3-phosphate to form pyridoxine 5′-phosphate and inorganic phosphate (Franco et al., 2001).
Among the identified extracellular proteins, two were involved in carbohydrate transport across cellular membranes in bacteria. The protein spot vps2 matched a phosphocarrier protein HPr, an essential component of the multicomponent phosphotransferase system, by which many sugars are transported into bacteria across the cytoplasmic membrane, concomitantly phosphorylated and then fed into glycolysis (Postma et al., 1993). In addition, the vps12 was a periplasmic maltose-binding protein MalE of the maltose ATP binding cassette (ABC) transporter (Davidson and Maloney, 2007), which also contains integral cytoplasmic membrane proteins MalF and MalG, and two copies of the ATPase subunit MalK (Boos and Lucht, 1996). To our knowledge, none of the secreted HPr and MalE derived from any toxic and non-toxic bacteria has been described thus far. In this study, our data implied possible unknown regulation mechanisms underlying the key sugar transports in central carbohydrate metabolism, which was mediated by the membrane-linked HPr and MalE in V. parahaemolyticus.
Several identified extracellular proteins have been reported to be involved in the virulence of some pathogenic bacteria. For example, in this study, two protein spots (vps1 and vps7) were identified as a Sigma-54 (σ54) modulation protein of V. parahaemolyticus, while the vps6 was a dihydrolipoyl dehydrogenase (DLDH). In addition, transaldolase (vps11 and vps13) can be recruited on the cell surface, and secreted via a non-classical secretion mechanism or an uncharacterized translocation pathway (González-Rodríguez et al., 2012). Moreover, the vps14 was found as a phosphoglycerate kinase (PGK) that catalyzes the transfer of a phosphate group from 1,3-diphosphoglycerate to ADP to produce 3-phosphoglycerate and ATP (Smith et al., 2011). Taken together, in this study, our data suggested that the extracellular proteins of σ 54, DLDH, transaldolase and PGK perhaps also contributed to the pathogenecity of V. parahaemolyticus. Further studies on these potential virulence-associated factors will improve our understanding of the pathogenesis of the bacterium.
In addition, the protein spot vps10 matched a hypothetical protein (9% sequence coverage, 64 score) encoded by the VP1454 gene of V. parahaemolyticus RIMD2210633 with currently unknown functions in the public databases.
Discussion
In this study, approximately 2.9% of the V. parahaemolyticus strains isolated from aquatic products sampled from fish markets in Shanghai in 2011–2014 carried the toxic trh gene, but none had the toxic tdh gene, consistent with previous reports (Su and Liu, 2007; Martinez-Urtaza et al., 2008; Flores-Primo et al., 2014). Antimicrobial susceptibility and heavy metal resistance of the V. parahaemolyticus strains chosen for the comparative secretome analysis also correlated with those reported by Song et al. (2013), where ampicillin resistance was the most predominant amongst the Vibrios isolated from Shanghai fish markets in 2011. Moreover, compared with the prior work (Song et al., 2013), the data in this study revealed similar but narrow heavy metal resistance patterns, although based on a fairly small number of isolates analyzed here.
Distinct secretomic profiles derived from the seven V. parahaemolyticus strains of clinical and food origins as well as the LC-MS/MS analysis revealed approximately 43% of the common extracellular proteins involved in protein synthesis of the bacterium. Bacterial ribosome is a large protein-RNA complex that consists of two major subunits (a small 30S and a large 50S subunits), each of which is composed of a variety of proteins (Brodersen et al., 2002). The components S2 (vps3), S6 (vps4) and L9 (vps9) are associated with the formation of the translation initiation complex and possibly interact with mRNA and other components of the ribosome (Lieberman et al., 2000; Brodersen et al., 2002; Wei et al., 2014). The S2 and ribosomal protein L7/L12 were also detected as extracellular proteins of Haemophilus parasuis and Streptococcus suis by 2-DE analysis (Wu et al., 2008b; Wei et al., 2014). Despite highly conserved translational machinery with invariable rRNA and protein components, the formation of distinct ribosomal subpopulations has been reported in bacteria when encountered adverse conditions (Moll and Engelberg-Kulka, 2012). It will be interesting to investigate the biological function of the ribosomal proteins of V. parahaemolyticus in the future research. In addition, the RRF (vps5) is responsible for the dissociation of ribosomes from messenger RNA after the termination of translation (Li et al., 2010). It has been characterized as an essential protein for the survival of Escherichia coli (Selvaraj et al., 2013). In this study, two other translation-associated proteins, vps15 (EF-Tu) and vps 16 (EF-Ts), were also identified. These two proteins are important components of the multistep ribosomal decoding pathway that ensures rapid and accurate translation (Burnett et al., 2013; Yikilmaz et al., 2014). It has been reported that EF-Tu of Streptococcus pneumonia was a new virulence factor that binds human complement factors, aids in immune evasion and host tissue invasion (Sarkar et al., 2013). To our knowledge, no linking to secreted proteins of the RRF and EF-Ts has been described previously.
The PdxJ (vps8) is absent from humans, but exists in many human pathogens, such as E. coli, Cercospora spp. and Aspergillus nidulans. The enzyme has been suggested as a potential target for the development of novel drugs to control the pathogens (Garrido-Franco, 2003).
Previous studies have revealed that the σ54 (vps1 and vps7) played an important role in the virulence of Pseudomonas syringae, Erwinia carotovora and Pseudomonas aeruginosa (Hendrickson et al., 2000, 2001; Chatterjee et al., 2002). It was also essential for biofilm formation in Vibrio fischeri (Wolfe et al., 2004), and bacterial quorum sensing regulation and pilin and flagellin production in Vibrio cholerae (Prouty et al., 2001; Ishikawa et al., 2009). In this study, the protein vps6 was identified as a DLDH. It was also detected as an extracellular enzyme secreted by Bacillus subtilis WY34 (Wu et al., 2008a), and identified as a virulence-associated determinant of Streptococcus pneumoniae and Mycoplasma gallisepticum (Smith et al., 2002; Hudson et al., 2006). DLDH could be recognized by anti-sera as one of the cell wall-associated proteins isolated from Staphylococcus epidermidis (Sellman et al., 2005). Recently, western blot analyses of whole cell lysates of Vibrio harveyi and Vibrio alginolyticus suggested that DLDH was a novel immunogenic protein (Pang et al., 2010, 2013). In addition, the extracellular transaldolase [vps11 (6% sequence coverage, 69 score) and vps13] of Bifidobacterium bifidum A8 was demonstrated in vitro as a specific aggregation factor contributing to the strong auto-aggregation phenotype of the bacterium. When exposed on the cell surface, it could act as an important colonization factor for the survival of B. bifidum in host intestinal tract (González-Rodríguez et al., 2012). In this study, the protein vps14 was identified as a PGK. In Streptococcus agalactiae, PGK is a major outer surface protein that can be recognized by host immune system, and the recombinant PGK has been used as an antigen in a neonatal-animal model against S. agalactiae infection (Hughes, 2002; Wang et al., 2014). PGK has also been suggested as a potential vaccine candidate against Lactococcus garvieae infection and Streptococcus suis biofilm formation (Shin et al., 2007; Valette et al., 2012).
Overall, this study constitutes the first investigation of the secretomes of seven V. parahaemolyticus strains with toxic and non-toxic genotypes. Fourteen common extracellular proteins were identified from the distinct secretomic profiles derived from the strains by 2-DE and LC-MS/MS analysis. Of these, half were involved in protein synthesis and sugar transport of V. parahaemolyticus. Notably, six identified proteins were virulence-associated factors involved in the pathogenicity of some other pathogenic bacteria, including the EF-Tu, PdxJ, σ54, DLDH, transaldolase and PGK. In addition, comparative secretomics also uncovered several extracellular proteins that have not been described in any bacteria, such as the ribosome-recycling factor, EF-Ts, HPr and MalE. Our secretome data, coupled with the previous studies, indicated that bacterial virulence-associated factors fall into three distinct categories. One of these exhibits strong strain-specificity and plays a crucial role in the virulence of the pathogens, e.g., the TDH and TRH in V. parahaemolyticus, as well as the cholera toxin (CTX) in V. cholerae. The EF-Tu, PdxJ, σ54, DLDH, transaldolase and PGK fall into the second category, the members of which are likely essential for the colonization, invasion and survival of the pathogens in host environments, such as human gastrointestinal and respiratory tracts. The third category may contain the secreted proteins involved in the key cellular synthesis and metabolism, e.g., EF-Ts, HPr and MalE in V. parahaemolyticus. Given that the members belonging to the latter two categories were also identified from the non-toxic V. parahaemolyticus strains, in the future research, it will be interesting to further address their functions in the virulence of the bacterium, particularly in the complex gastrointestinal flora. The results in this study will facilitate the better elucidation of the pathogenecity of V. parahaemolyticus, and also provide data in support of the development of novel vaccine against the leading seafood-borne pathogen worldwide.
Author Contributions
YH, HW, LC participated in the design and or discussion of the study. YH carried out the major experiments. YH, LC analyzed the data. LC wrote the article. HW revised it for important intellectual improvement. All authors read and approved the final version to be published.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a grant No. B-9500-10-0004 from Shanghai Municipal Education Commission and a grant No. 31271830 from National Natural Science Foundation of China. The authors thank Y Lin for some help to the experiments of susceptibility to antimicrobial agents and heavy metals in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00707
Figure S1. Growth curves of the V. parahaemolyticus strains. The strains were cultured in LB broth (pH 8.5, 3% NaCl) at 37°C without shaking, and growth curves were determined using Synergy 2 Multi-Mode Microplate Reader.
References
Baumann, P., Baumann, L., and Reichelt, J. L. (1973). Taxonomy of marine bacteria: Beneckea parahaemolytica and Beneckea alginolytica. J. Bacteriol. 113, 1144–1155.
Bhattacharjee, R. N., Park, K. S., Kumagai, Y., Okada, K., Yamamoto, M., Uematsu, S., et al. (2006). VP1686, a Vibrio type III secretion protein, induces toll-like receptor-independent apoptosis in macrophage through NF-κB inhibition. J. Biol. Chem. 281, 36897–36904. doi: 10.1074/jbc.M605493200
Boos, W., and Lucht, J. (1996). “Periplasmic binding protein-dependent ABC transporters,” in Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd Edn., eds F. C. Neidhardt, R. Curtiss, III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (Washington, DC: ASM Press), 1175–1209.
Boyd, E. F., Cohen, A. L., Naughton, L. M., Ussery, D. W., Binnewies, T. T., Stine, O. C., et al. (2008). Molecular analysis of the emergence of pandemic Vibrio parahaemolyticus. BMC Microbiol. 8:110. doi: 10.1186/1471-2180-8-110
Broberg, C. A., Calder, T. J., and Orth, K. (2011). Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 13, 992–1001. doi: 10.1016/j.micinf.2011.06.013
Brodersen, D. E., Clemons, W. M. Jr. Carter, A. P., Wimberly, B. T., and Ramakrishnan, V. (2002). Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 316, 725–768. doi: 10.1006/jmbi.2001.5359
Burdette, D. L., Seemann, J., and Orth, K. (2009). Vibrio VopQ induces PI3-kinase-independent autophagy and antagonizes phagocytosis. Mol. Microbiol. 73, 639–649. doi: 10.1111/j.1365-2958.2009.06798.x
Burnett, B. J., Altman, R. B., Ferrao, R., Alejo, J. L., Kaur, N., Kanji, J., et al. (2013). Elongation factor Ts directly facilitates the formation and disassembly of the Escherichia coli elongation factor Tu-GTP-aminoacyl-tRNA ternary complex. J. Biol. Chem. 288, 13917–13928. doi: 10.1074/jbc.M113.460014
Ceccarelli, D., Hasan, N. A., Huq, A., and Colwell, R. R. (2013). Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front. Cell Infect. Microbiol. 3:97. doi: 10.3389/fcimb.2013.00097
Chatterjee, A., Cui, Y., and Chatterjee, A. K. (2002). Regulation of Erwinia carotovora hrpLEcc (sigma-LEcc), which encodes an extracytoplasmic function subfamily of sigma factor required for expression of the HRP regulon. Mol. Plant Microbe Interact. 15, 971–980. doi: 10.1094/MPMI.2002.15.9.971
Chen, L., Xiong, Z., Sun, L., Yang, J., and Jin, Q. (2012). VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 40, D641–D645. doi: 10.1093/nar/gkr989
Davidson, A. L., and Maloney, P. C. (2007). ABC transporters: how small machines do a big job. Trends Microbiol. 15, 448–455. doi: 10.1016/j.tim.2007.09.005
Flores-Primo, A., Pardío-Sedas, V., Lizárraga-Partida, L., López-Hernández, K., Uscanga-Serrano, R., and Flores-Hernández, R. (2014). Seasonal abundance of total and pathogenic Vibrio parahaemolyticus isolated from American oysters harvested in the mandinga lagoon system, Veracruz, Mexico: implications for food safety. J. Food Prot. 77, 1069–1077. doi: 10.4315/0362-028X.JFP-13-482
Franco, M. G., Laber, B., Huber, R., and Clausen, T. (2001). Structural basis for the function of pyridoxine 5′-phosphate synthase. Structure 9, 245–253. doi: 10.1016/S0969-2126(01)00584-6
Fujino, T., Miwatani, T., Yasuda, J., Kondo, M., Takeda, Y., Akita, Y., et al. (1965). Taxonomic studies on the bacterial strains isolated from cases of “shirasu” food-poisoning (Pasteurella parahaemolytica) and related microorganisms. Biken J. 8, 63–71.
Garrido-Franco, M. (2003). Pyridoxine 5′-phosphate synthase: de novo synthesis of vitamin B6 and beyond. Biochim. Biophys. Acta 1647, 92–97. doi: 10.1016/S1570-9639(03)00065-7
González-Rodríguez, I., Sánchez, B., Ruiz, L., Turroni, F., Ventura, M., Ruas-Madiedo, P., et al. (2012). Role of extracellular transaldolase from Bifidobacterium bifidum in mucin adhesion and aggregation. Appl. Environ. Microbiol. 78, 3992–3998. doi: 10.1128/AEM.08024-11
Haley, B. J., Kokashvili, T., Tskshvediani, A., Janelidze, N., Mitaishvili, N., Grim, C. J., et al. (2014). Molecular diversity and predictability of Vibrio parahaemolyticus along the Georgian coastal zone of the Black Sea. Front. Microbiol. 5:45. doi: 10.3389/fmicb.2014.00045
Hendrickson, E. L., Guevera, P., Penaloza-Vazquez, A., Shao, J., Bender, C., and Ausubel, F. M. (2000). Virulence of the phytopathogen Pseudomonas syringae pv. maculicola is rpoN dependent. J. Bacteriol. 182, 3498–3507. doi: 10.1128/JB.182.12.3498-3507.2000
Hendrickson, E. L., Plotnikova, J., Mahajan-Miklos, S., Rahme, L. G., and Ausubel, F. M. (2001). Differential roles of the Pseudomonas aeruginosa PA14 rpoN gene in pathogenicity in plants, nematodes, insects, and mice. J. Bacteriol. 183, 7126–7134. doi: 10.1128/JB.183.24.7126-7134.2001
Higa, N., Toma, C., Koizumi, Y., Nakasone, N., Nohara, T., Masumoto, J., et al. (2013). Vibrio parahaemolyticus effector proteins suppress inflammasome activation by interfering with host autophagy signaling. PLoS Pathog. 9:e1003142. doi: 10.1371/journal.ppat.1003142
Hiyoshi, H., Kodama, T., Saito, K., Gotoh, K., Matsuda, S., Akeda, Y., et al. (2011). VopV, an F-actin-binding type III secretion effector, is required for Vibrio parahaemolyticus-induced enterotoxicity. Cell Host Microbe. 10, 401–409. doi: 10.1016/j.chom.2011.08.014
Hudson, P., Gorton, T. S., Papazisi, L., Cecchini, K., Frasca, S. Jr., and Geary, S. J. (2006). Identification of a virulence-associated determinant, dihydrolipoamide dehydrogenase (lpd), in Mycoplasma gallisepticum through in vivo screening of transposon mutants. Infect Immun. 74, 931–939. doi: 10.1128/IAI.74.2.931-939.2006
Hughes, M. J. G. (2002). Identification of major outer surface proteins of Streptococcus agalactiae. Infect Immun. 70, 1254–1259. doi: 10.1128/IAI.70.3.1254-1259.2002
Ishikawa, T., Rompikuntal, P. K., Lindmark, B., Milton, D. L., and Wai, S. N. (2009). Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. PLoS ONE 4:e6734. doi: 10.1371/journal.pone.0006734
Letchumanan, V., Chan, K.-G., and Lee, L.-H. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 5:705. doi: 10.3389/fmicb.2014.00705
Letchumanan, V., Yin, W.-F., Lee, L.-H., and Chan, K.-G. (2015). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 6:33. doi: 10.3389/fmicb.2015.00033
Li, L., Guo, J., Wen, Y., Zhi, C., Song, Y., and Li, J. (2010). Overexpression of ribosome recycling factor causes increased production of avermectin in Streptomyces avermitilis strains. J. Ind. Microbiol. Biotechnol. 37, 673–679. doi: 10.1007/s10295-010-0710-0
Lieberman, K. R., Firpo, M. A., Herr, A. J., Nguyenle, T., Atkins, J. F., Gesteland, R. F., et al. (2000). The 23 S rRNA environment of ribosomal protein L9 in the 50 S ribosomal subunit. J. Mol. Biol. 297, 1129–1143. doi: 10.1006/jmbi.2000.3621
Makino, K., Oshima, K., Kurokawa, K., Yokoyama, K., Uda, T., Tagomori, K., et al. (2003). Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholera. Lancet 361, 743–749. doi: 10.1016/S0140-6736(03)12659-1
Martinez-Urtaza, J., Lozano-Leon, A., Varela-Pet, J., Trinanes, J., Pazos, Y., and Garcia-Martin, O. (2008). Environmental determinants of the occurrence and distribution of Vibrio parahaemolyticus in the rias of Galicia, Spain. Appl. Environ. Microbiol. 74, 265–274. doi: 10.1128/AEM.01307-07
Matlawska-Wasowska, K., Finn, R., Mustel, A., O'Byrne, C. P., Baird, A. W., Coffey, E. T., et al. (2010). The Vibrio parahaemolyticus type III secretion systems manipulate host cell MAPK for critical steps in pathogenesis. BMC Microbiol. 10:329. doi: 10.1186/1471-2180-10-329
Moll, I., and Engelberg-Kulka, H. (2012). Selective translation during stress in Escherichia coli. Trends Biochem. Sci. 37, 493–498. doi: 10.1016/j.tibs.2012.07.007
Ono, T., Park, K. S., Ueta, M., Iida, T., and Honda, T. (2006). Identification of proteins secreted via Vibrio parahaemolyticus type III secretion system 1. Infect Immun. 74, 1032–1042. doi: 10.1128/IAI.74.2.1032-1042.2006
Pang, H. Y., Li, Y., Wu, Z. H., Jian, J. C., Lu, Y. S., and Cai, S. H. (2010). Immunoproteomic analysis and identification of novel immunogenic proteins from Vibrio harveyi. J. Appl. Microbiol. 109, 1800–1809. doi: 10.1111/j.1365-2672.2010.04808.x
Pang, H.-Y., Zhang, X.-Z., Wu, Z.-H., Jian, J.-C., Cai, S.-H., and Liang, J. (2013). Identification of novel immunogenic proteins of Vibrio alginolyticusby immunoproteomic methodologies. Aquacult. Res. 44, 472–484. doi: 10.1111/j.1365-2109.2012.03150.x
Postma, P., Lengeler, J., and Jacobson, G. (1993). Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57, 543.
Prouty, M. G., Correa, N. E., and Klose, K. E. (2001). The novel σ54−and σ28−dependent flagellar gene transcription hierarchy of Vibrio cholera. Mol. Microbiol. 39, 1595–1609. doi: 10.1046/j.1365-2958.2001.02348.x
Raghunath, P. (2014). Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front. Microbiol. 5:805. doi: 10.3389/fmicb.2014.00805
Sarkar, S., Hallström, T., Hammerschmidt, S., Skerka, C., and Zipfel, P. F. (2013). Elongation factor Tu (Tuf) is a new virulence factor of Streptococcus pneumoniae that binds human complement factors, aids in immune evasion and host tissue invasion. Mol. Immunol. 56, 312. doi: 10.1016/j.molimm.2013.05.202
Sellman, B. R., Howell, A. P., Kelly-Boyd, C., and Baker, S. M. (2005). Identification of immunogenic and serum binding proteins of Staphylococcus epidermidis. Infect Immun. 73, 6591–6600. doi: 10.1128/IAI.73.10.6591-6600.2005
Selvaraj, M., Govindan, A., Seshadri, A., Dubey, B., Varshney, U., and Vijayan, M. (2013). Molecular flexibility of Mycobacterium tuberculosis ribosome recycling factor and its functional consequences: an exploration involving mutants. J. Biosci. 38, 845–855. doi: 10.1007/s12038-013-9381-0
Shi, Y., Pan, Y., Li, B., He, W., She, Q., and Chen, L. (2013). Molecular cloning of a novel bioH gene from an environmental metagenome encoding a carboxylesterase with exceptional tolerance to organic solvents. BMC Biotechnol. 13:13. doi: 10.1186/1472-6750-13-13
Shin, G. W., Palaksha, K. J., Kim, Y. R., Nho, S. W., Cho, J. H., Heo, N. E., et al. (2007). Immunoproteomic analysis of capsulate and non-capsulate strains of Lactococcus garvieae. Vet. Microbiol. 119, 205–212. doi: 10.1016/j.vetmic.2006.08.021
Smith, A. W., Roche, H., Trombe, M. C., Briles, D. E., and Håkansson, A. (2002). Characterization of the dihydrolipoamide dehydrogenase from Streptococcus pneumoniae and its role in pneumococcal infection. Mol. Microbiol. 44, 431–448. doi: 10.1046/j.1365-2958.2002.02883.x
Smith, C. D., Chattopadhyay, D., and Pal, B. (2011). Crystal structure of Plasmodium falciparum phosphoglycerate kinase: evidence for anion binding in the basic patch. Biochem. Biophys. Res. Commun. 412, 203–206. doi: 10.1016/j.bbrc.2011.07.045
Song, Y., Yu, P., Li, B., Pan, Y., Zhang, X., Cong, J., et al. (2013). The mosaic accessory gene structures of the SXT/R391-like integrative conjugative elements derived from Vibrio spp. isolated from aquatic products and environment in the Yangtze River Estuary, China. BMC Microbiol. 12:214. doi: 10.1186/1471-2180-13-214
Su, Y.-C., and Liu, C. (2007). Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24, 549–558. doi: 10.1016/j.fm.2007.01.005
Sun, X., Liu, T., Peng, X., and Chen, L. (2014). Insights into Vibrio parahaemolyticus CHN25 response to artificial gastric fluid stress by transcriptomic analysis. Int. J. Mol. Sci. 15, 22539–22562. doi: 10.3390/ijms151222539
Valette, M., Poitou, C., Le Beyec, J., Bouillot, J.-L., Clement, K., and Czernichow, S. (2012). Melanocortin-4 receptor mutations and polymorphisms do not affect weight loss after bariatric surgery. PLoS ONE 7:e48221. doi: 10.1371/journal.pone.0048221
Wang, L., Zhou, D., Mao, P., Zhang, Y., Hou, J., Hu, Y., et al. (2013). Cell density-and quorum sensing-dependent expression of type VI secretion system 2 in Vibrio parahaemolyticus. PLoS ONE 8:e73363. doi: 10.1371/journal.pone.0073363
Wang, Y. T., Huang, H. Y., Tsai, M. A., Wang, P. C., Jiang, B. H., and Chen, S. C. (2014). Phosphoglycerate kinase enhanced immunity of the whole cell of Streptococcus agalactiae in tilapia, Oreochromis niloticus. Fish Shellfish Immunol. 41, 250–259. doi: 10.1016/j.fsi.2014.09.008
Wei, X., Cao, S., Zhang, L., Wen, X., Wen, Y., Wu, R., et al. (2014). Comparative proteome analysis of the extracellular proteins of two Haemophilus parasuis strains Nagasaki and SW114. Biochem. Biophys. Res. Commun. 446, 997–1001. doi: 10.1016/j.bbrc.2014.03.049
Wolfe, A. J., Millikan, D. S., Campbell, J. M., and Visick, K. L. (2004). Vibrio fischeri σ54 controls motility, biofilm formation, luminescence, and colonization. Appl. Environ. Microbiol. 70, 2520–2524. doi: 10.1128/AEM.70.4.2520-2524.2004
Wooldridge, K. (ed.). (2009). Bacterial Secreted Proteins: Secretory Mechanisms and Role in Pathogenesis. Poole: Caister Academic Press.
Wu, A. M., Jiang, Z. Q., Wei, Y., and Sun, C. Y. (2008a). Purification of an extracellular dihydrolipoamide dehydrogenase from Bacillus subtilis. J. China Agricultural Universit. 3, 16–19.
Wu, Z., Zhang, W., and Lu, C. (2008b). Comparative proteome analysis of secreted proteins of Streptococcus suis serotype 9 isolates from diseased and healthy pigs. Microb. Pathog. 45, 159–166. doi: 10.1016/j.micpath.2008.04.009
Yarbrough, M. L., Li, Y., Kinch, L. N., Grishin, N. V., Ball, H. L., and Orth, K. (2009). AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science 323, 269–272. doi: 10.1126/science.1166382
Yikilmaz, E., Chapman, S. J., Schrader, J. M., and Uhlenbeck, O. C. (2014). The interface between Escherichia coli elongation factor Tu and aminoacyl-Trna. Biochemistry 53, 5710–5720. doi: 10.1021/bi500533x
Zhou, X. H., Gewurz, B. E., Ritchie, J. M., Takasaki, K., Greenfeld, H., Kieff, E., et al. (2013). A Vibrio parahaemolyticus T3SS effector mediates pathogenesis by independently enabling intestinal colonization and inhibiting TAK1 activation. Cell Reports. 3, 1690–1702. doi: 10.1016/j.celrep.2013.03.039
Keywords: Vibrio parahaemolyticus, virulence factor, secretome, aquatic products, vaccine candidate
Citation: He Y, Wang H and Chen L (2015) Comparative secretomics reveals novel virulence-associated factors of Vibrio parahaemolyticus. Front. Microbiol. 6:707. doi: 10.3389/fmicb.2015.00707
Received: 28 April 2015; Accepted: 26 June 2015;
Published: 17 July 2015.
Edited by:
Sahdeo Prasad, The University of Texas MD Anderson Cancer Center, USAReviewed by:
Pendru Raghunath, Dr. VRK Women's Medical College, IndiaLearn-Han Lee, Monash University Malaysia, Malaysia
Parag A. Parekh, The University of Texas MD Anderson Cancer Center, USA
Narayan Sastri Palla, Baylor College of Medicine, USA
Copyright © 2015 He, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lanming Chen, Key Laboratory of Quality and Safety Risk Assessment for Aquatic Products on Storage and Preservation (Shanghai), China Ministry of Agriculture, College of Food Science and Technology, Shanghai Ocean University, 999 Hu Cheng Huan Road, Shanghai 201306, China,bG1jaGVuQHNob3UuZWR1LmNu
 Yu He
Yu He Hua Wang
Hua Wang Lanming Chen
Lanming Chen