- 1Department of Molecular Genetics and Microbiology, Stony Brook University, Stony Brook, NY, USA
- 2Department of Medicine, Stony Brook University, Stony Brook, NY, USA
- 3Department of Biomedical Science and Human Oncology, Hygiene Section, University of Bari, Bari, Italy
- 4Department of Physiology and Biophysics, Stony Brook University, Stony Brook, NY, USA
Cryptococcosis caused by Cryptococcus neoformans and Cryptococcus gattii affects a large population and is a cause of significant morbidity and mortality. Despite its public health burden, there are currently no vaccines against cryptococcosis and new strategies against such infections are needed. In this study, we demonstrate that C. neoformans has the biochemical ability to metabolize sterylglucosides (SGs), a class of immunomodulatory glycolipids. Genetic manipulations that eliminate cryptococccal sterylglucosidase lead to the accumulation of SGs and generate a mutant strain (Δsgl1) that is non-pathogenic in the mouse models of cryptococcosis. Interestingly, this mutant strain acts as a vaccine strain and protects mice against cryptococcosis following infection with C. neoformans or C. gattii. The immunity induced by the Δsgl1 strain is not CD4+ T-cells dependent. Immunocompromised mice, which lack CD4+ T-cells, are able to control the infection by Δsgl1 and acquire immunity against the challenge by wild-type C. neoformans following vaccination with the Δsgl1 strain. These findings are particularly important in the context of HIV/AIDS immune deficiency and suggest that the Δsgl1 strain might provide a potential vaccination strategy against cryptococcosis.
Introduction
Cryptococcus is an opportunistic fungal pathogen and the causative agent of the disease cryptococcosis. Infections caused by Cryptococcus neoformans and Cryptococcus gattii lead to more than 600000 deaths per year (Park et al., 2009), especially among immunocompromised individuals. Despite its significant public health burden, no vaccines currently exist in the clinic for cryptococcosis (or other fungal infections Nanjappa and Klein, 2014). Although experimental vaccines have been developed using the glucuronoxylomannan (GXM) capsule bound to tetanus toxoid (Devi et al., 1991; Casadevall et al., 1992; Devi, 1996), these formulations have not been translated to the clinic and have suffered from drawbacks such as inducing detrimental antibodies in mice (Casadevall and Pirofski, 2005; Datta and Pirofski, 2006). Recent attempts in the mouse models of cryptococcosis have been focused on the use of genetically engineered C. neoformans strains that generate cytokines (Wormley et al., 2007; Wozniak et al., 2011) or protein preparations from C. gattii administered prior to infection (Chaturvedi et al., 2014). Although these attempts have provided valuable insights, studies are still limited and shortcomings exist. For example, complete immunity against C. gattii (responsible for severe infections in the USA Datta et al., 2009; Walraven et al., 2011) has not been achieved (Chaturvedi et al., 2014) demonstrating the need for the development of more effective vaccines.
Sterylglucosides (SGs) are a class of glycolipids produced by animals, plants, bacteria and various fungi including C. neoformans (Warnecke et al., 1999; Watanabe et al., 2015). SGs show immunomodulatory properties as reported by various studies. For example, SGs have been shown to increase proliferation of lymphocytes and eosinophils in vivo (Donald et al., 1997) and cytokine secretion in vitro (Bouic et al., 1996; Lee and Han, 2006; Lee et al., 2007). In the realm of fungal infections, it has been observed that SGs administration increases the survival of mice infected with a lethal dose of Candida albicans by inducing a Th1 immune response (Lee et al., 2007).
A recent report has identified a endoglycoceramidase-related protein (named EGCrP2), capable of hydrolyzing SGs in C. neoformans (Watanabe et al., 2015). We hypothesized that the lack of this enzyme should lead to a lack of SG catabolism and subsequent SG accumulation, allowing for the immunomodulatory properties of SGs to manifest. Thus, we sought to utilize the immunomodulatory properties of fungal SGs by engineering a C. neoformans strain that lacks the sterylglucosidase enzyme. This strain was non-pathogenic in the mouse models of cryptococcosis and conferred complete protection against both C. neoformans and C. gattii, suggesting that it might be a suitable candidate for vaccine development against cryptococcosis.
Materials and Methods
Strains, Plasmid, and Culture Conditions
The fungal strains used in this study were C. neoformans (Cn) var. grubii strain H99 wild-type (WT) and C. gattii strain R265 and Saccharomyces cerevisiae ΔYIR007W mutant (ScΔYIR) derived from BY4741. Bacterial strain Escherichia coli DH5-α (Life Technologies, Carlsbad, CA, USA) was used as competent cells. The plasmid pCR II-TOPO 4.0 kb was used for cloning, and pYES2/CT was used for expression of Cn 5607 in ScΔYIR.
Bacterial strains were grown at 37°C in Luria-Bertani medium with 75 mg/L of ampicillin. Cn strains were routinely grown in YPD broth at 30°C and 0.04% atmospheric CO2 for 20–22 h with shaking at 250 rpm. Dulbecco’s modified eagle media (DMEM) buffered with 25 mM HEPES and 2% glucose at pH 7.4 or pH 4 was used for growing Cn at 37°C and 5% CO2 (i.e., physiologically relevant conditions). ScΔYIR strain transformed with pYES2/CT or pYES2/CT- Cn 5607, was grown in yeast nitrogen base (YNB) without amino acids, 1.2 g/L amino acid mixture lacking uracil (ura-), and 2% glucose or 2% galactose (48 h) at 30°C to induce Cn 5607 expression.
Expression of Cn 5607 in S. cerevisiae
Cn 5607 was identified by blasting the Cn WT endoglycoceramidase-related Protein 1 (EGCrP1) sequence in Cn WT Broad Institute genome database [http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/MultiHome.html]. Two sequences were found: one 100% identical to EGCrP1 and another one with an E-value of 9e-57, located on chromosome 14 (Cn 5607). To express Cn 5607 in S. cerevisiae strains, total RNA was extracted from Cn and the cDNA was synthesized from 1 μg of the total RNA using SuperScript III RNase H-Reverse Transcriptase (Life Technologies). PCR was performed using the cDNA as a template and the following expression primers: PRSGL1-5′ forward (5′-GAGCTCATGCCTCCTCCACCAGAAGT-3′) and PRSGL1-5′ reverse (5′-TCTAGAAGCAATAACGCATTCAGGACA-3′) carrying the restriction enzyme sites for SacI and XbaI respectively. A 2,556-pb fragment was cloned into pCR II-TOPO vector generating plasmid pCR- Cn 5607 and sequenced. After digestion with SacI and XbaI, Cn 5607 was inserted into pYES2/CT vector generating pYES2/CT-Cn 5607. ScΔYIR was grown in YNB medium overnight at 30°C and transformed with pYES2/CT empty-vector or pYES2/CT-Cn 5607 using lithium acetate transformation, as previously described (Kawai et al., 2010). After transformation, cells were plated on YNB ura- plates and incubated for 2–3 days in 30°C incubator. Then, S. cerevisiae colonies were patched with sterile toothpicks to fresh YNB ura- plates. To verify the expression of Cn 5607, one single colony containing pYES2/CT or pYES2/CT-Cn 5607 construct was inoculated into 10 ml of YNB ura- containing 2% of glucose and was grown overnight at 30°C with shaking. The cells were washed twice with PBS 1X and a suitable amount of overnight culture necessary to obtain an OD600 of 0.4 was transferred to a fresh YNB ura- medium containing 2% of galactose (induction medium) and incubated for 48 h at 30°C with shaking. After 48 h, the cells were washed and harvested by centrifugation at 3000g for 5 min at 4°C, and the cell pellets was stored at -80°C until ready to be used.
Total proteins were extracted from S. cerevisiae strains as previously described (Singh et al., 2011). Protein content was assessed by the method of Bradford using bovine serum albumin (Sigma-Aldrich, St. Louis, MO, USA) as a standard. Fifty micrograms of S. cerevisiae proteins were loaded onto SDS-PAGE and stained with Coomassie Brilliant Blue. Western Blot was used to detect the expression of recombinant fusion protein using Anti-His (C-term)-HRP antibody.
In Vitro Activity Assay of Cn 5607 Using Standard Plants SGs and Purified Cn SGs
The in vitro Cn 5607 enzymatic assay was performed using 20 μg of standard SGs (purified from plants and commercially available) or 10 μg of endogenously purified SGs as substrate and ScΔYIR + empty vector or ScΔYIR-Cn 5607 as source of enzyme. The decrease in the intensity of the SGs band was monitored by thin layer chromatography (TLC). Plants SGs were incubated with 200 μg of ScΔYIR + empty vector or 50, 100, and 200 μg of ScΔYIR-Cn 5607 at 30°C for 1 h. Endogenously purified Cn SGs were incubated with 200 μg of ScΔYIR + empty vector or 100 μg of ScΔYIR-Cn 5607 at 30°C for 1 h. The reactions were terminated by the addition of 300 μl of CHCl3/MeOH (1:1 ratio), the lower phases were dried down, resuspended in 50 μl of CHCl3/MeOH (2:1 ratio) and analyzed by TLC. Cn 5607 specifically cleaves SGs, thus we called Cn 5607 sterylglucosidase1 (Sgl1).
The pH dependence of Sgl1 was determined using as substrate SGs in a pH range of 4.5–8 using the following buffers at the final concentration of 50 mM: sodium acetate (pH 4.5–5.0), MES (pH 5.5–6.0), sodium phosphate (pH 6.5–7.0), and HEPES (pH 7.5–8.0). The optimal temperature of Sgl1 was determined in the range from 25 to 37°C. The effect of detergents was assessed using Triton X-100, Sodium Deoxycholate and CHAPS at the concentration of 0.05, 0.15, and 0.3%.
In Vitro Activity Assay of Cn 5607 Using NBD-C6-Glucosylceramide and Cn Long Chain GlcCer
To verify the enzymatic activity of Cn 5607, we used different substrates: NBD-C6-glucosylceramide (NBD-C6-GlcCer; Matreya, LLC, State-College, PA, USA) and Cn long chain GlcCer. Briefly, 200 μg of yeast proteins from ScΔYIR + empty vector or ScΔYIR-Cn 5607 were incubated first with 20 μM of NBD-C6-Glucosylceramide and at 30°C for 1 h in a final reaction volume of 100 μl. The production of NBD-C6-Ceramide was identified as a fluorescent band using a PhosphorImagerTM 860 STORM unit and ImageQuant analysis (GE Healthcare, Rahway, NJ, USA) as previously described (Rittershaus et al., 2006).
The in vitro Cn 5607 activity was also valuated using 10 μg of Cn long chain GlcCer and 200 μg of ScΔYIR + empty vector or ScΔYIR-Cn 5607 cell extracts. Cerezyme (10 μg), generously provided by the Genzyme Corporation (Cambridge, MA, USA), was used as positive control for the catalytic reaction. The reactions were terminated by the addition of 300 μl of CHCl3/MeOH (1:1 ratio), the samples were mixed and the phases were separated by centrifugation at 3000g for 5 min. The lower phases were dried down using a SPD 2010 SpeedVac vacuum dryer (Thermo Electron Corp.). The dried samples were resuspended in 50 μl of CHCl3/MeOH (2:1 ratio) and analyzed by TLC on silica gel plate (EMD Millipore, Billerica, MA, USA) developed with chloroform/methanol/water (65:25:4, v/v/v) and stained with iodine and resorcinol. The in vitro activity assay using Cn long chain GlcCer and ScΔYIR + empty vector or ScΔYIR-Cn 5607 was also repeated with a longer incubation time (4 h) and the results were evaluated by liquid chromatography–mass spectrometry (LC–MS).
Disruption and Reconstitution of SGL1 Gene in Cn
The SGL1 gene (locus number CNAG_05607 in C. neoformans var. grubii serotype A genome database) was deleted using NAT1 (Nourseothricin Acetyl transferase1) split marker. A knockout cassette was generated containing a 1.035 bp of the 5′ untranslated region (5′UTR) upstream of the ATG start codon of the SGL1 gene and a 1.059 bp of the 3′UTR. The 5′UTR was amplified by PCR using H99 genomic DNA as a template and the following primers: 5′UTR-F (5′-GTCAAGCTAAGAGCTCCATTTGATCAGCGGGATTCT-3′) and 5′UTR-R (5′-TCCACTCCGAACTAGTATCGCGTAAACGAAGAGGTG-3′), containing SacI and SpeI sites, respectively (underlined). The 3′UTR was amplified by PCR using H99 genomic DNA as a template and the following primers: 3′UTR-F (5′-GTCAAGCTAATCTAGAAGCCCATTCTGGTTGTTCTG-3′) and 3′UTR-R (5′-ACATCACACTTCTAGATTTAGCGAGCCACGTTTTCT-3′). The amplified fragments were cloned in pCR II-TOPO and sequenced, generating plasmid pCR-5′UTR-TOPO and pCR-3′UTR-TOPO. NAT1 gene, which confers resistance to the antibiotic nourseothricin (Werner BioAgents, Jena, Germany), under the control of Cn actin promoter was digested from the plasmid pCR-NAT1-TOPO by SacI and SpeI and ligated with 5′UTR digested with the same restriction enzyme generating pCR-5′UTR-NAT1-TOPO. Finally, pCR-5′UTR-NAT-TOPO was digested by EcoRV and ligated with 3′UTR generating the disruption cassette pCR-5′UTR-NAT1-3′UTR-TOPO that was named pΔsgl1. The deletion scheme is illustrated in Supplementary Figure S3A. Cn WT was transformed with the plasmid pΔsgl1 using biolistic DNA delivery device, as described previously (Toffaletti et al., 1993). Stable transformants were grown on YPD plates containing 100 μg/ml of nourseothricin. Colonies were chosen randomly and genomic DNA was isolated and digested with EcoRV and KpnI for Southern blot analysis. The DNA fragments were screened by probing with a fragment of 5′UTR. Transformant #106 showing deletion of the SGL1 gene by insertion of the NAT1 was chosen and designated Δsgl1 mutant strain. SGL1 gene was reintroduced back into the Δsgl1 using the reconstitution cassette pSK-SGL1-HYG, which had the Hygromycin B allele as selectable marker. The reconstitution scheme is illustrated in Supplementary Figure S3B. The plasmid pSK-SGL1-HYG was biolistically delivered into Δsgl1. Homologous recombinants were screened by Southern hybridization using a 800 bp fragment of the SGL1 open reading frame as probes. Transformant #21 showing reconstitution of SGL1 gene was designated Δsgl1+SGL1 reconstituted strain.
Wild-type, mutant, and reconstituted strains were characterized for their growth profile, capsule formation, stress response, and intracellular growth. For growth profile studies, WT, Δsgl1, and Δsgl1+SGL1 reconstituted strains were grown overnight in YPD at 30°C, the cells were washed three times with PBS, counted, and diluted to a final density of 104 cells/ml in DMEM at pH 7.4 or pH 4 and incubated at 37°C in the presence of 5% CO2. Aliquots were taken at different time points, diluted, and plated in duplicates onto YPD agar plates for assessment of CFUs. Capsule thickness and melanin production were determined as previously described (Wang et al., 1995; Shea et al., 2006). For oxidative stress studies, strains were spotted in serial dilution (107, 106, 105, 104, 103) on YPD agar plates with 25 mM HEPES (pH 7 or pH 4) supplemented with 5 mM H2O2, cells growth was assessed after incubation at 30°C for 96 h. Nitrosative stress response was studied by spotting the strains in serial dilution (107, 106, 105, 104, 103) on YNB agar plates with 25 mM succinate acid (pH 4) supplemented with 0.1 mM NaNO2. Cell growth was assessed after 96 h of incubation at 30, 37°C in atmospheric environment or 37°C in the presence of 5% CO2.
Phagocytosis and intracellular killing studies were performed in J774.16 macrophage-like cells as previously described (Tripathi et al., 2012). Briefly, for phagocytosis experiments cells were plated in a 96 well plate in Dulbecco’s minimal essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS). C. neoformans cells were grown overnight in YPD at 30°C. Cells were washed twice in PBS and counted. Approximately 105 cells in DMEM + FBS medium were opsonized with 10 μg/ml of anti-GXM monoclonal antibody 18B7 (kindly provided by Dr. Arturo Casadevall) and added to macrophage-like cells activated with 50 units/ml of recombinant murine gamma interferon and 0.3 μg/ml of lipopolysaccharide at an effector-to-target ratio of 1:1. After incubation for 2 h, extracellular C. neoformans cells were washed with three changes of warm DMEM medium and fresh medium. Then, 200 μl of sterile water was added to each well and the macrophage-like cells were lysed by pipetting several times. CFUs were analyzed by plating them on YPD agar plates and the numbers of internalized fungal cells were reported. Intracellular killing were performed in the same way with the following change: extracellular C. neoformans cells were washed off once 2 h after the initial incubation and another time after 24 h of incubation. Macrophage-like cells were lysed after 24 h by pipetting several times and CFUs were analyzed by plating them on YPD agar plates.
Lipid Analysis of Cryptococcus Strains by TLC
Total lipids from Cryptococcus strains were extracted, as described previously (Singh et al., 2012). Briefly, a single colony of Cryptococcus strains was grown in 15 ml of YPD broth at 30°C for 20 h at 250 rpm. Cryptococcus cells (5 × 108) were placed in a single glass tube to which the Mandala extraction buffer was added (Mandala et al., 1995). Lipid extraction was performed according to the methods of Bligh and Dyer (Bligh and Dyer, 1959) followed by base hydrolysis. One set of dried samples was resuspended in 50 μl of CHCl3/MeOH (2:1 ratio) and analyzed by TLC developed with chloroform/methanol/water (65:25:4, v/v/v) and stained with iodine and resorcinol, the other set was used for gas chromatography–mass spectrometry (GC–MS).
Lipid Profiling by Mass Spectrometry
Total lipids were extracted from Cryptococcus strains, using the methods described previously (Singh et al., 2012). For sterylglucosides analyses, extracted lipid samples were derivatized using N, O-bis (trimethylsilyl) trifluoroacetamide/trimethylchlorosilane reagent (Sigma-Aldrich) and then analyzed using 30 mt (0.25 μm) DB5-MS column on Agilent 7890 GC–MS (Agilent Technologies, Santa Clara, CA, USA). The retention time and mass spectral patterns of plant SGs standard (Avanti Polar Lipids, Inc., Alabaster, AL, USA) were used as a reference (Gutierrez and del Rio, 2001). Cholesterol was added as an internal standard for these analyses prior to lipid extraction. Ceramide and glucosylceramide species were analyzed by multiple reactions monitoring (MRM) as described previously (Singh et al., 2012) using TSQ Quantum UltraTM Triple Quadrupole Mass Spectrometer (Thermo Scientific, USA). Samples were delivered by Accela pump (Thermo Finnigan, USA) to the HPLC fitted with 3 μm C8SR, 150 mm × 3.0 mm column (Peeke Scientific, Sommerset, NJ, USA). C17 sphingosine and C17 ceramide were added as an internal standard for these analyses prior to lipid extraction. Determination of plant sterols and sterylglucosides for enzymatic activity assay was performed using MRM monitoring on LC–MS (Wewer et al., 2011). Standard plant sterols and sterylglucosides (Avanti Polar Lipids, Inc.) were used as the external standards in these measurements.
Animal Studies
Four weeks old female CBA/J mice (Harlan Laboratories, Indianapolis, IN, USA) were used for all studies. Mice were anesthetized with an intraperitoneal injection of 60 μl xylazine/ketamine mixture containing 95 mg ketamine and 5 mg xylazine per kilogram of body weight and infected. For the infection studies, 24 mice (eight for each group) were infected intranasally with 5 × 105 cells/20 μl of WT, Δsgl1 or Δsgl1+SGL1 reconstituted strain. Mice were inspected twice a day and those that appeared moribund or in pain were sacrificed with CO2 inhalation followed by cervical dislocation. All animal procedures were approved by Stony Brook University Institutional Animal Care and Use Committee and followed the guidelines of American Veterinary Medical Association. For tissue burden analysis, four mice per strain were used. Lung, brain, liver, kidney and spleen were excised and homogenized in 10 ml of PBS using Stomacher 80 (Seward, UK) for 2 min at high speed. Several dilutions were plated in duplicate onto YPD agar plates and incubated for 48–72 h at 30°C. The CFUs per organ were counted. For histopathology analysis, three mice per strain were used. Mice organs were fixed in 3.7% of formaldehyde in paraffin and stained with haematoxylin and eosin and mucicarmine.
For in vivo vaccination studies, mice were pre-treated with vehicle (PBS), Δsgl1 (5 × 105 cells), and Δgcs1 (5 × 105 cells). After 30 days, mice pre-treated with vehicle or Δsgl1 were challenged with 5 × 105 cells of Cn WT or Cg R265. Mice pre-treated with Δgcs1 were challenged with 5 × 105 cells of Cn WT. Mouse survival was monitored for 80 days after post-challenge. CD4+ T-cell depletion was achieved by weekly intraperitoneal administration of anti-CD4+ (GK1.5, rat IgG2b, 200 μg in 200 μL of PBS; National Cell Culture Center, Minneapolis, MN, USA). A rat IgG2b (eBioscience, Inc., San Diego, CA, USA) was used as control. T-cell depletion was assessed by flow cytometry in the spleens. For vaccination studies, mice (eight for each group) were pre-treated with vehicle (PBS) or Δsgl1 strain after 48 h from the first round of T-cell depletion and after 30 days were challenged with a lethal dose of Cn WT (5 × 105 cells) and their survival was monitored for 80 days.
Results
CNAG_05607 has Sterylglucosidase and not Glucosylceramidase Activity
A S. cerevisiae expression system was used for characterizing the activity of the CNAG_05607 enzyme. The blast search of CNAG_05607 in Saccharomyces genome database revealed a gene YIR007W with an identity of 41% (expect = 1.8e-129) to CNAG_05607 (Supplementary Figure S1). Therefore, a ScΔYIR mutant strain lacking of YIR007W gene was used for the studies. CNAG_05607 was cloned in pYES/CT vector and overexpressed in S. cerevisiae YIR007W mutant strain (ScΔYIR + Cn 5607). As a negative control, ScΔYIR mutant was transformed with pYES/CT empty vector (ScΔYIR + empty vector). Total proteins were extracted from S. cerevisiae strains, which contained the empty vector (control) or overexpressed the CNAG_05607 enzyme, and were incubated with plant (Figure 1A) or cryptococcal sterylglucosides (SGs; Figure 1B). With either substrate, 100 μg of total protein extract was enough to significantly degrade the SGs as evidenced by the disappearance of the SGs band on the TLC. No difference in the intensity of the SGs band was detected compared to the SG control when ScΔYIR mutant strain carrying the empty vector was incubated with plant or cryptococcal SGs. The activity of the enzyme was dependent on pH (Figure 1C) and temperature (data not shown), with the maximum activity observed at a pH 4.5 in sodium acetate buffer and a temperature of 37°C. In addition to cryptococcal SGs, the CNAG_05607 enzyme was also able to degrade cholesterol glucoside, the mammalian form of SGs (Supplementary Figure S2).
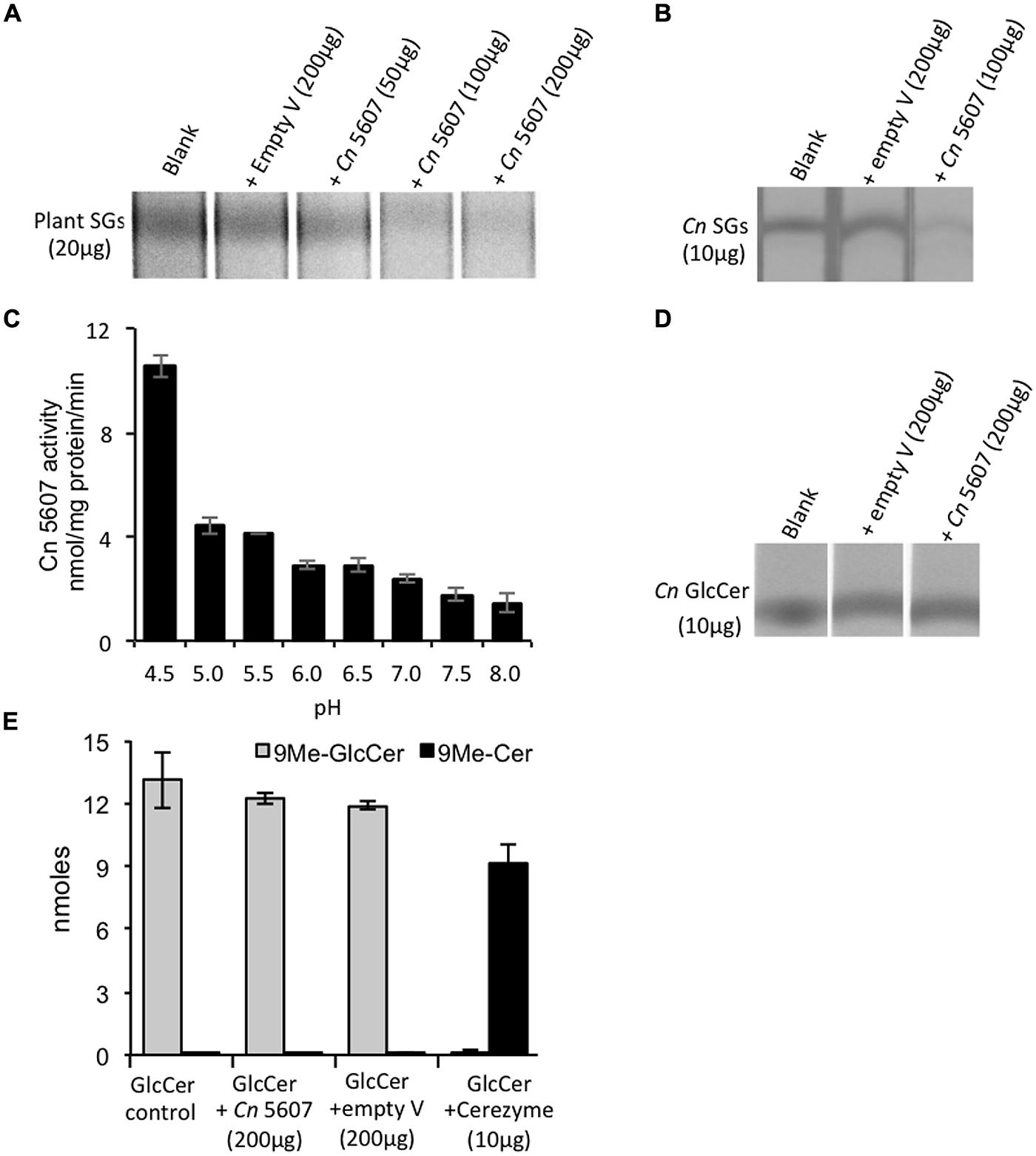
FIGURE 1. CNAG_05607 enzyme has sterylglucosidase and not glucosylceramidase activity. (A) Cryptococcus neoformans CNAG_05607 (Cn 5607) metabolizes plants SGs in a dose dependent manner and (B) metabolizes SGs extracted from Cn cells (Cn SGs). (C) pH dependence of Cn 5607 activity, as measured based on its ability to cleave plants SGs and produce free sterols, showed maximal activity at pH 4.5. Results are based on the measurement of free sterols performed by liquid chromatography–mass spectrometry (LC–MS). Cn 5607 does not metabolize C. neoformans GlcCer (Cn GlcCer) as analyzed by (D) thin layer chromatography (TLC) or by (E) LC–MS (Cerezyme is an analog of the human enzyme β-glucocerebrosidases, and was used as a positive control). Cn 5607 is the endoglycoceramidase-related protein 2 (EGCrP2) also identified by Watanabe et al. (2015). Empty V, empty vector.
The CNAG_05607 enzyme has recently been characterized as a glucosylceramidase due to its ability to hydrolyze short-chain glucosylceramides (Watanabe et al., 2015). Our initial biochemical characterization also showed that CNAG_05607 metabolizes short chain glucosylceramide (data not shown) similarly to what was observed by Watanabe et al. (2015). However, CNAG_05607 did not metabolize long-chain, physiologically relevant, Δ8-C9 methyl glucosylceramides (Figures 1D,E), which is the form of glucosylceramide found in C. neoformans. To the best of our knowledge, glucosylceramide synthase and glucosylcerebrosidase, do not need a co-factor or activator to exert their activity on long chain GlcCer (i.e., C16 GlcCer; Akiyama et al., 2013). Cerezyme, a human recombinant glucosylcerebrosidase, was used as control. This enzyme metabolized NBD-C6-GlcCer (data not shown) as well as long-chain CryptococcusΔ8-C9 methyl glucosylceramides resulting in ceramide production (Figure 1E). Cerezyme did not exhibit activity on plants or cryptococcal SGs (data not shown). Thus, these results demonstrate that CNAG_05607 has specific activity toward sterylglucosides, therefore we re-named this enzyme Sterylglucosidase 1 (Sgl1).
Deletion of SGL1 Causes Accumulation of SGs and not GlcCer
Since the Sgl1 enzyme acts to metabolize cryptococcal SGs, deletion of this enzyme in C. neoformans should lead to a SGs accumulating strain. This hypothesis was tested by genetically eliminating the sterylglucosidase enzyme (Supplementary Figure S3) in C. neoformans and monitoring the lipid profile by performing TLC and GC–MC on the total lipids extracted from the WT and the mutant strain. It was found that while the WT C. neoformans produces very little SGs, genetic elimination of sterylglucosidase (the Δsgl1 mutant) leads to a dramatic SGs accumulation; a phenomenon that is restored in the reconstituted strain (Δsgl1+SGL1; Figures 2A,B). In agreement with the in vitro activity studies, elimination of sterylglucosidase did not affect glucosylceramide levels in the cell (Figure 2C), further confirming the sterylglucosides-specific activity of this enzyme.
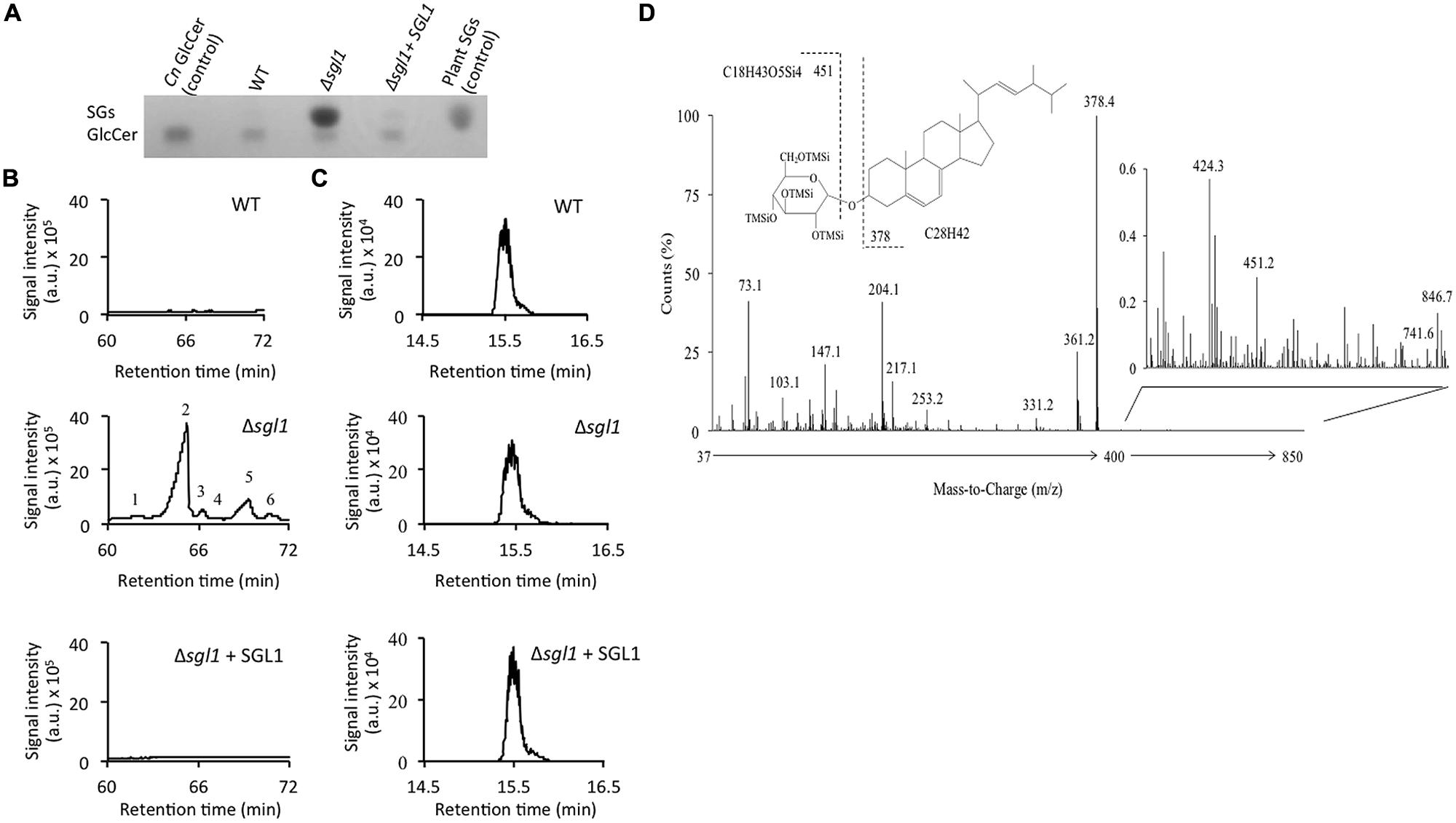
FIGURE 2. Deletion of SGL1 causes accumulation of SGs and not GlcCer. Analysis of sterylglucosides (SGs) and glucosylceramide (GlcCer) was performed by (A) TLC and (B,C) gas chromatography-mass spectrometry. Results show that the Δsgl1 mutant dramatically accumulates SGs (A,B), which are normally undetectable in wild-type (WT) or reconstituted strain (Δsgl1+ SGL1). In contrast, the level of GlcCer (A,C) in the Δsgl1 mutant is identical to the one observed in the WT or Δsgl1+SGL1 reconstituted strain. Chromatograms are representatives of three separate experiments showing similar results. Peaks denote: (1) Dehydroergosteryl-β-D-glucoside∗; (2) Ergosteryl-β-D-glucoside; (3) Ergosta-7,22-dien-3-oyl-β-D-glucoside∗; (4) Fecosteryl-β-D-glucoside∗; (5) Episteryl-β-D-glucoside∗; (6) Lanosteryl-β-D-glucoside∗ (∗: putative structures). (D) Structure and electron-impact (EI) mass spectrum of peak number 2.
In depth analysis of the MS spectrum of Δsgl1 strain showed the accumulation of 9 structures (Figure 2B) with ion fragments of m/z 147, 204, 217, 305, 361, 451. These structures were characteristic of tetramethylsilyl (TMSi) glucose ion fragments resulting from cleavage of C–O bonds. The fragments with m/z 361 and 451 are representative of TMSi derivative of hexoses. Ion fragments with m/z 129 and 255, characteristic of steroid moiety, were also present. Ion fragments with m/z of 73 and 147 represent the cleavage of 1 TMSi and 2 TMSsi groups respectively. The signal intensity of ion m/z 204 was greater than 217, which represented pyranoside configuration of the O-glycosidic linkage. These ion fragments resembled the fragmentation pattern generated during the MS analysis of plant sterylglucosides (Gutierrez and del Rio, 2001). Altogether, the ion fragments analysis confirmed that the structure possessed all characteristics of sterylglucosides. One of the most accumulated structures in the Δsgl1 mutant was ergosterolglucoside (Peak 2, Figure 2B). Apart from other characteristic ion fragments of sterolglucoside, MS fragmentation of peak 2 showed an ion fragment of m/z 378, which results from the cleavage C–O linkage of O-linked glucose moiety and is characteristic to ergosterol, suggesting that ergosterolglucoside was the structure with the highest concentration in the Δsgl1 strain. The chemical structure and the electron-impact mass spectrum of this molecule is presented in Figure 2D.
Sgl1 is a Virulence Factor of C. neoformans
Alterations in sphingolipid metabolism have been shown to attenuate cryptococcal virulence (Rittershaus et al., 2006; Singh et al., 2012). Thus, the virulence of the Δsgl1 strain in the mouse model of cryptococcosis was tested. Mice were infected with a lethal dose of fungal cells (5 × 105 cells) to establish cryptococcosis and monitored for their survival. The average survival of mice infected with the WT C. neoformans was 24 ± 6 days whereas all mice infected with Δsgl1 strain remained alive during the course of the experiment (i.e., 90 days post-infection). Mice infected with the Δsgl1+SGL1 strain showed a survival pattern similar to that observed in the WT (average survival of 21 ± 7 days; Figure 3A). During the course of infection, lungs and brains were removed from the mice infected with the three strains and analysis of tissue burden was performed at days 0, 3, 6, 9, and 14 post-infection.
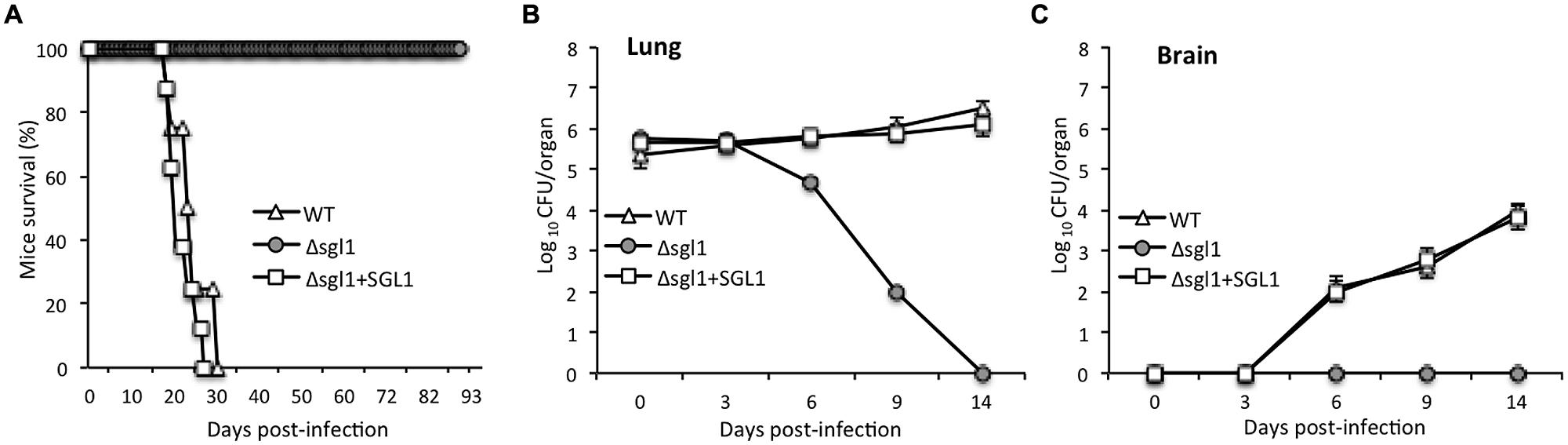
FIGURE 3. Deletion of the SGL1 gene in C. neoformans abolishes virulence. (A) Virulence studies showed that 100% of mice infected with Δsgl1 survived the infection whereas mice infected with C. neoformans WT H99 or with Δsgl1+SGL1 reconstituted strain succumbed to infection within 24 ± 6 and 21 ± 7 days, respectively; n = 8 mice in each group. (B) Lung tissue burden analysis showed that the Δsgl1 mutant is eliminated from the lungs after 14 days of inoculation; n = 3 mice each at time point. (C) Brain tissue burden analysis showed that Δsgl1 is not found in the brain at anytime during the course of experiment; n = 3 mice at each time point.
Interestingly, the number of Δsgl1 cells in the lungs decreased starting at day 3 and continued until day 14, at which point the lungs were completely clear of fungal cells (Figure 3B). Furthermore, no Δsgl1 cells were observed in the brain (Figure 3C), suggesting that fungal cells did not disseminate to the brain in the Δsgl1-infected mice. In contrast, a significant number of fungal cells were found in the lungs and brains of mice infected with the WT or the Δsgl1+SGL1 strain (Figures 3B,C). In both cases, the number of fungal cells in the brain increased as a function of time, demonstrating the occurrence of extrapulmonary dissemination and progression of the disease. The findings of the tissue burden studies were confirmed by lung and brain histology observations, which showed no fungal cells in the organs isolated from the Δsgl1-infected mice at the end of the experiment, but significant tissue damage and presence of fungal cells in the WT or Δsgl1+SGL1 strains (Supplementary Figure S4). These experiments reveal that sterylglucosidase is a virulence factor in C. neoformans, as the loss of this enzyme leads to loss of virulence in the mouse model.
The Δsgl1 Strain Acts as a Vaccine Against Cryptococcosis in the Mouse Model
Cryptococcus neoformans cells possess a number of virulence factors that contribute to their survival inside the host, resistance to immune response, and detrimental activity against the host (Coelho et al., 2014). To gain more insight into the loss of virulence of the Δsgl1 strain, a number of virulence factors in this strain were evaluated and compared to the WT. In comparison to the WT, the Δsgl1 strain showed similar growth in acidic and neutral pH (at 37°C and in the presence of 5% CO2), similar melanin production and capsule thickness, and no major difference in growth under oxidative or nitrosative stress. In addition, the WT and mutant strains showed similar intracellular growth during in vitro infection of the J774.16 macrophage-like cells (Supplementary Figure S5). These analyses suggest that the most common virulence factors are similar between the Δsgl1 strain and the WT denoting a different mechanism for the loss of virulence.
Given that the Δsgl1 strain was non-pathogenic and that SGs are known immunostimulators (Lee et al., 2007; Grille et al., 2010), the potential use of the Δsgl1 strain as a vaccine against cryptococcosis was investigated. Two controls were used for these studies: a vehicle (sterile PBS) and the C. neoformans Δgcs1 strain (Rittershaus et al., 2006), which is avirulent, but does not accumulate SGs. Mice were infected with the vehicle, or 5 × 105 cells of the Δgcs1 or the Δsgl1 strains and after 30 days were challenged with a lethal dose of the virulent WT C. neoformans or C. gattii R265 strains. Interestingly, the mice that were pre-treated with the Δsgl1 strain were completely protected against the subsequent infection. However, the mice that were pre-treated with the vehicle or the Δgcs1 strain succumbed to infection within 35 days (Figure 4A). These results suggest that the Δsgl1 strain may stimulate a host immune response that successfully kills Δsgl1 and makes the host resistant to subsequent cryptococcosis.
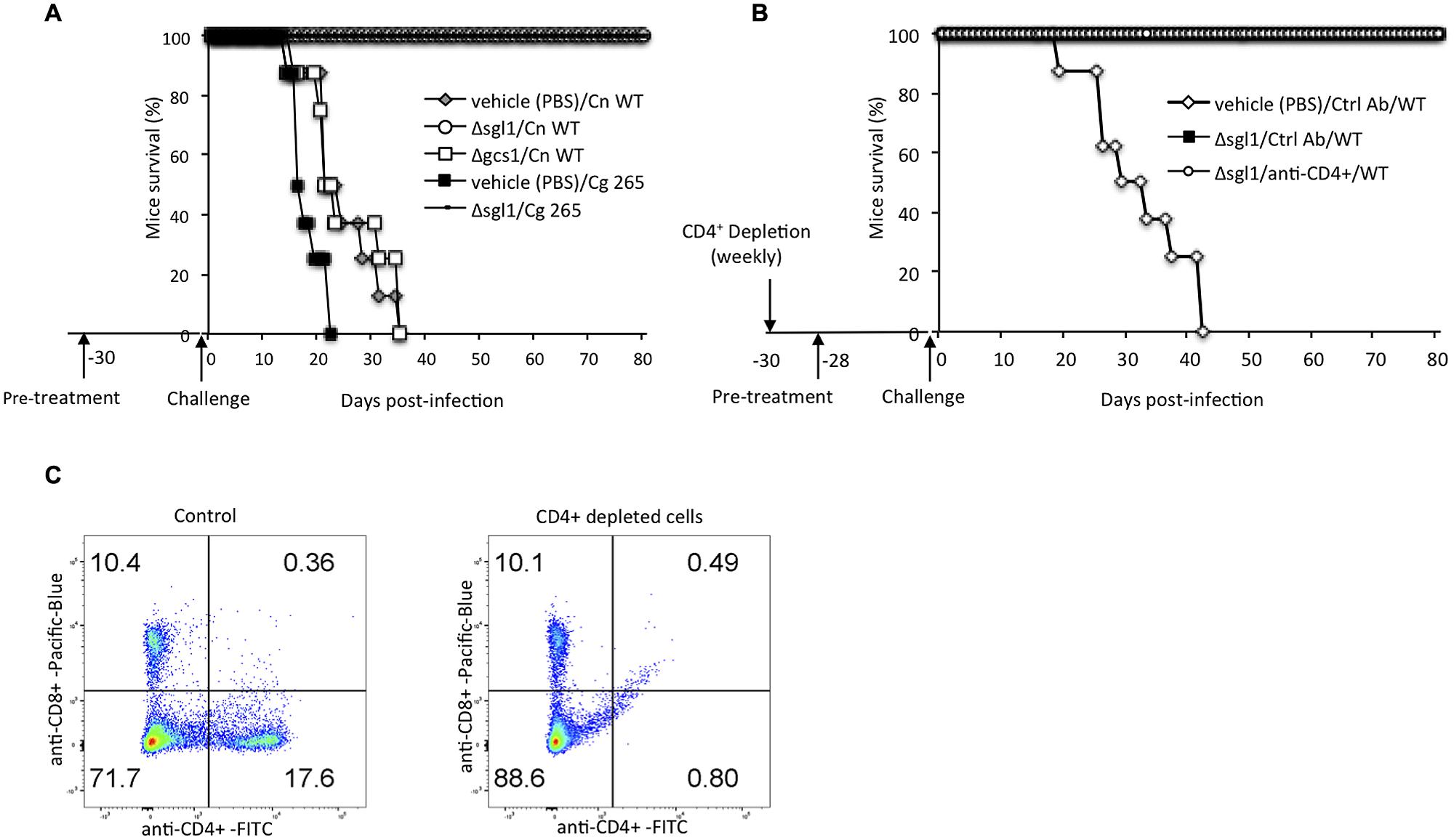
FIGURE 4. Vaccination studies. (A) Mice were “pre-treated” with Δsgl1 and, after 30 days (time 0), challenged with a lethal dose of C. neoformans wild-type H99 (Cn WT) or Cryptococcus gattii WT R265 (Cg 265). Mice exposed to Δsgl1 remained alive during the course of experiment (80 days post-infection) whereas all mice that were not exposed to Δsgl1 but to vehicle (PBS) or to the Δgcs1 mutant strain and then challenged with Cn WT succumbed to infection within 35 days; n = 8 mice in each group. (B) Pre-treatment with Δsgl1 completely protected CD4+ T-cell depleted mice from a subsequent lethal challenge with C. neoformans wild-type H99 (WT); n = 8 mice in each group. Depletion of CD4+ was achieved by administering anti-CD4 (Ab) weekly during the entire course of the experiment. The depletion was confirmed by flow cytometry performed at the day of Cn challenge (C). Results are representative of three separate experiments showing similar results.
Although Cryptococcus infections can afflict immunocompetent individuals, the majority of the population at risk, are those suffering from immune suppression, such as HIV/AIDS patients. A reduction in CD4+ T-cells in this population results in aggressive cryptococcosis, which can be life threatening (Jarvis et al., 2013). The efficiency of the Δsgl1 strain as a vaccine against cryptococcosis during immune suppression was examined by its administration in CD4+ T-cells depleted mice prior to infection with the WT C. neoformans. For these studies, mice were depleted of CD4+ by weekly administration of anti-CD4+ antibody or control antibody (rat IgG2b) starting a month prior to infection with WT C. neoformans (Figure 4B). A 94.3% percent reduction in CD4+ T-cells was achieved as confirmed by flow cytometry (Figure 4C). The Δsgl1 strain or control (PBS) were also administered to mice a month prior to infection. Mice were then infected with 5 × 105 cells of the virulent WT C. neoformans. All mice that received the PBS and the antibody control succumbed to infection in 41 days, while all the CD4+ T-cells depleted mice that were vaccinated with the Δsgl1 strain survived the infection, demonstrating that this strain is not infectious and can protect immune suppressed mice against a subsequent cryptococcal infection (Figure 4B).
Discussion
In the current study, a mutant C. neoformans strain lacking the enzyme sterylglucosidase 1 (Sgl1) was engineered and characterized. This strain accumulated SGs and was not virulent in the mouse model, despite not demonstrating any deficiency in commonly known cryptococcal virulence factors. Importantly, this strain acted as a vaccine and was able to protect CD4+ T-cells depleted and immunocompetent mice against subsequent infections with lethal doses of C. neoformans or C. gattii.
Deletion of the Sgl1 enzyme was used as a strategy to accumulate SGs in C. neoformans. The presence of this enzyme was recently reported in C. neoformans and found to hydrolyze both SGs and short chain glucosylceramide (Watanabe et al., 2015). However, our results provided a more thorough characterization because it included physiologically relevant glucosylceramide species, which are produced by C. neoformans cells. We found that Sgl1 does not metabolize long chain glucosylceramide but only SGs.
The Δsgl1 mutant was not virulent in the mouse model of cryptococcosis. In the Δsgl1 strain, fungal cells were killed in the lungs within 14 days post-infection. No fungal cells were found in the brain tissue suggesting that the infection is completely controlled by the host in the lungs. It is important to note that the reduction in the fungal burden in the lungs starts at 3 days post-infection. This is due to the fact that the Δsgl1 strain does not show an appreciable growth defect compared to the WT and exhibits a similar resistance profile to stresses. Thus, the delay in the reduction of fungal burden is due to the time it takes for this strain to stimulate the immune response, which then kills the strain in vivo. Given that the loss of Sgl1 does not affect the growth profile or the common virulence factors of C. neoformans, this enzyme appears to be a stand-alone factor in controlling cryptococcal virulence. This is confirmed by the observation that the virulence phenotype was restored in the reconstituted strain (Δsgl1+SGL1). The Sgl1 enzyme was active at physiological temperature and in the pH range of 4.5 to 8. The maximum activity was observed at pH = 4.5 after which the activity was sharply reduced with increased alkalinity. These activity profiles suggest that Sgl1 is highly active in the acidic environment inside alveolar macrophages; the activity is also present, although significantly reduced, in slightly acidic/neutral environment within the alveolar spaces.
Administration of the SGs accumulating Δsgl1 strain proved to be an effective vaccination strategy in the mouse models of cryptococcosis. When administered 30 days prior to the challenge, this strain was able to protect mice against subsequent infections with either WT C. neoformans H99 and C. gattii R265. These findings suggest that protection by the Δsgl1 strain is not serotype specific. Protection against the C. gattii R265 is of particular interest. The immune response against this strain is different from C. neoformans (Dong and Murphy, 1995; Wright et al., 2002; Cheng et al., 2009) and it has been the subject of a recent study for vaccination (Chaturvedi et al., 2014). It is important to note that the Sgl1 enzyme is capable of hydrolyzing cholesterol glucoside as well. Thus, although not experimentally tested, it is also possible that infection with this strain might lead to the uptake and accumulation of cholesterol glucoside from the host, which would be otherwise metabolized by the WT C. neoformans. However, it appears unlikely that cholesterol glucoside accumulation would contribute to immunity against the infection. The bacterium Helicobacter pylori routinely accumulates cholesterol glucosides during the infection of the mammalian host without eliciting any form of lipid-induced immunity (Wunder et al., 2006).
The Δsgl1 strain was also able to protect CD4+ T-cell depleted mice against cryptococcosis. SGs have been shown to enhance T-cell proliferation (Bouic et al., 1996) and promote Th1 immune response (Lee et al., 2007). A switch to Th1 cell-mediated immunity is the predominant mode of immune response against cryptococcosis (Decken et al., 1998; Koguchi and Kawakami, 2002; Wormley et al., 2007). In fact, promotion of Th1 response with an engineered interferon-gamma producing C. neoformans has been shown to protect mice against subsequent C. neoformans infections. Our results suggest that the immune stimulation by SGs goes beyond the Th1 immune response because our vaccination strategy is still effective in condition of CD4+ T cell deficiency. This is particularly advantageous as cryptococcosis mostly occurs in patients with this type of immune deficiency. It should be noted that while the Δsgl1 strain provides complete immunity in the mouse model of cryptococcosis, the administration of a live attenuated vaccine is associated with challenges such as undesired secondary mutations, which might revert the virulence (Teng et al., 2013). However, this strain can be used as a valuable starting point for the development of other vaccine formulations such heat-killed strains or vesicle formulations containing SGs, which would eliminate the dangers of using a live vaccine.
The immunomodulatory properties of SGs have been documented not only in the context of fungal infections (Lee et al., 2007), but also in other human diseases such as rhinitis, sinusitis, and tuberculosis (Bouic et al., 1996; Donald et al., 1997; Bouic, 2001). Although the underlying molecular mechanisms remain largely unknown; an increase in the level of T cells and cytokines induced by SG administration has been reported (Bouic et al., 1996; Donald et al., 1997). Although our data clearly show that the Δsgl1 strain accumulates SGs, we cannot rule out the presence of parallel/consequential mechanisms for the Δsgl1-induced immunity. It is possible that other factors such as stimulation of CD4+ and CD8+ T cells are involved. The immunological basis of protection provided by vaccination with the Δsgl1 strain is an interesting topic for future studies.
Conclusion
We have provided a comprehensive biochemical study of Sgl1, characterized the C. neoformansΔsgl1 strain and reported that the Sgl1 enzyme is a virulence factor in C. neoformans. The Δsgl1 strain is non-pathogenic and can protect immunocompromised mice against cryptococcosis. These results could have important implications for future vaccine development for cryptococcosis and other fungal infections.
Author Contributions
AR and MDP conceived the study. AR designed and performed the majority of the experiments, analyzed the results and helped with writing the paper. VM helped in the design of the study and the experimental work on animals. AF contributed to data analysis and the design of experiments and wrote the paper. AS performed and analyzed the mass spectrometry experiments. AAS performed and analyzed the flow cytometry experiments. EI, NC, and MM provided critical help with the study design. CL helped with the experiments and provided a critical review of the study design. MDP contributed to the design of the experiments, analysis of data, and writing and reviewing the paper. All authors reviewed the results and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Dr. Bidyut K. Mohanty for kindly providing the S. cerevisiae strains and Arielle Bryan for help with animal studies. This work was supported by NIH grants AI56168, AI71142, AI87541, AI100631 and by a Merit Review grant I01BX002624 from the Veterans Affairs Program in Biomedical Laboratory Research and Development to MDP. AR is an American Heart Association post-doctoral fellow. MDP is Burroughs Welcome Investigator in Infectious Diseases.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00836
References
Akiyama, H., Kobayashi, S., Hirabayashi, Y., and Murakami-Murofushi, K. (2013). Cholesterol glucosylation is catalyzed by transglucosylation reaction of beta-glucosidase 1. Biochem. Biophys. Res. Commun. 441, 838–843. doi: 10.1016/j.bbrc.2013.10.145
Bligh, E. G., and Dyer, W. J. A. (1959). rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. doi: 10.1139/o59-099
Bouic, P. J. (2001). The role of phytosterols and phytosterolins in immune modulation: a review of the past 10 years. Curr. Opin. Clin. Nutr. Metab. Care 4, 471–475.
Bouic, P. J., Etsebeth, S., Liebenberg, R. W., Albrecht, C. F., Pegel, K., and Van Jaarsveld, P. P. (1996). beta-Sitosterol and beta-sitosterol glucoside stimulate human peripheral blood lymphocyte proliferation: implications for their use as an immunomodulatory vitamin combination. Int. J. Immunopharmacol. 18, 693–700. doi: 10.1016/S0192-0561(97)85551-8
Casadevall, A., Mukherjee, J., Devi, S. J., Schneerson, R., Robbins, J. B., and Scharff, M. D. (1992). Antibodies elicited by a Cryptococcus neoformans-tetanus toxoid conjugate vaccine have the same specificity as those elicited in infection. J. Infect. Dis. 165, 1086–1093. doi: 10.1093/infdis/165.6.1086
Casadevall, A., and Pirofski, L. (2005). Insights into mechanisms of antibody-mediated immunity from studies with Cryptococcus neoformans. Curr. Mol. Med. 5, 421–433. doi: 10.2174/1566524054022567
Chaturvedi, A. K., Hameed, R. S., Wozniak, K. L., Hole, C. R., Leopold Wager, C. M., Weintraub, S. T., et al. (2014). Vaccine-mediated immune responses to experimental pulmonary Cryptococcus gattii infection in mice. PLoS ONE 9:e104316. doi: 10.1371/journal.pone.0104316
Cheng, P. Y., Sham, A., and Kronstad, J. W. (2009). Cryptococcus gattii isolates from the British Columbia cryptococcosis outbreak induce less protective inflammation in a murine model of infection than Cryptococcus neoformans. Infect. Immun. 77, 4284–4294. doi: 10.1128/IAI.00628-09
Coelho, C., Bocca, A. L., and Casadevall, A. (2014). The tools for virulence of Cryptococcus neoformans. Adv. Appl. Microbiol. 87, 1–41. doi: 10.1016/B978-0-12-800261-2.00001-3
Datta, K., Bartlett, K. H., Baer, R., Byrnes, E., Galanis, E., Heitman, J., et al. (2009). Spread of Cryptococcus gattii into Pacific Northwest region of the United States. Emerg. Infect. Dis. 15, 1185–1191. doi: 10.3201/eid1508.081384
Datta, K., and Pirofski, L. A. (2006)Towards a vaccine for Cryptococcus neoformans: principles and caveats. FEMS Yeast Res. 6, 525–536. doi: 10.1111/j.1567-1364.2006.00073.x
Decken, K., Kohler, G., Palmer-Lehmann, K., Wunderlin, A., Mattner, F., Magram, J., et al. (1998). Interleukin-12 is essential for a protective Th1 response in mice infected with Cryptococcus neoformans. Infect. Immun. 66, 4994–5000.
Devi, S. J. (1996). Preclinical efficacy of a glucuronoxylomannan-tetanus toxoid conjugate vaccine of Cryptococcus neoformans in a murine model. Vaccine 14, 841–844. doi: 10.1016/0264-410X(95)00256-Z
Devi, S. J., Schneerson, R., Egan, W., Ulrich, T. J., Bryla, D., Robbins, J. B., et al. (1991). Cryptococcus neoformans serotype A glucuronoxylomannan-protein conjugate vaccines: synthesis, characterization, and immunogenicity. Infect. Immun. 59, 3700–3707.
Donald, P. R., Lamprecht, J. H., Freestone, M., Albrecht, C. F., Bouic, P. J., Kotze, D., et al. (1997). randomised placebo-controlled trial of the efficacy of beta-sitosterol and its glucoside as adjuvants in the treatment of pulmonary tuberculosis. Int. J. Tuberc. Lung. Dis. 1, 518–522.
Dong, Z. M., and Murphy, J. W. (1995)Effects of the two varieties of Cryptococcus neoformans cells and culture filtrate antigens on neutrophil locomotion. Infect. Immun. 63, 2632–2644.
Grille, S., Zaslawski, A., Thiele, S., and Plat, J. (2010). Warnecke D The functions of steryl glycosides come to those who wait: recent advances in plants, fungi, bacteria and animals. Prog. Lipid Res. 49, 262–288. doi: 10.1016/j.plipres.2010.02.001
Gutierrez, A., and del Rio, J. C. (2001). Gas chromatography/mass spectrometry demonstration of steryl glycosides in eucalypt wood, Kraft pulp and process liquids. Rapid Commun. Mass Spectrom. 15, 2515–2520. doi: 10.1002/rcm.537
Jarvis, J. N., Casazza, J. P., Stone, H. H., Meintjes, G., Lawn, S. D., Levitz, S. M., et al. (2013). The phenotype of the Cryptococcus-specific CD4+ memory T-cell response is associated with disease severity and outcome in HIV-associated cryptococcal meningitis. J. Infect. Dis. 207, 1817–1828. doi: 10.1093/infdis/jit099
Kawai, S., Hashimoto, W., and Murata, K. (2010). Transformation of Saccharomyces cerevisiae and other fungi: methods and possible underlying mechanism. Bioeng Bugs 1, 395–403. doi: 10.4161/bbug.1.6.13257
Koguchi, Y., and Kawakami, K. (2002). Cryptococcal infection and Th1-Th2 cytokine balance. Int. Rev. Immunol. 21, 423–438. doi: 10.1080/08830180213274
Lee, J. H., and Han, Y. (2006). Ginsenoside Rg1 helps mice resist to disseminated candidiasis by Th1 type differentiation of CD4+ T cell. Int. Immunopharmacol. 6, 1424–1430. doi: 10.1016/j.intimp.2006.04.009
Lee, J. H., Lee, J. Y., Park, J. H., Jung, H. S., Kim, J. S., Kang, S. S., et al. (2007). Immunoregulatory activity by daucosterol, a beta-sitosterol glycoside, induces protective Th1 immune response against disseminated Candidiasis in mice. Vaccine 25, 3834–3840. doi: 10.1016/j.vaccine.2007.01.108
Mandala, S. M., Thornton, R. A., Frommer, B. R., Curotto, J. E., Rozdilsky, W., Kurtz, M. B., et al. (1995). The discovery of australifungin, a novel inhibitor of sphinganine N-acyltransferase from Sporormiella australis. Producing organism, fermentation, isolation, and biological activity. J. Antibiot. (Tokyo) 48, 349–356. doi: 10.7164/antibiotics.48.349
Nanjappa, S. G., and Klein, B. S. (2014). Vaccine immunity against fungal infections. Curr. Opin. Immunol. 28, 27–33. doi: 10.1016/j.coi.2014.01.014
Park, B. J., Wannemuehler, K. A., Marston, B. J., Govender, N., Pappas, P. G., and Chiller, T. M. (2009). Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23, 525–530. doi: 10.1097/QAD.0b013e328322ffac
Rittershaus, P. C., Kechichian, T. B., Allegood, J. C., Merrill, A. H. Jr., Hennig, M., Luberto, C., et al. (2006). Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. J. Clin. Invest. 116, 1651–1659. doi: 10.1172/JCI27890
Shea, J. M., Kechichian, T. B., Luberto, C., and Del Poeta, M. (2006). The cryptococcal enzyme inositol phosphosphingolipid-phospholipase C confers resistance to the antifungal effects of macrophages and promotes fungal dissemination to the central nervous system. Infect. Immun. 74, 5977–5988. doi: 10.1128/IAI.00768-06
Singh, A., Qureshi, A., and Del Poeta, M. (2011). Quantitation of cellular components in Cryptococcus neoformans for system biology analysis. Methods Mol. Biol. 734, 317–333. doi: 10.1007/978-1-61779-086-7_16
Singh, A., Wang, H., Silva, L. C., Na, C., Prieto, M., Futerman, A. H., et al. (2012). Methylation of glycosylated sphingolipid modulates membrane lipid topography and pathogenicity of Cryptococcus neoformans. Cell. Microbiol. 14, 500–516. doi: 10.1111/j.1462-5822.2011.01735.x
Teng, X., Dayhoff-Brannigan, M., Cheng, W. C., Gilbert, C. E., Sing, C. N., Diny, N. L., et al. (2013). Genome-wide consequences of deleting any single gene. Mol. Cell. 52, 485–494. doi: 10.1016/j.molcel.2013.09.026
Toffaletti, D. L., Rude, T. H., Johnston, S. A., Durack, D. T., and Perfect, J. R. (1993). Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175, 1405–1411.
Tripathi, K., Mor, V., Bairwa, N. K., Del Poeta, M., and Mohanty, B. K. (2012). Hydroxyurea treatment inhibits proliferation of Cryptococcus neoformans in mice. Front. Microbiol. 3:187. doi: 10.3389/fmicb.2012.00187
Walraven, C. J., Gerstein, W., Hardison, S. E., Wormley, F., Lockhart, S. R., Harris, J. R., et al. (2011). Fatal disseminated Cryptococcus gattii infection in New Mexico. PLoS ONE 6:e28625. doi: 10.1371/journal.pone.0028625
Wang, Y., Aisen, P., and Casadevall, A. (1995). Cryptococcus neoformans melanin and virulence: mechanism of action. Infect. Immun. 63, 3131–3136.
Warnecke, D., Erdmann, R., Fahl, A., Hube, B., Muller, F., Zank, T., et al. (1999). Cloning and functional expression of UGT genes encoding sterol glucosyltransferases from Saccharomyces cerevisiae, Candida albicans, Pichia pastoris, and Dictyostelium discoideum. J. Biol. Chem. 274, 13048–13059. doi: 10.1074/jbc.274.19.13048
Watanabe, T., Ito, T., Goda, H. M., Ishibashi, Y., Miyamoto, T., Ikeda, K., et al. (2015). Sterylglucoside Catabolism in Cryptococcus neoformans with endoglycoceramidase-related protein 2 (EGCrP2), the first steryl-beta-glucosidase identified in fungi. J. Biol. Chem. 290, 1005–1019. doi: 10.1074/jbc.M114.616300
Wewer, V., Dombrink, I., Vom Dorp, K., and Dormann, P. (2011). Quantification of sterol lipids in plants by quadrupole time-of-flight mass spectrometry. J. Lipid Res. 52, 1039–1054. doi: 10.1194/jlr.D013987
Wormley, F. L. Jr., Perfect, J. R., Steele, C., and Cox, G. M. (2007). Protection against cryptococcosis by using a murine gamma interferon-producing Cryptococcus neoformans strain. Infect. Immun. 75, 1453–1462. doi: 10.1128/IAI.00274-06
Wozniak, K. L., Young, M. L., and Wormley, F. L. Jr. (2011). Protective immunity against experimental pulmonary cryptococcosis in T cell-depleted mice. Clin. Vaccine Immunol. 18, 717–723. doi: 10.1128/CVI.00036-11
Wright, L., Bubb, W., Davidson, J., Santangelo, R., Krockenberger, M., Himmelreich, U., et al. (2002). Metabolites released by Cryptococcus neoformans var. neoformans and var. gattii differentially affect human neutrophil function. Microbes Infect. 4, 1427–1438. doi: 10.1016/S1286-4579(02)00024-2
Keywords: yeast genetics, gene expression, glycolipid, immunosuppression, vaccine development
Citation: Rella A, Mor V, Farnoud AM, Singh A, Shamseddine AA, Ivanova E, Carpino N, Montagna MT, Luberto C and Del Poeta M (2015) Role of Sterylglucosidase 1 (Sgl1) on the pathogenicity of Cryptococcus neoformans: potential applications for vaccine development. Front. Microbiol. 6:836. doi: 10.3389/fmicb.2015.00836
Received: 04 July 2015; Accepted: 29 July 2015;
Published: 11 August 2015.
Edited by:
Luis R. Martinez, New York Institute of Technology College of Osteopathic Medicine, USAReviewed by:
Radames J. B. Cordero, Johns Hopkins Bloomberg School of Public Health, USAAllan Guimaraes, AECOM, USA
Copyright © 2015 Rella, Mor, Farnoud, Singh, Shamseddine, Ivanova, Carpino, Montagna, Luberto and Del Poeta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maurizio Del Poeta, Department of Molecular Genetics and Microbiology, Stony Brook University, 145 Life Sciences Building, Stony Brook, NY 11794, USA,bWF1cml6aW8uZGVscG9ldGFAc3Rvbnlicm9vay5lZHU=
 Antonella Rella1
Antonella Rella1 Visesato Mor
Visesato Mor Ashutosh Singh
Ashutosh Singh Maria T. Montagna
Maria T. Montagna Maurizio Del Poeta
Maurizio Del Poeta