- 1Department of Clinical Laboratory, The First Hospital Affiliated to Henan University, Kaifeng, China
- 2State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing, China
- 3Department of Clinical Laboratory, The Second Affiliated Hospital of Soochow University, Suzhou, China
- 4Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing, China
A carbapenem-resistant Klebsiella pneumoniae strain 628 was isolated from a human case of intracranial infection in a Chinese teaching hospital. Strain 628 produces KPC-2 and CTX-M-55 encoded by two different conjugative plasmids, i.e., the IncFIIK plasmid p628-KPC and the IncI1 plasmid p628-CTXM respectively. blaKPC−2 is captured by a Tn1722-based unit transposon with a linear structure. ΔTn3-ISKpn27-blaKPC−2-ΔISKpn6-ΔTn1722 and this transposon together with a mercury resistance (mer) gene locus constitutes a 34 kb acquired drug-resistance region. blaKPC−2 has two transcription starts (nucleotides G and C located at 39 and 250 bp upstream of its coding region respectively) which correspond to two promoters, i.e., the intrinsic P1 and the upstream ISKpn27/Tn3-provided P2 with the core −35/−10 elements TAATCC/TTACAT and TTGACA/AATAAT respectively. blaCTX−M−55 is mobilized in an ISEcp1-blaCTX−M−55-Δorf477 transposition unit and appears to be the sole drug-resistant determinant in p628-CTXM. blaCTX−M−55 possesses a single transcription start (nucleotides G located at 116 bp upstream of its coding region) corresponding to the ISEcp1-provided P1 promoter with the core −35/−10 element TTGAAA/TACAAT. All the above detected promoters display a characteristic of constitutive expression. Coexistence of blaKPC and blaCTX−M in K. pneumoniae has been reported many times but this is the first report to gain deep insights into genetic platforms, promoters, and expression of the two coexisting bla genes with determination of entire nucleotide sequences of the two corresponding plasmids.
Introduction
KPC-producing Klebsiella pneumoniae has spread worldwide and became an emerging pathogen with serious clinical and infection control implications (Tzouvelekis et al., 2012; Munoz-Price et al., 2013). Coexistence of blaKPC and blaCTX−M in K. pneumoniae has been reported in several countries, such as blaKPC−2/blaCTX−M−1 group, blaKPC−2/blaCTX−M−2 group, and blaKPC−2/blaCTX−M−8 group in Brazil (Peirano et al., 2009), blaKPC−2/blaCTX−M−10, blaKPC−2/blaCTX−M−15, and blaKPC−3/blaCTX−M−2 in Israel (Leavitt et al., 2007, 2010), blaKPC−2/blaCTX−M−14 in China (Cai et al., 2008), and blaKPC−2/blaCTX−M−15 in Greece (Souli et al., 2010). However, all these studies are confined to PCR detection and sequencing of bla genes, lacking deeper characterization of mechanisms of drug resistance. This study describes co-production of KPC-2 and CTX-M-55 in a clinical K. pneumoniae strain 628 from China. The blaKPC−2 and blaCTX−M−55genes are encoded by two different conjugative plasmids, p628-KPC and p628-CTXM respectively. The complete nucleotide sequences of p628-KPC and p628-CTXM are determined and then compared with other genetically closely related plasmids to gain deep insights into genetic structures of relevant plasmids and resistance gene loci. In addition, the promoters and their expression characteristics of these two plasmid-borne bla genes are dissected experimentally.
Materials and Methods
Bacterial Strains and Identification
K. pneumoniae strain 628 was isolated from the cerebrospinal fluid specimen of a 64-year-old male with intracranial infection in a Chinese teaching hospital in October 2010. Bacterial species identification was performed using Bruker MALDI Biotyper (Bruker Daltonics, Bremen, Germany) and 16s rRNA gene sequencing (Frank et al., 2008). The major carbapenemase and extended-spectrum beta-lactamase (ESBL) genes were detected by PCR, followed by sequencing on an ABI Sequencer (Applied Biosystems, Foster City, CA, USA) (Chen et al., 2015). Bacterial antimicrobial susceptibility was tested by using VITEK 2 and judged by CLSI standard (CLSI, 2012).
Plasmid Transfer
Plasmid conjugal transfer experiments were carried out with Escherichia coli EC600 (LacZ−, NalR, RifR) being used as recipient and strain 628 as donor. Three milliliter of overnight culture of each of donor and recipient bacteria were mixed together, harvested and resuspended in 80 μl of Brain Heart Infusion broth (BD Biosciences, San Jose, CA, USA). The mixture was spotted on a 1 cm2 filter membrane that was placed on Brain Heart Infusion agar (BD Biosciences, San Jose, CA, USA) plate, and then incubated for mating at 37°C for 12 to 18 h. Bacteria were washed from filter membrane and spotted on Muller-Hinton agar (BD Biosciences, San Jose, CA, USA) plate containing 1000 mg/L rifampin (Merck, Darmstadt, Germany) and 200 mg/L ampicillin (Merck, Darmstadt, Germany) for selection of blaCTX−M- or blaKPC-positive E. coli transconjugants.
Determination of Plasmid DNA Sequence
Plasmid DNA was isolated from the cell culture of E. coli transconjugant using Qiagen large construct kit (Qiagen, Hilden, Germany) and then sequenced by using whole-genome shotgun strategy in combination with Illumina HiSeq 2500 (Illumina, San Diego, CA, USA) sequencing technology. The contigs were assembled with Velvet and the gaps were filled through combinatorial PCR and Sanger Sequencing on an ABI Sequencer. The genes were predicted with GeneMarkS™ and further annotated by BLASTP and BLASTN against UniProt and NR databases.
RNA Isolation and Primer Extension Assay
Bacteria were cultured overnight in Mueller-Hinton broth (BD Biosciences, San Jose, CA, USA). Total RNAs were extracted from harvested bacterial cells using TRIzol Reagent (Life Technologies, Carlsbad, CA, USA). RNA quality was monitored by agarose gel electrophoresis, and RNA quantity was determined by spectrophotometry. Each of the [γ-32P] ATP end-labeled primers GCTCAGTGGAACGAAAAC, AGCCGCCAAAGTCCTGTTCG, and CATGGGATTCCTTATTCTG, which corresponded to blaKPC−2 promoter P2, blaKPC−2 promoter P1, and blaCTX−M−55 promoter P1 respectively, was annealed with total RNA sample for primer extension assay as described previously (Zhang et al., 2011). For different cell cultures in a single experiment, equal amounts of total RNA were used as starting materials. The corresponding end-labeled primers were also used for sequencing the PCR amplicons generated by the primer pairs TCAGCGACATCGTCAACC/GGTCGTGTTTCCCTTTAGCC, TCAGGTGGCACTTTTCGG/GGTCGTGTTTCCCTTTAGCC, and AGACCTTTCGTTTGAAGTATG/AGCTTATTCATCGCCACGTT for blaKPC−2 promoter P2, blaKPC−2 promoter P1, and blaCTX−M−55 promoter P1 respectively. DNA sequencing was carried out using AccuPower & Top DNA Sequencing Kit (Bioneer, Daejeon, Korea). Primer extension products and sequencing materials were analyzed on 8 M urea-6% polyacrylamide gel electrophoresis. Radioactive species were detected by autoradiography.
Nucleotide Sequence Accession Numbers
The complete sequences of plasmids p628-KPC and p628-CTXM were submitted to GenBank under accession numbers KP987218 and KP987217 respectively.
Results and Discussion
Characterization of K. pneumoniae Strain 628
Strain 628 harbors blaKPC−2, blaCTX−M−55, blaSHV, and blaTEM. blaKPC−2 and blaCTX−M−55are located plasmids p628-KPC and p628-CTXM respectively. Conjugative transfer of p628-KPC or p628-CTXM into EC600 generates the transconjugant 628-KPC-EC600 (bla+KPC−2, bla−CTX−M−55, bla−SHV, and bla−TEM) or 628-CTXM-EC600 (bla−KPC−2, bla+CTX−M−55, bla−SHV, and bla−TEM) respectively. All of 628, 628-KPC-EC600 and 628-CTXM-EC600 are resistant to ampicillin, ampicillin/sulbactam, penicillin, monobactam, and cephalosporins tested (Table 1). 628 and 628-KPC-EC600 (but not 628-CTXM-EC600) are resistant to piperacillin/tazobactam. 628 and 628-KPC-EC600 (but not 628-CTXM-EC600) are carbapenem-resistant.
Complete Nucleotide Sequence of p628-KPC
The entire nucleotide sequence of p628-KPC is 105,008 bp in length, forming a circular plasmid with an average G+C content of 53.22 and a total of 127 open reading frames (ORFs) annotated (Figure 1A). p628-KPC belongs to the IncFIIK incompatibility group and harbors IncFIIK repA and the second IncFIB-like repA2, both of which encode replication initiation proteins. The p628-KPC backbone, 67,515 bp in length, is composed of DNA regions for plasmid replication (repA and repA2) and stability (parAB, stbAB, ssb, etc), and conjugal transfer (tra, trb, etc), which show >98% sequence identity to the corresponding regions of the IncFIIK plasmids pKPN4 (GenBank accession number CP000649), pKP048 (Jiang et al., 2010), and pKPHS2 (CP003224) (Figure 2A). The overall structure of p628-KPC is most similar to that of pKPHS2 (91% query coverage and 98% maximum nucleotide identity) (Figure 2A). pKPN4 is recovered from clinical K. pneumoniae MGH 78578 and represents the reference IncFIIK plasmid, carrying blaSHV−12 (cephalosporin resistance), and aac(6′) and aadA (aminoglycoside resistance). pKP048 and pKPHS2 are from two KPC-2-producing clinical K. pneumoniae isolates from China.
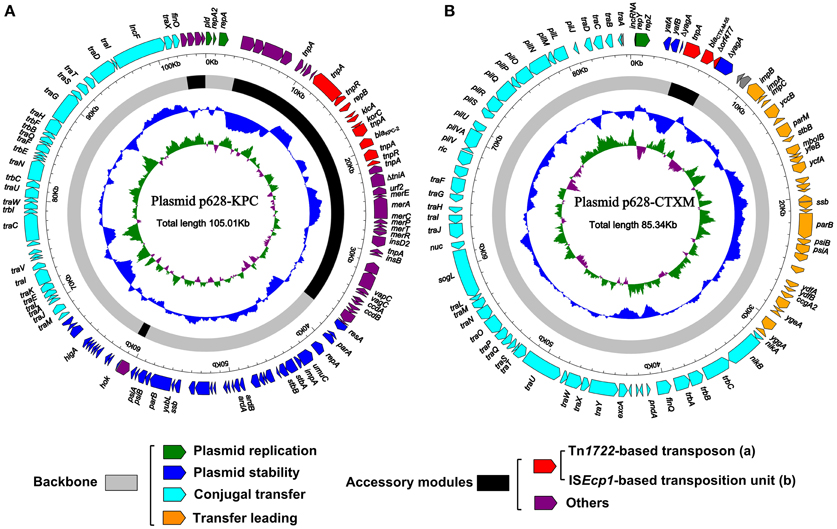
Figure 1. Schematic maps of p628-KPC (A) and p628-CTXM (B). Genes are denoted by arrows and colored based on gene function classification. The innermost circle presents GC-Skew [(G-C)/(G+C)] with a window size of 500 bp and a step size of 20 bp. The blue circle presents GC content. Shown also are backbone and accessory module regions.
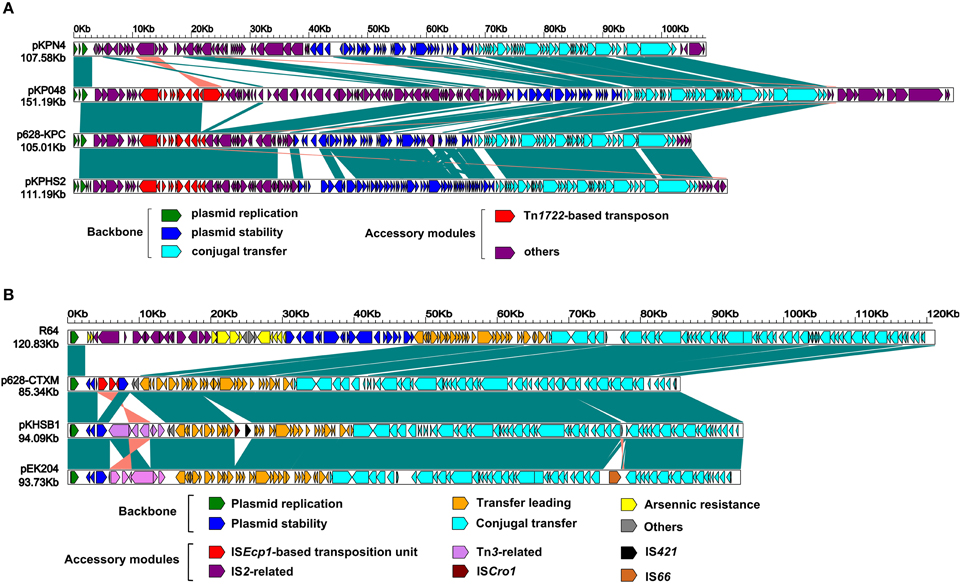
Figure 2. Linear comparison of sequenced plasmids. Genes are denoted by arrows and colored based on gene function classification. Shading regions denote regions of homology (>98% nucleotide similarity). Included are p628-KPC (A) and p628-CTXM (B) and their closely related plasmids.
As shown in Figures 1A,2A, p628-KPC contains three distinct accessory modules: a 34 kb drug-resistance region, a 1038 bp ISKpn28-based element, and a 2302 bp region of unknown function [identical sequences can be found in blaKPC−2-carrying plasmid pKPCAPSS (KP008371) and qnrS1-harboring pE66An (HF545433)]. The 34 kb region harbors two drug-resistance loci, the mer locus (mercury resistance) and the blaKPC−2 locus, and it is almost the same as the counterpart of pKPHS2 (Figure 2A). A 71 kb multi-drug-resistance region in pKP048 (Jiang et al., 2010) is composed of the 34 kb region of p628-KPC and the extra part (carrying blaDHA−1, qnrB4, and armA encoding resistance to cephalosporins, fluoroquinolones, and aminoglycosides respectively) absent from p628-KPC (Figure 2A).
Complete Nucleotide Sequence of p628-CTXM
The p628-CTXM genome consists of an 85,338 bp circular DNA molecule with an average G+C content of 49.71 and harbors a total of 92 ORFs annotated (Figure 1B). p628-KPC belongs to the IncI1 incompatibility group expressing the replication initiation protein RepZ. The p628-CTXM backbone, 82,357 bp in length, contains DNA regions for plasmid replication (repY, repZ, and inc), conjugal transfer (tra, trb, pil, etc) and transfer leading (imp, yfa to yfh, yga to ygg, etc), which show >98% sequence identity to the corresponding regions of the IncI1 plasmids R64 from Salmonella enterica serovar Typhimurium (Sampei et al., 2010), pKHSB1 from Shigella sonnei (Holt et al., 2013) and pEK204 from E. coli O25:H4-ST131 clone (Woodford et al., 2009) (Figure 2B). Another backbone component is the plasmid stability region, composed of three genes yafA, yafB, and yagA, which is highly conserved among p628-CTXM, pKHSB1, and pEK204; by contrast, the corresponding segment of R64 is a 17.8 kb region which harbors at least 18 genes and especially include those encoding site-specific recombination (resD and yefA) and partition (parAB) of replicated DNA into daughter cells during cell division (Sampei et al., 2010) (Figure 2B).
R64 carries a single accessory module, a 17 kb IS2-based transposon, which interrupts arsA1 (a member of the arsR1-arsD1-arsA1-arsB-arsC operon) (Sampei et al., 2010). p628-CTXM harbors a single accessory module, a 2980 bp ISEcp1-related element, which interrupts yagA (a member of the plasmid stability region) (Figures 1B, 2B). Two distinct Tn3-related elements, 7935 and 8014 bp in length, are inserted downstream of yagA in pKHSB1 and pEK204 respectively. There are still additional accessory modules including ISCro1 and IS421 for pKHSB1, and IS66 for pEK204.
Genetic Surroundings of blaKPC-2
As characterized in European and American countries, the blaKPC genes are located in a Tn3-family transposon named Tn4401, which is present on a wide variety of plasmids varying in size, structure and replicon (Naas et al., 2008; Kitchel et al., 2009, 2010; Chen et al., 2012; Bryant et al., 2013; Chmelnitsky et al., 2014). At least eight isoforms of Tn4401 have been named, i.e., Tn4401a to Tn4401g and a separate Tn4401d (Table S1 in Supplementary Material). Several unnamed Tn4401 isoforms have been also reported recently (Cuzon et al., 2011; Li et al., 2011; Ho et al., 2013b; Naas et al., 2013; Perez-Chaparro et al., 2014). Tn4401b is considered as the prototype one, and the other isoforms result from occurrence of distinct deletion or insertion events at different sites.
As shown in Table 2 and Figure 3, the blaKPC−2 genetic environments from China can be assigned into three main categories: Tn4401 with the ISKpn7-blaKPC−2-ISKpn6 core structure (pKPC-NY79), the Tn1722-based unit transposons with the ISKpn27-blaKPC−2-ΔISKpn6 core structure [pKP048, p628-KPC, pHS062105-3, pKPHS2, pKPC-LK30, and pHS102707; ISKpn27 is initially named in the ISfinder database (Siguier et al., 2006)], and the IS26-based composite transposons with the ISKpn27-blaKPC−2-ΔISKpn6 core structure (pKPC-LKEc, pECN580, and pKo6). The Tn4401 of pKPC-NY79 is a novel isoform of Tn4401a with tnpR truncated. The prototype Tn1722-based transposon as observed in pKP048 has a linear structure Δmcp-Tn3-ISKpn27-blaKPC−2-ΔISKpn6-korC-klcA-unkown ORF-ΔrepB- Tn1722. Various truncations within the 5′ terminal Δmcp-Tn3 region can be identified for different KPC-encoding plasmids from China; in p628-KPC, a truncation within Δmcp-Tn3 leaves only a 402 bp remnant of the Tn3 tnpR gene at the 5′ end of Tn1722-based transposon. Interestingly, an IS26-based composite transposon, which is almost identical to the counterpart in pHK23 (recovered from pig-derived E. coli in China) and harbors the fosfomycin resistance gene fosA3 (Ho et al., 2013a), is inserted into the tnpRA locus of Tn1722 in pHS102707, leaving tnpR and tnpA truncated. The IS26-based blaKPC−2-carrying transposons have a basic linear structure IS26-ΔTn3-ISKpn27-blaKPC−2-ΔISKpn6-IS26, for which presence of two IS26 elements at both ends truncates ISKpn6 and Tn3; notably, different lengths of truncated ISKpn6 can be observed for these IS26-based transposons from different plasmids.
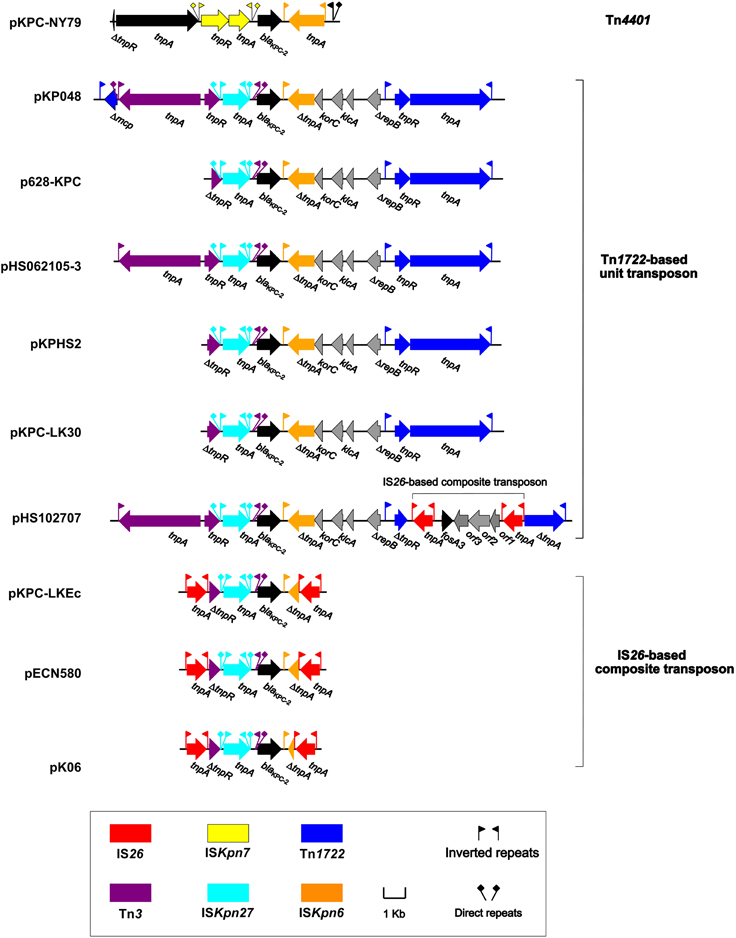
Figure 3. Linear comparison of blaKPC−2 genetic surroundings. Genes are denoted by arrows and colored based on gene function classification.
Genetic Surroundings of blaCTX−M−55
R64, p628-CTXM, pKHSB1, and pEK204 carry a 17 kb IS2-based mobile element, a 2980 bp ISEcp1-based transposition unit, a 7935 bp Tn3-based element, and an 8014 bp Tn3-based element respectively; each of them is the sole determinant for antibiotics resistance of the corresponding plasmid (Figure 4). For R64, stepwise insertions occur to eventually assemble the IS2-based element: insertion of IS2 into arsA1, that of Tn6082 into IS2, that of IS1133 into Tn6082, and finally that of Tn10 into IS1133; the tet locus carried by Tn10 and the strAB operon carried by Tn6082 account for resistance to tetracycline and streptomycin respectively.
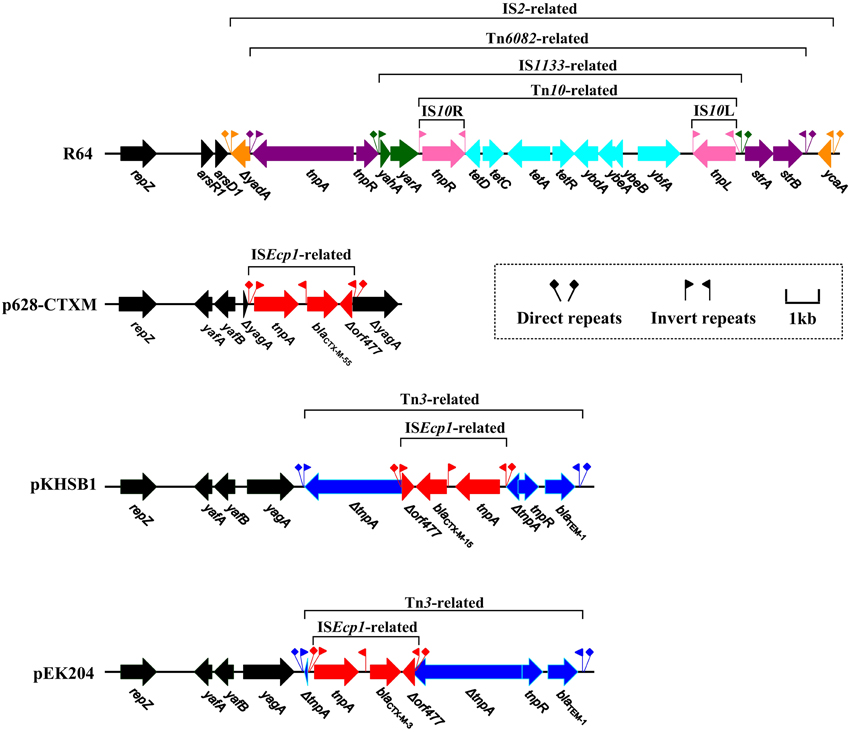
Figure 4. Linear comparison of blaCTX−M genetic surroundings. Genes are denoted by arrows and colored based on gene function classification.
A lot of blaCTX−M−1 group genes such as blaCTX−M−55, blaCTX−M−15 and blaCTX−M−3 are often connected with ISEcp1 (upstream; responsible for capture and mobilization of blaCTX−M) and Δorf477 (downstream), constituting an ISEcp1-blaCTX−M-Δorf477 transposition unit (Lartigue et al., 2006; Zong et al., 2010). In p628-KPC, the plasmid backbone gene yagA is disrupted by ISEcp1-blaCTX−M−55-Δorf477. In pKHSB1 and pEK204, a blaTEM−1-carrying Tn3 transposon is inserted at the site downstream of yagA and the Tn3 tnpA gene is further disrupted by ISEcp1-blaCTX−M−15-Δorf477 and ISEcp1-blaCTX−M−3-Δorf477, respectively. In addition, these two inserted ISEcp1-based structures differ from each other with respect to targeting sites and oriented directions (Figure 4).
Expression of blaKPC−2 and blaCTX−M−55
Each of the blaKPC−2 genes in Tn4401a, b, d, f, and g has two transcription starts, i.e., nucleotides G and C located at 39 and 289 bp upstream of blaKPC−2, which correspond to the two promoters P1 and P2 (re-designated P2ISKpn7 herein) with core −35/−10 elements TAATCC/TTACAT and TTGACA/TATCTT respectively (Naas et al., 2012). By contrast, blaKPC−2 from Tn4401c or e has only P1, while P2ISKpn7 is absent due to presence of 215 or 255 bp deletion within blaKPC−2 upstream region respectively (Naas et al., 2012).
In this work, the primer extension assay detected two transcription starts, i.e., nucleotides G and C located at 39 and 250 bp upstream of blaTn1722−basedKPC−2 from p628-KPC respectively; the corresponding two promoters were designated P1 and P2ISKpn27∕Tn3 with the core −35/−10 elements TAATCC/TTACAT and TTGACA/AATAAT respectively (Figures 5, 6). The first 74 bp fragments upstream of blaTn4401bKPC−2 and blaTn1722−basedKPC−2 are essentially identical; the P1 promoter is located within this 74 bp region and thereby shared by blaTn4401bKPC−2 and blaTn1722−basedKPC−2 (Figure 6). The next 280 bp region upstream of the above 74 bp fragment for blaTn4401bKPC−2 is dramatically divergent at nucleotide level from the counterpart for blaTn1722−basedKPC−2; these two distinct 280 bp regions contain P2ISKpn7 and P2ISKpn27∕Tn3 respectively. The −35 element of P2ISKpn7 is provided by ISKpn7 inserted at 319 bp upstream of blaTn4401bKPC−2, while the −35 and −10 elements of P2ISKpn27∕Tn3 are provided by ISKpn27 and Tn3 inserted at 281 and 75 bp upstream of blaTn1722−basedKPC−2 respectively (Figure 6).
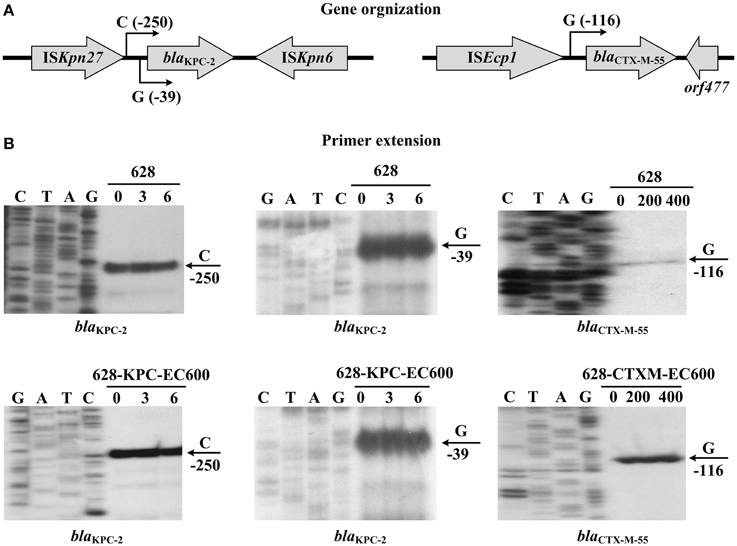
Figure 5. Organization and expression of bla genes. (A) Gene organization. Boxed arrows stand for length/direction of indicated genes. Broken-line arrows represent transcription starts for indicated bla genes. (B) Primer extension. Primer extension assay of the RNA transcript of each bla gene was performed for strain 628 cultured with addition of increasing amounts of imipenem or ampicillin. Lanes C, T, A, and G represent Sanger sequencing reactions. Lanes 0, 3, and 6 stand for 0, 3, and 6 mg/L imipenem. Lanes 0, 200, and 400 stand for 0, 200, and 400 mg/L ampicillin. Transcription start of each bla gene is indicated by arrow with nucleotide, and minus number under arrow indicates nucleotide position upstream of indicated bla gene. Representative data from at least two independent biological replicates are shown.

Figure 6. Alignment of blaKPC−2 upstream sequences. The 357 bp upstream sequences together with the start codon of the blaKPC−2 genes were aligned by CLUSTALW (http://clustalw.ddbj.nig.ac.jp/). Shown were core promoter regions, −35 and −10 elements, transcription starts, Shine-Dalgarno (SD) sequences for ribosome recognition, and translation starts. Asterisks indicate the identical nucleotides.
Spacer regions between ISEcp1 and blaCTX−M−55 from different ISEcp1-blaCTX−M−55 isoforms display three different lengths, namely 45 bp (e.g., blap1081−CTXMCTX−M−55) (Qu et al., 2014), 48 bp (e.g. blap628−CTXMCTX−M−55), and 127 bp (e.g., blaJQ343851CTX−M−55). Two promoters, TTGAAA-N18-TACAAT-N6-G (organized as -35 element/-10 element/transcription start; named P1) and TTGACT-N18-TTTCGT-N6-C (P2), are experimentally identified for blaAF550415CTX−M−3 with a 127 bp spacer and moreover, the ISEcp1-provided promoter P1 is stronger and more important than the intrinsic P2 promoter in the 127 bp spacer (Ma et al., 2011). The above result is applicable to the blaCTX−M−55 genes with the 127 bp spacer (Figure 7), because their ISEcp1+spacer region is identical to the counterpart of blaAF550415CTX−M−3.
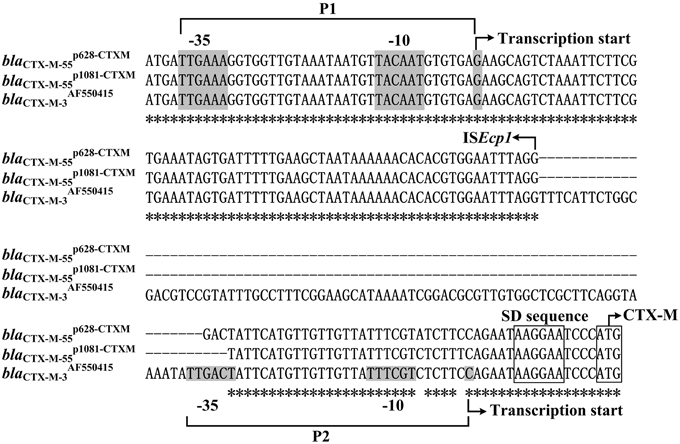
Figure 7. Alignment of blaCTX−M upstream sequences. The 153–235 bp upstream sequences together with the start codon of the blaCTX−M genes were aligned by CLUSTALW. Shown were core promoter regions, −35 and −10 elements, transcription starts, SD sequences for ribosome recognition, and translation starts. Asterisks indicate the identical nucleotides
In the present study, the primer extension assays detected a transcription start, i.e., nucleotides G located at 116 bp upstream of blaCTX−M−55 (Figure 5), which corresponded to the P1 promoter shared by blap1081−CTXMCTX−M−55 (Qu et al., 2014) and blap628−CTXMCTX−M−55 (Figure 7). Compared with the 127 bp spacer, the 45 or 48 bp spacer for blap1081−CTXMCTX−M−55 or blap628−CTXMCTX−M−55 is a truncated form due to absence of a 82 or 79 bp region respectively. The deletion event impairs the −35 element of P2, most likely making the P2 activity undetectable for blap1081−CTXMCTX−M−55 and blap628−CTXMCTX−M−55 (Figure 7).
In addition, the primer extension assay showed that addition of increasing amounts of imipenem or ampicillin during cultivation of indicated strains 628, 628-KPC-EC600 and 628-CTXM-EC600 had no effect on activity of all the above promoters detected for blaKPC−2 or blaCTX−M−55, denoting constitutive expression of the above two resistance genes (Figure 5).
Concluding Remarks
KPC-2 and CTX-M-55 enzymes are produced by two different conjugative plasmids, p628-KPC and p628-CTXM respectively, in K. pneumoniae strain 628, and the sequences of these two plasmids are >98% identical to other relevant plasmids carrying the same resistance determinants previously sequenced. The detected blaKPC−2 gene is captured by a Tn1722-based unit transposon carried by an IncFIIK-type multi-drug-resistant plasmid p628-KPC, and this gene has two different promoters, the intrinsic P1 and the ISKpn27/Tn3-provided P2, both characteristic of constitutive expression. The detected blaCTX−M−55 gene, being the sole drug-resistant determinant in the plasmid, is mobilized in an ISEcp1-based transposition unit carried by an IncI1 plasmid p628-CTXM, and this gene has a single ISEcp1-provided promoter driving blaCTX−M−55 expression in a constitutive manner. Coexistence of blaKPC and blaCTX−M in K. pneumoniae has been reported many times, but this is the first report to gain deep insights into genetic platforms, promoters, and expression of the two coexisted bla genes. The IncFIIK and IncI1 plasmids have been frequently identified to carry horizontally acquired drug-resistant gene modules and could be transmitted across a number of bacterial species (Woodford et al., 2009; Jiang et al., 2010; Sampei et al., 2010; Holt et al., 2013), and increased surveillance of these drug-resistant plasmids is needed.
Funding
This work is funded by National High-Tech Research and Development Program (2014AA021402), National Key Program for Infectious Disease of China (2013ZX10004216), and National Natural Science Foundation of China (31471184 and 31170127).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00838
References
Bryant, K. A., Van Schooneveld, T. C., Thapa, I., Bastola, D., Williams, L. O., Safranek, T. J., et al. (2013). KPC-4 Is encoded within a truncated Tn4401 in an IncL/M plasmid, pNE1280, isolated from Enterobacter cloacae and Serratia marcescens. Antimicrob. Agents Chemother. 57, 37–41. doi: 10.1128/AAC.01062-12
Cai, J. C., Zhou, H. W., Zhang, R., and Chen, G. X. (2008). Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli isolates possessing the plasmid-mediated carbapenem-hydrolyzing beta-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob. Agents Chemother. 52, 2014–2018. doi: 10.1128/AAC.01539-07
Chen, L., Chavda, K. D., Mediavilla, J. R., Jacobs, M. R., Levi, M. H., Bonomo, R. A., et al. (2012). Partial excision of blaKPC from Tn4401 in Carbapenem-resistant Klebsiella pneumoniae. Antimicrob. Agents Chemother. 56, 1635–1638. doi: 10.1128/AAC.06182-11
Chen, L., Hu, H., Chavda, K. D., Zhao, S., Liu, R., Liang, H., et al. (2014a). Complete sequence of a KPC-producing IncN multidrug-resistant plasmid from an epidemic Escherichia coli sequence type 131 strain in China. Antimicrob. Agents Chemother. 58, 2422–2425. doi: 10.1128/AAC.02587-13
Chen, Y. T., Lin, J. C., Fung, C. P., Lu, P. L., Chuang, Y. C., Wu, T. L., et al. (2014b). KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J. Antimicrob. Chemother. 69, 628–631. doi: 10.1093/jac/dkt409
Chen, Z., Li, H., Feng, J., Li, Y., Chen, X., Guo, X., et al. (2015). NDM-1 encoded by a pNDM-BJ01-like plasmid p3SP-NDM in clinical Enterobacter aerogenes. Front. Microbiol. 6:294. doi: 10.3389/fmicb.2015.00294
Chmelnitsky, I., Shklyar, M., Leavitt, A., Sadovsky, E., Navon-Venezia, S., Ben Dalak, M., et al. (2014). Mix and match of KPC-2 encoding plasmids in Enterobacteriaceae-comparative genomics. Diagn. Microbiol. Infect. Dis. 79, 255–260. doi: 10.1016/j.diagmicrobio.2014.03.008
CLSI (2012). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Second Informational Supplement M10-S22. Wayne, PA: CLSI.
Cuzon, G., Naas, T., Villegas, M. V., Correa, A., Quinn, J. P., and Nordmann, P. (2011). Wide dissemination of Pseudomonas aeruginosa producing beta-lactamase blaKPC−2 gene in Colombia. Antimicrob. Agents Chemother. 55, 5350–5353. doi: 10.1128/AAC.00297-11
Frank, J. A., Reich, C. I., Sharma, S., Weisbaum, J. S., Wilson, B. A., and Olsen, G. J. (2008). Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74, 2461–2470. doi: 10.1128/AEM.02272-07
Ho, P. L., Chan, J., Lo, W. U., Law, P. Y., and Chow, K. H. (2013a). Plasmid-mediated fosfomycin resistance in Escherichia coli isolated from pig. Vet. Microbiol. 162, 964–967. doi: 10.1016/j.vetmic.2012.09.023
Ho, P. L., Cheung, Y. Y., Lo, W. U., Li, Z., Chow, K. H., Lin, C. H., et al. (2013b). Molecular characterization of an atypical IncX3 Plasmid pKPC-NY79 carrying bla KPC-2 in a Klebsiella pneumoniae. Curr. Microbiol. 67, 493–498. doi: 10.1007/s00284-013-0398-2
Holt, K. E., Thieu Nga, T. V., Thanh, D. P., Vinh, H., Kim, D. W., Vu Tra, M. P., et al. (2013). Tracking the establishment of local endemic populations of an emergent enteric pathogen. Proc. Natl. Acad. Sci. U.S.A. 110, 17522–17527. doi: 10.1073/pnas.1308632110
Jiang, Y., Yu, D., Wei, Z., Shen, P., Zhou, Z., and Yu, Y. (2010). Complete nucleotide sequence of Klebsiella pneumoniae multidrug resistance plasmid pKP048, carrying blaKPC-2, blaDHA-1, qnrB4, and armA. Antimicrob. Agents Chemother. 54, 3967–3969. doi: 10.1128/AAC.00137-10
Kitchel, B., Rasheed, J. K., Endimiani, A., Hujer, A. M., Anderson, K. F., Bonomo, R. A., et al. (2010). Genetic factors associated with elevated carbapenem resistance in KPC-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 54, 4201–4207. doi: 10.1128/AAC.00008-10
Kitchel, B., Rasheed, J. K., Patel, J. B., Srinivasan, A., Navon-Venezia, S., Carmeli, Y., et al. (2009). Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 53, 3365–3370. doi: 10.1128/AAC.00126-09
Lartigue, M. F., Poirel, L., Aubert, D., and Nordmann, P. (2006). In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring beta-lactamase gene blaCTX-M of Kluyvera ascorbata. Antimicrob. Agents Chemother. 50, 1282–1286. doi: 10.1128/AAC.50.4.1282-1286.2006
Leavitt, A., Carmeli, Y., Chmelnitsky, I., Goren, M. G., Ofek, I., and Navon-Venezia, S. (2010). Molecular epidemiology, sequence types, and plasmid analyses of KPC-producing Klebsiella pneumoniae strains in Israel. Antimicrob. Agents Chemother. 54, 3002–3006. doi: 10.1128/AAC.01818-09
Leavitt, A., Navon-Venezia, S., Chmelnitsky, I., Schwaber, M. J., and Carmeli, Y. (2007). Emergence of KPC-2 and KPC-3 in carbapenem-resistant Klebsiella pneumoniae strains in an Israeli hospital. Antimicrob. Agents Chemother. 51, 3026–3029. doi: 10.1128/AAC.00299-07
Li, G., Wei, Q., Wang, Y., Du, X., Zhao, Y., and Jiang, X. (2011). Novel genetic environment of the plasmid-mediated KPC-3 gene detected in Escherichia coli and Citrobacter freundii isolates from China. Eur. J. Clin. Microbiol. Infect. Dis. 30, 575–580. doi: 10.1007/s10096-010-1124-7
Li, G., Zhang, Y., Bi, D., Shen, P., Ai, F., Liu, H., et al. (2015). First report of a clinical, multidrug-resistant Enterobacteriaceae isolate coharboring fosfomycin resistance gene fosA3 and carbapenemase gene blaKPC−2 on the same transposon, Tn1721. Antimicrob. Agents Chemother. 59, 338–343. doi: 10.1128/AAC.03061-14
Ma, L., Siu, L. K., and Lu, P. L. (2011). Effect of spacer sequences between bla(CTX-M) and ISEcp1 on bla(CTX-M) expression. J. Med. Microbiol. 60, 1787–1792. doi: 10.1099/jmm.0.033910-0
Munoz-Price, L. S., Poirel, L., Bonomo, R. A., Schwaber, M. J., Daikos, G. L., Cormican, M., et al. (2013). Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13, 785–796. doi: 10.1016/S1473-3099(13)70190-7
Naas, T., Bonnin, R. A., Cuzon, G., Villegas, M. V., and Nordmann, P. (2013). Complete sequence of two KPC-harbouring plasmids from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 68, 1757–1762. doi: 10.1093/jac/dkt094
Naas, T., Cuzon, G., Truong, H. V., and Nordmann, P. (2012). Role of ISKpn7 and deletions in blaKPC gene expression. Antimicrob. Agents Chemother. 56, 4753–4759. doi: 10.1128/AAC.00334-12
Naas, T., Cuzon, G., Villegas, M. V., Lartigue, M. F., Quinn, J. P., and Nordmann, P. (2008). Genetic structures at the origin of acquisition of the beta-lactamase bla KPC gene. Antimicrob. Agents Chemother. 52, 1257–1263. doi: 10.1128/AAC.01451-07
Peirano, G., Seki, L. M., Val Passos, V. L., Pinto, M. C., Guerra, L. R., and Asensi, M. D. (2009). Carbapenem-hydrolysing beta-lactamase KPC-2 in Klebsiella pneumoniae isolated in Rio de Janeiro, Brazil. J. Antimicrob. Chemother. 63, 265–268. doi: 10.1093/jac/dkn484
Pérez-Chaparro, P. J., Cerdeira, L. T., Queiroz, M. G., De Lima, C. P., Levy, C. E., Pavez, M., et al. (2014). Complete nucleotide sequences of two blaKPC-2-bearing IncN Plasmids isolated from sequence type 442 Klebsiella pneumoniae clinical strains four years apart. Antimicrob. Agents Chemother. 58, 2958–2960. doi: 10.1128/AAC.02341-13
Qu, F., Ying, Z., Zhang, C., Chen, Z., Chen, S., Cui, E., et al. (2014). Plasmid-encoding extended-spectrum beta-lactamase CTX-M-55 in a clinical Shigella sonnei strain, China. Future Microbiol. 9, 1143–1150. doi: 10.2217/fmb.14.53
Roberts, A. P., Chandler, M., Courvalin, P., Guédon, G., Mullany, P., Pembroke, T., et al. (2008). Revised nomenclature for transposable genetic elements. Plasmid 60, 167–173. doi: 10.1016/j.plasmid.2008.08.001
Sampei, G., Furuya, N., Tachibana, K., Saitou, Y., Suzuki, T., Mizobuchi, K., et al. (2010). Complete genome sequence of the incompatibility group I1 plasmid R64. Plasmid 64, 92–103. doi: 10.1016/j.plasmid.2010.05.005
Shen, P., Wei, Z., Jiang, Y., Du, X., Ji, S., Yu, Y., et al. (2009). Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob. Agents Chemother. 53, 4333–4338. doi: 10.1128/AAC.00260-09
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Souli, M., Galani, I., Antoniadou, A., Papadomichelakis, E., Poulakou, G., Panagea, T., et al. (2010). An outbreak of infection due to beta-Lactamase Klebsiella pneumoniae Carbapenemase 2-producing K. pneumoniae in a Greek University Hospital: molecular characterization, epidemiology, and outcomes. Clin. Infect. Dis. 50, 364–373. doi: 10.1086/649865
Tzouvelekis, L. S., Markogiannakis, A., Psichogiou, M., Tassios, P. T., and Daikos, G. L. (2012). Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25, 682–707. doi: 10.1128/CMR.05035-11
Woodford, N., Carattoli, A., Karisik, E., Underwood, A., Ellington, M. J., and Livermore, D. M. (2009). Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob. Agents Chemother. 53, 4472–4482. doi: 10.1128/AAC.00688-09
Zhang, Y., Gao, H., Wang, L., Xiao, X., Tan, Y., Guo, Z., et al. (2011). Molecular characterization of transcriptional regulation of rovA by PhoP and RovA in Yersinia pestis. PLoS ONE 6:e25484. doi: 10.1371/journal.pone.0025484
Keywords: Klebsiella pneumoniae, KPC-2, CTX-M-55, p628-KPC, p628-CTXM, promoter
Citation: Wang L, Fang H, Feng J, Yin Z, Xie X, Zhu X, Wang J, Chen W, Yang R, Du H and Zhou D (2015) Complete sequences of KPC-2-encoding plasmid p628-KPC and CTX-M-55-encoding p628-CTXM coexisted in Klebsiella pneumoniae. Front. Microbiol. 6:838. doi: 10.3389/fmicb.2015.00838
Received: 16 May 2015; Accepted: 31 July 2015;
Published: 19 August 2015.
Edited by:
Patrick Rik Butaye, Ghent University, BelgiumReviewed by:
Yoshikazu Ishii, Toho University School of Medicine, JapanAurora García-Fernández, Istituto Superiore di Sanità, Italy
Copyright © 2015 Wang, Fang, Feng, Yin, Xie, Zhu, Wang, Chen, Yang, Du and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Du, Department of Clinical Laboratory, The Second Affiliated Hospital of Soochow University, No. 181, Sanxiang Road, Suzhou, Jiangsu 215004, China,aG9uZ19kdUAxMjYuY29t;
Dongsheng Zhou, State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, No. 20, Dongda Street, Fengtai District, Beijing 100071, China,ZG9uZ3NoZW5nemhvdTE5NzdAZ21haWwuY29t
 Li Wang
Li Wang Haihong Fang
Haihong Fang Jiao Feng2
Jiao Feng2 Zhe Yin
Zhe Yin Jie Wang
Jie Wang Dongsheng Zhou
Dongsheng Zhou