- 1State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing, China
- 2Animal Husbandry Base Teaching and Research Section, College of Animal Science and Technology, Hebei North University, Zhangjiakou, China
Yersinia pestis, which is the causative agent of plague, has acquired exceptional pathogenicity potential during its evolution from Y. pseudotuberculosis. Two laterally acquired plasmids, namely, pMT1 and pPCP1, are specific to Y. pestis and are critical for pathogenesis and flea transmission. Small regulatory RNAs (sRNAs) commonly function as regulators of gene expression in bacteria. MicF, is a paradigmatic sRNA that acts as a post-transcriptional repressor through imperfect base pairing with the 5′-UTR of its target mRNA, ompF, in Escherichia coli. The high sequence conservation and minor variation in the RNA duplex of MicF-ompF has been reported in Yersinia. In this study, we utilized super-folder GFP reporter gene fusion to validate the post-transcriptional MicF-mediated regulation of target mRNA ompF in Y. pestis. Unexpectedly, upon MicF overexpression, the slightly upregulated expression of OmpF were found in the wild-type strain, which contradicted the previously established model. Interestingly, the translational repression of ompF target fusions was restored in the intrinsic plasmids-cured Y. pestis strain, suggesting intrinsic plasmids influence the MicF-mediated translational repression of ompF in Y. pestis. Further examination showed that plasmid pPCP1 is likely the main contributor to the abolishment of MicF-mediated translational repression of endogenous or plasmid-borne ompF. It represents that the possible roles of intrinsic plasmids should be considered upon investigating sRNA-mediated gene regulation, at least in Y. pestis, even if the exact mechanism is not fully understood.
Introduction
The human pathogenic species of Yersiniae include Yersinia pestis, Y. pseudotuberculosis, and Y. enterocolitica (Perry and Fetherston, 1997). Y. pestis evolved from its ancestor Y. pseudotuberculosis several thousands of years ago (Achtman et al., 1999). Interestingly, the two species induce remarkably different diseases by distinct transmission routes. Y. pestis causes pandemics of bubonic or pneumonic plague, which is a fatal disease to rodents and humans, and is transmitted by flea biting, whereas Y. pseudotuberculosis only causes a chronic and relatively mild disease as an enteric pathogen (Zhou et al., 2006). Although different plasmid combinations exist among Y. pestis strains, typical strains of Y. pestis harbor three virulence plasmids (pCD1, pMT1, and pPCP1), which encode various virulence determinants. The plasmid pCD1 is commonly shared in all three pathogenic species of Yersiniae. The other two plasmids (pPCP1 and pMT1) are laterally acquired by Y. pestis (Hu et al., 1998). These three plasmids in Y. pestis possess different replication systems. The replicon of plasmid pCD1 belongs to IncFIIA replication system (Perry et al., 1998). Plasmid pPCP1 has a ColE1-like replicon (Hu et al., 1998). The replication region of the pMT1 consists of a structural gene (repA) and accessorial elements of replication (Lindler et al., 1998). Although the plasmid-encoded virulence determinants are well documented, the way by which the laterally acquired plasmids have complicated the regulatory networks of Y. pestis during its evolution remains ambiguous.
Small RNAs (sRNAs) are involved in regulatory networks as regulators of gene expression to facilitate the quick adjustment of bacterial cells to environmental stresses (Waters and Storz, 2009). Trans-encoded sRNAs, which represent a major class of sRNAs in bacteria generally activate or repress mRNA translation by limited base-pairing with mRNA (Desnoyers et al., 2013). The RNA-binding protein Hfq is usually required to help the sRNA-mRNA interaction and RNA stability (Vogel and Luisi, 2011). Hfq has been implicated in stress adaptation and virulence in many bacterial pathogens such as Y. pestis (Sittka et al., 2007; Kulesus et al., 2008; Geng et al., 2009; Chiang et al., 2011). MicF is defined as a canonical trans-encoded sRNA that regulates outer membrane protein F (OmpF) synthesis in Escherichia coli and other related bacteria (Andersen et al., 1989). Approximately 20 nt of MicF forms a perfect RNA duplex by directly pairing with the Shine-Dalgarno region of the ompF mRNA. This process occludes the initiation of the 30S ribosome; thus translation is inhibited and the cleavage of mRNA is possibly induced (Andersen et al., 1989). MicF is a highly conserved sRNA in closely related Enterobacteriaceae genomes (Delihas, 1997). A strong phylogenetic relationship is also found in MicF/ompF interacting sites and RNA duplex in Yersiniae (Delihas, 2003). Thus, the regulatory outcome is expected to fit the generalized model, in which MicF forms a duplex with the 5′ UTR of the ompF mRNA thereby inhibiting the translation and promoting degradation of the ompF transcript. In this study we utilized super-folder GFP reporter gene fusion to validate the MicF-mediated regulation of the ompF 5′ UTR in Y. pestis. Unexpectedly, the OmpF translation was slightly induced by overexpressed MicF instead of being inhibited in Y. pestis Microtus strain 201. This phenomenon is paradoxical to the previous prediction. Interestingly, the translational repression was restored in the intrinsic plasmids-cured strain. Further investigations were conducted to determine which plasmid(s) are responsible for the abrogation of MicF-mediated ompF regulation.
Materials and Methods
Bacterial Strains and Growth
The bacterial strains in this study are listed in Table 1. Y. pestis strain 201 (F1+, VW+, Pst+, and Pgm+) isolated from Microtus brandti in Inner Mongolia, China, belongs to biovar Microtus. This strain is avirulent to humans but highly lethal to mice (Zhou et al., 2004). Strain 201 has gene content that is almost identical to that of Y. pestis strain 91001, which possesses four plasmids, namely, pCD1, pMT1, pPCP1, and pCRY1 (Song et al., 2004). All the combinations of plasmid(s)-cured strains derived from strain 201 were constructed by Bin et al. based on the plasmid incompatibility in our laboratory (Ni et al., 2008). E. coli and Y. pestis were grown to exponential phase on LB (Luria–Bertani) agar with 0.1% arabinose at 37°C or 26°C. Approximate concentrations of antibiotics (100 μg/mL ampicillin and 34 μg/mL chloramphenicol) were added to LB agar throughout this study.
Construction of gfp Reporter Fusions Plasmid
Plasmid pXG10-SF, which is an improved gfp-based translational fusion vector with the constitutive PLtetO promoter (Corcoran et al., 2012), was used to construct the GFP reporter fusion to ompF. An amplicon containing the 5′ UTR of the AUG start codon (92 nt) and 16 amino acids of OmpF was obtained by using the primer pair ompF-F/R. The resulting fragment was fused to sfGFP by inserting it into the NsiI/NheI-digested pXG10-SF, which yielded the plasmid pXG-ompF::gfp.
Construction of sRNA Overexpression Plasmid and pXG-1
QuikChange® Lightning Site-Directed Mutagenesis Kit (Stratagene) was used to construct an inducible transcriptional fusion vector by modifying an expression vector, pBAD/HisA. According to the manufacturer's instructions, the pBAD/HisA plasmid was amplified with pBAD-F/R primer pair by PCR followed by DpnI treatment. The resulting plasmid pBAD-TF removed the 327-nt fragment containing several elements (RBS, AUG, and polyclonal sites) and introduced EcoRI and HindIII restriction sites upstream of the rrnB terminator sequence. The modified vector pBAD-TF was used as pBAD control. To construct the MicF overexpression vector, a DNA fragment spanning full-length (85 nt) and 58 nt downstream of MicF was ligated to modified pBAD vector and pBAD-MicF was obtained.
The similar protocol was also used to construct a control plasmid designated as pXG-1 with sfGFP expression controlled by PtetO by modifying pXG10-SF. The pXG10-SF plasmid was amplified with primer pair pXG-1-F/R. The resulting plasmid removed the 725-nt lacZ-containing fragment between AatII and BheI and introduced RBS and AUG sequence (GAGGGGAAAUCUGAUG) upstream of lacZ from the transcriptional fusion vector pRW50 into the same site.
Imaging and Quantitative Measurements of GFP Fluorescence
Images of bacteria expressing plasmid-borne gfp fusions and grown overnight on LB plates were taken using a CCD camera in Gel Doc XR+ image analyzer (Bio-Rad) under the SYBR Green mode. The aliquots of each strain were scraped from the plates and resuspended into PBS buffer. Cell densities were adjusted to OD620nm ≈ 1.5 and 200 μL of bacterial suspensions were placed into 96-well microtiter plates. Two independent cultures as biological replicates and three aliquots as technical replicates were used throughout the study for each strain. Green fluorescence images were captured by SpectraMax M2 MicroplateReaders (Molecular Devices) with an excitation/emission wavelength of 485/525 nm. Fold changes in the MicF-mediated OmpF expression were calculated by dividing the specific fluorescence of strains with MicF overexpression by that of strains with negative control plasmid. The data were analyzed using One-Way analysis of variance (ANOVA) and Sidak's multiple comparisons test, where P-values of < 0.01 were considered significant.
OmpF Detection by Using Western Blot
Y. pestis were grown in LB agar plate at 26°C for 36 h. Equal amounts of bacterial aliquots were collected and lysed by ultrasonication. Total cellular proteins were separated on SDS-PAGE and immunoblotted with anti-OmpF multiclonal antibody and DyLight 680–labeled goat anti-rabbits antibody followed by detection using the Odyssey Infrared Imaging System. The abundance values were calculated as the expression level of the derivatives divided by that of strain 201 using the Quantity One software. The GroEL protein was detected in parallel as control.
RNA Detection by Using Northern Blot
Total RNA was then extracted from various bacterial strains grown in LB agar plate in the presence of 0.1% arabinose at 26°C for 36 h using the TRIzol Reagent (Invitrogen). Total RNA samples (1 μg) were denatured at 70°C for 5 min, separated on 6% polyacrylamide 7 M urea gel, and transferred onto Hybond N+ membranes (GE) via electroblot. The membranes were UV-crosslinked and pre-hybridized for 1 h. Northern hybridization was performed by adding the DIG-labeled MicF-specific RNA probe synthesized by in vitro transcription using T7 RNA polymerase. The RNAs were immunologically detected according to the instructions on the DIG Northern Starter Kit (Roche). Band intensities on the Northern blots were quantified by Quantity One software. The 5s rRNA species was monitored in parallel as control.
Results and Discussion
In this study, the MicF-mediated ompF regulation was validated using a two-compatible-plasmid reporter system established by (Corcoran et al., 2012). The sRNA plasmid is a high-copy vector (pBAD), which ensures the high level of expressed MicF and minimizes the inference of chromosome-encoded MicF. The target plasmid is a low-copy vector (pXG10), where transcription of the ompF::gfp gene is driven by the constitutive promoter PLtetO, thereby uncouples the translation from ompF transcription. In this system MicF is overexpressed and ompF-gfp fusion represents the OmpF abundance. We found that the intrinsic plasmid(s) have an effect on sRNA-mediated regulation, at least on the MicF-mediated ompF translation.
Intrinsic Plasmids Accounted for the Abolishment of the MicF-mediated Translational Repression of ompF in Y. pestis
Y. pestis-specific ompF-gfp fusion and sRNA MicF were cloned into low- and high-copy vectors, respectively. Both plasmids were transformed into E. coli and Y. pestis. All the plasmids were checked throughout all the tested strains by PCR. The results showed that all the plasmids (pMT1, pCD1, pPCP1, and MicF-expressing plasmid pBAD-MicF) were present as expected (Figure S1). The reporter GFP fluorescence activity was monitored to evaluate the regulatory roles in various bacterial strains.
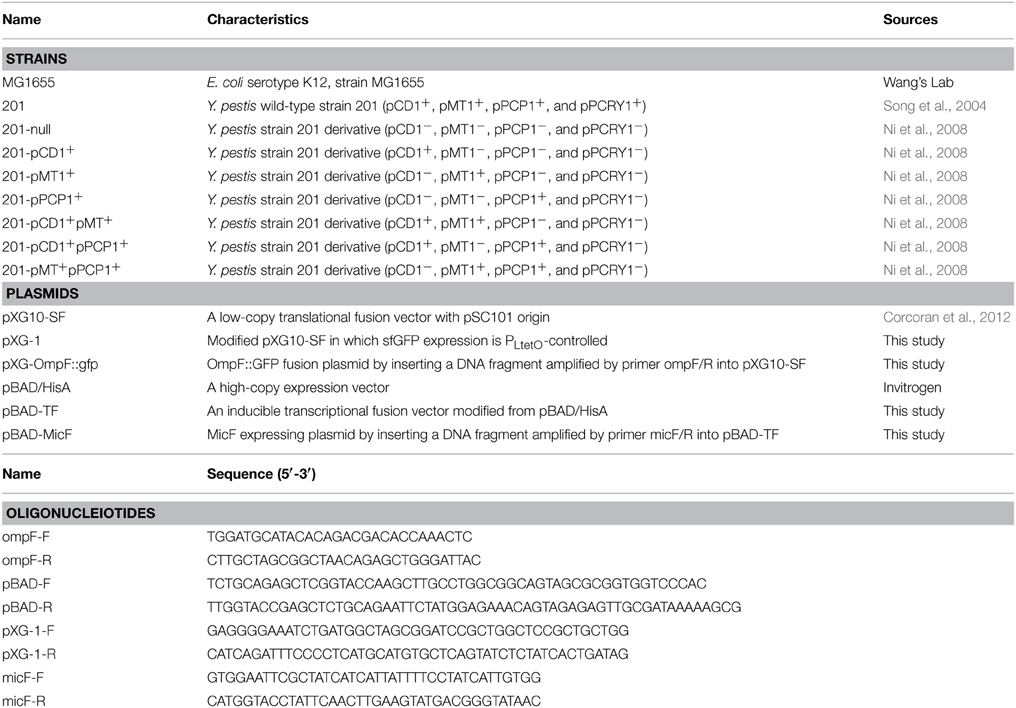
The inhibitory translation of ompF-gfp fusion upon MicF overexpression (approximately five-fold repression) was observed in E. coli strain MG1655. Only the 1.8-fold repression was observed in the Y. pestis strain 201 that is cured of all the endogenous plasmids (Figure 1). This finding is consistent with the results previously reported on E. coli (Mizuno et al., 1984) and also confirms that MicF-mediated ompF repression occurs in the 5′-UTR. Interestingly, the inhibitory effect was not found in the Y. pestis WT strain 201. Instead, more than three-fold upregulation of OmpF-GFP expression was observed under the same conditions (Figure 1). The relative quantification of fluorescence value was also measured in Y. pestis strains with different combinatorial plasmids grown to exponential phase in LB medium followed by arabinose induction for 1 h. Similar tendency was found in Y. pestis strains grown in liquid medium as that found in solid medium (Figures 1, 2 and Figure S2). To exclude the possibility that the effect was caused by the translational reporter system, we construct plasmid pXG-1 which sfGFP expression is constitutively PLtetO-controlled. No changes in fluorescence intensity were found between pBAD-MicF and pBAD-vector groups in Y. pestis strain 201 and 201-null carrying pXG-1 (Figure S3). The different regulatory consequences in Y. pestis strains carrying or cured of plasmids indicated that the virulence-associated plasmids might have been recruited to sRNA-mediated regulatory networks in the chromosome during evolution.
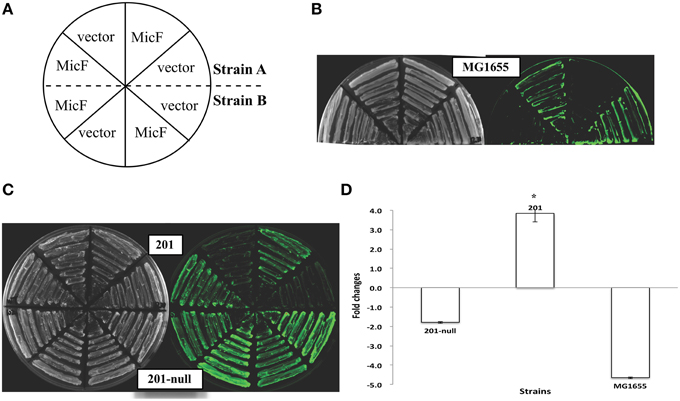
Figure 1. Expression of the ompF::gfp fusions upon MicF overexpression in Escherichia coli and Yersinia pestis strains. (A) Layout map of bacterial strains carrying the control plasmid without sRNA overexpression (denoted as “vector”) or MicF-overexpression plasmid (denoted as “MicF”). Duplicate images of representative strains are shown in parallel on plates. (B) Representative fluorescence images of E. coli strain MG1655. The image obtained under visible light mode is shown at the left panel and that of the same plate under the fluorescence mode at the right panel. (C) Representative fluorescence images of Y. pestis WT strain (201) and its plasmid-cured derivative strain (201-null). (D) Quantitative measurements of fluorescence produced by the tested strains. Fold changes are provided as the ratio of fluorescence values detected in MicF-overexpressed bacterial strains divided by those detected in strains carrying pBAD blank vectors. Values presented are means ± standard deviations of two independent experiments. The asterisks indicate statistically significant differences compared to the values detected in strain MG1655.
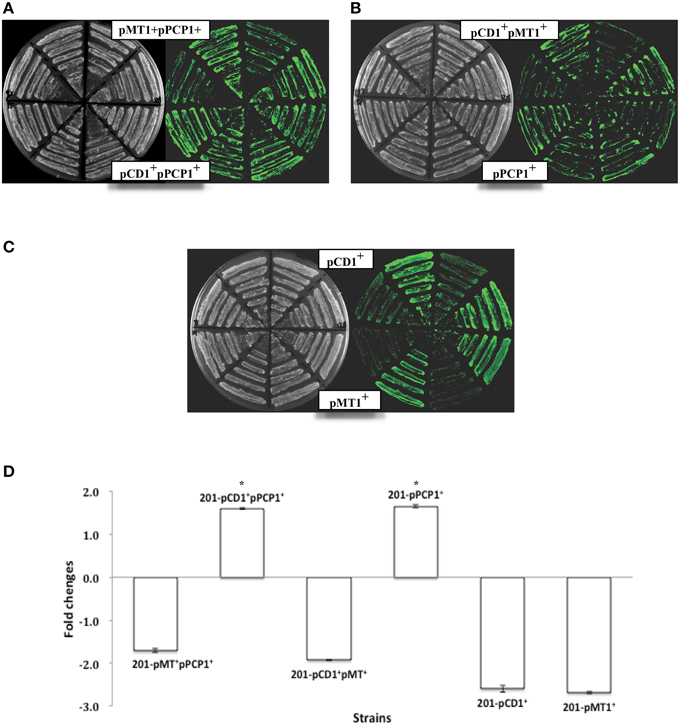
Figure 2. Expression of the ompF::gfp fusions upon MicF overexpression in the Yersinia pestis strains with different plasmid combinations. Representative fluorescence images of Y. pestis derivative strains are shown (A–C). The corresponding quantitative results are also shown (D), in which the asterisks indicate statistically significant differences compared to the values detected in strain MG1655 shown in Figure 1.
Plasmid pPCP1 Alone Abolished the MicF-mediated Translational Repression of OmpF
Y. pestis strains carrying different combinations of the three plasmids (pMT1, pPCP1, and pCD1) were used to determine the probable plasmid(s) responsible for the abolished MicF-mediated regulation of ompF. Upon MicF overexpression, ompF translation remained repressed in the pCD1- or pMT1-containing strains, as observed in the plasmid-null strain. However, the expression level of OmpF-GFP fusion protein was even slightly upregulated about 1.6-fold in the Y. pestis strain carrying plasmid pPCP1. Such activation was also observed in the strain carrying both plasmids pPCP1 and pCD1 (Figure 2). Plasmid pPCP1 alone could overwhelm the translational repression in the plasmid-cured strain, thereby suggesting that pPCP1 may mainly contribute to the abolishment of sRNA-mediated regulation. Paradoxically, translational repression was still found in the strain with pPCP1 and pMT1. We speculated that addition of pMT1 might interfere with the effect of pPCP1 on sRNA-mediated regulation because of the potential interactions between these two plasmids.
We also monitored MicF in the different bacterial strains under the same conditions as those shown in Figures 1, 2. No MicF were found expressed in the pBAD control of various Y. pestis strains by using Northern Blot, which might be mainly due to huge disparity in copy number of MicF between plasmid pBAD and chromosome and/or low expression levels of endogeneous MicF under our experimental conditions. Additionally, no significant differences in MicF overexpression were found among Y. pestis strains (Figure 3).
Figure 3. Detection of MicF expression in various strains of Y. pestis. MicF expression detected by Northern Blot, in which 5S rRNA images from each tested strain were provided as control.
Validation of MicF-mediated Regulation of Endogenous OmpF Elicited by Intrinsic Plasmids
To test whether the intrinsic plasmids interfere with the MicF-mediated regulation of the chromosome-encoded target gene ompF, the abundances of endogenous ompF transcript and OmpF protein were validated in various strains of Y. pestis by Northern Blot and Western Blot, respectively (Figure 4). In agreement with the results of gene fusion reporter systems, the abundance of ompF transcript or OmpF protein were found decreased 2.0–5.0 fold in four pPCP1-cured strains (122-null, 122-pCD1+pMT1+, 201-pCD1+, and 201-pMT1+) upon MicF overexpression relative to that in the control strains. Strikingly, the ompF transcript was stable, but approximate three-fold decrease was found in the expression level of OmpF in the strain 122-pMT1+pPCP1+, which is also consistent with the findings presented in Figure 2. However, only the comparable levels of ompF and OmpF were found among three groups of pPCP-containing strains (122, 122-pPCP1, and 122-pCD1+pPCP1+). The downregulation phenomenon is roughly consistent with that of plasmid-borne gfp fusion. The discrepancy is likely due to the different sensitivity between translational fusion assay and Western blot. However, no obvious upregulation was found in strain 201. Maybe the actual activation was magnified by translational fusion assay or veiled by detection threshold of Western blot or Northern Blot. Taken together, this observation further confirmed the conclusion that intrinsic plasmids have the potential impacts on abolishment of MicF-mediated ompF regulation in Y. pestis.
Figure 4. Abundance detection of endogenous ompF transcript and OmpF protein in various strains of Y. pestis. Northern Blot was used to detect the chromosome-encoded ompF transcript in various strains of Y. pestis grown under the same conditions as those shown in Figure 3. Meanwhile, the anti-OmpF rabbit multiclonal antibody was used in Western Blot to detect the endogenous OmpF protein in the indicated strains, in which GroEL protein images from each strain were provided as control. The numbers indicated below each panel represent the fold changes of mRNA or protein abundance detected in strains carrying pBAD-MicF divided by that of the corresponding strains carrying pBAD control vector.
Our study demonstrated that the mobile elements affect sRNA-mediated regulation in Y. pestis. Distinct conclusions may be drawn if various strains carrying different plasmids are used to investigate the sRNA-mediated regulation. For example, Hfq reportedly represses the biofilm formation in Y. pestis KIM6+ (an avirulent derivative of the fully virulent strain KIM, which was cured of the pCD1 plasmid) grown in BHI medium (Bellows et al., 2012). Opposite effects on biofilm formation were observed in the hfq mutant of Y. pestis wild-type strain 201 and its derivative strain lacked the plasmid pCD1 (unpublished data). Therefore, the background bacteria to be used as control strain should be carefully selected. The possible roles of plasmids in gene regulation should be considered even if the exact mechanism is not fully understood. Although a relationship exists between the intrinsic plasmids and sRNA-mediated regulation, the presence or absence of any plasmid did not cause the clear-cut effects on MicF-mediated ompF regulation. This phenomenon may indicate that mutual interactions exist among intrinsic plasmids in Y. pestis, and such interactions further influence sRNA-mediated regulation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We greatly thank Jörg Vogel's lab for kindly providing the pXG superfolder GFP series of plasmids and Prof. Hengliang Wang from Beijing Institute of Biotechnology for offering E. coli K12 strain MG1655. This study was funded by the National Basic Research Program of China (2014CB744405), the National Natural Science Foundation of China (31171248 and 31430006), and the State Key of Pathogen and Biosecurity (Academy of Military Medical Science, SKLPBS1418).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00862
References
Achtman, M., Zurth, K., Morelli, G., Torrea, G., Guiyoule, A., and Carniel, E. (1999). Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U.S.A. 96, 14043–14048. doi: 10.1073/pnas.96.24.14043
Andersen, J., Forst, S. A., Zhao, K., Inouye, M., and Delihas, N. (1989). The function of micF RNA. micF RNA is a major factor in the thermal regulation of OmpF protein in Escherichia coli. J. Biol. Chem. 264, 17961–17970.
Bellows, L. E., Koestler, B. J., Karaba, S. M., Waters, C. M., and Lathem, W. W. (2012). Hfq-dependent, co-ordinate control of cyclic diguanylate synthesis and catabolism in the plague pathogen Yersinia pestis. Mol. Microbiol. 86, 661–674. doi: 10.1111/mmi.12011
Chiang, M. K., Lu, M. C., Liu, L. C., Lin, C. T., and Lai, Y. C. (2011). Impact of Hfq on global gene expression and virulence in Klebsiella pneumoniae. PLoS ONE 6:e22248. doi: 10.1371/journal.pone.0022248
Corcoran, C. P., Podkaminski, D., Papenfort, K., Urban, J. H., Hinton, J. C., and Vogel, J. (2012). Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol. Microbiol. 84, 428–445. doi: 10.1111/j.1365-2958.2012.08031.x
Delihas, N. (1997). Antisense micF RNA and 5′-UTR of the target ompF RNA: phylogenetic conservation of primary and secondary structures. Nucleic Acids Symp. Ser. 36, 33–35.
Delihas, N. (2003). Annotation and evolutionary relationships of a small regulatory RNA gene micF and its target ompF in Yersinia species. BMC Microbiol. 3:13. doi: 10.1186/1471-2180-3-13
Desnoyers, G., Bouchard, M. P., and Massé, E. (2013). New insights into small RNA-dependent translational regulation in prokaryotes. Trends Genet. 29, 92–98. doi: 10.1016/j.tig.2012.10.004
Geng, J., Song, Y., Yang, L., Feng, Y., Qiu, Y., Li, G., et al. (2009). Involvement of the post-transcriptional regulator Hfq in Yersinia pestis virulence. PLoS ONE 4:e6213. doi: 10.1371/journal.pone.0006213
Hu, P., Elliott, J., McCready, P., Skowronski, E., Garnes, J., Kobayashi, A., et al. (1998). Structural organization of virulence-associated plasmids of Yersinia pestis. J. Bacteriol. 180, 5192–5202.
Kulesus, R. R., Diaz-Perez, K., Slechta, E. S., Eto, D. S., and Mulvey, M. A. (2008). Impact of the RNA chaperone Hfq on the fitness and virulence potential of uropathogenic Escherichia coli. Infect. Immun. 76, 3019–3026. doi: 10.1128/IAI.00022-08
Lindler, L. E., Plano, G. V., Burland, V., Mayhew, G. F., and Blattner, F. R. (1998). Complete DNA sequence and detailed analysis of the Yersinia pestis KIM5 plasmid encoding murine toxin and capsular antigen. Infect. Immun. 66, 5731–5742.
Mizuno, T., Chou, M. Y., and Inouye, M. (1984). A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA). Proc. Natl. Acad. Sci. U.S.A. 81, 1966–1970. doi: 10.1073/pnas.81.7.1966
Ni, B., Du, Z., Guo, Z., Zhang, Y., and Yang, R. (2008). Curing of four different plasmids in Yersinia pestis using plasmid incompatibility. Lett. Appl. Microbiol. 47, 235–240. doi: 10.1111/j.1472-765X.2008.02426.x
Perry, R. D., and Fetherston, J. D. (1997). Yersinia pestis–etiologic agent of plague. Clin. Microbiol. Rev. 10, 35–66.
Perry, R. D., Straley, S. C., Fetherston, J. D., Rose, D. J., Gregor, J., and Blattner, F. R. (1998). DNA sequencing and analysis of the low-Ca2+-response plasmid pCD1 of Yersinia pestis KIM5. Infect. Immun. 66, 4611–4623.
Sittka, A., Pfeiffer, V., Tedin, K., and Vogel, J. (2007). The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 63, 193–217. doi: 10.1111/j.1365-2958.2006.05489.x
Song, Y., Tong, Z., Wang, J., Wang, L., Guo, Z., Han, Y., et al. (2004). Complete genome sequence of Yersinia pestis strain 91001, an isolate avirulent to humans. DNA Res. 11, 179–197. doi: 10.1093/dnares/11.3.179
Vogel, J., and Luisi, B. F. (2011). Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9, 578–589. doi: 10.1038/nrmicro2615
Waters, L. S., and Storz, G. (2009). Regulatory RNAs in bacteria. Cell 136, 615–628. doi: 10.1016/j.cell.2009.01.043
Zhou, D., Han, Y., and Yang, R. (2006). Molecular and physiological insights into plague transmission, virulence and etiology. Microbes Infect. 8, 273–284. doi: 10.1016/j.micinf.2005.06.006
Keywords: Yersinia pestis, sRNA regulation, MicF-ompF, intrinsic plasmid, translational fusion
Citation: Liu Z, Wang H, Wang H, Wang J, Bi Y, Wang X, Yang R and Han Y (2015) Intrinsic plasmids influence MicF-mediated translational repression of ompF in Yersinia pestis. Front. Microbiol. 6:862. doi: 10.3389/fmicb.2015.00862
Received: 12 March 2015; Accepted: 07 August 2015;
Published: 21 August 2015.
Edited by:
Yi-Cheng Sun, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaReviewed by:
Jeff Shannon, National Institutes of Health, USAWeili Liang, Chinese Center for Disease Control and Prevention, China
Copyright © 2015 Liu, Wang, Wang, Wang, Bi, Wang, Yang and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruifu Yang and Yanping Han, 20, Dongdajie, Fengtai, Beijing 100071, China,cnVpZnV5YW5nQGdtYWlsLmNvbQ==;eWFucGluZ2hhbkBnbWFpbC5jb20=
 Zizhong Liu
Zizhong Liu Haili Wang
Haili Wang Hongduo Wang1
Hongduo Wang1 Yujing Bi
Yujing Bi Xiaoyi Wang
Xiaoyi Wang Yanping Han
Yanping Han