- 1Department of Horticulture, College of Horticulture, Northeast Agricultural University, Harbin, China
- 2Department of Horticulture, College of Life Science and Agroforestry, Qiqihar University, Qiqihar, China
- 3Heilongjiang Provincial Key University Laboratory of Cold Area Vegetable Biology, Northeast Agricultural University, Harbin, China
- 4Ministry of Agriculture Key Laboratory of Biology and Germplasm Enhancement of Horticultural Crops in Northeast China, Northeast Agricultural University, Harbin, China
The growth of watermelon is often threatened by Fusarium oxysporum f. sp. niveum (Fon) in successively monocultured soil, which results in economic loss. The objective of this study was to investigate the effect of D123 wheat as a companion crop on soil enzyme activities, microbial biomass and microbial communities in the rhizosphere of watermelon and to explore the relationship between the effect and the incidence of wilt caused by Fon. The results showed that the activities of soil polyphenol oxidase, urease and invertase were increased, the microbial biomass nitrogen (MBN) and microbial biomass phosphorus (MBP) were significantly increased, and the ratio of MBC/MBN was decreased (P < 0.05). Real-time PCR analysis showed that the Fon population declined significantly in the watermelon/wheat companion system compared with the monoculture system (P < 0.05). The analysis of microbial communities showed that the relative abundance of microbial communities was changed in the rhizosphere of watermelon. Compared with the monoculture system, the relative abundances of Alphaproteobacteria, Actinobacteria, Gemmatimonadetes and Sordariomycetes were increased, and the relative abundances of Gammaproteobacteria, Sphingobacteria, Cytophagia, Pezizomycetes, and Eurotiomycetes were decreased in the rhizosphere of watermelon in the watermelon/wheat companion system; importantly, the incidence of Fusarium wilt was also decreased in the watermelon/wheat companion system. In conclusion, this study indicated that D123 wheat as a companion crop increased soil enzyme activities and microbial biomass, decreased the Fon population, and changed the relative abundance of microbial communities in the rhizosphere of watermelon, which may be related to the reduction of Fusarium wilt in the watermelon/wheat companion system.
Introduction
Watermelon [Citrullus lanatus (Thunb.) Matsum and Nakai] is a widely cultivated fruit that is consumed globally. However, under continuous cropping patterns, the growth of watermelon is often threatened by the pathogen Fusarium oxysporum f. sp. niveum (Fon). The continuous cropping of the same crop in the same land can negatively affect the yield and quality of crops (Yu et al., 2000; Yao et al., 2006; Wang et al., 2014a) because it eliminates biological diversity (Blanco-Canqui and Lal, 2008). To overcome this problem, we proposed to increase the diversification of cultivated species and suggest that intercropping is the most efficient practice to reduce the incidence of soilborne diseases (Ren et al., 2008; Zhang et al., 2013).
Soil enzyme activities are indicators of soil health and have been applied to analyze soil quality and ecosystems (Ndiaye et al., 2000). Most commonly, urease is used to monitor the soil nitrogen cycle and nitrogen utilization because it can hydrolyze urea to ammonia. Invertase is used to monitor the change of soluble nutrients in the soil (Li et al., 2012). Phenol oxidase may be involved in the decomposition of toxic compounds in soils (Sinsabaugh, 2010). These enzymes, as well as others, have been used to evaluate the effects generated by agricultural practice (Zhou et al., 2011; Wang et al., 2014a,b).
The microbial biomass reflects the turnover of soil microorganisms and acts as both a source and a pool for nutrients (Irshad et al., 2012). Considering the species-specific effect of plants on the microbial community of a rhizosphere in an intercropping system, investigating the microbial biomass in the rhizosphere of intercrops compared with monocultures can provide meaningful data (Tang et al., 2014).
The soil microbiome is thought to be responsible for biological processes that are necessary for maintaining a healthy soil and suppressing plant diseases (Garbeva et al., 2004; Pedersen and Mills, 2004; Andreote et al., 2014). Mazzola (2004) reported that a decrease in soil microbial diversity was related to the development of soil-borne plant diseases. Soils with a higher fungal diversity exhibited a higher potential of disease suppression (Penton et al., 2014). Intercropping can improve soil microbial diversity and change microbial communities; therefore, it plays an important role in controlling plant disease. For example, tomato blight disease can be controlled by using a tomato–marigold intercropping system (Gómez-Rodrígueza et al., 2003). Ren et al. (2008) demonstrated that intercropping with aerobic rice alleviated Fusarium wilt in watermelon by changing the microbial communities in the rhizosphere.
In a previous study, D123 wheat was used as a companion crop to reduce the incidence of watermelon Fusarium wilt (Xu et al., 2015). However, little information is known about how D123 wheat, as a companion crop, exerts this effect. Therefore, the aim of this study was to evaluate the effect of D123 wheat on soil enzyme activities, microbial biomass, and microbial communities in the rhizosphere of watermelon and explore the relationship between the effect and incidence of watermelon Fusarium wilt.
Materials and Methods
Plant Material
The D123 wheat seeds were provided by the Vegetable Physiological Ecology Laboratory College of Horticulture at Northeast Agricultural University in Harbin, Heilongjiang Province. Seeds of the watermelon cultivar Jingxin No. 1, which is moderately susceptible to Fon, were bought from the Golden Seed Company, Beijing, China.
Greenhouse Experiment
This study was performed in a greenhouse located in the experimental center of Northeast Agricultural University in Harbin, China (45°41′N, 126°37′E). The soil used in pot experiments was collected from the surface of locations on Xiangfang farm in Harbin, China, where watermelon was cultivated continuously for 3 years and was infected by Fon. The soil contained 35.90 g·kg−1 of organic matter, 357.00 g·kg−1 of alkaline hydrolytic N, 378.80 g·kg−1 of available P, and 107.50 g·kg−1 of available K. The electrolytic conductivity was 1.27 ms·cm−1, and the pH was 7.23 (1:5, soil:water). Two cropping systems were included in these experiments: (I) watermelon monoculture (CK2) and (II) watermelon/wheat companion system, in which D123 wheat (D123) was used as a companion crop because a previous study reported that the root exudates from D123 wheat can inhibit the mycelial growth of Fon (Xu et al., 2015). In addition, control pots were designed that did not contain any plants (CK1). All of the pots were arranged randomly with three repeats per treatment and 10 pots per repeat.
Watermelon seedlings with five leaves were transplanted into plastic pots (20 cm in diameter and 17 cm in height). Each pot was filled with 3 kg fresh soil from the infected field mentioned previously. No fertilizers were added to the soil. In the watermelon/wheat companion system, D123 wheat seeds were surface sterilized with 5% (v/v) H2O2 for 30 min, rinsed four times with distilled water, and then directly sown on the side of the watermelon plant; each pot had 30 wheat seedlings and one watermelon seedling, and watermelon and wheat seedlings were kept apart 5–7 cm from each other. To ensure good aeration and avoid the shading of watermelon, the wheat seedlings were cut several times and kept to a 15 cm height during the experimental period. In the watermelon monoculture system, each pot contained one watermelon seedling only. Hand weeding was performed during the experiment. The water content of the soil was maintained by weight. No pesticides were sprayed. When the watermelon plants began to develop Fusarium wilt, the samples from the rhizosphere of the watermelon plant were collected from five plants in each repeat as described by Song et al. (2007). Briefly, five plants were excavated randomly, and the loosely adhered soil was shaken off; the tightly adhered soil was removed and mixed as the rhizosphere sample. Part of the sample was used to determine microbial biomass and soil enzyme activities, and the other part was stored at −70°C for DNA extraction.
Assessment of Disease Incidence
The incidence of wilt was expressed as a percentage that was calculated by diseased plants over the total number of plants (Wu et al., 2009).
Soil Enzyme Activities Analysis
Soil urease (EC 3.5.1.5) was measured by incubating 10 g of soil with 10 ml of 10% urea solution for 24 h at 37°C. The suspension was filtered, and the ammonia content in the filtrates was determined colorimetrically at 578 nm by mixing the filtrates with reagent (sodium phenolate, sodium hypochlorite). The activity was expressed as NH4-N mg·g−1 soil·24 h−1 (Kandeler and Gerber, 1988). Soil invertase (EC 3.2.1.26) activity was measured by incubating 5 g of soil with 15 ml of 8% sucrose solution for 24 h at 37°C. The suspension was filtered, and 1 ml filtrate was treated with 3 ml of 3,5-dinitrosalicylic acid; the absorbance was then detected at 508 nm. Enzyme activity is expressed as mg glucose·g−1 soil·24 h−1 (Frankeberger and Johanson, 1983). Soil catalase (EC 1.11.1.6) was measured by incubating 5 g of soil with 5 ml of 0.3% H2O2 at 30°C for 30 min. The suspension was titrated with 0.1 M KMnO4 solution. Enzyme activity was expressed as 0.1 M KMnO4 ml·g−1 soil·30 min−1 (Johnson and Temple, 1964). Polyphenol oxidase (EC 1.10.3.1) activity was determined as described by Peruccia et al. (2000) and expressed as purpurogallin mg·g−1 soil·3 h−1.
Estimation of Microbial Biomass in the Rhizosphere
Fumigation extraction methods were used to measure microbial biomass carbon (MBC), microbial biomass nitrogen (MBN), and microbial biomass phosphorus (MBP) in the watermelon rhizosphere as described by Brookes et al. (1985) and Vance et al. (1987). The 15 g fresh soil samples were placed in a 50 ml beaker, and another beaker was filled with 50 ml alcohol-free chloroform; both beakers were kept in a vacuum desiccator. As a control, soil that was not treated with chloroform was kept in another desiccator. Then, the two desiccators were kept in the dark for 24 h at room temperature. Subsequently, the two desiccators were evacuated using a vacuum pump, and the fumigated and non-fumigated samples were transferred to a 100 ml conical flask, respectively, and extracted with 0.5 M K2SO4 by shaking for 30 min on a rotator at 300 rpm. The extracts were filtered through filter paper. The contents of organic carbon from fumigated and non-fumigated soil were measured using the dichromate digestion method. An extractability factor of 0.45 was used to calculate the MBC (Brookes et al., 1982). For MBN, the filtrates (20 ml) were digested with sulfuric acid (96%) in a 50 ml Kjeldahl bottle. The digested filtrates were team distilled using a semi-micro Kjeldahl bottle and were titrated against hydrochloric acid (0.05 N). MBP was determined using the chloroform fumigation-extraction method (Brookes et al., 1982).
Soil DNA Extraction
The genomic DNA of rhizosphere microorganisms was extracted with the PowerSoil® DNA Isolation Kit (MO BIO, USA) according to the manufacturer's instructions. Extracted DNA was stored at −20°C until use.
PCR Amplification and MiSeq Sequencing
The protocol used for PCR amplification was described by (Magoč and Salzberg, 2011). Each treatment (CK1, CK2, and D123) was repeated three times, PCR reactions were performed in triplicate. The primer set of V338f (5′-ACTCCTACGGGAGG CAGCA-3′) and V806R (5′-ATGCAGGGACTACHVGGGT WTCTAAT-3′) was used to amplify the V3-V4 region of the bacterial 16S rDNA, and the primer set of ITS1-737F (5′-GGAAGTAAAAGTCGT AACAAGG-3′) /ITS2-2043R (5′-ATGCAGGCTGCG TTCTTCATCGATGC-3′) was used to amplify the ITS2 (internal transcribed spacer 2) region of the fungi rDNA (White et al., 1990; Bazzicalupo et al., 2013). Because the entire ITS region is too long for high-throughput sequencing methods, recent high-throughput sequencing studies have selected either the ITS1 or ITS2 region (Mello et al., 2011; Orgiazzi et al., 2012). Due to systematic length differences in the ITS2 region as well as the entire ITS, Bellemain et al. (2010) found that ascomycetes will more easily amplify than basidiomycetes using ITS2 regions as targets. Ascomycota represents the largest phylum of fungi (Bellemain et al., 2010). Amplicon pyrosequencing was performed by Majorbio Bioinformatics Technology Co., Ltd. (Shanghai, China). The protocol used to determine the composition of the microbial communities in three treatments was described by Caporaso et al. (2010). All data sets were deposited into the NCBI Sequence Read Archive database (http://www.ncbi.nlm.nih.gov/Traces/sra).
Sequences were analyzed using the QIIME (Wang et al., 2007) software and UPARSE pipeline (Edgar, 2013). Pairs of reads from the original DNA fragments were merged using FLASH (Caporaso et al., 2010), which is a very fast and accurate software tool that was designed to merge pairs of reads when the original DNA fragments are shorter than twice the length of the reads. Sequencing reads were assigned to each sample according to the unique barcode of each sample. Briefly, the reads were filtered by QIIME (version 1.17) quality filters. Then, we used the UPARSE (version 7.1 http://drive5.com/uparse/) pipeline to pick operational taxonomic units (OTUs) to make an OTU table. Sequences were assigned to OTUs at 97% similarity. The chimeric sequences were identified and removed using UCHIME. We picked a representative sequences for each OTU and used the ribosomal database project (RDP) classifier (Caporaso et al., 2011) to assign taxonomic data to each representative sequence.
The Analysis of Diversity
The Shannon index (Hshannon) was determined as follows (Schloss et al., 2009):
where Sobs is the number of observed OTUS, ni is the number of individuals in the ith OTU, and N is the total number of individuals in the community. Additionally, the principal co-ordinate analysis (PCoA) was performed to estimate the community diversity between samples (Lozupone and Knight, 2005) based on the Illumina-MiSeq sequencing data with unweighted UniFrac distance matrix (Peiffer et al., 2013).
Real-time PCR Assay
The population of Fon in the soil samples was analyzed by real-time PCR. The primer pair of fn-1 (5′-TACCACTTGTTG CCTCGGC-3′) and fn-2 (5′-TTGAGGAAC GCGAATTAAC-3′) was used to identify Fon (Zhang et al., 2005). The real-time PCR assays were performed with an IQ5 Real-Time PCR System (Bio-Rad Lab, LA, USA). The reaction volume was 20 μl, which contained 10 μl of 2 × Real SYBR Mixture (TIANGEN Biotech, China), 0.5 μl of each primer, and 2 μl of template DNA. The PCR process was 94°C for 5 min; 95°C for 30 s, 54°C for 30 s, 72°C for 30 s for 35 cycles in total; and a final elongation at 72°C for 7 min. The signal threshold was set automatically by the system. To evaluate amplification specificity, melt curve analysis was performed at the end of the PCR run (see Supplementary Material).
The plasmid standard for the quantification of Fon was generated from a cloned target gene from Fon genomic DNA. The amplicon was purified using the PCR Purification Kit (BioTeke Corporation, China) and ligated into a pMD18 T vector (Takara, Dalian) according to the manufacturer's instructions. Plasmid from the insert-positive clones was extracted with a Plasmid Extraction Kit (BioTeke Corporation, China). The concentration of plasmid DNA was measured and converted to copy concentration using the following equation as described by Whelan et al. (2003):
Standard curves (see Supplementary Material) were performed with 10-fold dilution series of plasmids. Sterile water was used as a negative control to replace the template. All real-time PCR reactions were done in technical triplicates such that each treatment was analyzed nine times.
Statistical Analyses
Differences were calculated with One-Way analysis of variance (ANOVA) at the end of each assay. The data were analyzed by Duncan's multiple range test and expressed as the means ± standard error, and the incidence of watermelon Fusarium wilt was analyzed by independent samples T-test using the SAS statistical software.
Results
Incidence of Watermelon Fusarium Wilt was Affected by Companion with Wheat
The incidence of watermelon Fusarium wilt was investigated. The rate was 63.3% (P < 0.05) in the monoculture system but was significantly lower at 21.1% (P < 0.05) in the watermelon/wheat companion system (Figure 1).

Figure 1. The effect of D123 wheat as a companion crop on Fusarium wilt in watermelon seedlings. CK2, monoculture of watermelon; D123, D123 wheat as a companion crop. Significant differences between treatments are indicated by different letters (P < 0.05, independent samples T-test).
Soil Enzyme Activities
In this study, the activities of four soil enzymes in the watermelon rhizosphere (including control soils) were compared. The invertase activity increased significantly in the watermelon/wheat companion system compared with that in the monoculture system; however, the invertase activity in the control soil (without plants) was lower than that in the monoculture (P < 0.05; Figure 2A). The catalase activity in the control soil was lower than that in both the watermelon/wheat companion system and the monoculture system, but no significant difference was found between the watermelon/wheat companion system and the watermelon monoculture system (P < 0.05; Figure 2B). The polyphenol oxidase activity was higher in the watermelon/wheat companion system than that in the monoculture and control (without plants, P < 0.05; Figure 2C). The urease activity was highest in the watermelon/wheat companion system, lower in the monoculture system, and lowest in the control (without plants), and significant differences in urease activity existed between these treatments (Figure 2D).
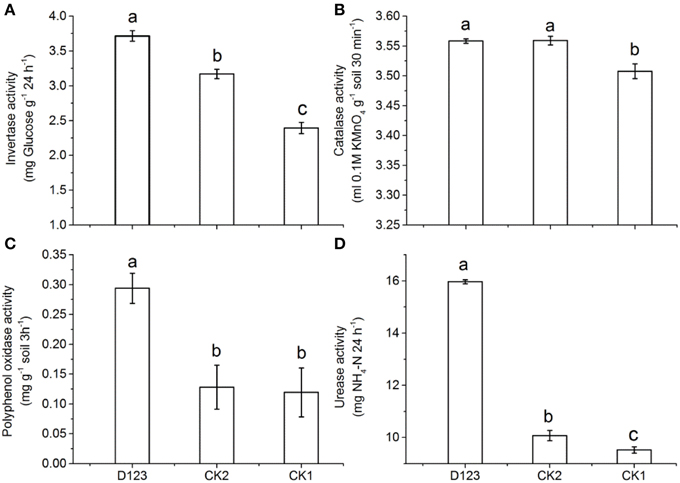
Figure 2. The effects of D123 wheat as a companion crop on invertase (A), catalase (B), polyphenol oxidase (C), urease (D) activities in the watermelon rhizosphere. CK1, control soil, without plants; CK2, monoculture of watermelon; D123, D123 wheat as a companion crop; Significant differences between treatments are indicated by different letters (Duncan's multiple range test, P < 0.05).
Microbial Biomass
No significant differences in MBC were detected among the three treatment soils (Table 1). However, the soil MBN and MBP were significantly higher in the watermelon/wheat companion system and the control (CK1) than in the monoculture system. Compared with the monoculture (CK2), the MBC/MBN ratio was significantly decreased in the watermelon/wheat companion system and control (CK1) (Table 1, P < 0.05).
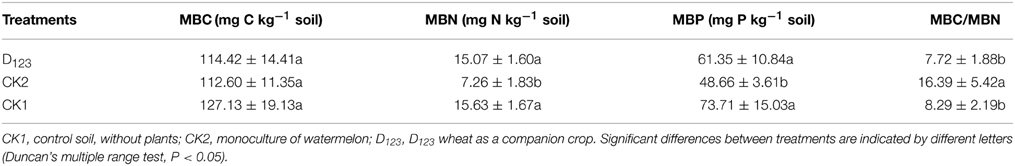
Table 1. The effect of wheat as companion crop on the MBC, MBN, MBP, and the MBC/MBN ratio in the rhizosphere of watermelon.
Fon Population
Real-time PCR analysis was performed to determine the copy numbers of the target DNA in the rhizosphere of watermelon collected from the watermelon/wheat companion system, monoculture system, and control soil, respectively. A significant difference was found between the watermelon/wheat companion system and the monoculture system (Figure 3). For the three treatment soils, the highest population of Fon was detected in the rhizosphere of watermelon in the monoculture system (up to 49.2 × 108 copies·g−1 soil). The population was lower in the control soil, at 31.1 × 108 copies·g−1 soil. However, the lowest Fon population was found in the rhizosphere of watermelon in the watermelon/wheat companion system, at 13.6 × 108 copies·g−1 soil.
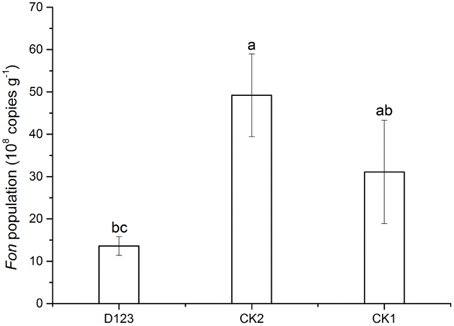
Figure 3. The abundance of Fusarium oxysporum f. sp. niveum in the rhizosphere of watermelon in companion cropping and monoculture systems. Significant differences between treatments are indicated by different letters (Duncan's multiple range test, P < 0.05). CK1, control soil, without plants; CK2, monoculture of watermelon; D123, D123 wheat as a companion crop.
Pyrosequencing and Sequence Analysis
The number of species detected in the samples, or the number of organisms at a given phylogenetic level, relies largely on the amount of analyzed sequences (Schloss and Handelsman, 2005). In the present study, the average sequence lengths for 16S rDNA and ITS2 were approximately 430 and 270 bp, respectively. After removing the low-quality sequence reads, a total of 174,251 sequences were obtained, in which 76,545 sequences were classified as bacterial and 97,706 were classified as fungal. They were used to evaluate the microbial richness and diversity and the differences among the treatments.
Microbial Community Composition Analysis
The maximum bacteria OTUs detected were 1854, 1763, and 1649 for the rhizosphere of the monoculture (CK2), the rhizosphere of the companion system (D123), and the control soil (CK1), respectively (Figure 4A). The cut-off for the analysis was at 97% sequence similarity. For fungi, 927 OTUs were found in the rhizosphere of the companion system (D123), 829 OTUs were found in the rhizosphere of the monoculture watermelon (CK2), and 678 OTUs were found in the control soil (CK1) (Figure 4B). The rarefaction curves of both bacteria and fungi showed that the sequencing capability was not sufficiently large to capture the complete diversity of communities because the curves did not reach a plateau by increasing sample size. However, the data were sufficient to show differences among the treatments.
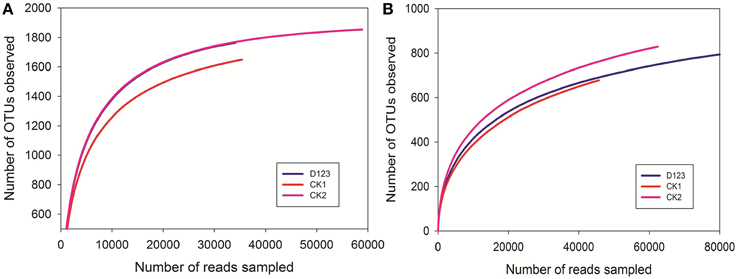
Figure 4. Rarefaction curves of both bacteria (A) and fungi (B) depicting the effect of 3% dissimilarity on the number of OTUs identified in the three soil treatments. The vertical axis shows the average number of OTUs that would be expected after sampling the number of sequences shown on the horizontal axis. CK1, control soil, without plants; CK2, monoculture of watermelon; D123, D123 wheat as a companion crop.
All of the sequences were classified into 19 known classes by the mother program, “Others,” if labeled in data, meant that the sequences could not be classified into any known group. The results showed that the overall bacterial composition was similar for each treatment, whereas the relative abundance of each class varied in different treatments (Figures 5A–C). Alphaproteobacteria, Gammaproteobacteria, and Actinobacteria were the top three classes among all bacterial classes, which comprised 19.06, 18.39, and 13%, respectively, in CK1 soil; 13.3, 17.99, and 10.59%, respectively, in CK2 soil; and 16.35, 16.2, and 10.83%, respectively, in D123 soil. Sphingobacteria comprised 4.55, 8.45, and 7.25% of the total bacterial communities in CK1, CK2, and D123 soil, respectively. Gemmatimonadetes comprised 7.29, 6.23, and 7.92% of the total bacterial communities in CK1, CK2, and D123 soil, respectively. Cytophagia comprised 3.37, 5.82, and 5.69% of the total bacterial communities in CK1, CK2, and D123 soil, respectively. The overall fungal composition of the different soil samples was similar, whereas the distribution of each class was varied (Figures 6A–C). Compared with CK2, the relative abundance of Sordariomycetes was higher in the watermelon/wheat companion system, and Pezizomycetes was lower in the watermelon/wheat companion system.
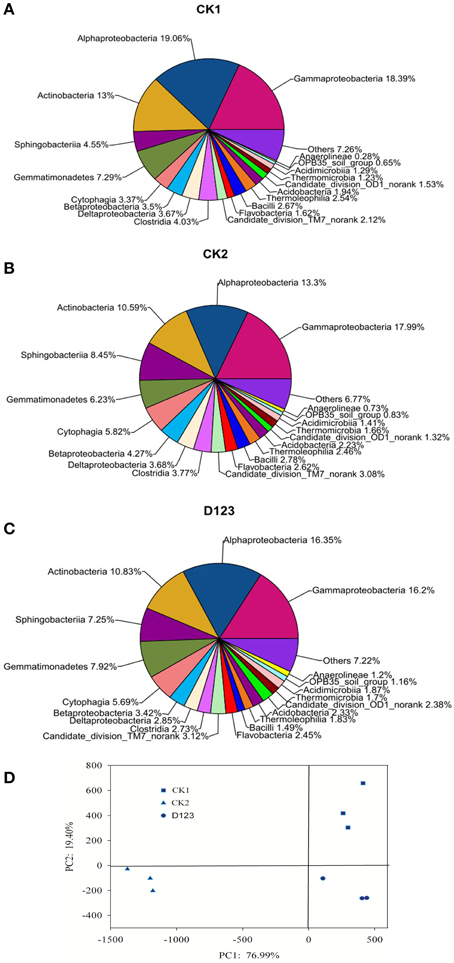
Figure 5. Comparison of the bacterial communities at the class level (A–C) and β-diversity analysis of the bacterial community across each genus by PCoA for the different cropping treatments (D). Sequences that could not be classified into any known group are labeled “Others.” CK1, control soil, without plants; CK2, monoculture of watermelon; D123, D123 wheat as a companion crop.
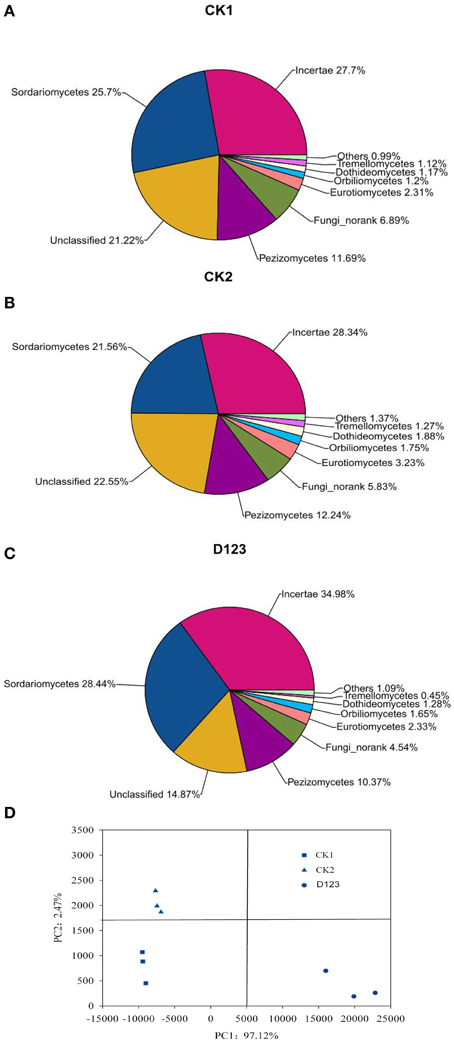
Figure 6. Comparison of the fungal communities at the class level (A–C) and β-diversity analysis of the fungal community across each genus by PCoA for the different cropping treatments (D). Sequences that could not be classified into any known group are labeled “Others.” CK1, control soil, without plants; CK2, monoculture of watermelon; D123, D123 wheat as a companion crop.
The PCoA analysis of the bacteria and fungi revealed three separated clusters (Figures 5D, 6D). Each cluster was distinguished from each other, which indicated that differences in the community diversity exist between the different cropping systems. The Hshannon indices of soil bacterial and fungal community were also calculated. Compared with monoculture (CK2), the Hshannon indices of soil bacterial and fungal community were increased in the watermelon/wheat companion system (Figure 7).
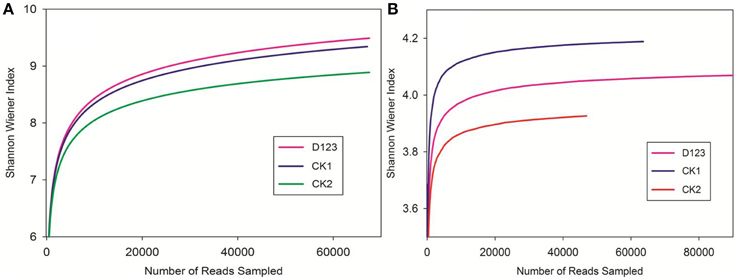
Figure 7. The Shannon index of both bacterial (A) and fungal (B) communities in three soil treatments. CK1, control soil, without plants; CK2, monoculture of watermelon; D123, D123 wheat as a companion crop.
Discussion
This study showed that the incidence of watermelon Fusarium wilt was decreased in the watermelon/wheat companion system compared with a monoculture system (Figure 1). The result was consistent with the decline of the Fon population (Figure 3). These results suggest that the Fusarium wilt may be suppressed in the companion system, in accordance with the results reported by other studies (Larkin et al., 1993a,b).
We also found that the activities of soil polyphenol oxidase, urease and invertase were increased significantly in the watermelon/wheat companion system compared with the monoculture system (Figure 2). Soil urease plays a vital role in utilization of soil nitrogen and the nitrogen cycle by decomposing urea into ammonia, carbon dioxide and water, which are beneficial for plant absorption (Wang et al., 2014a). The urease activity was sensitive to changes in cropping systems. The result is supported by other studies (Zhou et al., 2011; Xu et al., 2013). Extracellular phenol oxidases are deployed by both fungi and bacteria to mitigate the toxicity of phenolic molecules and aid in antimicrobial defense (Sinsabaugh, 2010). In our experiments, we observed that the polyphenol oxidase activity was higher in the watermelon/wheat companion system than in both the monoculture system and the control (without plants), which implied that the watermelon/wheat companion system was more beneficial in reducing the toxicity of phenolic acids in the rhizosphere of watermelon (Figure 2). Invertase widely exists in the soil and plays an important role in the transformation of carbon in the soil (Eivazi and Bayan, 1996). Dai et al. (2013) found that the intercropping of peanut with Atractylodes lancea effectively increased soil invertase activities. Additionally, Ahmad et al. (2013) found that pepper intercropping with green garlic significantly increased the activities of invertase in soil. The result obtained from this study is in agreement with their conclusion. In sum, the watermelon/wheat companion system (D123 wheat as companion crop) changed the activities of soil enzymes, and these changes may reflect changes in the soil micro-environment (Iyyemperumal and Shi, 2008) and result in the inhibition of the growth of Fon, which exhibits a decreased incidence of the Fusarium wilt.
Disease suppression is generally thought to be related to a global increase in soil microbial biomass (Janvier et al., 2007). In the present study, the watermelon/wheat companion system increased the total contents of MBN and MBP (Table 1). The increase in MBN may be attributed to the increase in root exudates because the root exudates could come from both watermelon and D123 wheat, which can lead to changes in the soil microbial community structure (Figures 5, 6). The MBP was also increased, which could be due to the watermelon/wheat companion system stimulating the transformation of P mediated by rhizosphere microorganisms. The ratio of MBC/MBN is often used to describe the structure and status of the microbial community. A high MBC/MBN ratio indicates that the microbial biomass contains a higher proportion of fungi, which means that the status of soil health is worse. In contrast, a lower ratio suggests that bacteria predominate in the microbial population, which represents good soil health (Campbell et al., 1992). In this study, the MBC/MBN ratio was decreased in the watermelon/wheat companion system compared with the monoculture system, which implied that the soil was healthier in the watermelon/wheat companion system. This may be another reason for the reduction of Fon wilt incidence.
Sturz and Christie (2003) confirmed that the optimum microbial community can promote soil defense capability. In successively mono-cropped soil, specific microbial communities were formed due to the accumulation of specific exudates (Brussaard et al., 2007; Janvier et al., 2007), which could promote the increase of soil-borne pathogens. In this study, we found that the composition of microbial communities was changed (Figures 3, 5, 6), which suggests that Fusarium wilt may be suppressed in this system and is in concordance with previous studies (Larkin et al., 1993a,b). However, the mechanism by which the Fusarium pathogen is reduced remains unclear. We believe that the decline of the Fon population in the rhizosphere of watermelon in a wheat-based companion system is associated with the D123 wheat root exudates because D123 wheat root exudates may be inhibiting the mycelial growth of Fon (Xu et al., 2015). Furthermore, the MiSeq Illumina technology was applied to analyze the differences of bacterial and fungal communities. The different cropping system could affect the soil microbial diversity and composition (Larkin and Honeycutt, 2006; Acosta-Martínez et al., 2010). Our results indicated that, the relative abundance and diversity of the soil bacterial and fungal community were differed in different cropping system (Figures 5–7). The relative abundances of bacterial classes Alphaproteobacteria, Actinobacteria, and Gemmatimonadetes were increased, but the relative abundances of Gammaproteobacteria, Sphingobacteria, and Cytophagia were decreased in the watermelon/wheat companion system compared with the monoculture system (Figures 5B,C). The relative abundance of the fungal class Sordariomycetes was increased, but the relative abundances of Pezizomycetes and Eurotiomycetes were decreased in the watermelon/wheat companion system compared with the monoculture system (Figures 6B,C). In addition, the diversity of soil microbiota was increased in the watermelon/wheat companion system compared with the monoculture system (Figure 7). These differences may be due to the interactions between the soil microorganisms and different plant exudates (Singh et al., 2007; Song et al., 2007). Plant root exudates may play a strong role in shaping the rhizospheric community structure and function (Huang et al., 2014). The changes in the relative abundance and diversity of soil microbiota can also be used to explain the decrease in Fusarium wilt incidence.
Conclusion
In this study, the incidence of Fusarium wilt was decreased and the Fon population was reduced in a watermelon/wheat companion system. However, the soil polyphenol oxidase, urease and invertase activities were increased, MBN and MBP were increased, and the ratio of MBC/MBN was decreased in the rhizosphere of watermelon in the watermelon/wheat companion system compared with a monoculture system. These results suggest that the decrease in the incidence of Fusarium wilt may be related to changes in soil enzyme activities, microbial biomass, microbial relative abundance and diversity.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge the National Nature Science Foundation of China (31471917), the Natural Science Foundation of Heilongjiang Province, China (C2015048) and the Program for Young Teachers Scientific Research at Qiqihar University, China (2014k-Z14) for their funding of this project.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00899
Abbreviations
MBC, Microbial biomass carbon; MBN, Microbial biomass nitrogen; MBP, Microbial biomass phosphorus; Fon, Fusarium oxysporum f. sp. niveum.
References
Acosta-Martínez, V., Burow, G., Zobeck, T. M., and Allen, V. G. (2010). Soil microbial communities and function in alternative systems to continuous cotton. Soil Sci. Soc. Am. J. 74, 1181–1192. doi: 10.2136/sssaj2008.0065
Ahmad, I., Cheng, Z., Meng, H., Liu, T., Wang, M., Ejaz, M., et al. (2013). Effect of pepper-garlic intercropping system on soil microbial and bio-chemical properties. Pak. J. Bot. 45, 695–702.
Andreote, F. D., Gumiere, T., and Durrer, A. (2014). Exploring interactions of plant microbiomes. Sci. Agric. 71, 528–539. doi: 10.1590/0103-9016-2014-0195
Bazzicalupo, A. L., Bálint, M., and Schmitt, I. (2013). Comparison of ITS1 and ITS2 rDNA in 454 sequencing of hyperdiverse fungal communities. Fungal Ecol. 6, 102–109. doi: 10.1016/j.funeco.2012.09.003
Bellemain, E., Carlsen, T., Brochmann, C., Coissac, E., Taverlet, P., and Kauserud, H. (2010). ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol. 10:189. doi: 10.1186/1471-2180-10-189
Blanco-Canqui, H., and Lal, R. (2008). Principles of Soil Conservation and Management. Heidelberg: Springer.
Brookes, P. C., Landman, A., Pruden, G., and Jenkinson, D. S. (1985). Chloroform fumigation and the release of soil nitrogen: a rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biol. Biochem. 17, 837–842. doi: 10.1016/0038-0717(85)90144-0
Brookes, P. C., Powlson, D. S., and Jenkinson, D. S. (1982). Measurement of microbial biomass phosphorus in soil. Soil Biol. Biochem. 14, 319–329. doi: 10.1016/0038-0717(82)90001-3
Brussaard, L., Ruiter, P. C., and Brown, G. G. (2007). Soil biodiversity for agricultural sustainability. Agric. Ecosyst. Environ. 121, 233–244. doi: 10.1016/j.agee.2006.12.013
Campbell, C. A., Moulin, A. P., Bowren, K. E., Janzen, H. H., Townley-Smith, L., and Biederbeck, V. O. (1992). Effect of crop rotations on microbial biomass, specific respiratory activity and mineralizable nitrogen in a black chernozemic soil. Can. J. Soil Sci. 72, 417–427. doi: 10.4141/cjss92-035
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Tumbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. 108 Suppl. 1, 4516–4522. doi: 10.1073/pnas.1000080107
Dai, C., Chen, Y., Wang, X., and Li, P. (2013). Effects of intercropping of peanut with the medicinal plant Atractylodes lancea on soil microecology and peanut yield in subtropical China. Agroforest Syst. 87, 417–426. doi: 10.1007/s10457-012-9563-z
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Eivazi, F., and Bayan, M. R. (1996). Effects of long-term prescribed burning on the activity of select soil enzymes in an oak hickory forest. Can. J. For. Res. 26, 1799–1804. doi: 10.1139/x26-204
Frankeberger, W. T., and Johanson, J. B. (1983). Method of measuring invertase activity in soils. Plant Soil 74, 301–311. doi: 10.1007/BF02181348
Garbeva, P., Veen, J. A. V., and Elsas, J. D. V. (2004). Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu. Rev. Phytopathol. 42, 243–270. doi: 10.1146/annurev.phyto.42.012604.135455
Gómez-Rodrígueza, O., Zavaleta-Mejía, E., and González-Hernándezb, V. A. (2003). Allelopathy and microclimatic modification of intercropping with marigold on tomato early blight disease development. Field Crops Res. 83, 27–34. doi: 10.1016/S0378-4290(03)00053-4
Huang, X. F., Chaparro, J. M., Reardon, K. F., Zhang, R., and Shen, Q. (2014). Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany 92, 267–275. doi: 10.1139/cjb-2013-0225
Irshad, U., Brauman, A., Villenave, C., and Plassard, C. (2012). Phosphorus acquisition from phytate depends on efficient bacterial grazing, irrespective of the mycorrhizal status of Pinus pinaster. Plant Soil 358, 155–168. doi: 10.1007/s11104-012-1161-3
Iyyemperumal, K., and Shi, W. (2008). Soil enzyme activities in two forage systems following application of different rates of swine lagoon effuent or ammonium nitrate. Appl. Soil Ecol. 38, 128–136. doi: 10.1016/j.apsoil.2007.10.001
Janvier, C., Villeneuve, F., Alabouvette, C., Edel-Hermann, V., Mateille, T., and Steinberg, C. (2007). Soil health through soil disease suppression: which strategy from descriptors to indicators? Soil Biol. Biochem. 39, 1–23. doi: 10.1016/j.soilbio.2006.07.001
Johnson, J. L., and Temple, K. L. (1964). Some variables affecting the measurement of “catalase activity” in soil. Soil Sci. Soc. Am. J. 28, 207–209. doi: 10.2136/sssaj1964.03615995002800020024x
Kandeler, E., and Gerber, H. (1988). Short-term assay of soil urease activity using colorimetric determination of ammonium. Biol. Fert Soils 6, 68–72. doi: 10.1007/BF00257924
Larkin, R. P., and Honeycutt, C. W. (2006). Effects of different 3-year cropping systems on soil microbial communities and Rhizoctonia diseases of potato. Phytopathology 96, 68–79. doi: 10.1094/PHYTO-96-0068
Larkin, R. P., Hopkins, D. L., and Martin, F. N. (1993a). Ecology of Fusarium oxysporum f. sp. niveum in soils suppressive and conducive to Fusarium wilt of watermelon. Phytopathology 83, 1105–1116. doi: 10.1094/Phyto-83-1105
Larkin, R. P., Hopkins, D. L., and Martin, F. N. (1993b). Effect of successive watermelon plantings on Fusarium oxysporum and other microorganisms in soils suppressive and conducive to Fusarium wilt of watermelon. Phytopathology 83, 1097–1105. doi: 10.1094/Phyto-83-1097
Li, H., Shao, H., Li, W., Bi, R., and Bai, Z. (2012). Improving soil enzyme activities and related quality properties of reclaimed soil by applying weathered coal in opencast-mining areas of the Chinese Loess Plateau. Clean Soil Air Water 40, 233–238. doi: 10.1002/clen.201000579
Lozupone, C., and Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mazzola, M. (2004). Assessment and management of soil community structure for disease suppression. Annu. Rev. Phytopathol. 42, 35–59. doi: 10.1146/annurev.phyto.42.040803.140408
Mello, A., Napoli, C., Murat, C., Morin, E., Marceddu, G., and Bonfante, P. (2011). ITS-1 versus ITS-2 pyrosequencing: a comparison of fungal population in truffle grounds. Mycologia 103, 1184–1193. doi: 10.3852/11-027
Ndiaye, E. L., Sandeno, J. M., McGrath, D., and Dick, R. P. (2000). Integrative biological indicators for detecting change in soil quality. Am. J. Alternative Agric. 15, 26–36. doi: 10.1017/S0889189300008432
Orgiazzi, A., Lumini, E., Nilsson, R. H., Girlanda, M., Vizzini, A., Bonfante, P., et al. (2012). Unravelling soil fungal communities from different Mediterranean land-use backgrounds. PLoS ONE 7:e34847. doi: 10.1371/journal.pone.0034847
Pedersen, B. S., and Mills, N. J. (2004). Single vs. multiple introduction in biological control: the roles of parasitoid efficiency, antagonism and niche overlap. J. Appl. Ecol. 41, 973–984. doi: 10.1111/j.0021-8901.2004.00953.x
Peiffer, J. A., Spor, A., Koren, O., Jin, Z., Tringe, S. G., Dangl, J. L., et al. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. U.S.A. 110, 6548–6553. doi: 10.1073/pnas.1302837110
Penton, C. R., Gupta, V. V., Tiedje, J. M., Neate, S. M., Ophel-Keller, K., Gillings, M., et al. (2014). Fungal community structure in disease suppressive soils assessed by 28S LSU gene sequencing. PLoS ONE 9:e93893. doi: 10.1371/journal.pone.0093893
Peruccia, P., Casucci, C., and Dumontet, S. (2000). An improved method to evaluate the odiphenol oxidase activity of soil. Soil Biol. Biochem. 32, 1927–1933. doi: 10.1016/S0038-0717(00)00168-1
Ren, L. X., Su, S. M., Yang, X. M., Xu, Y. C., Huang, Q. W., and Shen, Q. R. (2008). Intercropping with aerobic rice suppressed Fusarium wilt in watermelon. Soil Biol. Biochem. 40, 834–844. doi: 10.1016/j.soilbio.2007.11.003
Schloss, P. D., and Handelsman, J. (2005). Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71, 1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: opensource, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Singh, B. K., Munro, S., Potts, J. M., and Millard, P. (2007). Influence of grass species and soil type on rhizosphere microbial community structure in grassland soils. Appl. Soil Ecol. 36, 147–155. doi: 10.1016/j.apsoil.2007.01.004
Sinsabaugh, R. L. (2010). Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biol. Biochem. 42, 391–404. doi: 10.1016/j.soilbio.2009.10.014
Song, Y. N., Zhang, F. S., Marschner, P., Fan, F. L., Gao, H. M., Bao, X. G., et al. (2007). Effect of intercropping on crop yield and chemical and microbiological properties in rhizosphere of wheat (Triticum aestivum L.), maize (Zea mays L.), and faba bean (Vicia faba L.). Biol. Fert. soils. 43, 565–574. doi: 10.1007/s00374-006-0139-9
Sturz, A. V., and Christie, B. R. (2003). Beneficial microbial allelopathies in the root zone: the management of soil quality and plant disease with rhizobacteria. Soil Till. Res. 72, 107–123. doi: 10.1016/S0167-1987(03)00082-5
Tang, X. Y., Bernard, L., Brauman, A., Daufresne, T., Deleporte, P., Desclaux, D., et al. (2014). Increase in microbial biomass and phosphorus availability in the rhizosphere of intercropped cereal and legumes under field conditions. Soil Biol. Biochem. 75, 86–93. doi: 10.1016/j.soilbio.2014.04.001
Vance, E. D., Brookes, P. C., and Jenkinson, D. S. (1987). An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 19, 703–707. doi: 10.1016/0038-0717(87)90052-6
Wang, M. Y., Wu, C. N., Cheng, Z. H., Meng, H. W., Zhang, M. R., and Zhang, H. J. (2014a). Soil chemical property changes in eggplant/garlic relay intercropping systems under continuous cropping. PLoS ONE 9:e111040. doi: 10.1371/journal.pone.0111040
Wang, Q., Garrity, M. G., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, Z. G., Jin, X., Bao, X. G., Li, X. F., Zhao, J. H., Sun, J. H., et al. (2014b). Intercropping enhances productivity and maintains the most soil fertility properties relative to sole cropping. PLoS ONE 9:e113984. doi: 10.1371/journal.pone.0113984
Whelan, J. A., Russell, N. B., and Whelan, M. A. (2003). A method for the absolute quantification of cDNA using real-time PCR. J. Immunol. Methods 278, 261–269. doi: 10.1016/S0022-1759(03)00223-0
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky and T. J. White (New York, NY: Academic Press), 315–322.
Wu, H. S., Yang, X. N., Fan, J. Q., Miao, W. G., Ling, N., Xu, Y. C., et al. (2009). Suppression of Fusarium wilt of watermelon by a bio-organic fertilizer containing combinations of antagonistic microorganisms. BioControl 54, 287–300. doi: 10.1007/s10526-008-9168-7
Xu, W. H., Liu, D., Wu, F. Z., and Liu, S. W. (2015). Root exudates of wheat are involved in suppression of Fusarium wilt in watermelon in watermelon-wheat companion cropping. Eur. J. Plant Pathol. 141, 209–216. doi: 10.1007/s10658-014-0528-0
Xu, W. H., Wu, F. Z., Chang, C. L., Liu, S. W., and Zhou, Y. (2013). Effects of wheat as companion cropping on growth, soil enzymes and disease resistance of watermelon. Allelopathy J. 32, 267–278.
Yao, H. Y., Jiao, X. D., and Wu, F. Z. (2006). Effects of continuous cucumber cropping and alternative rotations under protected cultivation on soil microbial community diversity. Plant Soil 284, 195–203. doi: 10.1007/s11104-006-0023-2
Yu, J. Q., Shou, S. Y., Qian, Y. R., Zhu, Z. Z., and Hu, W. H. (2000). Autotoxic potential of cucurbit crops. Plant Soil 223, 147–151. doi: 10.1023/A:1004829512147
Zhang, H., Mallik, A., and Zeng, R. S. (2013). Control of panama disease of banana by rotating and intercropping with Chinese chive (Allium Tuberosum Rottler): role of plant volatiles. J. Chem. Ecol. 39, 243–252. doi: 10.1007/s10886-013-0243-x
Zhang, Z. G., Zhang, J. G., Wang, Y. C., and Zheng, X. B. (2005). Molecular detection of Fusarium oxysporum f. sp. niveum and Mycosphaerella melonis in infected plant tissue and soil. FEMS. Microbiol. Lett. 249, 39–47. doi: 10.1016/j.femsle.2005.05.057
Keywords: watermelon, D123 wheat, companion crop, Fusarium oxysporum f. sp. niveum, microbial biomass, soil enzyme activity, microbial community
Citation: Xu W, Wang Z and Wu F (2015) The effect of D123 wheat as a companion crop on soil enzyme activities, microbial biomass and microbial communities in the rhizosphere of watermelon. Front. Microbiol. 6:899. doi: 10.3389/fmicb.2015.00899
Received: 07 June 2015; Accepted: 17 August 2015;
Published: 01 September 2015.
Edited by:
Essaid Ait Barka, Reims University, FranceReviewed by:
Muthu Venkateshwaran, University of Wisconsin-Platteville, USARaffaella Balestrini, Consiglio Nazionale delle Ricerche, Italy
Copyright © 2015 Xu, Wang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fengzhi Wu, Department of Horticulture, College of Horticulture, Northeast Agricultural University, Harbin, China,Znp3dTIwMDZAYWxpeXVuLmNvbQ==
†These authors have contributed equally to this work and are co-first authors.
 Weihui Xu
Weihui Xu Zhigang Wang
Zhigang Wang Fengzhi Wu
Fengzhi Wu