- Institute of Technical Biochemistry, Faculty of Biotechnology and Food Sciences, Lodz University of Technology, Lodz, Poland
Multiple studies confirm laccase role in fungal pathogenicity and lignocellulose degradation. In spite of broad genomic research, laccases from plant wilt pathogen Fusarium oxysporum are still not characterized. The study aimed to identify F. oxysporum genes that may encode laccases sensu stricto and to characterize the proteins in silico in order to facilitate further research on their impact on the mentioned processes. Twelve sequenced F. oxysporum genomes available on Broad Institute of Harvard and MIT (2015) website were analyzed and three genes that may encode laccases sensu stricto were found. Their amino acid sequences possess all features essential for their catalytic activity, moreover, the homology models proved the characteristic 3D laccase structures. The study shades light on F. oxysporum as a new source of multicopper oxidases, enzymes with possible high redox potential and broad perspective in biotechnological applications.
Introduction
Fusarium oxysporum is a known plant wilt pathogen that attacks various crops such as corn, tomato, or banana. Most of the research involving these ascomycetes is focused on understanding of its virulence and prevention of plants’ devastation. The Broad Institute proceeds with Fusarium Comparative project and now 12 genomes of sequenced F. oxysporum strains are available on their website. Despite of broad genomic studies, still not much is understood about its proteome and in particular about its secreted enzymes. Extracellular laccases are one of the factors pointed out to be responsible for fungal virulence, however, so far only single genes were investigated, but now, with so many genomes available, broader analysis is possible (Ma et al., 2010; Broad Institute of Harvard and MIT, 2015).
Laccases (EC 1.10.3.2) are oxidoreductases from multicopper oxidases family that catalyze 4-electron reduction of O2 to water with simultaneous oxidation of organic substrates. These multicopper oxidases possess wide range of specificity and are able to oxidize both phenolic substances and non-phenolic ones with presence of mediators. Nowadays laccases are one of the most desired biocatalysts – they catalyze wide range of chemical reactions, thus can be applied in various industrial areas – pulp and paper, textile, food industries, diagnostics, waste water treatment, and chemical compounds synthesis. Their environmental friendly nature continuously focuses attention on the enzyme (Alcalde, 2007; Kunamneni et al., 2008; Madhavi and Lele, 2009; Giardina et al., 2010; Pezzella et al., 2015).
There are certain characteristics that distinguish laccases from other multicopper oxidases. Firstly, they must contain multicopper oxidase domains (Pfam: PF00394, PF07731, and PF07732), in fungi most of them contain all three domains. Moreover, signatures of laccases were distinguished – four 8–24 amino acid sequences characteristic for these proteins, called L1, L2, L3, and L4. These signatures comprise amino acids that coordinate copper atoms in T1 and T2/T3 centers. Two serines and one arginine in the signatures form SDS gate, a channel responsible for proton transfer. What is more, it is said that the laccases from ascomycetes should contain DSG[ILV] on the C-terminus (Kumar et al., 2003; Cázares-García et al., 2013).
In the past, 15 potential laccase genes were discovered in F. oxysporum f. sp. lycopersici 4287 and few of them studied in the aspect of pathogenicity (Cañero and Roncero, 2008; Reyes-Medina and Macías-Sánchez, 2015). Our study aimed to identify and analyze potential laccases sensu stricto in all sequenced F. oxysporum strains. Redundancy analysis of all multicopper oxidases was done and sequence, structure, and phylogenetic studies were performed on the chosen proteins.
Materials and Methods
Redundancy Analysis
All DNA and protein sequences used in the study were obtained from “Fusarium Comparative Sequencing Project, Broad Institute of Harvard and MIT (2015). The proteins that possess Cu-oxidase, Cu-oxidase2, or Cu-oxidase3 (Pfam: PF00394, PF07731, and PF07732) were selected using tools available on the website. The protein sequences were downloaded and subjected to further analysis (Step 1). ScanProsite was used to select proteins with certain sequence features (de Castro et al., 2006). In the following step (Step 2), Prosite patterns and L1 and L3 signatures were found. In the third step, only those proteins who possess all four L1–L4 signatures and MCO1 and MCO2 patterns were chosen (Step 3). Table 1 gathers the patterns used for our search.
All protein sequences in the sequential steps were divided into groups on the basis of sequence identity. The member of a given group must have been identical in more than 95%. It was assessed by multiple sequence alignment, tree building and identity analysis. The following tools were used: ClustalW2, Mega6, sequence identity and similarity (SIAS), interactive Tree Of Life (iTOL), and CLC Sequence Viewer.
Analysis of Gene and Protein Sequences
Gene localization on chromosomes were visualized by Ensembl website tools.
Subcellular localization of proteins were predicted by SignalP 4.0, TMHMM2.0, and Phobius (default settings; Käll et al., 2004; Petersen et al., 2011; TMHMM Server, v. 2.0, 2015). Isoelectric points and molecular weight were analyzed by ExPASy – Compute pI/Mw Tool (2015). NetNGlyc 1.0 Server (2015; ExPASy – Compute pI/Mw Tool, 2015) was used to find possible glycosylation sites. The localization of the domains were found by Pfam and the logo of L1–L4 sequences were obtained in Meme (Bailey et al., 2009; Finn et al., 2014). The dendrograms were created in Mega6 and edited in iTOL (Letunic and Bork, 2007; Tamura et al., 2013). The sequences were aligned by ClustalW2 program and subjected to the tree building process with Maximum Likelihood method by Mega6. The comparison of four laccase-encoding genes was done with K2+G model (Mega6 model test), while the strain tree was done on nucleotide sequence of laccase orthologs (genes encoding for Gr1 and Gr5 proteins) with T92+G model (1000 bootstrap). The orthologs were found by Proteinortho v.5.11 (Lechner et al., 2011) and the chosen genes had the algebraic connectivity of 0.915. Sequence similarities were assessed by SIAS (2015).
Modeling
Homology models were obtained by I-Tasser (Yang et al., 2015), the analysis of the models were done in pyMOL, ModFOLD4, and WinCoot (McGuffin et al., 2013).
Results and Discussion
Redundancy Analysis
Over 300 protein sequences that possess Cu-oxidase domains were downloaded from Broad Institute website. In the first step of the analysis, 214 sequences with MCO1 and MCO2 were retained. The sequences were divided into 20 groups, additionally there were 9 proteins that could not be included in neither of the groups as their identity was much below 95%. In the next step of the analysis, only those proteins were taken into account in which L1 and L3 could be found. Those 80 proteins were divided into 5 groups. Next, the group which does not have the L2 and L4 signature was excluded (Gr3). Deeper analysis was done with proteins from the Step 2.
Molecular Characteristics
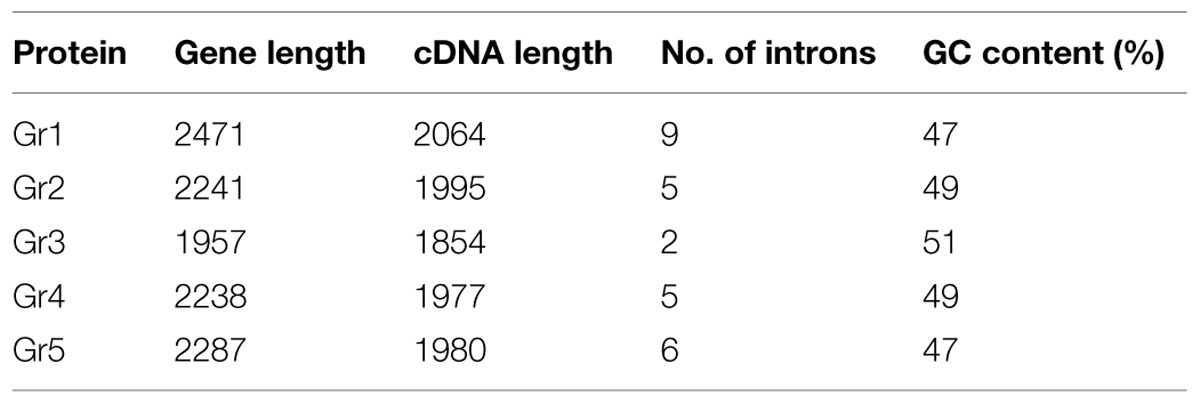
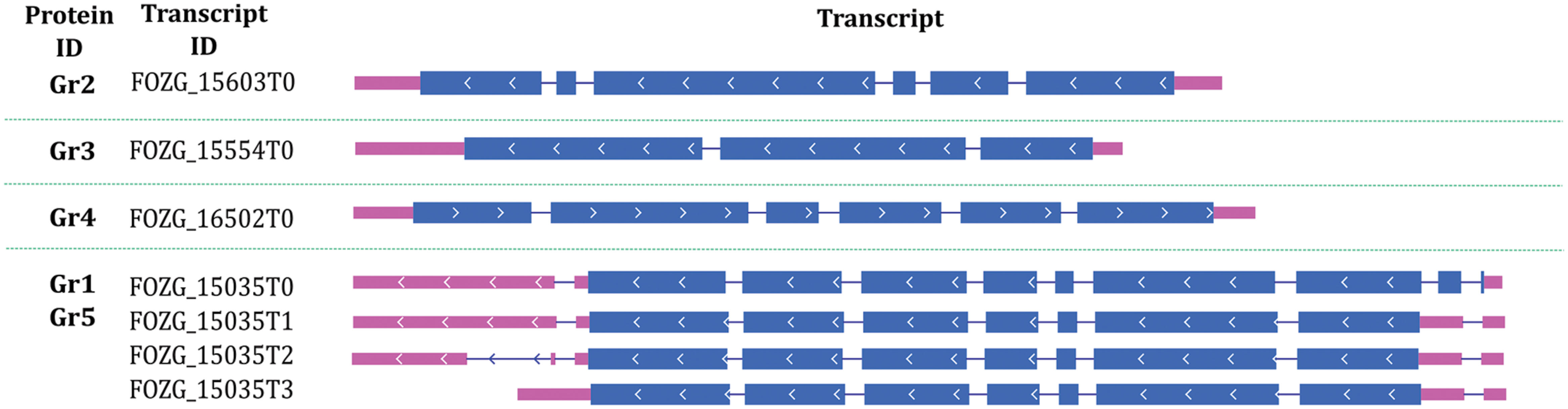
The length of F. oxysporum potential laccase genes varies between 1957 and 2471 nucleotides, while GC content of the genes ranges between 47 and 51% (Table 2). There are 2–9 introns in individual genes, which are not distributed similarly, thus division into subgroups within the species cannot be done as proposed in earlier scientific work (Kilaru et al., 2006; Cázares-García et al., 2013). We may observe that Gr3 gene is far distinct from other, which explains the exclusion of Gr3 in the last step of the analysis. Gene encoding for potential laccase Gr1 is the same gene that encodes for Gr5, although with an additional sequence on 5′ end (Figure 1).
Annotations of F. oxysporum strains genomes are in progress, so far one of 12 genomes is assembled in chromosomes. All potential multicopper oxidase-encoding genes are far from each other and are located on different chromosomes (Figure 2). Similar situation was observed before, for example in the genome of Trichoderma species or Laccaria bicolor; however, clusters of laccase genes were also discovered before (Courty et al., 2009).
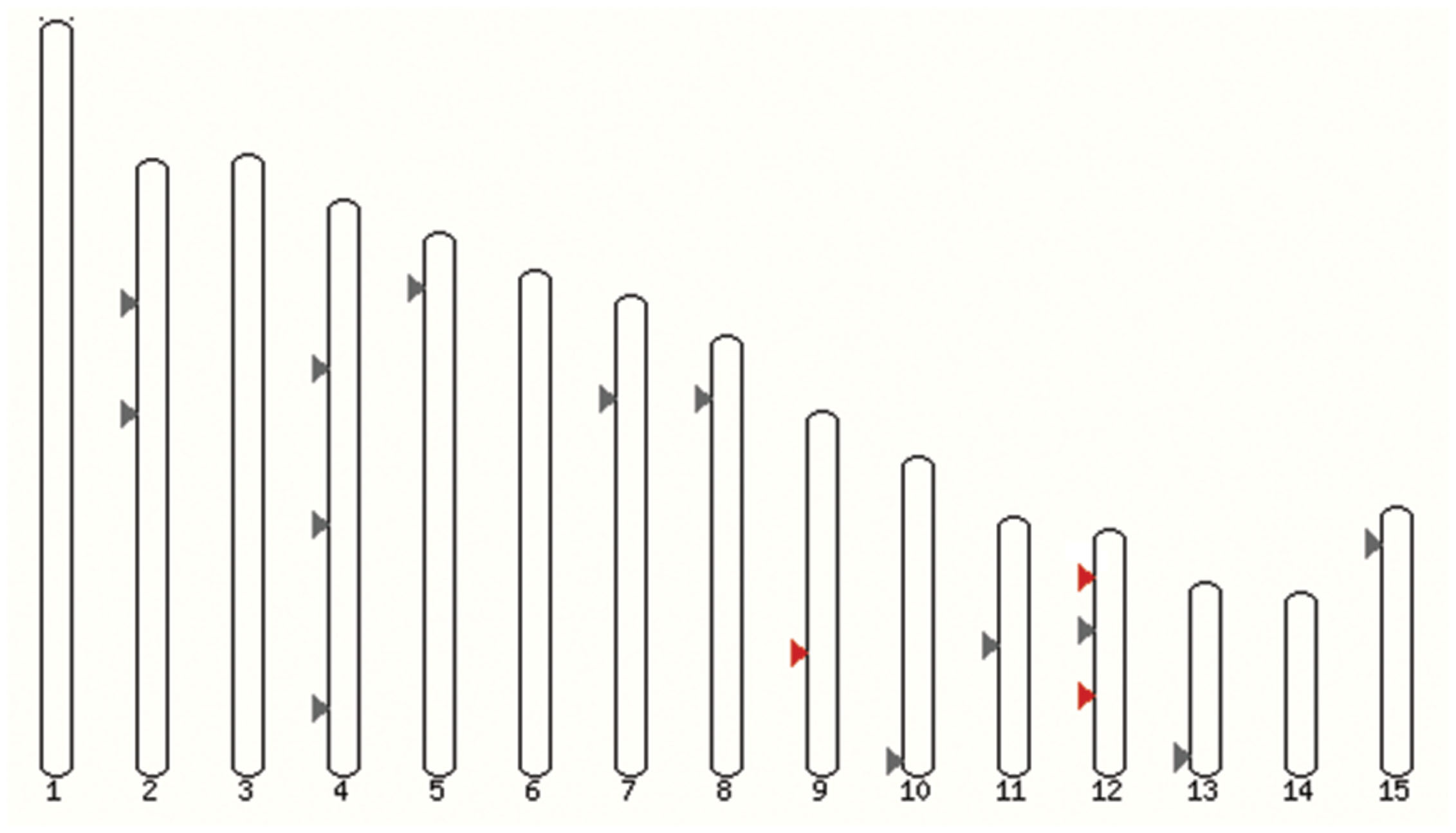
FIGURE 2. Localization of laccase genes in F. oxysporum f. sp. lycopersici 4287 genome. Grey, all multicopper oxidases genes; red, FOXG_12706 (Chr9), FOXG_13227 (Chr12), and FOXG_14565 (Chr12) – laccases sensu stricto (Ensembl).
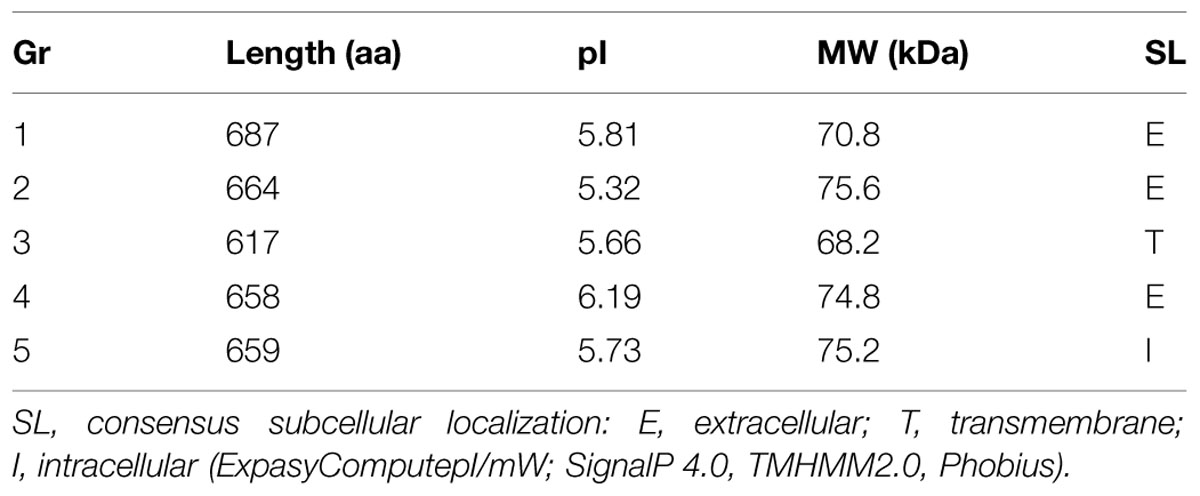
The length of chosen proteins varies between 617 and 687 amino acids (Table 3), which is atypical for fungal laccases (normally 500–600 amino acids). Putative laccases from F. oxysporum seem to have 68–76 kDa, while the typical weight of these protein ranges between 60 and 70 kDa; however, laccases of 70–80 kDa were also reported (Slomczynski et al., 1995; Eggert et al., 1996; Chefetz et al., 1998; Madhavi and Lele, 2009).
The consensus subcellular localization was determined for all five groups of laccases using SignalP 4.0, TMHMM2.0, and Phobius. Gr1, 2 are considered to be extracellular laccases (0.480 and 0.514, respectively, according to SignalP); moreover, Gr4 could also be an extracellular protein (0.439). Gr5 must be an intracellular laccase, this protein possess identical amino acid sequences to Gr1, however, without the first 28 amino acids, which is a further sign of the extracellular nature of Gr1 laccase. The putative signal sequence of extracellular laccases were about 25–26 amino acid long. N-glycosylation were predicted by NetNGlyc 1.0. All of them seems to have 11–13 Asn–Xaa–Ser/Thr sequons.
Three Cu-oxidase domains are present in proteins from Gr1–5. The first domain from N-terminus is Cu-oxidase3, then Cu-oxidase and the closest to C-termini Cu-oxidase2, which is characteristic for ascolaccases. Although the proteins from Step 1 possess all three Cu-oxidase domains, their localization on the sequence varies.
All proteins from Step 2 and 3 possess L1–L4 laccase signatures; however, Gr3 is relatively distinct from the others. The signatures of laccases from Step 3 are far more conservative than those proposed earlier (Kumar et al., 2003; Cázares-García et al., 2013), in L1 17/24aa are conserved in L2 20/22, in L3 7/9, and in L4 15/22 (Figure 3). The signatures are composed of the most important amino acids of laccases, those that coordinates copper atoms. Multiple sequence alignment of chosen protein further reveals differences between Gr3 and other sequences (Figure 4). Gr3 somehow lacks the C-termini characteristic for GR1, 2, 4, 5, which signifies that either the D residue forming SDS gate is located in different position or the proton transfer in this protein would function distinctly.
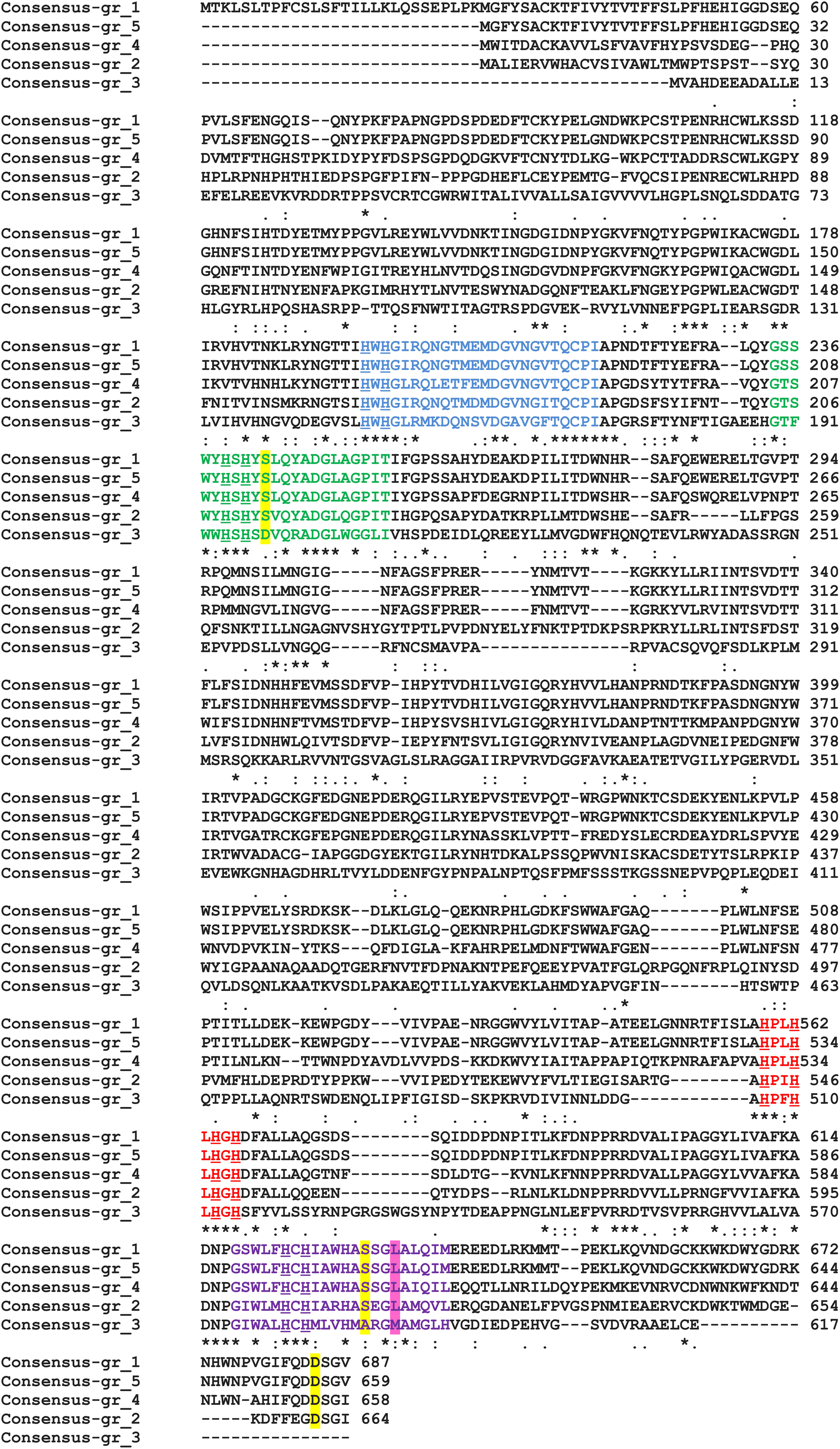
FIGURE 4. MSA of five chosen enzymes performed by ClustalW2. L1, blue; L2, green; L3, red; and L4, violet. H residues involved in copper binding – underlined, SDS-gate – yellow shaded, axial coordination – pink shaded. An asterisk indicates that the residus in a given position are identical, the colon indicates conserved substitutions while a dot signifies semiconserved substitutions.
Axial coordination is an important feature that affects redox potential of laccases (Hakulinen et al., 2002; Kumar et al., 2003; Garavaglia et al., 2004; Hakulinen and Rouvinen, 2015; Jones and Solomon, 2015; Mate and Alcalde, 2015). It was suggested that if laccase has Leu or Phe in around tenth position downstream the conserved Cys residue in L4, then the enzyme has a higher E0 –redox potential (700–800V), whereas the lower E0 is observed when Met is in this position. Laccases are classified into Lac1 (Met), Lac2 (Leu), and Lac3(Phe) on the basis of axial coordination. This would mean that the chosen putative laccases should be classified as Lac2 (Gr1, 2, 4, 5) proteins and should exhibit higher E0, while Gr3 should display a lower E0 (Lac1).
A classification with LccED database was performed with usage of a blast tool available on the website (default settings). With high probability Gr1, 2, 4, and 5 belong to H Family B1 (Ascomycetes-like MCO), while Gr4 is classified as H Family A2 (Basidiomycete laccases) (Sirim et al., 2011).
Multiple sequence alignments and dendrogram formation were essential for the redundancy analysis. The Figure 5 presents a dendrogram of all genes from Step 2 analysis, which visualize clearly the relationship between all four chosen genes. The identities between the 20 groups from Step 1 ranges between 20 and 97%. The groups with the highest identity are exactly Gr1 and Gr5 from Step 2 which differ only with the signal peptide in Gr1 protein.
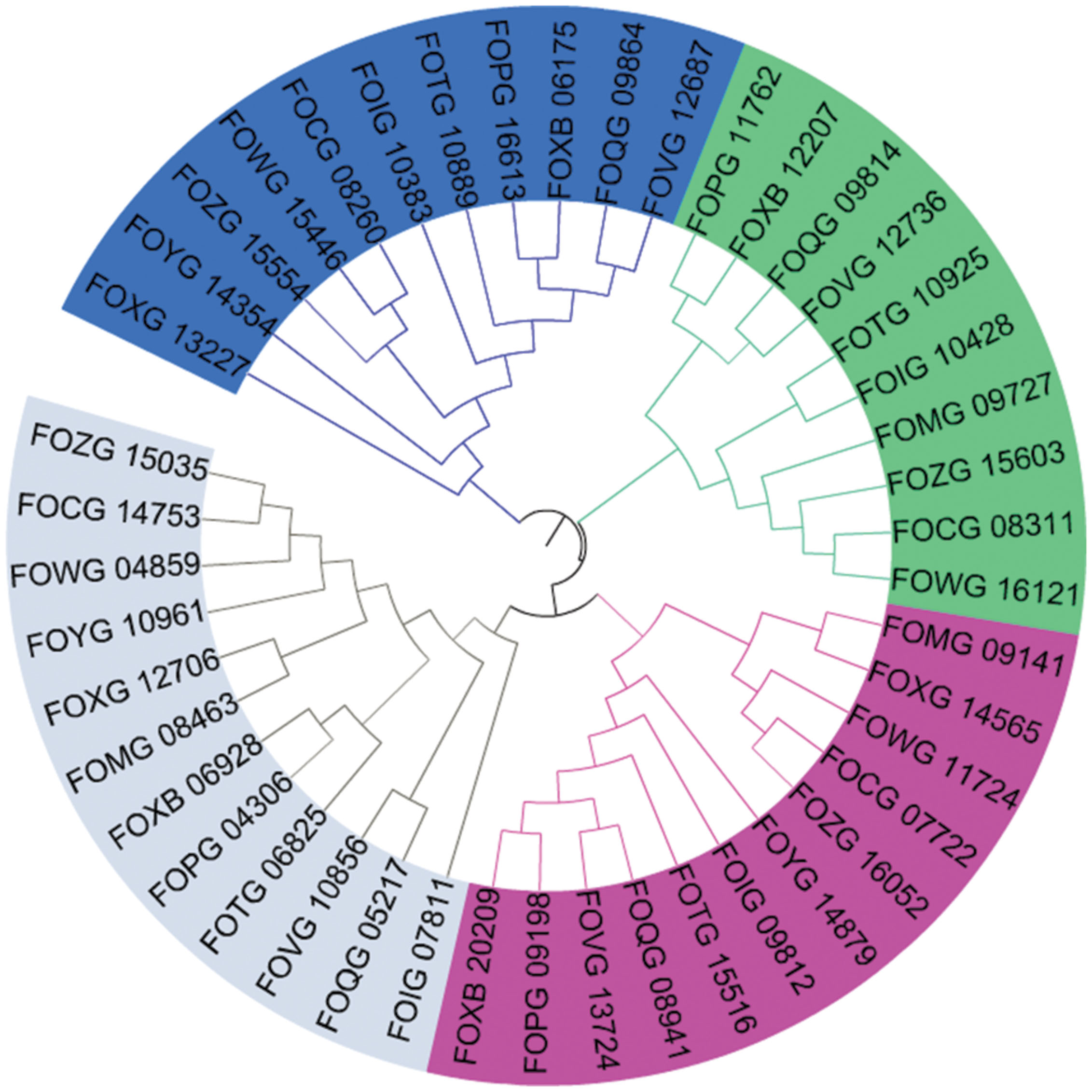
FIGURE 5. Phylogenetic tree of all 45 genes that may encode for laccases. The colors signify division to four groups: grey, Gr1 and Gr5; green, Gr2; blue, Gr3; and pink, Gr4 (Mega6, iTOL).
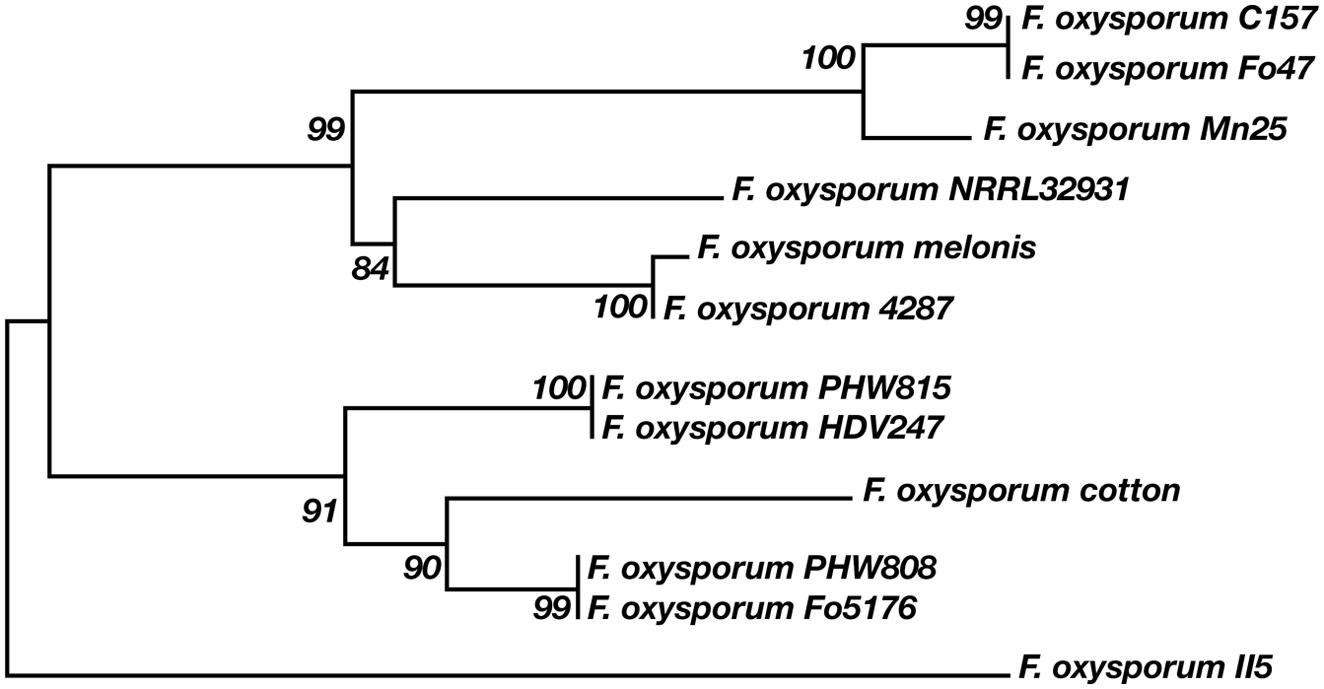
According to our analysis, the 12 F. oxysporum strains possess in minimum 16–21 potential multicopper oxidase genes and at least two putative laccase genes (Figure 6). Twelve F. oxysporum strains are similar in context of laccase genes; however, three of them seem to diverge evolutionary from the rest. F. oxysporum melonis does not contain a gene encoding for the putative transmembrane Gr3, while F. oxysporum f. sp. lycopersici4287 lost three putative laccase genes. The human pathogen F. oxysporum NRRL32931 lacks Gr1- and Gr2-encoding genes, those that may encode for extracellular proteins, probably due to the fact that it does not have to deal with lignocellulose during its virulence. These three F. oxysporum strains form a subgroup on a strain tree presented in the Figure 7. The mentioned subgroup is a part of one of three clades that forms the strain tree. Interestingly, the gene from F. oxysporum II5, pathogenic toward banana, has the highest number of mutations and was not included in a clade with other strains. F. oxysporum II5 as the only strain that was isolated in Indonesia, which climate differ significantly from weather conditions in Europe (Fo47, PHW815), America (Cl57, Mn25, melonis, PHW808), or Australia (HDV247). The climate may be an important factor for strain evolution.
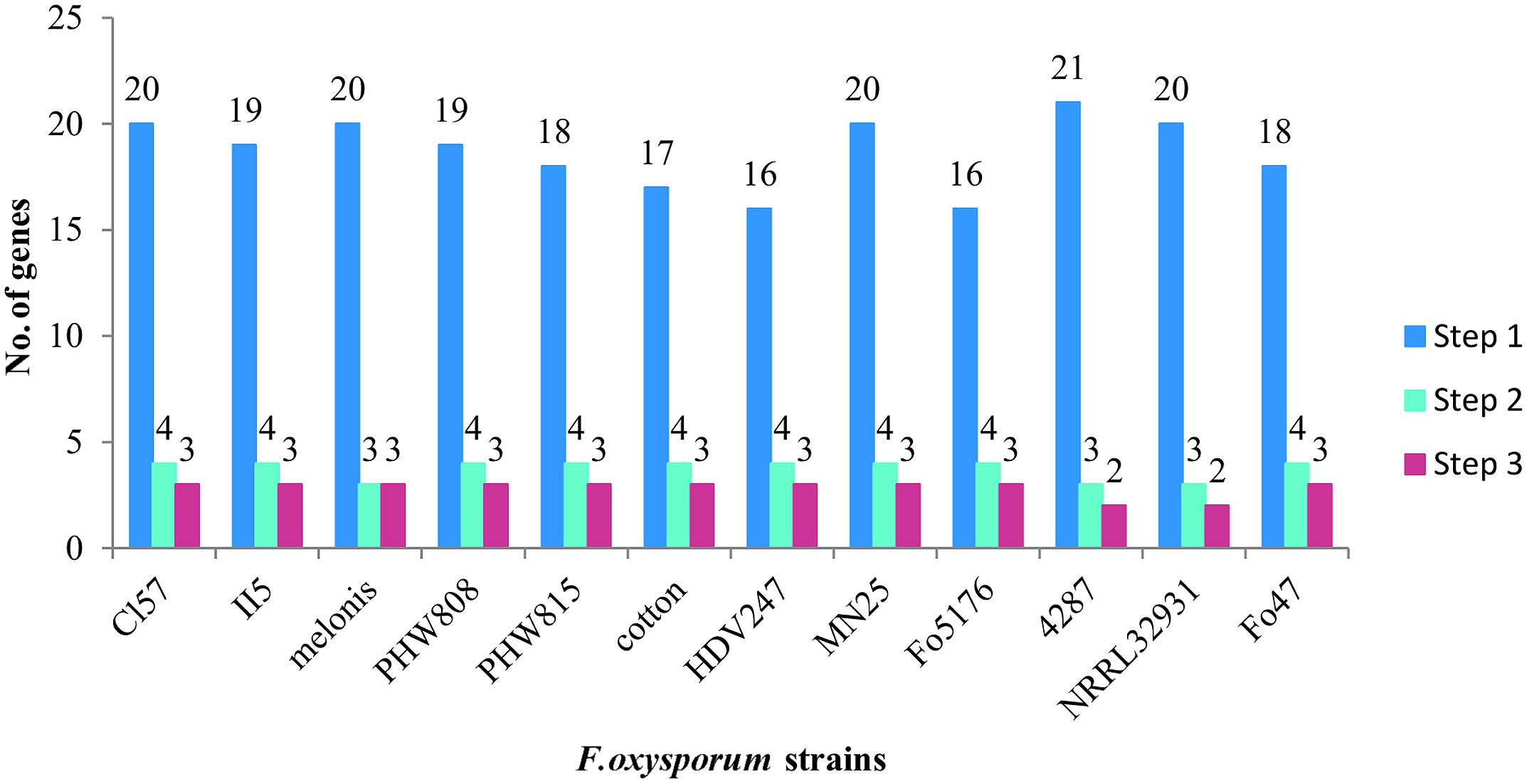
FIGURE 6. Number of genes coding for multicopper oxidases (Step 1), multicopper oxidases with certain features of laccases (Step 2), and laccases sensu stricto (Step 3).
Modeling
Three homologous models of potential laccases were successfully created by I-Tasser server for Gr2, 3, and 4; Gr1 and 5 models were built with an unsatisfactory effect. For protein Gr2 and 4 a potential signal peptides were removed and the modeling was done again which led to better results. This is a further sign of the extracellular nature of Gr2 and 4 proteins. The models are assessed by ModFOLD4 server as good models with less that 1:1000 chance that the models are incorrect. All three models have problematic region on their N-termini. It may suggest that this region may be cleft off during maturation of the protein (pro-peptide) and that is why the I-Tasser server could not find a match in the PDB database. Such pro-sequences important in protein expression were detected for example in Melanocarpus albomyces laccase (Bulter et al., 2003; Kiiskinen and Saloheimo, 2004; Kiiskinen et al., 2004). However, ProP1 server did not discover pro-peptide cleavage sites in these proteins. The Ramachandran plots for the models show a relatively high number of amino acids in disallowed regions (∼17%), which is an indication for further model optimizations.
It was suggested that the 3D position of C-terminus of laccase is important for its activity. C-terminus may act as a plug that obturates the trinuclear (T2/T3) channel, thus preventing oxygen to enter the channel and water to exit it. The trinuclear channel is widely regarded as the oxygen channel, however, a mutant of M. albomyces laccase (PDB code 3QPK) suggest a different oxygen route. The models of Gr2 and 4 reveals similar location of C-terminus as observed in M. albomyces laccase (2Q90; Kiiskinen and Saloheimo, 2004; Bleve et al., 2013, 2014). Such a conformation was proposed to be a common feature of ascolaccases. However, the amino acid sequences of Gr2 and 4 lacks the additional 14 amino acids upstream DSG[LIV] motif which is present in M. albomyces laccase. It is hypothesized that incorrect processing of this sequence leads to lack of enzyme activity. Among ascomycetes many laccases share similar C-terminal sequence with M. albomyces, which is cleft on a conserved site, while other are similar to our proteins and their sequence is finished with DSG[LIV] motif (Botrytis cinerea, Trichoderma reesei). It was suggested many times that DSG[LIV] is a common conservative motif for all ascomycetes laccase, which is a further reason for excluding Gr3 protein from laccases sensu stricto group (Andberg et al., 2009; Cázares-García et al., 2013). The studies on M. albomyces laccase proved that the deletion of DSG[LIV] inactivates the enzyme, whereas the point mutation in this region leads to lower thermostability, turnover number and the structural changes in T2 centre.
All modeled proteins hold characteristic 3D architecture. The domains are arranged in a typical greek-key motifs, the L1–L4 signatures encircle Cu atoms. A mononuclear site is located in Cu-oxidase2 domain (third domain from N-terminus), while trinuclear site is between Cu-oxidase3 and Cu-oxidase2 domains (first and third domains from N-terminus) (Hakulinen and Rouvinen, 2015; Pardo and Camarero, 2015). The architecture of Cu centers is similar to those in other studied laccases. T1 copper ion is coordinated by two His and one Cys residue, while T2/T3 center by eight His residues (Figures 8 and 9). The substrate binding site is located close to the mononuclear site, close to Cu-oxidase3 domain, whereas oxygen binding pocket is situated at the trinuclear copper site.
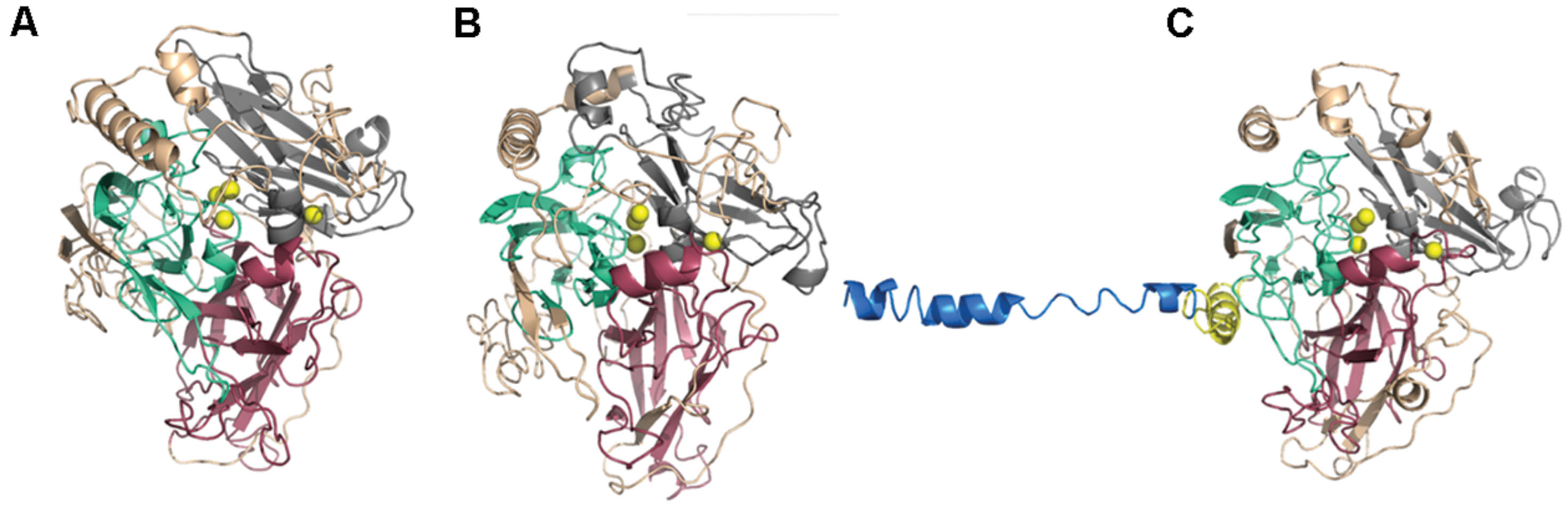
FIGURE 8. Gr2 (A), Gr4 (B), and Gr3 (C) models created by I-Tasser. Pink, Cu-oxidase domain; grey, Cu-oxidase2 domain; cyan, Cu-oxidase3 domain; and Cu atoms (yellow) in an approximate location. Yellow cartoon in (C) represents transmembrane region of the protein, blue cartoon represents the outside part.
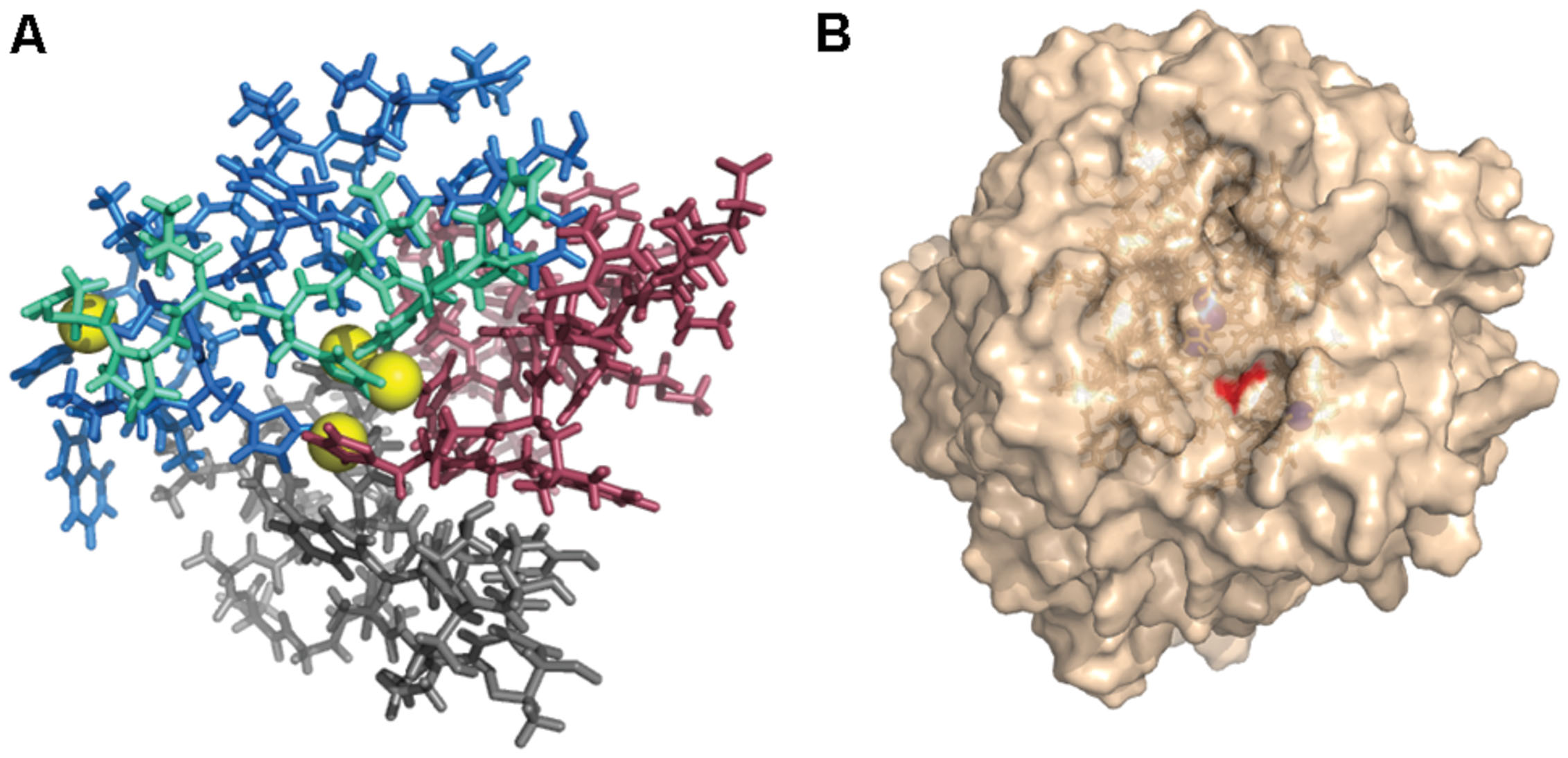
FIGURE 9. (A) L1–L4 signatures encirculating Cu atoms (yellow) (Gr4 model). Pink, L1; grey, L2; cyan, L3; and blue, L4. (B) Surface of Gr4 model with DSGI C-terminus sequence shown in red. Cu atoms shown in blue (approximate location).
Summary
Four proteins extracted in in silico analysis possess all characteristics of laccases sensu stricto. Their sequences contain L1–L4 signatures, three Pfam domains and conserved C-terminus. Their structure, according to homologous models, is similar to structures of known laccases. The rest of over 300 genes were not assigned as laccase-encoding genes due to the lack of mentioned sequences. However, further analysis of the proteins is important in their assessment to specific enzyme groups.
Genome-wide structural and phylogenetic in silico analysis of laccase genes in a plant pathogen is essential for further research on the virulence of the fungi. The determination of laccases sensu stricto may help, among others, in planning of knock-out studies in order to confirm or deny the role of laccase in pathogenicity. Six potential laccase gene were studied by a Spanish team for its role in virulence in tomato (Cañero and Roncero, 2008). Three of them were detected by RT-PCR as expressed during plant infection. Lcc1(EF990894.1), lcc3(EF990899.1), and lcc5(EF990897.1) lacking mutants were created and checked for virulence in root infection assays, leading to the same results as for the wild type strain. However, this negative results does not mean that laccases do not take part in the pathogenicity of F. oxysporum. The mentioned putative laccases were not included in our Step 2 and Step 3 analysis because of the lack of as conservative L1–L4 signatures as proposed earlier, thus they may not be laccases sensu stricto. Further research is essential for full understanding of laccase role in pathogenicity.
Laccase is nowadays a popular enzyme in many biotechnological applications, scientists and industrial partners still work hard to find laccases with better catalytic efficiencies, hence discovery of novel enzymes is needed. The presented putative laccases, to our knowledge, do not have very close homologs already used in industrial applications. Blastp (default settings) analysis of consensus sequences did not reveal any studied laccases that are identical at least in 50%. This and the fact that these enzymes may have high redox potential due to leucine in axial coordination make the effort of their study more meaningful in the perspective of biotechnological applications.
Funding
Project realized at Institute of Technical Biochemistry and founded by Ph.D. Funds from the Ministry of Sciene and Higher Education Government of RP and Institute of Technical Biochemistry funds.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Katarzyna Kubiak, Ph.D. and Marzena Jȩdrzejczak-Krzepkowska, Ph.D. for helpful discussions. Moreover, we would like to acknowledge Klaudia Jadczak for assistance in the analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00933
References
Alcalde, M. (2007). “Laccases: biological functions, molecular structure and industrial applications,” in Industrial Enzymes, eds J. Polaina and A. P. MacCabe (Berlin: Springer), 461–476. Available at: http://link.springer.com/chapter/10.1007/1-4020-5377-0_26 [Accessed April 16, 2015].
Andberg, M., Hakulinen, N., Auer, S., Saloheimo, M., Koivula, A., Rouvinen, J., et al. (2009). Essential role of the C-terminus in Melanocarpus albomyces laccase for enzyme production, catalytic properties and structure. FEBS J. 276, 6285–6300. doi: 10.1111/j.1742-4658.2009.07336.x
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bleve, G., Lezzi, C., Spagnolo, S., Rampino, P., Perrotta, C., Mita, G., et al. (2014). Construction of a laccase chimerical gene: recombinant protein characterization and gene expression via yeast surface display. Appl. Biochem. Biotechnol. 172, 2916–2931. doi: 10.1007/s12010-014-0734-4
Bleve, G., Lezzi, C., Spagnolo, S., Tasco, G., Tufariello, M., Casadio, R., et al. (2013). Role of the C-terminus of Pleurotus eryngii Ery4 laccase in determining enzyme structure, catalytic properties and stability. Protein Eng. Des. Sel. PEDS 26, 1–13. doi: 10.1093/protein/gzs056
Broad Institute of Harvard and MIT. (2015). Fusarium Comparative Sequencing Project. Available at: http://www.broadinstitute.org/.
Bulter, T., Alcalde, M., Sieber, V., Meinhold, P., Schlachtbauer, C., and Arnold, F. H. (2003). Functional expression of a fungal laccase in Saccharomyces cerevisiae by directed evolution. Appl. Environ. 69, 987. doi: 10.1128/AEM.69.2.987-995.2003
Cañero, D. C., and Roncero, M. I. G. (2008). Functional analyses of laccase genes from Fusarium oxysporum. Phytopathology 98, 509–518. doi: 10.1094/PHYTO-98-5-0509
Cázares-García, S. V., Vázquez-Garcidueñas, S., and Vázquez-Marrufo, G. (2013). Structural and phylogenetic analysis of laccases from Trichoderma: a bioinformatic approach. PLoS ONE 8:e55295. doi: 10.1371/journal.pone.0055295
Chefetz, B., Chen, Y., and Hadar, Y. (1998). Purification and characterization of laccase from Chaetomium thermophilium and its role in humification. Appl. Environ. Microbiol. 64, 3175–3179.
Courty, P. E., Hoegger, P. J., Kilaru, S., Kohler, A., Buée, M., Garbaye, J., et al. (2009). Phylogenetic analysis, genomic organization, and expression analysis of multi-copper oxidases in the ectomycorrhizal basidiomycete Laccaria bicolor. New Phytol. 182, 736–750. doi: 10.1111/j.1469-8137.2009.02774.x
de Castro, E., Sigrist, C. J. A., Gattiker, A., Bulliard, V., Langendijk-Genevaux, P. S., Gasteiger, E., et al. (2006). ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–W365. doi: 10.1093/nar/gkl124
Eggert, C., Temp, U., and Eriksson, K. E. (1996). The ligninolytic system of the white rot fungus Pycnoporus cinnabarinus: purification and characterization of the laccase. Appl. Environ. Microbiol. 62, 1151–1158.
ExPASy – Compute pI/Mw Tool. (2015). Available at: http://web.expasy.org/compute_pi/ [Accessed July 19, 2015].
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Garavaglia, S., Cambria, M. T., Miglio, M., Ragusa, S., Iacobazzi, V., Palmieri, F., et al. (2004). The structure of Rigidoporus lignosus Laccase containing a full complement of copper ions, reveals an asymmetrical arrangement for the T3 copper pair. J. Mol. Biol. 342, 1519–1531. doi: 10.1016/j.jmb.2004.07.100
Giardina, P., Faraco, V., Pezzella, C., Piscitelli, A., Vanhulle, S., and Sannia, G. (2010). Laccases: a never-ending story. Cell. Mol. Life Sci. CMLS 67, 369–385. doi: 10.1007/s00018-009-0169-161
Hakulinen, N., Kiiskinen, L.-L., Kruus, K., Saloheimo, M., Paananen, A., Koivula, A., et al. (2002). Crystal structure of a laccase from Melanocarpus albomyces with an intact trinuclear copper site. Nat. Struct. Biol. 9, 601–605. doi: 10.1038/nsb823
Hakulinen, N., and Rouvinen, J. (2015). Three-dimensional structures of laccases. Cell. Mol. Life Sci. CMLS 72, 857–868. doi: 10.1007/s00018-014-1827-1825
Jones, S. M., and Solomon, E. I. (2015). Electron transfer and reaction mechanism of laccases. Cell. Mol. Life Sci. CMLS 72, 869–883. doi: 10.1007/s00018-014-1826-1826
Käll, L., Krogh, A., and Sonnhammer, E. L. L. (2004). A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036. doi: 10.1016/j.jmb.2004.03.016
Kiiskinen, L.-L., Kruus, K., Bailey, M., Ylösmäki, E., Siika-Aho, M., and Saloheimo, M. (2004). Expression of Melanocarpus albomyces laccase in Trichoderma reesei and characterization of the purified enzyme. Microbiol. Read. Engl. 150, 3065–3074. doi: 10.1099/mic.0.27147-27140
Kiiskinen, L.-L., and Saloheimo, M. (2004). Molecular Cloning and expression in Saccharomyces cerevisiae of a laccase gene from the ascomycete Melanocarpus albomyces. Appl. Environ. Microbiol. 70, 137–144. doi: 10.1128/AEM.70.1.137-144.2004
Kilaru, S., Hoegger, P. J., and Kües, U. (2006). The laccase multi-gene family in Coprinopsis cinerea has seventeen different members that divide into two distinct subfamilies. Curr. Genet. 50, 45–60. doi: 10.1007/s00294-006-0074-71
Kumar, S. V. S., Phale, P. S., Durani, S., and Wangikar, P. P. (2003). Combined sequence and structure analysis of the fungal laccase family. Biotechnol. Bioeng. 83, 386–394. doi: 10.1002/bit.10681
Kunamneni, A., Plou, F. J., Ballesteros, A., and Alcalde, M. (2008). Laccases and their applications: a patent review. Recent Pat. Biotechnol. 2, 10–24. doi: 10.2174/187220808783330965
Lechner, M., Findeiß, S., Steiner, L., Marz, M., Stadler, P. F., and Prohaska, S. J. (2011). Proteinortho: detection of (Co-)orthologs in large-scale analysis. BMC Bioinform. 12:124. doi: 10.1186/1471-2105-12-124
Letunic, I., and Bork, P. (2007). Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinforma. Oxf. Engl. 23, 127–128. doi: 10.1093/bioinformatics/btl529
Ma, L.-J., van der Does, H. C., Borkovich, K. A., Coleman, J. J., Daboussi, M.-J., Di Pietro, A., et al. (2010). Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464, 367–373. doi: 10.1038/nature08850
Madhavi, V., and Lele, S. S. (2009). Laccase: properties and applications. Bioresources 4, 1694–1717. doi: 10.15376/biores.4.4.1694-1717
Mate, D. M., and Alcalde, M. (2015). Laccase engineering: from rational design to directed evolution. Biotechnol. Adv. 33, 25–40. doi: 10.1016/j.biotechadv.2014.12.007
McGuffin, L. J., Buenavista, M. T., and Roche, D. B. (2013). The ModFOLD4 server for the quality assessment of 3D protein models. Nucleic Acids Res. 41, W368–W372. doi: 10.1093/nar/gkt294
NetNGlyc 1.0 Server. (2015). Available at: http://www.cbs.dtu.dk/services/NetNGlyc/ [Accessed July 19, 2015].
Pardo, I., and Camarero, S. (2015). Laccase engineering by rational and evolutionary design. Cell. Mol. Life Sci. CMLS 72, 897–910. doi: 10.1007/s00018-014-1824-1828
Petersen, T. N., Brunak, S., von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Pezzella, C., Guarino, L., and Piscitelli, A. (2015). How to enjoy laccases. Cell. Mol. Life Sci. CMLS 72, 923–940. doi: 10.1007/s00018-014-1823-1829
Reyes-Medina, M. A., and Macías-Sánchez, K. L. (2015). GTPase Rho1 regulates the expression of xyl3 and laccase genes in Fusarium oxysporum. Biotechnol. Lett. 37, 679–683. doi: 10.1007/s10529-014-1709-1709
SIAS. (2015). SEQUENCE IDENTITY AND SIMILARITY. Available at: http://imed.med.ucm.es/Tools/sias.html [Accessed July 19, 2015].
Sirim, D., Wagner, F., Wang, L., Schmid, R. D., and Pleiss, J. (2011). The laccase engineering database: a classification and analysis system for laccases and related multicopper oxidases. Database (Oxford) 2011:bar006. doi: 10.1093/database/bar006
Slomczynski, D., Nakas, J. P., and Tanenbaum, S. W. (1995). Production and characterization of laccase from Botrytis cinerea 61-34. Appl. Environ. Microbiol. 61, 907–912.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
TMHMM Server, v. 2.0. (2015). Available at: http://www.cbs.dtu.dk/services/TMHMM/ [Accessed July 19, 2015].
Keywords: laccase, multicopper oxidase, Fusarium oxysporum, pathogen, in silico modeling
Citation: Kwiatos N, Ryngajłło M and Bielecki S (2015) Diversity of laccase-coding genes in Fusarium oxysporum genomes. Front. Microbiol. 6:933. doi: 10.3389/fmicb.2015.00933
Received: 22 July 2015; Accepted: 24 August 2015;
Published: 10 September 2015.
Edited by:
Helio K. Takahashi, Universidade Federal de São Paulo, BrazilReviewed by:
Luciana Lopes Guimaraes, Universidade Santa Cecília, BrazilAparecida S. Tanaka, Universidade Federal de São Paulo, Brazil
Copyright © 2015 Kwiatos, Ryngajłło and Bielecki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Natalia Kwiatos, Institute of Technical Biochemistry, Faculty of Biotechnology and Food Sciences, Lodz University of Technology, Stefanowskiego 4/10, 90-924 Lodz, Poland,bmF0YWxpYS5rd2lhdG9zQGRva3QucC5sb2R6LnBs
 Natalia Kwiatos
Natalia Kwiatos Małgorzata Ryngajłło
Małgorzata Ryngajłło Stanisław Bielecki
Stanisław Bielecki