- 1Department of Medical, Oral and Biotechnological Sciences, “G. d'Annunzio” University of Chieti-Pescara, Chieti, Italy
- 2Aging Research Center (Ce.S.I.), “G. d'Annunzio” University Foundation, Chieti, Italy
- 3Department of Pharmacy, “G. d'Annunzio” University of Chieti-Pescara, Chieti, Italy
- 4Children's Hospital and Research Institute “Bambino Gesù,” Rome, Italy
The present study was undertaken in order to understand more about the interaction occurring between S. maltophilia and P. aeruginosa, which are frequently co-isolated from CF airways. For this purpose, S. maltophilia RR7 and P. aeruginosa RR8 strains, co-isolated from the lung of a chronically infected CF patient during a pulmonary exacerbation episode, were evaluated for reciprocal effect during planktonic growth, adhesion and biofilm formation onto both polystyrene and CF bronchial cell monolayer, motility, as well as for gene expression in mixed biofilms. P. aeruginosa significantly affected S. maltophilia growth in both planktonic and biofilm cultures, due to an inhibitory activity probably requiring direct contact. Conversely, no effect was observed on P. aeruginosa by S. maltophilia. Compared with monocultures, the adhesiveness of P. aeruginosa on CFBE41o- cells was significantly reduced by S. maltophilia, which probably acts by reducing P. aeruginosa's swimming motility. An opposite trend was observed for biofilm formation, confirming the findings obtained using polystyrene. When grown in mixed biofilm with S. maltophilia, P. aeruginosa significantly over-expressed aprA, and algD—codifying for protease and alginate, respectively—while the quorum sensing related rhlR and lasI genes were down-regulated. The induced alginate expression by P. aeruginosa might be responsible for the protection of S. maltophilia against tobramycin activity we observed in mixed biofilms. Taken together, our results suggest that the existence of reciprocal interference of S. maltophilia and P. aeruginosa in CF lung is plausible. In particular, S. maltophilia might confer some selective “fitness advantage” to P. aeruginosa under the specific conditions of chronic infection or, alternatively, increase the virulence of P. aeruginosa thus leading to pulmonary exacerbation.
Introduction
Pulmonary disease is the leading cause of morbidity and mortality in cystic fibrosis (CF) patients, in whom defective mucociliary clearance and impaired innate immunity lead to chronic pulmonary infections (Lyczak et al., 2002). During CF airway disease periods of stability are punctuated by acute pulmonary exacerbations (PEs) in which overt immunological responses are the main causes of irreversible lung damage (Amadori et al., 2009; Sanders et al., 2011). Recurrent PEs are associated with a shortened survival (Emerson et al., 2002; Marshall, 2004).
The pathophysiology of PEs is not yet completely understood. Aside from acute viral infections which, especially in children, have been associated with up to one-third of PEs (Armstrong et al., 1998; Clifton et al., 2008), Pseudomonas aeruginosa has been considered the primary cause of PEs and related long-term decline in lung function (Goss and Burns, 2007; Sanders et al., 2010).
The pathogenesis of P. aeruginosa infection depends on several cell-associated and extracellular virulence factors, including proteases and toxins, whose expression is mainly regulated by hierarchically organized inter-bacterial communication LasRI and RhIRI quorum sensing (QS) systems, which monitor population size using various diffusible N-acylhomoserine lactones as signal molecules (Goodman and Lory, 2004).
However, this view is not convincingly supported by either the clinical or the microbiological evidence available.
Several studies have reported that in adult CF patients the anti-pseudomonal antibiotic therapy frequently did not reduce P. aeruginosa load and airways inflammation (Wolter et al., 1999; Reid et al., 2004), and that about 25% of patients experiencing a PE did not recover their baseline lung function after treatment, leading to a progressive deterioration in their clinical status over time (Sanders et al., 2010, 2011). Furthermore, in other studies no increase in P. aeruginosa concentration was observed immediately prior to, or at the time of, PE (Stressmann et al., 2011; Reid et al., 2013). Recent epidemiological data indicate that coinfections involving different species of bacteria are common, and probably represent the norm, in CF lung (Harrison, 2007; Bittar et al., 2008; Sibley et al., 2008a; Rogers et al., 2010a). P. aeruginosa is the most common species found in CF airways, but other species are frequently co-isolated in CF lung (Harrison, 2007).
The pathophysiology of PEs in CF could be, therefore, directly related to changes in microbial behavior and/or to the dynamics of the interactions between the constituents of the complex microbial communities present. Several studies have recently highlighted the potential role of interspecies interactions in influencing infection status, clinical outcomes or response to therapy in CF patients (Harrison, 2007; Ryan et al., 2008; Sibley et al., 2008b, 2009; Shank and Kolter, 2009; Rogers et al., 2010b). Taken together, these findings suggest that the role of microbial species other than P. aeruginosa needs to be considered.
Stenotrophomonas maltophilia is one of the most common emerging multi-drug resistant organisms found in the lungs of people with CF where its prevalence is increasing (Ciofu et al., 2013). Nevertheless, the role of S. maltophilia in the pathogenesis of CF lung disease is not yet clear because of conflicting results from clinical studies which focused on the correlation between the presence of this microorganism and lung damage (Karpati et al., 1994; Goss et al., 2002). In a series of studies, we found evidence highly suggestive of the pathogenic role of S. maltophilia in CF patients (Di Bonaventura et al., 2004, 2007a,b, 2010; Pompilio et al., 2008, 2010, 2011). This microorganism can grow as biofilms—sessile communities inherently resistant to antibiotics and host immune response—not only on abiotic surfaces (Di Bonaventura et al., 2004, 2007a,b; Pompilio et al., 2008), but also on CF-derived epithelial monolayer (Pompilio et al., 2010), probably because of a selective adaptation to CF airways (Pompilio et al., 2011). Furthermore, in a murine model of acute respiratory infection we observed that S. maltophilia significantly contributes to the inflammatory process resulting in compromised respiratory function and death (Di Bonaventura et al., 2010).
S. maltophilia is often co-isolated with P. aeruginosa, with a frequency ranging from 10 to 60% of CF patients (Spicuzza et al., 2009; Blau et al., 2014; Zemanick et al., 2015); however, no data are available on this at the onset of PE. It is, therefore, plausible to hypothesize that these species interact and that this could potentially affect the virulence and persistence of P. aeruginosa. While several studies have focused on the interaction of P. aeruginosa with other bacterial species (Qin et al., 2009; Pihl et al., 2010; Baldan et al., 2014), very little has been published on the interaction between P. aeruginosa and S. maltophilia. In this regard, Ryan et al. (2008) found that the presence of S. maltophilia increases P. aeruginosa resistance to polymyxin by a diffusible signal factor, while Kataoka et al. (2003) observed that β-lactamases leaking from S. maltophilia can encourage the growth of P. aeruginosa in the presence of imipenem or ceftazidime.
Recently, we observed that the pre-incubation of the CF bronchial epithelial IB3-1 cells with S. maltophilia decreases the adherence of P. aeruginosa, while previous P. aeruginosa infection may increase the likelihood of S. maltophilia colonizing a damaged CF pulmonary mucosa (Pompilio et al., 2010).
Our hypothesis, based on these clinical and experimental findings, was that in CF lung S. maltophilia may indirectly contribute to disease development by providing a favorable growth environment for P. aeruginosa and/or by modulating some virulence traits exhibited by P. aeruginosa. In order to test this hypothesis, S. maltophilia RR7 and P. aeruginosa RR8—two strains co-isolated at the same time from the lung of a CF patient during a PE episode—were evaluated, both in combined and monomicrobial cultures, with respect to planktonic growth, adhesion and biofilm formation onto both polystyrene and CF bronchial cell monolayer, and motility. The effect of S. maltophilia RR7 on the expression of selected P. aeruginosa RR8 virulence genes was also assessed in mixed biofilms.
We show that S. maltophilia has the potential to significantly affect P. aeruginosa virulence, which suggests that interspecies interactions are operative in promoting bacterial pathogenicity in CF lung infections.
Materials and Methods
Bacterial Strains and Growth Conditions
P. aeruginosa RR8 and S. maltophilia RR7 strains were co-isolated from the same sputum sample collected, during a PE, from a 15-year-old CF patient (named “RR”) attending Children Hospital “Bambino Gesù” of Rome. The patient was selected because he was chronically infected by both P. aeruginosa and S. maltophilia, according to the definition proposed by the EuroCareCF Working Group (Pressler et al., 2011). The strains were identified by the API 20-NE system (bioMérieux, Marcy-L'Etoile, France) and stored at −80°C until use. Fresh bacterial stocks were thawed and grown twice on Mueller-Hinton agar (MHA; Oxoid S.p.A., Garbagnate M.se, Italy) to check for purity and to regain the original phenotype: (i) P. aeruginosa RR8 strain was mucoid and resistant to amikacin and gentamicin only; (ii) S. maltophilia RR7 strain, not mucoid, showed resistance to ceftazidime, piperacillin/tazobactam, amikacin, gentamicin, ciprofloxacin, and levofloxacin.
All assays were carried out by using a standardized bacterial inoculum. Briefly, an overnight culture in tryptone soya broth (TSB), grew under agitation (130 rpm) at 37°C, was adjusted with sterile broth to an OD550 corresponding to 1–3 × 108 CFU/ml for both strains, then diluted 1:10 in cation-adjusted Mueller-Hinton broth (CAMHB; Becton, Dickinson and Company; Milan, Italy) (pH 7.2–7.4).
Mono- and Co-culture Planktonic Growth
Each well of a 96-well polystyrene, flat bottom, tissue culture-treated microtiter (BD Falcon, Milan, Italy) was seeded with 100 μl of each standardized inoculum (dual cultures) or 100 μl of standardized inoculum +100 μl sterile medium (single cultures). Microtiters were then statically incubated at 37°C, under aerobic atmosphere, for 24 h. At different time points (2, 4, 6, 8, 10, and 24 h) samples were taken, serially diluted in sterile phosphate buffered saline (PBS; Sigma-Aldrich S.p.A.; Milan, Italy) (pH 7.2), then plated onto MHA + imipenem at 32 μg/ml or Pseudomonas cetrimide agar (Oxoid) to discriminate S. maltophilia, and P. aeruginosa growth, respectively. The agar plates were incubated at 37°C for 24 h when the CFU number was recorded.
Kinetics of Adhesion and Biofilm Formation by Single and Mixed Cultures
Each well of a 96-well polystyrene, flat bottom, tissue culture-treated microtiter was seeded as described above, and statically incubated at 37°C, under aerobic atmosphere, for 3 h (adhesion) or up to 7 days (biofilm formation). During biofilm formation assay, CAMHB was replaced daily with fresh broth. At each time-point considered (3 h, and 1–7 days), samples were washed twice with sterile PBS, and then adhesion or biofilm were measured for both biomass and viability. Biomass was measured in terms of optical density read at 492 nm (OD492) following crystal violet staining, as previously described (Pompilio et al., 2008), considering a low cut-off of ODc + 3 × SDs, where ODc is OD492 of control wells (containing medium alone without bacteria). For viability assessment, samples were exposed to trypsin-ethylenediaminetetraacetic acid (EDTA) 0.25% (Sigma-Aldrich) to allow detachment from the polystyrene, and then the viable count was carried out as described above.
Preparation of Cell-free Culture Supernatant
For each strain, some colonies from an overnight MHA growth were suspended in 20 ml of TSB and incubated overnight at 37°C, under agitation (220 rpm). Supernatants were obtained after centrifugation at 12,000 × g, for 10 min at 4°C, followed by sterile filtration using a 0.20 μm-pore-size filter (Corning; Tewksbury, MA, USA), and storage at −80°C until use.
Evaluation of Antibacterial Activity by S. maltophilia RR7 and P. aeruginosa RR8
The antibacterial activity of each strain was evaluated by the drop test assay. Briefly, 100 μl of the standardized inoculum containing the indicator strain were streaked onto an MHA surface using a cotton swab, and then the plate was dried at 30°C for 30 min. The test strain was then evaluated for antimicrobial activity, both as cell suspension and supernatant. Drops containing 10 μl of the supernatant or standardized suspension, and a control, were placed on the agar surface and the inhibition zone diameter was measured after incubation at 37°C for 24 h.
To evaluate whether the inhibitory activity of P. aeruginosa RR8 on S. maltophilia RR7 required direct cell-cell interactions, a transwell-based assay was performed. Briefly, P. aeruginosa RR8 was inoculated at 1–5 × 108 CFU/ml in the upper insert (Cell Culture Insert; BD Falcon), while S. maltophilia RR7 was inoculated at the same concentration in the bottom of the culture well. Following 24 h-incubation at 37°C, suspensions from both compartments underwent a viable count. A sample where the upper insert contained TSB without P. aeruginosa RR8 was used as control.
Adhesion to, and Biofilm Formation on CF Bronchial Cells
CFBE41o- bronchial cells, derived from a CF patient homozygous for the F508del CFTR mutation, were obtained from Dr. Dieter C. Gruenert (University of California, San Francisco, USA). Briefly, confluent CFBEo- cells were grown in tissue polystyrene culture flasks (BD Falcon) in Minimum Essential Medium (GIBCO, Life-technologies; Monza, Italy) supplemented with 10% fetal bovine serum (GIBCO), 50 U/ml penicillin, 50 μg/ml streptomycin (both from Sigma-Aldrich), and 2 mM L-glutamine (GIBCO). Monolayers were simultaneously infected with S. maltophilia RR7 and P. aeruginosa RR8 strains (ratio 1:1, each at 106 CFU/ml; multiplicity of infection, MOI: 10), then incubated at 37°C for 3 h (adhesion assay) or 24 h (biofilm formation assay). CFBE41o- cell monolayers infected with a single strain (at 106 CFU/ml) were prepared as control. At the end of incubation, infected monolayers were washed twice with PBS, then detached by 0.25% trypsin/EDTA, and finally plated for colony count.
Tobramycin Activity Against Planktonic and Biofilm Cells
MICs of tobramycin (Sigma-Aldrich) for S. maltophilia RR7 and P. aeruginosa RR8 were respectively 128 and 4 μg/ml, as assessed by microdilution technique in accordance with the M100-S20 protocol [Clinical and Laboratory Standards Institute, 2010]. Tobramycin was also tested against preformed single species and mixed biofilms. Briefly, biofilms were allowed to form at 37°C for 24 h as described in “Kinetics of adhesion and biofilm formation by single and mixed cultures.” Samples were washed once with sterile CAMHB, and then exposed to 200 μl of tobramycin at 128 μg/ml. After incubation at 37°C for 24 h, non-adherent bacteria were removed by washing twice with sterile PBS, and biofilm cells were scraped with a pipette tip following 5 min-exposure to 100 μl trypsin-EDTA 0.25%. Cell suspension was vortexed at high speed for 1 min to break up the clumps, and then bacterial counts were assessed by plating serial 10-fold dilutions of the biofilm cell suspension onto selective media MHA plates.
Motility Assays
Swimming, swarming, and twitching motilities were evaluated using dedicated agar media, as previously described (Pompilio et al., 2008). The relative motility of two strains was assessed by using agar prepared with and without 1:2 diluted culture supernatant of the other strain.
Gene Expression Assay in Mixed Biofilm
The effect of S. maltophilia RR7 on the transcription levels of several virulence factors (Table 1) of P. aeruginosa RR8 was assessed in mixed biofilms by real-time PCR (RT-PCR). P. aeruginosa RR8 was cultured as biofilm, both alone and with S. maltophilia RR7, in 24-well polystyrene, flat bottom, tissue culture-treated microtiter (BD Falcon), as described above. Following 24 h-incubation at 37°C, biofilms were washed, and then harvested by scraping in Qiazol (Qiagen; Milan, Italy). RNA was then extracted by the phenol-chloroform technique (Kang et al., 2009), treated with DNase I (Applied Biosystems Italia; Monza, Italy), and checked for purity by NanoDrop-2000 spectrophotometer (Thermo Scientific Italia; Milan, Italy). First strand cDNA was synthesized using a High Capacity cDNA reverse transcription kit (Applied Biosystems) according to the manufacturer's protocol. Gene expression was evaluated using a SYBR green (Applied Biosystems) RT-PCR assay. The primers' specificity was assessed both in silico with BLAST and by PCR endpoint under the same RT-PCR conditions. Each amplification assay was also tested for S. maltophilia RR7 as an additional negative control in each RT-PCR. The ΔΔCt method was used to determine the relative gene expression of each gene in co-culture vs. monoculture biofilms normalized to the expression of the housekeeping gene proC.
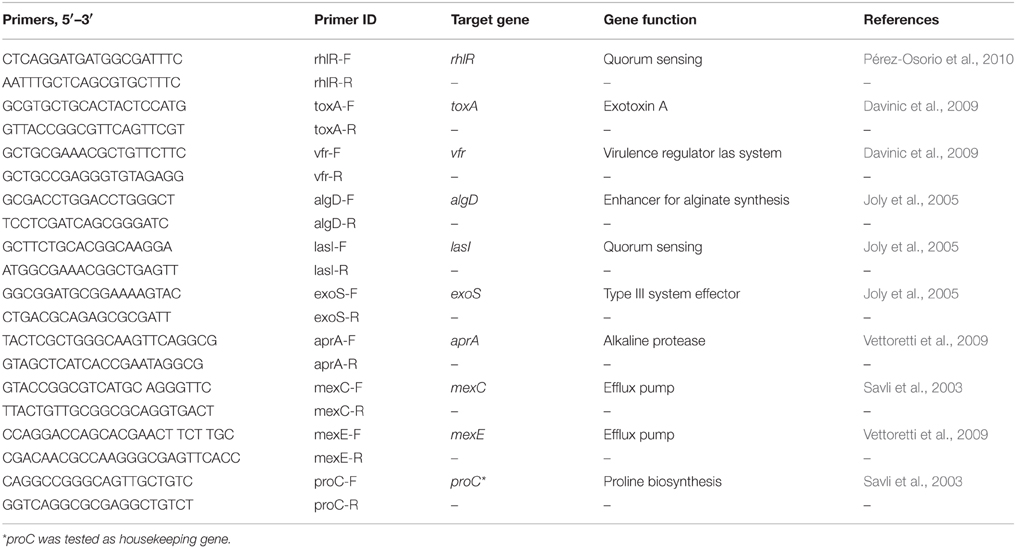
Table 1. List of primer sequences used in RT-PCR for expression analysis of virulence genes by P. aeruginosa RR8 grown as biofilm, both in solo and co-cultured with S. maltophilia RR7.
Interpretative Criteria and Statistical Analysis
Each experiment was performed in triplicate and repeated on two different occasions. All statistical analyses were performed by GraphPad Prism software (ver. 4.0; GraphPad Inc, San Diego, USA), considering as statistically significant a p-value less than 0.05. Differences were assessed by ANOVA-test followed by Newman-Keuls multiple comparison post-test (kinetics of planktonic growth, adhesion and biofilm formation onto both polystyrene and CFBE41o- cells, and tobramycin activity against preformed biofilms), chi-square test (adhesion and biofilm formation efficiency) or paired Student's t-test (motility and gene expression). The efficiency of adhesion and biofilm formation onto CFBE41o- cells was calculated for each strain, both in single and mixed infections, as a percentage referred to the initial inoculum: (A/B) × 100, where A is the number of adhered or biofilm bacteria, and B is the number of bacteria in the initial inoculum.
In mixed cultures, the Competitive Index (CI) was defined as the S. maltophilia/P. aeruginosa ratio within the output sample divided by the corresponding ratio in the inoculum (input): CI = (S. maltophilia/P. aeruginosa)output/(S. maltophilia/P. aeruginosa)input, where output and input samples were assessed after plating onto MHA serial dilutions of the sample taken at fixed times or the inoculum (t = 0), respectively (Macho et al., 2007). For statistical analyses, CI values were first subjected to a Log transformation for normal distribution, then interpreted as follows: a CI value equal to 0 indicates equal competition of the two species; a positive CI value indicates a competitive advantage for S. maltophilia; a negative CI value indicates a competitive advantage for P. aeruginosa.
Similarly to CI, the Relative Increase Ratio (RIR) was calculated based on the growth results obtained from monocultures of each strain (Macho et al., 2007). Each CI and RIR was analyzed using the Student's t-test and the null hypothesis: the mean index was not significantly different from 1.0. When appropriate, CI and RIR from a given experiment were compared using unpaired Student's t-test, and significant differences are suggestive of a meaningful competition between the species (Macho et al., 2007).
Results
P. aeruginosa Significantly Affects S. maltophilia Growth in Planktonic Culture
The growth curve kinetics of P. aeruginosa RR8 and S. maltophilia RR7 strains, tested as alone or in mixed culture, were assessed by colony count over 24 h, and the results are shown in Figure 1A. When grown in mixed cultures, the growth kinetics of the two strains were comparable. However, compared with single cultures, the growth of both S. maltophilia RR7 and P. aeruginosa RR8 in mixed cultures was negatively affected during both log and stationary phases.
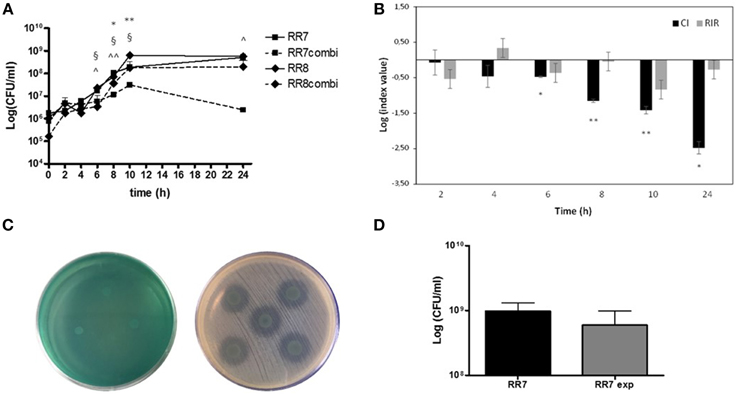
Figure 1. Kinetics of planktonic growth exhibited by S. maltophilia RR7 and P. aeruginosa RR8, considered both as alone and in combination. S. maltophilia RR7 and P. aeruginosa RR8 were grown for 24 h in CAMHB in single culture and in co-culture after inoculation at equal ratio from mid-exponential phase pure cultures. Growth rate was monitored by colony count after plating on selective media for both species. (A) Growth curves of S. maltophilia RR7 and P. aeruginosa RR8 strains in pure culture (RR7, RR8) and in co-culture (RR7combi, RR8combi). The results are shown as mean + SD (n = 6). *p < 0.05, **p < 0.01, RR7 vs. RR8; §p < 0.05, RR8 vs. RR8combi; ∧p < 0.05, ∧∧p < 0.01, RR7 vs. RR7combi; ANOVA + Newman-Keuls post-test. (B) Competitive index (CI; black bars) and Relative Increase Ratio (RIR; gray bars) calculated from single and dual planktonic cultures of S. maltophilia RR7 and P. aeruginosa RR8 strains. The results are shown as mean ± SD (n = 6). *p < 0.05, **p < 0.01, CI vs. RIR, unpaired t-test. (C) Evaluation of antibacterial activity using agar spot assay. S. maltophilia RR7 vs. P. aeruginosa RR8 (left): no antibacterial activity. P. aeruginosa RR8 vs. S. maltophilia RR7 (right): partial antibacterial activity, as suggested by the regrowth observed within the inhibition zone. (D) Transwell assay: S. maltophilia RR7 growth without or with (exp) P. aeruginosa RR8. The results are expressed as mean + SD (n = 6).
To assess further the meaning of the differences between the variations observed in single vs. mixed cultures, CI and RIR were calculated and compared, as shown in Figure 1B. The CI of S. maltophilia RR7 vs. P. aeruginosa RR8 was significantly different from the respective RIR values between 6 h- and 24 h-incubation, suggesting that P. aeruginosa RR8 exerts an inhibitory effect on S. maltophilia RR8 growth during both log and stationary growth phases.
P. aeruginosa Exhibits Antimicrobial Activity against S. maltophilia in a Contact-dependent Manner
The agar spot assay results showed that P. aeruginosa RR8 is active against S. maltophilia RR7, although only when tested as cell suspension (diameter of inhibition zone, mean ± SD: 12.5 ± 1.4 mm), and not as supernatant (Figure 1C). However, the inhibition halo showed a partial regrowth, suggesting that a partial inhibition occurred. Conversely, S. maltophilia RR7 showed no activity against P. aeruginosa RR8, as supernatant or suspension (Figure 1C).
The inhibitory effect of P. aeruginosa RR8 on S. maltophilia RR7 was also assessed using a transwell-based assay, and the results are shown in Figure 1D. S. maltophilia growth was not significantly affected by the presence of P. aeruginosa (9.6 ± 3.4 × 108 CFU/ml vs. 5.9 ± 3.9 × 108 CFU/ml for unexposed and exposed S. maltophilia, respectively; p > 0.05), which suggests that P. aeruginosa inhibits S. maltophilia growth in a contact-dependent manner.
Kinetics of S. maltophilia and P. aeruginosa Interaction during Adhesion and Biofilm Formation onto Polystyrene
The interaction during adhesion phase between S. maltophilia RR7 and P. aeruginosa RR8 strains was assessed, tested as single and mixed cultures, after 3 h-incubation, both by crystal violet and the viable count assays; the results are reported in Figure 2.
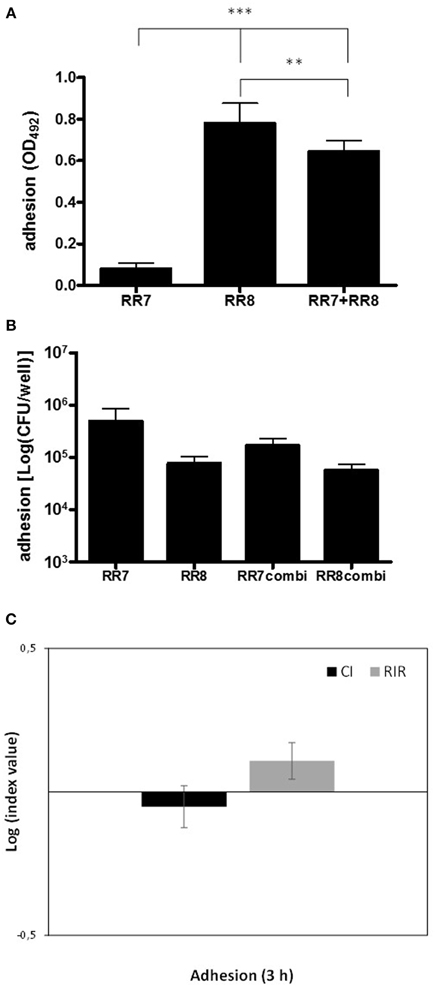
Figure 2. Kinetics of adhesion onto polystyrene by S. maltophilia RR7 and P. aeruginosa RR8, considered both as alone and in combination. Adhesion was assessed, after 3 h of incubation at 37°C, both by crystal violet and viable count assays. (A) Crystal violet assay. S. maltophilia RR7, and P. aeruginosa RR8 were tested both alone (RR7, RR8), and in mixed infection (RR7 + RR8), and the results are shown as mean + SD (n = 6). **p < 0.01, ***p < 0.001, ANOVA + Newman-Keuls post-test. (B) Viable count assay. S. maltophilia RR7 and P. aeruginosa RR8 strains were tested both alone (RR7, RR8), and in mixed infection (RR7combi, RR8combi), and the results are shown as mean + SD (n = 6). No statistically significant differences were found among groups by ANOVA + Newman-Keuls post-test. (C) Competitive index (CI; black bars) and Relative Increase Ratio (RIR; gray bars), calculated from single and dual cultures of S. maltophilia RR7 and P. aeruginosa RR8. The results are shown as mean ± SD (n = 6). CI vs. RIR, no statistically significant difference, unpaired t-test.
The crystal violet assay showed that P. aeruginosa RR8, when cultured alone, exhibited a significantly higher adhesiveness than S. maltophilia RR7 (OD492, mean ± SD: 0.781 ± 0.095 vs. 0.079 ± 0.026, respectively; p < 0.001) (Figure 2A). No differences, however, were found between the strains in terms of cell viability (Figure 2B), which suggests that extracellular polymeric substance (EPS) contributes substantially to P. aeruginosa RR8 adhesion.
In mixed infections, the numbers of viable cells of each strain that adhered to polystyrene were not affected by the presence of the other (Figure 2B), as was confirmed by comparable CI and RIR values (Figure 2C). However, in mixed culture the biomass adhesion level of S. maltophilia RR7 was significantly lower than that of P. aeruginosa RR8 (OD492, mean ± SD: 0.645 ± 0.051 vs. 0.781 ± 0.095, respectively; p < 0.01), which indicates that the presence of S. maltophilia RR7 leads to a reduction in P. aeruginosa RR8 EPS.
The interaction in biofilm growth between S. maltophilia RR7 and P. aeruginosa RR8, tested as single and mixed cultures, was monitored over 7 days, by both crystal violet and viable count assays; results are shown in Figure 3.
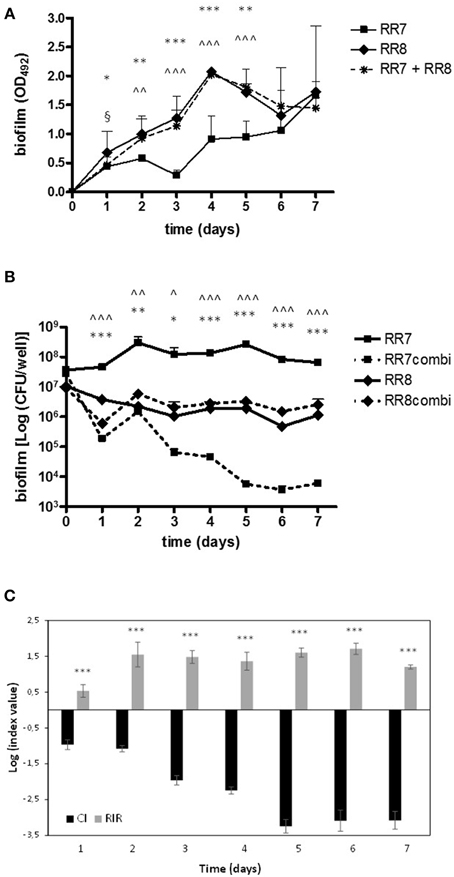
Figure 3. Kinetics of biofilm formation onto polystyrene by S. maltophilia RR7 and P. aeruginosa RR8, both considered as alone and in combination. Biofilm formation was assessed, over 7 days, both by (A) crystal violet and (B) viable count assays. S. maltophilia RR7 and P. aeruginosa RR8 were tested both alone (RR7, RR8), and in mixed infection (RR7 + RR8), and the results are shown as mean + SD (n = 6). *p < 0.05, **p < 0.01, ***p < 0.001, RR7 vs. RR8; ∧p < 0.05, ∧∧p < 0.01, ∧∧∧p < 0.001, RR7 vs. RR7combi; §p < 0.05, RR8 vs. RR8combi; ANOVA + Newman-Keuls post-test. (C) Competitive index (CI; black bars) and Relative Increase Ratio (RIR; gray bars) calculated from single and dual biofilm cultures of S. maltophilia RR7 and P. aeruginosa RR8. The results are shown as mean ± SD (n = 6). ***p < 0.001, CI vs. RIR, unpaired t-test.
The kinetics of biofilm formation, as assessed by crystal violet staining, are summarized in Figure 3A. In monoculture, from day 1 until day 5 of incubation P. aeruginosa RR8 formed significantly higher biofilm biomass amount than S. maltophilia RR7. The kinetics of biofilm formation by P. aeruginosa RR8 and RR7+RR8 mixed infection followed a comparable trend throughout the 7 day period. From day 2 until day 5 of incubation, the mean biofilm biomass amount formed by both P. aeruginosa RR8 and mixed infection was significantly higher than when S. maltophilia RR7 was cultured alone. On day 6, all conditions tested produced comparable mean biofilm biomass values.
The biofilm formation kinetics evaluated by the viable count are shown in Figure 3B. In monoculture, the viability of biofilm formed by S. maltophilia RR7 was higher than that of P. aeruginosa RR8 over the entire period (Figure 3B). Given the different endpoints measured by the crystal violet (biofilm biomass, consisting of both EPS and cells) and the viable count (biofilm viability) assays, our results indicate that only in P. aeruginosa does the amount of EPS increase over time during biofilm formation, as is also confirmed by the mucoid appearance of the biofilm samples observed during macroscopic analysis of the 96-well plate.
When S. maltophilia RR7 was co-cultured with P. aeruginosa RR8 in mixed biofilms, its viability was significantly decreased throughout the period. Conversely, the biofilm viability of P. aeruginosa RR8 was not affected by the presence of S. maltophilia RR7.
The CI values of S. maltophilia vs. P. aeruginosa were always significantly different from the RIR values (p < 0.001) throughout the 7 days of incubation, which suggests that P. aeruginosa outcompetes S. maltophilia affecting its growth in mixed biofilms as well as in planktonic cultures (Figure 3C).
Adhesion to and Biofilm Formation on CFBE41o- cells
P. aeruginosa RR8 and S. maltophilia RR7 were evaluated, both alone and in mixed culture, for adhesion to, and biofilm formation onto, CFBE41o- CF cell monolayer. The results of the viable count assay are summarized in Figure 4.
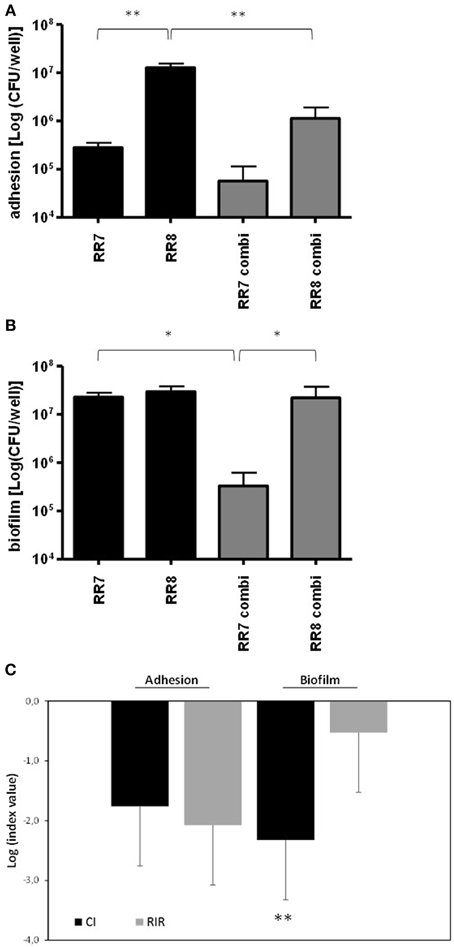
Figure 4. Adhesion and biofilm formation by S. maltophilia RR7 and P. aeruginosa RR8 onto CFBE41o- CF bronchial cells. CFBE41o- cell monolayers were exposed for (A) 3 h (adhesion assay) or (B) 24 h (biofilm formation assay) to S. maltophilia RR7 and P. aeruginosa RR8 (each at 106 CFU/ml; MOI: 10), tested as alone (RR7, RR8) or in mixed cultures (RR7combi, RR8combi). *p < 0.05, **p < 0.01, ANOVA + Newman-Keuls post-test. (C) In mixed cultures, the Competitive Index (CI) and the Relative Increase Ratio (RIR) were calculated as described in Materials and Methods. The results are shown as mean + SD (n = 6). **p < 0.01; unpaired t-test.
When monolayers were separately infected, the adhesiveness of P. aeruginosa RR8 to CFBE41o- cells was significantly higher than that of S. maltophilia RR7 (1.3 ± 0.3 × 107 vs. 2.8 ± 0.7 × 105 CFU/well, respectively; p < 0.01) (Figure 4A). Conversely, when CFBE41o- cell monolayer was concomitantly challenged by both strains their degrees of adhesiveness were comparable. However, the adhesiveness of P. aeruginosa RR8 was significantly less than that observed in monoculture (1.1 ± 0.7 × 106 vs. 1.3 ± 0.3 × 107 CFU/well, respectively; p < 0.05), unlike S. maltophilia RR7, whose adhesiveness was not affected by the presence of P. aeruginosa RR8. The comparative analysis of the CI and the RIR values showed no statistically significant difference (Figure 4C), although a comparison of efficiency values, from single and mixed cultures, showed a significant decrease for P. aeruginosa RR8 (from 12.9 to 4.9%, respectively; p < 0.05), whereas a significant increase was found for S. maltophilia RR7 (from 0.08 to 0.3%, respectively; p < 0.001).
Both S. maltophilia RR7 and P. aeruginosa RR8 showed comparable biofilm viability when tested in monoculture (Figure 4B). However, when CFBE41o- cell monolayer was simultaneously infected by both strains, the concentration of P. aeruginosa RR8 in mixed biofilm was significantly higher than that of S. maltophilia RR7 (2.2 ± 1.5 × 107 vs. 3.2 ± 2.9 × 105 CFU/well, respectively; p < 0.05), whose concentration was significantly lower than that observed in monomicrobial biofilm (2.3 ± 0.5 × 107 CFU/well; p < 0.05) (Figure 4B).
The comparative analysis of the CI and the RIR values obtained for biofilm formation assay showed statistically different values (−2.3 ± 0.4 vs. −0.5 ± 0.4, respectively; p < 0.01) (Figure 4C). Relative to the biofilm cells/initial inoculum ratio calculated for each strain, the values obtained from mono- and mixed cultures were significantly elevated for P. aeruginosa RR8 (from 31.8 to 96.2%, respectively; p < 0.001), while a significant decrease was observed for S. maltophilia RR7 (from 6.9 to 0.5%, respectively; p < 0.001).
Taken together, these results suggest the existence of a reciprocal interference between these two species in the adhesion step. S. maltophilia RR7 negatively affects P. aeruginosa RR8 adhesiveness, whereas later, during biofilm formation onto CFBE41o- cells, P. aeruginosa RR8 outcompetes S. maltophilia RR7, probably by inhibiting its growth.
Exposure to S. maltophilia Culture Supernatant Significantly Reduces Swimming Motility in P. aeruginosa
The effect of the presence of one strain, tested as culture supernatant, on swimming, swarming and twitching motilities of the other strain was evaluated and the results are summarized in Figures 5A,B.
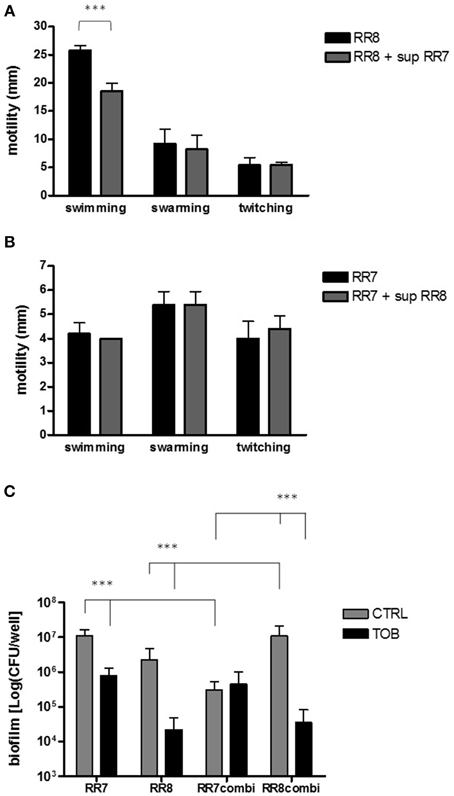
Figure 5. (A,B) Effect of culture supernatants on motility. Swimming, swarming, and twitching motilities of each strain were assessed as previously described (Pompilio et al., 2008). (A) P. aeruginosa RR8 motility was evaluated both in the presence and absence of S. maltophilia RR7 culture supernatant (sup RR7). (B) S. maltophilia RR7 motility was evaluated both in the presence and absence of P. aeruginosa RR8 culture supernatant (sup RR8). The results are shown as mean + SD (n = 6). ***p < 0.001, paired-t test. (C) Activity of tobramycin against preformed biofilms. Monomicrobial and mixed biofilms were allowed to grow for 24 h, and then they were exposed to tobramycin 128 μg/ml (TOB) or CAMHB only (control, CTRL) for further 24 h. Biofilm's viability was then assessed by viable count. The results are shown as mean + SD (n = 6). ***p < 0.001, ANOVA + Newman-Keuls post-test.
The swimming motility exhibited by P. aeruginosa RR8 was significantly reduced by exposure to S. maltophilia RR7 culture supernatant (mean ± SD: 18.6 ± 1.3 vs. 25.8 ± 0.8 mm, with or without supernatant, respectively; p < 0.001) (Figure 5A). No significant variations were observed between groups for swarming and twitching motilities. P. aeruginosa RR8 supernatant did not affect any of S. maltophilia RR7 motility types tested (Figure 5B).
S. maltophilia is Less Susceptible than P. aeruginosa to Tobramycin in Mixed Biofilms
The activity of tobramycin at 128 μg/ml against single and mixed preformed biofilm was assessed by the viable count and the results are summarized in Figure 5C. Exposure to tobramycin significantly reduced the viability of both S. maltophilia RR7 and P. aeruginosa RR8 monomicrobial biofilms (S. maltophilia RR7: 1.1 ± 0.5 × 107 vs. 8.0 ± 5.0 × 105 CFU/well; P. aeruginosa RR8: 2.2 ± 2.5 × 106 vs. 2.2 ± 2.4 × 104 CFU/well; unexposed and tobramycin-treated biofilms, respectively; p < 0.001). However, when mixed biofilms were tested, tobramycin resulted to be effective against P. aeruginosa RR8 only (1.1 ± 0.9 × 107 vs. 3.4 ± 4.7 × 104 CFU/well; unexposed and tobramycin-treated biofilms, respectively; p < 0.001).
S. maltophilia Significantly Affects P. aeruginosa Virulence in Mixed Biofilm
The effect of exposure to S. maltophilia RR7 on the expression of nine selected virulence genes (rhlR, lasI, aprA, vfr, exoS, toxA, algD, mexC, mexE) of P. aeruginosa RR8 was assayed, by RT-PCR, both in mono- and co-cultured biofilms; the results are summarized in Figure 6. When grown in mixed biofilm with S. maltophilia RR7, P. aeruginosa RR8 significantly overexpressed aprA, and algD genes (p < 0.05), codifying for protease and alginate, respectively, whereas the QS-related rhlR and lasI genes were down-regulated (p < 0.05). The presence of S. maltophilia RR7 also caused a considerable increase of efflux pump systems-related mexC and mexE genes, as well as of toxA, codifying for exotoxin A, expression by P. aeruginosa RR8, although this trend resulted not to be statistically significant, probably due to high SD values.
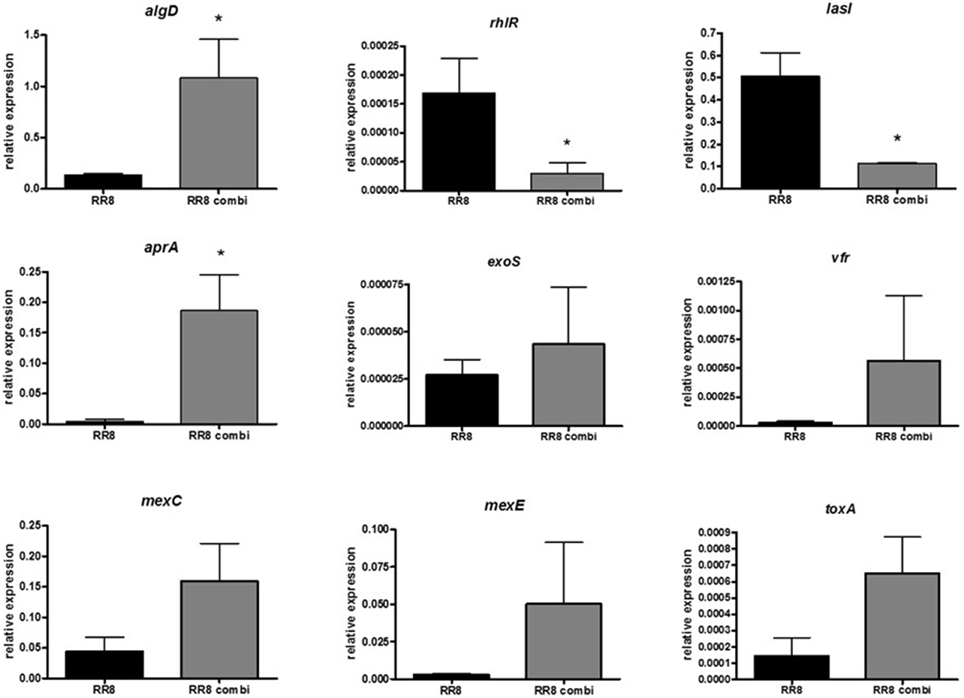
Figure 6. Effect of S. maltophilia RR7 on P. aeruginosa RR8 virulence-related gene expression in mixed biofilm. S. maltophilia RR7 and P. aeruginosa RR8 were allowed to grow, as mixed biofilm, in a 24-well microtiter polystyrene plate at 37°C for 24 h (RR8 combi, gray bars). Relative expression of nine selected P. aeruginosa virulence genes (algD, rhlR, lasI, aprA, exoS, vfr, mexC, mexE, toxA) were then measured by RT-PCR. P. aeruginosa grown alone as biofilm was considered as control (RR8, black bars). The results are shown as mean + SD (n = 6). *p < 0.05, paired t-test.
Discussion
The originality of this study is two-fold. Firstly, it evaluates the nature of interactions occurring between P. aeruginosa and S. maltophilia under both planktonic and biofilm growth, and specifically whether these interactions offer enhanced fitness compared to single cultures. Secondly, it considers S. maltophilia and P. aeruginosa strains isolated at the same time from the same CF patient during a PE episode, whereas previous works were conducted using strains isolated from different, and non-CF, patients (Kataoka et al., 2003; Ryan et al., 2008; Varposhti et al., 2014).
Following examination of the interaction between S. maltophilia and P. aeruginosa under planktonic growth conditions, the CI and RIR values suggested that P. aeruginosa outcompetes S. maltophilia, during both exponential and stationary growth phases. This microbial antagonism could be due to competition for both nutrients and space or, more directly, to an antibacterial effect of one species toward the other (Harrison, 2007; Hibbing et al., 2010). Our findings from the agar spot assay in fact clearly showed, for the first time, that P. aeruginosa exerts antibacterial activity against S. maltophilia. As suggested by the transwell-based assay results, this effect is probably not mediated by exoproducts, but requires direct contact between cells, which suggests that P. aeruginosa, in order to achieve new niches, might kill S. maltophilia by injecting specific proteins via the type VI secretion system (Tashiro et al., 2013).
Despite the increasing interest in the crucial role played by biofilm in determining CF infections, interspecies interactions in mixed biofilms are still poorly understood. Using a polystyrene microtiter assay, the reciprocal interaction between S. maltophilia and P. aeruginosa was initially evaluated at the adhesion phase, the first step of biofilm formation. Our findings confirmed that EPS plays a crucial role during P. aeruginosa adhesion, and interestingly showed that the presence of S. maltophilia reduces the amount of EPS, probably secondary to loss or a reduced synthesis, therefore affecting P. aeruginosa adhesion to the substratum.
The kinetics of biofilm formation were then monitored throughout the 7 day-incubation period with daily medium replacement, in order to simulate a chronic infection. When both species were cultured alone, we found that in P. aeruginosa, unlike S. maltophilia, EPS amount increases over time during biofilm formation. When simultaneously cultured in mixed biofilms, we observed that S. maltophilia and P. aeruginosa cannot coexist in a dynamic equilibrium in mixed biofilm: S. maltophilia will always be outcompeted. The results from the viable count in fact indicated that the contribution of S. maltophilia to the biomass of mixed biofilm is negligible, being at least 100-fold lower than P. aeruginosa concentration. The comparative evaluation between single and dual specie biofilms showed that the capacity of P. aeruginosa to form biofilms is unaffected by the presence of S. maltophilia, while that of S. maltophilia is significantly reduced in the presence of P. aeruginosa. The antibacterial activity exhibited in planktonic cultures by P. aeruginosa is probably one of the mechanisms that inhibits S. maltophilia biofilm formation.
In contrast with our findings, Varposhti et al. (2014) found that S. maltophilia, in mixed cultures, induces P. aeruginosa biofilm biomass formation on Foley catheter. The discrepancy could be explained by the fact that strains tested in combinations by Varposhti et al. were from different patients with polymicrobial lower respiratory infections. This might be particularly relevant in the case of CF chronic infections, where P. aeruginosa undergoes adaptation to the CF lung leading to patho-adaptative lineages genotypically and phenotypically different (Huse et al., 2010), which has been observed not only when comparing isolates from different CF patients, but also when comparing multiple isolates from the same CF respiratory specimen (Lee et al., 2005).
The polystyrene microtiter assay is classically used for biofilm studies due to its ease of use and ability for high-throughput testing. However, since not all variables can be controlled, the conditions still differ significantly from those observed during natural infections and, therefore, the results might be of limited clinical relevance. The interaction between P. aeruginosa and S. maltophilia during adhesion and biofilm formation was therefore also assessed using a CF-derived bronchial epithelial cell monolayer.
In single infections, P. aeruginosa was more adhesive than S. maltophilia, which disagrees with the polystyrene microtiter assay. However, when the cell monolayer was simultaneously challenged with mixed cultures, the adhesiveness of P. aeruginosa was significantly reduced relative to monoculture, while that of S. maltophilia remained unaffected. The results from motility assays suggested that S. maltophilia might reduce P. aeruginosa adhesiveness to bronchial cells by affecting its ability to reach the substratum via swimming motility, probably as a consequence of flagellar loss or a reduced expression of flagella-associated genes.
In accordance with the polystyrene microtiter assay, the viable count showed that P. aeruginosa outcompetes S. maltophilia in mixed biofilms formed onto CF bronchial cells. Accordingly, we also observed that S. maltophilia can not affect P. aeruginosa biofilm formation by modulating its swarming or twitching motility.
Recent studies on the behavior of multispecies communities have shown that the virulence and the gene expression of pathogens can be modulated by the presence of other bacterial species (Duan et al., 2003). Therefore, we performed a comparative analysis of transcript levels of several P. aeruginosa virulence factors both in single- and dual-species biofilms. The quantitative PCR results suggested that S. maltophilia changes the physiology and, consequently, the virulence of P. aeruginosa. Specifically, both aprA and algD operon—codifying for the frank P. aeruginosa virulence factors alkaline protease and alginate, respectively—were up-regulated in the presence of S. maltophilia, whereas the QS-related rhlR and lasI genes were down-regulated. These differences may be the expression of a defensive reaction by P. aeruginosa to the presence of S. maltophilia. Alternatively, since both bacterial species exist ubiquitously in various environments, including the rhizosphere of plants (Berg et al., 2005), the lack of significant differences in the expression of other genes observed in co-culture may be highly suggestive of their relative acclimatization to each other.
P. aeruginosa causes severe tissue damage by the expression of the alkaline protease AprA, especially in CF patients where it has been associated with increased infectivity and virulence, suggesting a role in processes related to bacterial colonization and/or exacerbation in the CF lung (Burke et al., 1991; Kim et al., 2006). Our findings suggested that in mixed infections S. maltophilia might indirectly facilitate the onset of PEs in CF patients by inducing increased proteolytic activity in P. aeruginosa, probably via a Las/RhI independent pathway.
On the other hand, S. maltophilia might also promote P. aeruginosa adaptation to the airways of CF patients by favoring phenotypic traits acquired by P. aeruginosa only during chronic infections, which has the result of promoting long-term survival in CF lungs. Particularly, we observed in mixed biofilms the loss of LasR function and overproduction of alginate. Loss of QS regulation is generally considered a hallmark of chronic virulence and has been described for several P. aeruginosa CF isolates (Geisenberger et al., 2000; D'Argenio et al., 2007). Specifically, loss of LasR function has clinical implications for disease progression and antibiotic resistance since it produces an increase in β-lactamase activity, which augments tolerance to ceftazidime, a widely used β-lactam antibiotic in CF patients (D'Argenio et al., 2007). In addition, mucoid strains overproducing alginate have been found to be dominant in chronic lung infections, and therefore correlated with a poor prognosis (Yang et al., 2011). Furthermore, by increasing algD expression S. maltophilia might favor the protection of P. aeruginosa against oxygen radicals from activated PMNs (Govan and Deretic, 1996), and improve its ability to form more robust biofilms (Hoffmann et al., 2005).
Understanding the interspecies interactions is essential, not only because they can potentially modulate the virulence and the persistence of pathogens such as P. aeruginosa, but also because it might lead to the development of new, more effective, therapeutic strategies. We found that the exposure of a mixed biofilm to tobramycin reduced the viability of P. aeruginosa but not of S. maltophilia. It is, therefore, plausible that the S. maltophilia-induced alginate expression by P. aeruginosa might ensure added protection both to S. maltophilia against tobramycin, and to P. aeruginosa against other antimicrobial agents. This might explain why about 25% of patients do not recover their baseline lung function after treatment for a PE episode (Sanders et al., 2010, 2011), therefore suggesting that a suitable CF treatment should take into consideration the threat of such nosocomial indirect pathogens as S. maltophilia.
In conclusion, for the first time our findings show that a reciprocal interference between S. maltophilia and P. aeruginosa in CF lung is plausible. P. aeruginosa exhibits an aggressive lifestyle dominating S. maltophilia during competitive planktonic growth and biofilm development. However, in CF lung S. maltophilia cannot be considered as simply a “by-stander.” In fact, growing in a biofilm together with S. maltophilia changes the physiology of P. aeruginosa and, therefore, modulates its virulence profile. This might confer some selective “fitness advantage” to P. aeruginosa under the specific conditions of chronic infection or, alternatively, cause its hypervirulentation thus leading to PE.
Our findings, however, need to be confirmed by in vitro and in vivo investigations focused on other CF S. maltophilia/P. aeruginosa combinations since, in infections where both species are present, the outcome over time could be highly influenced by the phenotype of the strains involved.
Author Contributions
AP performed analyses, statistically evaluated results and, together with GDB, drafted the manuscript and assisted in the study design. VC and SDN performed analyses. FV designed and supervised the analyses of gene expression. EF collected and processed clinical specimens, and provided clinical expertise for the discussion of the results. GDB supervised analyses, defined the study design, discussed the results and drafted the manuscript. All authors read, reviewed, and approved the final manuscript.
Conflict of Interest Statement
The Reviewer Jose L. Martinez declares that, despite co-hosting a Research Topic with Giovanni Di Bonaventura, the review process was handled objectively and no conflict of interest exists. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was partly supported by the grant “ex-60%, 2014-2015” from the “G. d'Annunzio” University of Chieti-Pescara, Italy.
Abbreviations
CF, cystic fibrosis; PE, pulmonary exacerbation; QS, quorum sensing; MHA, Mueller-Hinton agar; TSB, tryptone soya broth; CAMHB, cation-adjusted Mueller-Hinton broth; PBS, phosphate buffered saline; OD, optical density; EDTA, ethylenediaminetetraacetic acid; MOI, multiplicity of infection; RT-PCR, real-time PCR; CI, competitive index; RIR, relative increase ratio; EPS, extracellular polymeric substance.
References
Amadori, A., Antonelli, A., Balteri, I., Schreiber, A., Bugiani, M., and De Rose, V. (2009). Recurrent exacerbations affect FEV(1) decline in adult patients with cystic fibrosis. Respir. Med. 103, 407–413. doi: 10.1016/j.rmed.2008.09.024
Armstrong, D., Grimwood, K., Carlin, J. B., Carzino, R., Hull, J., Olinsky, A., et al. (1998). Severe viral respiratory infections in infants with cystic fibrosis. Pediatr. Pulmonol. 26, 371–379.
Baldan, R., Cigana, C., Testa, F., Bianconi, I., De Simone, M., Pellin, D., et al. (2014). Adaptation of Pseudomonas aeruginosa in cystic fibrosis airways influences virulence of Staphylococcus aureus in vitro and murine models of co-infection. PLoS ONE 9:e89614. doi: 10.1371/journal.pone.0089614
Berg, G., Eberl, L., and Hartmann, A. (2005). The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 7, 1673–1685. doi: 10.1111/j.1462-2920.2005.00891.x
Bittar, F., Richet, H., Dubus, J. C., Reynaud-Gaubert, M., Stremler, N., Sarles, J., et al. (2008). Molecular detection of multiple emerging pathogens in sputa from cystic fibrosis patients. PLoS ONE 3:e2908. doi: 10.1371/journal.pone.0002908
Blau, H., Linnane, B., Carzino, R., Tannenbaum, E. L., Skoric, B., Robinson, P. J., et al. (2014). Induced sputum compared to bronchoalveolar lavage in young, non-expectorating cystic fibrosis children. J. Cyst. Fibros. 13, 106–110. doi: 10.1016/j.jcf.2013.05.013
Burke, V., Robinson, J. O., Richardson, C. J., and Bundell, C. S. (1991). Longitudinal studies of virulence factors of Pseudomonas aeruginosa in cystic fibrosis. Pathology 23, 145–148. doi: 10.3109/00313029109060814
Ciofu, O., Hansen, C. R., and Høiby, N. (2013). Respiratory bacterial infections in cystic fibrosis. Curr. Opin. Pulm. Med. 19, 251–258. doi: 10.1097/MCP.0b013e32835f1afc
Clifton, I. J., Kastelik, J. A., Peckham, D. G., Hale, A., Denton, M., Etherington, C., et al. (2008). Ten years of viral and non-bacterial serology in adults with cystic fibrosis. Epidemiol. Infect. 136, 128–134. doi: 10.1017/S0950268807008278
Clinical Laboratory Standards Institute (CLSI). (2010). Performance Standards for Antimicrobial Susceptibility Testing; Sixteenth Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute.
D'Argenio, D. A., Wu, M., Hoffman, L. R., Kulasekara, H. D., Déziel, E., Smith, E. E., et al. (2007). Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 2, 512–533. doi: 10.1111/j.1365-2958.2007.05678.x
Davinic, M., Carty, N. L., Colmer-Hamood, J. A., San Francisco, M., and Hamood, A. N. (2009). Role of Vfr in regulating exotoxin A production by Pseudomonas aeruginosa. Microbiology 155(Pt 7), 2265–2273. doi: 10.1099/mic.0.028373-0
Di Bonaventura, G., Pompilio, A., Zappacosta, R., Petrucci, F., Fiscarelli, E., Rossi, C., et al. (2010). Role of excessive inflammatory response to Stenotrophomonas maltophilia lung infection in DBA/2 mice and implications for cystic fibrosis. Infect. Immun. 78, 2466–2476. doi: 10.1128/IAI.01391-09
Di Bonaventura, G., Prosseda, G., Del Chierico, F., Cannavacciuolo, S., Cipriani, P., Petrucca, A., et al. (2007b). Molecular characterization of virulence determinants of Stenotrophomonas maltophilia strains isolated from patients affected by cystic fibrosis. Int. J. Immunopathol. Pharmacol. 20, 529–537.
Di Bonaventura, G., Spedicato, I., D'Antonio, D., Robuffo, I., and Piccolomini, R. (2004). Biofilm formation by Stenotrophomonas maltophilia: modulation by quinolones, trimethoprim-sulfamethoxazole, and ceftazidime. Antimicrob. Agents Chemother. 48, 151–160. doi: 10.1128/AAC.48.1.151-160.2004
Di Bonaventura, G., Stepanović, S., Picciani, C., Pompilio, A., and Piccolomini, R. (2007a). Effect of environmental factors on biofilm formation by clinical Stenotrophomonas maltophilia isolates. Folia Microbiol. (Praha) 52, 86–90. doi: 10.1007/BF02932144
Duan, K., Dammel, C., Stein, J., Rabin, H., and Surette, M. G. (2003). Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 50, 1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x
Emerson, J., Rosenfeld, M., McNamara, S., Ramsey, B., and Gibson, R. L. (2002). Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatr. Pulmonol. 34, 91–100. doi: 10.1002/ppul.10127
Geisenberger, O., Givskov, M., Riedel, K., Høiby, N., Tümmler, B., and Eberl, L. (2000). Production of N-acyl-L-homoserine lactones by Pseudomonas aeruginosa isolates from chronic lung infections associated with cystic fibrosis. FEMS Microbiol. Lett. 2, 273–278.
Goodman, A. L., and Lory, S. (2004). Analysis of regulatory networks in Pseudomonas aeruginosa by genomewide transcriptional profiling. Curr. Opin. Microbiol. 7, 39–44. doi: 10.1016/j.mib.2003.12.009
Goss, C. H., and Burns, J. L. (2007). Exacerbations in cystic fibrosis. 1: epidemiology and pathogenesis. Thorax 62, 360–367. doi: 10.1136/thx.2006.060889
Goss, C. H., Ott, K., Aitken, M. L., and Rubenfeld, G. D. (2002). Detecting Stenotrophomonas maltophilia does not reduce survival of patients with cystic fibrosis. Am. J. Respir. Crit. Care Med. 166, 356–361. doi: 10.1164/rccm.2109078
Govan, J. R., and Deretic, V. (1996). Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 3, 539–574.
Harrison, F. (2007). Microbial ecology of the cystic fibrosis lung. Microbiology 153, 917–923. doi: 10.1099/mic.0.2006/004077-0
Hibbing, M. E., Fuqua, C., Parsek, M. R., and Peterson, S. B. (2010). Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25. doi: 10.1038/nrmicro2259
Hoffmann, N., Rasmussen, T. B., Jensen, P. Ø., Stub, C., Hentzer, M., Molin, S., et al. (2005). Novel mouse model of chronic Pseudomonas aeruginosa lung infection mimicking cystic fibrosis. Infect. Immun. 4, 2504–2514. doi: 10.1128/IAI.73.4.2504-2514.2005
Huse, H. K., Kwon, T., Zlosnik, J. E., Speert, D. P., Marcotte, E. M., and Whiteley, M. (2010). Parallel evolution in Pseudomonas aeruginosa over 39,000 generations in vivo. MBio 1:e00199–e00110. doi: 10.1128/mBio.00199-10
Joly, B., Pierre, M., Auvin, S., Colin, F., Gottrand, F., Guery, B., et al. (2005). Relative expression of Pseudomonas aeruginosa virulence genes analyzed by a real time RT-PCR method during lung infection in rats. FEMS Microbiol. Lett. 243, 271–278. doi: 10.1016/j.femsle.2004.12.012
Kang, S., Denman, S. E., Morrison, M., Yu, Z., and McSweeney, C. S. (2009). An efficient RNA extraction method for estimating gut microbial diversity by polymerase chain reaction. Curr. Microbiol. 58, 464–471. doi: 10.1007/s00284-008-9345-z
Karpati, F., Malmborg, A. S., Alfredsson, H., Hjelte, L., and Strandvik, B. (1994). Bacterial colonisation with Xanthomonas maltophilia–a retrospective study in a cystic fibrosis patient population. Infection 22, 258–263. doi: 10.1007/BF01739911
Kataoka, D., Fujiwara, H., Kawakami, T., Tanaka, Y., Tanimoto, A., Ikawa, S., et al. (2003). The indirect pathogenicity of Stenotrophomonas maltophilia. Int. J. Antimicrob. Agents 22, 601–606. doi: 10.1016/S0924-8579(03)00244-9
Kim, S. J., Park, R. Y., Kang, S. M., Choi, M. H., Kim, C. M., and Shin, S. H. (2006). Pseudomonas aeruginosa alkaline protease can facilitate siderophore-mediated iron-uptake via the proteolytic cleavage of transferrins. Biol. Pharm. Bull. 29, 2295–2300. doi: 10.1248/bpb.29.2295
Lee, B., Haagensen, J. A., Ciofu, O., Andersen, J. B., Høiby, N., and Molin, S. (2005). Heterogeneity of biofilms formed by nonmucoid Pseudomonas aeruginosa isolates from patients with cystic fibrosis. J. Clin. Microbiol. 43, 5247–5255. doi: 10.1128/JCM.43.10.5247-5255.2005
Lyczak, J. B., Cannon, C. L., and Pier, G. B. (2002). Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15, 194–222. doi: 10.1128/CMR.15.2.194-222.2002
Macho, A. P., Zumaquero, A., Ortiz-Martín, I., and Beuzón, C. R. (2007). Competitive index in mixed infections: a sensitive and accurate assay for the genetic analysis of Pseudomonas syringae-plant interactions. Mol. Plant. Pathol. 8, 437–450. doi: 10.1111/j.1364-3703.2007.00404.x
Marshall, B. C. (2004). Pulmonary exacerbations in cystic fibrosis: it's time to be explicit! Am. J. Respir. Crit. Care Med. 169, 781–782. doi: 10.1164/rccm.2401009
Pérez-Osorio, A. C., Williamson, K. S., and Franklin, M. J. (2010). Heterogeneous rpoS and rhlR mRNA levels and 16S RNA/rDNA (rRNA gene) ratios within Pseudomonas aeruginosa biofilms, sampled by laser capture microdissection. J. Bacteriol. 192, 2991–3000. doi: 10.1128/JB.01598-09
Pihl, M., Chávez de Paz, L. E., Schmidtchen, A., Svensäter, G., and Davies, J. R. (2010). Effects of clinical isolates of Pseudomonas aeruginosa on Staphylococcus epidermidis biofilm formation. FEMS Immunol. Med. Microbiol. 59, 504–512. doi: 10.1111/j.1574-695x.2010.00707.x
Pompilio, A., Crocetta, V., Confalone, P., Nicoletti, M., Petrucca, A., Guarnieri, S., et al. (2010). Adhesion to and biofilm formation on IB3-1 bronchial cells by Stenotrophomonas maltophilia isolates from cystic fibrosis patients. BMC Microbiol. 10:102. doi: 10.1186/1471-2180-10-102
Pompilio, A., Piccolomini, R., Picciani, C., D'Antonio, D., Savini, V., and Di Bonaventura, G. (2008). Factors associated with adherence to and biofilm formation on polystyrene by Stenotrophomonas maltophilia: the role of cell surface hydrophobicity and motility. FEMS Microbiol. Lett. 287, 41–47. doi: 10.1111/j.1574-6968.2008.01292.x
Pompilio, A., Pomponio, S., Crocetta, V., Gherardi, G., Verginelli, F., Fiscarelli, E., et al. (2011). Phenotypic and genotipic characterization of Stenotrophomonas maltophilia isolates from patients with cystic fibrosis: genome diversity, biofilm formation, and virulence. BMC Microbiol. 11:159. doi: 10.1186/1471-2180-11-159
Pressler, T., Bohmova, C., Conway, S., Dumcius, S., Hjelte, L., Høiby, N., et al. (2011). Chronic Pseudomonas aeruginosa infection definition: EuroCareCF Working Group report. J. Cyst. Fibros. 10, S75–S78. doi: 10.1016/s1569-1993(11)60011-8
Qin, Z., Yang, L., Qu, D., Molin, S., and Tolker-Nielsen, T. (2009). Pseudomonas aeruginosa extracellular products inhibit staphylococcal growth and disrupt established biofilms by Staphylococcus epidermidis. Microbiology 155, 2148–2156. doi: 10.1099/mic.0.028001-0
Reid, D. W., Lam, Q. T., Schneider, H., and Walters, E. H. (2004). Airway iron and iron regulatory cytokines in cystic fibrosis. Eur. Respir. J. 24, 286–291. doi: 10.1183/09031936.04.00104803
Reid, D. W., Latham, R., Lamont, I. L., Camara, M., and Roddam, L. F. (2013). Molecular analysis of changes in Pseudomonas aeruginosa load during treatment of a pulmonary exacerbation in cystic fibrosis. J. Cyst. Fibros. 12, 688–699. doi: 10.1016/j.jcf.2013.03.008
Rogers, G. B., Hoffman, L. R., Whiteley, M., Daniels, T. W., Carroll, M. P., and Bruce, K. D. (2010b). Revealing the dynamics of polymicrobial infections: implications for antibiotic therapy. Trends Microbiol. 18, 357–364. doi: 10.1016/j.tim.2010.04.005
Rogers, G. B., Stressmann, F. A., Walker, A. W., Carroll, M. P., and Bruce, K. D. (2010a). Lung infections in cystic fibrosis: deriving clinical insight from microbial complexity. Expert. Rev. Mol. Diagn. 10, 187–196. doi: 10.1586/erm.09.81
Ryan, R. P., Fouhy, Y., Garcia, B. F., Watt, S. A., Niehaus, K., Yang, L., et al. (2008). Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol. Microbiol. 68, 75–86. doi: 10.1111/j.1365-2958.2008.06132.x
Sanders, D. B., Bittner, R. C., Rosenfeld, M., Hoffman, L. R., Redding, G. J., and Goss, C. H. (2010). Failure to recover to baseline pulmonary function after cystic fibrosis pulmonary exacerbation. Am. J. Respir. Crit. Care Med. 182, 627–632. doi: 10.1164/rccm.200909-1421OC
Sanders, D. B., Bittner, R. C., Rosenfeld, M., Redding, G. J., and Goss, C. H. (2011). Pulmonary exacerbations are associated with subsequent FEV1 decline in both adults and children with cystic fibrosis. Pediatr. Pulmonol. 46, 393–400. doi: 10.1002/ppul.21374
Savli, H., Karadenizli, A., Kolayli, F., Gundes, S., Ozbek, U., and Vahaboglu, H. (2003). Expression stability of six housekeeping genes: a proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J. Med. Microbiol. 52(Pt 5), 403–408. doi: 10.1099/jmm.0.05132-0
Shank, E. A., and Kolter, R. (2009). New developments in microbial interspecies signaling. Curr. Opin. Microbiol. 12, 205–214. doi: 10.1016/j.mib.2009.01.003
Sibley, C. D., Duan, K., Fischer, C., Parkins, M. D., Storey, D. G., Rabin, H. R., et al. (2008b). Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 4:e1000184. doi: 10.1371/journal.ppat.1000184
Sibley, C. D., Parkins, M. D., Rabin, H. R., Duan, K., Norgaard, J. C., and Surette, M. G. (2008a). A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc. Natl. Acad. Sci. U.S.A. 105, 15070–15075. doi: 10.1073/pnas.0804326105
Sibley, C. D., Parkins, M. D., Rabin, H. R., and Surette, M. G. (2009). The relevance of the polymicrobial nature of airway infection in the acute and chronic management of patients with cystic fibrosis. Curr. Opin. Investig. Drugs 10, 787–794.
Spicuzza, L., Sciuto, C., Vitaliti, G., Di Dio, G., Leonardi, S., and La Rosa, M. (2009). Emerging pathogens in cystic fibrosis: ten years of follow-up in a cohort of patients. Eur. J. Clin. Microbiol. Infect. Dis. 28, 191–195. doi: 10.1007/s10096-008-0605-4
Stressmann, F. A., Rogers, G. B., Marsh, P., Lilley, A. K., Daniels, T. W., Carroll, M. P., et al. (2011). Does bacterial density in cystic fibrosis sputum increase prior to pulmonary exacerbation? J. Cyst. Fibros. 10, 357–365. doi: 10.1016/j.jcf.2011.05.002
Tashiro, Y., Yawata, Y., Toyofuku, M., Uchiyama, H., and Nomura, N. (2013). Interspecies interaction between Pseudomonas aeruginosa and other microorganisms. Microbes Environ. 28, 13–24. doi: 10.1264/jsme2.ME12167
Varposhti, M., Entezari, F., and Feizabadi, M. M. (2014). Synergistic interactions in mixed-species biofilms of pathogenic bacteria from the respiratory tract. Rev. Soc. Bras. Med. Trop. 47, 649–652. doi: 10.1590/0037-8682-0262-2013
Vettoretti, L., Plésiat, P., Muller, C., El Garch, F., Phan, G., Attrée, I., et al. (2009). Efflux unbalance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 53, 1987–1997. doi: 10.1128/AAC.01024-08
Wolter, J. M., Bowler, S. D., and McCormack, J. G. (1999). Are antipseudomonal antibiotics really beneficial in acute respiratory exacerbations of cystic fibrosis? Aust. N.Z. J. Med. 29, 15–21. doi: 10.1111/j.1445-5994.1999.tb01583.x
Yang, L., Rau, M. H., Hoiby, N., Molin, S., and Jelsbak, L. (2011). Bacterial adaptation during chronic infection revealed by independent component analysis of transcriptomic data. BMC Microbiol. 11:184. doi: 10.1186/1471-2180-11-184
Keywords: cystic fibrosis, lung infections, Stenotrophomonas maltophilia, Pseudomonas aeruginosa, microbial interactions
Citation: Pompilio A, Crocetta V, De Nicola S, Verginelli F, Fiscarelli E and Di Bonaventura G (2015) Cooperative pathogenicity in cystic fibrosis: Stenotrophomonas maltophilia modulates Pseudomonas aeruginosa virulence in mixed biofilm. Front. Microbiol. 6:951. doi: 10.3389/fmicb.2015.00951
Received: 17 April 2015; Accepted: 27 August 2015;
Published: 16 September 2015.
Edited by:
Evangelos Giamarellos-Bourboulis, University of Athens Medical School, GreeceReviewed by:
Malgorzata Anna Mikaszewska-Sokolewicz, The Medical University of Warsaw, PolandJose L. Martinez, Centro Nacional de Biotecnología, Spain
Athina Savva, Centre Hospitalier Universitaire Vaudois and University of Lausanne, Switzerland
Copyright © 2015 Pompilio, Crocetta, De Nicola, Verginelli, Fiscarelli and Di Bonaventura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giovanni Di Bonaventura, Laboratory of Clinical Microbiology, Department of Medical, Oral and Biotechnological Sciences, “G. d'Annunzio” University of Chieti-Pescara, Via dei Vestini 31, 66100 Chieti, Italy,Z2RpYm9uYXZlbnR1cmFAdW5pY2guaXQ=
 Arianna Pompilio
Arianna Pompilio Valentina Crocetta
Valentina Crocetta Serena De Nicola
Serena De Nicola Fabio Verginelli
Fabio Verginelli Ersilia Fiscarelli
Ersilia Fiscarelli Giovanni Di Bonaventura
Giovanni Di Bonaventura