- Pacific Northwest National Laboratory, Richland, WA, USA
The tandem gene clusters orfR-ombB-omaB-omcB and orfS-ombC-omaC-omcC of the metal-reducing bacterium Geobacter sulfurreducens PCA are responsible for trans-outer membrane electron transfer during extracellular reduction of Fe(III)-citrate and ferrihydrite [a poorly crystalline Fe(III) oxide]. Each gene cluster encodes a putative transcriptional factor (OrfR/OrfS), a porin-like outer-membrane protein (OmbB/OmbC), a periplasmic c-type cytochrome (c-Cyt, OmaB/OmaC) and an outer-membrane c-Cyt (OmcB/OmcC). The individual roles of OmbB, OmaB and OmcB in extracellular reduction of Fe(III), however, have remained either uninvestigated or controversial. Here, we showed that replacements of ombB, omaB, omcB, and ombB-omaB with an antibiotic gene in the presence of ombC-omaC-omcC had no impact on reduction of Fe(III)-citrate by G. sulfurreducens PCA. Disruption of ombB, omaB, omcB, and ombB-omaB in the absence of ombC-omaC-omcC, however, severely impaired the bacterial ability to reduce Fe(III)-citrate as well as ferrihydrite. These results unequivocally demonstrate an overlapping role of ombB-omaB-omcB and ombC-omaC-omcC in extracellular Fe(III) reduction by G. sulfurreducens PCA. Involvement of both ombB-omaB-omcB and ombC-omaC-omcC in extracellular Fe(III) reduction reflects the importance of these trans-outer membrane protein complexes in the physiology of this bacterium. Moreover, the kinetics of Fe(III)-citrate and ferrihydrite reduction by these mutants in the absence of ombC-omaC-omcC were nearly identical, which suggests that absence of any protein subunit eliminates function of OmaB/OmbB/OmcB protein complex. Finally, orfS was found to have a negative impact on the extracellular reduction of Fe(III)-citrate and ferrihydrite in G. sulfurreducens PCA probably by serving as a transcriptional repressor.
Introduction
Geobacter sp. play critical roles in global cycling of carbon, iron (Fe), manganese (Mn), and other elements. They can oxidize organic matter intracellularly and then transfer the released electrons to the terminal electron acceptors, such as Fe(III) and Mn(IV) oxides that are external to the bacterial cells. Because of their extracellular electron transfer capability, Geobacter sp. have been harnessed for the bioremediation of metal and organic contaminants in the subsurface sediments and for bioenergy production (Lovley et al., 2004, 2011).
To reduce extracellular Fe(III) and Mn(IV) oxides, Geobacter sp. transfer electrons from the quinone/quinol pool in the cytoplasmic or inner membrane, through the periplasm and across the outer membrane to the oxide surfaces directly via Geobacter nanowires and/or redox proteins, such as c-type cytochromes (c-Cyt; Weber et al., 2006; Shi et al., 2007, 2009; Bird et al., 2011; Strycharz-Glaven et al., 2011; Bond et al., 2012; Snider et al., 2012). The outer membrane of Gram-negative bacteria, however, is a physical barrier for electron conductance (Shi et al., 2012a). To overcome this barrier, Geobacter sulfurreducens PCA employ the porin-cytochrome (Pcc) trans-outer membrane protein complexes for electron conductance across the outer membrane (Liu et al., 2014; Shi et al., 2014). The characterized Pcc protein complexes consist of a porin-like outer-membrane protein (OmbB or OmbC), a periplasmic 8-heme c-Cyt (OmaB or OmaC) and an outer-membrane 12-heme c-Cyt (OmcB or OmcC). The Pcc protein complexes have been isolated from the membrane fraction of G. sulfurreducens PCA cultured with Fe(III)-citrate as the terminal electron acceptor. After they were reconstituted in proteoliposomes, the isolated Pcc protein complexes transferred electrons from methyl viologen inside the proteoliposomes, across the lipid-bilayer to the external Fe(III)-citrate and ferrihydrite [a poorly crystalline Fe(III) oxide] (Liu et al., 2014). It is proposed that OmbB or OmbC serves as a scaffold through which OmaB or OmaC and OmcB or OmcC are inserted to form a heme-based electron conduit of sufficient length to span the entire width of outer membrane. This model is similar to that proposed for MtrABC trans-outer membrane protein complex of the metal-reducing bacterium Shewanella oneidensis MR-1(Hartshorne et al., 2009; Richardson et al., 2012; White et al., 2013; Liu et al., 2014). In S. oneidensis MR-1, MtrABC complex is responsible for trans-outer membrane electron transfer during extracellular reduction of Fe(III) oxides and can transfer electrons directly to Fe(III) oxides at rates sufficient to support in vivo anaerobic respiration of S. oneidensis MR-1 (Hartshorne et al., 2009; White et al., 2013). It should be pointed out that the Pcc proteins and Mtr proteins are phylogenetically unrelated and they appear to have evolved independently to the similar functions (Liu et al., 2014; Shi et al., 2014).
The genes encoding Pcc proteins are clustered in the same region (i.e., pcc gene cluster) of bacterial genomes. The pcc gene clusters are found in the genomes of all sequenced Geobacter sp. and 11 other bacteria from six different phyla, which reflect the importance of Pcc protein complexes in trans-outer membrane electrons transfer by the Gram-negative bacteria (Shi et al., 2014). G. sulfurreducens PCA possesses four pcc gene clusters and two of which, ombB-omaB-omcB and ombC-omaC-omcC, are directly involved in extracellular reduction of Fe(III) (Liu et al., 2014; Shi et al., 2014). The ombB-omaB-omcB and ombC-omaC-omcC are part of tandem gene clusters, which also include orfR and orfS that encode putative transcriptional factors (Figure 1; Leang et al., 2003; Leang and Lovley, 2005; Liu et al., 2014; Shi et al., 2014). At the amino acid sequence level, OmbB/OmbC and OmaB/OmaC are 100% identical, respectively; while OrfR/OrfS and OmcB/OmcC are 99 and 71% identical, respectively. Thus, orfR-ombB-omaB-omcB and orfS-ombC-omaC-omcC are a result of gene duplication (Leang et al., 2003; Liu et al., 2014; Shi et al., 2014).

FIGURE 1. The orfR gene clusters of Geobacter sulfurreducens PCA. The genes encoding transcriptional factors, porin-like outer-membrane proteins, the periplasmic c-type cytochromes and the outer-membrane c-type cytochromes are labeled in black, green, red, and purple, respectively.
Previously, we found that replacement of ombB-omaB-omcB or ombC-omaC-omcC with an antibiotic resistant gene had little or no impact on Fe(III)-citrate reduction and limited impacts on ferrihydrite reduction by G. sulfurreducens PCA, while disruption of ombB-omaB-omcB-orfS-ombC-omaC-omcC significantly impaired the ability of G. sulfurreducens PCA to reduce Fe(III)-citrate and ferrihydrite. All these results demonstrate the direct involvement of both ombB-omaB-omcB and ombC-omaC-omcC in extracellular reduction of Fe(III)-citrate and ferrihydrite (Liu et al., 2014). However, the roles of individual genes of these clusters, such as ombB and omaB, in extracellular reduction of Fe(III) had remained largely uninvestigated. Moreover, the role of omcB in Fe(III) reduction had remained controversial (Leang et al., 2003; Liu et al., 2014). In contrast to our previous results in G. sulfurreducens PCA (Liu et al., 2014), disruption of omcB in the presence of omcC greatly impaired the bacterial ability to reduce Fe(III)-citrate by G. sulfurreducens DL-1, which suggests that OmcC is not involved in Fe(III)-citrate reduction (Leang et al., 2003). Moreover, OmcB was once believed to mediate electron conductance across the outer membrane by itself because it was deeply embedded in the outer membrane (Qian et al., 2007; Butler et al., 2009). To further clarify their roles, we conducted detailed characterizations of ombB, omaB, and omcB with regards to extracellular reduction of Fe(III)-citrate and ferrihydrite by G. sulfurreducens PCA.
Materials and Methods
Bacterial Growth, Mutant Construction, and Gene Cloning
Geobacter sulfurreducens PCA (ATCC® 51573TM) was routinely cultured in the medium with 10 mM acetate as an electron donor and 40 mM fumarate as an electron acceptor prior to construction of gene replacement mutants. The gene replacement mutants and related complement strains were constructed by using established protocols (Coppi et al., 2001; Leang et al., 2003; Lloyd et al., 2003; Rollefson et al., 2009; Liu et al., 2014). Briefly, the genomic DNA of G. sulfurreducens PCA was purchased from ATCC (Manassas, VA, USA), which served as a template for PCR amplification of respective PCR fragments that flanked the target genes. The kanamycin and chloramphenicol resistance genes were amplified with pBBR1-MCS2 and pACYC184 as templates, respectively (Chang and Cohen, 1978; Kovach et al., 1995). The PCR fragments that flanked the targeted genes and PCR-amplified kanamycin or chloramphenicol resistance gene were mixed and then served as the templates for the second round PCR amplification of the fragments that contained respective gene replacement mutants. These fragments from the second round PCR were electroporated into the target cells separately to make individual gene replacement mutant via double-homologous recombination between the flanking regions of the PCR fragments and the flanking regions of the target genes on the chromosome (Coppi et al., 2001; Lloyd et al., 2003).
For cloning, the target genes were separately amplified by PCR with their respective primers. After treatment with restriction enzymes, such as BamHI, EcoRI, HindIII, SpeI, and XhoI (New England Biolabs, Ipswich, MA, USA), the PCR fragments were cloned into pBBR1-MCS5 by using the Fast-link DNA ligation kit (Epicenter, Madison, WI, USA; Kovach et al., 1995). After verification by sequencing, the cloned genes were introduced to their respective mutants by conjugation via Escherichia coli strain WM3064 (Coppi et al., 2001). All gene replacement mutants and complement strains were confirmed by PCR amplifications of the disrupted regions of bacterial genome and the cloned genes in a plasmid, respectively. Bacterial strains, plasmids, and oligonucleotide primers used in this study are listed in Supplementary Table S1. The procedures for SDS-PAGE and heme staining were described previously (Thomas et al., 1976; Shi et al., 2006).
Fe(III) Reduction
Amorphous 2-line ferrihydrite was synthesized by hydrolysis of Fe(NO3)3 solution at pH 7 at room temperature. The synthesis was conducted inside an anaerobic chamber to prevent any CO2 contamination (100% N2, Innovative Technology, Inc., Amesbury, MA, USA). Briefly, 140 g of Fe(NO3)3⋅9H2O was dissolved in 700 mL of ddH2O in a 1 L Teflon bottle. The pH of the Fe(NO3)3 solution was slowly adjusted with 2 M NaOH solution until pH 7, which yielded a dark brown suspension. After overnight equilibration under a N2 atmosphere, the suspension pH was still 7. The suspension was centrifuged and resuspended in ddH2O, and this washing procedure was repeated for eight times (Schwertman and Cornell, 2000; Shi et al., 2012b). The synthesized two-line ferrihydrite was characterized using transmission electron microscopy (TEM, Jeol JEM 2010 high-resolution TEM, Peabody, MA, USA) and powder X-ray diffraction (XRD, Philips PW 3040/00 X’pert MPD system, Westborough, MA, USA). Fe(III)-citrate was prepared as described previously (Wang et al., 2008). For Fe(III)-reduction assays, all Geobacter strains were pre-cultured in the medium with fumarate as an electrons acceptor. Antibiotics were used at 200 μg/ml for kanamycin and 10 μg/ml for chloramphenicol and gentamicin. Reduction of 50 mM of Fe(III)-citrate or two-line ferrihydrite was carried out at 30°C with Geobacter cells at starting OD600 of 0.05 in the absence of antibiotic (Leang et al., 2003; Rollefson et al., 2009; Liu et al., 2014). All procedures were performed in an anaerobic chamber (Coy Laboratory Products Inc., Grass Lake, MI, USA) that was filled with 5% H2, 20% CO2, and 75% N2. The reduced Fe(II) was measured with a ferrozine assay (Stookey, 1970), and total Fe was determined with inductively coupled plasma emission spectroscopy (Perkin-Elmer, Waltham, MA, USA).
Results
Characterization of ombB, omaB, and omcB in the Presence of ombC-omaC-omcC
To characterize their roles, we first replaced ombB, omaB, omcB, and ombB-omaB with the kanamycin or chloramphenicol resistance gene. Following verification (Supplementary Figure S1A), the resulting mutants were tested for their growth with fumarate or Fe(III)-citrate as the terminal electron acceptor. For comparison, a previously made ΔombB-omaB-omcB was included. As shown in Figure 2, replacements of ombB, omaB, omcB, and ombB-omaB had no impact on bacterial growth with fumarate or reduction of Fe(III)-citrate. Consistent with our previous results, even replacement of the entire ombB-omaB-omcB gene cluster only slightly decreased the ability of G. sulfurreducens PCA to reduce Fe(III)-citrate at 24 h. Given that both ombB-omaB-omcB and ombC-omaC-omcC are involved in Fe(III)-citrate reduction (Liu et al., 2014), the lack of apparent phenotype of these mutants in reducing Fe(III)-citrate suggests an overlap function between ombB-omaB-omcB and ombC-omaC-omcC in Fe(III) reduction, which makes it difficult to accurately evaluate the functions of ombB, omaB, and omcB in the presence of ombC-omaC-omcC.
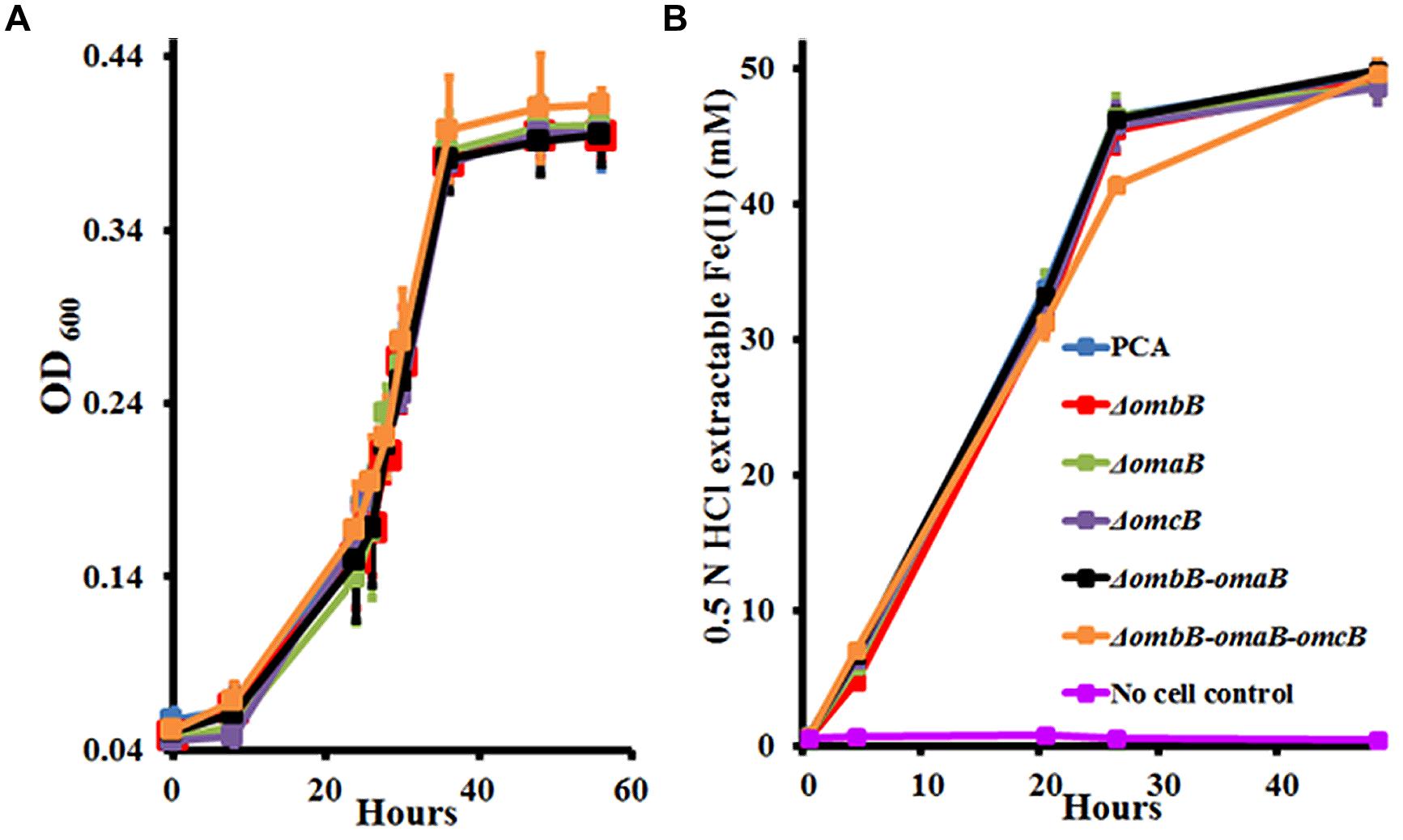
FIGURE 2. Characterization of ombB, omaB, and omcB in the presence of ombC-omaC-omcC. (A) Growth on fumarate. (B) Fe(III)-citrate reduction. The curves are labeled in the same way in (A,B), except that no cell control is omitted in (A). The values plotted at each time point are the average OD600 (A) and 0.5 N HCl extractable Fe(II) (B) measured for each strain from triplicate assays, respectively, and error bars are standard deviations. For points without error bar, the error was smaller than the symbol.
Characterization of ombB, omaB, and omcB in the Absence of ombC-omaC-omcC
To avoid any interference of ombC-omaC-omcC, we replaced ombB, omaB, omcB, ombB-omaB, and ombB-omaB-omcB with a chloramphenicol resistance gene in a previously constructed ΔombC-omaC-omcC (Supplementary Figure S1B) (Liu et al., 2014). Replacements of these genes had no impact on bacterial growth with fumarate as the terminal electron acceptor (Figure 3A), but significantly lowered the bacterial reduction of Fe(III)-citrate (Figure 3B). At 26 h, G. sulfurreducens PCA and ΔombC-omaC-omcC reduced 43.3 ± 1.4 and 42.4 ± 1.5 mM Fe(III)-citrate (n = 3), respectively, while ΔombB/ΔombC-omaC-omcC (or ΔC cluster), ΔomaB/ΔC cluster, ΔomcB/ΔC cluster, ΔombB-omaB/ΔC cluster, and ΔB/ΔC clusters had a decrease of at least 89% of the wild type’s ability to use Fe(III)-citrate as the terminal electron acceptor (Figure 3B). Moreover, the kinetics of Fe(III)-citrate reduction by these newly constructed mutants were nearly identical.
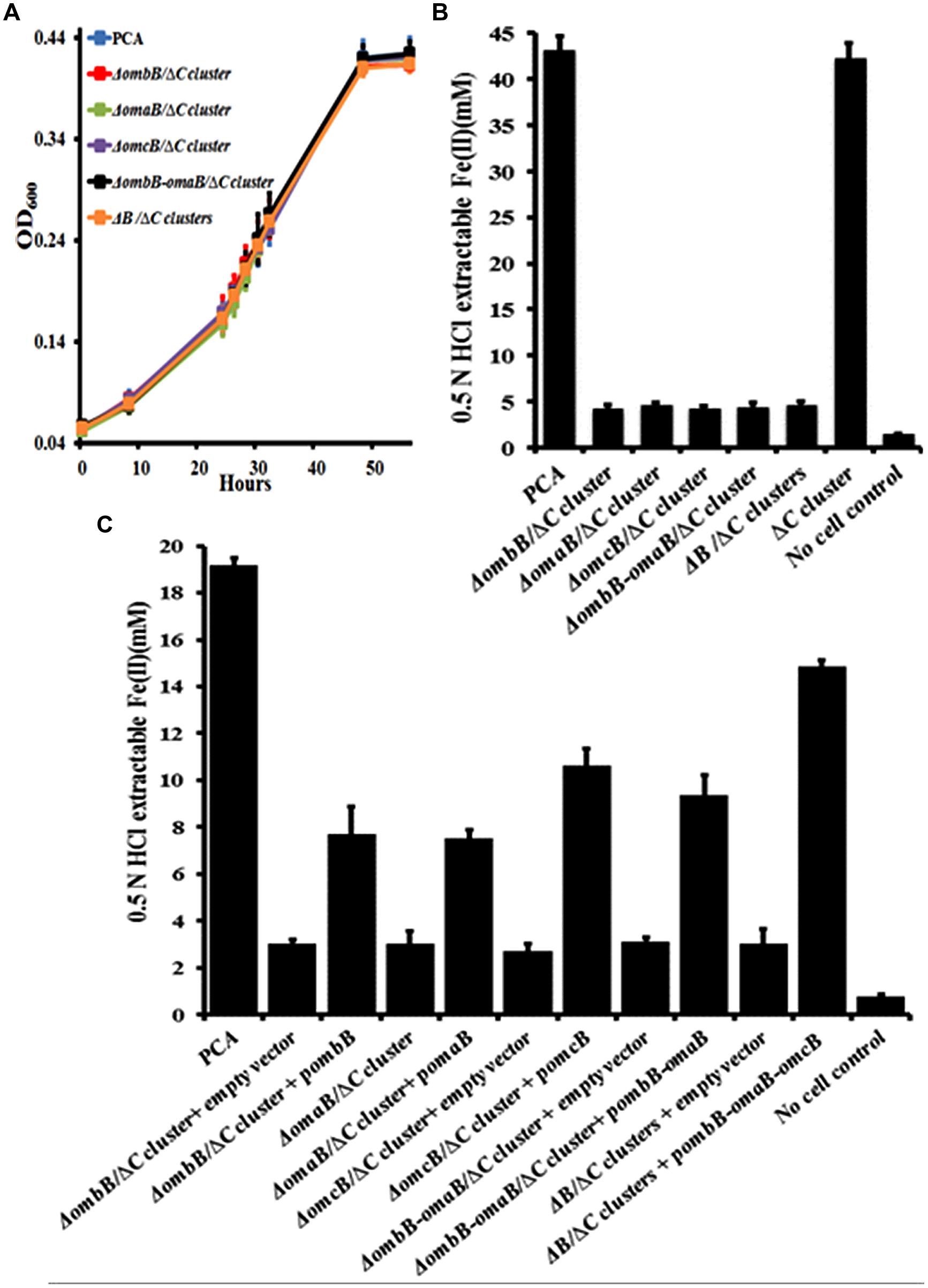
FIGURE 3. Characterization of ombB, omaB, and omcB in the absence of ombC-omaC-omcC. (A) Growth on fumarate. (B) Fe(III)-citrate reduction at 26 h. (C) Ferrihydrite reduction at 360 h. The values plotted at each time point are the average OD600 (A) and 0.5 N HCl extractable Fe(II) (B,C) measured for each strain from triplicate assays, respectively, and error bars are standard deviations. For points without error bar, the error was smaller than the symbol (A).
Apparently varied expression levels of heme-containing of OmcB or OmaB were observed in different gene disruption mutants (Supplementary Figures S1 and S2). To test any polar effect of gene-replacement, we cloned ombB, omaB, omcB, ombB-omaB, and ombB-omaB-omcB and introduced the cloned genes into their respective mutants. Empty vector was also introduced into the mutants and resulting strains served as controls. Addition of empty vector had little impact on Fe(III) reduction as the rates for reducing Fe(III)-citrate were nearly identical for those with or without empty vector (Figure 3B; Supplementary Figure S2). Compared to the controls that were introduced with the empty vector, complemented strains exhibited 2- to 5-fold increase in reducing Fe(III)-citrate at 48 h (Supplementary Figure S2). Because complement omaB and omcB displayed varied levels of expressed proteins (Supplementary Figure S3), the observed difference in Fe(III)-citrate reduction by the complement strains is most likely due to different expression levels of complement genes. All these results consistently show that the phenotypes of Fe(III)-citrate reduction exhibited by these mutants are unlikely attributable to any secondary effect of disrupting these genes. We then tested the impacts of disrupting ombB, omaB, omcB, ombB-omaB, and ombB-omaB-omcB on the ability of G. sulfurreducens PCA to reduce ferrihydrite with these strains.
As shown in Figure 3C, at 360 h, G. sulfurreducens PCA reduced 19.2 ± 1.2 mM ferrihydrite (n = 3), however, ΔombB/ΔC cluster, ΔomaB/ΔC cluster, ΔomcB/ΔC cluster, ΔombB-omaB/ΔC cluster, and ΔB/ΔC cluster, which all contained the empty vector, only reduced 2.7 ± 0.3 to 3.1 ± 0.2 mM ferrihydrite (n = 3). Similar to the results of Fe(III)-citrate reduction, the kinetics of ferrihydrite reduction by these mutants were nearly identical and complements with their respective genes increased the extent of ferrihydrite reduction from 2.5- to 4.9-fold.
Negative Role of orfS in Fe(III) Reduction
During the measurements, we noticed that the ΔB/ΔC clusters constructed in this study reduced much less Fe(III)-citrate than the ΔombB-omaB-omcB-orfS-ombC-omaC-omcC prepared previously (Figure 4B). The only difference between these two mutants is the presence of orfS in the ΔB/ΔC clusters, suggesting that the difference is related to the function of orfS.
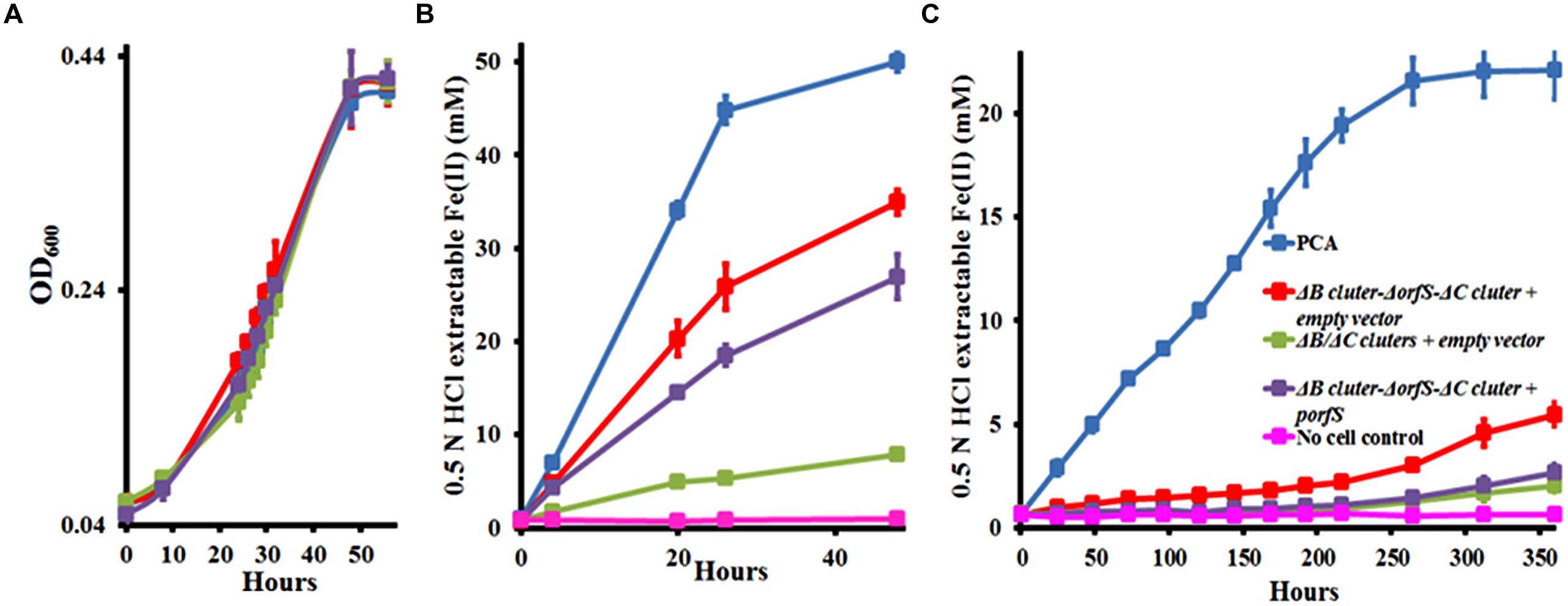
FIGURE 4. Characterization of orfS. (A) Growth on fumarate. (B) Fe(III)-citrate reduction. (C) Ferrihydrite reduction. The curves are labeled in the same way in (A–C), except that no cell control is omitted in (A). The values plotted at each time point are the average OD600 (A) and 0.5 N HCl extractable Fe(II) (B,C) measured for each strain from triplicate assays, respectively, and error bars are standard deviations. For points without error bar, the error was smaller than the symbol.
To further explore the orfS function, we complemented ΔombB-omaB-omcB-orfS-ombC-omaC-omcC with the cloned orfS. An empty vector was also introduced into ΔB/ΔC clusters and ΔombB-omaB-omcB-orfS-ombC-omaC-omcC and resulting strains served as controls. All these strains and G. sulfurreducens PCA showed the nearly identical growth pattern with fumarate as the terminal electron acceptor (Figure 4A). However, they reduced Fe(III)-citrate differently. At 48 h, G. sulfurreducens PCA and ΔB/ΔC clusters with the empty vector reduced 50.0 ± 1.0 and 7.8 ± 0.4 mM Fe(III)-citrate (n = 3), respectively, while ΔombB-omaB-omcB-orfS-ombC-omaC-omcC with the empty vector and ΔombB-omaB-omcB-orfS-ombC-omaC-omcC with orfS in trans reduced 34.0 ± 1.3 and 26.9 ± 2.4 mM Fe(III)-citrate (n = 3), respectively. Thus, complement of ΔombB-omaB-omcB-orfS-ombC-omaC-omcC with orfS further decreased Fe(III)-citrate reduction by 23% (Figure 4B). Similarly, complement of ΔombB-omaB-omcB-orfS-ombC-omaC-omcC with orfS further decreased ferrihydrite reduction. At 360 h, G. sulfurreducens PCA and ΔombB-omaB-omcB-orfS-ombC-omaC-omcC with the empty vector reduced 22.1 ± 1.4 and 5.5 ± 0.5 mM ferrihydrite (n = 3), respectively. Complement of ΔombB-omaB-omcB-orfS-ombC-omaC-omcC with orfS in trans reduced only 2.7 ± 0.4 mM ferrihydrite (n = 3), which was very close to 2.0 ± 0.3 mM ferrihydrite (n = 3) reduced by ΔB/ΔC clusters with the empty vector (Figure 4C). Thus, the presence of orfS appears to negatively impact Fe(III) reduction by G. sulfurreducens.
Discussion
In the presence of ombC-omaC-omcC, disruptions of ombB, omaB, omcB, or ombB-omaB had no impact on Fe(III)-citrate reduction by G. sulfurreducens PCA. These results are consistent with our previous finding that replacement of ombB-omaB-omcB with an antibiotic gene only slightly decreased Fe(III)-citrate reduction by G. sulfurreducens PCA (Liu et al., 2014). However, no observed phenotype for ΔomcB in Fe(III)-citrate reduction by G. sulfurreducens PCA contrasts with previous results for G. sulfurreducens DL-1, in which replacement of omcB significantly impaired this microorganism’s ability to reduce Fe(III)-citrate (Leang et al., 2003). This difference between G. sulfurreducens PCA and G. sulfurreducens DL-1 regarding the role of omcB in Fe(III)-citrate reduction was hypothesized to be attributed to the different ability of these two bacterial strains to compensate for the loss of omcB (Liu et al., 2014). In sharp contrast to that in the presence of ombC-omaC-omcC, disruptions of ombB, omaB, omcB, or ombB-omaB in the absence of ombC-omaC-omcC greatly impaired the bacterial ability to reduce Fe(III)-citrate as well as ferrihydrite. Together, these results unequivocally demonstrate that ombC, omaC, and omcC of G. sulfurreducens PCA can quickly compensate for the loss of their respective counterparts in ombB-omaB-omcB. These results are consistent with our previous findings that both ombB-omaB-omcB and ombC-omaC-omcC contributed to the extracellular reduction of Fe(III)-citrate and ferrihydrite by G. sulfurreducens PCA (Liu et al., 2014). They are also consistent with the findings that both ombB-omaB-omcB and ombC-omaC-omcC of G. sulfurreducens PCA are expressed under the conditions tested and they are highly identical (71–100% identical at the amino acid sequence level; Leang et al., 2003; Liu et al., 2014; Shi et al., 2014).
The observed overlapping role of ombB-omaB-omcB and ombC-omaC-omcC in Fe(III)-citrate and ferrihydrite reduction by G. sulfurreducens PCA is, however, different from that of mtrD-mtrE-mtrF-omcA-mtrC-mtrA-mtrB of S. oneidensis MR-1. Although mtrA/mtrD, mtrB/mtrD, and mtrC/mtrF are paralogs, respectively, only mtrC-mtrA-mtrB have been implicated in extracellular reduction of Fe(III) by S. oneidensis MR-1and the role of mtrD-mtrE-mtrF remains unclear (Hartshorne et al., 2009; Coursolle and Gralnick, 2010).
The reduction kinetics of ΔombB/ΔC cluster, ΔomaB/ΔC cluster, ΔomcB/ΔC cluster, ΔombB-omaB/ΔC cluster and ΔB/ΔC clusters are nearly identical. These results support previous findings that OmbB, OmaB, and OmcB form a functional protein complex, which is a 20-heme trimer, for transferring electrons across the outer membrane (Liu et al., 2014). Thus, loss of any subunit of this protein complex eliminates its function. This is, however, different from the MtrABC protein complex of S. oneidensis MR-1 in which MtrAB can still transfer electrons across the outer membrane in the absence of MtrC (Hartshorne et al., 2009; White et al., 2013). This difference between OmbB/OmaB/OmcB of G. sulfurreducens PCA and MtrABC of S. oneidensis MR-1 may be attributed to the fact that the periplasmic c-Cyt OmaB contains eight hemes and the outer-membrane c-Cyt OmcB has 12 hemes, while each of the periplasmic c-Cyt MtrA and outer-membrane c-Cyt MtrC possesses 10 hemes (Shi et al., 2005, 2006; Hartshorne et al., 2007; Liu et al., 2014). The eight hemes of OmaB may not form a heme-based conduit that is long enough to span the entire width of the outer membrane and may require the two hemes of OmcB for transferring electrons across the outer membrane. Consistent with this suggestion, OmcB is only partially exposed to the bacterial surface (Qian et al., 2007). It is, thus, possible that a substantial portion of OmcB is inserted into the porin-like outer-membrane protein OmbB where it interfaces with OmaB to facilitate transfer of electrons across the outer membrane, which is similar in principle to the MtrABC protein complex of S. oneidensis MR-1 (Hartshorne et al., 2009; Richardson et al., 2012). Alternatively, in S. oneidensis MR-1, Mtr-associated proteins, such as OmcA and MtrF, can compensate for the loss of MtrC (Coursolle and Gralnick, 2012), while in the absence of ombC-omaC-omcC, no additional outer membrane c-Cyt is available.
Our previous results showed that ΔombB-omaB-omcB-orfS-ombC-omaC-omcC could still reduce Fe(III)-citrate and ferrihydrite although at a significantly decreased rate compared to the wild type. Because G. sulfurreducens PCA possessed two more pcc gene clusters in addition to ombB-omaB-omcB and ombC-omaC-omcC, we suggested that the residual ability of reducing Fe(III)-citrate and ferrihydrite by ΔombB-omaB-omcB-orfS-ombC-omaC-omcC could be attributed to the functions of remaining pcc gene clusters or other redox proteins with trans-outer membrane electron transfer capabilities (Liu et al., 2014; Shi et al., 2014). The observed negative role of orfS in reducing Fe(III)-citrate and ferrihydrite by the mutant without ombB-omaB-omcB and ombC-omaC-omcC indeed supports our previous suggestion. OrfS is predicted to be a transcriptional factor of the TetR family. Previous results indicated that OrfS was not involved in regulating expression of ombB-omaB-omcB or ombC-omaC-omcC clusters and its functional role was unclear (Leang and Lovley, 2005). The results reported from this study clearly show that presence of orfS decreases Fe(III)-citrate and ferrihydrite reduction. Given that most members of the TetR family are transcriptional repressors (Ramos et al., 2005), we hypothesize that OrfS may also negatively regulate the expression of other genes with functions similar to that of ombB-omaB-omcB and ombC-omaC-omcC when Fe(III) serves as the terminal electron acceptor.
In summary, the results from this study clearly demonstrate the direct involvements of ombB, ombB, and omcB in extracellular reduction of Fe(III)-citrate and ferrihydrite by G. sulfurreducens PCA. They also show a negative role of orfS in extracellular reduction of Fe(III) by G. sulfurreducens PCA. Moreover, the results from this as well as previous investigations collectively demonstrate the involvement of ombB-omaB-omcB, ombC-omaC-omcC and probably other proteins in electron conductance across the outer membrane during extracellular reduction of Fe(III) by G. sulfurreducens PCA. Existence of multiple and parallel trans-outer membrane extracellular electron transfer pathways critical to extracellular reduction of Fe(III) not only reflects the importance of extracellular reduction of Fe(III) in the physiology of G. sulfurreducens, but also is probably one of the main reasons that the results of Fe(III) reduction by this bacterium from different research groups are sometimes not comparable.
Author Contributions
JF, JZ, and LS designed the study. YL conducted the research. YL, JF, JZ, and LS wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Subsurface Biogeochemical Research program (SBR)/Office of Biological and Environmental Research (BER), U.S. Department of Energy (DOE), and is a contribution of the Pacific Northwest National Laboratory (PNNL) Scientific Focus Area. YL was supported by the Genome Science Program (GSP)/BER (DE-SC0007229). A portion of the research was performed at the Environmental Molecular Sciences Laboratory (EMSL), a national scientific user facility sponsored by DOE-BER and located at PNNL. PNNL is operated for the DOE by Battelle under contract DE-AC05-76RLO 1830.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01075
References
Bird, L. J., Bonnefoy, V., and Newman, D. K. (2011). Bioenergetic challenges of microbial iron metabolisms. Trends Microbiol. 19, 330–340. doi: 10.1016/j.tim.2011.05.001
Bond, D. R., Strycharz-Glaven, S. M., Tender, L. M., and Torres, C. I. (2012). On electon transport through Geobacter biofilms. ChemSumChem 5, 1099–1105. doi: 10.1002/cssc.201100748
Butler, J. E., Young, N. D., and Lovley, D. R. (2009). Evolution from a respiratory ancestor to fill syntrophic and fermentative niches: comparative genomics of six Geobacteraceae species. BMC Genomics 10:103. doi: 10.1186/1471-2164-10-103
Chang, A. C., and Cohen, S. N. (1978). Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134, 1141–1156.
Coppi, M. V., Leang, C., Sandler, S. J., and Lovley, D. R. (2001). Development of a genetic system for Geobacter sulfurreducens. Appl. Environ. Microbiol. 67, 3180–3187. doi: 10.1128/AEM.67.7.3180-3187.2001
Coursolle, D., and Gralnick, J. A. (2010). Modularity of the Mtr respiratory pathway of Shewanella oneidensis strain MR-1. Mol. Microbiol. 77, 995–1008. doi: 10.1111/j.1365-2958.2010.07266.x
Coursolle, D., and Gralnick, J. A. (2012). Reconstruction of extracellular respiration pathways for iron(III) reduction in Shewanella oneidensis strain MR-1. Front. Microbiol. 3:56. doi: 10.3389/fmicb.2012.00056
Hartshorne, R. S., Jepson, B. N., Clarke, T. A., Field, S. J., Fredrickson, J., and Zachara, J. (2007). Characterization of Shewanella oneidensis MtrC: a cell-surface decaheme cytochrome involved in respiratory electron transport to extracellular electron acceptors. J. Biol. Inorg. Chem. 12, 1083–1094. doi: 10.1007/s00775-007-0278-y
Hartshorne, R. S., Reardon, C. L., Ross, D., Nuester, J., Clarke, T. A., and Gates, A. J. (2009). Characterization of an electron conduit between bacteria and the extracellular environment. Proc. Natl. Acad. Sci. U.S.A. 106, 22169–22174. doi: 10.1073/pnas.0900086106
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M., et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Leang, C., Coppi, M. V., and Lovley, D. R. (2003). OmcB, a c-type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens. J. Bacteriol. 185, 2096–2103. doi: 10.1128/JB.185.7.2096-2103.2003
Leang, C., and Lovley, D. R. (2005). Regulation of two highly similar genes, omcB and omcC, in a 10 kb chromosomal duplication in Geobacter sulfurreducens. Microbiology 151, 1761–1767. doi: 10.1099/mic.0.27870-0
Liu, Y., Wang, Z., Liu, J., Levar, C., Edwards, M. J., Babauta, J. T., et al. (2014). A trans-outer membrane porin-cytochrome protein complex for extracellular electron transfer by Geobactersul furreducens PCA. Environ. Microbiol. Rep. 6, 776–785. doi: 10.1111/1758-2229.12204
Lloyd, J. R., Leang, C., Hodges Myerson, A. L., Coppi, M. V., and Cuifo, S. (2003). Biochemical and genetic characterization of PpcA, a periplasmic c-type cytochrome in Geobacter sulfurreducens. Biochem. J. 369, 153–161. doi: 10.1042/bj20020597
Lovley, D. R., Holmes, D. E., and Nevin, K. P. (2004). Dissimilatory Fe(III) and Mn(IV) reduction. Adv. Microb. Physiol. 49, 219–286. doi: 10.1016/S0065-2911(04)49005-5
Lovley, D. R., Ueki, T., Zhang, T., Malvankar, N. S., Shrestha, P. M., and Flanagan, K. A. (2011). Geobacter: the microbe electric’s physiology, ecology, and practical applications. Adv. Microb. Physiol. 59, 1–100. doi: 10.1016/B978-0-12-387661-4.00004-5
Qian, X., Reguera, G., Mester, T., and Lovley, D. R. (2007). Evidence that OmcB and OmpB of Geobacter sulfurreducens are outer membrane surface proteins. FEMS Microbiol. Lett. 277, 21–27. doi: 10.1111/j.1574-6968.2007.00915.x
Ramos, J. L., Martinez-Bueno, M., Molina-Henares, A. J., Teran, W., Watanabe, K., and Zhang, X. (2005). The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69, 326–356. doi: 10.1128/MMBR.69.2.326-356.2005
Richardson, D. J., Butt, J. N., Fredrickson, J. K., Zachara, J. M., Shi, L., and Edwards, M. J. (2012). The ‘porin-cytochrome’ model for microbe-to-mineral electron transfer. Mol. Microbiol. 85, 201–212. doi: 10.1111/j.1365-2958.2012.08088.x
Rollefson, J. B., Levar, C. E., and Bond, D. R. (2009). Identification of genes involved in biofilm formation and respiration via mini-Himar transposon mutagenesis of Geobacter sulfurreducens. J. Bacteriol. 191, 4207–4217. doi: 10.1128/JB.00057-09
Schwertman, U., and Cornell, R. M. (2000). Iron Oxides in the Laboratory: Preparation and Characterization. Weinheim: Wiley-VCH.
Shi, L., Chen, B., Wang, Z., Elias, D. A., Mayer, M. U., and Gorby, Y. A. (2006). Isolation of a high-affinity functional protein complex between OmcA and MtrC: Two outer membrane decaheme c-type cytochromes of Shewanella oneidensis MR-1. J. Bacteriol. 188, 4705–4714. doi: 10.1128/JB.01966-05
Shi, L., Fredrickson, J., and Zachara, J. (2014). Genomic analyses of bacterial porin-cytochrome gene clusters. Front. Microbiol. 5:657. doi: 10.3389/fmicb.2014.00657
Shi, L., Lin, J. T., Markillie, L. M., Squier, T. C., and Hooker, B. S. (2005). Overexpression of multi-heme C-type cytochromes. BioTechniques 38, 297–299. doi: 10.2144/05382PT01
Shi, L., Richardson, D. J., Wang, Z., Kerisit, S. N., Rosso, K. M., and Zachara, J. M. (2009). The roles of outer membrane cytochromes of Shewanella and Geobacter in extracellular electron transfer. Environ. Microbiol. Rep. 1, 220–227. doi: 10.1111/j.1758-2229.2009.00035.x
Shi, L., Rosso, K. M., Clarke, T. A., Richardson, D. J., Zachara, J. M., and Fredrickson, J. K. (2012a). Molecular underpinnings of Fe(III) oxide reduction by Shewanella oneidensis MR-1. Front. Microbiol. 3:50. doi: 10.3389/fmicb.2012.00050
Shi, Z., Zachara, J. M., Shi, L., Wang, Z., Moore, D. A., and Kennedy, D. W. (2012b). Redox reactions of reduced flavin mononucleotide (FMN), riboflavin (RBF), and anthraquinone-2,6-disulfonate (AQDS) with ferrihydrite and lepidocrocite. Environ. Sci. Technol. 46, 11644–11652. doi: 10.1021/es301544b
Shi, L., Squier, T. C., Zachara, J. M., and Fredrickson, J. K. (2007). Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes. Mol. Microbiol. 65, 12–20. doi: 10.1111/j.1365-2958.2007.05783.x
Snider, R. M., Strycharz-Glaven, S. M., Tsoi, S. D., Erickson, J. S., and Tender, L. M. (2012). Long-range electron transport in Geobacter sulfurreducens biofilms is redox gradient-driven. Proc. Natl. Acad. Sci. U.S.A. 109, 15467–15472. doi: 10.1073/pnas.1209829109
Stookey, L. (1970). Ferrozine-a new spectrophotometric reagent for iron. Anal. Chem. 42, 779–781. doi: 10.1021/ac60289a016
Strycharz-Glaven, S. M., Snider, R. M., Guiseppi-Elie, A., and Tender, L. M. (2011). On the electrical conductivity of microbial nanowires and biofilms. Energy Environ. Sci. 4, 4366–4379. doi: 10.1039/c1ee01753e
Thomas, P. E., Ryan, D., and Levin, W. (1976). An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75, 168–176. doi: 10.1016/0003-2697(76)90067-1
Wang, Z., Liu, C., Wang, X., Marshall, M. J., Zachara, J. M., Rosso, K. M., et al. (2008). Kinetics of reduction of Fe(III) complexes by outer membrane cytochromes MtrC and OmcA of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 74, 6746–6755. doi: 10.1128/AEM.01454-08
Weber, K. A., Achenbach, L. A., and Coates, J. D. (2006). Microorganisms pumping iron: anaerobic microbial iron oxidation and reduction. Nat. Rev. Microbiol. 4, 752–764. doi: 10.1038/nrmicro1490
Keywords: Fe(III) reduction, Geobacter, porin-cytochrome, trans-outer membrane protein complex, extracellular electron transfer
Citation: Liu Y, Fredrickson JK, Zachara JM and Shi L (2015) Direct involvement of ombB, omaB, and omcB genes in extracellular reduction of Fe(III) by Geobacter sulfurreducens PCA. Front. Microbiol. 6:1075. doi: 10.3389/fmicb.2015.01075
Received: 06 June 2015; Accepted: 18 September 2015;
Published: 01 October 2015.
Edited by:
Jennifer Glass, Georgia Institute of Technology, USAReviewed by:
Caroline Ajo-Franklin, Lawrence Berkeley National Laboratory, USANadia Szeinbaum, Georgia Institute of Technology, USA
Copyright © 2015 Liu, Fredrickson, Zachara and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Shi, Pacific Northwest National Laboratory, 902 Battelle Boulevard, P.O. Box 999, Richland, WA 99352, USA, bGlhbmcuc2hpQHBubmwuZ292
 Yimo Liu
Yimo Liu James K. Fredrickson
James K. Fredrickson John M. Zachara
John M. Zachara Liang Shi
Liang Shi