- 1Department of Microbiology, Central Institute for Food and Nutrition Research (Zentralinstitut für Ernährungs- und Lebensmittelforschung), Technische Universität München, Freising, Germany
- 2Functional Microbiology, Institute of Microbiology, Department of Pathobiology, University of Veterinary Medicine Vienna, Vienna, Austria
- 3Chair of Food Chemistry and Molecular Sensory Science, Technische Universität München, Freising, Germany
- 4Lehrstuhl für Mikrobielle Ökologie, Wissenschaftszentrum Weihenstephan, Technische Universität München, Freising, Germany
The emetic toxin cereulide produced by Bacillus cereus is synthesized by the modular enzyme complex Ces that is encoded on a pXO1-like megaplasmid. To decipher the role of the genes adjacent to the structural genes cesA/cesB, coding for the non-ribosomal peptide synthetase (NRPS), gene inactivation- and overexpression mutants of the emetic strain F4810/72 were constructed and their impact on cereulide biosynthesis was assessed. The hydrolase CesH turned out to be a part of the complex regulatory network controlling cereulide synthesis on a transcriptional level, while the ABC transporter CesCD was found to be essential for post-translational control of cereulide synthesis. Using a gene inactivation approach, we show that the NRPS activating function of the phosphopantetheinyl transferase (PPtase) embedded in the ces locus was complemented by a chromosomally encoded Sfp-like PPtase, representing an interesting example for the functional interaction between a plasmid encoded NRPS and a chromosomally encoded activation enzyme. In summary, our results highlight the complexity of cereulide biosynthesis and reveal multiple levels of toxin formation control. ces operon internal genes were shown to play a pivotal role by acting at different levels of toxin production, thus complementing the action of the chromosomal key transcriptional regulators AbrB and CodY.
Introduction
The cyclic dodecadepsipeptide cereulide, a heat-, acid-, and proteolytically stable toxin, is responsible for the emetic type of food borne illness caused by a specific subgroup of Bacillus cereus. Intoxication with cereulide, which is preformed during vegetative growth of B. cereus in foods, causes nausea and heavy vomiting around 0.5–6 h after consumption of contaminated food (Ehling-Schulz et al., 2004). These symptoms are presumably induced by the interaction of cereulide with 5-HT3 serotonin receptors leading to the stimulation of the afferent vagus nerve (Agata et al., 1995). Usually, these symptoms decline after 24 h, but more severe foodborne intoxications requiring hospitalization or even including fatalities are reported increasingly (Dierick et al., 2005; Naranjo et al., 2011; Messelhäusser et al., 2014; Tschiedel et al., 2015).
In agreement with its chemical structure [D-O-Leu-D-Ala-L-O-Val-L-Val]3, cereulide is produced enzymatically by the non-ribosomal cereulide peptide synthetase Ces (Ces-NRPS; Ehling-Schulz et al., 2005b). NRPSs are large multifunctional enzyme complexes consisting of repetitive modules which selectively incorporate amino acid, α-hydroxy acid or carboxylic acid monomers in the peptide product (Marahiel et al., 1997). The order of the modules usually corresponds directly to that of the monomers in the assembled peptide chain, although strict co-linearity is not always reinforced in nature (Wenzel and Müller, 2005; Lane and Moore, 2011). Very recently we could show that the enzymatic activity of the Ces-NRPS does not follow the canonical NRPS biosynthesis logic, but represents a novel mechanism of non-ribosomal peptide assembly, by using dipeptides rather than monomers as basic units (Marxen et al., 2015a,b).
The cereulide biosynthetic genes were found to be located on a pXO1-like megaplasmid, organized in a 24-kb cluster comprising the seven ces genes cesH, cesP, cesT, cesA, cesB, cesC, and cesD (Ehling-Schulz et al., 2006). The cesPTABCD genes represent an operon being transcribed as a single 23-kb polycistronic mRNA, whereas the adjacently located cesH is transcribed from its own promoter (Dommel et al., 2010; Figure 1). The structural NRPS genes cesA and cesB encode the modules that are responsible for the activation and incorporation of each two monomers: The CesA2 and CesB2 submodules install D-alanine and L-valine, respectively, whereas CesA1 and CesB1 bind α-keto acids of leucine and valine (α-ketoisocaproic acid and α-ketoisovaleric acid; Magarvey et al., 2006; Alonzo et al., 2015). A central condensation domain in the C-terminal part of CesA together with a type I thioesterase (TE) domain located in the C-terminal part of CesB is suggested to act as esterification and elongation center before the final dodecadepsipeptide cereulide is released from the TE domain by macrocyclization (Marxen et al., 2015b).
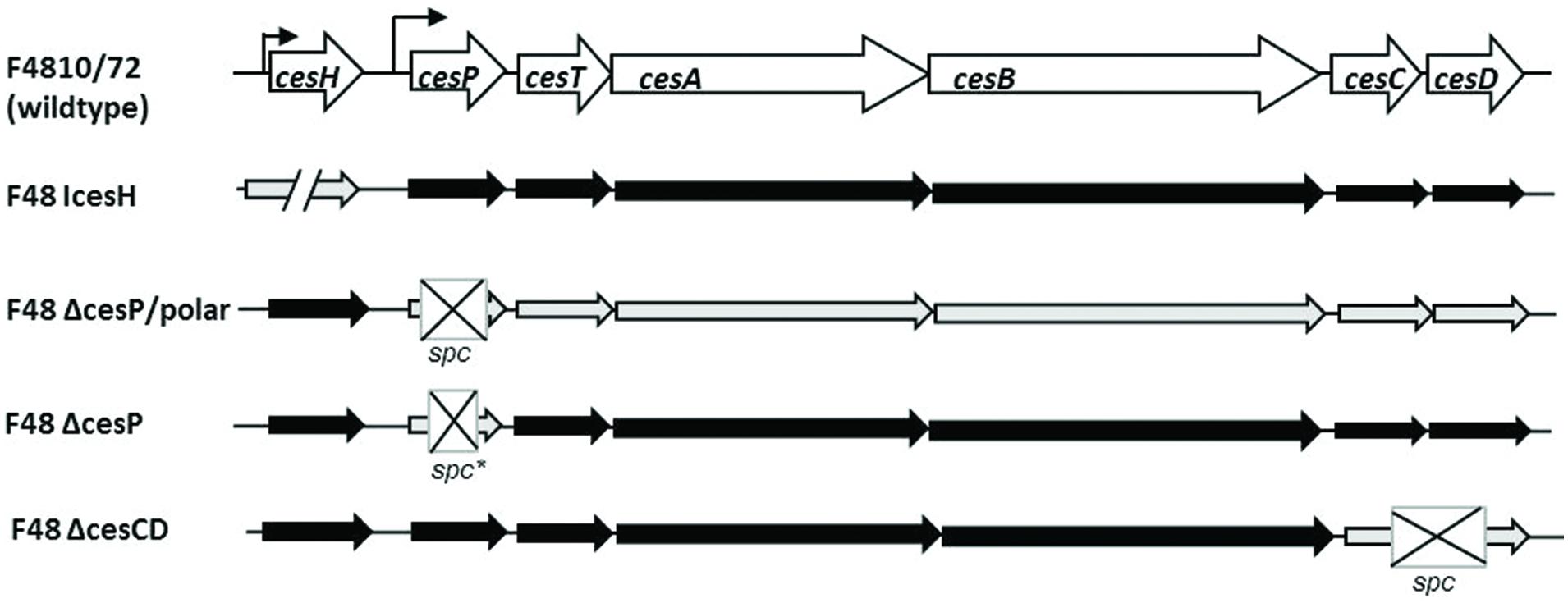
FIGURE 1. Genetic organization of the cereulide biosynthetic gene cluster in F4810/72 wildtype and ces mutant strains used in this study. The locus consists of seven open reading frames encoding a putative hydrolase/acetyltransferase (cesH), a phosphopantetheinyl transferase (cesP), a type II thioesterase (cesT), two non-ribosomal peptide synthetase modules (cesA and cesB) and a putative ABC transporter (cesC and cesD) (according to Ehling-Schulz et al., 2006; Dommel et al., 2010). Identified promoter regions are marked by bent arrows. Gene disruption is indicated by slashes, gene deletion, and introduction of a spectinomycin resistance cassette (spc) by crossed boxes. spc∗ represents a non-polar mutagenic spc cassette lacking promoter- and terminator sequences and followed by a ribosome binding site and a start codon.
The adjacent to cesAB located gene cesT encodes a putative type II thioesterase (TEII), an enzyme which is often found in association with NRPSs. In contrast to the type I thioesterases, which are an integrated part of the NRPS catalyzing the product release, external TEIIs reactivate the catalytic NRPS domains by removing misprimed monomers (Schwarzer et al., 2002). The cesP gene, located upstream of cesT, encodes a putative 4′-phosphopanthetheinyl transferase (PPTase). Such enzymes are detected frequently in association with NRPS enzymes, since they are crucial for their activation. PPTases catalyze the transfer of a 4′-phosphopantheteine (Ppant) moiety of coenzyme A to a conserved serine residue of the PCP domain, thereby converting the apo-carrier protein to its active holo-form (Lambalot et al., 1996).
Besides these typical NRPS-associated genes, three additional open reading frames with so far unknown functions are present in the ces gene cluster: cesH, at the 5′ end, encodes a putative hydrolase or acyltransferase, whose role regarding cereulide synthesis is unclear. cesC and cesD, encoding a putative ABC transporter, are located at the 3′ terminus of the ces operon (Ehling-Schulz et al., 2006).
The aim of this work was to assess the role of the ces genes flanking the structural cereulide synthetase genes in order to gain further insights into the unusual biosynthetic pathway, which catalyzes the production of the highly potent depsipeptide toxin cereulide. An essential prerequisite for translational studies was the availability of a Ces-NRPS specific antibody, which was raised successfully against the CesB1 module. Targeted gene inactivation- and overexpression mutants of the emetic reference strain F4810/72 were constructed and characterized regarding the different steps of toxin synthesis. The influence of the accessory ces genes on the different cereulide process levels is demonstrated and resulting functional predictions are discussed.
Materials and Methods
Bacterial Strains, Plasmids and Growth Conditions
Details on bacterial strains and plasmids used in this study are provided in Table 1 and Supplementary Table S1, respectively. Unless otherwise specified, all B. cereus, Bacillus subtilis and Bacillus megaterium were grown in LB broth (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) or on LB agar plates at 30°C. For liquid cultures, 100 ml LB media was inoculated with approximately 103 cfu/ml from a 14–16 h pre-culture and cultures were incubated in 500 ml baffled flasks with 150 rpm. Escherichia coli strains used for subcloning were cultured in LB at 37°C. Growth was monitored by optical density at 600 nm (OD600) using a GeneQuant pro spectrophotometer (Biochrom). Concentrations of antibiotics applied were 100 μg/ml for ampicillin, spectinomycin, and polymyxin B; 50 μg/ml for kanamycin; 5 μg/ml for erythromycin; 5 μg/ml for chloramphenicol; and 10 μg/ml for tetracycline.
Sequence Analysis
The genome information of the emetic reference strain F4810/72 (AH187), retrieved from the NCBI website (GenBank accession no. CP001177 and CP001179), was used for sequence analysis of the ces locus and homology searches. Amino acid sequences were retrieved from the NCBI database and protein homology search was performed using Blastp available at NCBI1. Multiple sequence alignments were carried out with Clustal W using default parameters (Thompson et al., 1994). Maximum-likelihood trees were constructed with MEGA 5 (Tamura et al., 2011) using the Jones–Thornton–Taylor (JTT) model. All positions containing gaps were eliminated. Numbers at nodes (≥50%) represent bootstrap support of 500 resamplings. Annotation of characteristic protein domains and membrane spanning motifs was achieved with the SMART database (Letunic et al., 2012) and the programs TMHMM2 and TMpred3.
Construction of B. cereus ΔcesP and ΔcesCD Deletion Mutants
Deletion and complementation mutants were constructed as described previously (Lücking et al., 2009). In brief, flanking regions of cesP and cesCD were amplified by PCR using the primer pairs listed in Supplementary Table S2. Digested PCR products, together with an excised spectinomycin resistance cassette (spc) from pUC1318spc (Murphy, 1985), were ligated into the TOPO pCR 2.1 vector (Invitrogen). For cesP, an additional construct with a non-polar mutagenic spectinomycin cassette from pSPCH+2 (Mesnage et al., 2000) was produced to avoid polar effects of the spectinomycin cassette on the ces operon genes located downstream of cesP. Constructs were excised from TOPO and inserted into the conjugative suicide vector pAT113, giving rise to pAT113ΔcesP/spc, pAT113ΔcesCD/spc, and pAT113ΔcesP/spcH+2 (see Supplementary Table S1 for details). Plasmids were then transformed into E. coli JM83/pRK24 by heat shock and the resulting strains were used for transconjugal transfer into B. cereus F4810/72 using a mating procedure previously described (Pezard et al., 1991). For complementation of F48ΔcesCD and F48ΔcesP/Ippt, the genes cesCD and cesP were amplified using the primers cesCD_F_Xba/cesCD_R_Pae and cesP_Pro_F2/cesP_Pro_R (Supplementary Table S2), respectively, and cloned into the shuttle vector pAD123 together with the ∼500 bp ces promoter region. For complementation of F48ΔcesP/Ippt with ppt (NCBI accession no. ACJ79141), the respective gene was amplified with ppt_chr_F and ppt_chr_R and ligated into the shuttle vector pMM1522, containing a xylose-inducible promoter. Constructs were propagated in the non-methylating E. coli strain INV110 cells and then introduced into the according B. cereus mutant by electroporation, giving rise to F48ΔcesCD/com_cesCD, F48ΔcesP/Ippt/com_cesP, and F48ΔcesP/Ippt/com_ppt.
Construction of B. cereus cesH and ppt Insertion- and Overexpression Mutants
Gene inactivation by pMAD integration was performed as described previously (Dommel et al., 2011). In brief, internal fragments (∼300 bp) of cesH and ppt (NCBI accession no. ACJ79141) were amplified by PCR using primers listed in Table S2 and ligated into the thermosensitive shuttle vector pMAD. Constructs were introduced into B. cereus F4810/72 or F48ΔcesP by electroporation and transformants were obtained after 2 days at 30°C on LB plates with erythromycin and X-Gal (20 μg/ml). Plasmid integration was enforced by re-cultivation of a blue colony in LB medium overnight at 42°C for several times. For further experiments, the resulting strains F48IcesH, F48Ippt, and F48ΔcesP/Ippt were cultured at high temperatures (37 or 42°C), which is essential to avoid the loss of the integrated plasmid.
For cesH overexpression, the promoterless cesH gene was amplified using the respective primers listed in Supplementary Table S2. The digested fragment was ligated into pMM1522, harboring a xylose-inducible promoter, and then introduced in B. cereus F4810/72 by electroporation. To induce gene overexpression, 0.1 % D-xylose (v/v) was added to the culture medium.
Production of a Monoclonal anti-CesB Antibody
For generation of a monoclonal antibody targeting the cereulide NRPS, the DNA fragment (3999 bp) encoding the first CesB submodule (CesB1: consisting of an adenylation-, ketoreductase-, and peptidyl carrier protein domain) including the native cesB ribosome binding site was amplified with Pfu polymerase (Promega) from genomic DNA of B. cereus F4810/72 using the primer pair cesB135Nco_for/cesB135Xho_rev (Supplementary Table S2). Cloning via the XhoI/NcoI sites into the pET28b(+) vector resulted in a transcriptional C-terminal His6 tag fusion. CesB1-His6 was amplified from pET28-cesB1 by PCR with cesBpETSpe_for2 and cesBpETSph_rev2 and cloned into the SpeI and SphI restriction sites of pWH1520, giving rise to the xylose-inducible overexpression plasmid pWHCesB1His. This construct was transferred into B. megaterium WH320 protoplasts by PEG-fusion according to the manufacturer’s protocol (MoBiTec, Göttingen). The CesB1His6 protein was overexpressed in recovered B. megaterium cells and purified using Ni-NTA affinity columns (Qiagen) according to the manufacturer’s instruction. The monoclonal antibody (mAB) CesB5-6 was raised by BioGenes (Berlin) using the purified CesB1His6 protein, NMRI mice (obtained from Janvier, France), and the myeloma cell line SP2/0-Ag14 (obtained from DSMZ, Germany). Purified CesB1 mAB with a concentration of 1.68 μg ml-1 in PBS solution was tested for its specificity by enzyme-linked immunosorbent assays (ELISA; data not shown) and by immunoblotting.
Reverse Transcription-qPCR
To analyze cesA transcript levels, 1 ml culture samples of B. cereus strains were harvested at OD600 8 by centrifugation (10000 g, 4°C, 2 min). Total RNA isolation, cDNA synthesis and RT-qPCR were carried out as described previously (Dommel et al., 2011). Transcript levels of the 16S rrn gene served as the reference control for data normalization and relative gene expression ratios were calculated with the REST software (Pfaffl et al., 2002).
Western- and Slot Blot Analysis of CesB
Twenty milliliter of B. cereus cultures, grown to different optical densities, were centrifuged (9500 rpm, 6 min, Sigma 3–18K with angle rotor 19776) and cell pellets resuspended in 2 ml lysis buffer [50 mM Tris-HCl pH 7.6, 2 mM EDTA pH 7.5, 1 mM Pefabloc (Merck)]. Cells were disrupted by two passages through a French Pressure cell press (1000 psi) and the soluble protein fraction was collected by centrifugation of the lysates (13000 rpm, 2x 30 min, 4°C). Protein concentration was determined according to Bradford using the Roti-Quant solution (Carl Roth GmbH). For Western blot analysis, 30 μg of total protein was separated by SDS-PAGE on 8% gels and the proteins were immobilized onto a PVDF membrane (Millipore) by semi-dry blotting for 2 h at 120 mA. A 1:2000 dilution of the monoclonal anti-CesB antibody was used to detect the CesB1 module. The second incubation was done with a 1:20000-dilution of HRP-conjugated goat anti-mouse IgG (Dianova) and blots were developed with the SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific). For slot blot analysis, 25 μg of total protein was blotted directly onto a PVDF membrane by vacuum filtration. Again, the 1:2000 dilution of the monoclonal anti-CesB antibody was used to detect the CesB module of the cereulide synthetase. In addition, blots of the same samples were performed with a 1:5000 dilution of a polyclonal rabbit anti-AtpB serum (Agrisera) detecting the beta subunit of the ATP synthase. As second antibodies served an alkaline phosphatase-conjugated goat anti-mouse IgG (1: 10000 dilution, Dianova) and an alkaline phosphatase-conjugated goat anti-rabbit IgG (1:3000, Dianova), respectively. Chromogenic detection of alkaline phosphatase activity was accomplished with BICP-p-toluidine salt- and NBT solutions (Carl Roth GmbH) according to the manufacturer’s instructions.
Sample Preparation and Cereulide Quantification by Means of UPLC-MS/MS
The biosynthetic production of cereulide in LB broth supplemented with 0.2% glucose and of 13C-labeled cereulide in MOD medium supplemented with 13C1-L-valine was carried out as described previously (Bauer et al., 2010). Extraction of cereulide from autoclaved cells of different B. cereus strains grown in 100 ml LB broth was carried out as described (Stark et al., 2013), using 1 ng of 13C6-cereulide as internal standard. Cereulide quantification via stable isotope dilution analysis (SIDA) was performed using a Xevo TQ-S Acquity i-class UPLC-MS/MS system and an UPLC BEH C18 column (2.1 mm × 50 mm, 1.7 μm; Waters, UK and USA). One microliter aliquots of ethanolic extracts (of B. cereus samples) were injected directly into the UPLC/MS-MS system. Operated with a flow rate of 1.0 ml/min at a temperature of 50°C, the following gradient was used for chromatography: starting with a mixture (90/10, v/v) of methanol and 10 mM ammonium formate (0.1% formic acid), the methanol content was increased to 100% within 0.5 min, kept constant for 0.3 min and decreased within 0.1 min to 90%. Measurements were performed using electrospray with positive ionization and the quantitative calibration mode consisting of the following ion source parameters: capillary voltage +3.5 kV, sampling cone 30 V, source offset 30 V, source temperature 150°C, desolvation temperature 600°C, cone gas 150 L/h, desolvation gas 1000 L/h, collision gas flow 0.15 ml/min and nebuliser gas flow 6.5 bar. Calibration of the TQ-S in the range from m/z 40-1963 was performed using a solution of phosphoric acid (0.1% in acetonitrile). The UPLC and Xevo TQ-S systems were operated with MassLnyxTM 4.1 SCN 813 software, data processing and analysis were performed using TargetLynx. By means of the multiple reaction monitoring (MRM) mode, the ammonium adducts of cereulide (qualifier: m/z 1170.7→172.2; 1170.7→314.2; quantifier: m/z 1170.7→357.2) and 13C6-cereulide (m/z 1176.7→173.2; 1176.7→316.3; 1176.7→358.3) were analyzed using the mass transitions (given in brackets) monitored for a duration of 25 ms. ESI+ mass and product ion spectra were acquired with direct flow infusion using IntelliStart. The MS/MS parameters were tuned for each individual compound, detecting the fragmentation of the [M+NH4]+ molecular ions into specific product ions after collision with argon. For quantitation, 10 ethanolic standard solutions of the analyte cereulide (0.05–10.0 ng/ml) and the internal standard 13C6-cereulide (1 ng/ml) were mixed and analyzed in triplicates by means of UPLC–MS/MS using the MRM mode. Calibration curve was prepared by plotting peak area ratios of analyte to internal standard against concentration ratios of each analyte to the internal standard using linear regression.
Cytotoxicity Test
Aliquots of B. cereus cultures were taken after 24 h of incubation and autoclaved (15 min at 121°C) to lyse cells and denature heat-labile toxins. Cereulide amounts of these samples were determined using the HEp-2 cell based cell culture assay as previously described (Lücking et al., 2009). Cereulide titres of the mutant strains were normalized to the value of the parental strain B. cereus F4810/72 (grown for 24 h at 30 or 37°C, respectively), which was defined as being 100%.
Results
Comparative Sequence Analysis of the Cereulide Synthetase Flanking Genes
Based on the genome information (NCBI accession no. CP001177 and CP001179) available for the emetic reference strain F4810/72 (AH187), a BLAST database search and extensive sequence analysis was carried out. This in silico approach reconfirmed the ces gene cluster boundaries suggested previously (Ehling-Schulz et al., 2006; Dommel et al., 2010). The ces gene locus comprises seven genes (cesHPTABCD), which are present in all emetic B. cereus and emetic Bacillus weihenstephanensis sequenced so far (data not shown). cesH represents the 5′ prime end of the ces locus while cesD, which is followed by a strong terminator sequence, marks the 3′ prime end.
CesH is a putative 31 kDa hydrolase belonging to the α/β-hydrolase fold superfamily of proteins. Besides the plasmid-borne cesH, F4810/72 carries 13 chromosomal genes annotated to code for members of this hydrolase subfamily, with none of them showing notable sequence similarity toward CesH. Comparison of the CesH with amino acid sequences retrieved from annotated genomes of the B. cereus group members revealed a large number of orthologs showing significant similarity. However, proteins with 100% identity (amino acids) were only found in emetic B. cereus strains possessing the ces gene cluster. The CesH sequences retrieved from the ces gene cluster in emetic B. weihenstephanensis strains showed a homology of 91–93% (Figure 2).
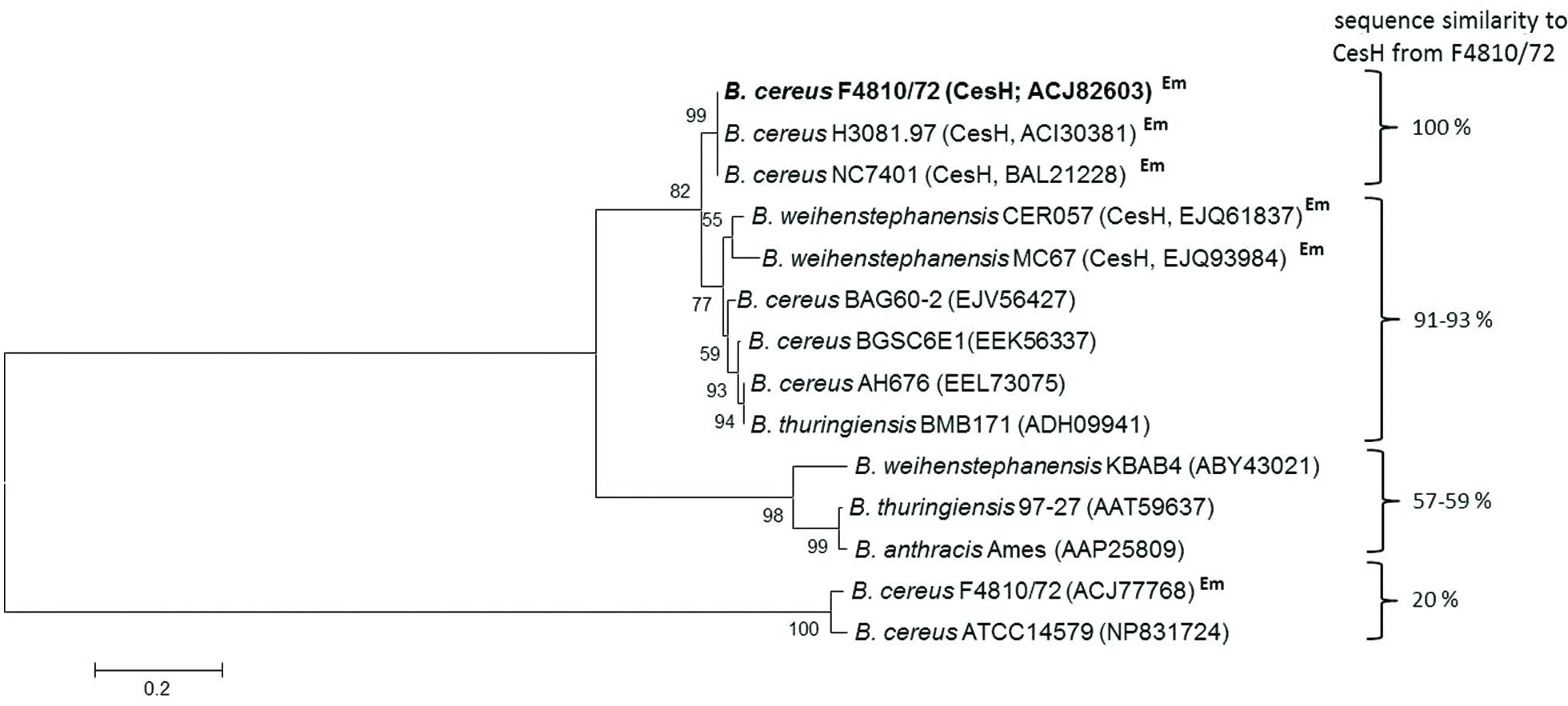
FIGURE 2. Maximum-likelihood protein similarity tree based on amino acid sequences of CesH from Bacillus cereus F4810/72 (AH187) and selected homologs of other B. cereus group members. NCBI accession numbers of proteins are given in brackets. Em indicates an emetic strain containing the ces gene cluster.
cesP codes for a putative 4′-phosphopantetheinyl transferase (PPtase) belonging to the Sfp-like subgroup of the PPTase superfamily. In silico analysis revealed the presence of an additional gene encoding a 4′-PPtase (ACJ79141; herein after referred to as ppt) in the chromosome of B. cereus F4810/72. A homolog of this protein with similarity above 80% is present in almost all B. cereus group members, while CesP homologs can only be found in emetic B. cereus and emetic B. weihenstephanensis strains. An alignment of the amino acid sequences of CesP and Ppt resulted in a rather low degree of homology (27% identity at 56% coverage), nevertheless, according to their size and three conserved motifs, both PPTases can be classified as typical members of the “Sfp-like” family (Copp and Neilan, 2006), showing 32% (CesP) and 26% (Ppt) identity to Sfp (P39135), the prototype PPTase of B. subtilis, respectively (Figure 3).

FIGURE 3. Amino acid sequence alignment of the PPTases CesP (ACJ82723), Ppt (ACJ79141) of B. cereus F4810/72, and Sfp (P39135) of B. subtilis. Three conserved motifs, which are typical for members of the “Sfp-like” PPTase subfamily, are shown in boxes. Identical residues are highlighted with asterisk, similar residues with colon.
The two open reading frames cesC and cesD at the 3′ terminus of the ces gene locus encode a putative ABC transporter. Protein domain annotation using the SMART database (Letunic et al., 2012) revealed the presence of characteristic motifs involved in ATP- binding and hydrolysis for CesC. The amino acid sequence of CesC was found to be homologous to several known ATP-binding proteins, e.g., BerA of B. thuringiensis required for β-exotoxin I production (Espinasse et al., 2002), or TnrB2 of Streptomyces longisporoflavus and BcrA of B. licheniformis, which have been shown to confer resistance to tetronasin and bacitracin, respectively (Linton et al., 1994; Podlesek et al., 1995; Supplementary Figure S1). For CesD, five hydrophobic transmembrane domains were predicted, which are typical for membrane-spanning proteins. CesD showed homology (33–34% identity) only toward the putative permease BerB of B. thuringiensis and its orthologs present in many B. cereus group members. Sequence alignment of CesCD with known ABC transporters of antimicrobial peptides confirmed that CesCD most closely resembles members of the BcrAB subgroup, which generally consist of one ATPase and one permease with six transmembrane helices (Gebhard, 2012; Figure 4).
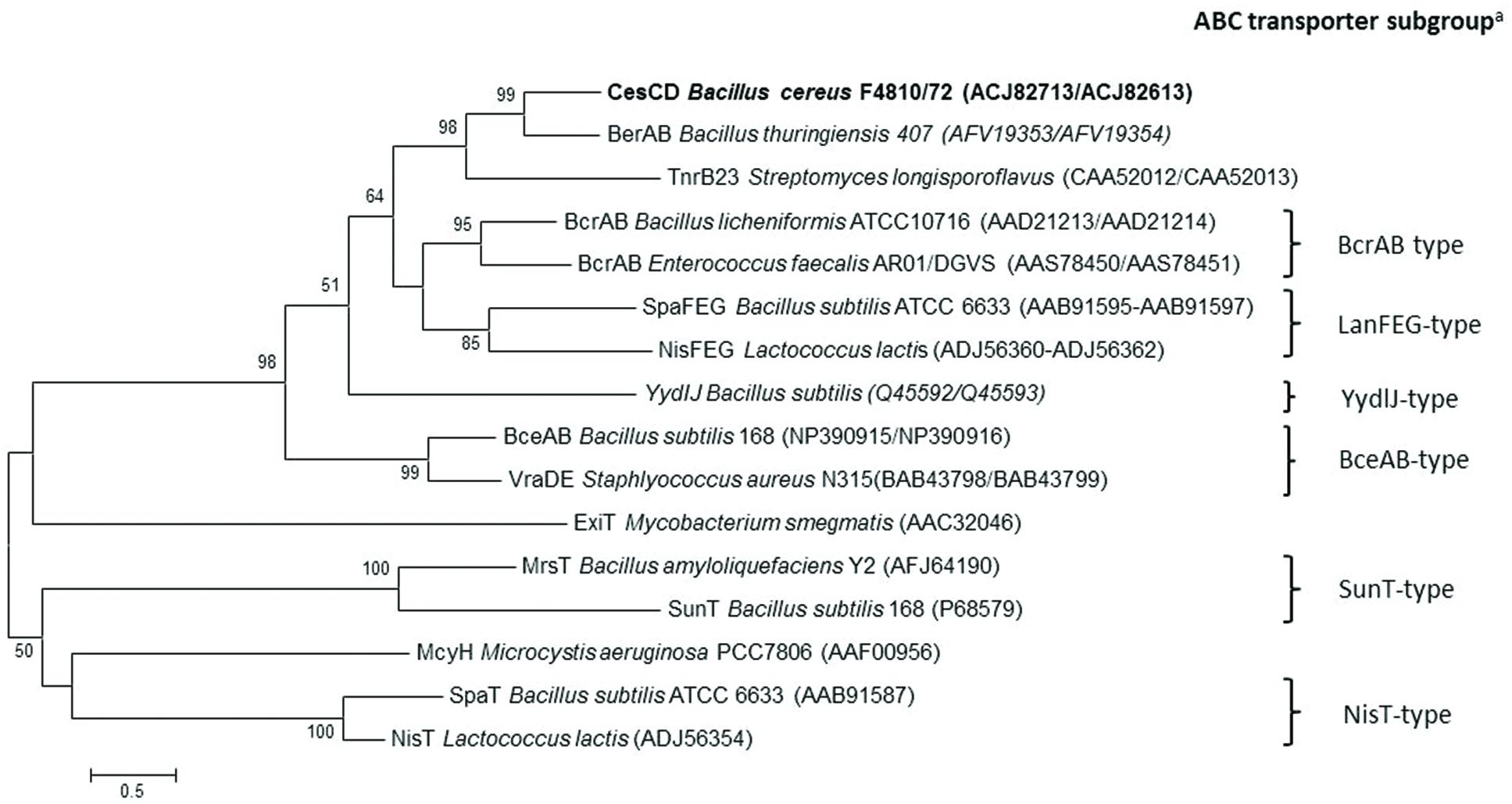
FIGURE 4. Protein similarity tree of the amino acid sequences of CesCD and previously characterized ABC transporters. aClassification of ABC transporters of antimicrobial peptides according to Gebhard (Gebhard, 2012).
Construction of Mutants and Determination of their Cereulide Production Capacities
To investigate the potential role of cesH, cesP, cesC, and cesD on cereulide production, various deletion-, insertion-, and overexpression mutants of B. cereus F4810/72 were constructed as described in the materials and methods section. An overview of mutants generated is provided in Figure 1 and the major genetic characteristics of mutant strains are summarized in Table 1, respectively. After growth in LB medium for 24 h at 30 or 37°C (insertion mutants), culture samples were taken and cells disrupted by autoclaving in order to obtain total cereulide amounts including extra- and intracellularly accumulated toxin. Cereulide concentrations of ethanolic extracts were measured by means of SIDA-UPLC-MS/MS. Furthermore, cytotoxicity was determined by the HEp-2 cell culture assay.
The cereulide production of the cesH inactivation mutant (F48IcesH) turned out to be almost twice as high compared to the wildtype strain. Interestingly, when cesH was overexpressed in the wildtype strain (F48pMM/cesH) with a xylose-inducible shuttle vector, almost no cereulide was detectable, while in the absence of the inducer xylose, cereulide production was comparable to the wildtype (Table 2). These results demonstrate that CesH inhibits cereulide synthesis.
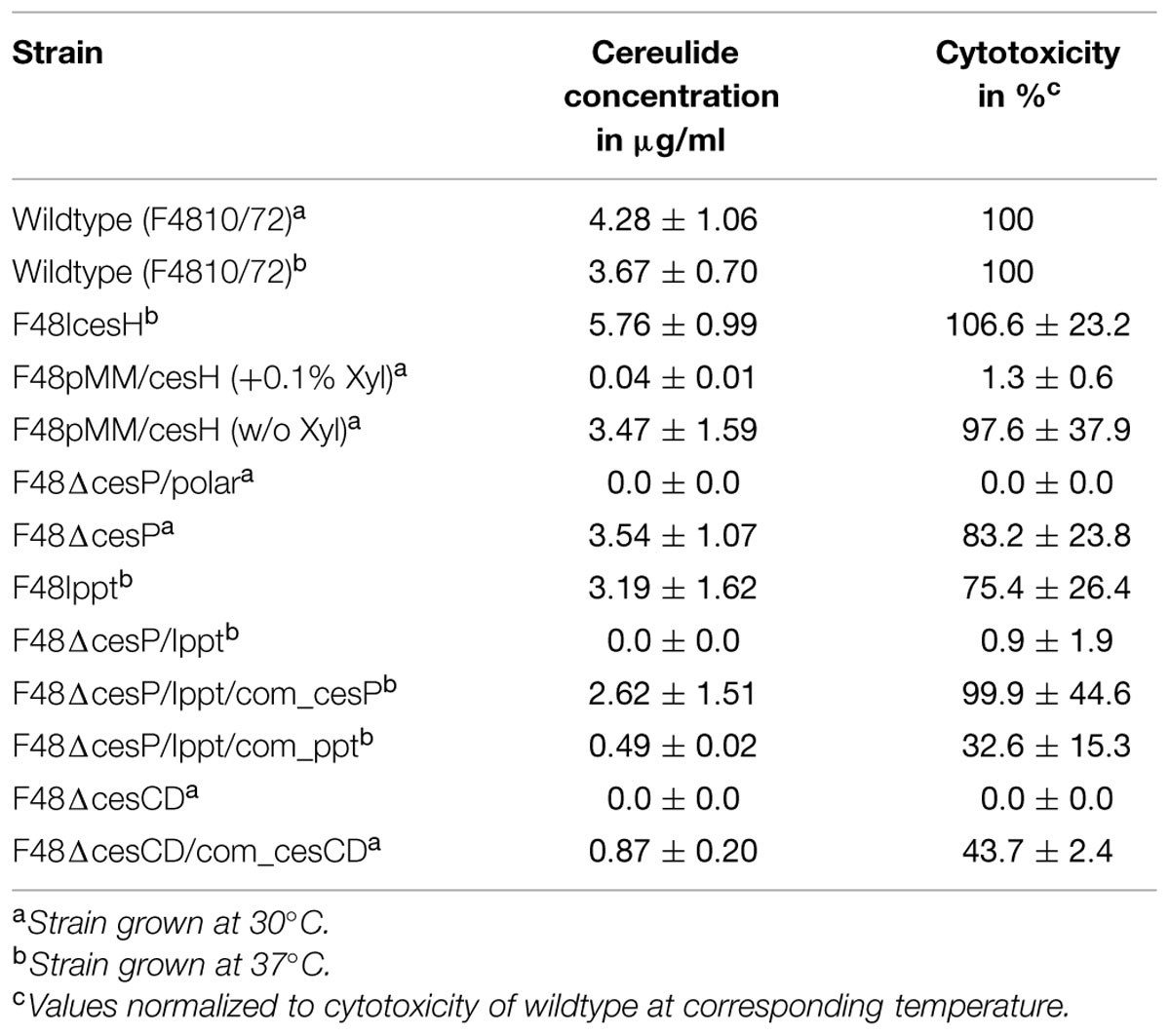
TABLE 2. Cereulide production of wildtype emetic B. cereus and ces gene mutants determined by SIDA-UPLC-MS/MS and HEp-2 bioassay.
For the analysis of cesP, two knockout strains were generated, since cesP is the first gene of the ces operon and disruption of this gene may affect the ces genes located downstream. Thus, in one strain a non-polar spectinomycin resistance cassette (spc) lacking promoter and terminator sequences (Mesnage et al., 2000) was used for mutagenesis (F48ΔcesP), while the other harbors a normal spc potentially leading to polar effects on downstream located genes (F48ΔcesP/polar; see also Figure 1). Indeed, SIDA-UPLC-MS/MS of the latter revealed no cereulide production and no toxicity could be detected toward HEp-2 cells. In contrast, F48ΔcesP produced toxin amounts similar to that in the wildtype, demonstrating cesP-independent cereulide synthesis (Table 2). Since in silico analysis exposed the presence of a second sfp-like PPTase in the chromosome of B. cereus F4810/72 (ppt), a respective gene inactivation mutant was constructed in the wildtype (F48Ippt) to test its possible involvement in toxin formation. UPLC-MS/MS showed that this mutant produces cereulide levels comparable to the wild type. However, when ppt was disrupted in F48ΔcesP, leading to a double knock out mutant in cesP and ppt (F48ΔcesP/Ippt), no cereulide or cytotoxicity was detected (Table 2). This effect was reversible by complementation of the mutant with cesP and – to a lesser extend – with ppt, indicating a redundant role of CesP and Ppt in cereulide production.
For the cesCD deletion mutant (F48ΔcesCD) neither cereulide by means of MS nor cytotoxicity in the HEp-2 bioassay could be determined. Both parameters could be restored to some extend by in trans complementation of F48ΔcesCD with a shuttle vector containing cesC and cesD under the control of the ces locus promoter (Table 2). These results demonstrate that the putative ABC transporter genes cesC and cesD are essential for cereulide formation in B. cereus F4810/72.
Generation of a Monoclonal Antibody Targeting the Cereulide NRPS
Next, we were interested to further dissect the level of regulation leading to the huge differences in cereulide quantities found in the ces mutants. To study the impact of the ces mutations on the expression of the Ces-NRPS, a specific antibody against the cereulide synthetase was generated. The CesB1 submodule of the cereulide synthetase, which was shown to present a ketoacid-binding module unique among Bacillus sp. NRPS (Magarvey et al., 2006), served as antigen for the generation of monoclonal antibodies (mAB) as outlined in the materials and methods section. The specificity of the resulting monoclonal anti-CesB1 antibody was assessed using two emetic-, two non-emetic B. cereus strains and one B. subtilis strain (possessing the surfactin NRPS genes) by ELISA (data not shown) and immunoblotting. Western blot analysis revealed the specificity of the antibody toward the two cereulide-positive strains B. cereus F4810/72 and A529, while no signal was detected for the cereulide-negative strains ATCC 10987 and ATCC 14579 or for B. subtilis 168 (Supplementary Figure S2). The CesB1 mAB reacted specifically with a large protein band migrating way above the range of the protein ladder used, which is consistent with the predicted molecular mass of CesB1 at 304 kDa. The appearance of an additional weak band at 170 kDa indicates fragmentation of the large CesB1 complex, which was repeatedly observed after denaturing PAGE.
Transcriptional and Translational Analysis of ces Mutant Strains
Quantitative RT-PCR detecting cesA mRNA levels as well as immunoblotting using the CesB mAB were carried out to investigate, whether the striking differences of cereulide synthesis of the B. cereus F4810/72 wildtype and the ces mutants (Table 2 and Figure 5C) were caused by variations in NRPS gene transcription or by translational regulation of the synthetase complex. Figure 5 combines the results of ces transcription-, translation-, and cereulide production analyses for the wildtype and seven mutant strains. For the cereulide-negative mutant F48ΔcesP/polar, no cesA transcripts or CesB expression was detectable (Figures 5A,B, lane 4), confirming the assumption that insertion of spc in cesP led to a transcriptional stop of the downstream located ces genes in the operon. In contrast, transcript levels and CesB protein signals for the other two cereulide-deficient mutants, F48ΔcesP/Ippt and F48ΔcesCD, resembled those of the parental strain (Figures 5A,B, lanes 6 and 7). These data suggest that cesP, ppt, cesCD and their respective gene products do not interfere with ces transcription or translation of the cereulide synthetase complex, but seem to play a role in the post-translational formation of active cereulide. Transcript- and CesB protein levels of the cesH insertion mutant (F48IcesH), which revealed higher toxin levels than the wildtype in the MS assay (Figure 5C, lane 2), were comparable to those of the wildtype. However, for the cesH overexpression mutant (F48pMM/cesH), cesA transcription was down-regulated significantly and only a very weak CesB band was detected by slot blot analysis (Figures 5A,B, lane 3). Therefore, the toxin deficiency of the latter strain may be due to cesH and its gene product, acting as a possible transcriptional repressor of the cereulide synthetase genes.
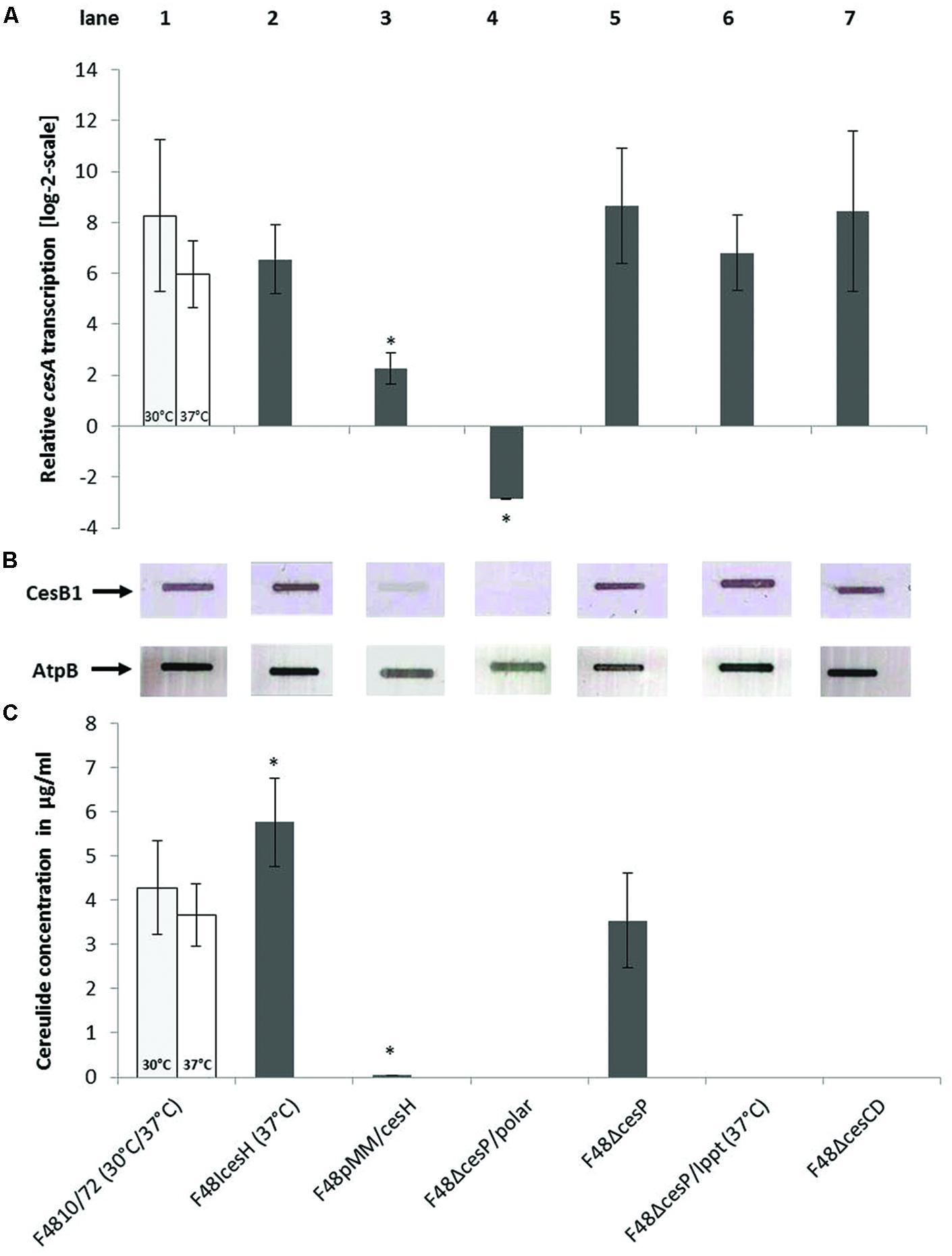
FIGURE 5. Comparison of ces gene transcription, cereulide synthetase expression, and cereulide production of B. cereus F4810/72 wildtype (WT; 1) and the mutant strains F48ΔcesP (5), F48ΔcesP/Ippt (6), F48ΔcesP/polar (4), F48ΔcesCD (7), F48IcesH (2), and F48pMM/cesH (3). (A) Relative cesA transcription of B. cereus strains grown in LB medium at 30 or 37°C (insertion mutants) and harvested at exponential growth phase (OD600 = 8) for RNA isolation. The asterisks denote statistically significant (P < 0.05) differences in cesA mRNA levels between wildtype and mutant strains grown under the same conditions. (B) Slot blot analysis of CesB and AtpB expression using 25 μg of total protein per slot from B. cereus strains harvested at OD600 = 8. The CesB1 module of the cereulide synthetase was detected with the CesB1 mAB and AtpB (beta subunit of ATP synthase) with a polyclonal anti-AtpB antibody, serving as a protein loading control. (C) Cereulide production of B. cereus strains determined by UPLC-MS/MS-SIDA. Strains were incubated in LB medium for 24 h (stationary growth phase) before cereulide was extracted for quantification. Statistically significant (P < 0.05) differences in cereulide concentration between wildtype and mutant strains are marked by asterisks.
Discussion
Hitherto, research on the cereulide biosynthetic ces operon focused on the molecular and biochemical characterization of the non-ribosomal cereulide peptide synthetase encoded by cesA and cesB (Ehling-Schulz et al., 2005b; Magarvey et al., 2006; Alonzo et al., 2015; Marxen et al., 2015b). However, little is known about the adjacent genes cesH, cesP, cesC, and cesD, which code for a putative hydrolase, a PPtase and a putative transport system of the ABC-type, respectively (Ehling-Schulz et al., 2006). So far it is unclear, why different strains of the emetic B. cereus group display highly variable toxicity (Carlin et al., 2006; Stark et al., 2013). Thus, it is important to dissect the role of ces locus genes and their gene products on cereulide formation. Since it was recently shown that the Ces-NRPS represents a novel mechanism for non-ribosomal depsipeptide assembly (Marxen et al., 2015b), information on the functional architecture of the ces gene locus would also improve our general understanding of the complex biochemical pathways involved in the production of natural, non-ribosomally synthesized peptides.
CesH, a Putative Repressor Involved in Timing of ces Gene Transcription
The first gene in the 5′ prime proximity of the ces cluster, cesH, codes for a putative hydrolase of the α/β-hydrolase fold superfamily. According to the ESTER classification database, cesH belongs to the 6_AlphaBeta_hydrolase subgroup, which to date consists of over 3800 members using various substrates (Hotelier et al., 2004). Although cesH is transcribed from its own promoter – while the remaining ces genes are co-transcribed – it is considered to be an integral part of the ces gene locus, since in silico analysis revealed a copy in all emetic B. cereus strains as well as in cereulide-producing B. weihenstephanensis strains. In general, the presence of hydrolase genes in NRPS gene clusters seems to be rare and their function is unknown. We found one gene coding for a CesH – homolog (presenting 59% identity) in close proximity of a 30 kb polyketide-NRPS locus of the non-emetic B. weihenstephanensis KBAB4. Surprisingly, our data revealed an inhibitory effect of CesH on cereulide formation, as CesH overexpression led to a non-toxic phenotype, while the cesH insertion mutant produced more cereulide than the wildtype (Table 2). Transcriptional analysis of the CesH overexpression mutant resulted in strongly down-regulated cesA mRNA levels, which is in line with the very weak CesB slot blot signal obtained for this mutant. These data indicate that CesH is an additional regulatory member of the cereulide pathway by acting directly or indirectly as a transcriptional repressor of the ces gene operon. Our previous work demonstrated that transcription of the polycistronic cesPTABCD genes is co-regulated in a complex manner by several key transcription factors of the chromosome (Ehling-Schulz et al., 2015), e.g., AbrB and CodY, leading to a tightly regulated transcription peak in late exponential phase (Lücking et al., 2009; Frenzel et al., 2012). While AbrB and CodY are responsible for the onset and strong increase of cesPTABCD transcription, CesH may function as a closing signal shutting down mRNA synthesis. Since a direct action of a hydrolase as a transcriptional regulator seems to be unlikely, another possibility may be that CesH acts indirectly by degrading metabolites or quorum sensing signaling molecules that influence ces gene transcription in later growth phases. This hypothesis is in line with transcriptional kinetic studies of cesH in B. cereus F4810/72, demonstrating highest cesH expression in stationary growth phase (Supplementary Figure S3B), while the other ces operon genes are known to be transcribed earlier in the growth cycle (Dommel et al., 2011). Further experiments including activity tests with purified CesH protein, which are clearly beyond the scope of the current study, are necessary to elucidate the regulatory role of cesH in cereulide production in detail.
Plasmid Encoded CesP and Chromosomally Encoded Ppt are Two Sfp Type- PPTases with Functionally Redundant Enzymatic Activities
In contrast to the frequency of proteins from the α/β-hydrolase fold superfamily, only very few members of the PPtase superfamily are present in Bacillus species. By post-translational modification and thereby activation of acyl-, aryl-, or peptidyl- carrier proteins, PPTases are essential enzymes for the synthesis of fatty acids, polyketides and non-ribosomal peptides (Walsh et al., 1997). Based on size, conserved sequence motifs and substrate selectivity, bacterial PPTases can be classified into two major groups, the Sfp-like PPTases and the acyl carrier protein synthases (AcpS; Finking and Marahiel, 2004). While PPTases of the Sfp-type are mostly found in association with NRPS, the enzymes of the AcpS type are linked to the activation of fatty acid and polyketide synthesis. Genome data analysis of B. cereus F4810/72 revealed the presence of two chromosomally encoded PPTases: an AcpS-type PPtase (ACJ81272) and a Sfp-like PPTase (ACJ79141, here named Ppt). The latter is encoded in the vicinity of the dhb operon, which has been shown to be responsible for the non-ribosomal synthesis of the siderophore bacillibactin in B. subtilis (May et al., 2001). The plasmid-encoded cesP gene codes for an additional Sfp-like PPTase and presents an integral part of the ces gene cluster, being co-transcribed with the NRPS genes (Dommel et al., 2010). Surprisingly, CesP was not essential for cereulide synthesis, as deletion of cesP did not significantly alter cereulide production or toxicity of the strain. Only an additionally disrupted ppt led to a toxin-negative phenotype, indicating that Ppt can function as a redundant CesP-PPTase in cereulide biosynthesis. Likewise, CesP seems to be able to functionally complement Ppt, as siderophore production (measured by a colorimetric assay) was detectable in low amounts in mutant strains concerning solely ppt or cesP, but not in the double knockout strain (data not shown). Transcriptional analysis of ppt in B. cereus F4810/72 revealed a weak, but constitutive expression throughout growth with a slight peak in stationary phase (Supplementary Figure S3A), similar to sfp transcription in B. subtilis, which was shown to be weak according to low promoter activity (Nakano et al., 1992). In contrast, transcription of cesP, which is linked to the ces operon, is strongly growth phase dependent, peaking highly in the late logarithmic phase (Dommel et al., 2011). Thus, we propose that the two PPtases encoded by cesP and ppt present functionally redundant enzymes with temporal different expression profiles in the growth cycle of B. cereus F4810/72.
ABC Transporter CesCD is Essential for Post-translational Cereulide Formation
Genes coding for transport systems of the ABC-type are frequently found part of, or in close proximity to, peptide synthetase operons of NRPS products, such as siderophores, antibiotics or lipopeptides. For example the production of lichenysin A of B. licheniformis, tyrocidine of B. brevis, syringomycin of Pseudomonas syringae or pyoverdine of P. aeruginosa are linked to different ABC transporters, which are thought to be involved in product secretion or self-resistance (Quigley et al., 1993; McMorran et al., 1996; Mootz and Marahiel, 1997; Yakimov et al., 1998). Gene disruption of the transporter genes cesC and cesD of B. cereus F4810/72 resulted in a complete cereulide deficient phenotype (Table 2), which could be restored partly by in trans complementation with cesCD, indicating that the putative ABC transporter is essential for toxin production. Sequence analysis of CesCD revealed homology to members of the BcrAB- or DRI- (drug resistance and immunity) subfamily of ABC systems (Dassa and Bouige, 2001; Davidson et al., 2008; Gebhard, 2012) with highest similarity toward the BerAB transporter of B. thuringiensis, which was shown to be essential for β-Exotoxin I production and suggested to cause toxin immunity by efflux of the molecule; but no experimental proof has been presented so far (Espinasse et al., 2002). Thus, it is tempting to speculate that CesCD is involved in the transport of the emetic toxin, maybe mediating its efflux and thereby conferring resistance. However, if CesCD were only responsible for cereulide export, an intracellular accumulation of toxin would be expected by inactivation of the transporter. Since no accumulation, but complete abolition of cereulide was observed in the CesCD knockout mutant, we propose that CesCD, besides having a potential transport function, plays a more direct role in the cereulide biosynthesis pathway without influencing ces gene transcription or NRPS translation. For the lantibiotics nisin and subtilin it was shown that the multimeric synthetase complexes, which are required for pre-peptide maturation, are associated to the membrane by interaction with ABC transporters linked to the enzyme gene clusters (Siegers et al., 1996; Kiesau et al., 1997). More recently, the NRPS/PKS enzymes responsible for the biosynthesis of the antibiotic bacillaene and the siderophore pyoverdine were found to form membrane-associated mega complexes (Straight et al., 2007; Imperi and Visca, 2013). Presuming that CesCD is a membrane-bound protein, it could be involved in membrane-anchoring of the cereulide synthetase complex or other cereulide-processing enzymes, which otherwise may be non-functional. So far, immunoblot analysis using our anti-CesB antibody detected the cereulide synthetase only in cytosolic but not in membrane cell fractions (Rütschle, unpublished data). Therefore, further experiments targeting the localisation of cereulide and its synthetase complex are in progress to define the exact function of CesCD in cereulide synthesis.
Conclusion
Taken together, our data demonstrate the importance of the cesAB- adjacent genes cesH, cesP, and cesCD in the regulation of cereulide biosynthesis. Interestingly, these genes exert their regulatory functions on very different levels of toxin production, ranging from the transcriptional to the post-translational level (Figure 6). To our knowledge this is the first report on the impact of genes located within an NRPS encoding operon, and their gene products, on the transcription, translation and synthesis of the NRPS product and its biosynthetic machinery. The fact that not only chromosomal encoded master transcriptional regulators, such as AbrB or CodY, but also genes embedded in the ces gene cluster itself, influence cereulide production, highlights the complexity of regulation of synthesis of NRPS products.
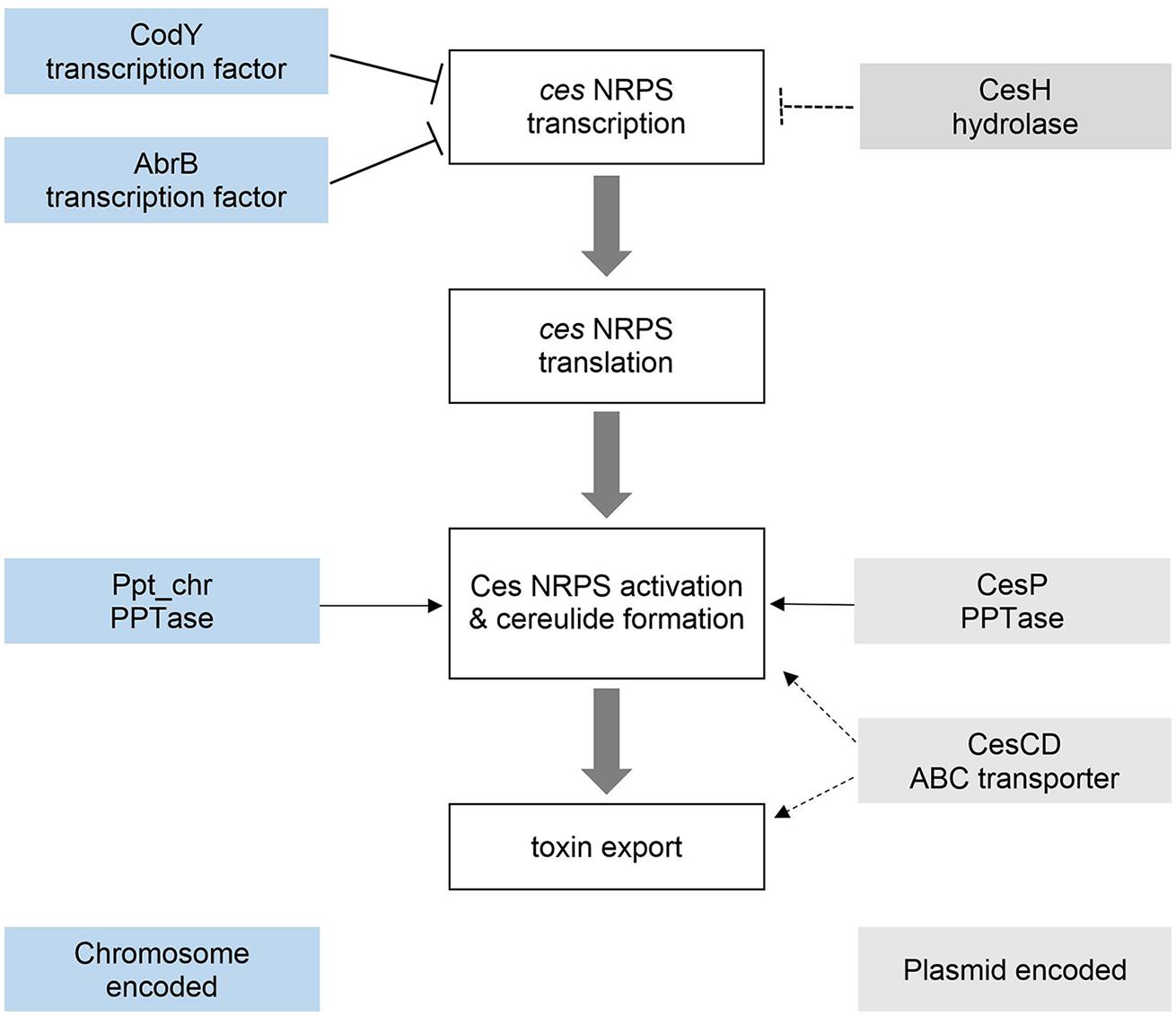
FIGURE 6. Regulation of cereulide synthesis. Interplay of chromosomally and plasmid encoded factors controlling cereulide synthesis at the transcriptional (AbrB, CodY, CesH) and post-translational (CesP, Ppt, cesCD) level.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Romy Wecko and Christine Braig for excellent technical support and Dietmar Hillman for advices concerning B. megaterium protoplast transformation procedures. Thanks to Daniel Zeigler (Bacillus Genetic Stock Center) for the gift of pAD123 and pMAD, and Wolfgang Hillen (University of Erlangen-Nürnberg) posthumously for the gift of pWH1520. We also thank Patrice Courvalin (Institut Pasteur) for kindly supplying us with E. coli JM83/pRK24, pAT113 and pSPCH+2. This research project was supported by the German Ministry of Economics and Technology (via AiF) and the FEI (Forschungskreis der Ernährungsindustrie e.V., Bonn), through projects AiF 15186 N and AiF 16845 N.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01101
Footnotes
- ^ http://blast.ncbi.nlm.nih.gov
- ^ http://www.cbs.dtu.dk/services/TMHMM/
- ^ http://www.ch.embnet.org/software/TMPRED_form.html
References
Agata, N., Ohta, M., Mori, M., and Isobe, M. (1995). A novel dodecadepsipeptide, cereulide, is an emetic toxin of Bacillus cereus. FEMS Microbiol. Lett. 129, 17–20. doi: 10.1111/j.1574-6968.1995.tb07550.x
Alonzo, D. A., Magarvey, N. A., and Schmeing, T. M. (2015). Characterization of cereulide synthetase, a toxin-producing macromolecular machine. PLoS ONE 10:e0128569. doi: 10.1371/journal.pone.0128569
Bauer, T., Stark, T., Hofmann, T., and Ehling-Schulz, M. (2010). Development of a stable isotope dilution analysis for the quantification of the Bacillus cereus toxin cereulide in foods. J. Agric. Food Chem. 58, 1420–1428. doi: 10.1021/jf9033046
Carlin, F., Fricker, M., Pielaat, A., Heisterkamp, S., Shaheen, R., Salonen, M. S., et al. (2006). Emetic toxin-producing strains of Bacillus cereus show distinct characteristics within the Bacillus cereus group. Int. J. Food Microbiol. 109, 132–138. doi: 10.1016/j.ijfoodmicro.2006.01.022
Copp, J. N., and Neilan, B. A. (2006). The phosphopantetheinyl transferase superfamily: phylogenetic analysis and functional implications in cyanobacteria. Appl. Environ. Microbiol. 72, 2298–2305. doi: 10.1128/AEM.72.4.2298-2305.2006
Dassa, E., and Bouige, P. (2001). The ABC of ABCS: a phylogenetic and functional classification of ABC systems in living organisms. Res. Microbiol. 152, 211–229. doi: 10.1016/S0923-2508(01)01194-9
Davidson, A. L., Dassa, E., Orelle, C., and Chen, J. (2008). Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–364. doi: 10.1128/MMBR.00031-07
Dierick, K., Van Coillie, E., Swiecicka, I., Meyfroidt, G., Devlieger, H., Meulemans, A., et al. (2005). Fatal family outbreak of Bacillus cereus-associated food poisoning. J. Clin. Microbiol. 43, 4277–4279. doi: 10.1128/JCM.43.8.4277-4279.2005
Dommel, M. K., Frenzel, E., Strasser, B., Blöchinger, C., Scherer, S., and Ehling-Schulz, M. (2010). Identification of the main promoter directing cereulide biosynthesis in emetic Bacillus cereus and its application for real-time monitoring of ces gene expression in foods. Appl. Environ. Microbiol. 76, 1232–1240. doi: 10.1128/AEM.02317-09
Dommel, M. K., Lücking, G., Scherer, S., and Ehling-Schulz, M. (2011). Transcriptional kinetic analyses of cereulide synthetase genes with respect to growth, sporulation and emetic toxin production in Bacillus cereus. Food Microbiol. 28, 284–290. doi: 10.1016/j.fm.2010.07.001
Ehling-Schulz, M., Frenzel, E., and Gohar, M. (2015). Food – bacteria interplay: pathometabolism of emetic Bacillus cereus. Front. Microbiol. 6:704. doi: 10.3389/fmicb.2015.00704
Ehling-Schulz, M., Fricker, M., Grallert, H., Rieck, P., Wagner, M., and Scherer, S. (2006). Cereulide synthetase gene cluster from emetic Bacillus cereus: structure and location on a mega virulence plasmid related to Bacillus anthracis toxin plasmid pXO1. BMC Microbiol. 6:20. doi: 10.1186/1471-2180-6-20
Ehling-Schulz, M., Fricker, M., and Scherer, S. (2004). Bacillus cereus, the causative agent of an emetic type of food-borne illness. Mol. Nutr. Food Res. 48, 479–487. doi: 10.1002/mnfr.200400055
Ehling-Schulz, M., Svensson, B., Guinebretiere, M. H., Lindback, T., Andersson, M., Schulz, A., et al. (2005a). Emetic toxin formation of Bacillus cereus is restricted to a single evolutionary lineage of closely related strains. Microbiology 151, 183–197. doi: 10.1099/mic.0.27607-0
Ehling-Schulz, M., Vukov, N., Schulz, A., Shaheen, R., Andersson, M., Martlbauer, E., et al. (2005b). Identification and partial characterization of the nonribosomal peptide synthetase gene responsible for cereulide production in emetic Bacillus cereus. Appl. Environ. Microbiol. 71, 105–113. doi: 10.1128/AEM.71.1.105-113.2005
Espinasse, S., Gohar, M., Lereclus, D., and Sanchis, V. (2002). An ABC transporter from Bacillus thuringiensis is essential for beta-exotoxin I production. J. Bacteriol. 184, 5848–5854. doi: 10.1128/JB.184.21.5848-5854.2002
Finking, R., and Marahiel, M. A. (2004). Biosynthesis of nonribosomal peptides1. Annu. Rev. Microbiol. 58, 453–488. doi: 10.1146/annurev.micro.58.030603.123615
Frenzel, E., Doll, V., Pauthner, M., Lücking, G., Scherer, S., and Ehling-Schulz, M. (2012). CodY orchestrates the expression of virulence determinants in emetic Bacillus cereus by impacting key regulatory circuits. Mol. Microbiol. 85, 67–88. doi: 10.1111/j.1365-2958.2012.08090.x
Gebhard, S. (2012). ABC transporters of antimicrobial peptides in Firmicutes bacteria – phylogeny, function and regulation. Mol. Microbiol. 86, 1295–1317. doi: 10.1111/mmi.12078
Hotelier, T., Renault, L., Cousin, X., Negre, V., Marchot, P., and Chatonnet, A. (2004). ESTHER, the database of the alpha/beta-hydrolase fold superfamily of proteins. Nucleic Acids Res. 32, D145–D147. doi: 10.1093/nar/gkh141
Imperi, F., and Visca, P. (2013). Subcellular localization of the pyoverdine biogenesis machinery of Pseudomonas aeruginosa: a membrane-associated “siderosome”. FEBS Lett. 587, 3387–3391. doi: 10.1016/j.febslet.2013.08.039
Ivanova, N., Sorokin, A., Anderson, I., Galleron, N., Candelon, B., Kapatral, V., et al. (2003). Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423, 87–91. doi: 10.1038/nature01582
Kiesau, P., Eikmanns, U., Gutowski-Eckel, Z., Weber, S., Hammelmann, M., and Entian, K. D. (1997). Evidence for a multimeric subtilin synthetase complex. J. Bacteriol. 179, 1475–1481.
Lambalot, R. H., Gehring, A. M., Flugel, R. S., Zuber, P., Lacelle, M., Marahiel, M. A., et al. (1996). A new enzyme superfamily – the phosphopantetheinyl transferases. Chem. Biol. 3, 923–936. doi: 10.1016/S1074-5521(96)90181-7
Lane, A. L., and Moore, B. S. (2011). A sea of biosynthesis: marine natural products meet the molecular age. Nat. Prod. Rep. 28, 411–428. doi: 10.1039/c0np90032j
Letunic, I., Doerks, T., and Bork, P. (2012). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–D305. doi: 10.1093/nar/gkr931
Linton, K. J., Cooper, H. N., Hunter, I. S., and Leadlay, P. F. (1994). An ABC-transporter from Streptomyces longisporoflavus confers resistance to the polyether-ionophore antibiotic tetronasin. Mol. Microbiol. 11, 777–785. doi: 10.1111/j.1365-2958.1994.tb00355.x
Lücking, G., Dommel, M. K., Scherer, S., Fouet, A., and Ehling-Schulz, M. (2009). Cereulide synthesis in emetic Bacillus cereus is controlled by the transition state regulator AbrB, but not by the virulence regulator PlcR. Microbiology 155, 922–931. doi: 10.1099/mic.0.024125-0
Magarvey, N. A., Ehling-Schulz, M., and Walsh, C. T. (2006). Characterization of the cereulide NRPS alpha-hydroxy acid specifying modules: activation of alpha-keto acids and chiral reduction on the assembly line. J. Am. Chem. Soc. 128, 10698–10699. doi: 10.1021/ja0640187
Marahiel, M. A., Stachelhaus, T., and Mootz, H. D. (1997). Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97, 2651–2674. doi: 10.1021/cr960029e
Marxen, S., Stark, T. D., Frenzel, E., Rütschle, A., Lücking, G., Purstinger, G., et al. (2015a). Chemodiversity of cereulide, the emetic toxin of Bacillus cereus. Anal. Bioanal. Chem. 407, 2439–2453. doi: 10.1007/s00216-015-8511-y
Marxen, S., Stark, T. D., Rütschle, A., Lücking, G., Frenzel, E., Scherer, S., et al. (2015b). Depsipeptide Intermediates Interrogate Proposed Biosynthesis of Cereulide, the Emetic Toxin of Bacillus cereus. Sci. Rep. 5, 10637. doi: 10.1038/srep10637
May, J. J., Wendrich, T. M., and Marahiel, M. A. (2001). The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem. 276, 7209–7217. doi: 10.1074/jbc.M009140200
McMorran, B. J., Merriman, M. E., Rombel, I. T., and Lamont, I. L. (1996). Characterisation of the pvdE gene which is required for pyoverdine synthesis in Pseudomonas aeruginosa. Gene 176, 55–59. doi: 10.1016/0378-1119(96)00209-0
Mesnage, S., Fontaine, T., Mignot, T., Delepierre, M., Mock, M., and Fouet, A. (2000). Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19, 4473–4484. doi: 10.1093/emboj/19.17.4473
Messelhäusser, U., Frenzel, E., Blöchinger, C., Zucker, R., Kämpf, P., and Ehling-Schulz, M. (2014). Emetic Bacillus cereus are more volatile than thought: recent foodborne outbreaks and prevalence studies in Bavaria (2007-2013). Biomed Res. Int. 2014, 465603. doi: 10.1155/2014/465603
Mootz, H. D., and Marahiel, M. A. (1997). The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J. Bacteriol. 179, 6843–6850.
Murphy, E. (1985). Nucleotide sequence of a spectinomycin adenyltransferase AAD(9) determinant from Staphylococcus aureus and its relationship to AAD(3”) (9). Mol. Gen. Genet. 200, 33–39. doi: 10.1007/BF00383309
Nakano, M. M., Corbell, N., Besson, J., and Zuber, P. (1992). Isolation and characterization of sfp: a gene that functions in the production of the lipopeptide biosurfactant, surfactin, in Bacillus subtilis. Mol. Gen. Genet. 232, 313–321.
Naranjo, M., Denayer, S., Botteldoorn, N., Delbrassinne, L., Veys, J., Waegenaere, J., et al. (2011). Sudden death of a young adult associated with Bacillus cereus food poisoning. J. Clin. Microbiol. 49, 4379–4381. doi: 10.1128/JCM.05129-11
Pezard, C., Berche, P., and Mock, M. (1991). Contribution of individual toxin components to virulence of Bacillus anthracis. Infect. Immun. 59, 3472–3477.
Pfaffl, M. W., Horgan, G. W., and Dempfle, L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30, e36. doi: 10.1093/nar/30.9.e36
Podlesek, Z., Comino, A., Herzog-Velikonja, B., Zgur-Bertok, D., Komel, R., and Grabnar, M. (1995). Bacillus licheniformis bacitracin-resistance ABC transporter: relationship to mammalian multidrug resistance. Mol. Microbiol. 16, 969–976. doi: 10.1111/j.1365-2958.1995.tb02322.x
Quigley, N. B., Mo, Y. Y., and Gross, D. C. (1993). SyrD is required for syringomycin production by Pseudomonas syringae pathovar syringae and is related to a family of ATP-binding secretion proteins. Mol. Microbiol. 9, 787–801. doi: 10.1111/j.1365-2958.1993.tb01738.x
Rasko, D. A., Ravel, J., Okstad, O. A., Helgason, E., Cer, R. Z., Jiang, L., et al. (2004). The genome sequence of Bacillus cereus ATCC 10987 reveals metabolic adaptations and a large plasmid related to Bacillus anthracis pXO1. Nucleic Acids Res. 32, 977–988. doi: 10.1093/nar/gkh258
Schwarzer, D., Mootz, H. D., Linne, U., and Marahiel, M. A. (2002). Regeneration of misprimed nonribosomal peptide synthetases by type II thioesterases. Proc. Natl. Acad. Sci. U.S.A. 99, 14083–14088. doi: 10.1073/pnas.212382199
Siegers, K., Heinzmann, S., and Entian, K. D. (1996). Biosynthesis of lantibiotic nisin. Posttranslational modification of its prepeptide occurs at a multimeric membrane-associated lanthionine synthetase complex. J. Biol. Chem. 271, 12294–12301. doi: 10.1074/jbc.271.21.12294
Spizizen, J. (1958). Transformation of biochemically deficient strains of Bacillus Subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. U.S.A. 44, 1072–1078. doi: 10.1073/pnas.44.10.1072
Stark, T., Marxen, S., Ruetschle, A., Luecking, G., Scherer, S., Ehling-Schulz, M., et al. (2013). Mass spectrometric profiling of Bacillus cereus strains and quantitation of the emetic toxin cereulide by means of stable isotope dilution analysis and HEp-2 bioassay. Anal. Bioanal. Chem. 405, 191–201. doi: 10.1007/s00216-012-6485-6
Straight, P. D., Fischbach, M. A., Walsh, C. T., Rudner, D. Z., and Kolter, R. (2007). A singular enzymatic megacomplex from Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 104, 305–310. doi: 10.1073/pnas.0609073103
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Trieu-Cuot, P., Carlier, C., Martin, P., and Courvalin, P. (1987). Plasmid transfer by conjugation from Escherichia coli to Gram-positive bacteria. FEMS Microbiol. Lett. 48, 289–294. doi: 10.1111/j.1574-6968.1987.tb02558.x
Tschiedel, E., Rath, P. M., Steinmann, J., Becker, H., Dietrich, R., Paul, A., et al. (2015). Lifesaving liver transplantation for multi-organ failure caused by Bacillus cereus food poisoning. Pediatr. Transplant. 19, E11–E14. doi: 10.1111/petr.12378
Walsh, C. T., Gehring, A. M., Weinreb, P. H., Quadri, L. E., and Flugel, R. S. (1997). Post-translational modification of polyketide and nonribosomal peptide synthases. Curr. Opin. Chem. Biol. 1, 309–315. doi: 10.1016/S1367-5931(97)80067-1
Wenzel, S. C., and Müller, R. (2005). Formation of novel secondary metabolites by bacterial multimodular assembly lines: deviations from textbook biosynthetic logic. Curr. Opin. Chem. Biol. 9, 447–458. doi: 10.1016/j.cbpa.2005.08.001
Keywords: Bacillus cereus, ces gene cluster, regulation, cesH, cesP, cesC, cesD, cereulide synthetase
Citation: Lücking G, Frenzel E, Rütschle A, Marxen S, Stark TD, Hofmann T, Scherer S and Ehling-Schulz M (2015) Ces locus embedded proteins control the non-ribosomal synthesis of the cereulide toxin in emetic Bacillus cereus on multiple levels. Front. Microbiol. 6:1101. doi: 10.3389/fmicb.2015.01101
Received: 27 July 2015; Accepted: 23 September 2015;
Published: 13 October 2015.
Edited by:
Michael Gänzle, University of Alberta, CanadaReviewed by:
Jinshui Zheng, Huazhong Agricultural University, ChinaAnne-Brit Kolstø, University of Oslo, Norway
Copyright © 2015 Lücking, Frenzel, Rütschle, Marxen, Stark, Hofmann, Scherer and Ehling-Schulz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Monika Ehling-Schulz, Functional Microbiology, Institute of Microbiology, Department of Pathobiology, University of Veterinary Medicine Vienna, Veterinärplatz 1, A-1210 Vienna, Austria, bW9uaWthLmVobGluZy1zY2h1bHpAdmV0bWVkdW5pLmFjLmF0
†Present address: Elrike Frenzel, Molecular Genetics Group, Groningen Biomolecular Sciences and Biotechnology Institute, Centre for Synthetic Biology, University of Groningen, Groningen, Netherlands
‡These authors have contributed equally to this work.
 Genia Lücking
Genia Lücking Elrike Frenzel
Elrike Frenzel Andrea Rütschle1‡
Andrea Rütschle1‡ Thomas Hofmann
Thomas Hofmann Siegfried Scherer
Siegfried Scherer Monika Ehling-Schulz
Monika Ehling-Schulz