- 1School of Biotechnology and Food Engineering, Hefei University of Technology, Hefei, China
- 2State Key Laboratory of Applied Microbiology Southern China, Guangzhou, China
- 3Guangdong Provincial Key Laboratory of Microbiology Culture Collection and Application, Guangzhou, China
- 4Guangdong Institute of Microbiology, Guangzhou, China
Cronobacter sakazakii is an opportunistic foodborne pathogen and the virulence differences were previously documented. However, information about membranous proteins involved in virulence differences was not available. In this study, virulent characterization such as biofilm formation and flagella motility between virulent C. sakazakii isolate G362 and attenuated L3101 were determined. Then, two-dimensional gel electrophoresis (2-DE) technology was used to preliminarily reveal differential expression of membranous proteins between G362 and L3101. On the mass spectrometry (MS) analysis and MASCOT research results, fourteen proteins with differential expression were successfully identified. At the threshold of twofold changes, five out of eight membranous proteins were up-regulated in G362. Using RT-PCR, the expression abundance of the protein (enzV, ompX, lptE, pstB, and OsmY) genes at mRNA levels was consistent with the results by 2-DE method. The findings presented here provided novel information and valuable knowledge for revealing pathogenic mechanism of C. sakazakii.
Introduction
The genus of Cronobacter is a group of opportunistic pathogens linked with life-threatening infections in neonates (Holý and Forsythe, 2014). Cronobacter sp. was isolated from various kinds of samples including food, soil, environments, clinical samples (Gurtler et al., 2005). In addition, these infections often occurred among poor immunity or low-birth weight newborn through consumption of contaminated powdered infant formula (Muytjens et al., 1983; Tall et al., 2014). The Cronobacter genus consists of Cronobacter sakazakii, C. malonaticus, C. turicensis, C. muytjensii, C. condiment, C. universalis, and C. dublinensis (Joseph et al., 2012a,b). Furthermore, C. sakazakii is the predominant species within Cronobacter isolated from powdered milk (Xu et al., 2014).
Membranous proteins were required for adhesion, invasion, and the release of toxin of pathogens, and further played important roles on host-pathogen interaction or host infections in foodborne pathogens (Allaoui et al., 1993; Monteville et al., 2003; Ellis and Kuehn, 2010). Transmembrane signal proteins of two-component regulatory system in gram-negative pathogens contributed to immune escape or biofilm formation (Krachler et al., 2011). In addition, outer membrane proteins A (OmpA) and X (OmpX) in C. sakazakii played critical roles in adhesion to and invading human cells (Mohan Nair and Venkitanarayanan, 2007; Mohan Nair et al., 2009; Kim et al., 2010). Kothary et al. (2007) reported that zpx gene encoding the cell-bound zinc-containing metalloprotease might play important roles in dissemination of Cronobacter into the systemic circulation. However, little information about membranous proteins involved in virulence differences among C. sakazakii isolates is available.
To determine the potential virulence factors of pathogenic bacteria, the use of global techniques such as proteomics was widely applied in exploring different factors between virulent and attenuated bacteria (Srinivasa Rao et al., 2004; Hecker et al., 2010; Niemann et al., 2011).
In this study, the virulent characterization including biofilm formation, flagella motility, and O serotypes between G362 and L3101 was determined. Then, the 2-DE technology coupled with mass spectrometry was applied to identify global molecular information about differentially expressed membranous proteins. Finally, Real-time PCR was used to further validate the expression abundance of these novel membranous protein genes between G362 and L3101.
Materials And Methods
Cronobacter sakazakii Isolates
The virulence differences between C. sakazakii G362 and L3101 from food samples were previously determined through intraperitoneal injection and histopathologic analysis (small intestine, kidney, and liver; Ye et al., 2015). The significant morphological change in small intestine, liver, and kidney further reveals the virulence differences between G362 and L3101.
Different Phenotypic Characterization between G362 and L3101
The biofilm formation between G362 and L3101 was determined using crystal violet staining (CVS) method as described by Hochbaum et al. (2011). The O serotypes were detected as described by Sun et al. (2012). In addition, the primers, PCR conditions, and PCR mixtures also referred to the description by Sun et al. (2012).
In addition, the motility of two isolates (G362 and L3101) was also determined. Media consisting of 10 g/L tryptose, 5 g/L NaCl, and 1.5 g/L agarose was used to test the swimming motility. Swimming plate was inoculated with C. sakazakii isolates from an overnight culture of TSA agar using a sterile toothpick and incubated at 37°C for 8 h. Media (8 g/L nutrient broth, 5 g/L D-glucose, and 5 g/L Difco bacto-agar) was made to determine the swarming motility. Swarming plate was inoculated with C. sakazakii isolates from swimming plate using a sterile toothpick and incubated at 37°C for 12 h. The diameter of motility at the agar dish interface was measured in triplicate.
Membranous Proteins Extraction and Quantification
Two C. sakazakii isolates (G362 and L3101) were inoculated into tryptic soy broth (TSB, Huankai, Guangzhou) for incubation at 37°C for 16 h. Two grams (wet weight) of G362 and L3101 were suspended in 5 ml of phosphate buffer saline (PBS, pH7.4) at 4°C. Then the cell suspension was sonicated in an ice bath (SONICS VC-505). Following the sonication process, Triton X-114 was applied for membranous proteins extraction using the method described by González de la Vara and Alfaro (2009). Then, two different phase collections and the insoluble pellet were performed to analyze SOD activity (Huang et al., 2006). Acetone precipitation was employed to remove the excess salts in the aqueous phase collection and detergent phase collection respectively. After lysis buffer (5 M urea, 2 M thiourea, 2% SB3-10, 2% CHAPS, 65 mM DTT, 40 mM Tris; Bio-Rad, USA) dissolution and centrifuged at 18, 000 × g for 1 h at 4°C, membrane proteins supernatant was stored at -80°C for use. Proteins qualification was measured by the bicinchoninic acid (BCA) assay as described by Krieg et al. (2005).
2-DE Analysis
For each sample, 100 μl (8 μg/μl) of membrane proteins extract, diluted with 200 μl of rehydration buffer (7 M urea, 2 M thiourea, 1% ASB-14, 65 mM DTT, 0.2% Bio-Lyte (w/v), 40 mM Tris, 0.001% Bromophenol blue; Bio-Rad, USA), was applied to a reswelling cassette with 17 cm immobiline dry strip (pH 3-10 NL, Bio-Rad, USA). Rehydration was allowed to proceed at 20°C to 16 h under silicone oil. The isoelectric focusing (IEF) was performed using the PROTEAN IEF system (Bio-Rad, USA). Each IPG strip current limit during IEF was set 50 μA at 20°C. The focusing was conducted in next five steps: (1) constant voltage at 250 V for 1 h; (2) constant voltage at 500 V for 30 min; (3) constant voltage at 1000 V for 2 h; (4) under linear ramping mode voltage increased from 1000 to 10, 000 V in 5 h; (5) at last constant voltage at 10, 000 V until the total volt-hours reached 68,000 V-h. The following steps including equilibration, SDS-PAGE electrophoresis, coomassie brilliant blue stain (CBB-stained), gel analysis, and subsequent MS analysis were performed as described by Cordwell et al. (2008) with modification. In brief, spots in gels between isolates corresponding to the same protein identifications were detected using PD-Quest (Bio-Rad) and the relative spot intensities calculated. Significantly changed spots were selected by rate increased/decreased twofold. Then Protein spots of interest were subjected to in-gel digestion, and then cut from the gels for mass spectrometric analysis. Protein identities were based on a combination of peptide fingerprint and MS/MS spectra. MS and MS/MS data were searched using MASCOT version 1.9.05 (Matrix Science) as search engine against the NCBI database. C. sakazakii species database was defined as a matching species. For each isolate sample, the 2-D gels in triplicate were analyzed.
Expression Abundance of Membranous Protein Genes by Real Time PCR
The overnight culture (1.0 ml) of C. sakazakii isolates was subject to extraction of RNA using bacterial RNA extraction kit (BIOMIGA, USA). Then, cDNA was obtained using first-strand cDNA synthesis kit (BIOMIGA, USA) according to the manufacturer’ instructions. The RT-PCR mixture consisted of 2 μl cDNA, 10 μl 2 × SYBR Green qPCR mix (Giagen, Beijing), 0.2 μM for each primer, and RNase-free water in a final volume of 20 μl. The PCR program was initiated at 95°C for 5 min, followed by 40 cycles at 95°C for 10 s, 59°C for 20 s, 72°C for 25 s using ABI Stepone plus (Applied Biosystems). The expression abundance of targeted genes between G362 and L3101 was determined in triplicate using ΔΔCt method.
Results And Discussion
Cronobacter sakazakii is an important foodborne pathogen associated with severe infections in poor immunity or low-birth weight newborn. The virulence differences of C. sakazakii G362 and L3101were previously documented by Ye et al. (2015). In this study, the virulent characterization including biofilm formation, O serotypes, and motility was further determined. In Figure 1A, both G362 and L3101 were O5 serotype by PCR assay (Sun et al., 2012). Xu et al. (2014) found that O2 serotype in C. sakazakii is the predominant serotype among isolates from powdered milk. However, the correlation between O serotypes and virulence was not revealed. Additionally, the significant differences of biofilm formation between G362 and L3101 were observed in Figure 1B, which might be involved in their adhesion to surfaces. The biofilm formation was associated with virulence in Candida (Hasan et al., 2009), Escherichia coli (Naves et al., 2008), and Pseudomonas (Ghadaksaz et al., 2015). Interestingly, the differences of motility (swimming and swarming) in Figure 1C were also founded between G362 and L3101 at 37°C for 8 h (swimming motility) and 12 h (swarming motility). Swimming ability of L3101 is stronger than that of G362, while the weakest swarming motility was also observed in L3101. This is the first report to determine the relationship between flagella motility and biofilm formation in C. sakazakii. Our result preliminarily indicated that swimming motility had negative effects on the biofilm formation, while swarming motility contributed to the biofilm formation. The biofilm formation required flagella motility in Pseudomonas aeruginosa (Barken et al., 2008), and motility and adhesion to surface was also connected in E. coli (Wood et al., 2006). However, flagella motility was not necessarily required for biofilm formation in P. aeruginosa (Caiazza et al., 2007) and Burkholderia pseudomallei (Lee et al., 2010). In C. sakazakii, the regulation of biofilm formation and flagella motility remains to be revealed.
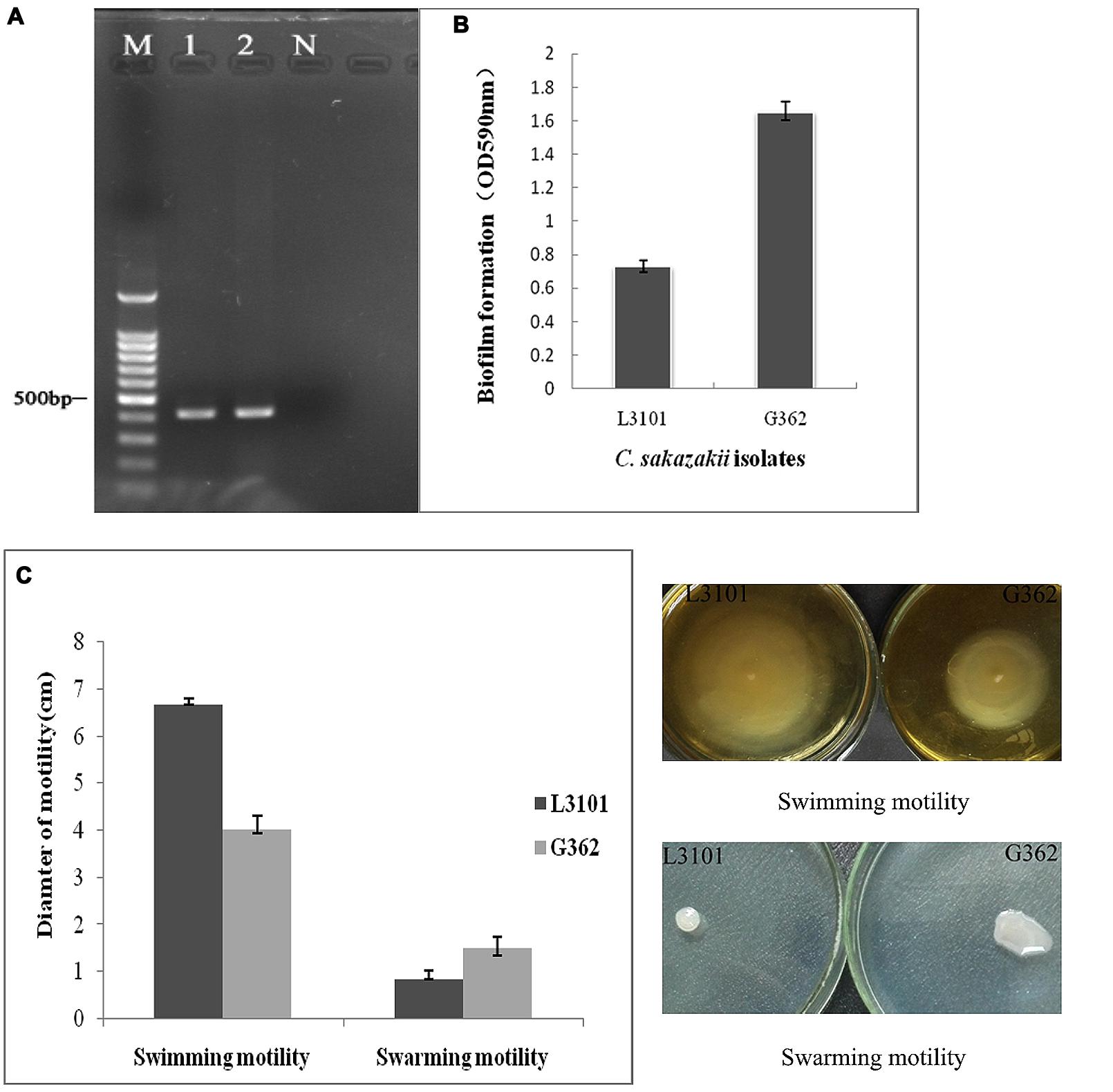
FIGURE 1. The phenotypic characterization (O serotypes, biofilm formation, and motility) of Cronobacter sakazakii virulent G362 and attenuated L3101 isolate. (A) Detection of O serotypes by PCR; (B) Biofilm formation using crystal violet staining (CVS); (C) Detection of swimming and swarming motility.
Thereafter, to reveal the membranous proteins involved in virulence differences among C. sakazakii isolates, the membranous proteomic profiles of G362 and L3101 were firstly constructed using the 2-DE technology (Figure 2). Changes of membranous proteins in intensity (>twofold) were considered as significant differences. The expression abundance of 14 proteins was successfully identified between G362 and L3101. In Table 1, 8 out of 14 differentially expressed proteins was confirmed as membrane-associated proteins and expression abundance of seven membranous proteins was increased in G362. The different expression abundance of genes of membranous proteins including enzV, ompX, lptE, pstB, and OsmY were detected using real time PCR. The primers were listed in Table 2 and 16S rRNA gene was used as internal control. From Figure 3, the expression of five membranous protein genes at mRNA levels using real-time PCR was consistent to results by 2-D method. The relative expression abundance of osmF, ompX, enzV, and lptE genes in G362 were 6.88, 4.49, 1.48, and 3.82 fold changes than those in L3101, respectively, while the expression of pstB in L3101 was 61.38 fold changes than that in G362. The relative expression of five factors by real-time PCR was consistent with results using 2-DE method.
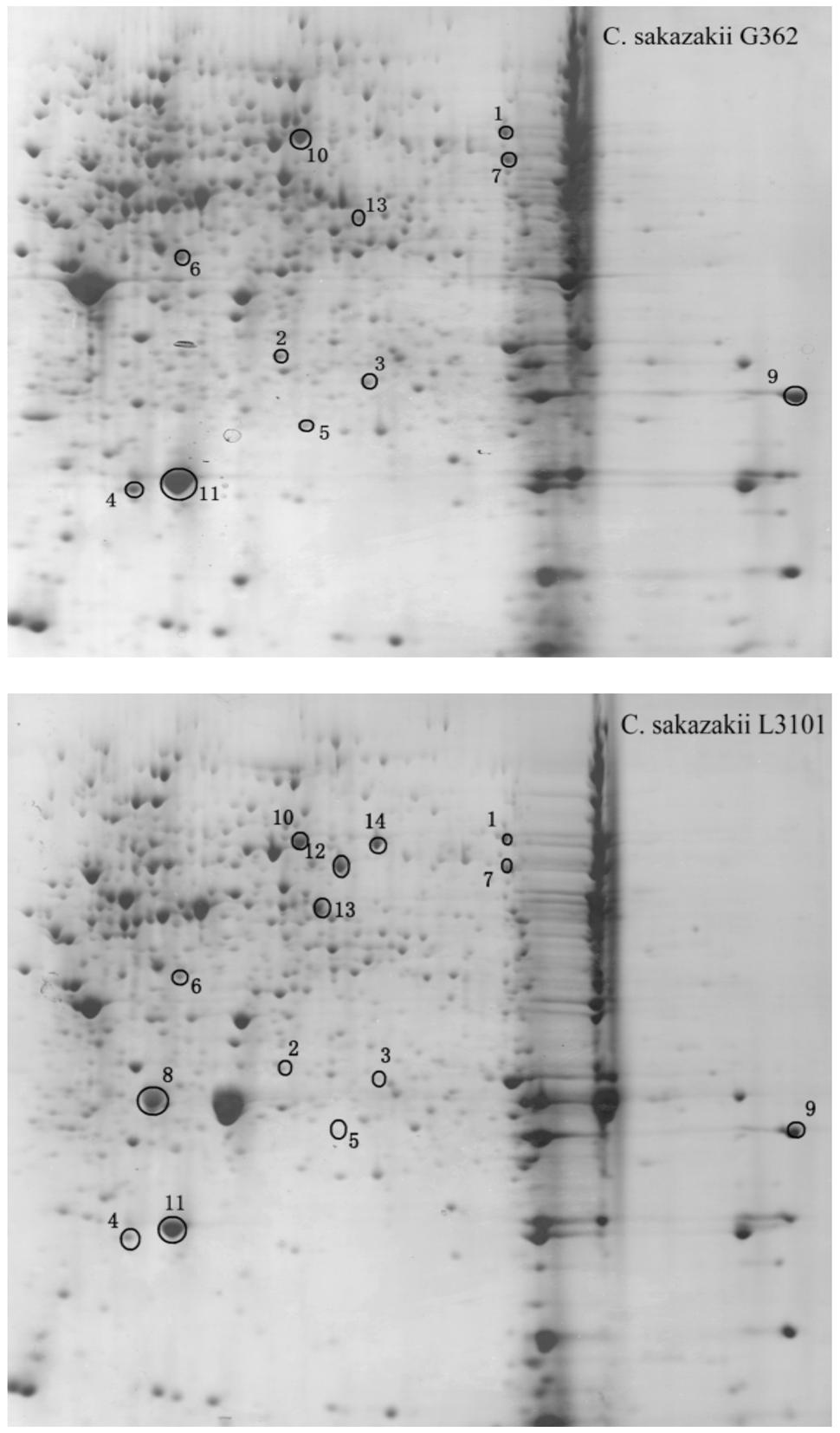
FIGURE 2. Membranous proteomics profiles of virulent (G362) and attenuated (L3101) isolates. pH 3-10 (from left to right).
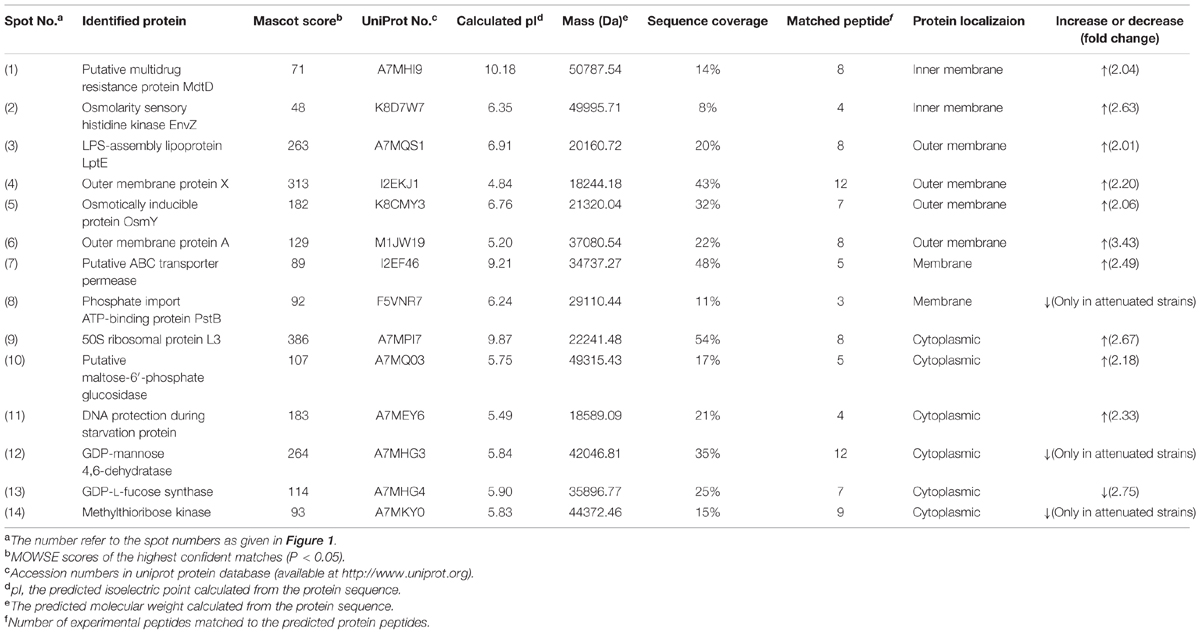
TABLE 1. Identification and relative quantification of differentially expressed membranous proteins by MALDI-TOF/TOF MS for virulent Cronobacter sakazakii G362 compared to L3101.
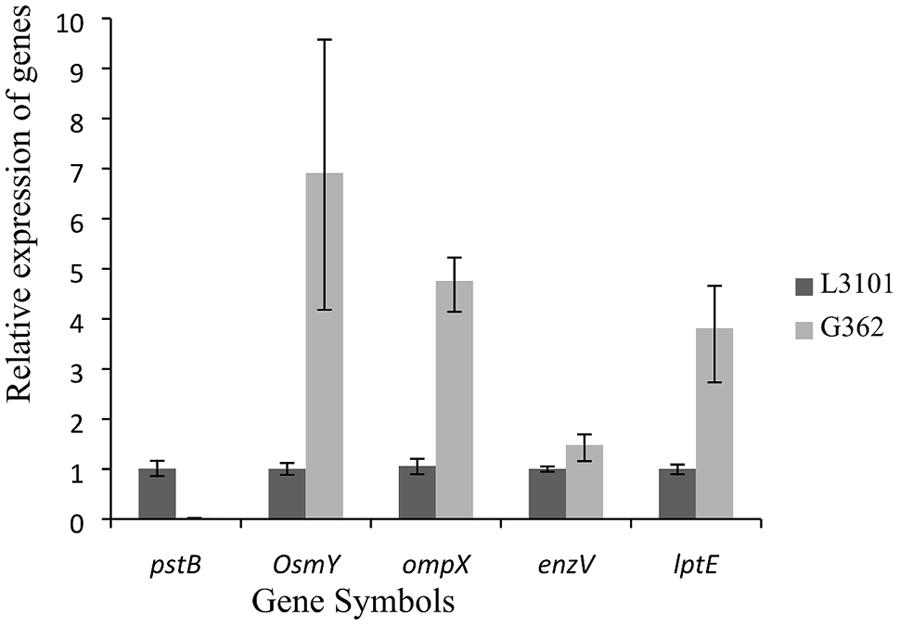
FIGURE 3. Relative fluorescence quantitative PCR (RT-PCR) of membrane protein genes involved in virulence differences between G362 and L3101.
Based on proteomic analysis, the outer membrane proteins (ompA and ompX) were increased in G362 compared to those in L3101. The ompA and ompX proteins were reported to enhance adherence and invasion of human cells (Mohan Nair and Venkitanarayanan, 2007; Mohan Nair et al., 2009; Liu et al., 2012). Outer membrane A of E. coli has been reported enhanced the invasion of brain microvascular endothelial (BMEC) by assisting through the blood-brain barrier (Prasadarao et al., 1996). Our findings also indicated that ompA and ompX might be involved in virulence differences of C. sakazakii at membranous protein levels.
Besides, the expression abundance of novel proteins of LPS-assembly lipoprotein LptE, Osmolarity sensory histidine kinase EnvZ, and Osmotically inducible protein OsmY in G362 were increased compared to those in L3101. The LPS-assembly lipoprotein LptE encoded by the gene lptE in E. coli is essential for the biosynthesis of lipopolysaccharide, resulting in strong immunogenicity with host cells (Chng et al., 2010). Inner membrane osmolarity sensory histidine kinase EnvZ-OmpR pair in C. sakazakii would be activated in response to environmental osmoic stress (Amalaradjou and Venkitanarayanan, 2011). The receptor ompR could influence bacterial biofilm formation and flagella motility by regulating genes ompF and ompC. Further findings indicated that virulence of ompR mutants decreased in Salmonella typhimurium (Bernardini et al., 1990; Raczkowska et al., 2011). Additionally, Osmotically inducible protein OsmY also played important roles in pathogenic bacteria. In S. typhimurium and E. coli, OsmY was indirectly associated with virulence behavior (Bader et al., 2003; Dong and Schellhorn, 2009). In addition, putative multidrug resistance protein MdtD and putative ABC transporter permease were also up-regulated in G362. Some authors reported mdtABCD together with BaeSR two-component system contributed to resisting to host defenses through membrane drug-efflux pump (Baranova and Nikaido, 2002; Zoetendal et al., 2008; Leblanc et al., 2011). ABC transporter permease, a multi-pass membrane protein, was responsible for oligogalacturonide transport, but its detailed mechanism involved in virulence of bacteria is not documented.
Interestingly, the expression abundance of some proteins like phosphate import ATP-binding protein PstB, GDP-L-fucose synthase and methylthioribose kinase were increased in L3101. To date, the correlation of these proteins and bacterial virulence was not documented.
In summary, the relationship between flagella motility and biofilm formation was preliminarily determined, which indicated that motility was not necessarily required for biofilm formation. Then, membranous proteomic profiles between virulent and attenuated C. sakazakii isolate were firstly constructed. In spite of ompA and ompX, some novel factors (envZ, LptE, MdtD, and OsmY) were successfully identified by proteomics which were potentially involved in virulence differences. The findings here provided novel insights and better knowledge for revealing pathogenic mechanisms of C. sakazakii. However, the detailed roles and regulation of these potential virulence factors of C. sakazakii, and study on interaction between host and bacteria are also urgent for exploration of pathogenic mechanism.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge the financial support of the National Natural Science Foundation of China (31201292) and Guangdong Province, Chinese Academy of comprehensive strategic cooperation project (2012B090400017).
References
Allaoui, A., Sansonetti, P. J., and Parsot, C. (1993). MxiD, an outer membrane protein necessary for the secretion of the Shigella flexneri Ipa invasions. Mol. Microbiol. 7, 59–68. doi: 10.1111/j.1365-2958.1993.tb01097.x
Amalaradjou, M. A. R., and Venkitanarayanan, K. (2011). Proteomic analysis of the mode of antibacterial action of trans-cinnamaldehyde against Cronobacter sakazakii 415. Foodborne Pathog. Dis. 8, 1095–1102. doi: 10.1089/fpd.2011.0841
Bader, M. W., Navarre, W. W., Shiau, W., Nikaido, H., Frye, J. G., McClelland, M., et al. (2003). Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol. Microbiol. 50, 219–230. doi: 10.1046/j.1365-2958.2003.03675.x
Baranova, N., and Nikaido, H. (2002). The BaeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J. Bacteriol. 184, 4168–4176. doi: 10.1128/JB.184.15.4168-4176.2002
Barken, K. B., Pamp, S. J., Yang, L., Gjermansen, M., Bertrand, J. J., Klausen, M., et al. (2008). Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 10, 2331–2343.1. doi: 10.1111/j.1462-2920.2008.01658.x
Bernardini, M., Fontaine, A., and Sansonetti, P. (1990). The two-component regulatory system ompR-envZ controls the virulence of Shigella flexneri. J. Bacteriol. 172, 6274–6281.
Caiazza, N. C., Merritt, J. H., and Brothers, K. M. (2007). Inverse regulation of biofilms formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189, 3603–3612. doi: 10.1128/JB.01685-06
Chng, S. S., Gronenberg, L. S., and Kahne, D. (2010). Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex. Biochemistry 49, 4565–4567. doi: 10.1021/bi100493e
Cordwell, S. J., Alice, C. L. L., Touma, R. G., Scott, N. E., Falconer, L., Jones, D., et al. (2008). Identification of membrane-associated proteins from Campylobacter jejuni strains using complementary proteomics technologies. Proteomics 8, 122–139. doi: 10.1002/pmic.200700561
Dong, T., and Schellhorn, H. E. (2009). Global effect of RpoS on gene expression in pathogenic Escherichia coli O157: H7 strain EDL933. BMC Genomics 10:349. doi: 10.1186/1471-2164-10-349
Ellis, T. N., and Kuehn, M. J. (2010). Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. 74, 81–94. doi: 10.1128/MMBR.00031-09
Ghadaksaz, A., Fooladi, A. A. I., Hosseini, H. M., and Amin, M. (2015). The prevalence of some Pseudomonas virulence genes related to biofilm formation and alginate production among clinical isolates. J. Appl. Biomed. 13, 61–68. doi: 10.1016/j.jab.2014.05.002
González de la Vara, L. E., and Alfaro, B. L. (2009). Separation of membrane proteins according to their hydropathy by serial phase partitioning with Triton X-114. Anal. Biochem. 387, 280–286. doi: 10.1016/j.ab.2009.01.035
Gurtler, J. B., Kornacki, J. L., and Beuchat, L. R. (2005). Enterobacter sakazakii: a coliform of increased concern to infant health. Int. J. Food Microbiol. 104, 1–34. doi: 10.1016/j.ijfoodmicro.2005.02.013
Hasan, F., Xess, I., Wang, X., Jain, N., and Fries, B. C. (2009). Biofilm formation in clinical Candida isolates and its association with virulence. Microb. Infect. 11, 753–761. doi: 10.1016/j.micinf.2009.04.018
Hecker, M., Becher, D., Fuchs, S., and Engelmann, S. (2010). A proteomic view of cell physiology and virulence of Staphylococcus aureus. Int. J. Med. Microbiol. 300, 76–87. doi: 10.1016/j.ijmm.2009.10.006
Hochbaum, A. I., Kolodkin-Gal, I., Foulston, L., Kolter, R., Aizenberg, J., and Losick, R. (2011). Inhibitory effects of D-amino acids on Staphylococcus aureus biofilm development. J. Bacteriol. 193, 5616–5622. doi: 10.1128/JB.05534-11
Holý, O., and Forsythe, S. (2014). Cronobacter spp. as emerging causes of healthcare-associated infection. J. Hosp. Infect. 86, 169–177. doi: 10.1016/j.jhin.2013.09.011
Huang, C. Z., Lin, X. M., Wu, L. N., Zhang, D. F., Liu, D., Wang, S. Y., et al. (2006). Systematic identification of the subproteome of Escherichia coli cell envelope reveals the interaction network of membrane proteins and membrane-associated peripheral proteins. J. Proteome Res. 5, 3268–3276. doi: 10.1021/pr060257h
Joseph, S., Cetinkaya, E., Drahovske, H., Levican, A., Figueras, M. J., and Forsythe, S. J. (2012a). Cronobacter condimenti sp. nov., isolated from spiced meat and Cronobacter universalis sp. nov., a novel species designation for Cronobacter sp. genomospecies 1, recovered from a leg infection, water, and food ingredients. Int. J. Syst. Evol. Microbiol. 62, 1277–1283. doi: 10.1099/ijs.0.032292-0
Joseph, S., Sonbol, H., Hariri, S., Desai, P., McClelland, M., and Forsythe, S. J. (2012b). Diversity of the Cronobacter genus as revealed by multilocus sequence typing. J. Clin. Microbiol. 50, 3031–3039. doi: 10.1128/JCM.00905-12
Kim, K., Kim, K.-P., Choi, J., Lim, J.-A., Lee, J., Hwang, S., et al. (2010). Outer membrane proteins A (OmpA) and X (OmpX) are essential for basolateral invasion of Cronobacter sakazakii. Appl. Environ. Microbiol. 76, 5188–5198. doi: 10.1128/AEM.02498-09
Krachler, A. M., Ham, H., and Orth, K. (2011). Outer membrane adhesion factor multivalent adhesion molecule 7 initiates host cell binding during infection by gram-negative pathogens. Proc. Natl. Aca. Sci. U.S.A. 108, 11614–11619. doi: 10.1073/pnas.1102360108
Kothary, M. H., McCardell, B. A., Frazar, C. D., Deer, D., and Tall, B. D. (2007). Characterization of the zinc-containing metalloprotease encoded by zpx and development of a species-specific detection method for Enterobacter sakazakii. Appl. Environ. Microbiol. 73, 4142–4151. doi: 10.1128/AEM.02729-06
Krieg, R. C., Dong, Y., Schwamborn, K., and Knuechel, R. (2005). Protein quantification and its tolerance for different interfering reagents using the BCA-method with regard to 2D-SDS PAGE. J. Biochem. Biophys. Methods 65, 13–19. doi: 10.1016/j.jbbm.2005.08.005
Leblanc, S. K., Oates, C. W., and Raivio, T. L. (2011). Characterization of the induction and cellular role of the BaeSR two-component envelope stress response of Escherichia coli. J. Bacteriol. 193, 3367–3375. doi: 10.1128/JB.01534-10
Lee, H. S., Gu, F., Ching, S. M., Lam, Y., and Chua, K. L. (2010). CdpA Is a Burkholderia pseudomallei cyclic di-GMP phosphodiesterase involved in autoaggregation, flagellum synthesis, motility, biofilm formation, cell invasion, and cytotoxicity. Infect. Immun. 78, 1832–1840. doi: 10.1128/IAI.00446-09
Liu, Q., Mittal, R., Emami, C. N., Iversen, C., Ford, H. R., and Prasadarao, N. V. (2012). Human isolates of Cronobacter sakazakii bind efficiently to intestinal epithelial cells in vitro to induce monolayer permeability and apoptosis. J. Surg. Res. 176, 437–447. doi: 10.1016/j.jss.2011.10.030
Mohan Nair, M. K., and Venkitanarayanan, K. (2007). Role of bacterial OmpA and host cytoskeleton in the invasion of human intestinal epithelial cells by Enterobacter sakazakii. Pediatr. Res. 62, 664–669. doi: 10.1203/PDR.0b013e3181587864
Mohan Nair, M. K., Venkitanarayanan, K., Silbart, L. K., and Kim, K. S. (2009). Outer membrane protein A (OmpA) of Cronobacter sakazakii binds fibronectin and contributes to invasion of human brain microvascular endothelial cells. Foodborne Pathog. Dis. 6, 495–501. doi: 10.1089/fpd.2008.0228
Monteville, M. R., Yoon, J. E., and Konkel, M. E. (2003). Maximal adherence and invasion of INT 407 cells by Campylobacter jejuni requires the CadF outer-membrane protein and microfilament reorganization. Microbiology 149, 153–165. doi: 10.1099/mic.0.25820-0
Muytjens, H. L., Zanen, H. C., Sonderkamp, H. J., Kolee, L. A., Wachsmuth, I. K., and Farmer, J. J. (1983). Analysis of eight cases of neonatal meningitis and sepsis due to Enterobacter sakazakii. J. Clin. Microbiol. 18, 115–120.
Naves, P., Prado, G., Huelves, L., Gracia, M., Ruiz, V., Blanco, J., et al. (2008). Correlation between virulence factors and in vitro biofilm formation by Escherichia coli strains. Microb. Pathog. 45, 86–91. doi: 10.1016/j.micpath.2008.03.003
Niemann, G. S., Brown, R. N., Gustin, J. K., Stufkens, A., Shaikh-Kidwai, A. S., Li, J., et al. (2011). Discovery of novel secreted virulence factors from Salmonella enterica serovar Typhimurium by proteomic analysis of culture supernatants. Infect. Immun. 79, 33–43. doi: 10.1128/IAI.00771-10
Prasadarao, N. V., Wass, C. A., Weiser, J. N., Stins, M. F., Huang, S. H., and Kim, K. S. (1996). Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect. Immun. 64, 146–153.
Raczkowska, A., Skorek, K., Bielecki, J., and Brzostek, K. (2011). OmpR controls Yersinia enterocolitica motility by positive regulation of flhDC expression. Antonie Van Leeuwenhoek 99, 381–394. doi: 10.1007/s10482-010-9503-8
Srinivasa Rao, P., Yamada, Y., Tan, Y. P., and Leung, K. Y. (2004). Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol. Microbiol. 53, 573–586. doi: 10.1111/j.1365-2958.2004.04123.x
Sun, Y. M., Wang, M., Wang, Q., Cao, B. Y., He, X., Li, K., et al. (2012). Genetic analysis of the Cronobacter sakazakii O4 to O7 O-antigen gene clusters and development of a PCR assay for identification of all C. sakazakii O serotypes. Appl. Environ. Microbiol. 78, 3966–3974. doi: 10.1128/AEM.07825-11
Tall, B. D., Chen, Y., Yan, Q., Gopinath, G. R., Grim, C. J., Jarvis, K. G., et al. (2014). Cronobacter: an emergent pathogen causing meningitis to neonates through their feeds. Sci. Prog. 97, 154–172. doi: 10.3184/003685014X13994743930498
Wood, T. K., Gonzalez Barrios, A. F., Herzberg, M., and Lee, J. (2006). Motility influences biofilm architecture in Escherichia coli. Appl. Microbiol. Biotechnol. 72, 361–367. doi: 10.1007/s00253-005-0263-8
Xu, X. K., Wu, Q. P., Zhang, J. M., Ye, Y. W., Yang, X. J., and Dong, X. H. (2014). Occurrence and characterization of Cronobacter spp. in powdered formula from chinese retail markets. Foodborne Pathog. Dis. 11, 307–312. doi: 10.1089/fpd.2013.1657
Ye, Y. W., Li, H., Ling, N., Han, Y. J., Wu, Q. P., Xu, X. K., et al. (2015). Identification of potential virulence factors of Cronobacter sakazakii isolates by comparative proteomic analysis. Int. J. Food Microbiol. doi: 10.1016/j.ijfoodmicro.2015.08.025
Keywords: membranous proteins, Cronobacter sakazakii, virulence factors, two-dimensional gel electrophoresis (2-DE)
Citation: Ye Y, Gao J, Jiao R, Li H, Wu Q, Zhang J and Zhong X (2015) The Membrane Proteins Involved in Virulence of Cronobacter sakazakii Virulent G362 and Attenuated L3101 Isolates. Front. Microbiol. 6:1238. doi: 10.3389/fmicb.2015.01238
Received: 23 August 2015; Accepted: 26 October 2015;
Published: 09 November 2015.
Edited by:
Giovanna Suzzi, Università degli Studi di Teramo, ItalyReviewed by:
Learn-Han Lee, Monash University Malaysia, MalaysiaMaria Schirone, Università degli Studi di Teramo, Italy
Copyright © 2015 Ye, Gao, Jiao, Li, Wu, Zhang and Zhong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingwang Ye, eWV5dzA0QG1haWxzLmd1Y2FzLmFjLmNu; Qingping Wu, d3VxcDIwM0AxNjMuY29t
†These authors have contributed equally to this work.
 YingWang Ye
YingWang Ye Jina Gao
Jina Gao Rui Jiao
Rui Jiao Hui Li1,2,3,4†
Hui Li1,2,3,4† Qingping Wu
Qingping Wu