- 1Department of Medical Microbiology and Parasitology, Faculty of Medicine and Health Sciences, Universiti Putra Malaysia, Serdang, Malaysia
- 2Department of Biomedical Sciences, Faculty of Medicine and Health Sciences, Universiti Putra Malaysia, Serdang, Malaysia
- 3Infectomics Cluster, Advanced Medical and Dental Institute, Universiti Sains Malaysia, Bertam, Malaysia
Candida glabrata is an emerging human fungal pathogen that has efficacious nutrient sensing and responsiveness ability. It can be seen through its ability to thrive in diverse range of nutrient limited-human anatomical sites. Therefore, nutrient sensing particularly glucose sensing is thought to be crucial in contributing to the development and fitness of the pathogen. This study aimed to elucidate the role of SNF3 (Sucrose Non Fermenting 3) as a glucose sensor and its possible role in contributing to the fitness and survivability of C. glabrata in glucose-limited environment. The SNF3 knockout strain was constructed and subjected to different glucose concentrations to evaluate its growth, biofilm formation, amphotericin B susceptibility, ex vivo survivability and effects on the transcriptional profiling of the sugar receptor repressor (SRR) pathway-related genes. The CgSNF3Δ strain showed a retarded growth in low glucose environments (0.01 and 0.1%) in both fermentation and respiration-preferred conditions but grew well in high glucose concentration environments (1 and 2%). It was also found to be more susceptible to amphotericin B in low glucose environment (0.1%) and macrophage engulfment but showed no difference in the biofilm formation capability. The deletion of SNF3 also resulted in the down-regulation of about half of hexose transporters genes (four out of nine). Overall, the deletion of SNF3 causes significant reduction in the ability of C. glabrata to sense limited surrounding glucose and consequently disrupts its competency to transport and perform the uptake of this critical nutrient. This study highlighted the role of SNF3 as a high affinity glucose sensor and its role in aiding the survivability of C. glabrata particularly in glucose limited environment.
Introduction
Glucose is commonly known as an important carbon source and energy for many organisms. Several studies have attempted to establish the linkage between glucose availability and physiological response of Candida species, including the biofilm formation, oxidative stress, and antifungal resistance (Rodaki et al., 2009; Uppuluri et al., 2010; Ene et al., 2012; Ng et al., 2015b). The regulatory effect by glucose found in these studies is suggestive of the importance of glucose sensing and uptake mechanism in contributing to the fitness of Candida species. Brown et al. (2006) have demonstrated that the loss of Hgt4, a high affinity glucose sensor resulted in a less virulent C. albicans type that failed to grow in low glucose and fermentation-preferred environments. In addition, the loss of Hgt4 also affects the ability of C. albicans to perform the yeast-hyphal morphological switch and therefore compromises its pathogenicity in mouse model of disseminated candidiasis (Brown et al., 2006). Apart from that, the ability to transport glucose by Cryptococcus neoformans is also diminished with the loss of Hxs1, a high affinity glucose sensor-like protein (Liu et al., 2013). The diminished glucose uptake activity leads to an attenuated strain of C. neoformans in which the strain demonstrated a delay in lethal infection in mice model. However, little is known about the role of high affinity glucose sensor in the emerging human fungal pathogen, Candida glabrata.
The ability of C. glabrata to thrive in several glucose-limited anatomical sites of host such as vaginal and blood (Ehrström et al., 2006) is suggestive of its sensitivity toward the low glucose availability in the niches with its superior glucose sensing ability. The primary mechanism for Saccharomyces cerevisiae to sense and transport surrounding glucose is through SNF3-RGT2 mediated sugar receptor repressor (SRR) pathway (Rolland et al., 2002; Santangelo, 2006; Gancedo, 2008). This pathway employs two glucose sensors with different affinity toward glucose: SNF3 (high affinity) and RGT2 (low affinity). They are located in the cell membrane and modulate the expression of the hexose transporters (HXTs) for the uptake of glucose through the interplay of transcription regulators (RGT1 and MIG1) and downstream component of SRR: YCK1 and YCK2 (casein kinase), GRR1 (Glucose Repression Resistant), STD1 (repressor of RGT1; summarized and illustrated in Figure 9; Schmidt et al., 1999; Kim and Johnston, 2006). The homologs of these key genes were found in the C. glabrata genome. The phylogenetic analysis conducted demonstrates the shared neighborhood between CgSNF3 (sequence ID: CAGL0J09020g) and ScSNF3 (Palma et al., 2009; Ng et al., 2015a). In addition, the key feature of glucose sensor as found in ScSNF3, for example the unusual long C-terminal segment amino acids and the signature Özcan motif were also found in the CgSNF3 (Palma et al., 2009; Ng et al., 2015a). Nevertheless, these studies have only managed to highlight the phylogenetic relatedness between ScSNF3 and CgSNF3, but the actual physiological role of SNF3 in C. glabrata is remains unknown. In respect to the importance of glucose in cell physiology, the disruption in SRR pathway may have negative impact on the fitness of C. glabrata. Therefore, this study aimed to explore the possible role of SNF3 in supporting the growth and fitness of C. glabrata under low glucose concentration environment. Its growth profile, biofilm formation, antifungal susceptibility, and capability to withstand phagocytosis of macrophage were assessed. In addition, its role in regulating the expression of the SRR pathway-related genes, which includes the hexose transporters (HXTs), was also deciphered.
Materials and Methods
Yeast Strain and Media Preparation
C. glabrata BG14 (gift from Brendan Cormack, John Hopkins University; Cormack and Falkow, 1999) and its parental strain C. glabrata BG2 (gift from Paul Fidel, Louisiana State University Health Sciences Center) were used in this study (Table 1). Three types of media were utilized: standard YPD (Becton, Dickinson and Company, USA; 20 g of peptone, 20 g of glucose, 10 g of yeast extract), synthetic minimal glucose medium, SD [0.67% of yeast nitrogen base (Becton, Dickinson and Company, USA) + glucose (Fisher Scientific, USA)] and synthetic complete media with uridine dropout [0.17% yeast nitrogen base without ammonium sulfate and amino acid (Becton, Dickinson and Company, USA) + 0.5% ammonium sulfate (Sigma Aldrich, USA) + 2% glucose (Fisher Scientific, USA) + complete supplement mixture with uridine dropout (ForMedium, UK)] (Sherman, 2002). All strains were maintained at 37°C in YPD, unless otherwise indicated.
Strain Construction
For the construction of the C. glabrata SNF3Δ strain, the C. glabrata BG14 was transformed to Ura3 + by replacing the SNF3 open reading frame (ORF) with SNF3::URA3 disruption cassette. SNF3::URA3 disruption cassette that consists of “upstream SNF3-URA3-downstream SNF3” was amplified by PCR (primers TS_SNF3_F and TS_SNF3_R; Table 2) and purified through Expin™ Combo GP purification kit (GeneAll, Korea). The purified cassette was then transformed into C. glabrata BG14 as described in Cormack and Falkow (1999). Transformants were selected on synthetic complete media with uridine dropout and insertion was confirmed by diagnostic PCR (primers CHK_F_1 CHK_R_1 and CHK_F_2 CHK_R_2; Table 2) for the absence of SNF3 and presence of URA3 at the correct locus. In order to eliminate the possible effect from secondary mutation of mutant constructed, three independently constructed SNF3Δ mutants were analyzed, and treated as three biological replicates in the subsequent assays (Odds et al., 2006).
Growth Profiling and Growth Rate Calculation
The capability of SNF3Δ strain and parental wild type BG2 to grow in different levels of surrounding glucose was assessed using a modified procedure as described in Brown et al. (2006) and Rodaki et al. (2009). C. glabrata cells were grown overnight in YPD medium at 37°C and washed with phosphate buffered saline (PBS, pH 7.4) for three times before re-inoculated into 50 mL fresh SD (OD600 = 0.1) with prepared glucose concentrations of 0.01 (extremely low), 0.1 (low), 0.2 (moderate), 1 (high), and 2% (extremely high), respectively. The cells were allowed to grow at 37°C, 200 rpm in shaking incubator (shaking). Another set of cells were incubated in the same manner but were left to be static. These two conditions served as respiration-preferred (shaking) and fermentation-preferred (static) atmosphere, respectively (Brown et al., 2006). The cells were harvested hourly for 10 h and the optical density of cells (OD600) for each hour were recorded and proceeded with the growth rate calculation as shown in Equation (1) (Widdel, 2010).
where,
μ = growth rate, h−1
OD1 = optical density obtained from the log phase of growth curve
OD2 = two times of OD1 obtained from the log phase of growth curve
t1 = time of OD1 obtained
t2 = time of OD2 obtained.
Biofilm Formation
The biofilm formation assay were performed with minor modification as described in Pierce et al. (2008) by replacing RPMI1640 medium with SD (0.01 and 0.1% glucose). Overnight grown C. glabrata cells were harvested and washed prior to re-inoculation into the defined SD (OD600 = 0.1). A volume of 100 μl of cell suspension was added to microtitre plate (U-shaped, tissue culture treated) (Becton, Dickinson and Company, USA). The microtitre plate was covered with lid and sealed with parafilm, followed by incubation for 24 h at 37°C. The media was aspirated and the plate was washed three times using 200 μl of PBS, pH 7.4. The plate was placed in an inverted position on a blotting paper to remove residual PBS. The biofilm activity was quantified via XTT 2,3-Bis (2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide reduction assay. A freshly prepared 100 μL mixture of 0.5 g/L XTT (Sigma Aldrich, USA) and 10 mM menadione (Sigma Aldrich, USA) (10000:1, v/v) was added to the washed-biofilm in the microtitre plate. The plate was wrapped with aluminum foil and incubated in dark at 37°C for 3 h. A volume of 80 μL of the supernatant was transferred to a new microtitre plate and the plate was read by using microtitre plate reader (Bio-Tek, USA) at wavelength of 490 nm.
Amphotericin B Susceptibility Assay
The inhibitory concentration of C. glabrata BG2 against amphotericin B was determined using the method as described in NCCLS (CLSI) M27-A2 by replacing the RPMI1460 with 0.1% glucose SD. The inhibitory concentration obtained was applied in the modified method from Rodaki et al. (2009) to elucidate the possible role of SNF3 in contributing to the anti-amphotericin B susceptibility. Briefly, overnight grown C. glabrata cells were harvested and washed prior to re-inoculation into the 0.1% glucose SD (OD600 = 0.1) and regrown to OD600 = 0.5. Cell suspension was added to 1.5 mL centrifuge tube (Axygen, USA) and microtitre plate (U-shaped, tissue culture-treated) (Becton, Dickinson and Company, USA) together with defined concentration of amphotericin B. The centrifuge tube and microtitre plate was covered and sealed with parafilm, followed by incubation for 24 h at 37°C. CFU (colony-forming unit) was determined from cell suspension in centrifuge tube for the calculation of survival percentage. In addition, the plate was read by using microtitre plate reader at wavelength of 600 nm to examine the cell density for the confirmation of C. glabrata viability. The survivability percentage of C. glabrata was calculated by applying the formula as below:
Candida-Macrophage Co-culture Assay
The capability of both C. glabrata strains to withstand engulfment of macrophage was analyzed as described by Kaur et al. (2006) and Collette et al. (2014) with minor modification. Murine macrophage cells, RAW264.7 (gift from Daud Ahmad Israf Ali, Universiti Putra Malaysia) were maintained and incubated in Dulbecco's Modified Eagle's Medium (DMEM; Life Technologies, USA), supplemented with 10% Fetal Bovine Serum (FBS; Life Technologies, USA) at 37°C/5% CO2. Prior to the co-culture step, 5 × 105 of RAW264.7 cells were seeded into 6-well plate (Becton, Dickinson and Company, USA) for 24 h at 37°C/5% CO2. After incubation, the washed cell was counted for the determination of cells number. For the preparation of C. glabrata cells, overnight grown C. glabrata cells were washed and regrown in fresh YPD (OD600 = 0.5). Harvested mid-log phase cells were washed and re-inoculated in fresh DMEM + 10% FBS for desired cell density to match the ratio of 1:1 (effector: target) prior to the co-culturing with RAW264.7 cells prepared. The mixed culture of C. glabrata and RAW264.7 was incubated at 37°C/5% CO2. In order to measure the growth of macrophage engulfed-yeast, non-engulfed yeast cell were washed away with DMEM after 2 h of incubation. The lysates of infected macrophages were scrapped and collected from wells at two time points (2 and 24 h) in ice-cold deionized water and plated on YPD agar. CFUs were determined after incubation of 24 h at 37°C and the growth ratio of engulfed cells were determined by applying the formula below:
RNA Extraction
Overnight-cultured C. glabrata cells in YPD medium were washed and regrown in fresh YPD (OD600 = 0.1) to mid-log phase (OD600 = 0.5). The mid-log phase cells were collected, washed and re-suspended in SD (0.01% glucose) and allowed to grow at 37°C for 2 h. The collected cell was washed and RNA extraction was performed based on the described protocol in Yeast Current Protocols in Molecular Biology (Collart and Oliviero, 1993). Verification of the RNA integrity and quality were performed by visualization on 1% Tris-acetate-EDTA gel and NanoPhotometer® (Implen, Germany). RNAs were treated with Maxima H minus first strand cDNA synthesis kit with dsDNase (Thermo Scientific, USA) as described in kit manual with minor modification where RNAs were reverse transcribed with the mixture of Oligo(dT) 18 and random hexamer for the generation of full length transcripts (Resuehr and Spiess, 2003). The RNAs were also reverse transcribed with reaction suspension lacking reverse transcriptase (Non Reverse Transcriptase, NRT) and without RNA template (Non Template Control, NTC) as controls, respectively. The RNA isolation and subsequent cDNA synthesis were performed in three biological independent experiments.
Quantitative Real-time PCR (qRT-PCR)
Two reference genes were employed as internal controls namely the ACT1 and UBC13 (Li et al., 2012) for a more reliable and accurate normalization output. All PCR primers (Table 2) were designed to amplify target genes based on the gene sequences sourced from the Candida Genome Database (http://www.candidagenome.org/). The PCR efficiency using each set of primers of the respective genes was determined in two independent experiments by running a series of five-fold dilution of C. glabrata DNA in MiniOpticon™ Real time PCR (Bio-Rad, USA) machine. The amplification efficiency for each respective gene was determined to be between 90 and 110%. For the expression analysis of the genes, all samples were performed in technical triplicate. The total volume of each reaction was 20 μl where it contained the cDNA template, 500–600 nM primers, 2X SensiFAST SYBR No-ROX (SYBR green) master mix (Bioline, UK) and type-1 ultrapure water (Milipore, USA). The reagents mixture was placed in low-profile white strip tube (Life Technologies, USA) and allowed to amplify in two-step cycling PCR amplification (polymerase activation: 95°C for 2 min, 40 cycles of denaturation: 95°C for 5 s and annealing/extension: 60°C for 30 s). Melting curve analysis was performed to ensure no non-specific PCR products were generated. A NRT and NTC were included for each gene during the qRT-PCR analysis. For post-experimental expression analysis, normalized expression ratios were calculated based on the mathematical equation developed by Pfaffl (2001). Wild type BG2 was chosen as the reference strain (baseline) when interpreting the result for the transcript profiling. Normalized expression ratio calculated was presented in logarithms based (log10).
Statistical Analysis
Statistical analyses were performed using SPSS Statistics (Version 17.0) software. All the experiments were performed at least three times and the data presented are mean of all experiments performed. Error bars represent standard error of the mean (SEM). Statistical significance was assessed by unpaired t-test to compare control (wild type) and sample (mutant). The relative expression software tool (REST©) version 2009 (Pfaffl et al., 2002) was employed to test the statistical significance in qRT-PCR analysis.
Results
Loss of SNF3 Resulted in the Failure of C. Glabrata to Thrive in Low Glucose Concentration Environments
The inabilities of SNF3Δ to grow in low glucose environments were demonstrated in both shaking and static condition (Figures 1–3). After 10 h of incubation, the growth rate of SNF3Δ strain was significantly reduced (p-value < 0.05) in 0.01 and 0.1% glucose environment for respiration preferred-condition (shaking) and in 0.01, 0.1, and 0.2% glucose environment for fermentation preferred-condition (static). However, deletion of SNF3 did not weaken the growth of SNF3Δ strain in higher glucose environment (1 and 2%; Figures 1–3). These observations highlighted the role of SNF3 in sustaining the growth of C. glabrata, particularly in low glucose for both respiration and fermentation-preferred environment. Furthermore, SNF3 is deemed to be more important in fermentation process where the growth defect of SNF3Δ was found to be more severe (extended up to 0.2% glucose) in fermentation-preferred condition. In respect of the data obtained, which suggested the deleterious effect of SNF3Δ is seen only in low glucose environment, the subsequent assays including biofilm formation and amphotericin B susceptibility assays were carried out in glucose limited environment (0.01 and 0.1%).
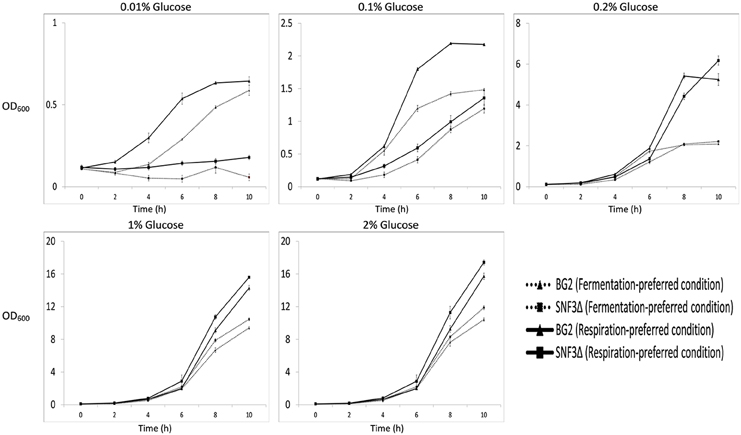
Figure 1. Growth profile of Candida glabrata BG2 and SNF3Δ in five difference glucose concentrations tested for both fermentation and respiration-preferred condition.
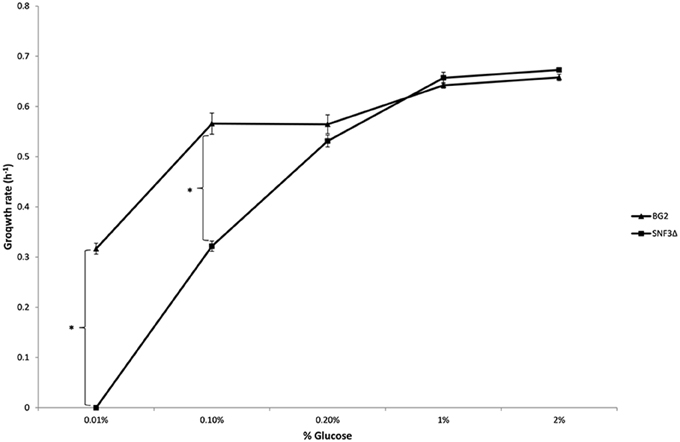
Figure 2. Growth rate of Candida glabrata BG2 and SNF3Δ in five differences glucose concentrations with respiration-preferred condition. Significant differences (indicated by *) were found between wild type and mutant under low glucose environments: 0.01 and 0.1% (p-value < 0.05).
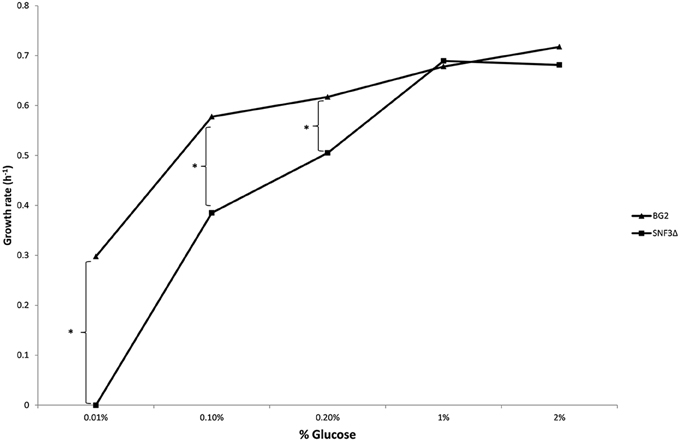
Figure 3. Growth rate of Candida glabrata BG2 and SNF3Δ in five differences glucose concentrations with fermentation-preferred condition. Significant differences (indicated by *) were found between wild type and mutant under low glucose environments: 0.01, 0.1, and 0.2% (p-value < 0.05).
Deletion of SNF3 Gives no Effect in the Biofilm Formation Capability of C. Glabrata in Glucose-Limited Environments
Previous study demonstrated the effects of glucose levels in directing C. albicans to form biofilm. Candida albicans tends to form biofilm in low glucose environment and lives in planktonic form in higher glucose environment (Uppuluri et al., 2010; Ng et al., 2015b). The sensitivity of SNF3 in responding to surrounding glucose leads to the thought whether this putative high affinity glucose sensor could contribute in detecting the flow of surrounding glucose and therefore orchestrates the biofilm/planktonic living form of Candida species in accordance to the availability of glucose. Result showed SNF3 did not participate in the biofilm formation of C. glabrata in low glucose environment as no significant differences were found between BG2 and SNF3Δ in the 0.01 and 0.1% glucose tested, respectively (Figure 4).
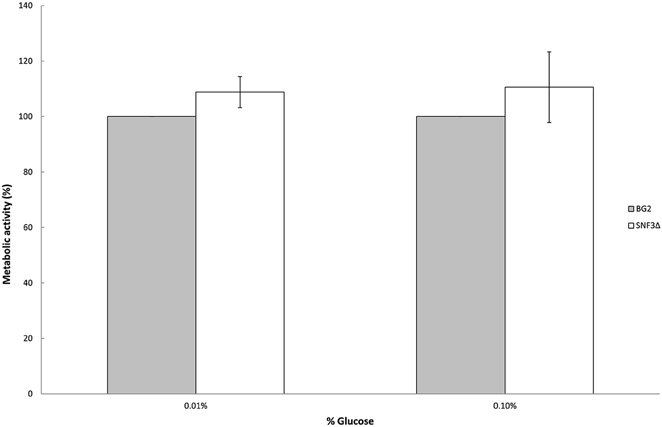
Figure 4. Biofilm formation activity of Candida glabrata BG2 and SNF3Δ strains under 0.01 and 0.1% glucose concentration. Unpaired T-test was carried out for the statistical analysis to examine the significant difference between BG2 and SNF3Δ and no significant difference was found.
The Loss of SNF3 Makes C. Glabrata More Vulnerable to Amphotericin B Treatment in Low Glucose Concentration Environment
Previous study demonstrated the ability of Candida species to withstand antifungal is affected by the type of carbon sources and levels (Ene et al., 2012; Mota et al., 2015; Ng et al., 2015b). With the aim to elucidate further the possible role of SNF3 in regulating the fitness of C. glabrata, the ability of both strains to withstand amphotericin B in low glucose environment was tested. However, the complete retarded growth of SNF3Δ in 0.01% glucose leads to the inability in the effort to set up an unstressed control for the calculation of survival percentage. Thus, only 0.1% glucose was tested in this assay. The growth of both wild type and SNF3Δ strain were arrested at 2 μg/mL of amphotericin B. The wild type strain was able to resist amphotericin B at 1 μg/mL while complete inhibition was observed in the SNF3Δ strain (p-value < 0.01). The wild type showed a better growth in comparison to SNF3Δ strain at 0.5 μg/mL amphotericin B (Figure 5). These data established the fact that glucose sensing by the SNF3 gene may contribute to the ability of C. glabrata in withstanding the effects of amphotericin B under low glucose environment.
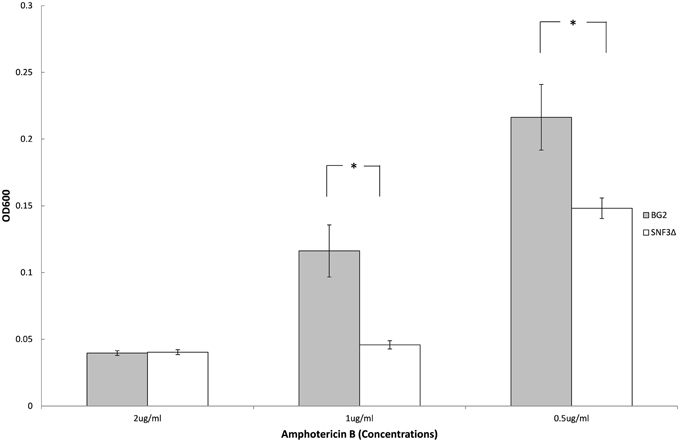
Figure 5. Survivability of Candida glabrata BG2 and SNF3Δ strains under treatment of three different concentrations of amphotericin B in 0.1% glucose. Unpaired T-test was carried out for the statistical analysis to examine the significant differences (indicated by *) between BG2 and SNF3Δ (p-value < 0.01).
SNF3Δ Strain Shows Reduced Growth in Macrophages
The microenvironment in macrophage is always linked to nutrient-limited environment, particularly in glucose availability. In order to validate the possible role of SNF3 in promoting the fitness of C. glabrata under glucose-limited environment, the survivability of macrophage trapped- C. glabrata was assayed in an ex vivo manner. Results demonstrated a significant reduced growth (p-value < 0.01) of the internalized mutant strain in comparison to wild type strain (Figure 6) and this suggests the essential role of SNF3 in supporting the survivability of C. glabrata upon macrophage engulfment.
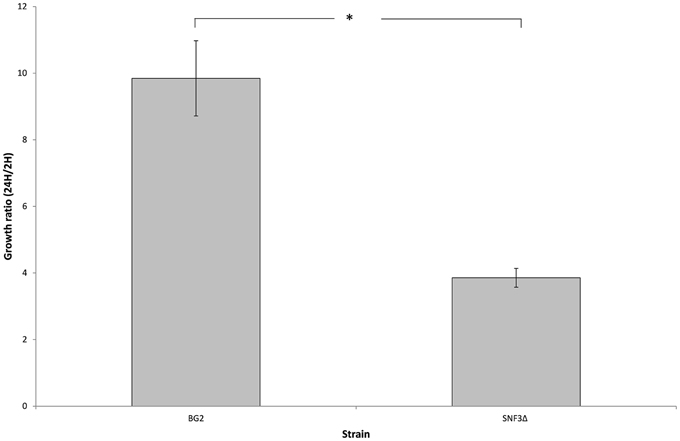
Figure 6. The survival ratio of Candida glabrata BG2 and SNF3Δ strains recovered from macrophages at 24 h vs. 2 h after co-cultivation. Unpaired T-test was carried out for the statistical analysis to examine the significant differences (indicated by *) between BG2 and SNF3Δ (p-value < 0.01).
Deletion of SNF3 Affects the Expression of Downstream Hexose Transporters (HXTs)
There are 11 hexose transporters found in C. glabrata. The expressions of these hexose transporters were examined and compared between wild type and SNF3Δ strain. Out of 11 hexose transporters, only nine hexose transporters were studied because the nucleotide sequences of putative hexose transporters CAGL0A02211 and CAGL0A02233 were found to be 96% similar while CAGL0A02662 and CAGL0A02640 displayed 100% similarity. The high similarity among these hexose transporters caused the inability in primer design for the expression study of those genes. Out of nine hexose transporters, six of them were affected with the deletion of SNF3, where four of them (CAGL0A1804_HXT1, CAGL0A01782_HXT4, CAGL0A02211/2233_HXT6/7, and CAGL0D02662/2640_HXT2/10) were down regulated while two (CAGL0A02321_HXT3 and CAGL0A01826_HXT5) were up regulated (Figure 7). In addition, deletion of SNF3 resulted in down-regulation of STD1, YCK1, and YCK2, which serve as the downstream messengers of SNF3 to modulate the expression of hexose transporters (Figure 8). Nevertheless, the expression of RGT2 was up regulated while expression of RGT1, GRR1, and MIG1 did not change significantly with the deletion of SNF3 (Figure 8). These observations suggest the significant role of SNF3 in regulating the signaling pathway of glucose uptake mechanism.
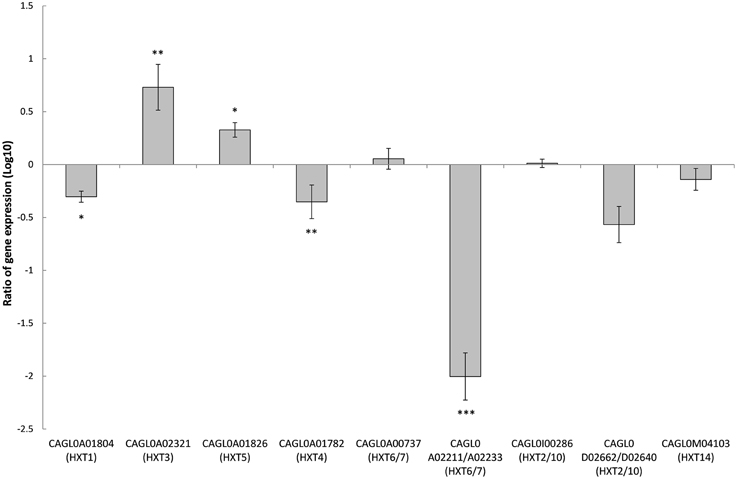
Figure 7. Comparison of expression ratios (Log10) for the Candida glabrata hexose transporters (HXTs) after the knockout of SNF3. *p < 0.1, **p < 0.05, ***p < 0.01.
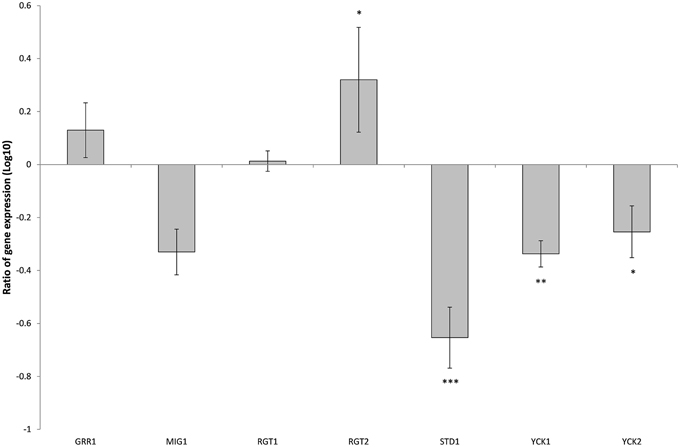
Figure 8. Comparison of expression ratios (Log10) for the Candida glabrata Sugar Receptor Repressor (SRR) related genes after the knockout of SNF3. *p < 0.1, **p < 0.05, ***p < 0.01.
Discussion
Data presented in present study is suggestive of the role of SNF3 as high affinity glucose sensor in C. glabrata, which is essential for it to grow in glucose-limited environment. SNF3 appeared to be important for the growth of C. glabrata in both respiration and fermentation preferred condition with low glucose environments (0.01 and 0.1%) and up to 0.2% glucose in fermentation preferred condition. Brown et al. (2006) demonstrated the deletion of glucose sensor, HGT4 in C. albicans attenuates its ability to grow only in the fermentation-preferred condition and low level of fermentable carbon source (0.2%). Data suggest high affinity glucose sensor appears to be more essential in C. glabrata than in C. albicans. The dissimilarities observed in both the Candida species could be due to the differences in their nature of glucose utilization. C. glabrata is identified as Crabtree-positive yeast or aerobic fermenter where, it prefers fermentation over respiration and produces ethanol even there is presence of oxygen (Van Urk et al., 1990), while C. albicans is known as Crabtree-negative yeast or respiratory yeast, which prefers respiration whenever there is presence of oxygen. The preferred-fermentation in Crabtree-positive yeast produces only two ATP per glucose, in comparison to 36/38 ATP per glucose produced in respiration. Owing to the preferred-inefficient mode of metabolism, Crabtree-positive yeast is found to exhibit higher glucose consumption rate than Crabtree-negative yeast (Van Urk et al., 1990; Fleck et al., 2011). Therefore, the presence of two specialized glucose sensors in C. glabrata (Palma et al., 2009; Ng et al., 2015a) may contribute to the higher glucose uptake sensitivity in order to fulfill its ATP demands and removal of this high affinity glucose sensor lead to detrimental effect on the growth of C. glabrata in low glucose environment (Figures 1–3). Unlike C. glabrata, there is only one high affinity glucose sensor (HGT4) found in C. albicans (Brown et al., 2006). The capability of C. albicans to assimilate both fermentable and non-fermentable carbon sources at the same time suggests C. albicans has evolved distinctively to adapt itself by not relying solely on glucose for its growth in hostile host niche with limited glucose availability (Sandai et al., 2012). Thus, a single glucose sensor is probably sufficient to support the life process of C. albicans. However, it is still unclear whether C. glabrata is equipped with the same metabolic flexibility. The presence of the two glucose sensors with different affinity in C. glabrata similar to the ones found in S. cerevisiae suggests it would behave more like S. cerevisiae. The baker's yeast is unable to assimilate both fermentable and non-fermentable carbon sources at the same time (Sandai et al., 2012). Further investigation on the carbon metabolic flexibility in C. glabrata is warranted as this could provide insight into the metabolic adaptation on the disease progression of this fungal pathogen.
Apart from glucose sensing and uptake mechanism, the expression of glucose sensor gene HGT4 in C. albicans is deemed to be regulated by macrophage engulfment, antifungal mechanism, and biofilm formation activity of yeast (Barker et al., 2004; Liu et al., 2005; Brown et al., 2006). The deletion of SNF3 indeed diminished the capability of C. glabrata to withstand the macrophage challenge and amphotericin B treatment but did not affect its biofilm formation activity. Data presented demonstrated the importance of SNF3 in supporting the growth of C. glabrata under low glucose environment in the growth profiling assay. This observation was extended further to the nutrient-limiting microenvironment of macrophage. Macrophage is critically important in building up an immunological barrier to counter infectious agents through its unique nutritional seal off and oxidative stress to destroy engulfed intruders (Kaur et al., 2006). Previous study demonstrated the capability of C. glabrata to perform autophagy for the nutritional scavenging and recycling in order to sustain its growth upon phagocytosis (Roetzer et al., 2010). Data (Figure 6) suggests in addition to autophagy mechanism, glucose sourcing and uptake are also important in aiding C. glabrata to sustain prolonged phagocytosis. The absence of SNF3 may result in the inability of C. glabrata to absorb sufficient glucose to perform basic physiological function or even to initiate autophagy mechanism and therefore lead to the diminished growth. On the other hand, deletion of SNF3 did not affect the capability of C. glabrata to form biofilm under low glucose condition as expected. Data presented suggest there is probably another sensor in C. glabrata but not SNF3 that assists in detecting the nutrient flow in environment. Further investigation is warranted for a clearer picture on how this pathogenic yeast senses and alters its lifestyle to adapt itself in such environment where abrupt change of nutrients takes place.
The transcriptional analysis on selected hexose transporters (HXTs) revealed that almost half (four out of nine) hexose transporters were down regulated with the removal of SNF3, together with the down-regulation of downstream casein kinase (YCK1 and YCK2) and STD1 (Figures 7, 8). The disruption of the signaling pathway for high affinity hexose transporters explained the compromised fitness of C. glabrata under low glucose environment (Figures 1–3) as this triggers the failure in transporting sufficient glucose to support its growth. In addition, data presented concurs with the view that the expression of transcription regulator, RGT1 is regulated by the glucose concentration but not affected by the signal generated from glucose sensors (Özcan and Johnston, 1999) as the expression of RGT1 remain unchanged even with the missing signal from SNF3. However, the direct regulation of glucose concentration on the expression level of RGT1 is still not fully understood. Nonetheless, the shutting down of these four hexose transporters did not diminish the growth of C. glabrata completely as there are two other hexose transporters that were still actively expressed namely the CAGL0A0232 (HXT3) and CAGL0A01826 (HXT5), together with the up regulation of RGT2. This could be a compensatory mechanism used by C. glabrata to compensate the loss of SNF3 with the activation of RGT2. Notably, these HXT3 and HXT5 were regarded as key hexose transporters for C. glabrata in low glucose environment from our previous work (Ng et al., 2015a). Nevertheless, this compensatory mechanism still failed to salvage C. glabrata from glucose uptake crisis as the growth defect is still significant (p-value < 0.05; Figures 1–3) in the absence of SNF3. We opine the compensation of glucose uptake by HXT3 and HXT5 is insufficient to provide the amount of glucose needed and this highlights the importance of four other repressed HXTs in supporting the growth of C. glabrata under low glucose environment. In addition, the capability of RGT2 to induce expression of HXT3 and HXT5 supports the view that SNF3 and RGT2 have separate but overlapping functions. Özcan et al. (1998) demonstrated the capability of SNF3 in S. cerevisiae to restore the expression of HXT1 (supposedly induce by RGT2) by 64%, in a RGT2 mutant. This observation suggests a complex and interconnected regulatory pathway of glucose sensing and uptake mechanism in yeast. From the data obtained, a model of glucose sensing in C. glabrata through the modulation of SNF3 is illustrated based on the understanding of the homolog and the inferred glucose sensing mechanism in S. cerevisiae (Figure 9). Further work is warranted, as the compensatory mechanism proposed here is still not fully deciphered. In addition, effort to study the transcriptional profile of the highly homologous HXTs genes using other approach should be carried out. With more complete information on the role of each hexose transporters present in C. glabrata, a clearer and more comprehensive picture on the role of SNF3 in SRR pathway will be achieved.
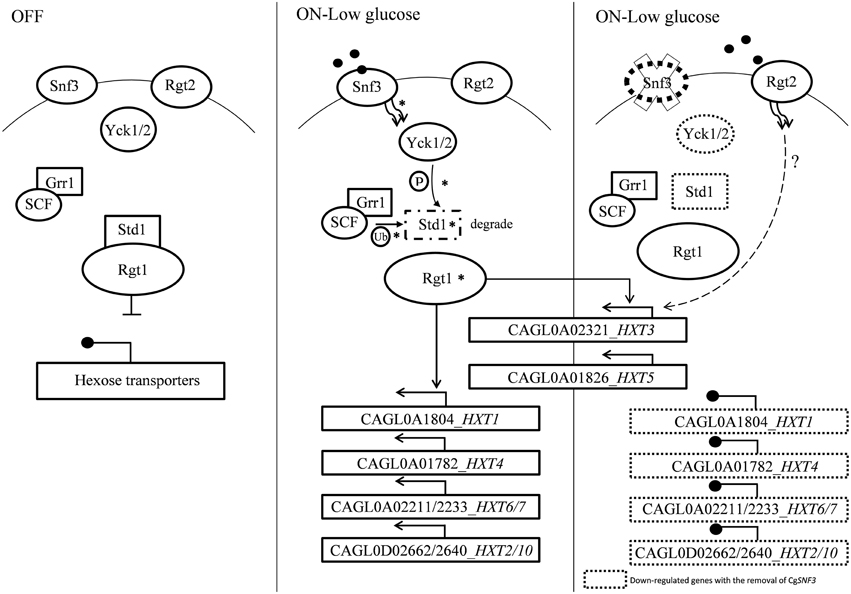
Figure 9. A model of glucose sensing in Candida glabrata under low glucose environment. The part of the pathway labeled with asteisks inferred from published works done on S. cerevisiae (Rolland et al., 2002; Santangelo, 2006; Gancedo, 2008). Hexose transporters are repressed by Std1-bounded-Rgt1 when there is no stimulation from glucose sensor located in the cell membrane. Presence of low concentration of glucose induced signal from high affinity glucose sensor, Snf3 to the phosphorylation of Std1 by the Yck kinase. Phosphorylated Std1 is then subjected to the SCFGrr1—mediated ubiquitination and degraded by proteasome. Degraded Std1 results in the activation of Rgt1, which then leads to derepression of downstream hexose transporters. Deletion of SNF3 gives rise to the disruption of hexose transporters expression and glucose uptake mechanism, therefore leads to the interference of Candida glabrata fitness under low glucose environment. However, the possible interaction between RGT2 and downstream HXT3/HXT5 (labeled with dotted line) is remains unclear and requires further investigation.
In conclusion, our results thus far suggest the important role of SNF3 in C. glabrata in the expression of hexose transporters under low glucose environment. We also highlight the vital role of SNF3 in promoting C. glabrata growth, resistant toward amphotericin B under glucose limited environment and macrophage engulfment by governing the glucose uptake mechanism. These results suggest SNF3 could be a potential factor for C. glabrata to survive and thrive in host niches with limited glucose availability. Further investigation such as RNA-sequencing and comparative proteomic study could be carried out for the analysis of global transcriptomes and validation of the obtained result. Owing to the essential role of glucose on metabolic network of organism, further exploration on the glucose sensing mechanism highlighted in current study could contribute in the discovery of novel drug target and help in controlling the emergence of C. glabrata.
Author Contributions
TS, LT, and DS designed the experiments. TS, SY, and PR carried out the experiments. TS analyzed and interpreted the data. TS and LT wrote the manuscript with critical revision for important intellectual content from MD, DS, and PP.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was financed by the Universiti Putra Malaysia (04-02-12-1799RU). TSN is a recipient of the Mybrain 15 Scholarship offered by the Ministry of Education (MOE) Malaysia. This work was presented, in part, at the 19th Congress of the International Society for Human and Animal Mycology (abstract number: 706), 4th-8th May 2015, Melbourne, Australia. We would like to acknowledge the generosity of Prof. Brendan Cormack and Prof. Paul Fidel for granting us the C. glabrata strains used in this study. In addition, we also would like to thank Dr. Iryna Bohovych for her advice in the mutant construction work and Ms Ley Juen Looi for her valuable comments on the manuscript.
References
Barker, K. S., Crisp, S., Wiederhold, H., Lewis, R. E., Bareither, B., Eckstien, J., et al. (2004). Genome-wide expression profiling reveals genes associated with amphotericin B and fluconazole resitance in experimentally induced antifungal resistant isolates of Candida albicans. J. Antimicrob. Chemoth. 54, 376–385. doi: 10.1093/jac/dkh336
Brown, V., Sexton, J. A., and Johnston, M. (2006). A glucose sensor in Candida albicans. Eukaryot. Cell 5, 1726–1737. doi: 10.1128/EC.00186-06
Collart, M. A., and Oliviero, S. (1993). Preparation of yeast RNA. Curr. Protoc. Mol. Biol. 23, 13.12.1–13.12.5.
Collette, J. R., Zhou, H. J., and Lorenz, M. C. (2014). Candida albicans suppresses nitric oxide generation from macrophage via a secreted molecule. PLoS ONE 9:e96203. doi: 10.1371/journal.pone.0096203
Cormack, B. P., and Falkow, S. (1999). Efficient homologous and illegitimate recombination in the opportunistic yeast pathogen Candida glabrata. Genetics 151, 979–987.
Ehrström, E., Yu, A., and Rylander, E. (2006). Glucose in vaginal secretions before and after oral glucose tolerance testing in women with and without recurrent vulvovaginal candidiasis. Obstet. Gynecol. 108, 1432–1437. doi: 10.1097/01.AOG.0000246800.38892.fc
Ene, I. V., Adya, A. K., Wehmeier, S., Brand, A. C., MacCallum, D. M., Gow, N. A. R., et al. (2012). Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen. Cell. Microbiol. 14, 1319–1335. doi: 10.1111/j.1462-5822.2012.01813.x
Fleck, C. B., Schöbel, F., and Brock, M. (2011). Nutrient acquisition by pathogenic fungi: nutrient availability, pathway regulation, and differences in substrate utilization. Int. J. Med. Microbiol. 301, 400–407. doi: 10.1016/j.ijmm.2011.04.007
Gancedo, J. M. (2008). The early steps of glucose signalling in yeast. FEMS Microbiol. Rev. 32, 673–704. doi: 10.1111/j.1574-6976.2008.00117.x
Kaur, R., Ma, B., and Cormack, B. P. (2006). A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc. Natl. Acad. Sci. U.S.A. 104, 7628–7633. doi: 10.1073/pnas.0611195104
Kim, J. H., and Johnston, M. (2006). Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transcporter genes in Saccharomyces cerevisiae. J. Biol. Chem. 281, 26144–26149. doi: 10.1074/jbc.M603636200
Li, Q. Q., Skinner, J., and Bennett, J. E. (2012). evalution of reference genes for real-time quantitative PCR studies in Candida glabrata following azole treatment. BMC Mol. Biol. 13:22. doi: 10.1186/1471-2199-13-22
Liu, T. B., Wang, Y., Baker, G. M., Fahmy, H., Jiang, L., and Xue, C. (2013). The glucose sensor-like protein Hxs1 is a high affinity glucose transporter and required for virulence in Cryptococcus neoformans. PLoS ONE 8:e64239. doi: 10.1371/journal.pone.0064239
Liu, T. T., Lee, R. E. B., Barker, K. S., Lee, R. E., Wei, L., Homayouni, R., et al. (2005). Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 49, 2226–2236. doi: 10.1128/AAC.49.6.2226-2236.2005
Mota, S., Alves, R., Carneiro, C., Silva, S., Brown, A. J., Istel, F., et al. (2015). Candida glabrata susceptibility to antifungals and phagocytosis is modulated by acetate. Front. Microbiol. 6:919. doi: 10.3389/fmicb.2015.00919
Ng, T. S., Desa, M. N. M., Sandai, D., Chong, P. P., and Than, L. T. L. (2015a). Phylogenetic and transcripts profiling of glucose sensing related genes in Candida glabrata. Jundishapur J. Microbiol. 8:e25177. doi: 10.5812/jjm.25177
Ng, T. S., Desa, M. N. M., Sandai, D., Chong, P. P., and Than, L. T. L. (2015b). Growth, biofilm formation, antifungal susceptibility and oxidative stress resistance of Candida glabrata are affected by different glucose concentrations. Infect. Genet. Evol. doi: 10.1016/j.meegid.2015.09.004. [Epub ahead of print].
Odds, F. C., Gow, N. A. R., and Brown, A. J. P. (2006). “Chapter 22: Toward a molecular understanding of Candida albicans virulence,” in Molecular Principles of Fungal Pathogenesis, eds J. Heitman, S. G. Filler, J. E. Edwards. Jr., and A. P. Mitchell (Washington, DC: American Society for Microbiology), 305–319. doi: 10.1128/9781555815776.ch22
Özcan, S., Dover, J., and Johnston, M. (1998). Glucose sensing and signaling by two glucsoe receptors in the yeast Saccharomyces cerevisiae. EMBO J. 17, 2566–2573. doi: 10.1093/emboj/17.9.2566
Özcan, S., and Johnston, M. (1999). Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 63, 554–569.
Palma, M., Seret, M. L., and Baret, P. V. (2009). Combined phylogenetic and neighbourhood analysis of the hexose transporters and glucose sensors in yeasts. FEMS Yeast Res. 9, 526–534. doi: 10.1111/j.1567-1364.2009.00511.x
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Pfaffl, M. W., Horgan, G. W., and Dempfle, L. (2002). Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. doi: 10.1093/nar/30.9.e36
Pierce, C. G., Uppuluri, P., Tristan, A. R., Wormley, F. L. Jr., Mowat, E., Ramage, G., et al. (2008). A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat. Protoc. 3, 1494–1500. doi: 10.1038/nprot.2008.141
Resuehr, D., and Spiess, A. N. (2003). A real-time polymerase chain reaction-based evaluation of cDNA synthesis priming methods. Anal. Biochem. 322, 287–291. doi: 10.1016/j.ab.2003.07.017
Rodaki, A., Bohovych, I. M., Enjalbert, B., Young, T., Odds, F. C., Gow, N. A. R., et al. (2009). Glucose promotes stress resistance in the fungal pathogen Candida albicans. Mol. Biol. Cell 20, 4845–4855. doi: 10.1091/mbc.E09-01-0002
Roetzer, A., Gratz, N., Kovarik, P., and Schüller, C. (2010). Autophagy supports Candida glabrata survival during phagocytosis. Cell. Microbiol. 12, 199–216. doi: 10.1111/j.1462-5822.2009.01391.x
Rolland, F., Winderickx, J., and Thevelein, J. M. (2002). Glucose-sensing and –signalling mechanism in yeast. FEMS Yeast Res. 2, 183–201. doi: 10.1111/j.1567-1364.2002.tb00084.x
Sandai, D., Yin, Z., Selway, L., Stead, D., Walker, J., Leach, M. D., et al. (2012). The evolutionary rewiring of ubiquitination targets has reprogrammed the regulation of carbon assimilation in pathogenic yeast Candida albicans. mBio 3, 1–12. doi: 10.1128/mBio.00495-12
Santangelo, G. M. (2006). Glucose signalling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70, 253–282. doi: 10.1128/MMBR.70.1.253-282.2006
Schmidt, M. C., McCartnet, R. R., and Zhang, X. D. (1999). Std1 and Mth1 proteins interacts with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 4561–4571. doi: 10.1128/MCB.19.7.4561
Sherman, F. (2002). Getting started with yeast. Method Enzymol. 350, 3–41. doi: 10.1016/S0076-6879(02)50954-X
Uppuluri, P., Chaturvedi, A. K., Srinivsan, A., Banerjee, M., Ramasubramaniam, A. K., and Köhler, J. R. (2010). Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 6:e1000828. doi: 10.1371/journal.ppat.1000828
Van Urk, H, Voll, W. S. L., Scheffers, W. A., and Van Dijken, J. P. (1990). Transient-state analysis of metabolic fluxes in crabtree-positive and crabtree-negative yeasts. Appl. Environ. Microbiol. 56, 281–287.
Widdel, F. (2010). Theory and Measurement of Bacterial Growth. University Bremen. Available online at: www.mpi-bremen.de/Binaries/Binary13037/Wachstumsversuch.pdf
Keywords: Candida glabrata, glucose sensor, SNF3, glucose-limited environment, hexose transporter
Citation: Ng TS, Chew SY, Rangasamy P, Mohd Desa MN, Sandai D, Chong PP and Than LTL (2015) SNF3 as High Affinity Glucose Sensor and Its Function in Supporting the Viability of Candida glabrata under Glucose-Limited Environment. Front. Microbiol. 6:1334. doi: 10.3389/fmicb.2015.01334
Received: 08 September 2015; Accepted: 16 November 2015;
Published: 01 December 2015.
Edited by:
Edvaldo Antonio Ribeiro Rosa, The Pontifical Catholic University of Paraná, BrazilReviewed by:
Patrick Van Dijck, KU Leuven, BelgiumVishvanath Tiwari, Central University of Rajasthan, India
Copyright © 2015 Ng, Chew, Rangasamy, Mohd Desa, Sandai, Chong and Than. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leslie Thian Lung Than, bGVzbGllQHVwbS5lZHUubXk=
 Tzu Shan Ng
Tzu Shan Ng Shu Yih Chew
Shu Yih Chew Premmala Rangasamy
Premmala Rangasamy Mohd N. Mohd Desa
Mohd N. Mohd Desa Doblin Sandai
Doblin Sandai Pei Pei Chong2
Pei Pei Chong2 Leslie Thian Lung Than
Leslie Thian Lung Than