- 1State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, China
- 2The Provincial Key Lab of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China
Microbial opsins play a crucial role in responses to various environmental signals. Here, we report that the microbial opsin homolog gene sop1 from the necrotrophic phytopathogenic fungus Sclerotinia sclerotiorum was dramatically up-regulated during infection and sclerotial development compared with the vegetative growth stage. Further, study showed that sop1 was essential for growth, sclerotial development and full virulence of S. sclerotiorum. Sop1-silenced transformants were more sensitive to high salt stress, fungicides and high osmotic stress. However, they were more tolerant to oxidative stress compared with the wild-type strain, suggesting that sop1 is involved in different stress responses and fungicide resistance, which plays a role in the environmental adaptability of S. sclerotiorum. Furthermore, a Delta blast search showed that microbial opsins are absent from the genomes of animals and most higher plants, indicating that sop1 is a potential drug target for disease control of S. sclerotiorum.
Introduction
The white mold fungus Sclerotinia sclerotiorum (Lib.) de Bary is a typical necrotrophic phytopathogen with a remarkably broad host range and worldwide distribution. At least 408 described species of plants from 278 genera in 75 families are susceptible to this pathogen (Boland and Hall, 1994), and it often leads to great losses of economically important crops such as rapeseed (Brassica spp.), sunflowers, soybeans, and peanuts. There are several key steps in the life cycle of S. sclerotiorum: vegetative growth, infection, sclerotial development, sclerotial myceliogenic germination, sclerotial carpogenic germination, and apothecium formation (stipe). The sclerotia produced by S. sclerotiorum are pigmented, hard, asexual resting structures capable of surviving for years in soil (Adams and Ayers, 1979; Le Tourneau, 1979; Willetts and Bullock, 1992). Sclerotia can either germinate myceliogenically to produce mycelia or carpogenically to form apothecia, which can produce abundant ascospores. Ascospores can undergo airborne dissemination, which is the most important means of S. sclerotiorum dispersal (Chet and Henis, 1975; Abawi and Grogan, 1979; Steadman, 1979). Previous research has demonstrated that oxalic acid and a wide array of cell wall degrading enzymes were key pathogenic factors in this pathogen (Riou et al., 1991; Cessna et al., 2000) and that signal transduction pathways also played a crucial role in pathogenesis and sclerotial development (Chen et al., 2004; Erental et al., 2008).
Sensing and responding to the environment and ensuring appropriate cellular responses are vitally important for the survival and proliferation of many organisms in a range of biological niches. Organisms receive and respond to diverse extracellular signals or stimuli through cell signal-transduction cascades. Genes involved in the signal-transduction cascades are designated as signaling genes, and play subtle, versatile and crucial roles in diverse biological processes (Krauss, 2006). High osmolarity is a common extracellular signal that reflects the adaptation of organisms to various extreme environments. During pathogen-host interactions, an oxidative burst is a conserved and nonspecific defense reaction of plants against invading pathogens. In this process, reactive oxygen species (ROS) can be produced by both plants and pathogens around the infection site (Tiedemann, 1997; Schouten et al., 2002; Tenberge et al., 2002; Egan et al., 2007). Large quantities of ROS can kill pathogens, hence, oxidative stress is also a common extracellular stimuli that is crucial for the survival of pathogens during infection. Many signaling networks specifically respond to an increase in extracellular osmolarity, among which the Hog1-MAPK signaling pathway plays a particularly important role in cellular adaptation to osmotic stress (Brewster et al., 1993; Xu, 2000). In filamentous fungi, Hog1 homologs response for both osmoregulation and oxidative stress (Heller et al., 2012). In addition to Hog1 homologs, many other signaling genes are involved in the oxidative stress response, such as AP-1-like transcription factors that have been characterized as regulators of ROS resistance in mammalian cells as well as in fungi (Temme and Tudzynski, 2009).
Some signaling genes produce specialized integral membrane proteins known as receptors that initiate cell signal transduction and trigger cascade changes in cell functions (Seger and Krebs, 1995). Proteinaceous light sensors are a class of receptors generally grouped into three categories in plants: phototropins, cryptochromes, and phytochromes. These light sensors are actually chromoprotein complexes. Both phototropins and cryptochromes use flavin as the chromophore for blue-light or near-UV-light sensing, and the phytochromes use bilin as the chromophore for red-light or far-red-light sensing (Purschwitz et al., 2006). Similar photosensory systems have been found in fungi and have been confirmed to play multiple roles in fungal phototropism, photoconidiation, photomorphogenesis, circadian rhythm, pathogenicity, or virulence, light-dependent differentiation, secondary metabolism, pigmentation and asexual and sexual development (Idnurm and Heitman, 2005b; Lee et al., 2006; Liu and Bell-Pedersen, 2006; Purschwitz et al., 2006; Herrera-Estrella and Horwitz, 2007; Cai et al., 2013). In addition to the photosensory systems described above, the photosensory opsin system has been found in microbes and animals. Typical opsins are seven-transmembrane (TM) helix receptors that use retinal as a chromophore for light sensing (Luecke et al., 1999; Palczewski et al., 2000); thus, this opsin-retinal complex is referred to as rhodopsin (Zhang et al., 2011). A conserved lysine residue is located on TM helix seven (TM7), which is necessary for the incorporation of retinal within the 7-TM helices (Zhang et al., 2011). However, there are some opsin-like or opsin-related proteins (ORPs, such as some bacteriorhodopsins) that do not contain the retinal binding lysine and hence cannot function as opsins (Oesterhelt and Tittor, 1989; Brown, 2004). The opsins are divided into two distinct types: type I microbial opsins and type II animal opsins (Spudich et al., 2000). Although, the biological functions of many opsins in animals, prokaryotes and algae have been confirmed (Sineshchekov and Govorunova, 2001; Nickle and Robinson, 2007; Zhang et al., 2011), their roles in many fungi remains to be determined. Although, many opsins have been identified in fungal genomes, only a few fungal opsins, such as in Neurospora crassa, Leptosphaeria maculans, Fusarium fujikuroi, Cryptococcus neoformans, and Botrytis cinerea, have been documented, and they as yet have no known function (Bieszke et al., 1999; Idnurm and Howlett, 2001; Brown, 2004; Idnurm and Heitman, 2005a; Estrada and Avalos, 2009; Heller et al., 2012). The aim of this study was to elucidate the biological functions of a microbial opsin homolog gene, sop1 in S. sclerotiorum. We found that sop1 had diverse roles and was involved in growth, sclerotial development and virulence of S. sclerotiorum. Sop1 was also involved in the osmotic stress and the oxidative stress responses and therefore partially contributed to the environmental adaptability of S. sclerotiorum.
Materials and Methods
Fungal and Bacterial Strains, Plants, and Culture Condition
S. sclerotiorum virulent wild-type strain Ep-1PNA367 (Xie et al., 2006) was used in this study. Fungal cultures were grown on potato dextrose agar (PDA, Difco, Detroit, MI, USA) at 20°C. S. sclerotiorum transformants were obtained and cultured on PDA amended with 80 μg/mL hygromycin B (EMD Biosciences, USA) at 20°C. Escherichia coli strain JM109 was used to propagate all plasmids. Agrobacterium tumefaciens strain EHA105 was used for fungal transformation. Seedlings of Arabidopsis thaliana (ecotype Columbia-0) were grown in greenhouse at 20 ± 2°C for 1 month under a 12 h light/dark cycle with 70% relative humidity.
Construction of RNAi Vectors and Transformation of S. sclerotiorum
The vector pCXDPH with PtrpC-ccdB-Pgpd fragment (the promoter PtrpC and Pgpd were placed in opposite directions) and the vector pCIT with PtrpC-intron-TtrpC fragment (two same exogenetic gene fragments were inserted into this region in opposite directions to make the transcripts form “hairpin structure”) (Nguyen et al., 2008; Yu et al., 2012) were used to construct S. sclerotiorum RNAi vectors. In detail, a 530 bp sop1 DNA fragment was amplified with a pair of specific primers RNAi-sop1-F (5′-CCATCGATGG ATCCCCAGTCCATCAGCCACTCCATCTGTT-3′) and RNAi-sop1-R (5′-CCCAAGCT TCTGCAGGAAAGCAACGCATCCCA TAGCATACCAA-3′) from the cDNA library of S. sclerotiorum and then ligated into pCXDPH digested by XcmI (New England Biolabs, Beverly, MA, USA) to replace the ccdB gene to produce the pRNAi-1 vector (Figure 3A). The same amplified sop1 DNA fragment was digested with two pairs of suitable enzymes, respectively, and then both of the digested DNA fragments were ligated into the pCIT vector. Afterwards, the fusion fragment PtrpC-sop1 fragment-intron-sop1 fragment-TtrpC was digested with Sac I and Xho I before it was ligated into the pCH vector (Yu et al., 2012) to produce the pRNAi-2 vector (Figure S2A). The Agrobacterium-mediated transformation (ATMT) with mycelial agar discs method was used to transform S. sclerotiorum as previously described (Yu et al., 2012). The knockdown transformants were validated by quantitative reverse transcription PCR (qRT-PCR).
Extraction and Manipulation of Nucleic Acids
To examine the sop1 expression patterns during infection, fresh hyphae of the wild-type strain without any medium were inoculated on A. thaliana (Col-0) leaves and cultured at 20°C. The hyphae and inoculated leaves were collected together at 0, 3, 6, 9, and 12 h post inoculation (hpi), respectively, for total RNA extraction. To examine the sop1 expression patterns during sclerotial development, mycelial agar discs of the wild-type strain were taken from active colony edge and inoculated on the center of cellophane over PDA plates, and cultured at 20°C for 1–7 days post inoculation (dpi) before the cultures were collected, respectively, for total RNA extraction. This period was selected because it represents the entire biological processes from vegetative growth, the initial stages of sclerotial development to the sclerotial formation and maturation stages. To evaluate the relative sop1 expression levels in the wild-type strain and different transformants, mycelial agar discs were taken from active colony edge and inoculated on the center of cellophane over PDA plates, and cultured at 20°C for 7 dpi and then the cultures (including both mycelium and sclerotia) were collected for total RNA extraction. To examine the sop1 expression patterns when the wild-type strain were cultured with oxidative stress, the mycelial agar discs were taken from active colony edge and inoculated on the center of cellophane over complete medium (CM) plates without any treatment and CM plates supplemented with different concentrations of H2O2 (0–12 mM), respectively, and cultured at 20°C for 24 h before the cultures were collected for total RNA extraction. The total RNA was isolated from each sample described above with TriZOL reagent (Invitrogen, USA) according to the manufacturer's protocols. First strand cDNA was generated with RevertAid™ First Strand cDNA Synthesis Kit (MBI Fermentas, Lithuania) after the total RNA samples were treated with DNase I (TaKaRa, Dalian). Gene expression was analyzed by qRT-PCR using a Bio-Rad CFX96 Real Time System (America) and Quantitect SYBR Green PCR master mix (Bio-Rad, USA), according to the manufacturer's instructions. The S. sclerotiorum β-tubulin gene (SS1G_04652) was used as housekeeping gene for qRT-PCR normalization (Harel et al., 2006). For qRT-PCR analysis, the primers qPCR-sop1-F (5′-TTCCAAGTGTTGTTCCT AATGC-3′) and qPCR-sop1-R (5′-GAGAGTGATGAATGCGGTAATAAC-3′) were used to amplify the sop1 gene fragment; the primers β-tubulin-F (5′-TTGGATTTGCTCC TTTGACCAG-3′) and β-tubulin-R (5′-AGCGGCCATCATGTTCTTAGG-3′) were used to amplify the β-tubulin gene fragment. For every experiment, three samples were harvested and independent qRT-PCR assays were repeated three times, with each repetition having three replicates.
Characterization of Sop1 Silenced Transformants
To examine colony morphology and assay growth rates, mycelial agar discs (0.8 mm) of the silenced transformants and the wild-type strain were taken from active colony edge and inoculated on the center of PDA Petri dishes, and cultured at 20°C. Colony diameters were measured at 24 hpi and the first day growth rate (semidiameter of the colony minus semidiameter of the mycelial agar disc) was calculated for statistical analysis. To compare the morphology of hyphal tips, mycelial agar discs of the silenced transformants and the wild-type strain were taken from active colony edge and inoculated on cellophane over PDA plates, and cultured for 48 h at 20°C before the morphology of hyphal tips were examined using a light microscope. To evaluate virulence, mycelial agar discs (diameter 6 mm) of the silenced transformants and the wild-type strain were taken from active colony edge and inoculated on detached Brassica napus and tomato leaves, and cultured at 20°C for 48 h before the induced lesions were measured. To calculate the inhibition rate of hyphal growth when the silenced transformants and the wild-type strain were cultured with high osmotic stress, mycelial agar discs (diameter 8 mm) were taken from active colony edge and inoculated on the center of CM plates without any treatment and CM plates supplemented with 1 M glucose, 1 M sorbitol and 1.2 M sucrose, respectively, and cultured for 24 h at 20°C before the colony diameters were measured. To measure the colony diameters of the silenced transformants and the wild-type strain cultured with membrane damage stress, high salt stress, fungicide stress, and oxidative stress, the mycelial agar discs (diameter 8 mm) of the silenced transformants and the wild-type strain were taken from active colony edge and inoculated on the center of CM plates without any treatment and CM plates supplemented with 0.02% sodium dodecylsulphate (SDS), 1 M NaCl, 1 M KCl, 0.25 μg/mL carbendazim, 0.40 μg/mL dimetachlone, 0–24 mM H2O2, respectively, and cultured at 20°C. Colony diameters were measured every day before the colony reached margins of the plates. In all experiments, at least three independent replications were performed.
Bioinformatics Analysis
The signaling genes were identified according to the genome annotation of S. sclerotiorum (Amselem et al., 2011). The Phyre2 server (Kelley and Sternberg, 2009) was used to predict transmembrane domain regions and construct 3D structural model which was displayed by PyMOL (DeLano, 2002). Sequences for sop1 homologs were collected from the non-redundant protein sequence database at the NCBI website1 using DELTA-BLAST (Boratyn et al., 2012) with sop1 as the query sequence. Datasets were assembled with a cut-off E-value < 1e-6. All sequences available for sop1 homologs form Volvox carteri f. nagariensis, Dunaliella salina, Acetabularia acetabulum, Chlorella vulgaris, Guillardia theta CCMP2712, Podospora anserina S mat+, L. maculans JN3, Haloarcula argentinensis, Halobacterium salinarum, Natronomonas pharaonis, Haloarcula marismortui, Chlamydomonas reinhardtii, Cryptomonas sp. S2, Cyanophora paradoxa, N. crassa OR74A, F. fujikuroi, C. neoformans var. grubii H99, C. neoformans var. neoformans JEC21, B. cinerea B05.10 and Oryza sativa Indica Group were used to perform amino acid alignment using COBALT (Papadopoulos and Agarwala, 2007). These species were selected because their opsin genes were experimentally characterized or reviewed previously (Bieszke et al., 1999; Idnurm and Howlett, 2001; Brown, 2004; Ruiz-Gonzalez and Marin, 2004; Idnurm and Heitman, 2005a; Estrada and Avalos, 2009; Zhang et al., 2011; Heller et al., 2012). Multiple protein sequence alignment without any truncation was viewed and edited by Jalview (Clamp et al., 2004). The entire protein sequence alignment was subjected to model testing with the ProtTest v.2.4, using the Aikake information criterion (AIC) and default settings (Abascal et al., 2005), before it was used for building a phylogenetic tree. Protein maximum likelihood (ML) tree was inferred with PhyML-mixtures (Guindon and Gascuel, 2003; Le et al., 2008), assuming the best-fit LG model (Le and Gascuel, 2008) according to the ProtTest and SPR tree topology search strategy (Hordijk and Gascuel, 2005). Gaps in alignment were systematically treated as unknown characters. The reliability of internal branches was evaluated based on aLRT SH-like branch support (Anisimova and Gascuel, 2006). The phylogenetic tree graphic was produced using the Interactive Tree of Life (Letunic and Bork, 2007).
Results
Characterization of the Microbial Opsins in S. sclerotiorum Genome
Our previous study on the transcriptomic analysis of the six key developmental stages (including vegetative growth, infection, sclerotial development, sclerotial myceliogenic germination, sclerotial carpogenic germination, and apothecium formation) of S. sclerotiorum based on digital gene expression (DGE) (Lyu et al., 2015) revealed the expression of a large number of signaling genes that were significantly regulated during corresponding biological processes, as summarized in Table S1. A microbial opsin homolog gene (SS1G_01614) is very special among these signaling genes because it was dramatically induced during all developmental stages compared with the vegetative growth stage (Table S1) and therefore was selected for further study. Two microbial opsin homologs were found in S. sclerotiorum genome: SS1G_01614 (GenBank accession: XP_001597420) and SS1G_04339 (GenBank accession: XP_001594532) and these were designated as sop1 and sop2, respectively, in this study. Sop1 and sop2 have high protein sequence similarity (E-value: 3e–33). Sop1 consists of 313 amino acid residues with a bacteriorhodopsin domain (pfam01036, E-value: 1.93e–25). It has seven predicted transmembrane helices and its initial N terminus and C terminus were predicted to be extracellular and cytoplasmic, respectively (Figure 1A). The predicted three dimensional (3D) structure of sop1 showed that it shared a strong structural similarity with the eukaryotic light-driven proton-pumping rhodopsin Acetabularia rhodopsin II (ARII) from the marine alga A. acetabulum (Wada et al., 2011; Figure 1B). The multiple protein sequence alignment of sop1 and sop2 with Mac in L. maculans (Idnurm and Howlett, 2001), nop1 in N. crassa OR74A (Bieszke et al., 1999), Pop in Podospora anserine S mat+ (Espagne et al., 2008), AaRh in A. acetabulum (Tsunoda et al., 2006), OpsCp in C. paradoxa (Frassanito et al., 2010), Gt1Rh in G. theta (Sineshchekov et al., 2005), CsRh in Cryptomonas sp. S2 (Sineshchekov et al., 2005), and Gt2Rh in G. theta (Sineshchekov et al., 2005) indicated that the lysine residues of these microbial opsins in the last transmembrane domains were conserved (Figure 1C). In addition, it also showed that the seven TM regions and their contiguous regions in these proteins were more conserved than other regions (Figure 1C).
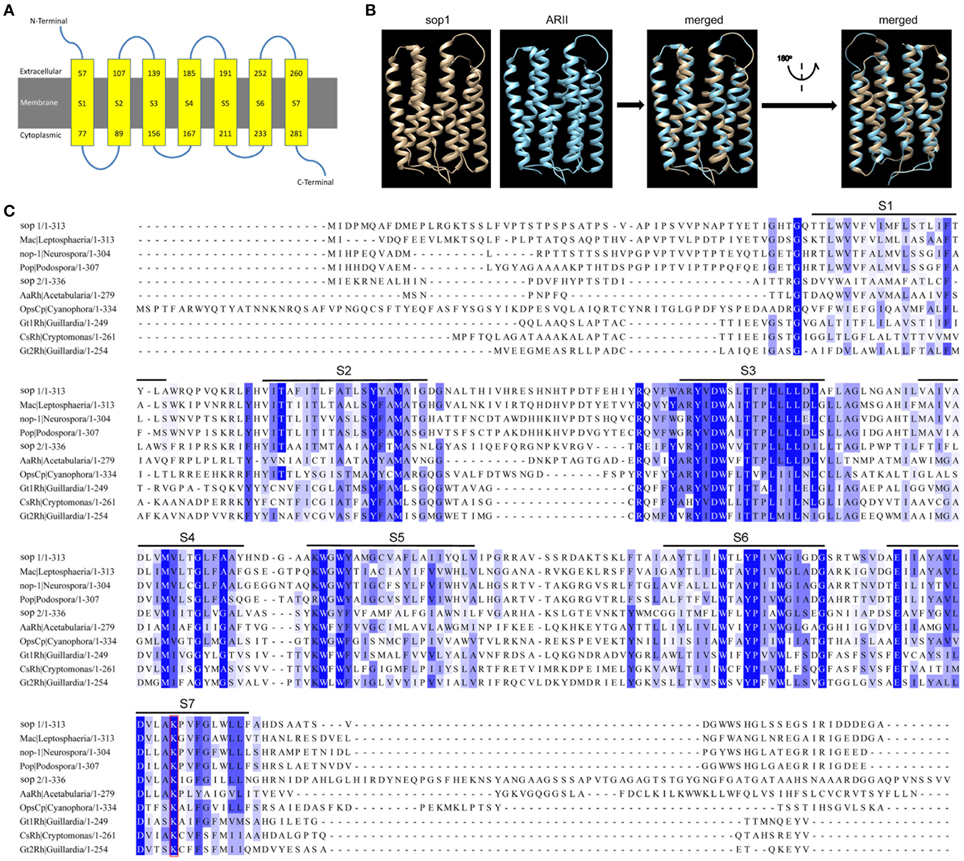
Figure 1. Characterization of sop1 in S. sclerotiorum. (A) A schematic diagram of seven transmembrane structures (S1–S7, yellow boxes) that were predicted with the Phyre2 program. Numbers in the yellow boxes indicate start and end sites of each transmembrane helix of sop1, respectively. (B) Comparison of 3D structural models of sop1 (from 50 aa to 287 aa) and eukaryotic light-driven proton pumping rhodopsin, Acetabularia rhodopsin II (arII), from marine alga A. acetabulum (Wada et al., 2011). The 3D structural models were generated with the Phyre2 program. 218 residues (70% of sop1) have been modeled with 100% confidence by the single highest scoring template c3am6C in the Phyre 2 database. (C) Multiple alignment of the protein sequences of sop1, sop2, Mac in L. maculans (GenBank accession: AAG01180), nop-1 in N. crassa OR74A (GenBank accession: XP_959421), Pop in P. anserine S mat+ (GenBank accession: XP_001904282), AaRh in A. acetabulum (GenBank accession: AAY82897), OpsCp in C. paradoxa (GenBank accession: ACV05065), Gt1Rh in G. theta (GenBank accession: ABA08437), CsRh in Cryptomonas sp. S2 (GenBank accession: ABA08439), and Gt2Rh in G. theta (GenBank accession: ABA08438) using COBALT program.
Expression Patterns of Sop1
Our DGE data indicated that sop1 was not expressed during the vegetative growth stage, but was dramatically up-regulated above 10-fold (log2Ratio) during all other developmental stages (Table S1). To examine the expression patterns of sop1 during infection and sclerotial developmental in more detail, qRT-PCR was used to detect sop1 expression levels at different times after inoculation. The results showed that sop1 was dramatically up-regulated during the very early stage of infection in A. thaliana (Col-0) leaves. The expression level of sop1 reached a peak at 3 hpi before it gradually decreased during the later stages of infection (6, 9, and 12 hpi, Figure 2A). In addition, the qRT-PCR results showed that the expression of sop1 was dramatically induced during the initial stage of sclerotial development (at 3 dpi) and up-regulated approximately 300,000-fold during the sclerotial maturation stage (at 7 dpi, Figure 2B). Taken together, these results were in accordance with the DGE data and suggested that sop1 was possibly involved in the infection and sclerotial developmental processes of S. sclerotiorum.
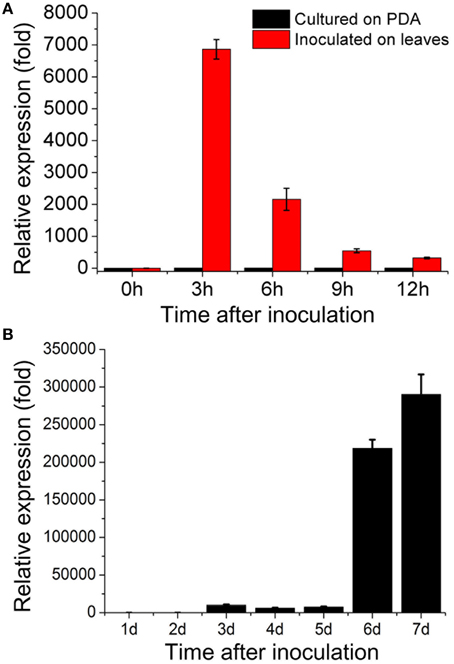
Figure 2. Expression patterns of sop1. (A) Relative expression of sop1 during infection and vegetative growth as determined by qRT-PCR analysis. The abundance of cDNA from cultures on PDA at 0 hpi was assigned a value of 1. (B) Relative expression of sop1 during sclerotial development as determined by qRT-PCR analysis. The abundance of cDNA from cultures on PDA at 1 dpi was assigned a value of 1. The gene expression levels of sop1 in the wild-type strain were normalized to that of the β-tubulin transcripts from each sample. Bars indicate the standard error.
RNA Interference-Mediated Down-Regulation of Sop1 Impaired Growth, Sclerotial Development, and Virulence of S. sclerotiorum
Because of multi-nucleated cells of S. sclerotiorum, the RNAi technique was used to study the biological functions of sop1. We had noted the “off-target” effect of the RNAi technique before silencing sop1. Although the protein sequence similarity between sop1 and sop2 is high, there was no sequence similarity between sop1 transcript and all other S. sclerotiorum transcripts, even between the transcripts of sop1 and sop2 according to a BLASTN search. Additionally, sop2 is almost not expressed during vegetative growth, sclerotial development and infection as well as all the other developmental stages according to our DGE data (Lyu et al., 2015). Two strategies (Figure 3A and Figure S2A) were used to silence the sop1 gene in the wild-type strain Ep-1PNA367. QRT-PCR analysis was used to examine the accumulation of sop1 transcripts in each transformant. The results showed that the silenced transformants with markedly reduced expression levels of sop1 exhibited abnormal sclerotial development and significantly reduced virulence. For example, the silenced transformants sop1-22 and sop1-45 showed abnormal sclerotial development when they were cultured on PDA at 20°C for 8 days, while the wild-type strain had already produced large amount of mature sclerotia under the same conditions (Figure 3B). Ten days later, sop1-22 and sop1-45 produced a small amount of immature sclerotia indicating that the sclerotial development in sop1-22 and sop1-45 was severely delayed. A similar result was obtained by the other silencing strategy (Figure S2B). Compared with the wild-type strain, the tip hyphae of sop1-22 and sop1-45 were denser and more likely to adhere to each other (Figure 3D), and the hyphal growth rate was also significantly reduced in sop1-22 and sop1-45 (Figure 3G and Figure S2F). The virulence of sop1-22 and sop1-45 were also significantly impaired (Figures 3C,F). Similar results were observed when sop1-22 and sop1-45 were inoculated on detached tomato leaves (Figure S1), indicating that the virulence reduction was not species-specific. Similar results were observed for the other silencing strategy (Figures S2C,E). In addition, the silencing efficiency was positively correlated with the virulence reduction (Figures 3E,F; Figures S1, S2D,E); for example, the lesions caused by another transformant (sop1-46) with lower silencing efficiency were larger than those caused by sop1-22 with higher silencing efficiency. This phenomenon indicated that the phenotypic change of silenced transformants was caused by the silencing of sop1. Taken together, our results showed that sop1 has pleiotropic effects on sclerotial development, virulence and hyphal growth of S. sclerotiorum.
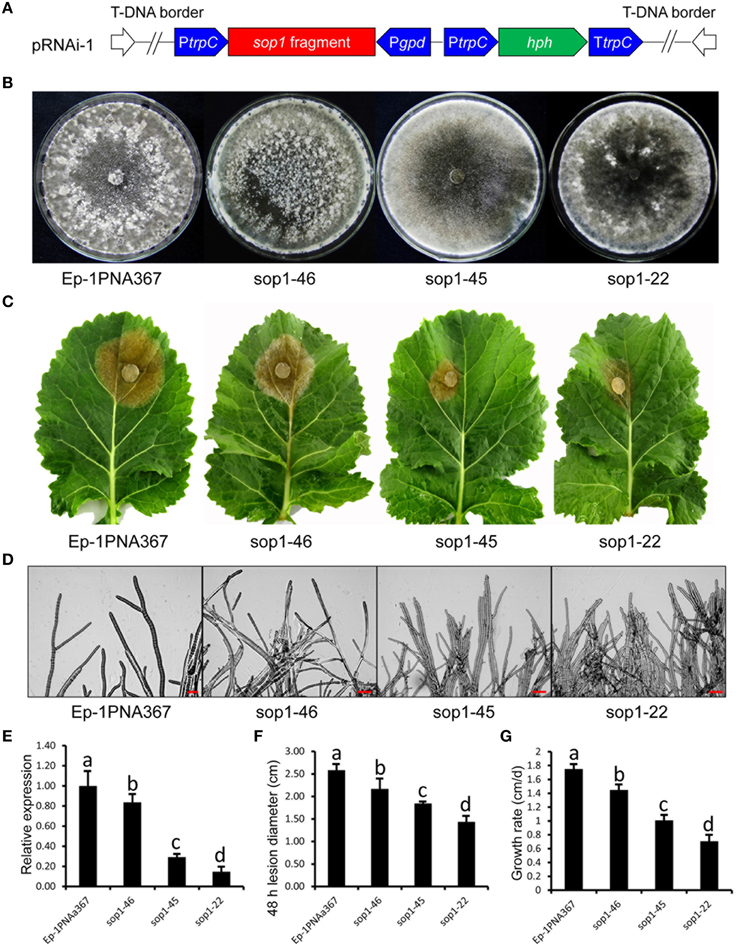
Figure 3. Biological characterization of the sop1 silenced transformants. (A) Construction of sop1 silencing vector pRNAi-1. (B) Comparison of the phenotypes of the sop1 silenced transformants and the wild-type strain. Photos were taken at 8 dpi. (C) Virulence assay of the silenced transformants and the wild-type strain on detached oilseed rape leaves. (D) Morphological comparison of the hyphal tips of the sop1 silenced transformants and the wild-type strain. (E) Relative sop1 expression level of the sop1 silenced transformants and the wild-type strain as determined by qRT-PCR analysis. A value of 1 was assigned to the abundance of cDNA from the wild-type strain. The gene expression levels of sop1 in the silenced transformants and the wild-type strain were normalized to that of the β-tubulin transcripts from each strain. Bars indicate the standard error. (F) Comparison of lesion diameter for the silenced transformants and the wild-type strain. (G) Comparison of the hyphal growth rate of the silenced transformants and the wild-type strain. Three independent replications were performed. Bars indicate the standard error. The values are presented as the mean ± s.d. Differentiation was evaluated by a t-test. Different letters on a graph indicate significant differences, while same letters on a graph indicate no significant differences, P = 0.05.
Sop1 is Involved in the Adaptability of S. sclerotiorum to Different Environmental Stressors
Because sop1 was predicted to be a transmembrane protein, the tolerance of sop1 silenced transformants to membrane damage stress was tested. In the presence of 0.02% SDS, which attacks the cell membrane (Temme et al., 2012), the growth rate of both the silenced transformants (sop1-22 and sop1-45) and the wild-type strain was dramatically suppressed during the entire growth process on CM. However, the wild-type strain could grow slowly at the later stages after inoculation on CM with SDS, while the growth of silenced transformants sop1-22 and sop1-45 was completely inhibited on CM with SDS even during the later stages after inoculation (Figures 4A,B), suggesting that the sop1 silenced transformants were more sensitive to SDS. This result indicated that sop1 might be involved in the tolerance to the cytomembrane damage stress, or sop1 might affect the homeostasis of S. sclerotiorum. To further explore the biological roles of sop1 under different environmental stresses, sop1 silenced transformants and the wild-type strain were cultured with high concentrations of sugars and salts. When the sop1 silenced transformants and the wild-type strain were cultivated with a high concentration of glucose (1 M), sorbitol (1 M), and sucrose (1.2 M), the growth of sop1-22 and sop1-45 was more significantly inhibited compared with the wild-type (Figures 4C,D), suggesting that the silencing of sop1 made S. sclerotiorum more sensitive to high osmotic stress. When sop1 silenced transformants and the wild-type strain were cultivated with a high concentration of salts, the growth rate of sop1-22, sop1-45 and the wild-type strain were all severely decreased during the initial stages (1-2 dpi, Figures 4F,G). For example, the colony diameter of untreated sop1-22, sop1-45, and the wild-type strain were 2.63 ± 0.12, 2.81 ± 0.10, and 3.52 ± 0.14 cm, respectively, at 1 dpi on CM without any treatment; but the colony diameter of sop1-22, sop1-45 and the wild-type strain were only 0.91 ± 0.08, 0.93 ± 0.07, and 0.87 ± 0.04 cm, respectively, at 1 dpi on CM with 1 M NaCl; and were only 0.98 ± 0.04, 0.98 ± 0.06, and 0.97 ± 0.03 cm, respectively, at 1 dpi on CM with 1 M KCl. However, the wild-type strain grew better than sop1-22 and sop1-45 after 4 dpi on CM with 1 M NaCl and 1 M KCl (Figures 4E–G). For example, the colony diameter of the wild-type strain was 5.59 ± 0.25 cm and 7.03 ± 0.13 cm at 5 dpi on CM with 1 M NaCl and 1 M KCl, respectively. But the colony diameter of sop1-22 and sop1-45 were only 2.01 ± 0.05 cm and 3.10 ± 0.09 cm, respectively, on CM with 1 M NaCl, and were only 2.15 ± 0.07 cm and 3.07 ± 0.11 cm, respectively, on CM with 1 M KCl. These results indicated that although the growth of both of the sop1 silenced transformants and the wild-type strain was dramatically depressed during the initial stages, the capacity to overcome high salt stress of the wild-type strain was greater than that of the sop1 silenced transformants, and therefore the silenced transformants were more sensitive to high salt stress during the later stages. Likewise, similar results were obtained when the silenced transformants and the wild-type strain were inoculated on CM supplemented with the two fungicides carbendazim and dimetachlone, respectively. The results showed that although the growth of all strains was severely depressed during the initial stage (1 dpi, Figures 4I,J), the sop1 silenced transformants sop1-22 and sop1-45 were more sensitive to the two fungicides than the wild-type during the later stages (3 dpi, Figures 4H–J). Additionally, our qRT-PCR result showed that the expression of sop1 was significantly down-regulated in the various stress conditions (Figure 4K). Taken together, these results indicated that sop1 was involved in the response to high osmotic stress, high salt stress and fungicides resistance, and might partially contribute to the capacity of S. sclerotiorum to overcome extreme environmental factors. Thus, sop1 is associated with the environmental adaptability of S. sclerotiorum.
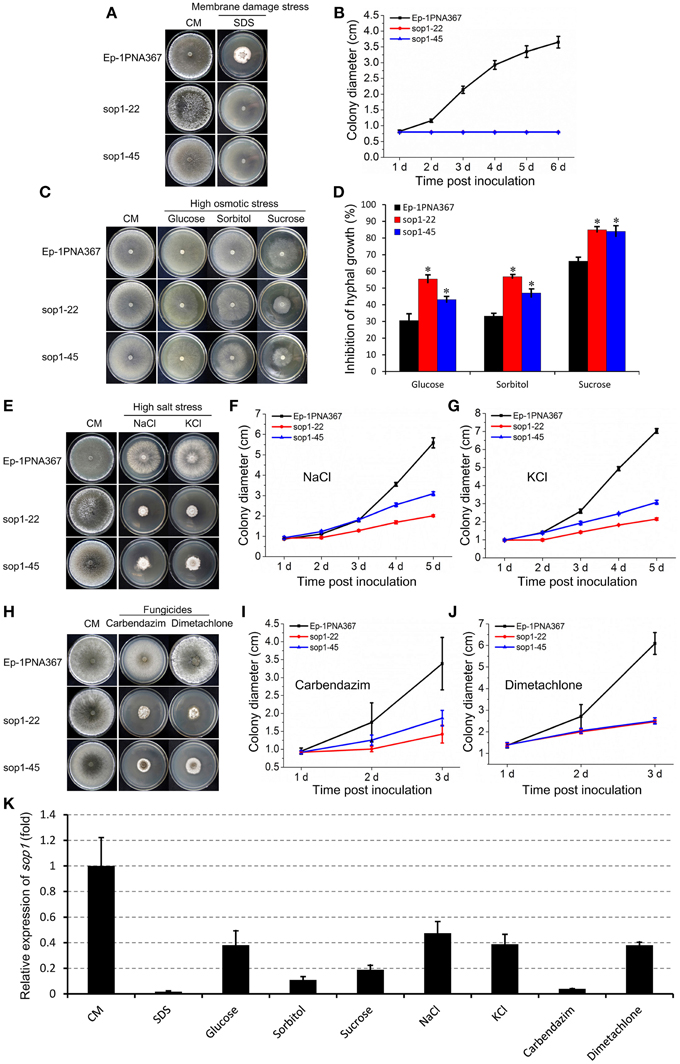
Figure 4. Growth of sop1 silenced transformants in the presence of various stressors. (A) Phenotypes of the sop1 RNAi-silenced transformants and the wild-type strain grown on CM supplemented with 0.02% SDS at 20°C. Photos were taken at 6 dpi. (B) The colony diameter of the sop1 RNAi-silenced transformants and the wild-type strain on CM supplemented with 0.02% SDS at different times. (C) Phenotypes of the sop1 RNAi-silenced transformants and the wild-type strain that grown on CM supplemented with glucose (1 M), sorbitol (1 M), and sucrose (1.2 M), respectively, at 20°C. Photos were taken at 3 dpi. (D) The inhibition rate of the hyphal growth of the sop1 RNAi-silenced transformants and the wild-type strain on stress-inducing media. * on the graph indicate significant differences, P = 0.05. (E) Phenotypes of the sop1 RNAi-silenced transformants and the wild-type strain grown on CM supplemented with NaCl (1 M) and KCl (1 M), respectively, at 20°C. Photos were taken at 6 dpi. (F,G) The colony diameter of the sop1 RNAi-silenced transformants and the wild-type strain on CM supplemented with 1 M NaCl and 1 M KCl, respectively, at different times. (H) Phenotypes of the sop1 RNAi-silenced transformants and the wild-type strain grown on CM supplemented with carbendazim (0.25 μg/mL) and dimetachlone (0.40 μg/mL), respectively, at 20°C. Photos were taken at 6 dpi. (I,J) The colony diameter of the sop1 RNAi-silenced transformants and the wild-type strain on CM supplemented with 0.25 μg/mL carbendazim and 0.40 μg/mL dimetachlone, respectively, at different times. (K) Relative expression of sop1 in the wild-type strain in the various stress conditions tested as determined by qRT-PCR analysis. Samples were collected for gene expression detection at 3 dpi on CM without any treatment and CM with 0.02% SDS, 1 M glucose, 1 M sorbitol, 1.2 M sucrose, 1 M NaCl, 1 M KCl, 0.25 μg/mL carbendazim, and 0.40 μg/mL dimetachlone, respectively. The abundance of cDNA from cultures on CM without any treatment was assigned a value of 1. The gene expression levels of sop1 were normalized to that of the β-tubulin transcripts from each sample. In all experiments at least three independent replications were performed, and the values are presented as the mean ± s.d. Bars indicate the standard error.
Sop1 is involved in the Tolerance of S. sclerotiorum to Oxidative Stress
Previous studies showed that the expression of bop1 (a sop1 homologous gene) in B. cinerea, a necrotrophic fungus with a close phylogenetic relationship to S. sclerotiorum, were induced upon H2O2 exposure (Temme and Tudzynski, 2009; Heller et al., 2012; Temme et al., 2012), suggesting that the opsins of filamentous fungi are involved in the oxidative stress response. To explore the role of sop1 in the oxidative stress response, the sop1 silenced transformants and the wild-type strain were cultured on CM supplemented with different concentrations of H2O2. Intriguingly, our results showed that the degree of the growth inhibition of the wild-type stain on CM supplemented with H2O2 was obviously greater than that of sop1-22 and sop1-45, indicating that silencing sop1 made S. sclerotiorum more tolerant to higher concentrations of H2O2 (Figures 5A,B). The qRT-PCR results showed that the expression of sop1 was induced when the wild-type strain was inoculated on CM with a low concentration of H2O2 (4 mM), however, it was down-regulated with a high concentration of H2O2 (8 and 12 mM, Figure 5C). This result indicated that S. sclerotiorum may adapt to oxidative stress partially through the regulation of sop1 expression, which is in accordance with a previous result (Temme and Tudzynski, 2009). Taken together, these results indicated that sop1 was involved in the response to oxidative stress, and it might play many roles in the response to various environmental signals.
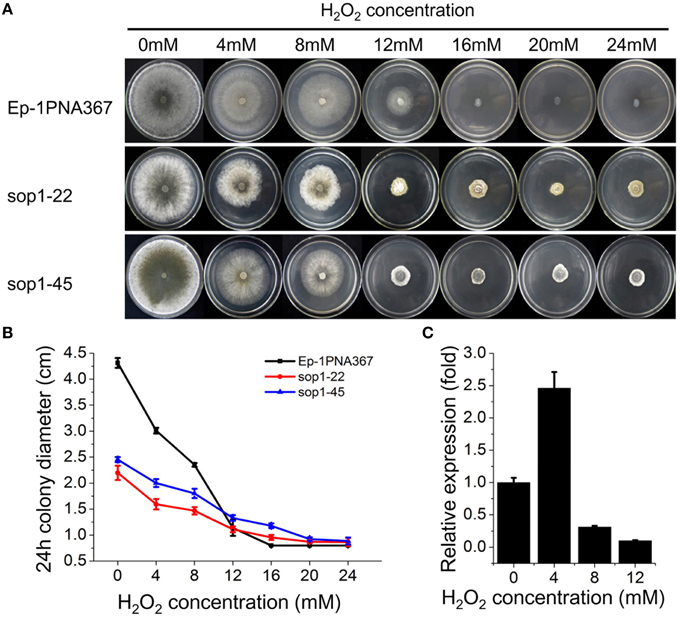
Figure 5. Tolerance of sop1 silenced transformants to oxidative stress. (A) Phenotypes of the sop1 silenced transformants and the wild-type strain on CM supplemented with different concentrations of H2O2. Photos were taken at 5 dpi. (B) The colony diameter of the sop1 silenced transformants and the wild-type strain on CM supplemented with different concentrations of H2O2 at 24 hpi. (C) The sop1 relative expression levels when the wild-type strain was inoculated on CM with different concentrations of H2O2 as indicated by qRT-PCR analysis. The abundance of cDNA from cultures on CM without H2O2 was assigned a value of 1. The gene expression levels of sop1 were normalized to that of the β-tubulin transcripts from each sample. In all experiments three independent replications were performed, and the values are presented as the mean ± s.d. Bars indicate the standard error.
A Phylogenetic Analysis of Microbial Opsins
An exhaustive search using DELTA-BLAST in the non-redundant protein sequence database at NCBI for sop1 showed that sop1 homologs were widespread in archaea, ascomycetes, basidiomycetes and lower plants such as algae (data not shown here), and the only exception was the presence of a putative rhodopsin (GenBank accession: AAQ75384.1) in the higher plant O. sativa indica group (Figure 6, see below), which was considered to be a horizontal gene transfer (HGT) event (Ruiz-Gonzalez and Marin, 2004). Additionally, not all sequenced species of bacteria, fungi or lower plants contain microbial opsins. This patchy distribution was thought to be caused by gene losses in multiple lineages in evolution (Ruiz-Gonzalez and Marin, 2004). All homology sequences of sop1 derived from the selected organisms (described in Materials and Methods) and S. sclerotiorum were used to perform a phylogenetic analysis, which showed that the microbial opsins had different copy numbers in different species (Figure 6). For example, there were 38 copies of microbial opsin homologs in G. theta, while most ascomycetes only had one or two copies (Figure 6). This phenomenon indicated that gene duplication events in microbial opsins occurred in some organisms in evolution. For the microbial opsins that have been experimentally studied or reviewed, sop1 appeared in the same branch with proton pumps: AaRh in A. acetabulum, Pop in P. Anserine S mat+ and Mac in L. maculans (Zhang et al., 2011), and had relatively distant phylogenetic relationship with the histidine kinase rhodopsin VcCop6 (GenBank accession: XP_002957065) in the multicellular green alga V. carteri f. nagariensis (Figure 6; Prochnik et al., 2010). Thus, the function of sop1 may have a closer phylogenetic relationship with the microbial proton-extruding pumps, that is accordance with the 3D structural analysis of sop1 (described as above).
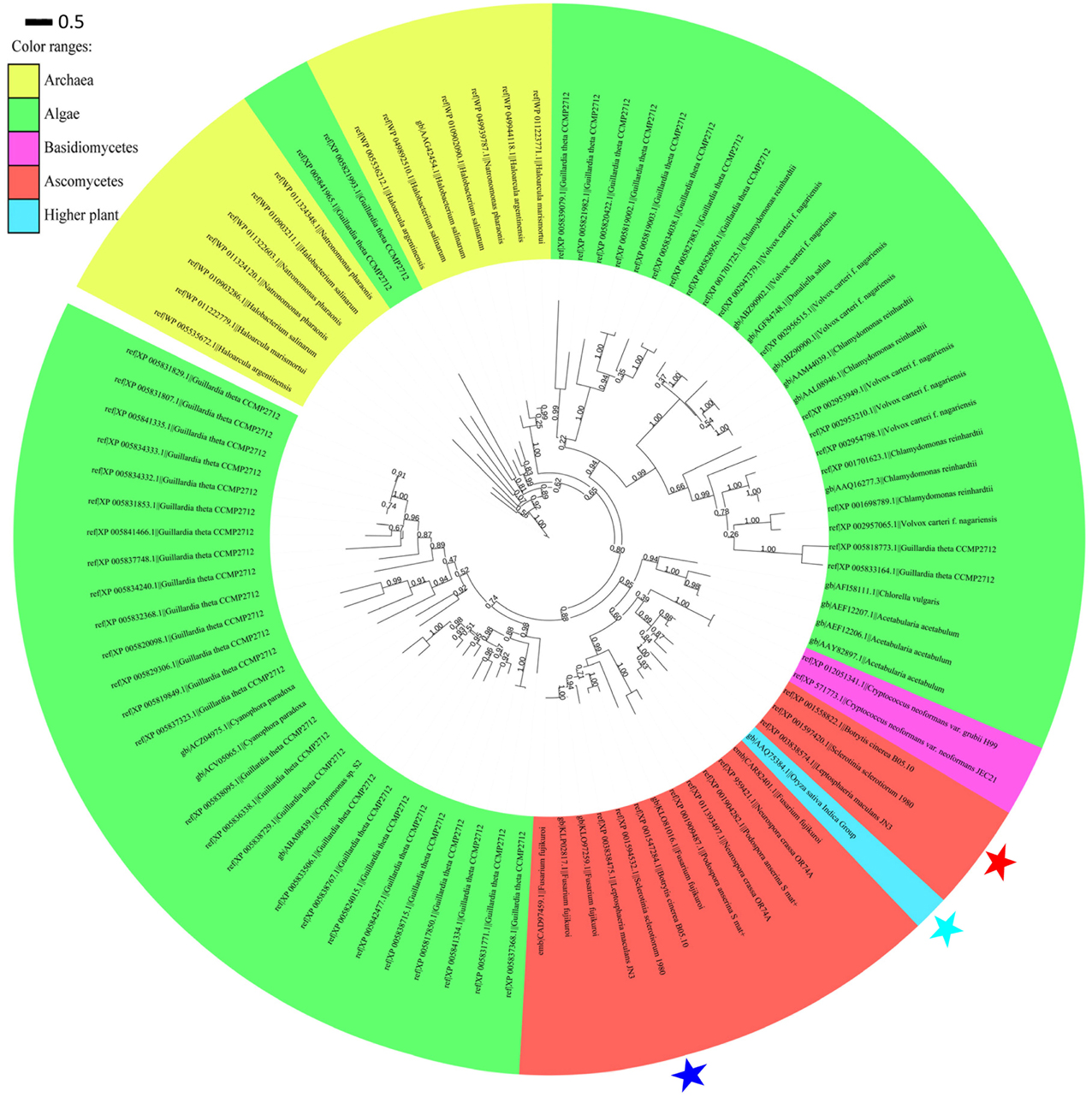
Figure 6. Phylogenic analysis of type I microbial opsin homologs. A phylogenetic tree was built using the PhyML-mixtures based on a multiple protein sequence alignment generated using COBALT with the Constraint E-value parameter setting to 0.003. The approximate likelihood ratios (SH-test) are indicated. Sop1, sop2, and the putative rhodopsin (GenBank accession: AAQ75384.1) in the higher plant O. sativa indica group are indicated by the red, blue, and sky blue stars, respectively. The characters before and after “||” indicate the GenBank accession numbers and corresponding species and strains, respectively. The scale bar corresponds to 0.5 amino acid substitutions per site.
Discussion
Many signaling genes were significantly regulated not only during infection but also during multiple developmental stages, indicating that these genes may play diverse roles during the corresponding biological processes and that defective mutants of these genes may have a pleiotropic phenotype. Previous research demonstrated that the signaling gene-related, pathogenicity-defective strains of S. sclerotiorum often exhibit abnormal sclerotial development (Rollins, 2003; Erental et al., 2007; Jurick and Rollins, 2007). In this study, the sop1 gene was also experimentally confirmed to be involved in growth, sclerotial development and virulence of S. sclerotiorum in accordance with its expression patterns (Table S1). These results indicated sop1 affected the basal development of S. sclerotiorum, and therefore indirectly affected its sclerotial development and virulence.
Undoubtedly, light is very important for the regulation of various physiological processes for almost all life on Earth. The type I opsins are photoreceptors that harvest light energy to carry out metabolic processes. Although this family also includes some bacteriorhodopsin-like proteins that do not contain the retinal binding lysine and cannot function as opsins (Oesterhelt and Tittor, 1989), sop1 is obviously not grouped with these proteins because it has a conserved lysine residue in the last transmembrane domains. Sop1 was found to be necessary for full virulence and sclerotial development of S. sclerotiorum. However, unlike the crucial roles of light-dependent regulation during sclerotial development and virulence in other fungi (Calvo et al., 2004; Duran et al., 2007; Canessa et al., 2013), the role of light-dependent regulation in sclerotial development and virulence seems not to be significant in S. sclerotiorum because our experience indicated no obvious differences when this fungus was cultured in the dark or in the light under normal laboratory conditions during sclerotial development and infection. This result indicated that the roles of sop1 in sclerotial development and infection and the inferred traditional molecular function of sop1 (light-driven proton-pumping) are independent because these biological functions of sop1 are independent on light while the molecular function of sop1 relies on light.
The expression patterns of sop1 indicated that the expression of sop1 has a intimate relationship with the development of S. sclerotiorum because the expression level of sop1 was very low during vegetative growth stage while the expression of sop1 was significantly up-regulated during later stages on PDA, especially during the mature period of sclerotial formation. Previous research showed that bop1 expression was induced when B. cinerea hyphae were treated with 10 mM H2O2 for 30 min (Temme and Tudzynski, 2009; Heller et al., 2012; Temme et al., 2012), indicating that the expression of opsin genes in filamentous fungi is regulated by oxidative stress. Our qRT-PCR and DGE data further showed that the expression of sop1 was regulated not only by oxidative stress but also by the developmental cycle itself because it was dramatically up-regulated during all developmental stages compared with the vegetative growth stage. However, our results indicated that sop1 expression was reduced when the mycelia were exposed to high concentrations of H2O2 (8 and 12 mM); this may be caused by a species difference or by the different treatment approaches. For example, to make sop1 expression correspond to the growth of the wild-type strain and the sop1 silenced transformants of S. sclerotiorum on CM medium supplemented with different concentrations of H2O2, the mycelia were cultured for 24 h for qRT-PCR analysis. The sop1 silenced transformants had a pleiotropic phenotype, indicating that sop1 has diverse biological functions, which is obviously different from its homologs in many filamentous fungi. Our results indicated that biological functions of microbial opsins may be not conserved in different organisms.
Previous research indicated obvious cross-talk between the osmotic stress signaling pathway and the oxidative stress pathway. For examples, the stress-activated MAP kinase BcSak1 could be activated by both oxidative and osmotic stress. Additionally, several genes were induced by oxidative stress treatment and most of them were also induced by osmotic stress (Heller et al., 2012). The Hog1 homologs are responsible not only for osmoregulation but also for oxidative stress response in many filamentous fungi (Dixon et al., 1999; Zhang et al., 2002; Du et al., 2006; Moriwaki et al., 2006). In this study, we noted the novel example that sop1 also responded to both oxidative and osmotic stress, although in a contrary manner: the presence of sop1 makes S. sclerotiorum more sensitive to oxidative stress, but more tolerant to osmotic stress. Large quantities of ROS are harmful for both pathogens and plant cells. In plants, the rapid accumulation of ROS can directly kill the cells surrounding the infection site. Local plant cell death caused by the hypersensitive response (HR) restricts biotrophic pathogens and inhibits their further expansion into neighbor cells and consequently protects the other parts of the plant (Talarczyk and Hennig, 2001). The oxidative burst is an effective process that protects the host against biotrophic pathogens that depend on living host cells but not necrotrophs that live on dead plant tissues (Rivas and Thomas, 2005). On the contrary, the HR reaction facilitates plant infection by necrotrophic pathogens (Govrin and Levine, 2000). For pathogens, higher levels of ROS evoke oxidative stress to harm the pathogen directly, so ROS tolerance is crucial for pathogens to eliminate ROS in their own cells with ROS-detoxification systems such as catalases, peroxidases or superoxide dismutase (SOD) (Temme and Tudzynski, 2009). A previous research showed that B. cinerea induces a significant oxidative burst in all host tissues analyzed (Lyon et al., 2007) but does not suffer H2O2-induced oxidative stress in plants (Temme and Tudzynski, 2009). In this study, our results showed that high concentration of H2O2 down-regulates sop1 expression in S. sclerotiorum and low sop1 expression level in turn increases oxidative stress tolerance. This is the first report of a novel positive feedback mechanism that allows S. sclerotiorum to strengthen its tolerance to oxidative stress through the regulation of sop1 expression.
Additionally, as a predicted conserved cytomembrane protein necessary for full virulence, normal growth and sclerotial development of S. sclerotiorum, sop1 homologs are only present in the microbes and lower plants, but not in higher plants and animals (with a single exception as previously noted). These properties make sop1 a potential drug target for S. sclerotiorum. Furthermore, our results indicated that the possibility of generation of fungicide-resistance or tolerance existed in the wild-type strain of S. sclerotiorum (Figures 4H–J), however disabling sop1 makes S. sclerotiorum more sensitive to fungicides and to various extreme environments, indicating that sop1 is a prospective, attractive drug target because it not necessary to consider fungicide-resistance or tolerance in the control of this pathogen.
Conclusion
The microbial opsin homolog sop1 was experimentally confirmed to be essential for growth, sclerotial development and full virulence of S. sclerotiorum. Sop1 plays many roles in responding to multiple environmental signals such as osmotic and oxidative stresses and is involved in fungicide resistance, which may be responsible for the environmental adaptability of S. sclerotiorum. Furthermore, our results showed that microbial opsins do not exist in animals and almost all higher plants indicating that sop1 is a potential novel drug target for disease control of S. sclerotiorum.
Author Contributions
XL and JC designed the research and wrote the paper; XL and CS executed the experiments. XL, JX, YF, DJ, GL, and JC performed the data and bioinformatics analyses. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the Special Fund for Agro-scientific Research in the Public Interest (201103016), the National Basic Research Program (2012CB114000), the Programme of Introducing Talents of Discipline to Universities in China (the 111 Project no. B14032), and the earmarked fund for China Agriculture Research System (CARS-13).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01504
Figure S1. Sop1 silenced transformants showing significantly reduced virulence on detached tomato leaves. Virulence of the sop1 silenced transformants and the wild-type strain was evaluated according to the lesion diameter at 20°C for 48 h. Three independent replications were performed. The values are presented as the mean ± s.d. Differentiation was evaluated by a t-test. Different letters on the graph indicate statistical significance, P = 0.05.
Figure S2. Biological characterization of the sop1 silenced transformants obtained by the other gene silencing strategy. (A) The construction of sop1 silencing vector pRNAi-2. (B) Phenotypes of the sop1 silenced transformants and the wild-type strain grown on PDA at 20°C. Photos were taken at 8 dpi. (C) Comparison of the virulence of the silenced transformants and the wild-type strain on detached oilseed rape leaves at 20°C for 48 h. (D) Relative sop1 expression level of the sop1 silenced transformants and the wild-type strain as determined by qRT-PCR analysis. A value of 1 was assigned to the abundance of cDNA from the wild-type strain. The sop1 expression levels in the silenced transformants and the wild-type strain were normalized to that of the β-tubulin transcripts from each strain. (E) Comparison of lesion diameter of the silenced transformants and the wild-type strain. (F) Comparison of the hyphal growth rate of the silenced transformants and the wild-type strain. Three independent replications were performed. Bars indicate the standard error. The values are presented as the mean ± s.d. Differentiation was evaluated by a t-test. Different letters on a graph indicate statistical significance, P = 0.05.
Table S1. Differentially expressed signaling genes identified by the DGE during different developmental stages of S. sclerotiorum compared with the vegetative growth stage. Red and green indicate significantly up-regulated and down-regulated gene expression, respectively. Blank cells without numbers indicate genes that were not detected in the DGE data or not differentially expressed during corresponding developmental stages.
Footnotes
1. ^http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome.
References
Abascal, F., Zardoya, R., and Posada, D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105. doi: 10.1093/bioinformatics/bti263
Abawi, G., and Grogan, R. (1979). Epidemiology of diseases caused by Sclerotinia species. Phytopathology 69, 899–904. doi: 10.1094/Phyto-69-899
Adams, P., and Ayers, W. (1979). Ecology of Sclerotinia species. Phytopathology 69, 896–898. doi: 10.1094/Phyto-69-896
Amselem, J., Cuomo, C. A., van Kan, J. A., Viaud, M., Benito, E. P., Couloux, A., et al. (2011). Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7:e1002230. doi: 10.1371/journal.pgen.1002230
Anisimova, M., and Gascuel, O. (2006). Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55, 539–552. doi: 10.1080/10635150600755453
Bieszke, J. A., Braun, E. L., Bean, L. E., Kang, S., Natvig, D. O., and Borkovich, K. A. (1999). The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsins. Proc. Natl. Acad. Sci. U.S.A. 96, 8034–8039. doi: 10.1073/pnas.96.14.8034
Boland, G., and Hall, R. (1994). Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 16, 93. doi: 10.1080/07060669409500766
Boratyn, G. M., Schäffer, A., Agarwala, R., Altschul, S. F., Lipman, D. J., and Madden, T. L. (2012). Domain enhanced lookup time accelerated BLAST. Biol. Direct 7:12. doi: 10.1186/1745-6150-7-12
Brewster, J. L., de Valoir, T., Dwyer, N. D., Winter, E., and Gustin, M. C. (1993). An osmosensing signal transduction pathway in yeast. Science 259, 1760–1763. doi: 10.1126/science.7681220
Brown, L. S. (2004). Fungal rhodopsins and opsin-related proteins: eukaryotic homologues of bacteriorhodopsin with unknown functions. Photochem. Photobiol. Sci. 3, 555–565. doi: 10.1039/b315527g
Cai, M., Fang, Z., Niu, C., Zhou, X., and Zhang, Y. (2013). Light regulation on growth, development, and secondary metabolism of marine-derived filamentous fungi. Folia Microbiol. (Praha). 58, 537–546. doi: 10.1007/s12223-013-0242-x
Calvo, A. M., Bok, J., Brooks, W., and Keller, N. P. (2004). veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl. Environ. Microbiol. 70, 4733–4739. doi: 10.1128/AEM.70.8.4733-4739.2004
Canessa, P., Schumacher, J., Hevia, M. A., Tudzynski, P., and Larrondo, L. F. (2013). Assessing the effects of light on differentiation and virulence of the plant pathogen Botrytis cinerea: characterization of the White Collar Complex. PLoS ONE 8:e84223. doi: 10.1371/journal.pone.0084223
Cessna, S. G., Sears, V. E., Dickman, M. B., and Low, P. S. (2000). Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell 12, 2191–2200. doi: 10.1105/tpc.12.11.2191
Chen, C., Harel, A., Gorovoits, R., Yarden, O., and Dickman, M. B. (2004). MAPK regulation of sclerotial development in Sclerotinia sclerotiorum is linked with pH and cAMP sensing. Mol. Plant Microbe Interact. 17, 404–413. doi: 10.1094/MPMI.2004.17.4.404
Chet, I., and Henis, Y. (1975). Sclerotial morphogenesis in fungi. Annu. Rev. Phytopathol. 13, 169–192. doi: 10.1146/annurev.py.13.090175.001125
Clamp, M., Cuff, J., Searle, S. M., and Barton, G. J. (2004). The Jalview Java alignment editor. Bioinformatics 20, 426–427. doi: 10.1093/bioinformatics/btg430
DeLano, W. L. (2002). Pymol: an open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 40, 82–92.
Dixon, K. P., Xu, J. R., Smirnoff, N., and Talbot, N. J. (1999). Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant Cell 11, 2045–2058. doi: 10.1105/tpc.11.10.2045
Du, C., Sarfati, J., Latge, J. P., and Calderone, R. (2006). The role of the sakA (Hog1) and tcsB (s1n1) genes in the oxidant adaptation of Aspergillus fumigatus. Med. Mycol. 44, 211–218. doi: 10.1080/13693780500338886
Duran, R. M., Cary, J. W., and Calvo, A. M. (2007). Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 73, 1158–1168. doi: 10.1007/s00253-006-0581-5
Egan, M. J., Wang, Z.-Y., Jones, M. A., Smirnoff, N., and Talbot, N. J. (2007). Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc. Natl. Acad. Sci. U.S.A. 104, 11772–11777. doi: 10.1073/pnas.0700574104
Erental, A., Dickman, M. B., and Yarden, O. (2008). Sclerotial development in Sclerotinia sclerotiorum: awakening molecular analysis of a “Dormant” structure. Fungal Biol. Rev. 22, 6–16. doi: 10.1016/j.fbr.2007.10.001
Erental, A., Harel, A., and Yarden, O. (2007). Type 2A phosphoprotein phosphatase is required for asexual development and pathogenesis of Sclerotinia sclerotiorum. Mol. Plant Microbe Interact. 20, 944–954. doi: 10.1094/MPMI-20-8-0944
Espagne, E., Lespinet, O., Malagnac, F., Da Silva, C., Jaillon, O., Porcel, B. M., et al. (2008). The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 9:R77. doi: 10.1186/gb-2008-9-5-r77
Estrada, A. F., and Avalos, J. (2009). Regulation and targeted mutation of opsA, coding for the NOP-1 opsin orthologue in Fusarium fujikuroi. J. Mol. Biol. 387, 59–73. doi: 10.1016/j.jmb.2009.01.057
Frassanito, A. M., Barsanti, L., Passarelli, V., Evangelista, V., and Gualtieri, P. (2010). A rhodopsin-like protein in Cyanophora paradoxa: gene sequence and protein immunolocalization. Cell. Mol. Life Sci. 67, 965–971. doi: 10.1007/s00018-009-0225-x
Govrin, E. M., and Levine, A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10, 751–757. doi: 10.1016/S0960-9822(00)00560-1
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Harel, A., Bercovich, S., and Yarden, O. (2006). Calcineurin is required for sclerotial development and pathogenicity of Sclerotinia sclerotiorum in an oxalic acid-independent manner. Mol. Plant Microbe Interact. 19, 682–693. doi: 10.1094/MPMI-19-0682
Heller, J., Ruhnke, N., Espino, J. J., Massaroli, M., Collado, I. G., and Tudzynski, P. (2012). The mitogen-activated protein kinase BcSak1 of Botrytis cinerea is required for pathogenic development and has broad regulatory functions beyond stress response. Mol. Plant Microbe Interact. 25, 802–816. doi: 10.1094/MPMI-11-11-0299
Herrera-Estrella, A., and Horwitz, B. A. (2007). Looking through the eyes of fungi: molecular genetics of photoreception. Mol. Microbiol. 64, 5–15. doi: 10.1111/j.1365-2958.2007.05632.x
Hordijk, W., and Gascuel, O. (2005). Improving the efficiency of SPR moves in phylogenetic tree search methods based on maximum likelihood. Bioinformatics 21, 4338–4347. doi: 10.1093/bioinformatics/bti713
Idnurm, A., and Heitman, J. (2005a). Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 3:e95. doi: 10.1371/journal.pbio.0030095
Idnurm, A., and Heitman, J. (2005b). Photosensing fungi: phytochrome in the spotlight. Curr. Biol. 15, R829–R832. doi: 10.1016/j.cub.2005.10.001
Idnurm, A., and Howlett, B. J. (2001). Characterization of an opsin gene from the ascomycete Leptosphaeria maculans. Genome 44, 167–171. doi: 10.1139/g00-113
Jurick, W. M. II., and Rollins, J. A. (2007). Deletion of the adenylate cyclase (sac1) gene affects multiple developmental pathways and pathogenicity in Sclerotinia sclerotiorum. Fungal Genet. Biol. 44, 521–530. doi: 10.1016/j.fgb.2006.11.005
Kelley, L. A., and Sternberg, M. J. (2009). Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371. doi: 10.1038/nprot.2009.2
Krauss, G. (2006). Biochemistry of Signal Transduction and Regulation. Manhattan: John Wiley & Sons.
Le, S. Q., and Gascuel, O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. doi: 10.1093/molbev/msn067
Le, S. Q., Lartillot, N., and Gascuel, O. (2008). Phylogenetic mixture models for proteins. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 363, 3965–3976. doi: 10.1098/rstb.2008.0180
Lee, K., Singh, P., Chung, W. C., Ash, J., Kim, T. S., Hang, L., et al. (2006). Light regulation of asexual development in the rice blast fungus, Magnaporthe oryzae. Fungal Genet. Biol. 43, 694–706. doi: 10.1016/j.fgb.2006.04.005
Le Tourneau, D. (1979). Morphology, cytology, and physiology of Sclerotinia species in culture. Phytopathology 69, 887–890. doi: 10.1094/Phyto-69-887
Letunic, I., and Bork, P. (2007). Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. doi: 10.1093/bioinformatics/btl529
Liu, Y., and Bell-Pedersen, D. (2006). Circadian rhythms in Neurospora crassa and other filamentous fungi. Eukaryotic Cell 5, 1184–1193. doi: 10.1128/EC.00133-06
Luecke, H., Schobert, B., Richter, H. T., Cartailler, J. P., and Lanyi, J. K. (1999). Structure of bacteriorhodopsin at 1.55 A resolution. J. Mol. Biol. 291, 899–911. doi: 10.1006/jmbi.1999.3027
Lyon, G. D., Goodman, B. A., and Williamson, B. (2007). “Botrytis cinerea perturbs redox processes as an attack strategy in plants,” in Botrytis: Biology, Pathology and Control (Springer), 119–141. doi: 10.1007/978-1-4020-2626-3_8
Lyu, X., Shen, C., Xie, J., Fu, Y., Jiang, D., Hu, Z., et al. (2015). A “footprint” of plant carbon fixation cycle functions during the development of a heterotrophic fungus. Sci. Rep. 5:12952. doi: 10.1038/srep12952
Moriwaki, A., Kubo, E., Arase, S., and Kihara, J. (2006). Disruption of SRM1, a mitogen-activated protein kinase gene, affects sensitivity to osmotic and ultraviolet stressors in the phytopathogenic fungus Bipolaris oryzae. FEMS Microbiol. Lett. 257, 253–261. doi: 10.1111/j.1574-6968.2006.00178.x
Nguyen, Q. B., Kadotani, N., Kasahara, S., Tosa, Y., Mayama, S., and Nakayashiki, H. (2008). Systematic functional analysis of calcium-signalling proteins in the genome of the rice-blast fungus, Magnaporthe oryzae, using a high-throughput RNA-silencing system. Mol. Microbiol. 68, 1348–1365. doi: 10.1111/j.1365-2958.2008.06242.x
Nickle, B., and Robinson, P. R. (2007). The opsins of the vertebrate retina: insights from structural, biochemical, and evolutionary studies. Cell. Mol. Life Sci. 64, 2917–2932. doi: 10.1007/s00018-007-7253-1
Oesterhelt, D., and Tittor, J. (1989). Two pumps, one principle: light-driven ion transport in halobacteria. Trends Biochem. Sci. 14, 57–61. doi: 10.1016/0968-0004(89)90044-3
Palczewski, K., Kumasaka, T., Hori, T., Behnke, C. A., Motoshima, H., Fox, B. A., et al. (2000). Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289, 739–745. doi: 10.1126/science.289.5480.739
Papadopoulos, J. S., and Agarwala, R. (2007). COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079. doi: 10.1093/bioinformatics/btm076
Prochnik, S. E., Umen, J., Nedelcu, A. M., Hallmann, A., Miller, S. M., Nishii, I., et al. (2010). Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329, 223–226. doi: 10.1126/science.1188800
Purschwitz, J., Müller, S., Kastner, C., and Fischer, R. (2006). Seeing the rainbow: light sensing in fungi. Curr. Opin. Microbiol. 9, 566–571. doi: 10.1016/j.mib.2006.10.011
Riou, C., Freyssinet, G., and Fevre, M. (1991). Production of cell wall-degrading enzymes by the phytopathogenic fungus Sclerotinia sclerotiorum. Appl. Environ. Microbiol. 57, 1478–1484.
Rivas, S., and Thomas, C. M. (2005). Molecular interactions between tomato and the leaf mold pathogen Cladosporium fulvum. Annu. Rev. Phytopathol. 43, 395–436. doi: 10.1146/annurev.phyto.43.040204.140224
Rollins, J. A. (2003). The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Mol. Plant Microbe Interact. 16, 785–795. doi: 10.1094/MPMI.2003.16.9.785
Ruiz-Gonzalez, M. X., and Marin, I. (2004). New insights into the evolutionary history of type 1 rhodopsins. J. Mol. Evol. 58, 348–358. doi: 10.1007/s00239-003-2557-8
Schouten, A., Tenberge, K. B., Vermeer, J., Stewart, J., Wagemakers, L., Williamson, B., et al. (2002). Functional analysis of an extracellular catalase of Botrytis cinerea. Mol. Plant Pathol. 3, 227–238. doi: 10.1046/j.1364-3703.2002.00114.x
Sineshchekov, O. A., and Govorunova, E. G. (2001). Rhodopsin receptors of phototaxis in green flagellate algae. Biochemistry (Mosc.) 66, 1300–1310. doi: 10.1023/A:1013191504508
Sineshchekov, O. A., Govorunova, E. G., Jung, K. H., Zauner, S., Maier, U. G., and Spudich, J. L. (2005). Rhodopsin-mediated photoreception in cryptophyte flagellates. Biophys. J. 89, 4310–4319. doi: 10.1529/biophysj.105.070920
Spudich, J. L., Yang, C. S., Jung, K. H., and Spudich, E. N. (2000). Retinylidene proteins: structures and functions from archaea to humans. Annu. Rev. Cell Dev. Biol. 16, 365–392. doi: 10.1146/annurev.cellbio.16.1.365
Steadman, J. (1979). Control of plant diseases caused by Sclerotinia species. Phytopathology 69, 904–907. doi: 10.1094/Phyto-69-904
Talarczyk, A., and Hennig, J. (2001). Early defence responses in plants infected with pathogenic organisms. Cell. Mol. Biol. Lett. 6, 955–970.
Temme, N., Oeser, B., Massaroli, M., Heller, J., Simon, A., Collado, I. G., et al. (2012). BcAtf1, a global regulator, controls various differentiation processes and phytotoxin production in Botrytis cinerea. Mol. Plant Pathol. 13, 704–718. doi: 10.1111/j.1364-3703.2011.00778.x
Temme, N., and Tudzynski, P. (2009). Does Botrytis cinerea ignore H2O2-induced oxidative stress during infection? characterization of Botrytis activator protein 1. Mol. Plant Microbe Interact. 22, 987–998. doi: 10.1094/MPMI-22-8-0987
Tenberge, K. B., Beckedorf, M., Hoppe, B., Schouten, A., Solf, M., and von den Driesch, M. (2002). In situ localization of AOS in host-pathogen interactions. Microsc. Microanal. 8, 250–251.
Tiedemann, A. (1997). Evidence for a primary role of active oxygen species in induction of host cell death during infection of bean leaves with Botrytis cinerea. Physiol. Mol. Plant P. 50, 151–166. doi: 10.1006/pmpp.1996.0076
Tsunoda, S. P., Ewers, D., Gazzarrini, S., Moroni, A., Gradmann, D., and Hegemann, P. (2006). H+-pumping rhodopsin from the marine alga Acetabularia. Biophys. J. 91, 1471–1479. doi: 10.1529/biophysj.106.086421
Wada, T., Shimono, K., Kikukawa, T., Hato, M., Shinya, N., Kim, S. Y., et al. (2011). Crystal structure of the eukaryotic light-driven proton-pumping rhodopsin, Acetabularia rhodopsin II, from marine alga. J. Mol. Biol. 411, 986–998. doi: 10.1016/j.jmb.2011.06.028
Willetts, H., and Bullock, S. (1992). Developmental biology of sclerotia. Mycol. Res. 96, 801–816. doi: 10.1016/S0953-7562(09)81027-7
Xie, J., Wei, D., Jiang, D., Fu, Y., Li, G., Ghabrial, S., et al. (2006). Characterization of debilitation-associated mycovirus infecting the plant-pathogenic fungus Sclerotinia sclerotiorum. J. Gen. Virol. 87, 241–249. doi: 10.1099/vir.0.81522-0
Xu, J. R. (2000). Map kinases in fungal pathogens. Fungal Genet. Biol. 31, 137–152. doi: 10.1006/fgbi.2000.1237
Yu, Y., Jiang, D., Xie, J., Cheng, J., Li, G., Yi, X., et al. (2012). Ss-Sl2, a novel cell wall protein with PAN modules, is essential for sclerotial development and cellular integrity of Sclerotinia sclerotiorum. PLoS ONE 7:e34962. doi: 10.1371/journal.pone.0034962
Zhang, F., Vierock, J., Yizhar, O., Fenno, L. E., Tsunoda, S., Kianianmomeni, A., et al. (2011). The microbial opsin family of optogenetic tools. Cell 147, 1446–1457. doi: 10.1016/j.cell.2011.12.004
Keywords: Sclerotinia sclerotiorum, microbial opsin, sclerotial development, virulence, stress response
Citation: Lyu X, Shen C, Fu Y, Xie J, Jiang D, Li G and Cheng J (2016) The Microbial Opsin Homolog Sop1 is involved in Sclerotinia sclerotiorum Development and Environmental Stress Response. Front. Microbiol. 6:1504. doi: 10.3389/fmicb.2015.01504
Received: 09 September 2015; Accepted: 14 December 2015;
Published: 07 January 2016.
Edited by:
Stéphane Hacquard, Max Planck Institute for Plant Breeding Research, GermanyReviewed by:
Biswapriya Biswavas Misra, University of Florida, USASylvain Raffaele, Institut National de la Recherche Agronomique, France
Copyright © 2016 Lyu, Shen, Fu, Xie, Jiang, Li and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiasen Cheng, amlhc2VuY2hlbmdAbWFpbC5oemF1LmVkdS5jbg==
 Xueliang Lyu
Xueliang Lyu Cuicui Shen
Cuicui Shen Yanping Fu
Yanping Fu Jiatao Xie
Jiatao Xie Daohong Jiang
Daohong Jiang Guoqing Li
Guoqing Li Jiasen Cheng
Jiasen Cheng