- 1Japan Agency for Marine-Earth Science Technology, Kochi Institute for Core Sample Research, Nankoku, Japan
- 2Japan Agency for Marine-Earth Science Technology, Research and Development Center for Submarine Resources, Nankoku, Japan
- 3Department Earth and Planetary Sciences, Kyushu University, Fukuoka, Japan
- 4Environmental Management Research Institute, National Institute of Advanced Industrial Science and Technology, Tsukuba, Japan
- 5Mishimamura Village, Kagoshima, Japan
Previous studies of microbial communities in deep-sea hydrothermal ferric deposits have demonstrated that members of Zetaproteobacteria play significant ecological roles in biogeochemical iron-cycling. However, the ecophysiological characteristics and interaction between other microbial members in the habitat still remain largely unknown. In this study, we investigated microbial communities in a core sample obtained from shallow hydrothermal iron-oxyhydroxide deposits at Nagahama Bay of Satsuma Iwo-Jima, Japan. Scanning electron microscopic observation showed numerous helical stalk structures, suggesting the occurrence of iron-oxidizing bacteria. Analysis of 16S rRNA gene sequences indicated the co-occurrence of iron-oxidizing Zetaproteobacteria and iron-reducing bacteria such as the genera Deferrisoma and Desulfobulbus with strong correlations on the sequence abundance. CARD-FISH indicated that the numbers of Zetaproteobacteria were not always consistent to the frequency of stalk structures. In the stalk-abundant layers with relatively small numbers of Zetaproteobacteria cells, accumulation of polyphosphate was observed inside Zetaproteobacteria cells, whereas no polyphosphate grains were observed in the topmost layers with fewer stalks and abundant Zetaproteobacteria cells. These results suggest that Zetaproteobacteria store intracellular polyphosphates during active iron oxidation that contributes to the mineralogical growth and biogeochemical iron cycling.
Introduction
Members of the class Zetaproteobacteria were first found at the Loihi Seamount, Hawaii (Moyer et al., 1995). Mariprofundus ferrooxydans is the only isolate characterized as a marine iron-oxidizing bacterium within the Zetaproteobacteria (Emerson et al., 2007). Previous studies of 16S rRNA genes in deep-sea hydrothermal fields showed that members of the Zetaproteobacteria are widely distributed in iron-oxyhydroxide deposits on the plate spreading centers, hot-spot seamounts, and island arcs (Davis et al., 2009; Kato et al., 2009; Forget et al., 2010; Edwards et al., 2011; McAllister et al., 2011; Fleming et al., 2013). These observations strongly suggest that microbial communities involving Zetaproteobacteria play significant ecological roles in biogeochemical iron and other elemental cycles.
In the deep-sea hydrothermal iron-oxyhydroxide deposits, it has been demonstrated that the dissolved oxygen is present but generally lower than that of the surface seawater, e.g., less than 50 μM of oxygen was observed in iron-rich mats around the Loihi Seamount (Glazer and Rouxel, 2009), suggesting that members of the Zetaproteobacteria preferentially inhabit and grow by oxidizing ferrous iron to ferric iron at the sub-oxic redox condition. Consistently, a kinetic model study using a pure culture supported the notion that the habitable zone of iron-oxidizing microorganisms is severely and sensitively constrained by in situ oxygen concentration, and the maximum value for the geochemical niche approaches ~50 μM (Druschel et al., 2008). In addition, a genomic study of Mariprofundus ferroxydans PV-1 (Singer et al., 2011), which was isolated from hydrothermal venting at Loihi Seamount, revealed that it has the complete TCA cycle, the ability to fix CO2, and genes encoding aerotaxis as well as antioxidant functionalities. Although strain PV-1 does not always represent metabolic pathways and functions of all iron-oxidizing Zetaproteobacteria, the genomic information suggest that this strain is capable of sensing and responding to the redox state of the iron-oxyhydroxide deposits. Comparative genomic analyses of single-amplified genomes also indicated the niche specialization of Zetaproteobacteria, most likely controlled by the oxygen tolerance (Field et al., 2015). However, ecophysiology and growth/survival strategy of Zetaproteobacteria correlated with other members in the iron-oxidizing microbial ecosystem are still largely unknown.
Satsuma Iwo-Jima is a small volcanic island located at ~40 km south of Kyushu Island, Japan. The volcanic activity provides a shallow hydrothermal field in the Nagahama Bay, where the formation of iron-oxyhydroxide deposits, including chimney-like structures was widely observed on the seafloor (Figure 1). Pilot geological and sedimentological studies of this environment showed that the depositional rates are exceptionally high, ranging from 2.8 to 4.9 cm per year (Kiyokawa and Ueshiba, 2015). Light microscopic observation of these deposits showed twisted stalk structures, suggesting the occurrence of iron-oxidizing microbial communities that mediate the formation process of iron-oxyhydroxide deposits.
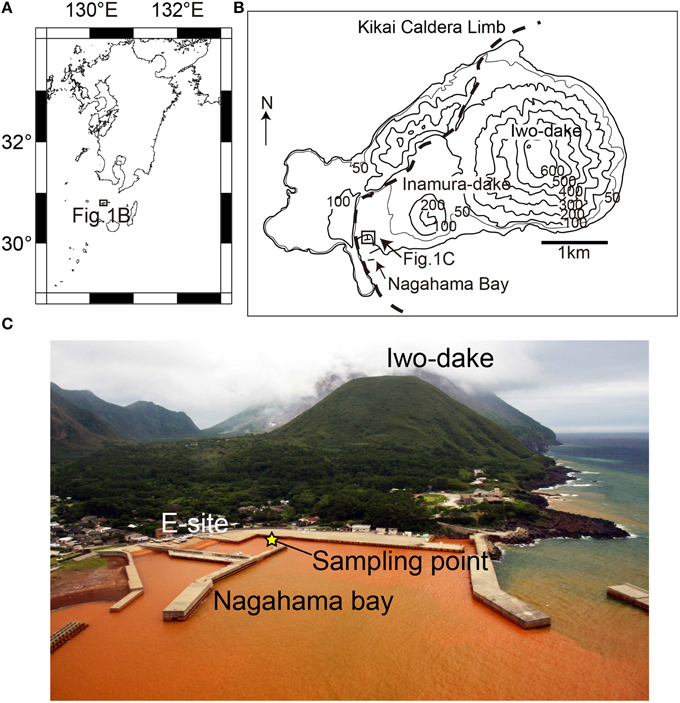
Figure 1. Regional (A) and local (B) maps of Satsuma Iwo-Jima. (C) An overview photo of the Nagahama Bay. The seawater is brownish-red due to the presence of iron oxyhydroxides. The yellow star indicates the sampling point in this study.
In this study, we investigated microbial communities in the shallow hydrothermal iron-oxyhydroxide deposits (water depth: ~3 m) in the Nagahama Bay. To understand the distribution and ecophysiological characteristics of Zetaproteobacteria cells in this iron-rich habitat, we obtained a 50 cm-long core sample and studied microbial communities using scanning electron microscopy (SEM), image-based cell count, and catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH) techniques as well as diversity and correlation analyses of 16S rRNA gene-tagged sequences.
Materials and Methods
Geologic Setting
Satsuma Iwo-Jima is a small volcanic island of the southern Kyushu, Japan. The volcanism is associated with iron-rich sedimentations at hydrothermal hot springs along the island coast. The Nagahama Bay is located on the southwest coast of the island and is one of the most active regions of hot water discharge (Figure 1). The seawater is reddish-brown because of the oxidation of ferrous iron in hydrothermal fluids as it mixes with seawater (Nogami et al., 1993; Kiyokawa et al., 2012). The hot waters discharged along the coast of Nagahama Bay (55–60°C and pH 5.5) is microaerobic (Eh = 69 mV) and contain high concentrations of ferrous iron (~191 ppm). The bottom seawater around iron deposits in the bay is anaerobic to micro-aerobic (Shikaura and Tazaki, 2001).
Sample Collection
A 50-cm-long sediment core was collected from an iron-rich hydrothermal mound in the Nagahama Bay in May 2013 by scuba divers using a clear acrylic tube (Figure 2). For DNA and SEM analyses, 10 cm3 subsamples were collected from the split core and stored at −20°C until further processing. There are hard and soft layers inside the core and the subsamples were only taken from relatively soft and undisturbed layers in this study (Figure 2B). For microscopic analyses such as cell count and fluorescence in situ hybridization (FISH), 10 cm3 of sediments were fixed with 4% paraformaldehyde, washed twice with 1 × PBS, re-suspended in 50% PBS-ethanol solution, and then stored at −20°C.
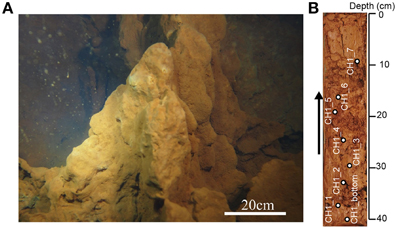
Figure 2. (A) The shallow hydrothermal iron-oxyhydroxide deposits off Satsuma Iwo-Jima. (B) A 50 cm-long core sample examined in this study. Arrow indicates the direction of the core, with the right side representing the seafloor. White circles indicate sampling points.
Scanning Electron Microscopy
Sample preparation and observation by a SEM was conducted as previously described (Kiyokawa and Ueshiba, 2015). The frozen iron-oxyhydroxide sample was dried in a freeze dryer (IWAKI, FRD-82M), and then subsequently fixed on a microscope slide using conductive tapes. The slide was coated with a 30-nm platinum film using an ion-sputtering device (JEOL, JFC-1600). The coated samples were analyzed by a field emission-SEM (JEOL, JSM-6500F).
Cell Count of SYBR Green I-Stained Cells
Cell concentration in sediment samples was determined based on a previously described procedure (Morono et al., 2013). Briefly, after washing fixed sediment using NaCl solution, a detergent mix and methanol were added and the sediment was shaken for 60 min at 500 rpm using a Shake Master (Bio Medical Science, Japan). Then, the sediment slurry was sonicated and carefully layered onto a high-density cushion consisting of multiple Nycodenz and sodium polytungstate solutions. Following centrifugation at 10,000 × g for 60 min, the supernatant was transferred to a separate vial. Subsequently, detergent mix and methanol were added to the remaining pellet, followed by sonication and centrifugation with the high-density cushion solution. The resulting supernatants were pooled, and 10% of the recovered suspension was filtered on a polycarbonate membrane. The number of cells stained by SYBR Green I was enumerated using an automated cell count system (Morono and Inagaki, 2010), with analysis of acquired images using MetaMorph software (Molecular Devices, USA).
CARD-FISH and DAPI-Stain of Zetaproteobacteria Cells
CARD-FISH was employed to detect and quantify Zetaproteobacteria in this study. A new probe, Zeta709 (5′-GCCTCAGGTGTTCCTCCG-3′), was designed in this study for the specific detection of Zetaproteobacteria. The optimal formamide (FA) concentration in the hybridization buffer was determined by clone-FISH (Schramm et al., 2002). Briefly, using ARB software (Ludwig et al., 2004), the specific probe was designed for targeting all Zetaproteobacteria in the public databases, including nearly full-length of Zetaproteobacteria sequences obtained in this study. To find the optimal FA concentration for CARD-FISH, the clones were tested at diffrent FA concentrations from 0 to 80%, resulting in 60%. Then, hybridization was carried out using microbial cells placed on a polycarbonate filter with 60% FA at 46°C for 2 h, followed by tyramide signal amplification using Alexa555-labeled tyramide at 37°C in the dark for 30 min (Hoshino et al., 2008). DNA was then counterstained using SYBR Green I. Polyphosphate inside cells was stained using a high concentration of DAPI solution (80 μg ml−1), if needed (Serafim et al., 2002).
DNA Extraction and PCR Amplification
DNA was extracted from 2 g of frozen sediment using a PowerMax® Soil DNA isolation kit (MO BIO Laboratories, USA) according to the manufacturer's instructions. The extracted DNA was further concentrated by isopropanol precipitation. For clone-FISH, nearly full-length 16S rRNA was amplified using a 27F-1492R primer set (Lane, 1991; Loy et al., 2002), whereas a 27F-927R primer set (Amann et al., 1995) was used for fine classification of Zetaproteobacteria. After purification, the amplicon was cloned, and then the sequence was determined by Sanger sequencing as described elsewhere. For analyzing community structure, the V4 hyper-variable region of 16S rRNA was amplified using the universal primer set 515F-806R (Lane, 1991; Caporaso et al., 2011). After indexing using the barcoded primers, illumina sequencing was performed using a MiSeq platform with MiSeq Reagent Kit v2 500 cycles (Illumina, USA).
Processing of Sequence Data and Statistical Analysis
Sequence data were processed using the Mothur software package (v.1.35.0) (Schloss et al., 2009). Spearman's rank correlation coefficients were calculated using the phylogenetic compositions by Mothur, and the data were illustrated using Cytoscape (v.3.2.0) (Shannon et al., 2003). For the data illustration, we used only statistically significant correlations (p < 0.05) with coefficients greater than 0.9. A phylogenetic tree was constructed using MEGA5 software (Tamura et al., 2011). ARB software was used for designing an oligonucleotide probe specific for Zetaproteobacteria.
DNA Sequences in the Databases
The sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession number LC074716 to LC074722 and DRA004034.
Results
Cell Concentrations and Abundance of Stalk Structures
The abundance of cells and stalk structure was determined in different depths (10–40 cm from the seafloor) of the 50 cm-long core dominated by red-brownish iron deposits (Figure 2). SEM observations showed the twisted stalk structures, which are morphologically similar to those previously reported as biogenic iron oxyhydroxides (Krepski et al., 2013; Figure 3). However, other morphotypes such as sheath-cased structures (Fleming et al., 2013) were not observed in this environment. Fine particulate precipitates were observed on the surface of the stalks, suggesting that the structures function as nuclei for the growth of iron minerals. Abundance of those helical stalk structures was also evaluated by using a light microscopy. In the topmost and bottom layers (CH1_7, CH1_1, and CH1_bottom), only a few stalk structures were observed, whereas the abundance increased in the middle layer sediments (CH1_2 to CH1_6, Figure 4, middle).
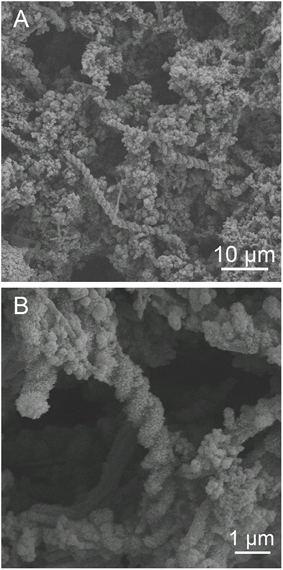
Figure 3. Scanning electron micrograph of iron oxyhydroxides. (A) Overview of iron-oxyhydroxide morphology. Helical stalk-like structures and particulate deposits on the surface were observed. (B) A higher-magnification image of (A). Fine particulate minerals adhere to the stalks. Behind the stalks, ribbon-like structures were observed.
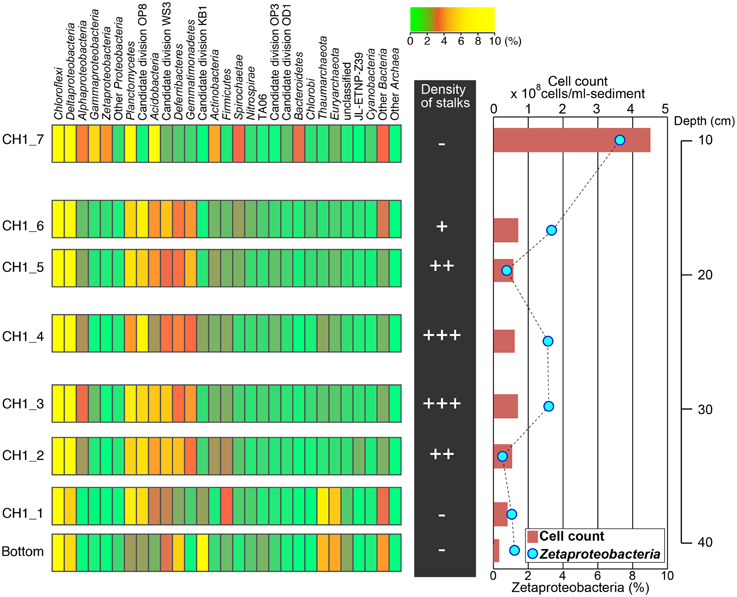
Figure 4. Left: Microbial community structures based on phylum-level classification using 16S rRNA gene-tagged sequences. Phyla comprising higher than 10% of the total sequence reads are shown in yellow. Middle: Relative abundance of stalks observed by light microscopy. Right: Cell concentrations estimated by SYBR Green I staining (red bars) and the relative abundance of Zetaproteobacteria detected by CARD-FISH (blue circles; also see Figure 5).
Fluorescent image-based cell count showed that the number of microbial cells was highest in the topmost CH1_7 at 4.23 × 108 cells cm−3 and declined sharply in CH1_6 to 7.26 × 107 cells cm−3 (Figure 4, right). Cell concentrations in iron-oxyhydroxide deposits below CH1_6 were relatively constant at around 6 to 7 × 107 cell cm−3 down to the sample CH1_2, and then decreased again to 1.65 × 107 cells cm−3 in the deepest sample CH1_bottom.
Taxonomic Composition and Microbial Community Structure
Illumina sequencing provided around 100,000 16 S rRNA sequences per sample after removing chimeras and low-quality sequences. Taxonomic analysis of the 16S rRNA sequences indicated that members within the phylum Chloroflexi are the most predominant microbial component throughout the 50 cm-core column, representing 35–40% of the total sequence reads in each sample (Figure 4). Sequences related to Deltaproteobacteria, including sulfate-reducers, were also abundant, accounting for 5–15% of the total sequence reads in all depth horizons. Sequences related to Zetaproteobacteria were prominent only in the topmost layer, composing 4% of the total sequence reads. The proportion of Zetaproteobacteria sequences decreased down to ~1% or less in the middle and bottom layers.
In the middle layers from CH1_2 to CH1_6 where the stalk structures were frequently observed, Deferribacteres represented 3–4% of the total sequence reads. Almost all of the Deferribacteres sequences were closely related to the genus Caldithrix, which are known as thermophilic nitrate-reducing bacteria (Miroshnichenko et al., 2003). Gemmatimonadetes and the candidate division OP8 were also prevalent only in the middle layer, representing 3–5% and 7–9% of the total sequence reads, respectively. In the lowest layer, members of the candidate division KB1 represent 11% of the total sequence reads, but its metabolic and physiological characteristics are not predictable because of no representative isolates in this group.
As for the archaeal 16S rRNA gene sequences, members within the Euryarchaeota and Thaumarchaeota were abundant only in the bottom layer. Sequences related to the Crenarchaeota were barely detected in our samples. Based on the sequence frequency of archaeal 16S rRNA genes, archaeal communities are relatively minor in this bacteria-dominated sedimentary habit. Nevertheless, the archaeal sequences we detected were similar to those found in an iron-rich bacterial mat at Pele's Vents on the summit of Loihi Seamount (Moyer et al., 1998).
Microscopic Observation and Quantification of Zetaproteobacteria
To determine the optimal formamide (FA) concentration for specific detection of Zetaproteobacteria, clone-FISH was performed using the nearly full-length of the 16 rRNA genes amplified from DNA extracted from the iron-oxyhydroxide samples. The optimal FA concentration was determined to be 60% when hybridization was carried out at 46°C. Analysis of CARD-FISH images for naturally occurring communities showed that all of the Zetaproteobacteria cells detected by CARD-FISH were small coccoids with the diameter of ~0.5 μm (Figures 5A,C), and they accounted for 7% of the total number of cells in the topmost layer. In the middle layer (CH1_2-CH1_6), the abundance of Zetaproteobacteria cells ranged only from ~1 to 3%. No clear correlations between the Zetaproteobacteria abundance and the frequency of helical stalk structures were observed (Figure 4, right). In the bottom layer of CH1_bottom and CH1_1, the Zetaproteobacteria abundance was as low as 1%, where only a few stalk-structures were observed.
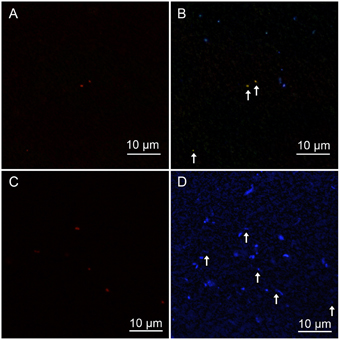
Figure 5. (A,C) Zetaproteobacteria detected by CARD-FISH using a Zeta709 probe. (B,D) High-concentration DAPI staining showed polyphosphate that emits yellowish fluorescence; blue indicates DNA inside cells. White arrows indicate Zetaproteobacteria cells detected by CARD-FISH. The contrast was enhanced in (D), because DAPI fluorescence was faint. (A,B) show CH1_5 sediment, whereas (C,D) show CH1_7 sediment.
Since the reported genome of M. ferroxydans PV-1 has genes related to polyphosphate metabolism (Singer et al., 2011), we tried to stain cells with a high concentration of DAPI, revealing that Zetaproteobacteria accumulate polyphosphate grains within the cells at the middle layer (Figure 5B). Interestingly, however, no polyphosphate grains were observed in Zetaproteobacteria cells at the top and bottom layers.
Beta Diversity of Microbial Communities
Beta-diversity of microbial communities in the iron-oxyhydroxide deposits was examined by Bray-Curtis dissimilarity analysis at the genus-level taxonomic classification (Figure 6). The result showed that microbial communities could be clustered into 3 groups corresponding to the sample depth, i.e., top, middle, and bottom. The top group (CH1_7) was markedly distinct from the other groups. The other 7 samples were more similar to each other and were categorized into two groups: the middle group (containing 4 samples) and the bottom group (containing 2 samples). In the middle group, CH_4 was relatively distinct from the other 4 samples. This zonation pattern is consistent with the occurrence of stalk structures (Figure 4, right), suggesting that there are at least 3 types of ecological niches in the cored 50 cm-long iron-oxyhydroxyide deposit.
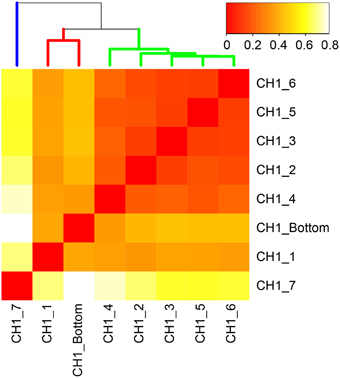
Figure 6. Bray-Curtis dissimilarity matrix of microbial communities based on 16S rRNA gene-tagged sequences at the genus-level classification. More red color indicates higher similarity. Three distinct clusters correspond to the top, middle, and bottom layers of the examined 50 cm-long core sample colored by blue, yellow, and red, respectively.
Phylogenetic Analysis of the Zetaproteobacteria Sequences
Because the length of the sequence obtained by Illumina MiSeq (254 bp) did not provide enough information for the genus-level classification, we additionally performed Sanger sequencing and obtained near-full-length sequences for the V1–V4 regions of the 16S rRNA gene, providing the finer and more precise taxonomic positions of the Zetaproteobacteria sequences from the examined environment. A total of 22 Zetaproteobacteria sequences were obtained from 331 bacteria clones in this study, although the proportions of Zetaproteobacteria in the Miseq sequence libraries were relatively low at around 1–5%. A phylogenetic tree was constructed based on the previously reported sequences (McAllister et al., 2011). The clone CH1_BAC3 composed 66% of the total Zetaproteobacteria sequences and formed a unique cluster with CH1_BAC21 (Figure 7). CH1_BAC80 and CH1_BAC2 were closely related to the sequence detected from the Kermadec Arc (Hodges and Olson, 2009) and the Loihi Seamount (McAllister et al., 2011), respectively. Other sequences such as CH1_BAC4 were closely related to sequences of Mariprofundus ferrooxydans and its relatives.
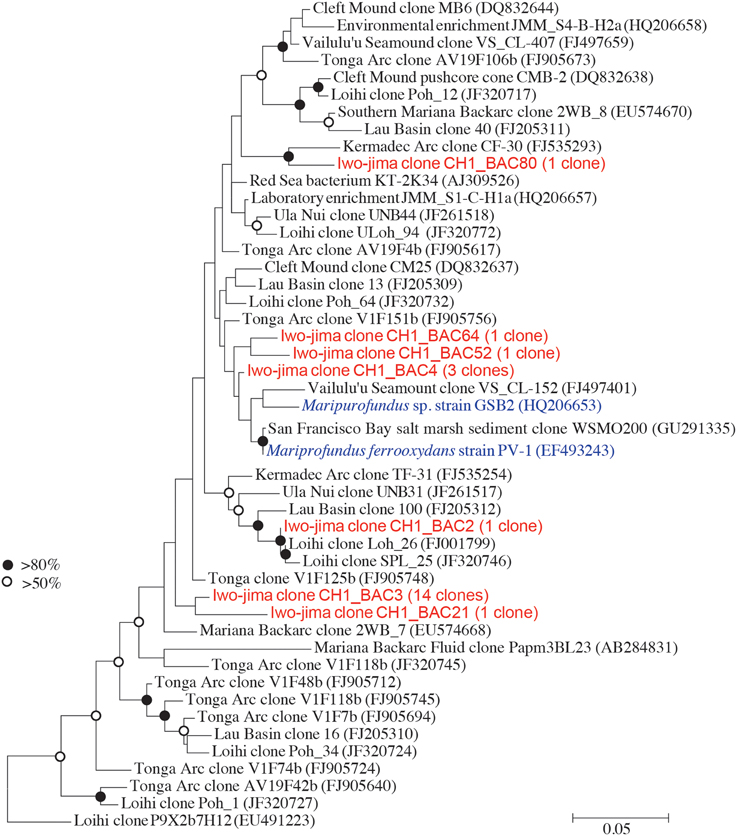
Figure 7. Molecular phylogenetic analysis of Zetaproteobacteria using the maximum-likelihood method with the Jukes-Cantor model. The V1-V4 region of 16 S rRNA sequences was used. Sequences marked in red indicate Zetaproteobacteria obtained in this study and in blue indicate pure cultures.
Correlation Between Zetaproteobacteria and other Microbial Members
We performed the community network analysis based on Spearman's rank correlation coefficient data at the genus-level classification of the sequences. Interestingly, the frequencies of iron-oxidizing Zetaproteobacteria strongly correlated with anaerobic or microaerophilic bacteria, such as the members of iron-reducing Deferisoma and sulfate-reducing Desulfobulbus (Figure 8). The members of Acidimicrobiales, which contains iron-oxidizing bacteria that can also reduce iron under reductive conditions, were also positively correlated with Zetaproteobacteria. The only exception was the members of Marinicella, which are aerobic heterotrophs positively correlated with the other microbial members. Negative correlation was observed with the candidate division NPL-UPA2, which sequences have been widely detected in both marine and terrestrial environments and their metabolic and physiological characteristics are still unknown.
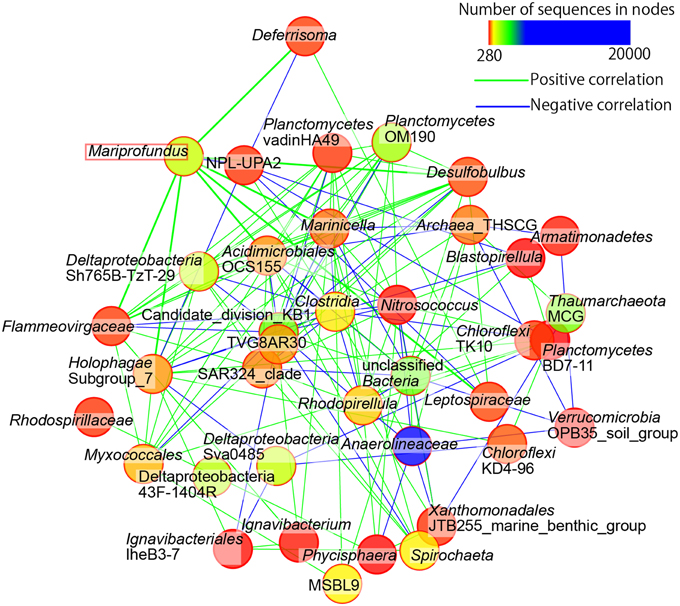
Figure 8. Community networks based on Spearman's correlation analyses. Only nodes representing first and second neighbors of Zetaproteobacteria (i.e., Mariprofundus sp.) and only strong correlations (rho values greater than 0.9) are shown. Green and blue lines indicate positive and negative correlations, respectively. Lines connected to Mariprofundus are drawn in bold.
Discussion
In the 50 cm-long core sample obtained from shallow hydrothermal iron deposits in the Nagahama Bay, members of the phylum Chloroflexi are the most predominant microbes throughout the core column. All the Chloroflexi sequences were most likely non-phototrophic, mesophilic to moderatory thermophilic anaerobes and no sequences related to phototrophic members are present, e.g., members of the class Anaerolineae are known to be strictly anaerobic heterotrophs that have been often detected from various organic-rich sedimentary environments, e.g., terrestrial soils, hydrothermal sediments, and marine subsurface sediments (Inagaki et al., 2006; Yamada et al., 2006). In the iron deposits that we examined, these Chloroflexi sequences present 8–20% of the total reads in all samples, indicating that, like other sedimentary microbial ecosystems, these members may play ecological roles in biogeochemical carbon cycling. It may also be possible that Anaerolineae ferment organic matter and produce fermented secondary products for iron-reducing bacteria (also iron-oxidizing members, if these are mixotrophs). In addition, the phylum Chloroflexi has been reported to be dominant in iron reducing enrichment culture (Hori et al., 2015) and Ardenticatena maritima that can reduce ferric iron was isolated from iron-rich coastal hydrothermal field (Kawaichi et al., 2013). Therefore, we cannot exclude the possibility that some of the Chloroflexi detected here might also associate with ferric iron reduction. In fact, we observed potential sulfate reducers such as members of the Deltaproteobacteria throughout the cored iron deposit we examined. If some of those members have multiple metabolic functions that can utilize either iron or sulfate as an electron acceptor, the presence of high concentration of iron-oxyhydroxides might be advantageous for iron reduction (Chapelle and Lovley, 1992) and hence they might contribute to iron cycling with Zetaproteobacteria.
It is not surprising in this iron-rich shallow sedimentary microbial ecosystem that many 16S rRNA sequences in all the examined depths are closely related to known sulfate-reducing bacteria (i.e., Deltaproteobacteria). Because the prevalence of sulfate-reducing conditions down to at least 40 cm below the seafloor and hence, even through the iron deposits are soft and highly permeable for discharging hydrothermal fluids and/or the overlying seawater, the redox state within the iron deposits are heterologous but generally sub-oxic to anaerobic conditions, providing habitable zones for both Chloroflexi and sulfate reducers with iron oxidizers. Beta-diversity analysis of microbial communities indicated the zonation pattern of three ecological niches in the iron deposits: the top (10 cm), middle (17–33cm), and bottom layers (~40 cm), suggesting the occurrence of redox-sensitive microbial habitats associated with iron and other elemental cycles. In fact, microscopic observations revealed that twisted stalk structures are most abundant in the middle layer. In addition, based on the visual observation, upward hydrothermal fluid flows seem to form complicated webs of the fluid passage in the middle zone (Kuratomi et al., 2015), which may create the patchily heterologous redox states that biogeochemical iron and other elemental cycles preferentially occur.
The twisted stalk structure is a typical morphological feature formed by iron-oxidizing Zetaproteobacteria under relatively low oxygen concentrations ranged from 3 to 23 μM (Krepski et al., 2013). Because we did not find any other known iron-oxidizers such as Marinobacter and Hyphomonas in the shallow hydrothermal iron deposits that we examined, the detected members of Zetaproteobacteria should play a primary role in the formation process of thick iron-oxyhydroxide deposits in the Nagahama Bay. Our previous observations indicated that the vertical formation rate of the iron deposit mound is ~12 mm y−1 (Kuratomi et al., 2015), which is similar to the formation rate of ~19 mm y−1 estimated by using a pure culture of M. ferroxydans (Chan et al., 2011). It is also conceivable that abiotic autocatalysis on the iron-oxyhydroxide stalks may accelerate the iron deposition (Rentz et al., 2007). On the other hand, we did not observed any stalk- or sheath-like structures in the topmost and bottom layers, implying that abiotic iron oxidation is dominant or other biotic oxidation process occurs without formation of the unique structures.
The community correlation network analysis based on Spearman's rank of SSU rRNA sequences at genus-level classification also exhibited the co-occurrence of iron-oxidizing Zetaproteobacteria and other anaerobic respirers in the iron-oxyhydroxide deposits. For example, Deferrisoma and Desulfobulbus showed a strong correlation to Zetaproteobacteria (Figure 8). Deferrisoma is mesophilic heterotrophs that can grow optimally at 50°C and use ferric iron or elemental sulfur as electron acceptors (Slobodkina et al., 2012), and Desulfobulbus is characterized as sulfate and iron reducers (Holmes et al., 2004). It has been reported that some sulfate-reducing bacteria are tolerant to relatively high concentrations of oxygen and have aerobic respiration systems (Muyzer and Stams, 2008). Although it still remains unknown if these members primarily utilize ferric iron for the anaerobic energy respiration instead of sulfur compounds or even both flexible to the in situ redox state, the strong correlations between iron oxidizers and reducers indicate the occurrence of iron cycling in this environment.
Another interesting finding in this study would be a discrepancy between the frequency of twisted stalk structures and the number of Zetaproteobacteria cells. Both deep sequencing analysis of 16S rRNA and CARD-FISH consistently showed that Zetaproteobacteria were most abundant in the top layer (i.e., CH1_7), although fewer stalk structures were observed (Figure 4). Although the taxonomic compositions of Zetaproteobacteria are varied in sediment depths, OTU1 was found to dominate all sedimentary horizons where we observed stalk-like structures (see Supplementary Figure 1). We might not be able to eliminate a possibility that minor Zetaproteobacteria or other iron-oxidizers may contribute to iron oxidation and mineral growth at top and bottom layers, however, we infer that OTU1-relatives play a significant ecological role in this habitat. In marked contrast, the occurrence of polyphosphate-accumulated cells was observed only in the middle layer, whereas no polyphosphate accumulations were observed in the top and bottom layers. It has been reported that a diverse array of microbes are capable of polyphosphate accumulation in natural and artificial environments (Hupfer et al., 2007), suggesting that intracellular polyphosphate grains act as a reservoir of energy and phosphate. In fact, M. ferroxydans PV-1 has genes encoding polyphosphate conversion and stores it via ATP under the aerobic conditions of excess carbon and energy substrates. Although the pure culture of M. ferroxydans PV-1 cannot grow using any organic carbon, some single amplified genomes obtained from the Loihi Seamount implicate the metabolic potential for heterotrophy or mixotrophy of Zetaproteobacteria (Field et al., 2015). Taken together, we interpret that Zeteproteobacteria in the middle layer oxidize ferrous iron, produce stalks, and store polyphosphate grains inside the cell. In the top and bottom layers, they may be different metabolic and/or physiological state(s), i.e., gaining energy from iron-oxidation and/or heterotrophy without forming stalk-like structures. This might be an adaption mechanism of iron-oxidizing Zetaproteobacteria; however, mechanisms for their redox-sensing and energetic response to the environmental change still remain largely elusive.
In conclusion, we show that Zetaproteobacteria play significant ecological roles in iron cycling with other iron reducers and construct a unique microbial ecosystem in shallow hydrothermal iron-oxyhydroxide deposits. In addition, we first demonstrated that Zetaproteobacteira in the natural ecosystem store polyphosphate grains inside cells for energy storage and a phophate source with iron-oxidation and stalk formation. We also observed that Zetaproteobacteria without pholyphosphate grains with fewer stalk formations have largest population, indicating a possibility that Zetaproteobacteria changed their metabolism depending on the environments. These results provide some new insights into ecophysiology of iron-oxidizing Zetaproteobacteria and the microbial ecosystem in marine hydrothermal environments.
Author Contributions
TK, HO, and SK sampled and pretreated a core sample. TTH, YM and TMH performed the molecular works as DNA extraction and sequencing of 16S rRNA. TTH and YM carried out microscopic works including cell count and FISH. TTH analyzed sequencing data. TTH, SK, and FI designed of the work and drafted the manuscript, which was critically riveted by all authors. The final manuscript was approved by all authors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful for the technical assistance of S. Hashimoto and T. Terada in microbiological analyses. This work is supported in part by the JSPS Grants-in-Aid for Science Research (no. 26251041 to FI, no. 15K14907 to TH).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01554
References
Amann, R. I., Ludwig, W., and Schleifer, K. H. (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143–169.
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4516–4522. doi: 10.1073/pnas.1000080107
Chan, C. S., Fakra, S. C., Emerson, D., Fleming, E. J., and Edwards, K. J. (2011). Lithotrophic iron-oxidizing bacteria produce organic stalks to control mineral growth: implications for biosignature formation. ISME J. 5, 717–727. doi: 10.1038/ismej.2010.173
Chapelle, F. H., and Lovley, D. R. (1992). Competitive-exclusion of sulfate reduction by Fe(Iii)-reducing bacteria - a mechanism for producing discrete zones of high-iron ground-water. Ground Water 30, 29–36. doi: 10.1111/j.1745-6584.1992.tb00808.x
Davis, R. E., Stakes, D. S., Wheat, C. G., and Moyer, C. L. (2009). Bacterial variability within an iron-silica-manganese-rich hydrothermal mound located off-axis at the cleft segment, Juan de Fuca Ridge. Geomicrobiol. J. 26, 570–580. doi: 10.1080/01490450902889080
Druschel, G. K., Emerson, D., Sutka, R., Suchecki, P., and Luther, G. W. (2008). Low-oxygen and chemical kinetic constraints on the geochemical niche of neutrophilic iron(II) oxidizing microorganisms. Geochim. Cosmochim. AC 72, 3358–3370. doi: 10.1016/j.gca.2008.04.035
Edwards, K. J., Glazer, B. T., Rouxel, O. J., Bach, W., Emerson, D., Davis, R. E., et al. (2011). Ultra-diffuse hydrothermal venting supports Fe-oxidizing bacteria and massive umber deposition at 5000 m off Hawaii. ISME J. 5, 1748–1758. doi: 10.1038/ismej.2011.48
Emerson, D., Rentz, J. A., Lilburn, T. G., Davis, R. E., Aldrich, H., Chan, C., et al. (2007). A novel lineage of proteobacteria involved in formation of marine Fe-oxidizing microbial mat communities. PLoS ONE 2:e667. doi: 10.1371/journal.pone.0000667
Field, E. K., Sczyrba, A., Lyman, A. E., Harris, C. C., Woyke, T., Stepanauskas, R., et al. (2015). Genomic insights into the uncultivated marine Zetaproteobacteria at Loihi Seamount. ISME J. 9, 857–870. doi: 10.1038/ismej.2014.183
Fleming, E. J., Davis, R. E., McAllister, S. M., Chan, C. S., Moyer, C. L., Tebo, B. M., et al. (2013). Hidden in plain sight: discovery of sheath-forming, iron-oxidizing Zetaproteobacteria at Loihi Seamount, Hawaii, USA. FEMS Microbiol. Ecol. 85, 116–127. doi: 10.1111/1574-6941.12104
Forget, N. L., Murdock, S. A., and Juniper, S. K. (2010). Bacterial diversity in Fe-rich hydrothermal sediments at two South Tonga Arc submarine volcanoes. Geobiology 8, 417–432. doi: 10.1111/j.1472-4669.2010.00247.x
Glazer, B. T., and Rouxel, O. J. (2009). Redox speciation and distribution within diverse iron-dominated microbial habitats at Loihi Seamount. Geomicrobiol. J. 26, 606–622. doi: 10.1080/01490450903263392
Hodges, T. W., and Olson, J. B. (2009). Molecular comparison of bacterial communities within iron-containing flocculent mats associated with submarine volcanoes along the Kermadec Arc. Appl. Environ. Microbiol. 75, 1650–1657. doi: 10.1128/AEM.01835-08
Holmes, D. E., Bond, D. R., and Lovley, D. R. (2004). Electron transfer by Desulfobulbus propionicus to Fe(III) and graphite electrodes. Appl. Environ. Microbiol. 70, 1234–1237. doi: 10.1128/AEM.70.2.1234-1237.2004
Hori, T., Aoyagi, T., Itoh, H., Narihiro, T., Oikawa, A., Suzuki, K., et al. (2015). Isolation of microorganisms involved in reduction of crystalline iron(III) oxides in natural environments. Front. Microbiol. 6:386. doi: 10.3389/fmicb.2015.00386
Hoshino, T., Yilmaz, L. S., Noguera, D. R., Daims, H., and Wagner, M. (2008). Quantification of target molecules needed to detect microorganisms by fluorescence in situ hybridization (FISH) and catalyzed reporter deposition-FISH. Appl. Environ. Microbiol. 74, 5068–5077. doi: 10.1128/AEM.00208-08
Hupfer, M., Gloess, S., and Grossart, H. P. (2007). Polyphosphate-accumulating microorganisms in aquatic sediments. Aquat. Microb. Ecol. 47, 299–311. doi: 10.3354/ame047299
Inagaki, F., Nunoura, T., Nakagawa, S., Teske, A., Lever, M., Lauer, A., et al. (2006). Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc. Natl. Acad. Sci. U.S.A. 103, 2815–2820. doi: 10.1073/pnas.0511033103
Kato, S., Yanagawa, K., Sunamura, M., Takano, Y., Ishibashi, J., Kakegawa, T., et al. (2009). Abundance of Zetaproteobacteria within crustal fluids in back-arc hydrothermal fields of the Southern Mariana Trough. Environ. Microbiol. 11, 3210–3222. doi: 10.1111/j.1462-2920.2009.02031.x
Kawaichi, S., Ito, N., Kamikawa, R., Sugawara, T., Yoshida, T., and Sako, Y. (2013). Ardenticatena maritima gen. nov., sp. nov., a ferric iron- and nitrate-reducing bacterium of the phylum 'Chloroflexi' isolated from an iron-rich coastal hydrothermal field, and description of Ardenticatenia classis nov. Int. J. Syst. Evol. Microbiol. 63, 2992–3002. doi: 10.1099/ijs.0.046532-0
Kiyokawa, S., Ninomiya, T., Nagata, T., Oguri, K., Ito, T., Ikehara, M., et al. (2012). Effects of tides and weather on sedimentation of iron-oxyhydroxides in a shallow-marine hydrothermal environment at Nagahama Bay, Satsuma Iwo-Jima Island, Kagoshima, southwest Japan. Isl. Arc. 21, 66–78. doi: 10.1111/j.1440-1738.2012.00808.x
Kiyokawa, S., and Ueshiba, T. (2015). Rapid sedimentation of iron oxyhydroxides in an active hydrothermal shallow semi-enclosed bay at Satsuma Iwo-Jima Island, Kagoshima, Japan. Sediment Geol. 319, 98–113. doi: 10.1016/j.sedgeo.2015.01.010
Krepski, S. T., Emerson, D., Hredzak-Showalter, P. L., Luther, G. W. III., and Chan, C. S. (2013). Morphology of biogenic iron oxides records microbial physiology and environmental conditions: toward interpreting iron microfossils. Geobiology 11, 457–471. doi: 10.1111/gbi.12043
Kuratomi, T., Kiyokawa, S., Ikehara, M., Goto, S., Hoshino, T., Ikegami, F., et al. (2015). “The structure of iron oxidized mounds at shallow marine hydrothermal environment in Satsuma Iwo-jima Island, Kagoshima,” in Japan Geoscience Union Meeting (Chiba).
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (New York, NY: John Wiley & Sons), 115–147.
Loy, A., Lehner, A., Lee, N., Adamczyk, J., Meier, H., Ernst, J., et al. (2002). Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68, 5064–5081. doi: 10.1128/AEM.68.10.5064-5081.2002
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371. doi: 10.1093/nar/gkh293
McAllister, S. M., Davis, R. E., McBeth, J. M., Tebo, B. M., Emerson, D., and Moyer, C. L. (2011). Biodiversity and emerging biogeography of the neutrophilic iron-oxidizing Zetaproteobacteria. Appl. Environ. Microbiol. 77, 5445–5457. doi: 10.1128/AEM.00533-11
Miroshnichenko, M. L., Kostrikina, N. A., Chernyh, N. A., Pimenov, N. V., Tourova, T. P., Antipov, A. N., et al. (2003). Caldithrix abyssi gen. nov., sp. nov., a nitrate-reducing, thermophilic, anaerobic bacterium isolated from a Mid-Atlantic Ridge hydrothermal vent, represents a novel bacterial lineage. Int. J. Syst. Evol. Microbiol. 53, 323–329. doi: 10.1099/ijs.0.02390-0
Morono, Y., and Inagaki, F. (2010). Automatic slide-loader fluorescence microscope for discriminative enumeration of subseafloor life. Sci. Drill. 9, 32–36. doi: 10.5194/sd-9-32-2010
Morono, Y., Terada, T., Kallmeyer, J., and Inagaki, F. (2013). An improved cell separation technique for marine subsurface sediments: applications for high-throughput analysis using flow cytometry and cell sorting. Environ. Microbiol. 15, 2841–2849. doi: 10.1111/1462-2920.12153
Moyer, C. L., Dobbs, F. C., and Karl, D. M. (1995). Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl. Environ. Microbiol. 61, 1555–1562.
Moyer, C. L., Tiedje, J. M., Dobbs, F. C., and Karl, D. M. (1998). Diversity of deep-sea hydrothermal vent Archaea from Loihi seamount, Hawaii. Deep-Sea Res. Part II-Top. Stud. Oceanogr. 45, 303–317. doi: 10.1016/S0967-0645(97)00081-7
Muyzer, G., and Stams, A. J. (2008). The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 6, 441–454. doi: 10.1038/nrmicro1892
Nogami, K., Yoshida, M., and Ossaka, J. (1993). Chemical composition of discolored seawater around Satuma-Iojima, Kagoshima, Japan. Volcanol. Soc. JPN 38, 71–77.
Rentz, J. A., Kraiya, C., Luther, G. W. III., and Emerson, D. (2007). Control of ferrous iron oxidation within circumneutral microbial iron mats by cellular activity and autocatalysis. Environ. Sci. Technol. 41, 6084–6089. doi: 10.1021/es062203e
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schramm, A., Fuchs, B. M., Nielsen, J. L., Tonolla, M., and Stahl, D. A. (2002). Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ. Microbiol. 4, 713–720. doi: 10.1046/j.1462-2920.2002.00364.x
Serafim, L. S., Lemos, P. C., Levantesi, C., Tandoi, V., Santos, H., and Reis, M. A. (2002). Methods for detection and visualization of intracellular polymers stored by polyphosphate-accumulating microorganisms. J. Microbiol. Methods 51, 1–18. doi: 10.1016/S0167-7012(02)00056-8
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shikaura, H., and Tazaki, K. (2001). Cementations of sand grains are accelerated by microbes. Clay Sci. Soc. JPN 40, 229–241. doi: 10.11362/jcssjnendokagaku1961.40.229
Singer, E., Emerson, D., Webb, E. A., Barco, R. A., Kuenen, J. G., Nelson, W. C., et al. (2011). Mariprofundus ferrooxydans PV-1 the first genome of a marine Fe(II) oxidizing Zetaproteobacterium. PLoS ONE 6:e25386. doi: 10.1371/journal.pone.0025386
Slobodkina, G. B., Reysenbach, A. L., Panteleeva, A. N., Kostrikina, N. A., Wagner, I. D., Bonch-Osmolovskaya, E. A., et al. (2012). Deferrisoma camini gen. nov., sp. nov., a moderately thermophilic, dissimilatory iron(III)-reducing bacterium from a deep-sea hydrothermal vent that forms a distinct phylogenetic branch in the Deltaproteobacteria. Int. J. Syst. Evol. Microbiol. 62, 2463–2468. doi: 10.1099/ijs.0.038372-0
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Yamada, T., Sekiguchi, Y., Hanada, S., Imachi, H., Ohashi, A., Harada, H., et al. (2006). Anaerolinea thermolimosa sp. nov., Levilinea saccharolytica gen. nov., sp. nov. and Leptolinea tardivitalis gen. nov., sp. nov., novel filamentous anaerobes, and description of the new classes Anaerolineae classis nov. and Caldilineae classis nov. in the bacterial phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 56, 1331–1340. doi: 10.1099/ijs.0.64169-0
Keywords: iron-oxidizing bacteria, zetaproteobacteria, shallow hydrothermal vents, iron cycle, network analysis, fluorescence in situ hybridization (FISH), MiSeq Illumina, polyphosphates granules
Citation: Hoshino T, Kuratomi T, Morono Y, Hori T, Oiwane H, Kiyokawa S and Inagaki F (2016) Ecophysiology of Zetaproteobacteria Associated with Shallow Hydrothermal Iron-Oxyhydroxide Deposits in Nagahama Bay of Satsuma Iwo-Jima, Japan. Front. Microbiol. 6:1554. doi: 10.3389/fmicb.2015.01554
Received: 05 October 2015; Accepted: 22 December 2015;
Published: 11 January 2016.
Edited by:
Michael Rappe, University of Hawaii at Manoa, USAReviewed by:
Craig Lee Moyer, Western Washington University, USALingling Wu, University of Wisconsin – Madison, USA
Copyright © 2016 Hoshino, Kuratomi, Morono, Hori, Oiwane, Kiyokawa and Inagaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tatsuhiko Hoshino, aG9zaGlub3RAamFtc3RlYy5nby5qcA==
 Tatsuhiko Hoshino
Tatsuhiko Hoshino Takashi Kuratomi3
Takashi Kuratomi3 Yuki Morono
Yuki Morono Tomoyuki Hori
Tomoyuki Hori Fumio Inagaki
Fumio Inagaki