- 1Department of Microbiology, Pondicherry University, Kalapet, India
- 2Department of Botany and Microbiology, Addiriyah Chair for Environmental Studies, College of Sciences, King Saud University, Riyadh, Saudi Arabia
- 3School of Life Sciences, Bharathidasan University, Tiruchirappalli, India
- 4Department of Food Science and Technology, Pondicherry University, Kalapet, India
The important biological macromolecules, such as lipopeptide and glycolipid biosurfactant producing marine actinobacteria were analyzed and their potential linkage between type II polyketide synthase (PKS) genes was explored. A unique feature of type II PKS genes is their high amino acid (AA) sequence homology and conserved gene organization. These enzymes mediate the biosynthesis of polyketide natural products with enormous structural complexity and chemical nature by combinatorial use of various domains. Therefore, deciphering the order of AA sequence encoded by PKS domains tailored the chemical structure of polyketide analogs still remains a great challenge. The present work deals with an in vitro and in silico analysis of PKS type II genes from five actinobacterial species to correlate KS domain architecture and structural features. Our present analysis reveals the unique protein domain organization of iterative type II PKS and KS domain of marine actinobacteria. The findings of this study would have implications in metabolic pathway reconstruction and design of semi-synthetic genomes to achieve rational design of novel natural products.
Introduction
Natural products of microorganisms are potential source of bioactives that have been extensively exploited to develop next generation anti-infective drugs proposed by pharmaceutical companies (De Carvalho and Fernandes, 2010). But in recent years, the exploration of marine microorganisms received greater attention due to their complex biosynthetic pathways and potential implications on the development of anti-cancer agents and anti-infectives to combat multi-resistant strains (De Carvalho and Fernandes, 2010). Past few decades the bioprospecting of natural resources and microbial isolates were tremendously increased, however, the leads transformed to drugs are very few (Watve et al., 2001). Perhaps this trend might have led to the exploration of pristine and unexplored bioresources including hydrothermal vents and extreme niches. Marine sponges are sedentary animals harboring more than 40% of microorganisms by volume. Among the marine fauna and flora, marine sponges are potential source of bioactive natural products (Faulkner and Ghiselin, 1983, 1994; Matsunaga and Fusetani, 2003). However, recent deliberations envisage that the sponge derived secondary metabolites are biosynthesized by the associated microorganisms. However, this hypothesis is being remained unproven as sponge-specific bacteria are uncultivable with conventional approaches. Exploration of sponge associated microbial diversity and symbiont-assisted complex biosynthetic pathway of bioactive leads have increased the scope of natural product discovery from marine sponges (Faulkner and Ghiselin, 1983, 1994; Hentschel et al., 2002). Recent developments in genome mining and metagenomics have widely used in the exploitation of such complex biosynthetic pathways of marine natural products. By and large the biosynthetic pathways of polyketides, non-ribosomal peptides, and their derivatives are useful to integrate sponges and their symbiotic biosynthetic machineries. Marine sponges are richest source of polyketide and peptide bioactive molecules. Unlike terrestrial counterparts, sponge-derived bioactive molecules are unique and having specific targeted activities expected for drug leads (Li et al., 2002; Matsunaga and Fusetani, 2003; Montalvo et al., 2005; Montalvo and Hill, 2011). The sponge-derived bioactive peptides are non-ribosomal origin and are modified with unusual amino acids (AAs; Matsunaga et al., 1985).
Polyketide synthases (PKSs) are modular proteins involved in the biosynthesis of complex bioactive molecules through sequential catalytic activities. These enzymes mediate biosynthesis of bioactive molecules with diverse structural complexities by combinatorial use of a specific sequential order of catalytic domains. The tailoring of catalytic domains and AA sequence of these domains are drastically changes with natural bioresources and therefore, the nature and chemical structure of end product is varied between/within the species (Yadav et al., 2009). The mechanism of sequential order and/or selection of catalytic domains remains a major challenge in chemical ecology of secondary metabolite synthesis. The fully dissociable complex of small, discrete mono-functional proteins that catalyze combinatorial synthesis of aromatic polyketides, which is in general termed as type II PKS. In the iterative PKSs, the active site of each catalytic module for tailoring of type II PKS is encoded by a single gene. There is only one set of a hetero-dimeric ketosynthase (KSα–KSβ) and an acyl carrier protein (ACP) that tailored the synthesis of polyketide molecule in a specific order and defined number of cycles to build a polyketide chain (He and Hertweck, 2003). The chain length is maintained through sequential iterative process including cyclisation, reduction, and aromatization steps which are catalyzed by cyclase (CYC), KR, and aromatase (ARO), respectively. In certain group of type II PKSs, the malonyl-CoA ACP acyl transferase (MAT), which catalyzes condensation of acyl transfer between malonyl-CoA and the ACP (Revill et al., 1995). The type II PKSs in general catalyze the biosynthesis of diverse range of multi-functional aromatic polyketides and are mostly restricted among bacteria (Shen et al., 2000). The type II PKSs, such as those responsible for the biosynthesis of the aromatic polyketides actinorhodin (ACT; Fernández-Moreno et al., 1992) and tetracenomycin (TCM), (Bibb et al., 1989; Summers et al., 1992) are composed of three to seven separate mono- or bi-functional proteins, the active sites of which are used iteratively for the assembly and early modification of the polyketide chain.
The KS domain of PKS gene was retrieved from marine actinobacteria producing biosurfactants and antimicrobial compounds. Therefore, this study was aimed to integrate PKS gene in biosurfactant production. Based on the literature, PKS gene can be expected from actinobacteria producing antimicrobial compounds, but PKS gene was not linked with biosurfactant production. A PKS gene possibly encodes biosynthesis of some biosurfactants, being considered as smart biomolecules having the ability to reduce surface and interfacial tension, wider bioactivities and possibly involved in bacterial quorum sensing (Kiran et al., 2015). Biosurfactant production has been reported by our research group in several actinobacteria (Selvin et al., 2009b; Kiran et al., 2010) and they were linked with non-ribosomal peptide synthases (NRPS), PKS (Kiran et al., 2010), and large multifunctional proteins with a modular organization. Biosynthetic pathway of biosurfactants in Bacillus and Pseudomonas was well-established. However, biosynthetic pathway of biosurfactants produced by marine actinobacteria, in general remains undisclosed. The biosurfactants invariably showed antibiofilm activity without inhibiting the biomass of pathogens tested. Based on in vitro experiments, it was found that the biosurfactants produced by marine actinobacteria is having antimicrobial and antibiofilm activity. The PCR amplified KS domain from these actinobacteria envisages the biosynthetic pathway of biosurfactants might have mediated through PKS biosynthetic gene clusters. Therefore, in this study, the in vitro findings are integrated with in silico analysis to substantiate the hypothesis that the biosynthesis of biosurfactants produced by marine actinobacteria might have mediated by PKS gene. To date, there are few reports about the interaction between PKS type II gene clusters and biosurfactant production (Kiran et al., 2010). There is no report on marine actinobacteria and their PKS structural diversity related with biosurfactant production. Hence we decided to focus on this aspect with three biosurfactants (MSA10, MSA13, and MSA21; Gandhimathi et al., 2009; Kiran et al., 2010, 2014) and two antagonistic compounds producing (MAD01 and MSI051; Selvin et al., 2009a,b) actinobacterial strains and they were isolated from marine sponges, Fasciospongia cavernosa and Dendrilla nigra, respectively. In silico analysis of PKS gene clusters and modular structure of iterative type II PKS are important tool for designing various experimental approaches toward the combinatorial synthesis of diverse aromatic polyketides. Therefore, present study was aimed to analyze and evaluate the KS domains of iterative PKS gene type II and ketosynthase genes retrieved from marine sponge-associated actinobacteria and their biosurfactant producing ability related to iterative type II PKS gene.
Materials and Methods
Microorganisms and PKS Type II Gene Amplification
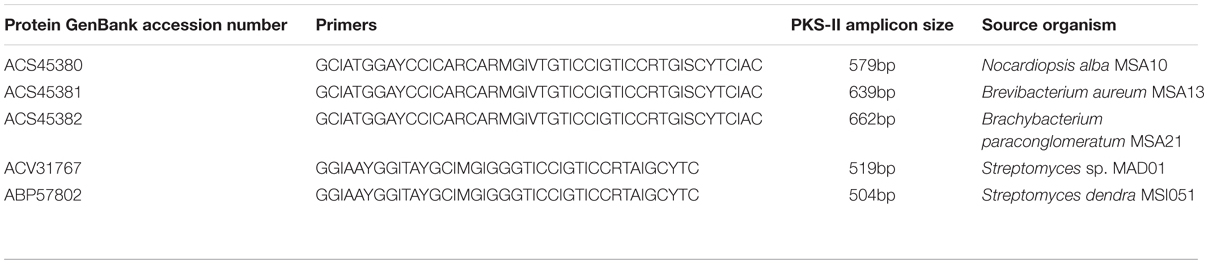
The actinobacterial strains used in this study were already been isolated from marine sponges, such as F. cavernosa (MSA10) and D. nigra (MSA13, MSA21, MAD01, and MSI051) collected from southwest cost of India. The 16S rRNA GenBank accession numbers as follows Nocardiopsis alba MSA10: EU563352 (Gandhimathi et al., 2009), Brevibacterium aureum MSA13: GQ153943 (Kiran et al., 2010), Brachybacterium paraconglomeratum MSA21: GQ153945 (Kiran et al., 2014), Streptomyces sp. MAD01: GQ246755 (Selvin et al., 2009b), and Streptomyces dendra MS1051: EF417875 (Selvin, 2009), respectively. The PKS type II gene was amplified from five actinobacterial strains (MSA10, MSA13, MSA21, MAD01, and MSI051) according to Selvin (2009). The genes encoding PKS were amplified using degenerate primers (Table 1). The PCR temperature profile used was 95°C for 3 min, and then followed by 30 cycles at 95°C for 30 s, 56°C for 30 s, and 72°C for 60 s and finally an extension step at 72°C for 10 min. The resultant amplified PCR products were purified and cloned using the TOPO TA cloning kit (Invitrogen) for sequencing.
Evaluation of Antibiofilm Effect
The culture supernatant obtained from actinobacterial strains were evaluated for biofilm inhibitory effect against Vibrio harveyi. The biofilm was allowed to develop on cover slips and treated with the actinobacterial extracts and incubated for 48 h at 37°C. After incubation the planktonic and spent media were discarded. The cells were washed twice with deionized water air dried and stained with 0.1% acridine orange and examined under confocal laser scanning microscopy (CLSM).
Determination of Bacterial Cell Viability in Biofilm
Cell viability of the bacteria in the biofilm was assessed using MTT assay as described by Traba and Liang (2011) with necessary modifications. Biofilm of V. harveyi was allowed to develop on 96-well plate and treated with 50 μl culture filtrates of the five actinobacterial strains and incubated for 24 h at 37°C. Untreated wells were set as control. After 24 h the bacterial suspension was collected and then treated with 100 μl of phosphate buffered saline and 50 μl of MTT at concentration of 0.3% were added and then incubated for 2 h at 37°C. The MTT solutions were removed and formazan crystals formed were dissolved in 150 μl of DMSO and 25 μl of 0.1 M glycine buffer of pH 10.2. The absorbance was recorded in a microplate reader at 550 nm.
KS Domain Protein Data Set, Phylogeny Construction, and Domain Structural Analysis
Type II KS domain sequences and ketosynthase gene sequences were translated using sequence manipulation suite1 and these deduced AA sequences of type PKS II and ketosynthase were deposited to NCBI-GenBank with the accession numbers of ACS45380–ACS45382 (type II PKS), and ketosynthase bearing following accession numbers ACV31767 and ABP57802. KS domain of type II PKS gene sequences and ketosynthase (Cds) sequences of sponge-associated actinobacteria were retrieved from National center for Biotechnology Information2. GenBank accession numbers of these KS domains and ketosynthase sequences were given as GQ153947 (N. alba MSA10), GQ153948 (B. aureum MSA13), GQ153949 (Brachybacterium sp. MSA21), and GQ246762 (Streptomyces sp. MAD01), EF520724 (Streptomyces dendra MS1051), respectively. The predicted KS domains of all retrieved actinobacterial gene sequences and the PKS type II protein sequences from reference actinobacteria were aligned by CLUSTAL W23 and translated deduced AA sequences were verified using the NCBI-BLAST4 search with expected value set to the default value of 10 was performed using the protein sequences of N. alba, B. aureum, Brachybacterium sp. MSA21, Streptomyces MAD01 and S. dendra, respectively, and the various sequences against 138 complete eubacterial and 20 complete archaebacterial genomes. Phylogenetic tree of the deduced AA sequences of PKS II segments and ketosynthase genes were generated using neighbor-joining method through MEGA programs (Kumar et al., 2004). KS domain phylogeny was based in the prediction of putative enzymes of identical or nearly identical biochemical function. The type of KS was identified based on the top BLAST match in the reference data set. NCBI CDD search, SEARCPKS and Motif scan were performed to derive the existence of significant domains and their organization. Comparative analyses of KS domains of five subject organisms were performed with known polyketide producers and with the structure of polyketides using NCBI CDD and SEARCHPKS, respectively. The AA composition was also predicted to substantiate the function of type II PKS and ketosynthase of our interest.
Profile Hidden Morkov Model (HMM) analysis was carried out by HMMER package. The available three (Nocardiopsis, Brevibacterium, Brachybacterium) actinobacterial KS dataset was analyzed, whether these domains are modular or iterative KS domains. All these three iterative KS domains of PKS type II gene clusters of actinomycetes were modeled using comparative modeling approach. Threading analysis was carried out using a local version of threader package5 to identify the structural templates for modeling of actinobacterial KS domains. The remaining two KS domains (from Streptomyces MAD01 and S. dendra MSI051) have been modeled using fatty acid KAS structure as template (Escherichia coli KAS I), which show only about 40% sequence identity with polyketide KS domains. Even the sequence identity was lesser between the target and template, the two KS proteins structures can be reliable and they adopt similar structure. The secondary structures of type II PKS and ketosynthase domains of 3D models were created using a (PS)2 is an automated homology modeling server (Chen et al., 2006). The (PS)2 combines PSI-BLAST, IMPALA, and T-Coffee in both template selection and target-template alignment. The final three dimensional structures were built using the modeling package MODELLER.
Results and Discussion
The Nature of KS Domains of Type II PKS and Ketosynthase
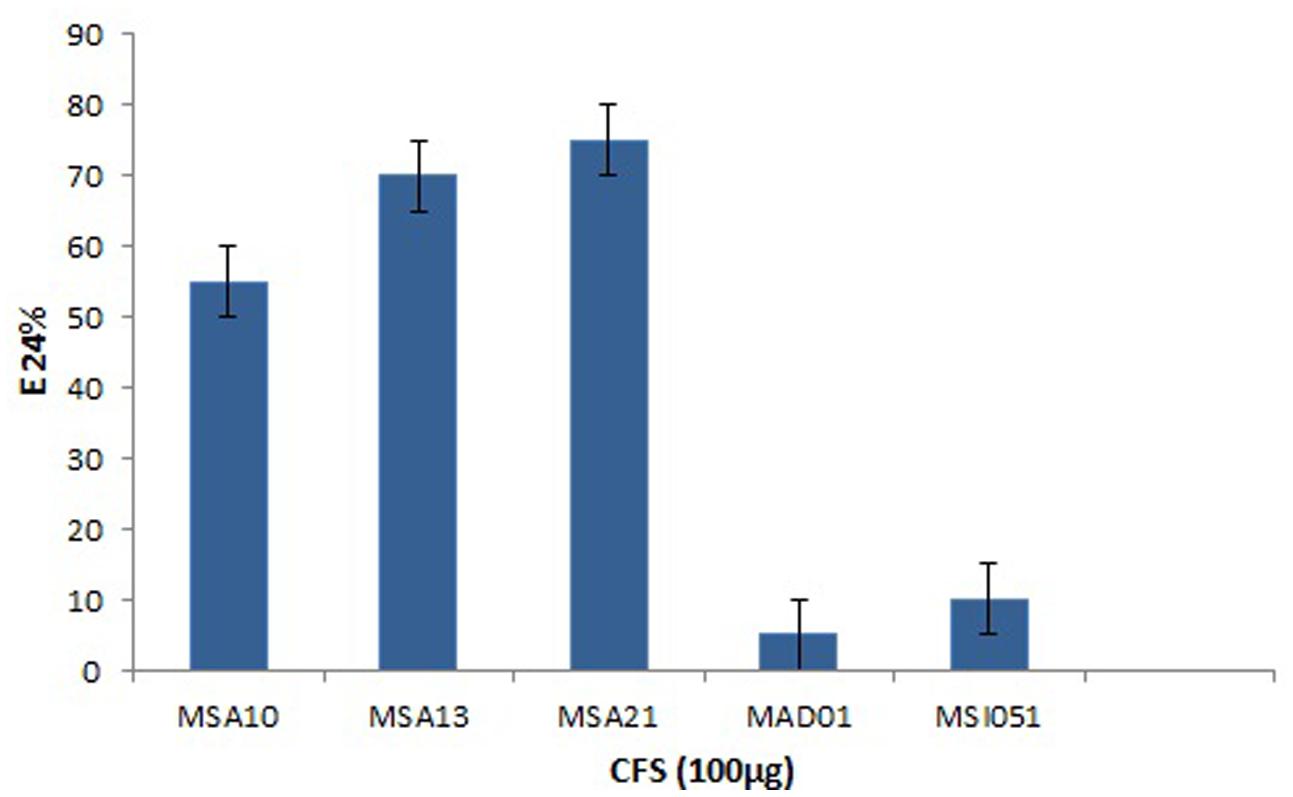
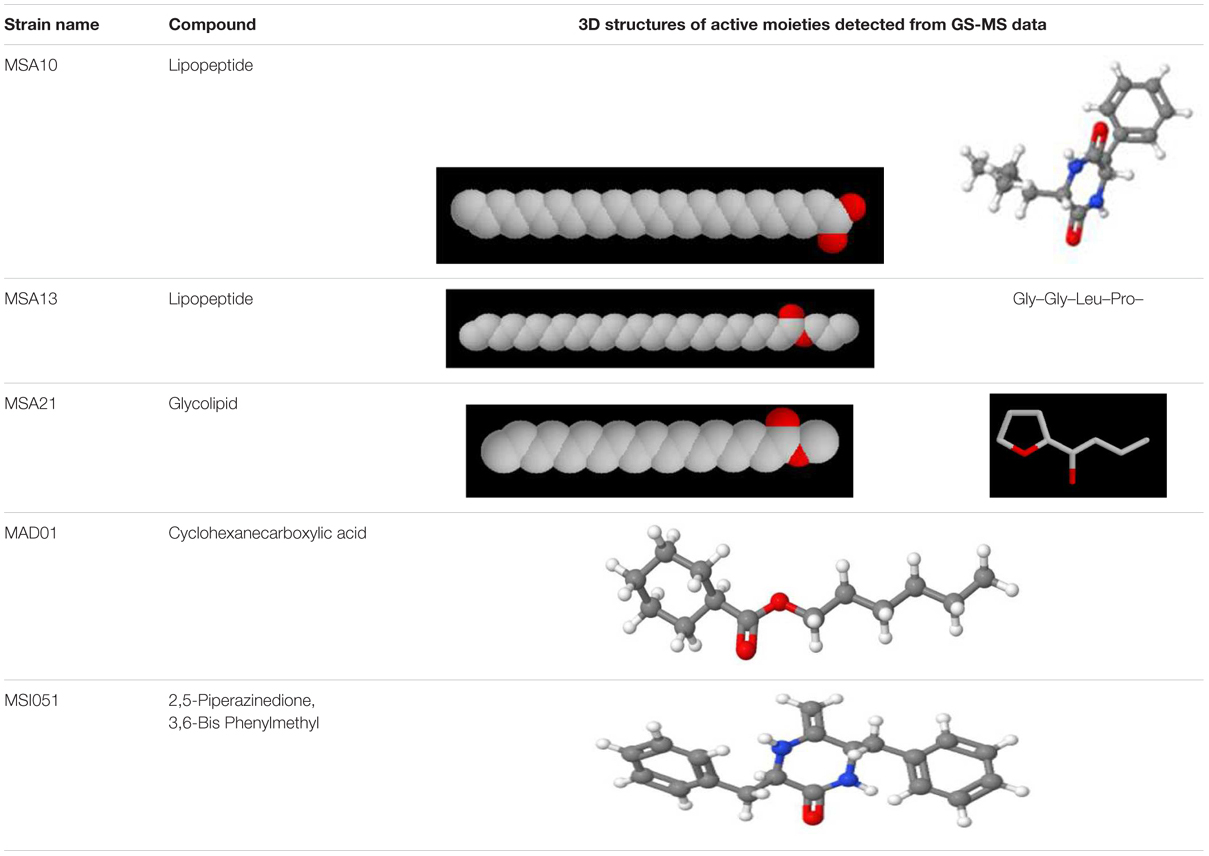
The actinobacterial isolates from marine sponges were screened for biosurfactant activity using emulsification index (E24) as per Kiran et al. (2010). Among the five actinobacteria MSA10, MSA13, and MSA21 were potential producer of biosurfactants (Figure 1). The active moieties were identified from GCMS data. The active moieties of MSA10, MSA13, and MSA21 were evidenced as biosurfactant molecules, but the moieties of MAD01 and MSI051 were not related with biosurfactants (Table 2). The antimicrobial moiety of MAD801 was identified as cyclohexane carboxylic acid hexyl ester. It was reported that cyclohexane carboxylic acid is a moiety of the antifungal polyketide ansatrienin A (Patton et al., 2000).
Sponge-associated actinobacteria: i.e., N. alba, B. aureum, and Brachybacterium paraconglomeratum beared the type II PKS (GQ153947, GQ153948, and GQ153949, Table 1). The KS domain of these gene segments were translated into AAs counts, viz; 191, 212 and 220, respectively. All these KS domains encodes the condensation enzymes (cds), which catalyzes (decarboxylation or non-decarboxylation) Claisen-like condensation reaction, and the KS domains sharing the strong structural similarities are involved in the synthesis and degradation of fatty acids.
KS domain of PKS gene is the most conserved catalytic domain and is involved in the tailoring PKS molecule by catalyzing the chain condensation step. We have performed in silico analysis to identify KS domain counterparts from modular and iterative PKSs and other PKS families. The analyzed domains are separated into distinct clusters in a phylogenetic tree (Figure 2). Based on HMM by the HMMEP package, three actinobacterial KS domain of type II PKS genes were analyzed and the results show that these three isolates contains iterative PKS gene and this outcome provides potential in genome sequencing efforts for the identification of novel PKS genes. Iterative condensation steps play a vital role in biosynthesis by PKS proteins and phylogenetic analysis of iterative KS domains inferred that the clustering of iterative PKS gene sequence is highly correlated with the number of iterations they perform. From this study, we suggest that marine sponge associated actinobacterial community predominantly possesses the iterative KS domain of type II PKS rather than modular type I PKS or NRPS-PKS hybrids. The type II PKS from three different genera is characterized to study and understand their function and diversity. The isolation and identification of PKS with different enzymatic activity in marine actinobacteria has been reported, as well as the occurrence of PKS gene families in a community (Kim and Fuerst, 2006). This is the first report on the in silico analysis of iterative type II PKS of sponge-associated actinobacteria. Recent literature (Kiran et al., 2010, 2015) evidenced that these actinobacteria are potent biosurfactant producers with antimicrobial activity (lipopeptide and glycolipid derivatives). The present in silico analysis revealed that these isolates possessing iterative domains (Figure 2) type II PKS genes and it can be hypothesized that the antimicrobial biosurfactants synthesis might be mediated by iterative type II PKS genes. Another group of actinobacterial antibiotics producers from the marine sponge D. nigra such as S. dendra MSI051 (Selvin, 2009) and Streptomyces sp. MAD01 (Selvin et al., 2009b) were included in the analysis. Their partial ketosynthase genes were retrieved from GenBank (GQ246762 and EF520724) with 519 and 504 bp encoding 173 and 168 aa, respectively (Table 1).
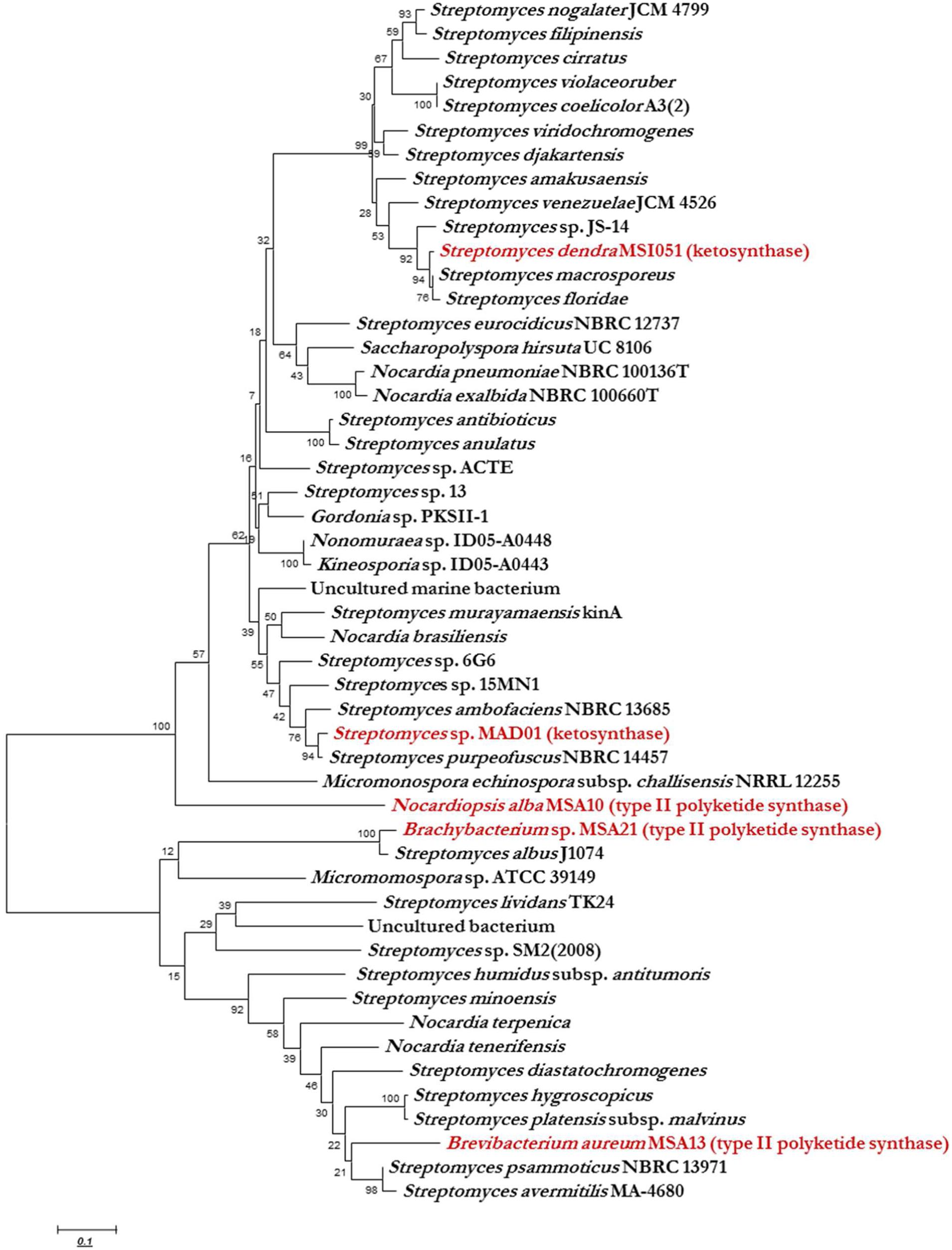
FIGURE 2. Phylogenetic analysis (MEGA 5.0) of ketosynthase regions with respect to the diverse range of ketosynthase domains, including iterative types II, modular PKS, and KS domain. The phylogenetic trees were constructed using bootstrapping and the neighbor-joining rules.
Antibiofilm Effect Against Vibrio
Antibiofilm effect of the culture supernatant was well noticed by CLSM. The culture supernatant inhibits the biofilm formation of V. harveyi. Among the extract used the lipopeptide producer MSA10 and MSA 13 inhibit the biofilm formation by 80% compared to the other actinobacterial extracts (Figure 3). The antibiofilm effect may be due to the biosurfactant production mediated by PKS gene.
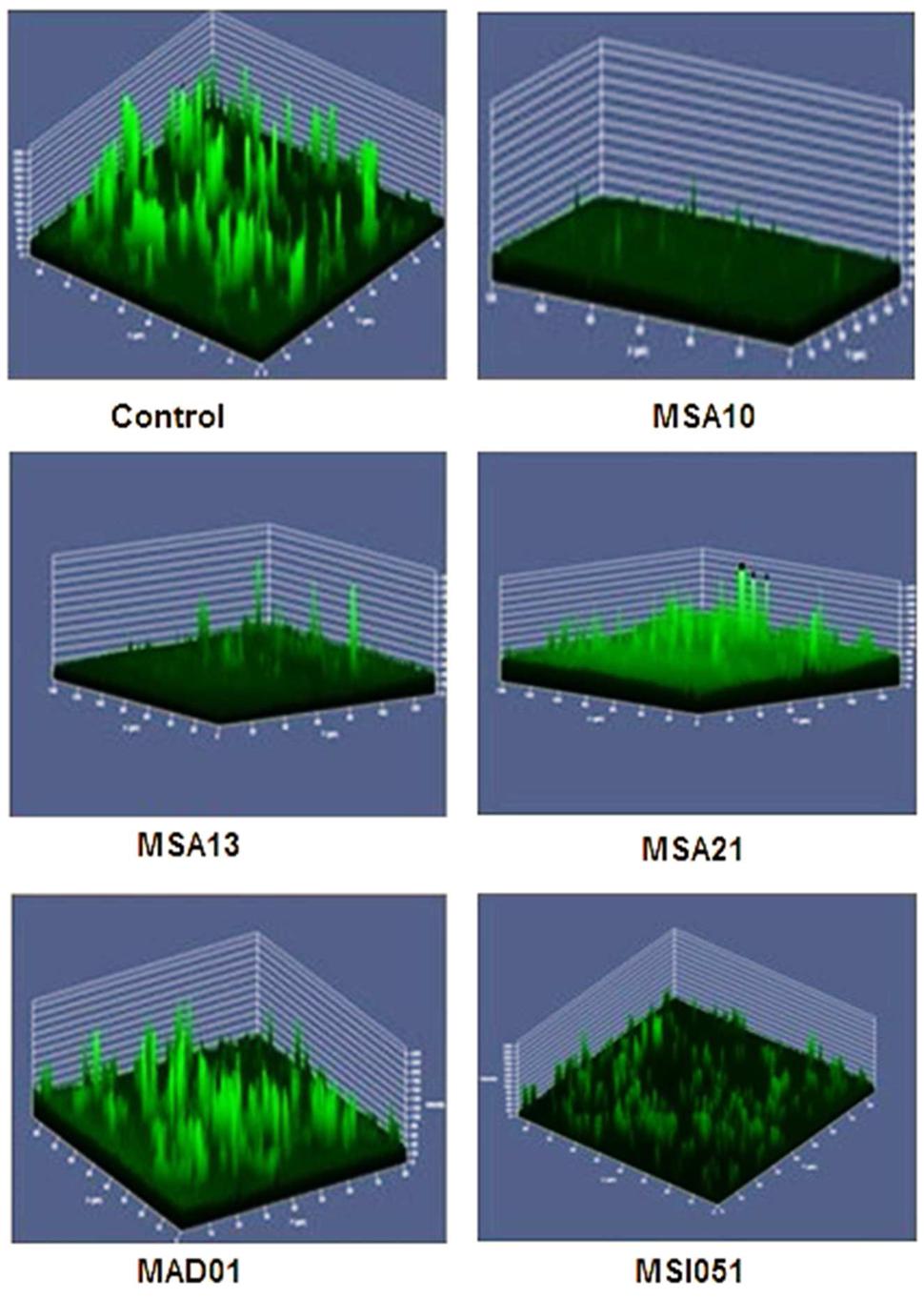
FIGURE 3. Antibiofilm activity of cell free supernatants from marine actinobacteria. The pre-formed biofilm on glass slides was treated for 24 h with 150 μg each of freeze-dried cell free supernatant (CFS). Untreated biofilm (control) and remaining biofilm of V. harveyi treated after CFS on glass slides were stained with 0.1% acridine orange and observed in CLSM (LSM 710, Carl Zeiss).
Cell Viability in Biofilm
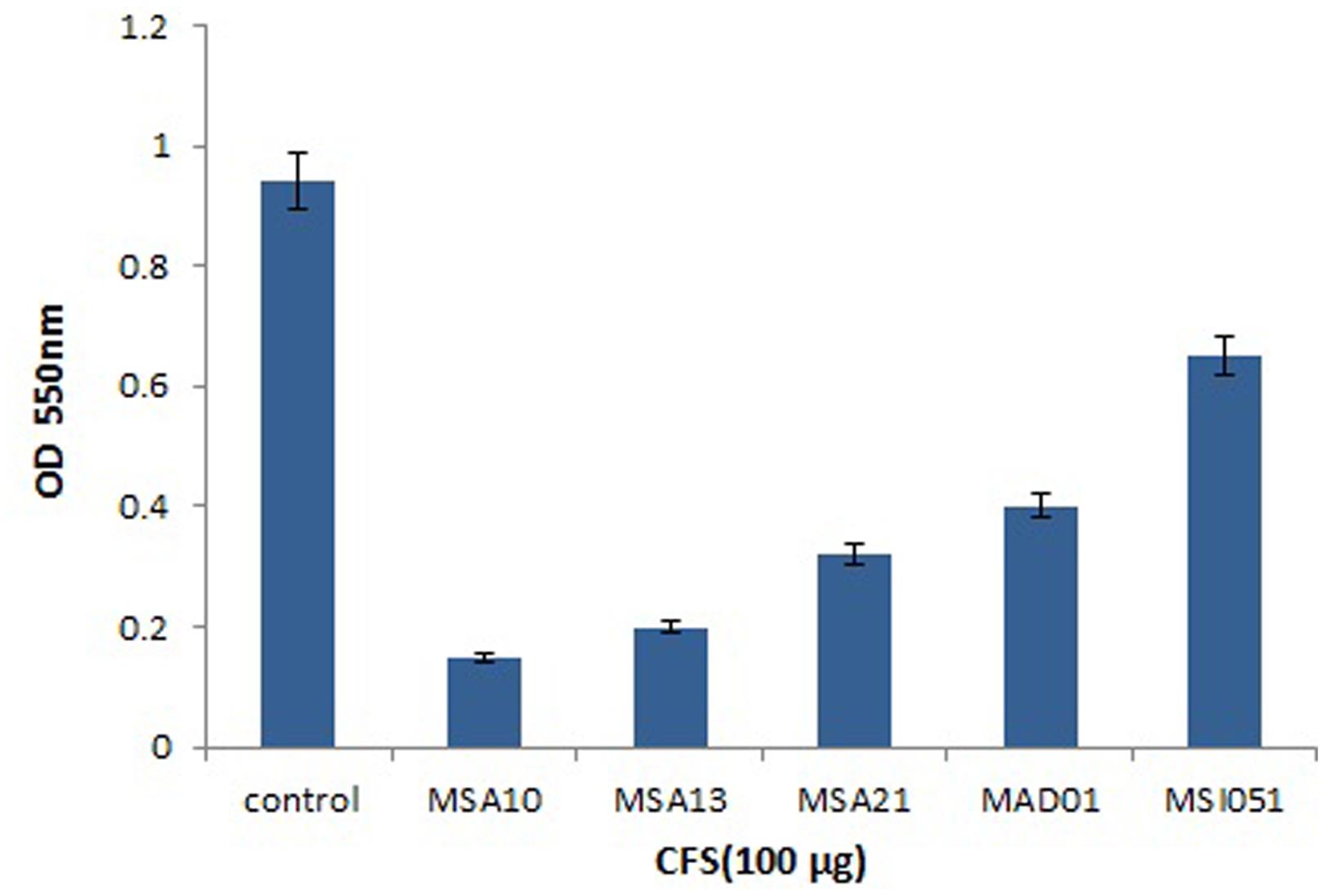
The viability of the cells were reduced by adding the biosurfactants as shown in Figure 4. When compared to the control the extracts from MSA 10 and MSA 13 inhibits the viability of Vibrio cells by more than 80%, followed by MSA 21 by 70%.
Domain Architecture and Homology Modeling of Iterative Type II PKS
In silico analysis of Type II PKS and ketosynthase unveiled an unprecedented organization of various domains encoding discrete ketoacyl synthase (KAS) and thiolase, PKC, CK2, and ACP some are lacking an ACP. Certain polyketides undergoes non-iterative biosynthesis which involves a novel type II PKS that acts directly on acyl CoA substrates. These results demonstrate the capability of nature’s in designing complex bioactive compounds and suggest new methods for PKS design and engineering through synthetic biology approaches to expand the scope and diversity of polyketide library. The structural diversity of PKS would ultimately help in searching for PKS with novel chemistry for combinatorial biosynthesis (Shen and Kwon, 2002). All the proteins studied here are found to have potential KS domains which catalyze the polyketide chain elongation step. In the beginning of chain elongation, an enzyme intermediate is formed between the growing polyketide chain and the thiol of its active site Cys. Then condensation reaction occurs with the methylmalonyl-ACP or malonyl-ACP co-substrate (Shen, 2003).
The phylogenetic trees were constructed using bootstrapping and the neighbor-joining rules.
Analysis evidenced that the PKS sequence retrieved from N. alba and B. aureum are having ACP domain, i.e., beta-ketoacyl-ACP synthase and beta-KAS (Figure 5). KASs are involved in the elongation steps in the pathway of fatty acid biosynthesis. KAS III is involved in the catalysis of the initial condensation and KAS I and II are responsible for elongation steps by Claisen condensation of malonyl-ACP with acyl-ACP. Remaining three protein sequences lack ACP, some non iterative type II PKSs lack ACP, utilize acyl CoAs as substrates for macrotetrolide biosynthesis. It was reported that the PKSs are using ACP to activate the acyl CoA substrate and channel the polyketide intermediates (Shen, 2003).
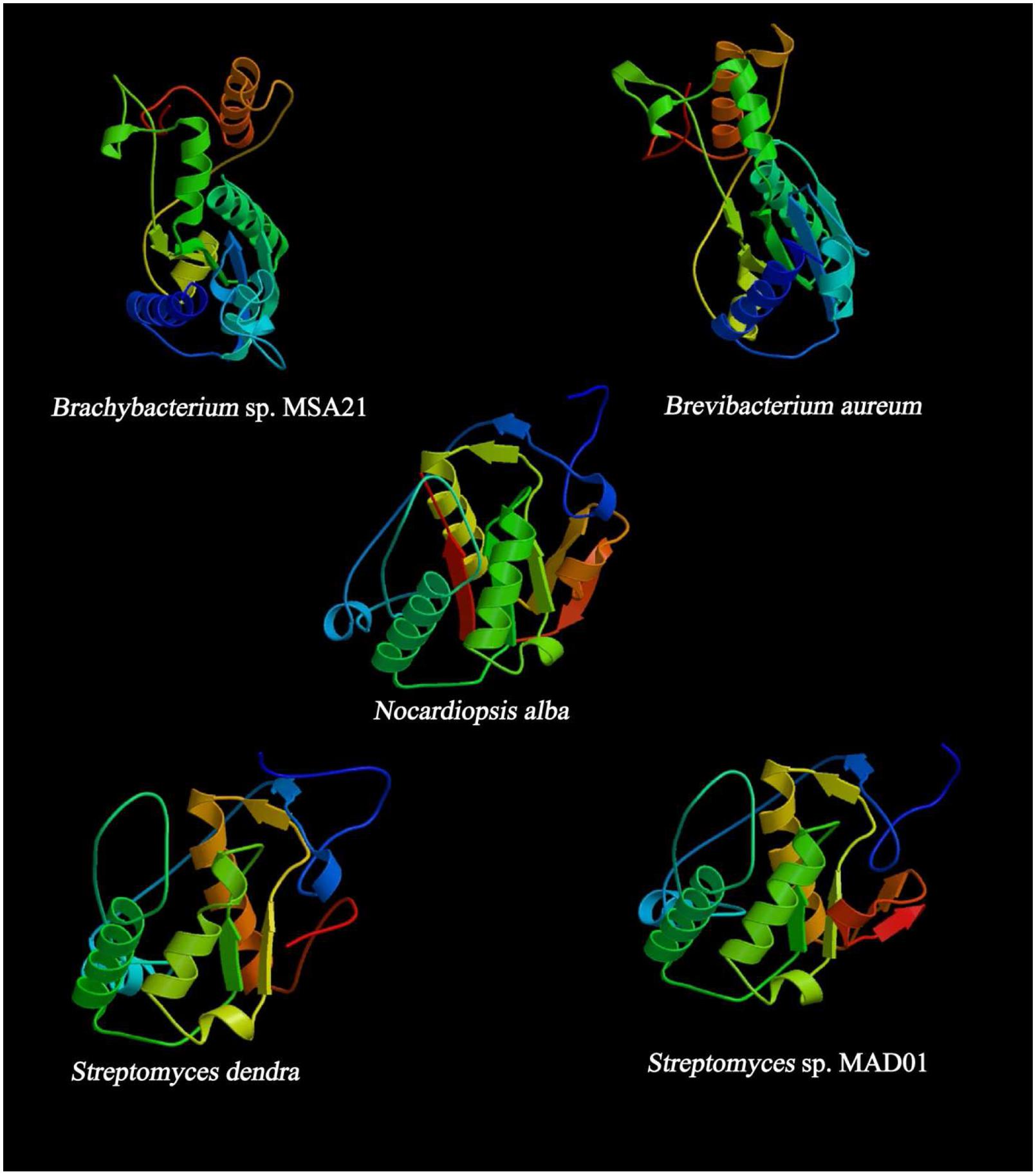
FIGURE 5. Predicted secondary structure of KS domain using (PS)2: Protein Structure Prediction Server.
Outside of the module, the beta-KAS domains are dimeric. However, the number of domains within the module is dimeric still remains to be established (Tsai et al., 2001, 2002; Broadhurst et al., 2003). Perhaps every enzyme within the module made contacts across the ser and cys, ACP suppose to diffuse farther than the peptide linkers on each side would permit (Keatinge-Clay and Stroud, 2006). The deduced quaternary structure of the proteins indicates a surprising configuration which is homologous to many PKS genes that are capable of synthesizing active polyketides. Even alignment of KS domains of our sequence of interest shows 53–62% of similarity with structures like amphotericin, ACT, epothilone, meagalomycin, myxalomycin, and rifamycin.
Most of the KS are dimeric with active site at the interface of dimer and type II PKS probably functions by making contact across the twofold axis and the active sites of KS are accessible to ACP (Keatinge-Clay and Stroud, 2006). In the present findings, we observed the PKC domain is common in all the protein sequences and lack AT domain. The analysis showed the chances of inactive enzymes within the modules may perform some important functions. The ACP module is bound by peptide linkers on both ends, and this module can pass between each enzyme in the module as well as the next KS or thiolase C and N terminal (Perham, 2000). The linkers helps to prevent a polyketide from interacting with enzymes and contribute little translational freedom to the polyketide compared to the peptide linkers on both ends of ACP. Thus helps in the biosynthesis of polyketides (Keatinge-Clay and Stroud, 2006). The interaction of ACP with the KS domain facilitate to docks in a deep groove which is formed by the interaction of the KS, PKC, and the other linker, thereby implicating both the PKC and the thioesterase linker in functional KS-ACP recognition (Liu et al., 2002). The KS domain of type II PKS (N. alba) ACS45380.1 was closely related to those of ACT PKS. Type II PKS (N. alba) consists of seven structural domains includes Asn Glycosylation, CK2 Phospho site, PKC, Tyr Phosphosite, ACP and KAS (49–161 AA residues) which shares 59% similarity with ACT polyketide putative beta-KAS 2 which contains eight chains, out of which two chains are homologous to our PKS protein which are chain A: beta-KAS/acyl transferase and chain B: ACT polyketide putative beta-KAS 2. The synthesis of aromatic polyketides are mostly begins with the formation of a polyketide chain (Keatinge-Clay and Stroud, 2006). The polymeric chains of type II PKS are tailored by the heterodimeric ketosynthase-chain length factor (KS-CLF). KS-CLF is the homolog of KS domain of type II PKS of N. alba which regulates chain length by catalyzing both chain initiation and elongation. Exploration of the mechanistic details of this central PKS polymerase may support designing and reconstruction of pathways being invented on synthetic biology platforms. This protein was structurally elucidated with four alpha helix and seven beta sheets. And it is slightly acidic composed of 39.79% of aliphatic (G,A,V,L,I), 18.32% of Acidic (B,D,E,N,Q,Z), 15.18% of basic (K,R,H), 3.66% of sulfur (C,M), 3.66% of aromatic (F,W,Y), and 12.04% of aliphatic hydroxyl (S,T).
Type II PKS of (B. aureum) ACS45381.1 is sequentially identical to type I ketosynthase (Streptomyces sp. T12-208) ACR61389.1. Structurally it is similar to the human fatty acid synthase (FAS), a modular enzyme involved in the metabolism of fatty acids and a drug target of antineoplastic and anti-obesity agents. Detailed structural study on human FAS has been limited due to its size and flexibility. Large part of human FAS that encompasses the tandem domain of beta-KAS is closely related to the KS domain of B. aureum. The KS domains are appear as the canonical dimer, and its substrate-binding site differs from that of bacterial homologs but is similar to type II PKS of B. aureum.
According to domain analysis, the PKS is a multi-domain protein consists of 14 domains includes ASN glycosylation, PKC, CK2, beta KAS, ACP synthase III, thiolase C and N terminal, and KAS C terminal domains. The position of KAS domain is 1–151 and 159–212 AA residues. The AA composition of the protein is predicted with 48.58% of aliphatic (G,A,V,L,I), 5.19% of aromatic (F,W,Y), 2.83% of sulfur (C,M), 9.43% of basic (K,R,H), 16.51% of acidic (B,D,E,N,Q,Z), and 14.15% of aliphatic hydroxyl (S,T). Type II PKS (Brachybacterium sp. MSA21) ACS45382.1 is identical to type I PKS of Streptomyces sp. and structurally proposed to contain five domains as follows KAS C and KAS N, myristyl site, PKC, and thiolase. The tertiary structure of the protein depicts six alpha helix and six beta sheets and composed of 50.45% of aliphatic (G,A,V,L,I), 5% of aromatic (F,W,Y), 2.73% of sulfur (C,M), 11.36% of basic (K,R,H), 13.18% of acidic (B,D,E,N,Q,Z), and 13.18% of aliphatic hydroxyl (S,T) AAs.
The position of the KS domain is 1–159 and 167–220 AA residues. In PSI- BLAST, PKS is predicted to have a structure similar to chain A, the ACT ketosynthase chain length factor since 73% identity, the E-value: 6.61e – 67, bit-score: 256, aligned-length: 173, this protein is structurally related to ACT ketosynthase and proposed to be rich in acidic and aliphatic AA residues, since the ligands may be acetyl group/magnesium ion/sodium ion. Six alpha helix and five beta sheets., and the composition is 41.04% of aliphatic (G,A,V,L,I), 6.94% of aromatic (F,W,Y), 3.47% of sulfur (C,M), 13.87% of basic (K,R,H), 19.08% of acidic (B,D,E,N,Q,Z), and 10.40% of aliphatic hydroxyl (S,T). The ketosynthase (Streptomyces dendra) ABP57802.1 found to have seven domains includes CKII phosphorylation site, PKC, KS C terminal and N terminal, phage tail fiber repeat and the AA composition is 42.26% of aliphatic (G,A,V,L,I), 4.76% of aromatic (F,W,Y), 4.17% of (sulfur C,M), 13.69% of basic (K,R,H), 18.45% of acidic (B,D,E,N,Q,Z) and 11.90% of aliphatic hydroxyl (S,T). The structural configuration presents six alpha helix and four beta sheets and mimics the structure of chain A. The ACT ketosynthase CLF with the values as follows, E-value: 1.00e – 72, bit-score: 258, aligned-length: 191, and identity to query: 67%. The PSI-BLAST shares 99% of similarity with doxorubicin PKS (E-value 4e – 75), 3-oxoacyl-ACP synthase I (Streptomyces avermitilis MA-4680), putative ketosynthase of Streptomyces antibioticus with (E-value 4e – 60), and granaticin polyketide putative beta-KAS 1 of Streptomyces hygroscopicus ATCC 53653 (E-value 3e – 60). KS domain analysis of type II PKS and ketosynthase was performed using PSI BLAST and MEGA (CLUSTAL W2) to highlight the unique conservative motif of each protein. The strain B. aureum shares specific motif with BAH67362.1 (PKS Streptomyces minoensis) denoted as “VDTACSSSLVALHLAAQALRSG.” Comparative analysis of KS domain of Brachybacterium sp. MSA21 exhibit the presence of unique motif PQQR(H)L in all the reference sequences which are capable of synthesizing cirramycin (BAH67190), minomycin (BAH67362), maridomycin (BAH67036), an anticancer compound (BAH67464), and platiomycin (BAH67144). This is the first report on the possible structural diversity mediated by type II PKS in Brachybacterium sp. MSA21.
KS Domain Phylogeny
Ketosynthase domain phylogeny was used to infer the phylogeny of type II PKS and ketosynthase. Phylogenetic analysis showed that the sponge associated actinobacterial sequences of PKS II genes and KS fragments were matched to conserved regions of previously characterized functional domains of other PKS I, II, and ketosynthase proteins. The KS domain of Brachybacterium and Brevibacterium showed a unique clustering, found KS domain of Brevibacterium clustering between two Streptomyces group and each group having two isolates and they possess high similarities among them like 100 and 98, respectively, but having less homology with B. aureum (Figure 2). The Brachybacterium was closely clustering with S. albus J1074 with 100% similarity. N. alba was not clustered with any actinobacterial KS domain since it was having the unique identity with KS domain of Streptomyces MAD01 and S. dendra. KS domain of Streptomyces MAD01 showing 97% of similarities with S. purpeofuces NBRC 14457 and S. dendra was clustering between Streptomyces sp. JS-14 and S. macrosporeus with 98 and 97% similarity, respectively. From the cluster analysis, we observed that two different marine sponge-associated actinobacteria possessing the identical KS domains of iterative PKS.
The major aim of this study was to find out the gene diversity of the PKS II and ketosynthase in two marine sponge associated actinobacterial polulation. The KS gene diversity could be useful to understand the evolution pattern of actinobacteria in the marine sponges, mode of interaction between sponge and associated microbes (Selvin, 2009) and chemical diversity of PKS II in marine sponge. Phylogenetic analysis of iterative PKS sequences is highly correlated with the number of iterations they performs. The PKS gene analysis provide a new insights that the poorly studied genera, such as Brevibacterium and Brachybacterium represent the KS genes which proves the unexplored resource for natural-product discovery. Conversely, the nearly ubiquitous detection of PKS genes in Streptomyces and Nocardiopsis envisages the possibility similar kind of natural products, but in reality the compounds are expected to be highly complex with diverse bioactivities (Selvin, 2009; Selvin et al., 2009b). To overcome these challenges, KS domain of PKS genes retrieved and analyzed in this study. KS domains tend to cluster phylogenetically based on the secondary metabolites of the actinobacterium from which the gene was retrieved. The active KS domains predictions could be based simply on the analysis of around 500 bp regions of KS domain from single PKS gene. The level of KS sequence domain in the iterative biosynthesis of natural products needs to be determined. The level of KS domain in strains may differ as it depends on the rate of sequence evolution, niche selectivity, host evolution pattern, and the time of pathways have been isolated in the respective genomes. It is also of interest that the three KS sequences associated with iterative type II PKS pathways were widely distributed among diverse taxonomic groups. The fact that all these KS domain sequences display relatively low levels of identity to the ketosynthase domain of Streptomyces MAD01 and S. dendra suggests that they are not associated with the production of iterative type of PKS domains. The mixed clustering of different sponge associated KS domains already been documented here for the first time we are reporting the evolutionary relatedness of KS domains of type II PKS and ketosynthase from D. nigra and F. cavernosa isolates. According to recent literatures, the PKS genes and their products exhibit novel insights in antimicrobial drug discovery (Selvin, 2009; Sasso et al., 2014; Wang et al., 2014). KS domain of type II PKS phylogeny is also highly need to know their relationship and structural diversity.
Potential Linkage Between Iterative Type II PKS Gene and Lipopeptide and Glycolipid Biosurfactant Production in Marine Sponge-Associated Actinobacteria
The marine sponge-associated bacteria have been recognized as rich source of biological macromolecules that are of potential interest to various industrial sectors (Kiran et al., 2015). Study reports evidenced that marine actinobacteria are unexplored resource for biosurfactant production. In this study, three actinobacterial strains (MSA10, MSA13, and MSA21) isolated from marine sponge were able to produce lipopeptide and glycolipid biosurfactants, respectively and showed positive for type II PKS gene. Two actinobacterial strains (MAD01 and MSI051) from D. nigra failed to produce biosurfactants but has the capability to synthesis polyketide based antagonistic compounds. There is an existing evidence for the synthesis of lipopeptide biosurfactant in Bacillus subtilis by NRPSs or hybrid PKS/NRPSs (Ongena and Jacques, 2008). These modular proteins in marine sponge associated microbes are responsible for the biosynthesis of several bioactive metabolites. They are mega-enzymes structured by iterative functional units called modules catalyzes various condensation, reduction, transferase reactions leading to polyketide and peptide transformation for the synthesis of biosurfactant. The positive strains display biosurfactant activity and, significantly, iterative type II PKS domain gene fragments, indicating the existence of a PKS gene cluster associated with biosurfactive compound biosynthesis. The present study reveals that the actinobacteria are a rich source of bioactive compounds and biosurfactant, and also represent the unrecognized group of organisms having type II PKS systems for polyketide biosynthesis. In this study, the bacterial motility was also checked for the surfactive compound production (data not shown). Bacterial motility mechanisms, comprising swimming, swarming, and twitching, are known to have significant roles in biofilm formation, colonization, and the subsequent expansion into complete organized surface populations. All the actinobacterial strains showed positive in swimming, swarming, and twitching motility assays which indicate that these strains possess biofilm forming ability. The strains with increased swimming motility also possess good swarming ability. Current research evidenced that the strain Streptomyces sp. MAD01 possess good biofilm forming capacity as well as antimicrobial activity against test organisms. It also proves that a ketosynthase type II PKS system is responsible for the biosynthesis of the antagonistic compounds in marine actinobacteria. Based on the present findings, the production of biosurfactants might be linked with type II iterative PKS gene cluster and the synthesis of biosurfactant by the sponge-associated actinobacteria might have significant role in the chemical ecology of host sponge (Kiran et al., 2015). However, the hypothesis has not been tested in controlled in vitro and in vivo experiments. Based on the functions of biosurfactants including antibacterial/antibiofilm activity, the biosurfactants may play a role in host defense fouling processes (Gandhimathi et al., 2009; Kiran et al., 2014). Therefore, explorations of marine sponge-associated actinobacteria for the lipopeptide and glycolipid biosurfactant production will have wider applications in industrial processes, bioremediation, and enhanced oil recovery.
Conclusion
In the current study, the iterative nature of actinobacterial type II PKS was proved by HMM profile. The domain architecture of N. alba and B. aureum have the potential of constructing “minimal PKS” and the later species share the specific motif “VDTACSSS” with S. minoensis. Both strains displayed PKS domains structurally similar with encoding ACT. S. dendra is found to have a unique repeat called phage tail fiber repeat which is responsible for altering the host specificity of secondary metabolites through protein–protein interaction. The other three actinobacterial strains Brachybacterium sp., Streptomyces sp., and S. dendra lack ACP results in inactive minimal PKS or may act non-iteratively. This study also provides a new insight on the KS genes of Brevibacterium and Brachybacterium proving that marine resources are still largely unexplored for natural-product discovery. In these regards, in silico gene mining is quite useful for prospecting novel metabolites produced by marine sponge endosymbionts. Further in vitro studies are needed to design novel natural products using a biosynthetic engineering approach.
Author Contributions
JS and GSK designed the experiments, GS performed in silico analysis, ANL performed in vitro assays, NAA and MV performed data analysis and interpretation, GSK, GS, and JS written the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
GK is thankful to the Department of Biotechnology (DBT), Govt. of India for a research grant. JS is thankful to Department of Science and Technology (DST). Authors of KSU acknowledged Deanship of Scientific Research at King Saud University for funding this Prolific Research Group (PRG-1437-28).
Footnotes
References
Bibb, M. J., Biró, S., Motamedi, H., Collins, J. F., and Hutchinson, C. R. (1989). Analysis of the nucleotide sequence of the Streptomyces glaucescens tcmI genes provides key information about the enzymology of polyketide antibiotic biosynthesis. EMBO J. 8, 2727–2736.
Broadhurst, R. W., Nietlispach, D., Wheatcroft, M. P., Leadlay, P. F., and Weissman, K. J. (2003). The structure of docking domains in modular polyketide synthases. Chem. Biol. 10, 723–731. doi: 10.1016/S1074-5521(03)00156-X
Chen, C.-C., Hwang, J.-K., and Yang, J.-M. (2006). (PS)(2):protein structure prediction server. Nucleic. Acids Res. 34, W152–W157. doi: 10.1093/nar/gkl187
De Carvalho, C. C. C. R., and Fernandes, P. (2010). Production of metabolites as bacterial responses to the marine environment. Mar. Drugs 8, 705–727. doi: 10.3390/md8030705
Faulkner, D. J., and Ghiselin, M. T. (1983). Chemical defense and evolutionary ecology of dorid nudibranchs and some other opisthobranch gastropods. Mar. Ecol. Prog. ser. 13, 295–301. doi: 10.3354/meps013295
Faulkner, D. J., Unson, M. D., and Bewley, C. A. (1994). The chemistry of some sponges and their symbionts. Pure Appl Chem. 66, 1983–1990. doi: 10.1351/pac199466101983
Fernández-Moreno, M. A., Martínez, E., Boto, L., Hopwood, D. A., and Malpartida, F. (1992). Nucleotide sequence and deduced functions of a set of cotranscribed genes of Streptomyces coelicolor A3(2) including the polyketide synthase for the antibiotic actinorhodin. J. Biol. Chem. 267, 19278–19290.
Gandhimathi, R., SeghalKiran, G., Hema, T. A., Selvin, T. A., RajeethaRaviji, T., and Shanmughapriya, S. (2009). Production and characterization of lipopeptide biosurfactant by a sponge-associated marine actinomycetes Nocardiopsis alba MSA10. Bioprocess. Biosyst. Eng. 32, 825–835. doi: 10.1007/s00449-009-0309-x
He, J., and Hertweck, C. (2003). Iteration as programmed event during polyketide assembly; molecular analysis of the aureothin biosynthesis gene cluster. Chem. Biol. 10, 1225–1232. doi: 10.1016/j.chembiol.2003.11.009
Hentschel, U., Hopke, J., Horn, M., Friedrich, A. B., Wagner, M., Hacker, J., et al. (2002). Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68, 4431–4440. doi: 10.1128/AEM.68.9.4431-4440.2002
Keatinge-Clay, A. T., and Stroud, R. M. (2006). The Structure of a ketoreductase determines the organization of the β-Carbon processing enzymes of modular polyketide synthases. Structure 14, 737–748. doi: 10.1016/j.str.2006.01.009
Kim, T. K., and Fuerst, J. A. (2006). Diversity of polyketide synthase genes from bacteria associated with the marine sponge Pseudoceratina clavata: culture-dependent and culture-independent approaches. Environ. Microbiol. 8, 1460–1470. doi: 10.1111/j.1462-2920.2006.01040.x
Kiran, G. S., Anto Thomas, T., Selvin, J., Sabarathnam, B., and Lipton, A. P. (2010). Optimization and characterization of a new lipopeptidebiosurfactant produced by marine Brevibacterium aureum MSA13 in solid state culture. Bioresour. Technol. 101, 2389–2396. doi: 10.1016/j.biortech.2009.11.023
Kiran, G. S., Ninawe, A. S., Lipton, A. N., Pandian, V., and Selvin, J. (2015). Rhamnolipid biosurfactants: evolutionary implications, applications and future prospects from untapped marine resource. Crit. Rev. Biotechnol. doi: 10.3109/07388551.2014.979758 [Epub ahead of print].
Kiran, G., Sabarathnam, B., Thajuddin, N., and Selvin, J. (2014). Production of glycolipid biosurfactant from sponge-associated marine actinobacterium brachybacterium paraconglomeratum MSA21. J. Surfactants. Deterg. 17, 531–542. doi: 10.1007/s11743-014-1564-7
Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5, 150–163. doi: 10.1093/bib/5.2.150
Li, W. L., Yi, Y. H., Wu, H. M., Xu, Q. Z., Tang, H. F., Zhou, D. Z., et al. (2002). Isolation and structure of the cytotoxic cycloheptapeptide phakellistatin 13. J. Nat. Prod. 66, 146–148. doi: 10.1021/np020223y
Liu, W., Christenson, S. D., Standage, S., and Shen, B. (2002). Biosynthesis of the enediyne antitumor antibiotic C-1027. Science 297, 1170–1173. doi: 10.1126/science.1072110
Matsunaga, S., and Fusetani, N. (2003). Nonribosomal peptides from marine sponges. Curr. Org. Chem. 7, 945–966. doi: 10.2174/1385272033486648
Matsunaga, S., Fusetani, N., and Konosu, S. (1985). Bioactive marine metabolites, IV. isolation and the amino acid composition of discodermin a, an antimicrobial peptide, from the marine sponge discodermiakiiensis. J. Nat. Prod. 48, 236–241. doi: 10.1021/np50038a006
Montalvo, N. F., and Hill, R. T. (2011). Sponge-associated bacteria are strictly maintained in two closely related but geographically distant sponge hosts. Appl. Environ. Microbiol. 77, 7207–7216. doi: 10.1128/AEM.05285-11
Montalvo, N. F., Mohamed, N. M., Enticknap, J. J., and Hill, R. T. (2005). Novel actinobacteria from marine sponges. Antonie Van Leeuwenhoek 87, 29–36. doi: 10.1007/s10482-004-6536-x
Ongena, M., and Jacques, P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trend. Microbiol. 16, 115–125. doi: 10.1016/j.tim.2007.12.009
Patton, S. M., Ashton, T., Cropp, A., and Reynolds, K. A. (2000). A novel Δ3,Δ2 enoyl CoA isomerase involved in the biosynthesis of the cyclohexanecarboxylic acid-derived moiety of the polyketide ansatrienin A. Biochemistry 39, 7595–7604. doi: 10.1021/bi0005714
Perham, R. N. (2000). Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu. Rev. Biochem. 69, 961–1004. doi: 10.1146/annurev.biochem.69.1.961
Revill, W. P., Bibb, M. J., and Hopwood, D. A. (1995). Purification of a malonyltransferase from Streptomyces coelicolor A3(2) and analysis of its genetic determinant. J. Bacteriol. 177, 3946–3952.
Sasso, S., Shelest, E., and Hoffmeister, D. (2014). Comments on the distribution and phylogeny of type I polyketide synthases and nonribosomal peptide synthetases in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 111, E3946. doi: 10.1073/pnas.1412766111
Selvin, J. (2009). Exploring the antagonistic producer Streptomyces MSI051: implications of polyketide synthase gene Type II and a ubiquitous defense enzyme phospholipase a2 in the host sponge Dendrilla nigra. Curr. Microbiol. 58, 459–463. doi: 10.1007/s00284-008-9343-1
Selvin, J., Gandhimathi, R., Seghal Kiran, G., Shanmughapriya, S., Rajeetha Ravji, T., and Hema, T. A. (2009a). Culturable heterotrophic bacteria from the marine sponge Dendrilla nigra: isolation and phylogenetic diversity of actinobacteria. Helgol. Mar. Res. 63, 239–247. doi: 10.1007/s10152-009-0153-z
Selvin, J., Shanmughapriya, S., Gandhimathi, R., Seghal Kiran, G., Rajeetha Ravji, T., Natarajaseenivasan, K., et al. (2009b). Optimization and production of novel antimicrobial agents from sponge associated marine actinomycetes Nocardiopsis dassonvillei MAD08. Appl. Microbiol. Biotechnol. 83, 435–445. doi: 10.1007/s00253-009-1878-y
Shen, B. (2003). Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr. Opin. Chem. Biol. 7, 285–295. doi: 10.1016/S1367-5931(03)00020-6
Shen, B., and Kwon, H.-J. (2002). Macrotetrolide biosynthesis: a novel type II polyketide synthase. Chem. Rec. 2, 389–396. doi: 10.1002/tcr.10042
Shen, B., Leeper, F., and Vederas, J. (eds) (2000). Biosynthesis of Aromatic Polyketides. In Biosynthesis. Berlin: Springer, 1–51.
Summers, R. G., Wendt-Pienkowski, E., Motamedi, H., and Hutchinson, C. R. (1992). Nucleotide sequence of the tcmII-tcmIV region of the tetracenomycin C biosynthetic gene cluster of Streptomyces glaucescens and evidence that the tcmN gene encodes a multifunctional cyclase-dehydratase-O-methyl transferase. J. Bacteriol. 174, 1810–1820.
Traba, C., and Liang, J. F. (2011). Susceptibility of Staphylococcus aureus biofilms to reactive discharge gases. Biofouling 27, 763–772. doi: 10.1080/08927014.2011.602188
Tsai, S.-C., Lu, H., Cane, D. E., Khosla, C., and Stroud, R. M. (2002). Insights into channel architecture and substrate specificity from crystal structures of two macrocycle-forming thioesterases of modular polyketide synthases. Biochemistry 41, 12598–12606. doi: 10.1021/bi0260177
Tsai, S.-C., Miercke, L. J. W., Krucinski, J., Gokhale, R., Chen, J. C. H., Foster, P. G., et al. (2001). Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: versatility from a unique substrate channel. Proc. Natl. Acad. Sci. U.S.A. 98, 14808–14813. doi: 10.1073/pnas.011399198
Wang, H., Zhang, H., Zou, Y., Mi, Y., Lin, S., Xie, Z., et al. (2014). Structural insight into the tetramerization of an iterative ketoreductasesiam through aromatic residues in the interfaces. PLoS ONE 9:e97996. doi: 10.1371/journal.pone.0097996
Watve, M., Tickoo, R., Jog, M., and Bhole, B. (2001). How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 176, 386–390. doi: 10.1007/s002030100345
Yadav, G., Gokhale, R. S., and Mohanty, D. (2009). Towards prediction of metabolic products of polyketide synthases: an in silico analysis. PLoS Comput. Biol. 5:e1000351. doi: 10.1371/journal.pcbi.1000351
Keywords: glycolipid, lipopeptide, biosurfactant, polyketide synthases, actinobacteria, three-dimensional structure
Citation: Selvin J, Sathiyanarayanan G, Lipton AN, Al-Dhabi NA, Valan Arasu M and Kiran GS (2016) Ketide Synthase (KS) Domain Prediction and Analysis of Iterative Type II PKS Gene in Marine Sponge-Associated Actinobacteria Producing Biosurfactants and Antimicrobial Agents. Front. Microbiol. 7:63. doi: 10.3389/fmicb.2016.00063
Received: 28 July 2015; Accepted: 14 January 2016;
Published: 12 February 2016.
Edited by:
Syed Gulam Dastager, CSIR-National Chemical Laboratory, IndiaReviewed by:
Virginia Helena Albarracín, Centro Integral de Microscopía Electrónica, ArgentinaKarthik Loganathan, Shanghai Jiao Tong University, China
Copyright © 2016 Selvin, Sathiyanarayanan, Lipton, Al-Dhabi, Valan Arasu and Kiran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: George S. Kiran, c2VnaGFsa2lyYW5AZ21haWwuY29t
†Present address: Ganesan Sathiyanarayanan, Department of Biological Engineering, College of Engineering, Konkuk University, Seoul 143-701, South Korea
 Joseph Selvin
Joseph Selvin Ganesan Sathiyanarayanan
Ganesan Sathiyanarayanan Anuj N. Lipton1,2
Anuj N. Lipton1,2 Mariadhas Valan Arasu
Mariadhas Valan Arasu George S. Kiran
George S. Kiran