- 1Unit of Microbiology, Belgian Nuclear Research Centre (SCK•CEN), Mol, Belgium
- 2Laboratory of Food Microbiology and Leuven Food Science and Nutrition Research Centre, Department of Microbial and Molecular Systems, Faculty of Bioscience Engineering, Katholieke Universiteit Leuven, Leuven, Belgium
Bacteria can respond to adverse environments by increasing their genomic variability and subsequently facilitating adaptive evolution. To demonstrate this, the contribution of Insertion Sequence (IS) elements to the genetic adaptation of Cupriavidus metallidurans AE126 to toxic zinc concentrations was determined. This derivative of type strain CH34, devoid of its main zinc resistance determinant, is still able to increase its zinc resistance level. Specifically, upon plating on medium supplemented with a toxic zinc concentration, resistant variants arose in which a compromised cnrYX regulatory locus caused derepression of CnrH sigma factor activity and concomitant induction of the corresponding RND-driven cnrCBA efflux system. Late-occurring zinc resistant variants likely arose in response to the selective conditions, as they were enriched in cnrYX disruptions caused by specific IS elements whose transposase expression was found to be zinc-responsive. Interestingly, deletion of cnrH, and consequently the CnrH-dependent adaptation potential, still enabled adaptation by transposition of IS elements (ISRme5 and IS1086) that provided outward-directed promoters driving cnrCBAT transcription. Finally, adaptation to zinc by IS reshuffling can also enhance the adaptation to subsequent environmental challenges. Thus, transposition of IS elements can be induced by stress conditions and play a multifaceted, pivotal role in the adaptation to these and subsequent stress conditions.
Introduction
Bacteria continuously evolve to survive different environmental challenges and the field of evolutionary biology has long been interested in the interplay between generation of genetic diversity and natural selection favoring the best adapted organism. The central dogma that evolution proceeds through natural selection of heritable mutations was first proposed by Darwin (Darwin, 1910). In addition, however, Darwin also suggested that environmental stress might generate a diversity of genetic variants, upon which subsequent natural selection works (Darwin, 1910). The claim that mutation rate might be influenced by stress is, however, still under debate (Rosenberg and Hastings, 2004; Roth, 2011). The classic experiments of Luria and Delbrück (1943) and Lederberg (1989) demonstrated for the first time that mutations can arise without the influence of selective stress. However, the lethal selections they used could likely not have detected mutations induced by selective conditions, and others have described genetic systems in which selective conditions indeed seemed to increase the mutation rate (Shapiro, 1984; Cairns et al., 1988; Hall, 1990; Foster, 2007; MacLean et al., 2013; Ram and Hadany, 2014). In addition, the coupling of potential mutation-generating systems to local events such as stress-induced transcription of a specific subset of genes (Wright, 2004) might form an example of stress-directed mutation where the mutation is specifically induced by the stress condition that the mutation relieves (Cairns et al., 1988). However, non-selected mutations readily occur during selective conditions disputing the existence of non-random mutations (Foster, 2007).
One frequently encountered type of mutation results from the hopping of transposable elements from the donor to the target site, potentially leading to important phenotypic changes (Mahillon and Chandler, 1998; Mahillon et al., 1999). Insertion sequence (IS) elements, which are widely distributed in the genomes of bacteria as well as higher organisms (Siguier et al., 2006, 2014), are of interest as they constitute an important driving force for genome plasticity (Kazazian, 2004). On the one hand, intra-genomic transposition of these small elements (<2.5 kb), which generally encode no other functions than those involved in their mobility, can both inactivate or activate adjacent genes by physically interrupting coding sequences or encoding regulatory sequences, respectively. The activation can be caused by a promoter encoded by the element and directed outward to the adjacent gene (Ciampi et al., 1982; Zhang and Saier, 2009; Wang and Wood, 2011; Saier and Zhang, 2014) or by the formation of a hybrid promoter in which the −35 region at the end of the IS element is correctly situated close to the −10 region of the adjacent gene (Jaurin and Normark, 1983; Prentki et al., 1986). On the other hand, homologous IS elements within the chromosome can become substrates of the cell's recombination machinery, which can result in complex chromosomal rearrangements (e.g., inversions, duplications, or deletions) depending on the orientation of the involved IS elements (Louarn et al., 1985; Schneider et al., 2000a; Parkhill et al., 2003).
Selective conditions can promote the activation and redistribution of IS elements that exist in the chromosome. Indeed, stress factors were found to induce the transposition of mobile genetic elements (MGEs) in plants (McClintock, 1984; Studer et al., 2011), flies (González et al., 2010), yeast (Stanley et al., 2010), and bacteria (Ohtsubo et al., 2005; Drevinek et al., 2010). The acquisition of phenotypic traits caused by transposition of IS elements has been well-documented, particularly in bacteria. Different conditions such as UV light, low frequency magnetic fields or sub-inhibitory concentrations of antibiotics were shown to promote transposition of IS elements in Escherichia coli (Eichenbaum and Livneh, 1998; Del Re et al., 2003; Lartigue et al., 2006). Transposition of IS elements in response to a selective condition has also been described in other gram-negative genera, e.g., Pseudomonas and Burkholderia in response to high temperature, conjugation or oxidative stress (Ohtsubo et al., 2005; Christie-Oleza et al., 2009; Drevinek et al., 2010). Active redistribution of IS elements was also observed in the gram-positive Deinococcus radiodurans after exposure to radiation (Pasternak et al., 2010) and in the gram-positive Bacillus subtilis under competence-inducing conditions (Takahashi et al., 2007).
Cupriavidus metallidurans strains are, next to metal-contaminated soils (Diels and Mergeay, 1990; Brim et al., 1999; Goris et al., 2001), being increasingly recovered from other anthropogenic environments not typified by metal contamination (Van Houdt et al., 2012; Mijnendonckx et al., 2013), including medically-relevant sources (Coenye et al., 2005; Langevin et al., 2011; D'Inzeo et al., 2015). They are studied for their resistance and adaptation to toxic levels of metal ions. Type strain C. metallidurans CH34 counteracts metal ion toxicity via a battery of resistance mechanisms, including transporters belonging to the Resistance Nodulation cell Division (RND), the cation diffusion facilitator (CDF) and the P-type ATPase families, with an important role for its two mega-plasmids as most of the heavy metal resistance mechanisms are encoded on either pMOL28 or pMOL30 (Mergeay et al., 1985, 2003; Nies and Silver, 1995; Nies, 1999, 2003; Monchy et al., 2007; von Rozycki and Nies, 2009; Janssen et al., 2010). In addition, C. metallidurans CH34 harbors 21 distinct IS elements belonging to 10 different IS families (Mijnendonckx et al., 2011). Out of the 21 distinct IS elements, active transposition has been observed for ISRme1, ISRme3, IS1086, IS1087B, IS1088, and IS1090 (Dong et al., 1992; Collard et al., 1993; Grass et al., 2000; Ma-e-Talat, 2000; Schneider et al., 2000b; Tibazarwa et al., 2000). In fact, active involvement of IS1087B in metal resistance has been shown for C. metallidurans AE126, which is a derivative of CH34 sensitive to zinc as it only carries plasmid pMOL28 and not plasmid pMOL30 (Collard et al., 1993; Tibazarwa et al., 2000). Spontaneous zinc-resistant AE126 mutants constitutively expressing cnrCBAT coding for the RND-driven efflux system CnrCBAT appear when AE126 is grown in the presence of 0.8 mM Zn2+ (Collard et al., 1993; Grass et al., 2000; Tibazarwa et al., 2000). The cnrCBAT operon is regulated by the upstream cnrYXH locus coding for the membrane-bound anti-sigma factor CnrY, the sensor protein CnrX, and the ECF (Extracytoplasmic Function) family sigma factor CnrH. The latter is released from the CnrYX transmembrane anti-sigma factor complex in the presence of inducers (Ni2+ or Co2+) and subsequently promotes transcription of both cnrYXH and cnrCBAT (Grosse et al., 2007; Trepreau et al., 2011, 2014; Maillard et al., 2015). Analysis of two of such derivatives indicated that the inactivation of the gene coding for the anti-sigma factor CnrY (by IS1087B and a frameshift mutation), and concomitant release of CnrH increased transcription of the structural cnrCBAT cluster and led to increased (non-specific) Zn2+ efflux (Collard et al., 1993; Grass et al., 2000; Tibazarwa et al., 2000).
These observations indicate that IS transposition may play a dynamic role in the genetic adaptation of C. metallidurans AE126 to toxic zinc concentrations. Therefore, in this study, we thoroughly scrutinized the mutations leading to, and the genetic determinants essential for increased zinc resistance in AE126. The involvement of IS elements was dissected via the identification of the concerned IS elements, their target sites, and the effect elicited by their transposition as well as via analysis of transposase expression upon metal ion exposure.
Materials and Methods
Strains, Media, and Culture Conditions
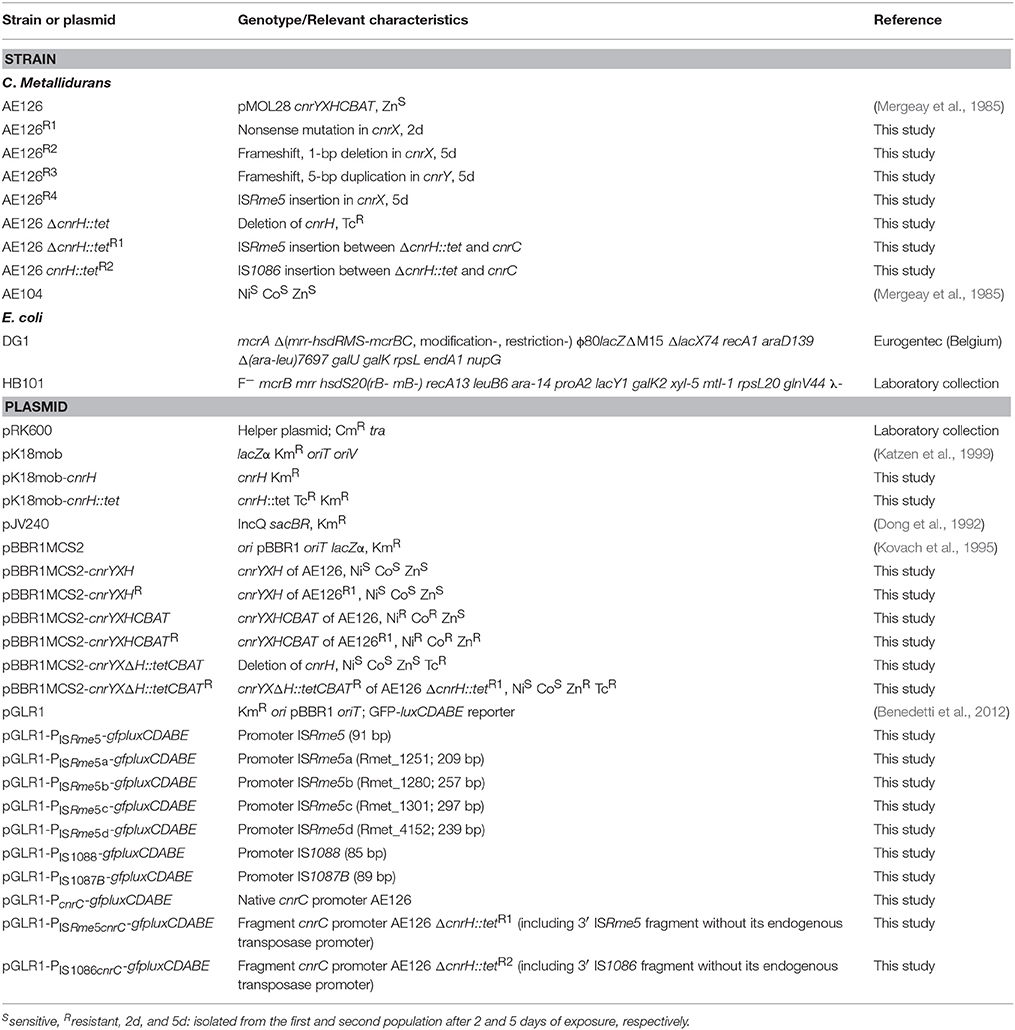
Bacterial strains and plasmids used in this study are listed in Table 1. C. metallidurans strains were routinely cultured at 30°C in Tris-buffered mineral medium supplemented with 0.2% (w/v) sodium gluconate (MM284) as described previously (Mergeay et al., 1985). E. coli strains were routinely cultured at 37°C in Lysogeny Broth (LB). Liquid cultures were grown in the dark on a rotary shaker at 150 rpm, for culturing on agar plates 2% agar (Thermo Scientific, Oxoid) was added. When appropriate, the following chemicals (Sigma-Aldrich or Thermo Scientific) were added to the growth medium at the indicated final concentrations: kanamycin [50 μg/ml for E. coli (Km50) or 1500 μg/ml for C. metallidurans (Km1500)], tetracycline (20 μg/ml), chloramphenicol (30 μg/ml), Zn2+ (0.1 or 0.8 mM; as zinc sulfate heptahydrate), Ni2+ (0.1 mM; as nickel chloride hexahydrate), Cd2+ (0.01 mM; as cadmium chloride hemipentahydrate), Co2+ (0.05 mM; as cobalt chloride hexahydrate), 5-bromo-4-chloro-3-indolyl-β-galactopyranoside (X-Gal; 40 μg/ml), and isopropyl β-D-1-thiogalactopyranoside (IPTG; 0.1 mM).
Determination of Growth and Minimal Inhibitory Concentration
The susceptibility of C. metallidurans strains to different heavy metals was determined via minimal inhibitory concentrations (MICs). The strains were cultivated in triplicate by inoculating 2 ml MM284 supplemented with different concentrations of heavy metals with 20 μL of a stationary phase culture at 30°C. The MIC is defined as the lowest concentration of heavy metal that will inhibit visible growth of the culture after 2 days of incubation. Growth experiments were performed by 1:100 dilution of stationary phase cultures in fresh MM284 and subsequently for each sample (biological replicates), 200 μL was added to a 96-well white cell culture plate (Nunc Flat-bottom, Thermo Scientific) which was placed into a CLARIOstar® (BMG LABTECH). Plates were incubated at 30°C with shaking for 48 h and at 30-min time intervals, the optical density at 600 nm was measured. The growth data were fitted by the model of Baranyi and Roberts (1994) using the Microsoft Excel add-in package DMFit (Institute of Food Research, Norwich, United Kingdom). Statistical comparison of the growth curve parameters was performed using a one-way ANOVA analysis, followed by a post-hoc Tukey test.
Isolation of Zinc-Resistant Mutants
C. metallidurans AE126 was cultivated in MM284 at 30°C up to stationary phase, 109 cells were pelleted and cell suspensions (100 μL) of a serial ten-fold dilution in saline were spread on MM284 agar plates containing a final concentration of 0.8 mM Zn2+ and incubated at 30°C. Colony forming units (CFU) were counted after day 2 up to day 5 and survival frequency was calculated as viable cell count on MM284 0.8 mM Zn2+ agar plates divided by viable cell count on MM284 agar plates. The variance-to-mean ratio and mutation rate in a fluctuation assay with 20 independent cultures was calculated according to Luria and Delbrück (1943) and using Fluctuation Analysis CalculatOR (FALCOR) (Hall et al., 2009), respectively.
Analysis of IS Transposition
Transposition of IS elements in the regulatory locus of the cnr operon (Supplementary Figure 1) was identified by colony PCR (DreamTaq DNA polymerase, Thermo Scientific) with primers CnrFw and CnrRv (Supplementary Table 1) and sequencing. A sacBR IS trap (Gay et al., 1985; Dong et al., 1992) was used to compare the adaptation potential of AE126R4, which carries an ISRme5 copy inserted in cnrX in the same transcriptional orientation as cnrYX, with AE126. Sucrose-resistant derivatives of C. metallidurans AE126 and AE126R4 carrying pJV240 (Gay et al., 1985; Dong et al., 1992) were selected on LB plates without NaCl and supplemented with 10% (w/v) sucrose and 1500 μg/ml kanamycin. The insertion of ISRme5 in pJV240 was identified by colony PCR with primer pairs ISRme5_Fw and SacR_Rv or pISRme5_Rv and SacR_Rv, respectively (Supplementary Table 1).
Whole Genome Gene Expression Microarrays
The whole-genome expression profile of AE126R2 and AE126R3 were compared to its parental strain AE126 by whole genome oligonucleotide microarrays. The strains were cultivated by inoculating 30 ml of MM284 in biological triplicates with 300 μL of a stationary phase culture at 30°C. These subcultures were allowed to grow until an OD600-value of around 0.6 was reached. Next, cells were harvested by centrifugation for 2 min at 10,000 rpm and the bacterial pellets were flash frozen by immersion into liquid nitrogen. The bacterial pellets were kept frozen at −80 °C until total RNA extraction was performed using the SV Total RNA Isolation system (Promega Corporation). The quantity of extracted RNA was measured using a NanoDrop™ 1000 spectrophotometer (Thermo Scientific). The RNA quality was determined with a Bioanalyzer (Agilent 2100 Electrophoresis Bioanalyzer Agilent Technologies). Only extracted RNA with a RNA integrity number (RIN number) of higher than eight was accepted for further analysis (Schroeder et al., 2006). Twenty micrograms of RNA were reverse transcribed following the instructions provided with the Pronto kit (Promega). Microarray slide spotting, RNA labeling, hybridization, and data analysis were performed as previously described (Monsieurs et al., 2011). The full description of the microarray data have been deposited at the Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE74091.
cnrH Mutant Deletion Construction
The cnrH gene of C. metallidurans AE126 was amplified by PCR (Phusion High-Fidelity DNA polymerase) with primer pairs cnrH_Fw-Rv (Supplementary Table 1), providing HindIII/EcoRI restriction sites. Afterwards, these PCR products were cloned as a HindIII/EcoRI fragment into the mobilizable suicide vector pK18mob. The resulting pK18mob cnrH plasmid from an E. coli DG1 transformant selected on LB Km50 was further confirmed by sequencing prior to amplifying of the flanking sequences of cnrH by inverse PCR (Phusion High-Fidelity DNA polymerase) with primer pair cnrH_tet_Fw-Rv (Supplementary Table 1), providing BcuI/XbaI restriction sites. At the same time the tet gene from pACYC184 (Table 1; Chang and Cohen, 1978) was amplified by PCR (Phusion High-Fidelity DNA polymerase) with primer pair Tet_Fw-Rv (Supplementary Table 1), providing BcuI/XbaI restriction sites. Afterwards, this PCR product was cloned as a BcuI/XbaI fragment into the former inverse cnrH PCR product. The resulting pK18mob-cnrH::tet plasmid from an E. coli DG1 transformant selected on LB Tc20 Km50 was further confirmed by sequencing prior to conjugation (triparental with E. coli HB101 pRK600 as helper) to C. metallidurans AE126. The resulting transformants selected on MM284 Km1500 were replica plated on MM284 Tc20 and MM284 Km1500. AE126ΔcnrH::tet cells resistant to Tc20 but sensitive to Km1500 were further confirmed by sequencing.
Identification of cnrCBAT Transcription Start Site
The transcription start site of cnrCBAT in wild type AE126 and in the two zinc-resistant derivatives AE126ΔcnrH::tetR1 and AE126ΔcnrH::tetR2 were identified using the cRACE method (Maruyama et al., 1995; Dallmeier and Neyts, 2013). The strains were cultivated by inoculating 30 ml of MM284 with 300 μL of a stationary phase C. metallidurans AE126, AE126ΔcnrH::tetR1, or AE126ΔcnrH::tetR2 culture at 30°C. The cultures were allowed to grow until an OD600-value of around 0.6 was reached. Next, cells were harvested by centrifugation for 2 min at 10,000 rpm and the bacterial pellets were flash frozen by immersion into liquid nitrogen. The bacterial pellets were kept frozen at −80°C until total RNA extraction was performed using the SV Total RNA Isolation system (Promega Corporation). The quantity of extracted RNA was measured using a NanoDrop™ 1000 spectrophotometer (Thermo Scientific). The RNA quality was determined with a Bioanalyzer (Agilent 2100 Electrophoresis Bioanalyzer Agilent Technologies). Only extracted RNA with a RNA integrity number (RIN number) of higher than eight was accepted for further analysis (Schroeder et al., 2006). One microgram of RNA was reverse transcribed following the instructions provided with the GoScript™ Reverse Transcription System (Promega) with a gene-specific 5′-phosphorylated oligonucleotide cnrC_R1 (Supplementary Table 1). Subsequently, three volumes of TE buffer (10 mM Tris-HCl and 1 mM ethylenediaminetetraacetic acid, pH 8.0) containing 4 μg/ml RNase A (Promega) was added to the reaction. Successively, 28 μL of the resulting cDNAs was circularized by 20 U of T4 RNA ligase (Thermo Scientific) in the presence of 15% (w/v) polyethylene glycol (PEG4000; Thermo Scientific) in a total volume of 50 μL at 37°C for 60 min. Next 1.5 U of Pfu DNA polymerase (Thermo Scientific) was added to the reaction at 37°C for 30 min employing its 3′-5′ exonuclease activity to remove unreacted residual first-strand primers and cDNAs. Finally, a 2 μL aliquot was directly used as template for the amplification by PCR (DreamTaq DNA polymerase) with primer pair cnrC_F1-R2 (Supplementary Table 1), the PCR product was cloned by TOPO cloning (Thermo Scientific) and the transcription start site was identified by sequencing.
Construction Of Plasmids
The cnr locus (cnrYXHCBAT) and its regulatory part (cnrYXH) from C. metallidurans AE126 and the zinc-resistant derivative AE126R1 were amplified by PCR (Phusion High-Fidelity DNA polymerase) with primer pairs CnrIns_Fw-CnrIns_Rv and CnrIns_Fw-CnrT_Rv (Supplementary Table 1), respectively, providing SacI/BcuI restriction sites. Afterwards, these PCR products were cloned as a SacI/BcuI fragment into pBBR1MCS2. The resulting pBBR1MCS2-cnrYXH, pBBR1MCS2-cnrYXHR, pBBR1MCS2-cnrYXHCBAT, and pBBR1MCS2-cnrYXHCBATR plasmids from E. coli DG1 transformants selected on LB Km50 were further confirmed by sequencing prior to transformation to C. metallidurans AE104. In addition, the cnr locus (cnrYXΔH::tetCBAT) from C. metallidurans AE126 ΔcnrH::tet (see above) and the zinc-resistant derivative AE126 ΔcnrH::tetR1 (Table 1) were amplified by PCR (Phusion High-Fidelity DNA polymerase) with primer pair Cnr_Fw-RvpBBR1MCS2 (Supplementary Table 1). At the same time, the pBBR1MCS2 (Table 1) plasmid was linearized by PCR (Phusion High-Fidelity DNA polymerase) with primer pair pBBR1MCS2_Fw-Rv (Supplementary Table 1), providing homologous ends with the former amplified cnr locus. Afterwards, these PCR products were cloned using the GeneArt® Seamless Cloning and Assembly Enzyme Mix (Thermo Scientific) and the resulting pBBR1MCS2-cnrYXΔH::tetCBAT and pBBR1MCS2-cnrYXΔH::tetCBATR plasmids from a E. coli DG1 transformant selected on LB Km50 were further confirmed by sequencing prior to conjugation (triparental with E. coli HB101 pRK600 as helper) to C. metallidurans AE104.
Monitoring of Transcription
For quantitatively assaying endogenous promoter activity, C. metallidurans AE126 carrying luminescence reporter constructs (Table 1) were constructed to analyze expression from the ISRme5, IS1088, and IS1087B transposase promoters. DNA fragments comprising the IS endogenous promoter (from the left inverted repeat until the transposase start codon) were amplified by PCR (Phusion High-Fidelity DNA polymerase) for ISRme5 (91 bp), IS1088 (85 bp), and IS1087B (89 bp) with primer pairs, pISRme5_Fw-Rv, pIS1088_Fw-Rv, and pIS1087B_Fw-Rv, respectively (Supplementary Table 1), providing EcoRI/XbaI restriction sites. Similarly, fragments comprised of the sequence upstream of the transposase start codon up to the adjacent gene sequence were amplified for all four ISRme5 copies [209 bp for ISRme5a (Rmet_1251), 257 bp for ISRme5b (Rmet_1280), 297 bp for ISRme5c (Rmet_1301), and 239 bp for ISRme5d (Rmet_4152)] with primer pairs pISRme5a/b/c/d_Fw and pISRme5_Rv, respectively (Supplementary Table 1). Afterwards, these PCR products were cloned as EcoRI/XbaI fragments into pGLR1. The resulting pGLR1-PISRme5∕a∕b∕c∕d -gfpluxCDABE, pGLR1-PIS1088-gfpluxCDABE, and pGLR1-PIS1087B-gfpluxCDABE plasmids from transformants selected on LB Km50 were further confirmed by sequencing prior to conjugation (triparental with E. coli HB101 pRK600 as helper) to C. metallidurans AE126 (Table 1). The luminescence reporter constructs were inoculated (in triplicate) into 4 ml of MM284 Km1500 and grown for 48 h. These stationary phase cells were diluted 1:100 (v/v) into fresh MM284 Km1500 and grown for 4 h. Next, exponential phase cell suspensions were divided and supplemented with different concentrations of Zn2+, Cd2+, Ni2+ or Co2+. For each sample, 200 μL was added to a 96-well black cell culture plate (Nunc Flat-bottom, Thermo Scientific) which was placed into a CLARIOstar® (BMG LABTECH). Plates were incubated at 30°C with shaking for several hours. At 30-min time intervals, the optical density at 600 nm and luminescence were measured. Data are shown as the ratio of the relative light unit to optical density (RLU/OD600) normalized to the MM284 control.
The expression of cnrCBAT from two zinc-resistant derivatives, AE126 ΔcnrH::tetR1 and AE126 ΔcnrH::tetR2 (Table 1), was quantitatively assayed via a transcriptional reporter fusion. A 953 and 853-bp DNA fragment comprising the cnrC promoter of AE126 ΔcnrH::tetR1 (harboring also a 3′ fragment of ISRme5 without its transposase promoter) and AE126 ΔcnrH::tetR2 harboring also a 3′ fragment of IS1086 without its transposase promoter) were amplified by PCR (Phusion High-Fidelity DNA polymerase) with primer pairs ISRme5_Internal-pCnrC_Rv and IS1086_Internal-pCnrC_Rv, respectively (Supplementary Table 1) In addition, as control, the native cnrC promoter of AE126 was amplified by PCR (Phusion High-Fidelity DNA polymerase) with primer pair pCnrC_Fw-Rv (Supplementary Table 1). At the same time, the pGLR1 (Table 1) plasmid was linearized by PCR (Phusion High-Fidelity DNA polymerase) with primer pair pGLR1_Fw-Rv (Supplementary Table 1), providing homologous ends with the former amplified fragments. Afterwards, these PCR products were cloned using the GeneArt® Seamless Cloning and Assembly Enzyme Mix (Thermo Scientific) and the resulting pGLR1-PISRme5cnrC-gfpluxCDABE, pGLR1-PIS1086cnrC-gfpluxCDABE, and pGLR1-PcnrC-gfpluxCDABE plasmids from an E. coli DG1 transformant selected on LB Km50 was further confirmed by sequencing prior to conjugation (triparental with E. coli HB101 pRK600 as helper) to C. metallidurans AE104 (Table 1). The luminescence reporter constructs were inoculated (in triplicate) into 4 ml of MM284 Km1500 and grown for 48 h. These stationary phase cells were diluted 1:100 (v/v) into fresh MM284 Km1500 and grown for 4 h. Next, 200 μL exponential phase cell suspensions were added to a 96-well black cell culture plate (Nunc Flat-bottom, Thermo Scientific) which was placed into a CLARIOstar® (BMG LABTECH). Plates were incubated at 30°C and the optical density at 600 nm and luminescence were measured.
Results
Isolation and Phenotypic Characterization of Zinc-Resistant AE126 Derivatives
The minimal inhibitory concentration (MIC) of Zn2+ for C. metallidurans AE126, which is type strain CH34 cured from its pMOL30 mega plasmid encoding the czc operon (cadmium/zinc/cobalt resistance), is much less than its parent, being 0.2 and 12 mM, respectively. However, AE126 has the capability to spontaneously acquire additional zinc resistance, i.e., the ability to grow in the presence of 0.8 mM Zn2+ at a frequency of ~10−6 (Table 2; Collard et al., 1993). Interestingly, using a zinc plate assay two populations of zinc-resistant AE126 derivatives were observed that appeared at different incubation times. The first population, which arose after 2 days of incubation, showed a high variance-to-mean ratio (26 CFU) in a fluctuation assay with 20 independent cultures and a calculated mutation rate to zinc resistance of 0.105 ± 0.022 × 10−7 per generation. However, new colonies emerged on the selective plate in the ensuing days and we defined derivatives specifically emerging at day 5 as the second population. A higher mutation rate to zinc resistance of 0.307 ± 0.056 × 10−7 per generation and lower variance-to-mean ratio (3 CFU) was calculated for the second population. The lower mutation rate and high variance-to-mean ratio of the first population, with most plates yielding a variable number of mutant colonies while occasional plates showed a large number of mutants (so-called “jackpots”), suggested that the first population was pre-existing while the second population emerged on the plate itself.
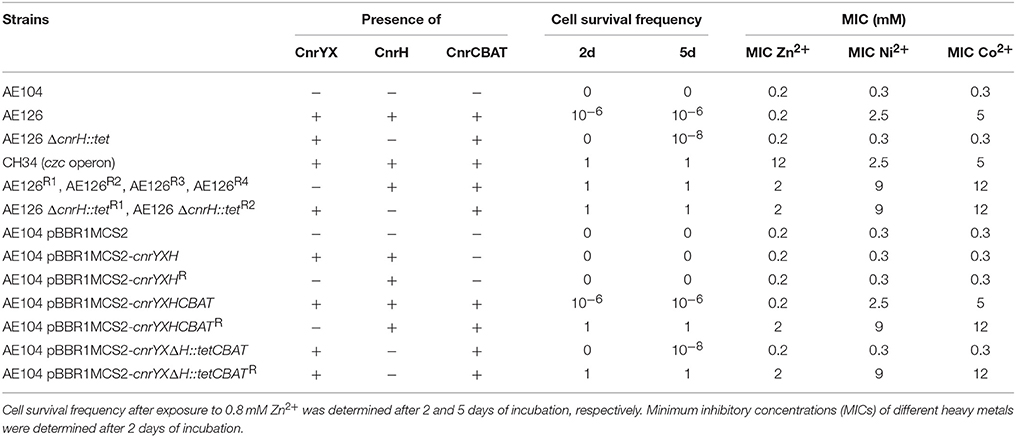
Table 2. Phenotypic characterization of C. metallidurans strains carrying different parts of the cnr operon and their relation with resistance to different heavy metals.
Cultured separately in liquid MM284 without zinc demonstrated similar growth rates for AE126 and zinc-resistant derivatives from the first (AE126R1) and second (AE126R3) population (Table 3). Moreover, the zinc-resistant derivatives exhibited a similar lag phase that was shorter than that of AE126 (Table 3). In addition, both derivatives grew equally fast in the presence of 0.8 mM Zn2+ but displayed a prolonged lag phase compared to MM284 without zinc (Table 3). Next to increased Zn2+ resistance (a 10-fold increase of the MIC), the characterized zinc-resistant AE126 derivatives also displayed an increased resistance to Ni2+, Co2+ and Cd2+ (3, 2.5 and two-fold increase of the MIC, respectively). However, the level of zinc and cadmium resistance of these mutants is still considerably lower than those from the type strain C. metallidurans CH34 (Table 2).
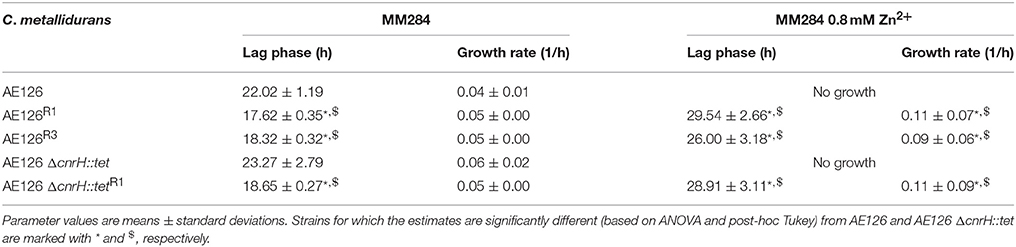
Table 3. Growth parameters of different C. metallidurans strains in MM284 medium with and without 0.8 mM Zn2+.
Genetic Characterization of Zinc-Resistant AE126 Derivatives
In previous work, only two zinc-resistant AE126 derivatives were analyzed (Collard et al., 1993; Grass et al., 2000). Therefore, we scrutinized cnrY and cnrX of 498 randomly selected zinc-resistant AE126 isolates, of which 296 belong to the first population and 202 belong to the second population, by PCR and sequencing to determine if cnrY and cnrX were indeed the sole targets leading to increased zinc resistance of AE126 derivatives (Supplementary Figure 1). This analysis confirmed that all 498 randomly selected isolates either had an IS element inserted or another mutation (point mutation, small insertions, or deletion) in cnrY or cnrX (Figure 1). Interestingly, however, the first population (46.3%) had a significantly lower contribution of IS elements than the second population (68.8%) as determined by a Fisher Exact test with a p < 0.05 (Figure 1), indicating that transposition of these IS elements might be promoted by the selective zinc condition.
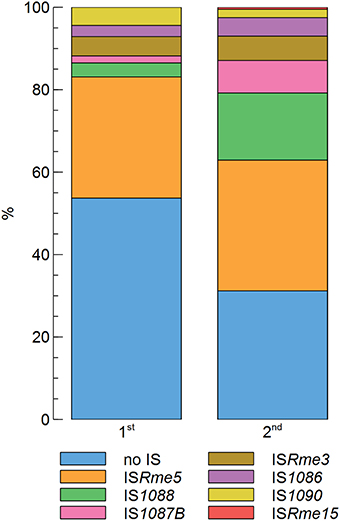
Figure 1. Contribution of different types of cnrYX mutations in the first and second population of zinc-resistant C. metallidurans AE126 derivatives comprising 296 and 202 mutants, respectively.
Characterization of IS-Independent Zinc-Resistant AE126 Mutants
Regarding the IS-independent mutations in cnrYX, it is clear that only mutations resulting in inactivation of either CnrY or CnrX are selected in the zinc plate assays. The IS-independent mutations are dispersed throughout the cnrYX locus and include frameshift mutations, non-sense mutations and larger deletions. For instance, a 360-nucleotide deletion was observed including the complete cnrY gene.
Two zinc-resistant IS-independent AE126 derivatives, AE126R2 (harboring a frameshift mutation caused by a single nucleotide deletion in cnrX) and AE126R3 (harboring a five nucleotide 5′-CGCGA-3′ duplication in cnrY), both belonging to the second population of zinc-resistant AE126 derivatives, were selected for whole-genome expression profiling to further confirm the cnr-dependency underlying toxic zinc stress and subsequent acquired zinc resistance. Although, IS-independent mutations are discussed here, the five nucleotide duplication in cnrY of AE126R3 might suggest an insertion and subsequent excision event of an IS element resulting in a direct target repeat in cnrY. Both AE126R2 and AE126R3 bear mutations leading to inactivation of the anti-sigma CnrYX complex, and whole-genome expression profiling showed 219 and 86 open reading frames (ORFs) that were significantly differentially expressed (>2-fold with an adjusted p < 0.05) in comparison with the parental AE126 strain, respectively (Supplementary Table 2). Twenty ORFs were commonly differentially expressed in both zinc-resistant derivatives. The mutated regulatory genes cnrY and cnrX were the most upregulated (11- and 15-fold in AE126R2, and 33- and 57-fold in AE126R3, respectively) followed by the structural locus cnrCBAT (nine-fold average for AE126R2 and 18-fold average for AE126R3). Transcription of cnrH was three- or six-fold upregulated in AE126R2 and AE126R3, respectively (Supplementary Table 2). As such, it seems that constitutive overexpression of the cnr pump is responsible for the acquired zinc resistance. The high amount of other differentially expressed genes and the fact that most of these genes (93%) were unique for one of both zinc-resistant derivatives indicate that probably different mutations, not involved in the acquired zinc resistance, occurred during the selective exposure to Zn2+, which thus may comprise a more general response to stress with mutations randomly happening, supporting zinc-induced mutagenesis for the second population of zinc-resistant derivatives.
To further and unambiguously confirm the essential role of the cnrYXHCBAT locus in the capability to spontaneously acquire zinc resistance, complementation assays were performed in the zinc-sensitive and plasmidless C. metallidurans CH34 derivative, i.e., strain AE104 (Table 2). The findings of Collard et al. (1993) that strain AE104 can never be evolved to withstand the toxic zinc concentration used in zinc plate assays (0.8 mM Zn2+) was confirmed. Complementation of C. metallidurans AE104 with the complete cnr locus, thus both the regulatory and structural genes, resulted in the ability to acquire zinc resistance (frequency of 0.96 ± 0.38 × 10−6 after 48 h exposure to 0.8 mM Zn2+), whereas complementation of AE104 with the complete but modified cnr locus from AE126R1 resulted in full resistance (frequency of 100%; Table 1). To test if the CnrH sigma factor could drive other potential resistance genes as well, the parental regulatory cnrYXH locus, and the mutated cnrYXH locus from AE126R1 were introduced in AE104 but no zinc resistance could be acquired. Thus, the presence of the cnrCBAT genes encoding the RND-driven efflux system was essential to acquire zinc resistance (Table 2). These complementation assays showed that the cnrYXHCBAT cluster is necessary for acquiring zinc resistance at high frequency in C. metallidurans AE104 or AE126 and that the inactivation of cnrYX and concomitant CnrH-mediated derepression of cnrCBAT is responsible for this zinc resistance.
Characterization of IS-Dependent Zinc-Resistant AE126 Mutants
Focusing further on the IS-dependent zinc-resistant mutants, ISRme5 appeared to be the main contributor to the obtained insertions both in the first (29.4%) and second (31.7%) population (Figure 1). Other identified IS elements included ISRme3, ISRme15, IS1090, IS1088, IS1086 and IS1087B. Comparing the contribution of specific IS elements in the zinc-resistant AE126 population to their abundance in the genome showed that some IS elements were clearly more represented than would be expected from their copy number (Figure 2). ISRme5 (four identical copies in the genome) had a significant higher contribution to both populations, and IS1087B (two identical copies in the genome) to the second population as determined by a Fisher Exact test with a p < 0.05. In addition, IS1088 (nine identical copies in the genome) was significantly more represented in the second than in the first population (Fisher Exact test; p < 0.05), but comparing its contribution to the second population to its abundance in the genome revealed no significant higher contribution than would be expected from its copy number (Figure 2).
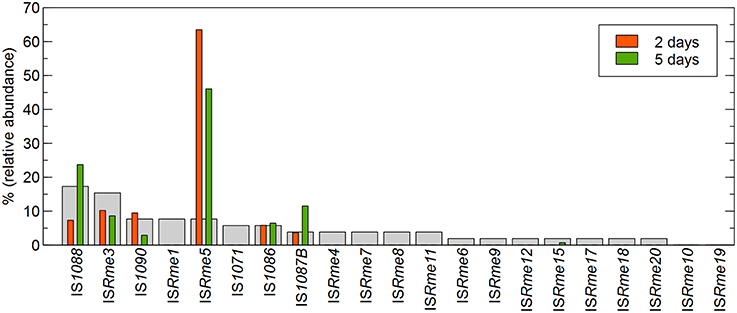
Figure 2. Correlation between the copy number and transposition of IS elements in the cnrYX locus identified in zinc-resistant AE126 derivatives. Red and green bars represent the relative abundance of a specific IS element identified in the pool of IS-mediated zinc-resistant AE126 derivatives isolated after 2 and 5 days of incubation, respectively. Light gray bars represent the relative abundance of a specific IS element in the genome of C. metallidurans AE126.
Next, the identified IS insertion sites were scrutinized. A different use of target genes (i.e., cnrY or cnrX) was detected for the identified IS elements, although this had no influence on zinc resistance as determined by cell survival at 0.8 mM Zn2+ and by MIC testing (data not shown). Most IS elements inactivated cnrY with the exception of ISRme5 and IS1086. In general, all IS elements, except IS1088, inserted exclusively in the same transcriptional orientation as the disrupted gene. No difference in target site use was detected between mutants from the first or second population. ISRme5 used six different insertion sites throughout the target locus and two were used preferentially (Figure 3).
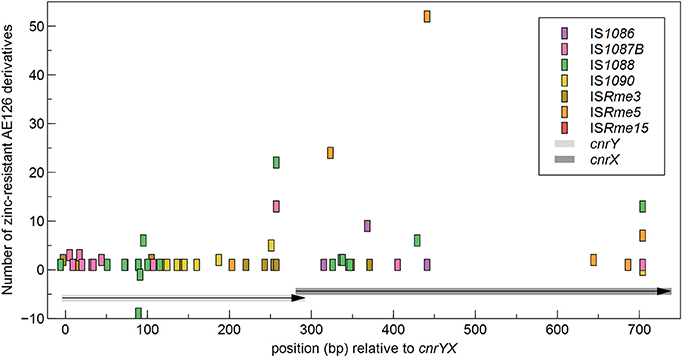
Figure 3. Distribution of IS elements in the cnrYX locus of zinc-resistant AE126 derivatives. Insertions in the same and opposite transcriptional orientation as the cnr operon are shown as positive and negative values, respectively.
A position-specific scoring matrix was derived from the multiple sequence alignment of all ISRme5 target sites (Figure 4). This position-specific scoring matrix was used to scan the genome sequence of C. metallidurans CH34 via the RSAT motif scan (Turatsinze et al., 2008) and resulted in numerous hits (Supplementary Table 3). Besides cnrX, other genes involved in regulation are found in the list composed of the top 100 hits, including genes encoding for transcriptional regulators, receptor proteins, a sensor histidine kinase, and a signal transduction protein. In addition, the family of transporters is with 21 genes also overrepresented in the motif scan. The other identified IS elements in cnrYX showed a more random insertion pattern although IS1087B preferentially inserted into the 5′-region of the regulatory locus.
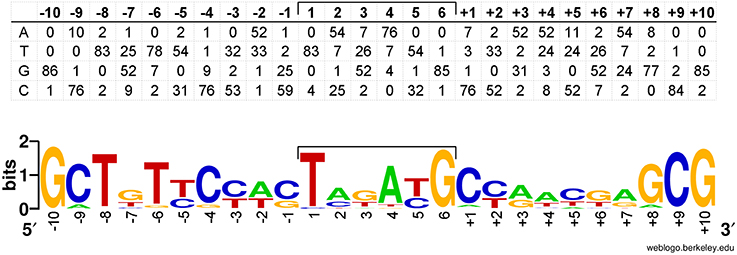
Figure 4. Position-specific scoring matrix derived from the multiple sequence alignment of all target sites used by ISRme5. Eighty-seven target sites and the flanking DNA sequences have been aligned to generate the matrix shown. The sequences compiled are those adjacent to the left (−1 to −10) and right end (+1 to +10) of ISRme5. The central six nucleotides are those that would be duplicated on insertion. A sequence logo was generated from this matrix with WebLogo (Crooks et al., 2004).
Characterization of cnrH-Independent Zinc-Resistant AE126 Mutants
To further analyze the adaptation capacities of C. metallidurans AE126 to toxic zinc concentrations, a cnrH deletion strain was created to assess if other resistance mechanisms than CnrH-mediated expression of cnrCBAT could result in increased zinc resistance. No difference in zinc resistance was observed between the parental and ΔcnrH deletion strain, however, as expected, the cnrH deletion resulted in a drastically decreased cobalt and nickel resistance (Table 2). Interestingly, zinc-resistant AE126 ΔcnrH::tet derivatives could be isolated after exposure to 0.8 mM Zn2+, but only after a minimum exposure time of 96 h and at a frequency 100-fold less compared with AE126 (Table 2). Cultured separately in liquid MM284 without zinc demonstrated similar growth rates for AE126, AE126 ΔcnrH::tet, and the zinc-resistant derivative AE126 ΔcnrH::tetR1 (Table 3). In addition, again, the zinc-resistant derivate exhibited a lag phase that was shorter than that of AE126 ΔcnrH::tet and AE126 (Table 3). Moreover, AE126 ΔcnrH::tetR1 displayed the same growth rate in the presence of 0.8 mM Zn2+ than the CnrH-dependent zinc-resistant AE126R1 and AE126R3 (Table 3).
Phenotypic characterization of zinc-resistant AE126 ΔcnrH::tet derivatives further indicated an increased resistance to Ni2+ (22-fold) and Co2+ (32-fold; Table 2), indicative for a role of the cnr locus. Genetic characterization of six independent zinc-resistant AE126 ΔcnrH::tet derivatives revealed for five derivatives an insertion of ISRme5 at the same target site 25 bp upstream the cnrC start codon (Figure 5). One representative was further used and designated AE126 ΔcnrH::tetR1. The other derivative (AE126 ΔcnrH::tetR2) carried an IS1086 insertion 29 bp upstream of the cnrC start codon (Figure 5). To exclude any other possible involvement of additional resistance determinants, complementation assays were performed where the complete but modified cnr operon of AE126 ΔcnrH::tet and AE126 ΔcnrH::tetR1 were transferred to C. metallidurans AE104, respectively. These complementation assays demonstrated that cnrYXΔcnrH::tetCBATR mediated the increased zinc resistance (Table 2). Furthermore, transcriptional reporter fusions with cnrC promoter fragments of AE126 ΔcnrH::tetR1 (953 bp fragment harboring also a 3′ fragment of ISRme5 without its transposase promoter) and AE126 ΔcnrH::tetR2 (853 bp fragment harboring also a 3′ fragment of IS1086 without its transposase promoter) confirmed the constitutive expression of cnrCBAT (Supplementary Figure 2). Therefore, the insertion of these IS elements in the intergenic region between ΔcnrH::tet and cnrC resulted in a constitutively increased transcription of cnrCBAT, which was also confirmed by microarray analysis of AE126 ΔcnrH::tetR1 (Supplementary Table 4). Since read-through expression from the transposase promoter did not account for this increased expression, the transcription start site of cnrCBAT in AE126 ΔcnrH::tetR1 (ISRme5 insertion) was identified and showed that expression is driven by an outward-directed promoter positioned at the 3′ extremity of ISRme5 (Figure 5). This could also be deduced for IS1086 based on the insertion site relative to the transcription start site of cnrC (Figure 5).
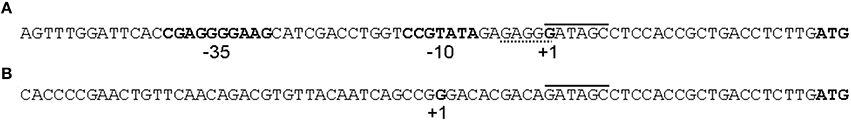
Figure 5. (A) Sequence of the intergenic region between cnrH and cnrC with the last three nucleotides (ATG) being the start codon of cnrC. The −35 and −10 box and transcription start of cnrC are highlighted in bold. Integration sites of ISRme5 (full line) and IS1086 (dashed line) are indicated above and below the sequence, respectively. (B) Sequence of the newly formed cnrC promoter via ISRme5 insertion with direct repeat at the point of insertion (full line) and transcription start.
Endogenous Promoter Activity of IS Elements
Based on their relative high abundance in the zinc-resistant derivatives of AE126, especially in the second population, ISRme5, IS1088 and IS1087B were selected for further investigation. To analyze the effect of metal ion stress on the endogenous promoter activity of the transposase of the three selected IS elements, transcriptional fusions between the promoterless tandem bi-cistronic gfp-luxCDABE reporter and fragments of ISRme5, IS1088, and IS1087B comprising the left inverted repeat up until the start codon of the transposase were constructed and analyzed. The promoters of all three IS elements were induced by Zn2+ and Cd2+, but not by Ni2+ and Co2+ (Figure 6). Of the three endogenous promoters analyzed, the ISRme5 promoter was induced the most by Zn2+, and in lesser extent by Cd2+ (1.7-fold for Zn2+ and 1.4-fold for Cd2+ after 4 h). A dose-dependent response was observed with the highest response for 100 μM Zn2+ and 10 μM Cd2+, respectively (Figure 7). Higher doses of Zn2+ or Cd2+ could not be tested as they are detrimental to the growth of C. metallidurans AE126 strains. The IS1088 and IS1087B endogenous transposase promoters were induced 1.14 and 1.25-fold by Zn2+ (after 4 h), respectively, and to a lesser extent by Cd2+.
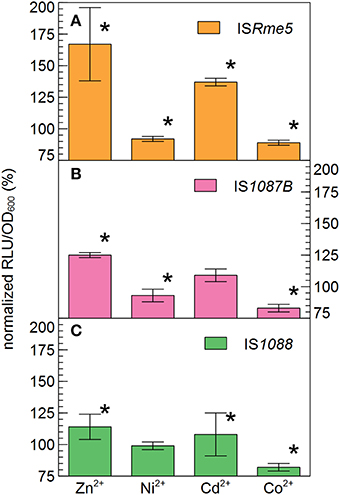
Figure 6. Endogenous promoter activity of the ISRme5 (A), IS1088 (C), and IS1087B (B) transposase in response to Zn2+ (100 μM), Ni2+ (100 μM), Cd2+ (10 μM) and Co2+ (50 μM) after 4 h of incubation, calculated by normalizing the reporter signal RLU to cell density ratio (RLU/OD600) to that of the control without added metals. *indicates a significant difference between the tested metal ion and the control, determined by a t-test with a p < 0.05. The average values of three independent experiments with standard deviations are shown.
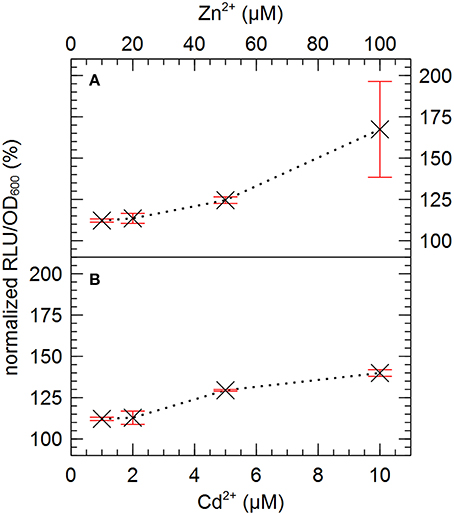
Figure 7. Normalized dose-response curve for the transcription of the endogenous promoter of ISRme5 in response to zinc (A) and cadmium (B) measured after 4 h of incubation. The average values of three independent experiments with standard deviations are shown.
Analysis of Upstream Flanking Genomic Sequence on ISRme5 Transposase Expression
For ISRme5, the most active element in response to zinc, the effect of the upstream flanking genomic sequence on transposase expression was scrutinized in more detail. C. metallidurans AE126 harbors three copies of the ISRme5 element on the chromosome and one copy on the chromid, which are identical at the sequence level. However, the transposase open reading frame contains no stop codon within the IS element, implying that the different ISRme5 transposases, encoded by the four copies, have different lengths generated by sequence specific insertion. This is not unique as more members of the IS481-family do not display a stop codon for the transposase open reading frame within their respective IS elements (Redenbach et al., 1996; Tauch et al., 1998). The transposase of the corresponding ISRme5a, b, c and d element consist of 333, 463, 321 and 321 amino acids, respectively. Two copies are located in genomic island CMGI-2, which is involved in hydrogenotrophy and the metabolism of aromatic compounds (Van Houdt et al., 2009; Mijnendonckx et al., 2011). Transcriptional fusions between the promoterless tandem bi-cistronic gfp-luxCDABE reporter and sequences starting 118, 166, 206 or 148 bp upstream the left inverted repeat up until the transposase start codon of all four ISRme5 copies were constructed and analyzed. Comparing the basic expression activity, it is clear that the genomic sequence upstream the left inverted repeat of all ISRme5 copies exerts a negative effect on ISRme5 endogenous promoter activity. This is especially true for ISRme5b where almost no expression is detected, i.e., < 0.5% (Figure 8). Furthermore, although the reporter construct composed of only the promoter region of ISRme5, thus without additional genomic sequences, is Zn2+-inducible, no Zn2+ induction was detected for the reporter constructs composed of upstream genomic content of ISRme5a, ISRme5b and ISRme5d. The reporter construct of ISRme5c, however, was more than two-fold induced for Zn2+ (Figure 8).
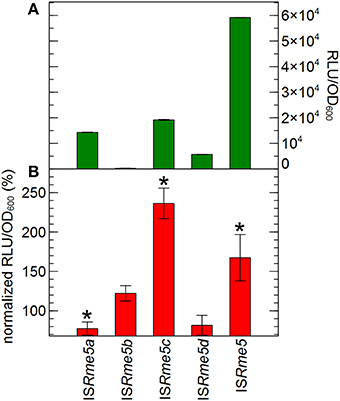
Figure 8. Transposase expression of the different ISRme5 copies. Reporter constructs contain the ISRme5 tranpsosase promoter (starting from the left inverted repeat) and for the four ISRme5 (a, b, c, and d) copies, in addition, the DNA sequence (~250 bp) upstream the integration site. (A) Basal expression of the reporter constructs at the start of the experiment, calculated by normalizing the reporter signal RLU to cell density (RLU/OD600). (B) Expression after 4 h of incubation in the presence of 100 μM Zn2+, calculated by normalizing the reporter signal RLU to cell density (RLU/OD600) to that of the control without added Zn2+. *indicates significant difference between Zn2+ and control, determined by a t-test with a p < 0.05. The average values of three independent experiments with standard deviations are shown.
Enhanced Adaptation Potential to Other Stress Challenges
The effect of IS transposition in the cnrYX genes on the general fitness was analyzed. In this test, the parental AE126 and the zinc-resistant C. metallidurans AE126R4 strain were used. AE126R4 carries an additional ISRme5 element inserted in cnrX in the same transcriptional orientation as the cnr operon, yielding a continuous read-through ISRme5 transposase expression as whole-genome expression verified that expression of cnrYX is induced in CnrH-dependent zinc-resistant AE126 derivatives (Supplementary Table 2). The pJV240 plasmid was introduced into AE126 and AE126R4, and biological triplicates were subsequently exposed to 10% sucrose. Sucrose-resistant derivatives were isolated and the pJV240-encoded sacBR locus was screened by PCR to determine the involvement of ISRme5. AE126R4 pJV240 displayed a higher survival frequency (4.78 ± 1.58 × 10−6) than AE126 pJV240 (1.42 ± 0.30 × 10−7) and ISRme5 was found to disrupt sacBR in sucrose-resistant AE126R4 pJV240 derivatives (3/20) opposed to sucrose-resistant AE126 pJV240 derivatives (0/20).
Discussion
MGEs have been shown to provide their hosts with a wide diversity of adaptive traits. Although, transposition of IS elements mostly results in detrimental mutations, a fraction of insertions can be beneficial and lead to increased survival. The simplest effect of IS transposition is the inactivation of a gene at a particular target site. In addition, IS transposition can also lead to altered expression of adjacently located genes by co-transcription or by providing a (hybrid) promoter or other protein binding elements located inside the IS element.
It was previously demonstrated that C. metallidurans AE126 could readily acquire increased zinc resistance (Collard et al., 1993). We observed that zinc-resistant AE126 derivatives arose at different time points after exposure to 0.8 mM Zn2+ and all 498 tested derivatives had mutations targeting the regulatory locus of the RND-driven CnrCBA efflux system. The first population arose after an exposure time of 48 h and new derivatives kept appearing in the ensuing days with a second population defined as derivatives that arose after an exposure time of 120 h. The first population displayed a high variance-to-mean ratio, with most plates yielding a variable number of mutant colonies while occasional plates showed a large number of mutants (so-called “jackpots”), indicating the involvement of pre-existing mutations (Luria and Delbrück, 1943). Interestingly, the second population displayed a higher mutation rate and lower variance-to-mean ratio. Although, the latter was still three times the theoretical value of one when only stress-induced mutagenesis is considered, the much lower value and higher mutation rate are indicative for the involvement of zinc-induced mutagenesis in the second population. Furthermore, mutants from both populations have the same growth rate on zinc medium suggesting once more that the second population may originate from selection on the selective zinc plate. Adaptive mutation and accumulation of colonies throughout incubation time has been described for the activation of a promoterless phenol degradation operon in starving Pseudomonas putida. Although, phenol-utilizing mutants already appeared on phenol-minimal plates on day 2, the majority of the colonies began to appear on day 3 and continued to accumulate during the next 7 days (Kasak et al., 1997). In addition, mutations to ciprofloxacin resistance also continually occurred in non-dividing E. coli cells during a 7-day exposure to ciprofloxacin in agar and the late-appearing resistant mutants had no growth rate defect compared to wild type or early-appearing mutants (Riesenfeld et al., 1997). Moreover, E. coli cells could utilize glycerol in the absence of Crp by activation of the silent glpFK glycerol utilization operon by transposition of IS5 upstream of glpFK. The transposed IS5 element harbors a permanent bend and IHF binding site at its 3′-end that promoted the expression of the adjacently located glpFK operon (Zhang and Saier, 2011). When crp− cells of E. coli are incubated on solid glycerol minimal medium, Glp+ colonies first appeared after 3 days of incubation and new colonies continued to appear in the ensuing days. However, wild type and crp Glp+ cells plated on the same solid medium formed already colonies in less than 2 days, indicating that the Glp+ mutants that arose from the crp cells on glycerol minimal medium plates were not present in the cell culture before plating (Zhang and Saier, 2011).
For acquiring zinc resistance at high frequency (10−6), inactivation of anti-sigma factor CnrYX complex is essential to liberate CnrH and increase cnrCBAT expression. Although the efflux pump CnrCBAT is necessary to mediate zinc efflux, the role of the sigma factor CnrH in promoting cnrCBAT expression is not. In the absence of cnrH, expression of cnrCBAT and acquisition of zinc resistance could be mediated by the correct insertion of ISRme5 or IS1086 carrying an outward-directed promoter positioned at their 3′ extremity, albeit to a lower frequency (10−8), and only after a minimum exposure time of 96 h. These mutants displayed the same growth rate on zinc medium than CnrH-dependent zinc-resistant AE126 derivatives from the first population, which were isolated after 48 h, suggesting that these CnrH-independent zinc-resistant derivatives may originate from selection on the selective zinc plate. In addition, using the cRACE method, the wild type promoter transcription start site of the cnrCBAT structural resistance genes was found to be located downstream of cnrH as previously reported by Grass et al. (2000) and not upstream as previously reported by Tibazarwa et al. (2000).
Exposure of AE126 to 0.8 mM Zn2+ revealed transposition of seven different IS elements in the cnrY or cnrX gene (occurrence of transposition ISRme5 > IS1088 > ISRme3 > IS1087B > IS1090 > IS1086 > ISRme15). Active IS transposition in C. metallidurans was already observed for ISRme1, ISRme3, IS1086, IS1087B, IS1088 and IS1090 (Dong et al., 1992; Collard et al., 1993; Grass et al., 2000; Ma-e-Talat, 2000; Schneider et al., 2000b; Tibazarwa et al., 2000; Mijnendonckx et al., 2011), but not for ISRme5 and ISRme15. Overall, IS elements contributed more to the second population that arose after five days of incubation on plate. ISRme5 was the main IS contributing to both populations, whereas IS1088 and IS1087B showed a higher contribution to the second compared with the first population. Most of the transposed IS elements (>95%) inserted exclusively in the same transcriptional orientation as cnrCBAT, although inactivation of CnrYX is sufficient to acquire zinc resistance. It is unclear why a given site should be used in only one orientation (Lampe et al., 1998). Most transposases did not show insertion site specificity, only for ISRme5 two hotspot loci were identified that were used preferentially and specifically by ISRme5. Scanning the genome of C. metallidurans CH34 with the generated target sequence matrix resulted in numerous hits, with various hits in genes involved in regulation and transport processes, indicating the evolutionary power that ISRme5 might exert as inactivation of those genes mostly results in significant phenotypic alterations. In some cases transposases prefer a particular DNA structure, most notably bent DNA, as target site (Craig, 1997). It seems that this was the case for the IS1087B transposase as it mostly inserted in the 5′-region of cnrY, indicating that the IS1087B transposase does not recognize target DNA in any sequence-specific manner but that certain sequences surrounding the insertion site may allow local structures to form that are conductive for IS1087B insertion. There is precedence for this phenomenon in the non-random use of target sequences of P transposable elements, where the local target preference depends more on DNA structure than on primary sequence (Liao et al., 2000).
Our study showed for the first time that the endogenous promoters of the ISRme5, IS1088 and IS1087B transposases can be induced by metal ions. This increased promoter induction was specific for zinc and cadmium but not nickel and cobalt. The dual zinc and cadmium induction could be explained by the fact that both metal ions have many similar (bio)chemical characteristics. Other examples of transposase transcriptional expression induced by environmental stimuli include Tn4652 and IS1246 in P. putida KT2440 by zinc, cadmium and nickel but not cobalt (Haritha et al., 2009), and ISFtu1 and ISFtu2 by spermine in Francisella tularensis (Carlson et al., 2009). Elevated transpositional activity of several IS elements was also observed at high temperature in Burkholderia multivorans (Ohtsubo et al., 2005). However, the induced transpositional activity in the examples above did not mediate enhanced tolerance to the inducer. Although, the ISRme5 transposase promoter internal to the element is inducible by zinc and cadmium, the genomic region upstream IS integration has a strong regulatory effect on the expression of the IS element. The upstream genomic region resulted in reduced promoter activity of all ISRme5 copies, especially for ISRme5b, and only the ISRme5c copy was found to be metal-inducible. In addition, the transposase of ISRme5 (IS481 family member) has no stop codon within the element. Consequently, the ISRme5b transposase has a prolonged carboxy-terminal end compared with the other copies, generated by its insertion site, which might influence its enzymatic stability and hence, could contribute to the possible diminished transposition activity of this copy compared to the other ISRme5 elements. Therefore, the evolutionary success of ISRme5, as probably for all IS elements, depends not only on the element itself, but also on its genomic integration site. Taking into account that many transposases preferentially act in cis (Mahillon and Chandler, 1998), which reduces the probability that transposase expression from a particular element would activate transposition of related copies elsewhere in the genome, the higher basal expression level and zinc-inducibility of the ISRme5c transpose indicate that most ISRme5-mediated CnrYX inactivation events are probably the result of ISRme5c transposition. Repression or enhancement of promoter activity by the upstream flanking sequence is also observed for other MGEs (Lavie et al., 2004).
Bacteria respond to environmental changes by adapting the expression of key genes, however, such reprogramming requires time and energy. Interestingly, all tested zinc-resistant derivatives showed a considerably shorter lag phase in liquid MM284 without zinc compared to wild type, rendering the evolved population better suited to quickly respond to other environmental challenges. For instance, in conditions where the lag phase is long, adaptations reducing the lag phase are highly beneficial as the evolved population can start their exponential growth earlier and consume most of the environmental nutrients before the wild type exits the lag phase (Oxman et al., 2008). Furthermore, adaptation to zinc by IS reshuffling can have an additional impact on the adaptation to subsequent stress challenges. Insertion of an IS element in cnrYX in the same transcriptional orientation as the cnr operon will probably result in read-through transposase expression as whole-genome expression indeed verified that expression of cnrYX is induced in CnrH-dependent zinc-resistant AE126 derivatives. Thus, redistribution of IS elements yielding IS-dependent zinc-resistant derivatives, hence harboring an additional copy of the IS element in a constitutively transcribed region, might result in an increased pool of the transposase in the cell and subsequent enhance the adaptation potential to new environmental challenges, such as was shown here for ISRme5 and SacB-mediated sucrose hypersensitivity. The higher survival frequency of the IS-dependent zinc-resistant derivative compared to wild type can be explained by (i) an increased pool of ISRme5 transposase resulting in ISRme5-mediated sacBR disruption, (ii) the possible insertion and subsequent excision of ISRme5, resulting in various possible frameshift deletions (Ooka et al., 2009) or duplications (Mahillon and Chandler, 1998) inactivating sacBR, however, the presence of these mutations was not investigated, and (iii) the uncharacterized genetic background of the IS-dependent zinc-resistant derivative, which likely, consistent with other derivatives from the second population, harbors more mutations.
To summarize, it was shown that zinc induced the expression of IS elements and that these elements mediate the adaptation of C. metallidurans to zinc stress by modulating its transcriptional network or by providing outward-directed mobile promoters.
Author Contributions
Conceived and designed the study: MM, AA, and RVH. Performed the experiments: JV. Analyzed the data: all authors. Wrote the paper: JV, AA, and RVH.
Funding
This work was supported through a Ph.D. grant for JV by SCK•CEN.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00359
Supplementary Figure 1. Schematic representation of the cnr operon of megaplasmid pMOL28 from C. metallidurans extracted from the MaGe platform (https://www.genoscope.cns.fr/agc/microscope/mage/) with cnrYX encoding two membrane-bound anti-sigma factors, cnrH a sigma factor, cnrCBA a RND efflux pump, and cnrT a cation diffusion facilitator. Black line represent the amplicon generated with the primer pair Cnr_Fw and Cnr_Rv used for screening the cnrYXH structural locus.
Supplementary Figure 2. Promoter activity of the cnrC promoter of AE126 ΔcnrH::tetR1 and AE126 ΔcnrH::tetR2. Data are represented as fold change of the report signal normalized to cell density (RLU/OD600) obtained for AE104 pGLR1-PISRme5cnrC-gfpluxCDABE (953 bp fragment harboring also a 3′ fragment of ISRme5 without its transposase promoter) and AE104 pGLR1-PIS1086cnrC-gfpluxCDABE (853 bp fragment harboring also a 3′ fragment of IS1086 without its transposase promoter) compared with the control AE104 pGLR1-PcnrC-gfpluxCDABE.
Supplementary Table 1. Primers used in this study.
Supplementary Table 2. Whole-genome microarray analysis of gene expression in two zinc-resistant AE126 derivatives (AE126R2 and AE126R3). Data are represented as fold change relative to the parental AE126. Only open reading frames (ORFs) that were significantly expressed (>2-fold with an adjusted p < 0.05) are shown. (A) ORFs differentially expressed in both derivatives, (B) ORFs differentially expressed in AE126R2 and (C) ORFs differentially expressed in AE126R3 with ND not determined. The full description of the microarray data have been deposited at the Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE74091.
Supplementary Table 3. Scan of the C. metallidurans CH34 genome sequence (+ for forward and – for reverse strand) with the position-specific scoring matrix obtained from the multiple sequence alignment of the ISRme5 target sites in 87 zinc-resistant AE126 derivatives using RSAT. The weight score Ws = log (P(S|M)/P(S|B)) is the probability for the sequence segment S to occur according to the motif model M, whereas P (S|B) is the probability for the same sequence S to occur under the background model B. The weight score is thus the log-ratio of the likelihood of S in these two respective models. Top 100 targets are shown.
Supplementary Table 4. Transcriptional expression of the cnrCBAT cluster, determined by whole-microarray analysis, in two zinc-resistant AE126 derivatives (AE126R2 and AE126R3) and in a zinc-resistant AE126 ΔcnrH::tet derivative (AE126ΔcnrH::tetR1). Data are represented as fold change relative to the parental AE126 and AE126 ΔcnrH::tet, respectively. The full description of the microarray data have been deposited at the Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE74091.
References
Baranyi, J., and Roberts, T. A. (1994). A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 23, 277–294. doi: 10.1016/0168-1605(94)90157-0
Benedetti, I. M., de Lorenzo, V., and Silva-Rocha, R. (2012). Quantitative, non-disruptive monitoring of transcription in single cells with a broad-host range GFP-luxCDABE dual reporter system. PLoS ONE 7:e52000. doi: 10.1371/journal.pone.0052000
Brim, H., Heyndrickx, M., de Vos, P., Wilmotte, A., Springael, D., Schlegel, H. G., et al. (1999). Amplified rDNA restriction analysis and further genotypic characterisation of metal-resistant soil bacteria and related facultative hydrogenotrophs. Syst. Appl. Microbiol. 22, 258–268. doi: 10.1016/S0723-2020(99)80073-3
Cairns, J., Overbaugh, J., and Miller, S. (1988). The origin of mutants. Nature 335, 142–145. doi: 10.1038/335142a0
Carlson, P. E. Jr., Horzempa, J., O'Dee, D. M., Robinson, C. M., Neophytou, P., Labrinidis, A., et al. (2009). Global transcriptional response to spermine, a component of the intramacrophage environment, reveals regulation of Francisella gene expression through insertion sequence elements. J. Bacteriol. 191, 6855–6864. doi: 10.1128/JB.00995-09
Chang, A. C., and Cohen, S. N. (1978). Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134, 1141–1156.
Christie-Oleza, J. A., Lanfranconi, M. P., Nogales, B., Lalucat, J., and Bosch, R. (2009). Conjugative interaction induces transposition of ISPst9 in Pseudomonas stutzeri AN10. J. Bacteriol. 191, 1239–1247. doi: 10.1128/JB.01071-08
Ciampi, M. S., Schmid, M. B., and Roth, J. R. (1982). Transposon Tn10 provides a promoter for transcription of adjacent sequences. Proc. Natl. Acad. Sci. U.S.A. 79, 5016–5020. doi: 10.1073/pnas.79.16.5016
Coenye, T., Spilker, T., Reik, R., Vandamme, P., and Lipuma, J. J. (2005). Use of PCR analyses to define the distribution of Ralstonia species recovered from patients with cystic fibrosis. J. Clin. Microbiol. 43, 3463–3466. doi: 10.1128/JCM.43.7.3463-3466.2005
Collard, J. M., Provoost, A., Taghavi, S., and Mergeay, M. (1993). A new type of Alcaligenes eutrophus CH34 zinc resistance generated by mutations affecting regulation of the cnr cobalt-nickel resistance system. J. Bacteriol. 175, 779–784.
Craig, N. L. (1997). Target site selection in transposition. Annu. Rev. Biochem. 66, 437–474. doi: 10.1146/annurev.biochem.66.1.437
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Dallmeier, K., and Neyts, J. (2013). Simple and inexpensive three-step rapid amplification of cDNA 5′ ends using 5′ phosphorylated primers. Anal. Biochem. 434, 1–3. doi: 10.1016/j.ab.2012.10.031
Del Re, B., Garoia, F., Mesirca, P., Agostini, C., Bersani, F., and Giorgi, G. (2003). Extremely low frequency magnetic fields affect transposition activity in Escherichia coli. Radiat. Environ. Biophys. 42, 113–118. doi: 10.1007/s00411-003-0192-9
Diels, L., and Mergeay, M. (1990). DNA probe-mediated detection of resistant bacteria from soils highly polluted by heavy metals. Appl. Environ. Microbiol. 56, 1485–1491.
D'Inzeo, T., Santangelo, R., Fiori, B., De Angelis, G., Conte, V., Giaquinto, A., et al. (2015). Catheter-related bacteremia by Cupriavidus metallidurans. Diagn. Microbiol. Infect. Dis. 81, 9–12. doi: 10.1016/j.diagmicrobio.2014.09.015
Dong, Q., Sadouk, A., van der Lelie, D., Taghavi, S., Ferhat, A., Nuyten, J. M., et al. (1992). Cloning and sequencing of IS1086, an Alcaligenes eutrophus insertion element related to IS30 and IS4351. J. Bacteriol. 174, 8133–8138.
Drevinek, P., Baldwin, A., Lindenburg, L., Joshi, L. T., Marchbank, A., Vosahlikova, S., et al. (2010). Oxidative stress of Burkholderia cenocepacia induces insertion sequence-mediated genomic rearrangements that interfere with macrorestriction-based genotyping. J. Clin. Microbiol. 48, 34–40. doi: 10.1128/JCM.01433-09
Eichenbaum, Z., and Livneh, Z. (1998). UV light induces IS10 transposition in Escherichia coli. Genetics 149, 1173–1181.
Foster, P. L. (2007). Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 42, 373–397. doi: 10.1080/10409230701648494
Gay, P., Le Coq, D., Steinmetz, M., Berkelman, T., and Kado, C. I. (1985). Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 164, 918–921.
González, J., Karasov, T. L., Messer, P. W., and Petrov, D. A. (2010). Genome-wide patterns of adaptation to temperate environments associated with transposable elements in Drosophila. PLoS Genet. 6:e1000905. doi: 10.1371/journal.pgen.1000905
Goris, J., De Vos, P., Coenye, T., Hoste, B., Janssens, D., Brim, et al., (2001). Classification of metal-resistant bacteria from industrial biotopes as Ralstonia campinensis sp nov., Ralstonia metallidurans sp nov and Ralstonia basilensis Steinle et al. 1998 emend. Int. J. Syst. Evol. Micr. 51, 1773–1782. doi: 10.1099/00207713-51-5-1773
Grass, G., Grosse, C., and Nies, D. H. (2000). Regulation of the cnr cobalt and nickel resistance determinant from Ralstonia sp. strain CH34. J. Bacteriol. 182, 1390–1398. doi: 10.1128/JB.182.5.1390-1398.2000
Grosse, C., Friedrich, S., and Nies, D. H. (2007). Contribution of extracytoplasmic function sigma factors to transition metal homeostasis in Cupriavidus metallidurans strain CH34. J. Mol. Microbiol. Biotechnol. 12, 227–240. doi: 10.1159/000099644
Hall, B. G. (1990). Directed evolution of a bacterial operon. Bioessays 12, 551–558. doi: 10.1002/bies.950121109
Hall, B. M., Ma, C. X., Liang, P., and Singh, K. K. (2009). Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics 25, 1564–1565. doi: 10.1093/bioinformatics/btp253
Haritha, A., Sagar, K. P., Tiwari, A., Kiranmayi, P., Rodrigue, A., Mohan, P. M., et al. (2009). MrdH, a novel metal resistance determinant of Pseudomonas putida KT2440, is flanked by metal-inducible mobile genetic elements. J. Bacteriol. 191, 5976–5987. doi: 10.1128/JB.00465-09
Janssen, P. J., Van Houdt, R., Moors, H., Monsieurs, P., Morin, N., Michaux, A., et al. (2010). The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS ONE 5:e10433. doi: 10.1371/journal.pone.0010433
Jaurin, B., and Normark, S. (1983). Insertion of IS2 creates a novel ampC promoter in Escherichia coli. Cell 32, 809–816. doi: 10.1016/0092-8674(83)90067-3
Kasak, L., Horak, R., and Kivisaar, M. (1997). Promoter-creating mutations in Pseudomonas putida: a model system for the study of mutation in starving bacteria. Proc. Natl. Acad. Sci. U.S.A. 94, 3134–3139. doi: 10.1073/pnas.94.7.3134
Katzen, F., Becker, A., Ielmini, M. V., Oddo, C. G., and Ielpi, L. (1999). New mobilizable vectors suitable for gene replacement in gram-negative bacteria and their use in mapping of the 3′ end of the Xanthomonas campestris pv. campestris gum operon. Appl Environ Microb 65, 278–282.
Kazazian, H. H. Jr. (2004). Mobile elements: drivers of genome evolution. Science 303, 1626–1632. doi: 10.1126/science.1089670
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M. II, et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Lampe, D. J., Grant, T. E., and Robertson, H. M. (1998). Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics 149, 179–187.
Langevin, S., Vincelette, J., Bekal, S., and Gaudreau, C. (2011). First case of invasive human infection caused by Cupriavidus metallidurans. J. Clin. Microbiol. 49, 744–745. doi: 10.1128/JCM.01947-10
Lartigue, M. F., Poirel, L., Aubert, D., and Nordmann, P. (2006). In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring beta-lactamase gene blaCTX−M of Kluyvera ascorbata. Antimicrob. Agents Chemother. 50, 1282–1286. doi: 10.1128/AAC.50.4.1282-1286.2006
Lavie, L., Maldener, E., Brouha, B., Meese, E. U., and Mayer, J. (2004). The human L1 promoter: variable transcription initiation sites and a major impact of upstream flanking sequence on promoter activity. Genome Res. 14, 2253–2260. doi: 10.1101/gr.2745804
Lederberg, J. (1989). Replica plating and indirect selection of bacterial mutants: isolation of preadaptive mutants in bacteria by sib selection. Genetics 121, 395–399.
Liao, G. C., Rehm, E. J., and Rubin, G. M. (2000). Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 97, 3347–3351. doi: 10.1073/pnas.97.7.3347
Louarn, J. M., Bouche, J. P., Legendre, F., Louarn, J., and Patte, J. (1985). Characterization and properties of very large inversions of the Escherichia coli chromosome along the origin-to-terminus axis. Mol. Gen. Genet. 201, 467–476. doi: 10.1007/BF00331341
Luria, S. E., and Delbrück, M. (1943). Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28, 491–511.
MacLean, R. C., Torres-Barceló, C., and Moxon, R. (2013). Evaluating evolutionary models of stress-induced mutagenesis in bacteria. Nat. Rev. Genet. 14, 221–227. doi: 10.1038/nrg3415
Ma-e-Talat (2000). Genetic Mechanism of Heavy Metal Resistance of Pseudomonas aeruginosa Cmg103. Karachi: University of Karachi.
Mahillon, J., Léonard, C., and Chandler, M. (1999). IS elements as constituents of bacterial genomes. Res. Microbiol. 150, 675–687. doi: 10.1016/S0923-2508(99)00124-2
Maillard, A. P., Künnemann, S., Grosse, C., Volbeda, A., Schleuder, G., Petit-Härtlein, I., et al. (2015). Response of CnrX from Cupriavidus metallidurans CH34 to nickel binding. Metallomics 7, 622–631. doi: 10.1039/C4MT00293H
Maruyama, I. N., Rakow, T. L., and Maruyama, H. I. (1995). Crace - a simple method for identification of the 5′-end of messenger-RNAs. Nucleic Acids Res. 23, 3796–3797. doi: 10.1093/nar/23.18.3796
McClintock, B. (1984). The significance of responses of the genome to challenge. Science 226, 792–801. doi: 10.1126/science.15739260
Mergeay, M., Monchy, S., Vallaeys, T., Auquier, V., Benotmane, A., Bertin, P., et al. (2003). Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal-responsive genes. FEMS Microbiol. Rev. 27, 385–410. doi: 10.1016/S0168-6445(03)00045-7
Mergeay, M., Nies, D., Schlegel, H. G., Gerits, J., Charles, P., and Vangijsegem, F. (1985). Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy-metals. J. Bacteriol. 162, 328–334.
Mijnendonckx, K., Provoost, A., Monsieurs, P., Leys, N., Mergeay, M., Mahillon, J., et al. (2011). Insertion sequence elements in Cupriavidus metallidurans CH34: distribution and role in adaptation. Plasmid 65, 193–203. doi: 10.1016/j.plasmid.2010.12.006
Mijnendonckx, K., Provoost, A., Ott, C. M., Venkateswaran, K., Mahillon, J., Leys, N., et al. (2013). Characterization of the survival ability of Cupriavidus metallidurans and Ralstonia pickettii from space-related environments. Microb. Ecol. 65, 347–360. doi: 10.1007/s00248-012-0139-2
Monchy, S., Benotmane, M. A., Janssen, P., Vallaeys, T., Taghavi, S., van der Lelie, D., et al. (2007). Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans are specialized in the maximal viable response to heavy metals. J. Bacteriol. 189, 7417–7425. doi: 10.1128/JB.00375-07
Monsieurs, P., Moors, H., Van Houdt, R., Janssen, P. J., Janssen, A., Coninx, I., et al. (2011). Heavy metal resistance in Cupriavidus metallidurans CH34 is governed by an intricate transcriptional network. Biometals 24, 1133–1151. doi: 10.1007/s10534-011-9473-y
Nies, D. H. (1999). Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51, 730–750. doi: 10.1007/s002530051457
Nies, D. H. (2003). Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27, 313–339. doi: 10.1016/S0168-6445(03)00048-2
Nies, D. H., and Silver, S. (1995). Ion efflux systems involved in bacterial metal resistances. J. Ind. Microbiol. 14, 186–199. doi: 10.1007/BF01569902
Ohtsubo, Y., Genka, H., Komatsu, H., Nagata, Y., and Tsuda, M. (2005). High-temperature-induced transposition of insertion elements in Burkholderia multivorans ATCC 17616. Appl. Environ. Microbiol. 71, 1822–1828. doi: 10.1128/AEM.71.4.1822-1828.2005
Ooka, T., Ogura, Y., Asadulghani, M., Ohnishi, M., Nakayama, K., Terajima, J., et al. (2009). Inference of the impact of insertion sequence (IS) elements on bacterial genome diversification through analysis of small-size structural polymorphisms in Escherichia coli O157 genomes. Genome Res. 19, 1809–1816. doi: 10.1101/gr.089615.108
Oxman, E., Alon, U., and Dekel, E. (2008). Defined order of evolutionary adaptations: experimental evidence. Evolution 62, 1547–1554. doi: 10.1111/j.1558-5646.2008.00397.x
Parkhill, J., Sebaihia, M., Preston, A., Murphy, L. D., Thomson, N., Harris, D. E., et al. (2003). Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35, 32–40. doi: 10.1038/ng1227
Pasternak, C., Ton-Hoang, B., Coste, G., Bailone, A., Chandler, M., and Sommer, S. (2010). Irradiation-induced Deinococcus radiodurans genome fragmentation triggers transposition of a single resident insertion sequence. PLoS Genet. 6:e1000799. doi: 10.1371/journal.pgen.1000799
Prentki, P., Teter, B., Chandler, M., and Galas, D. J. (1986). Functional promoters created by the insertion of transposable element IS1. J. Mol. Biol. 191, 383–393. doi: 10.1016/0022-2836(86)90134-8
Ram, Y., and Hadany, L. (2014). Stress-induced mutagenesis and complex adaptation. Proc. R. Soc. B 281:20141025. doi: 10.1098/rspb.2014.1025
Redenbach, M., Kieser, H. M., Denapaite, D., Eichner, A., Cullum, J., Kinashi, H., et al. (1996). A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21, 77–96. doi: 10.1046/j.1365-2958.1996.6191336.x
Riesenfeld, C., Everett, M., Piddock, L. J., and Hall, B. G. (1997). Adaptive mutations produce resistance to ciprofloxacin. Antimicrob. Agents Chemother. 41, 2059–2060.
Rosenberg, S. M., and Hastings, P. J. (2004). Adaptive point mutation and adaptive amplification pathways in the Escherichia coli Lac system: stress responses producing genetic change. J. Bacteriol. 186, 4838–4843. doi: 10.1128/JB.186.15.4838-4843.2004
Roth, J. R. (2011). The joys and terrors of fast adaptation: new findings elucidate antibiotic resistance and natural selection. Mol. Microbiol. 79, 279–282. doi: 10.1111/j.1365-2958.2010.07459.x
Saier, M. H. Jr., and Zhang, Z. (2014). Transposon-mediated directed mutation controlled by DNA binding proteins in Escherichia coli. Front. Microbiol. 5:390. doi: 10.3389/fmicb.2014.00390
Schneider, D., Duperchy, E., Coursange, E., Lenski, R. E., and Blot, M. (2000a). Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics 156, 477–488.
Schneider, D., Faure, D., Noirclerc-Savoye, M., Barrière, A. C., Coursange, E., and Blot, M. (2000b). A broad-host-range plasmid for isolating mobile genetic elements in gram-negative bacteria. Plasmid 44, 201–207. doi: 10.1006/plas.2000.1483
Schroeder, A., Mueller, O., Stocker, S., Salowsky, R., Leiber, M., Gassmann, M., et al. (2006). The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7:3. doi: 10.1186/1471-2199-7-3
Shapiro, J. A. (1984). Observations on the formation of clones containing araB-lacZ cistron fusions. Mol. Gen. Genet. 194, 79–90. doi: 10.1007/BF00383501
Siguier, P., Filée, J., and Chandler, M. (2006). Insertion sequences in prokaryotic genomes. Curr. Opin. Microbiol. 9, 526–531. doi: 10.1016/j.mib.2006.08.005
Siguier, P., Gourbeyre, E., and Chandler, M. (2014). Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol. Rev. 38, 865–891. doi: 10.1111/1574-6976.12067
Stanley, D., Fraser, S., Stanley, G. A., and Chambers, P. J. (2010). Retrotransposon expression in ethanol-stressed Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 87, 1447–1454. doi: 10.1007/s00253-010-2562-y
Studer, A., Zhao, Q., Ross-Ibarra, J., and Doebley, J. (2011). Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 43, 1160–1163. doi: 10.1038/ng.942
Takahashi, K., Sekine, Y., Chibazakura, T., and Yoshikawa, H. (2007). Development of an intermolecular transposition assay system in Bacillus subtilis 168 using IS4Bsu1 from Bacillus subtilis (natto). Microbiology 153, 2553–2559. doi: 10.1099/mic.0.2007/007104-0
Tauch, A., Zheng, Z., Pühler, A., and Kalinowski, J. (1998). Corynebacterium striatum chloramphenicol resistance transposon Tn5564: genetic organization and transposition in Corynebacterium glutamicum. Plasmid 40, 126–139. doi: 10.1006/plas.1998.1362
Tibazarwa, C., Wuertz, S., Mergeay, M., Wyns, L., and van Der Lelie, D. (2000). Regulation of the cnr cobalt and nickel resistance determinant of Ralstonia eutropha (Alcaligenes eutrophus) CH34. J. Bacteriol. 182, 1399–1409. doi: 10.1128/JB.182.5.1399-1409.2000
Trepreau, J., Girard, E., Maillard, A. P., de Rosny, E., Petit-Haertlein, I., Kahn, R., et al. (2011). Structural basis for metal sensing by CnrX. J. Mol. Biol. 408, 766–779. doi: 10.1016/j.jmb.2011.03.014
Trepreau, J., Grosse, C., Mouesca, J. M., Sarret, G., Girard, E., Petit-Haertlein, I., et al. (2014). Metal sensing and signal transduction by CnrX from Cupriavidus metallidurans CH34: role of the only methionine assessed by a functional, spectroscopic, and theoretical study. Metallomics 6, 263–273. doi: 10.1039/C3MT00248A
Turatsinze, J. V., Thomas-Chollier, M., Defrance, M., and van Helden, J. (2008). Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat. Protoc. 3, 1578–1588. doi: 10.1038/nprot.2008.97
Van Houdt, R., Monchy, S., Leys, N., and Mergeay, M. (2009). New mobile genetic elements in Cupriavidus metallidurans CH34, their possible roles and occurrence in other bacteria. Antonie Van Leeuwenhoek 96, 205–226. doi: 10.1007/s10482-009-9345-4
Van Houdt, R., Monsieurs, P., Mijnendonckx, K., Provoost, A., Janssen, A., Mergeay, M., et al. (2012). Variation in genomic islands contribute to genome plasticity in Cupriavidus metallidurans. BMC Genomics 13:111. doi: 10.1186/1471-2164-13-111
von Rozycki, T., and Nies, D. H. (2009). Cupriavidus metallidurans: evolution of a metal-resistant bacterium. Antonie Van Leeuwenhoek 96, 115–139. doi: 10.1007/s10482-008-9284-5
Wang, X., and Wood, T. K. (2011). IS5 inserts upstream of the master motility operon flhDC in a quasi-Lamarckian way. ISME J. 5, 1517–1525. doi: 10.1038/ismej.2011.27
Wright, B. E. (2004). Stress-directed adaptive mutations and evolution. Mol. Microbiol. 52, 643–650. doi: 10.1111/j.1365-2958.2004.04012.x
Zhang, Z., and Saier, M. H. Jr. (2009). A mechanism of transposon-mediated directed mutation. Mol. Microbiol. 74, 29–43. doi: 10.1111/j.1365-2958.2009.06831.x
Keywords: metal ions, resistance, Cupriavidus, insertion sequence element, adaptation
Citation: Vandecraen J, Monsieurs P, Mergeay M, Leys N, Aertsen A and Van Houdt R (2016) Zinc-Induced Transposition of Insertion Sequence Elements Contributes to Increased Adaptability of Cupriavidus metallidurans. Front. Microbiol. 7:359. doi: 10.3389/fmicb.2016.00359
Received: 10 November 2015; Accepted: 07 March 2016;
Published: 23 March 2016.
Edited by:
John R. Battista, Louisiana State University and A&M College, USAReviewed by:
Daniel Stoebel, Harvey Mudd College, USAChristophe Merlin, University of Lorraine, France
Copyright © 2016 Vandecraen, Monsieurs, Mergeay, Leys, Aertsen and Van Houdt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abram Aertsen, YWJyYW0uYWVydHNlbkBiaXcua3VsZXV2ZW4uYmU=;
Rob Van Houdt, cnZob3VkdG9Ac2NrY2VuLmJl
 Joachim Vandecraen1,2
Joachim Vandecraen1,2 Abram Aertsen
Abram Aertsen Rob Van Houdt
Rob Van Houdt