- Department of Microbiology, National Reference Centre for Respiratory Pathogens, University Hospital Antwerp, Antwerp, Belgium
Mycoplasma pneumoniae (M. pneumoniae) belongs to the class Mollicutes and has been recognized as a common cause of respiratory tract infections (RTIs), including community-acquired pneumonia (CAP), that occur worldwide and in all age groups. In addition, M. pneumoniae can simultaneously or sequentially lead to damage in the nervous system and has been associated with a wide variety of other acute and chronic diseases. During the past 10 years, the proportion of LRTI in children and adults, associated with M. pneumoniae infection has ranged from 0 to more than 50%. This variation is due to the age and the geographic location of the population examined but also due to the diagnostic methods used. The true role of M. pneumoniae in RTIs remains a challenge given the many limitations and lack of standardization of the applied diagnostic tool in most cases, with resultant wide variations in data from different studies. Correct and rapid diagnosis and/or management of M. pneumoniae infections is, however, critical to initiate appropriate antibiotic treatment and is nowadays usually done by PCR and/or serology. Several recent reviews, have summarized current methods for the detection and identification of M. pneumoniae. This review will therefore provide a look at the general principles, advantages, diagnostic value, and limitations of the most currently used detection techniques for the etiological diagnosis of a M. pneumoniae infection as they evolve from research to daily practice.
About 50 years ago, an outbreak of M. pneumoniae in a pediatric chronic care facility was described (Baernstein et al., 1965). Twenty years earlier, the organism had been identified by Eaton and since the early 1960s, it was clearly identified as a bacterium which was associated both in children and adults with community-acquired infections of the respiratory tract (Lambert, 1964). Since then, numerous reports have been published on the association of M. pneumoniae with community-acquired infections (Waites and Talkington, 2004). Given the wide variations of data from studies with equally wide variation of and lack of standardized diagnostic methods, the true role of M. pneumoniae in RTIs still remains a challenge.
Since its discovery, scientists have explored several strategies for an optimal diagnosis of a M. pneumoniae infection in the laboratory to initiate an appropriate treatment. Because of its fastidious nature, M. pneumoniae is not routinely cultured from respiratory specimens. Culture methods have been the gold standard for diagnosis but are too insensitive producing a result after several days or even several weeks and are therefore not relevant for the management of acute illness. Alternative diagnostic procedures were developed: Detection of IgM and/or IgG by ELISA, antigen detection by immunochromatography, and nucleic acid amplification techniques (NAATs), mainly PCR, although also isothermal amplification techniques such as LAMP (loop-mediated isothermal amplification method) have been developed. The utility of culture for M. pneumoniae was assessed by comparing it to PCR and IgM serology in a large study (She et al., 2010). Given the extremely low yield of culture and the wide availability of NAAT and serology, the authors concluded that culture for M. pneumoniae should be discontinued. Nowadays, most studies are serology and/or PCR-based. Different clinical specimens can be used as described in the review by Loens et al. (2009) for the latter.
Application of NAATs
PCR is accepted as a rapid diagnostic test. Few of the currently available NAATs have been extensively validated against culture. The sensitivity of NAATs is almost always superior to that of traditional procedures and they are more and more considered as the “new gold standard.”
An increasing body of literature describing the use of in-house NAATs for detection of M. pneumoniae DNA or RNA in various diseases is available with a great variation of methods used from study to study, including variability of target (P1 adhesin gene, 16S rRNA, ATPase gene, protease gene, CARDS toxin gene), NAAT (conventional, nested, real-time; monoplex vs. multiplex; PCR vs. isothermal amplification technologies), detection formats, and different platforms. An overview of the literature on the use of NAATs to detect M. pneumoniae since 1989 is given in two reviews (Loens et al., 2003, 2010a).
Lately, efforts have been mainly emphasized on the development of multiplex assays (Nummi et al., 2015; Shen et al., 2015) and on the evaluation of commercially available assays. Respiratory viruses and other so called “atypical bacteria” are all responsible for RTIs that may produce clinically similar manifestations. In order to reduce costs and hands-on-time, multiplex NAATs for the simultaneous detection of 2, 3, or up to more than 20 different respiratory pathogens in one tube with a mixture of primers have been developed by some groups. However, comparison between mono-and multiplex assays has been rarely performed. Findings and conclusions result frequently in contradictory and conflicting data concerning the sensitivity and specificity of the multiplex NAATs compared to the mono NAATs. This is not unexpected since the presence of several pairs of primers may increase the probability of mispairing resulting in non-specific amplification products and the formation of primer-dimers. Furthermore, enzymes, primers, and salt concentrations as well as temperature cyclings required for each target may be slightly different. The results of the proficiency panels (Loens et al., 2010b, 2012) described previously seem to confirm that multiplex assays are somewhat less sensitive than monoplex assays but until the number of organisms present in clinical specimens of diseased individuals is known, it is impossible to state whether the degree of sensitivity attained is clinically acceptable.
Since the previous review (Ieven and Loens, 2013) new NAATs became commercially available such as the Illumigene (Meridian Bioscience, USA) kit. It has been proposed that industry-produced assays in kit form result in better standardization. The analytical sensitivity of the Illumigene assay was evaluated by using 36 frozen stock cultures of M. pneumoniae reference strains, and a collection of other microorganisms and human DNA. (Ratliff et al., 2014). Serial dilutions of cultures with a known CFU/ml defined the analytical sensitivity at ≤88 CFU/ml. Based on the results obtained with 214 archived respiratory specimens, previously cultured for M. pneumoniae, the clinical sensitivity and specificity were found to be 100 and 99%, respectively, after resolving discrepancies by PCR and sequencing.
A second example of a test approved for the detection of a number of respiratory viruses by the US Food and Drug Administration is the Filmarray Respiratory panel (bioMérieux, France). The Filmarray is a small desktop closed single-piece flow real-time PCR system. It includes automation of nucleic acid extraction, an initial reverse transcription and multiplex PCR, followed by singleplex second stage PCR reactions for the detection of 15 viral agents including adenovirus, coronavirus HKU1, coronavirus NL63, human metapneumovirus, rhinovirus/enterovirus, influenza A/B, influenza A H1, AH1 2009, A H3, parainfluenza 1–4, and respiratory syncytial virus (Poritz et al., 2011). In May 2012, the US Food and Drug Administration expanded the use for the Filmarray respiratory panel with the addition of B. pertussis, M. pneumonia, and C. pneumoniae. The expanded panel detects now a total of 17 viruses and three bacteria. The test requires 5 min hands-on-time and 65 min instrumentation time. In 2013, a new version of the Filmarray (version 1.7) was released (Doern et al., 2013).
The Argene Respiratory MWSr-gene concept allows the detection of numerous pathogens (Influenza A/B, respiratory syncytial virus/human metapneumovirus, rhinovirus/enterovirus, adenovirus/bocavirus, Chlamydia/Mycoplasma pneumoniae, human coronavirus/parainfluenza virus, Bordetella, Bordetella parapertussis) in the same run. In addition, the diagnostic strategy can be adapted to the season: searching for the most likely pathogens can be considered in 1st stage, the remaining pathogens being searched for systematically in a 2nd stage.
Pillet et al. (2013) compared six commercially available multiplex assays for the diagnosis of respiratory pathogens. Two out of six were also capable of detecting M. pneumoniae: the RespiFinder SMART 22 (PathoFinder, The Netherlands) and the Seeplex RV15 OneStep ACE detection and Pneumobacter ACE detection (Seegene Inc, South Korea). Sensitivities and specificities were calculated against the ArgeneChla/Myco pneumo assay (bioMérieux, France). Sensitivity and specificity were 70.0 and 100%, respectively, for the RespiFinder assay and 80.0 and 98.73% for the Seegene assay.
Dumke et al. compared four commercially available real-time PCR assays recommended for use with the Roche LightCycler 1.5 and 2.0 instruments [Diagenode Mycoplasma/Chlamydophila pneumoniae real-time PCR (Diagenode, Belgium), GeneProof M. pneumoniae (GeneProof, Czech Republic), BactoReal M. pneumoniae (Ingenetix, Austria), LightMix kit M. pneumoniae (TIB MOLBIOL, Germany)] for the detection of M. pneumoniae to results obtained with an in-house approach (Dumke and Jacobs, 2014) by using serial dilutions of a cultured M. pneumoniae strain tested in eight parallel runs and 37 clinical specimens, previously found to be M. pneumoniae positive by the in-house assay. All NAATs detected 20 colony forming units (CFU)/5μl sample. Only the in-house-test (repMP1-based approach) was able to detect 0.2 CFU/5μl sample. 37/37,35/37, 35/37, 34/37 M. pneumoniae positive clinical specimens were confirmed by the Diagenode test, the Ingenetix and Lightmix assay, and the GeneProof assay respectively.
An overview of commercially available NAATS for the detection of M. pneumoniae is presented in Table 1.
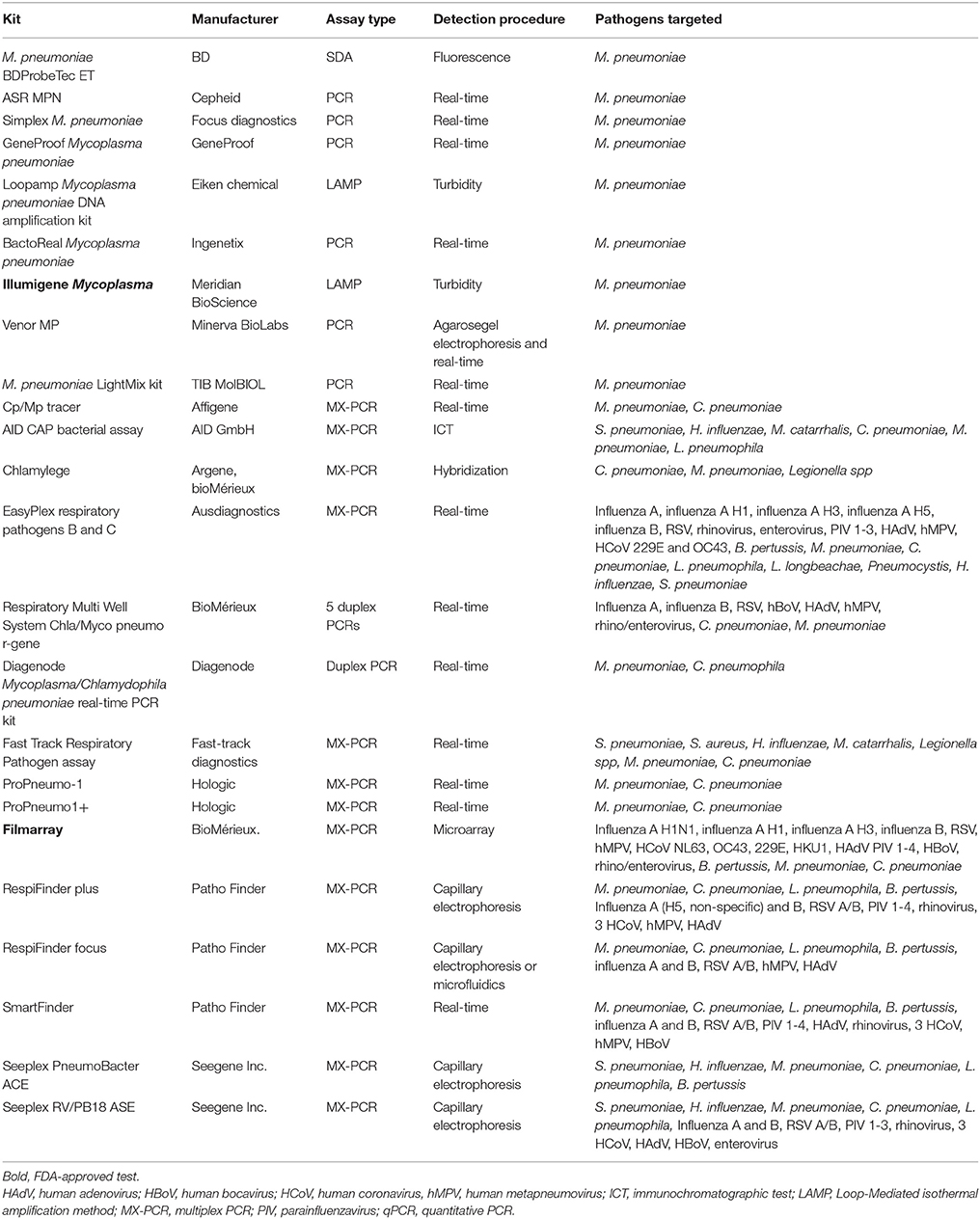
Table 1. Summary of commercially available single and multiplex PCR assays for detection of M. pneumonia.
Since the calculation of the sensitivities of the commercial multiplex assays was mainly dependent on DNA copy number, further evaluation and standardization using an extended number of clinical specimens that may have a low bacterial load are needed. The use of an international standard developed by the WHO for harmonization of Mycoplasma NAAT (Nübling et al., 2015) or the yearly participation in the external quality assessment (EQA) panel for M. pneumoniae and Chlamydophila pneumoniae available from Quality Control for Molecular Diagnostics (QCMD, United Kingdom) should be considered.
So far, it is unclear whether asymptomatic carriage of M. pneumoniae in adults and children exists and if colonization could be differentiated from infection by the current diagnostic methods. There are only few data on the relation between the bacterial load and the severity of infection. 405 asymptomatic children and 321 children with a RTI were enrolled in a cross-sectional study (Spuesens et al., 2013). Nasopharyngeal washings and pharyngeal swabs were investigated by culture and quantitative real-time PCR (qPCR). Serum was collected for IgM and IgG ELISA. Neither qPCR, serology nor culture was capable of differentiating colonization from infection. In 21.2 and 16.2% of the asymptomatic and symptomatic children, M. pneumoniae DNA was detected. In addition, persistence of M. pneumoniae in the upper respiratory tract was shown for up to 4 months by longitudinal sampling. A retrospective study investigated the clinical significance of the M. pneumoniae bacterial load in children with a M. pneumoniae pneumonia (Jiang et al., 2014). The authors concluded that a high bacterial load was indicative for a M. pneumoniae infection, whereas for a low bacterial load the etiologic role of M. pneumoniae remains to be determined.
Edin et al. developed a qPCR with duplex reactions targeting eight bacteria, including M. pneumoniae, and six viruses (Edin et al., 2015). Clinical specimens from the upper and lower respiratory tract were used to compare the qPCR assay with standard microbiological methods. The use of the qPCR assay resulted in 113 positive identifications in 94 respiratory specimens compared with 38 by using standard diagnostics. The authors conclude that in parallel qPCR detection of the targeted respiratory bacteria and viruses is feasible since a good technical performance of the assay in clinical specimens was obtained.
In contrast to the above mentioned studies, Jain et al. (2015) examined specimens from 2222 hospitalized children with community-acquired pneumonia and 521 asymptomatic controls for the detection of a variety of respiratory pathogens. M. pneumoniae was detected in 8%, and in 3% or less of controls.
Another trend is the simultaneous detection of M. pneumoniae and mutations associated with macrolide resistance directly in clinical specimens (Ji et al., 2014; Liu et al., 2014; Nummi et al., 2015; Zhao et al., 2015).
Application of Serology for the Detection of M. pneumoniae Infections
Serological methods, in particular enzyme-linked immunosorbent assays (ELISA), are most widely used to diagnose a M. pneumoniae infection. The complement fixation test (CFT) has been replaced by assays which allow for quantification of IgM, IgA, or IgG. However, the most convincing evidence of an ongoing infection is a significant increase in IgG or an IgG seroconversion in paired sera, collected 3–4 weeks apart (Nir-Paz et al., 2006). Although IgM antibodies appear earlier than IgG antibodies, and are thus an attractive alternative for diagnosis of a M. pneumoniae infection, one should realize that IgM is not often produced in very young children, in a proportion of primary infections and during re-infections (Waites et al., 2008; Loens et al., 2010a).
Ten serological assays for the diagnosis of a M. pneumoniae infection were recently evaluated by using 145 sera from 120 patients (Busson et al., 2013): SeroMP IgM and IgG (Savyon Diagnostics), SeroMP Recombinant IgM, IgA and IgG (Savyon Diagnostics), LIAISON M. pneumoniae IgM and IgG (Biotrin International Ltd), M. pneumoniae IgM, IgA and IgG Medac (Medac GmbH). A low IgM specificity and cross-reactivity was noticed for the SeroMP recombinant and Liaison assay. For IgA, the Medac assay tended to be less specific than the SeroMP Recombinant assay. All four tests showed discrepancies in the IgG measurements confirming results of previous studies (Talkington et al., 2004; Beersma et al., 2005). In conclusion, serology remains a diagnostic tool of choice but improvement and standardization of the assays are still needed, especially for the determination of IgG.
The clinical significance of a serologic test, both for IgM and IgG, should be defined by studies of patients with a documented infection and for whom detailed information concerning the time lapses between onset of disease and the collection of the serum specimens are known.
A promising blotting technique improving the performance of the M. pneumoniae serological assays has been described (Dumke et al., 2012).
Detection of M. pneumoniae by both NAATs and Serology
Data from recent studies using PCR based methods and serology published during the last decade in different patient populations from around the world are summarized in the recent reviews published by Ieven and Loens (Loens et al., 2010a; Ieven and Loens, 2013) and updated in Table 2.
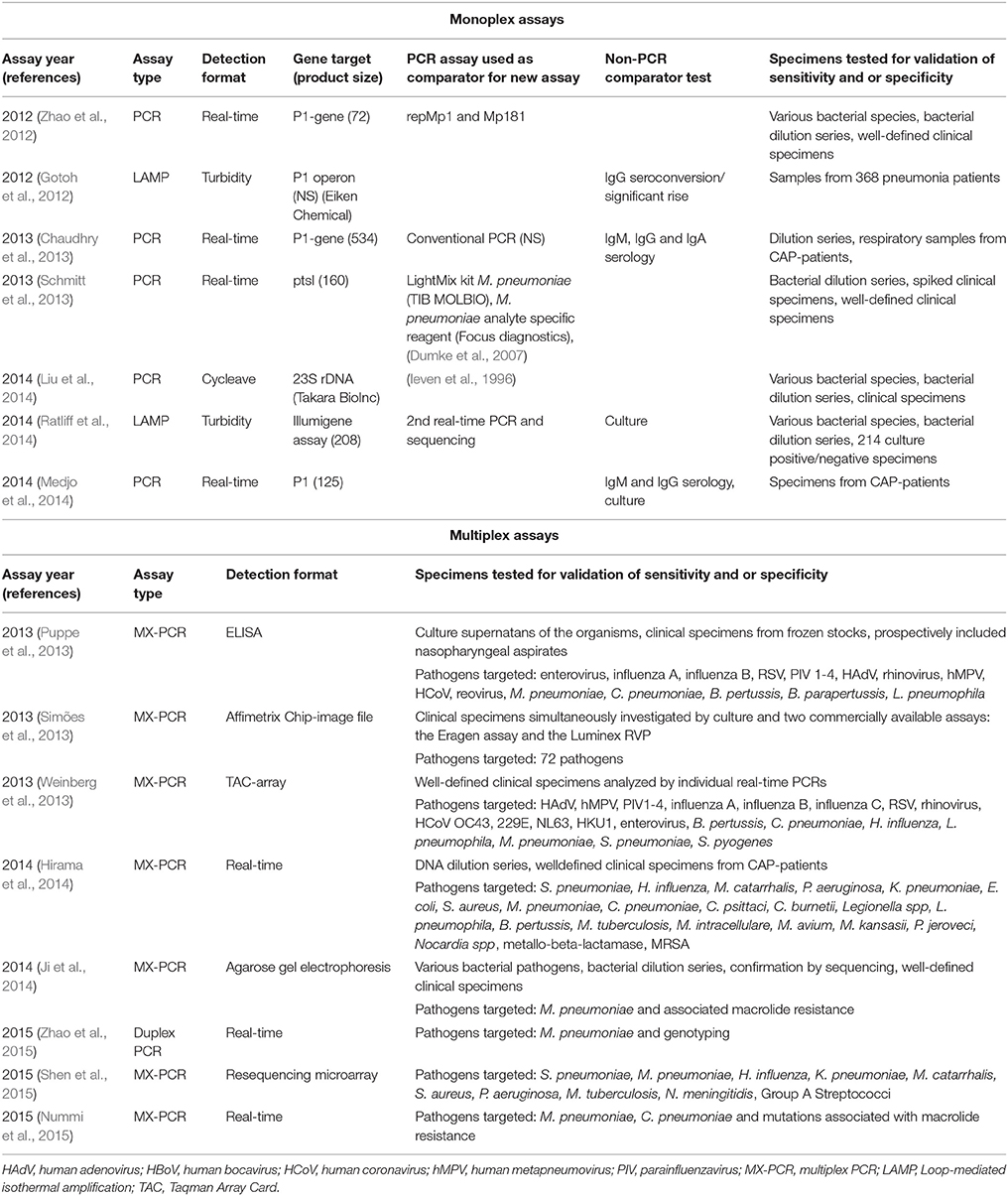
Table 2. Summary of recent single and multiplex NAATs for detection of Mycoplasma pneumoniae published since the previous review, and previously validated assays used as comparators.
The availability of the very sensitive NAATs has in recent years also put the often used serological tests in their right perspective and allow a better interpretation of the serological test results and their limitations such as the low sensitivity of IgM antibodies in acute phase specimens and importance of the delay between two serum samples. Studies in which also NAAT's are used on respiratory specimens should allow a better interpretation of the serological test results.
A rapid response report from the Canadian Agency for Drugs and Technologies in Health (Canadian Agency for Drugs and Technologies in Health, 2015) presents the results of a literature search in order to identify the diagnostic test accuracy, clinical effectiveness, and cost-effectiveness of serum IgM and molecular tests for the detection of M. pneumoniae in patients with a respiratory infection1. Six relevant studies were identified, but no evidence regarding the clinical effectiveness or cost-effectiveness of a serum IgM test compared with molecular tests was identified. Zhang et al. conducted a systematic review and meta-analysis on the diagnosis of M. pneumoniae by PCR and serology (Zhang et al., 2011) and reported a significant heterogeneity between the studies and inconsistent results as well.
Two studies compared the application of real-time PCR and serology in children with pneumonia. In 2011, 54/290 children were found to be positive by PCR (Chang et al., 2014), 44/182 were M. pneumoniae IgM positive. 12.6% of patients were found to be M. pneumoniae positive by both tests at the same time. Using PCR as gold standard, a sensitivity and specificity of resp. 62.2 and 85.5% were obtained. The specificity could be increased to 90.3% by increasing the cut-off without changing the sensitivity of the IgM assay. A study conducted by Medjo et al. (2014) applied PCR, culture, IgM and IgG in paired sera for the detection of a M. pneumoniae infection in 166 children. Using IgG serology as gold standard, the sensitivity of IgM, PCR, and culture was found to be equal (81.8%), specificity was found to be 100, 98.6, and 100% respectively. It was concluded that during the acute phase of disease, detection of IgM antibodies in combination with PCR allowed for a precise and reliable M. pneumoniae diagnosis. A prospective study in children with community-acquired CAP (Kakuya et al., 2014) compared loop-mediated isothermal amplification, (LAMP), culture and serology at first visit. Patients were defined positive if positive by culture and/or sero-conversion or a four-fold increase in IgG in paired sera. 31/191 patients met the criteria. Thirteen were positive by culture and serology, 17 on culture only, and one by serology only. A positive LAMP result was obtained for all patients that were culture positive. The sensitivity and specificity for LAMP, EIA, and the particle agglutination test, were 96.8, 38.7, 19.4, and 100%, 76.9 and 93.1%, respectively.
When establishing the etiology in 267 adult CAP-patients in Norway, 10 were found to be M. pneumoniae positive: two by serology, seven and one by PCR applied to a nasopharyngeal flocked swab and an oropharyngeal flocked swab, respectively (Holter et al., 2015).
Amplification-free and other Technological Developments
Newer technologies such as microfluidics and the application of nanotechnology offer the potential to an even more rapid detection of important pathogens allowing even near-patient testing. Since these technologies, as NAATs, do not require viable organisms, and thus avoid any adverse effect of longer specimen transport, they can be successfully applied to both the in- and outpatient settings. Several companies currently possess the technical expertise and research infrastructure to bring a useful diagnostic testing approach to the clinical trial stage shortly.
Li et al. (2015) developed a colloidal gold-based immune-chromatographic assay by using a pair of monoclonal antibodies targeting a region of the P1 gene. When applied to 303 clinical specimens from children suspected with a M. pneumoniae infection, the sensitivity and specificity against real-time PCR were 100 and 97.4%. This is in contrast to the results obtained with a commercially available rapid antigen test targeting the ribosomal protein L7/L12 (Ribotest Mycoplasma). Compared to real-time PCR, a sensitivity and specificity of respectively 62.5 and 90.9% were obtained when applied to clinical specimens (Miyashita et al., 2015). Based on these results, the authors concluded that treatment decisions should not be taken based on the Ribotest results alone.
Other amplification-free detection methodologies are currently being developed as biosensing detection strategies: A proto-type of an enzyme-free electrochemical genosensor on nanostructured screen-printed gold electrodes (Garcia-Gonzalez et al., 2015); A silver nanorod array-surface enhanced Raman Spectroscopy biosensing platform was successfully applied for the detection of M. pneumoniae in simulated and clinical throat swabs (Henderson et al., 2014, 2015).
Conclusions
With the use of tools such as NAATs a greater understanding of the etiology and epidemiology of M. pneumoniae is possible. Taken into account the results obtained in recent studies, there is more evidence that real-time NAATs are superior to other M. pneumoniae detection strategies during the early phase of infection. NAATs, however, cannot completely replace serology. In epidemiological studies, serology is certainly more useful than for the management of individual patients with LRTI or even CAP since results are often delayed by the need for paired sera to detect a seroconversion or a significant rise in titer; early in the course of an infection, false-negative results often occur.
In case a specific IgM test is used, serology should not completely be abolished despite the fact that IgM serology shows a moderate sensitivity. Nowadays, a combination of the detection of IgM antibodies and PCR may be the most optimal approach for early diagnosis of a M. pneumoniae infection, especially in children.
The implementation of quantitative tests could shed further light on the relation between bacterial load and the seriousness of the disease, produce useful prognostic information and help in the differentiation between colonization and infection. More information could be gathered on the length of the post infection carrier state as well as on the importance of subclinical infections and how prone these are for spreading infection.
It remains important to recognize the urgent need for the adoption of a more unified and consistent diagnostic approach for current and future investigations. Therefore, a common set of recommendations should be developed.
Author Contributions
KL drafted the manuscript. GI revised and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
KL is supported through The Belgian National Reference Centre for Respiratory Pathogens which is partially supported by the Belgian Ministry of Social affairs through a fund within the Health Insurance System. KL is also supported by the EU FP7 project PREPARE (602525).
Footnotes
1. ^(2015). Serum IgM and Molecular Tests for Mycoplasma pneumoniae Detection: A Review of Diagnostic Test Accuracy, Clinical Effectiveness, Cost-Effectiveness, and Guidelines, Ottawa, ON.
References
Baernstein, H. D. Jr. Trevisani, E., Axtell, S., and Quilligan, J. J. Jr (1965). Mycoplasma pneumoniae (eaton atypical pneumonia agent) in children's respiratory infections. J. Pediatr. 66, 829–837. doi: 10.1016/S0022-3476(65)80057-9
Beersma, M. F., Dirven, K., van Dam, A. P., Templeton, K. E., Claas, E. C., and Goossens, H. (2005). Evaluation of 12 commercial tests and the complement fixation test for Mycoplasma pneumoniae-specific immunoglobulin G (IgG) and IgM antibodies, with PCR used as the “gold standard.” J. Clin. Microbiol. 43, 2277–2285. doi: 10.1128/JCM.43.5.2277-2285.2005
Busson, L., Van den Wijngaert, S., Dahma, H., Decolvenaer, M., Di Cesare, L., Martin, A., et al. (2013). Evaluation of 10 serological assays for diagnosing Mycoplasma pneumoniae infection. Diagn. Microbiol. Infect. Dis. 76, 133–137. doi: 10.1016/j.diagmicrobio.2013.02.027
Canadian Agency for Drugs Technologies in Health (2015). Serum IgM and Molecular Tests for Mycoplasma pneumoniae Detection: A Review of Diagnostic Test Accuracy, Clinical Effectiveness, Cost-Effectiveness, and Guidelines [Internet]. CADTH Rapid Response Reports. Ottawa, ON: Canadian Agency for Drugs and Technologies in Health.
Chang, H. Y., Chang, L. Y., Shao, P. L., Lee, P. I., Chen, J. M., Lee, C. Y., et al. (2014). Comparison of real-time polymerase chain reaction and serological tests for the confirmation of Mycoplasma pneumoniae infection in children with clinical diagnosis of atypical pneumonia. J. Microbiol. Immunol. Infect. 47, 137–144. doi: 10.1016/j.jmii.2013.03.015
Chaudhry, R., Sharma, S., Javed, S., Passi, K., Dey, A. B., and Malhotra, P. (2013). Molecular detection of Mycoplasma pneumoniae by quantitative real-time PCR in patients with community acquired pneumonia. Indian J.Med. Res. 138, 244–251.
Doern, C. D., Lacey, D., Huang, R., and Haag, C. (2013). Evaluation and implementation of FilmArray version 1.7 for improved detection of adenovirus respiratory tract infection. J. Clin. Microbiol. 51, 4036–4039. doi: 10.1128/JCM.02546-13
Dumke, R., and Jacobs, E. (2014). Evaluation of five real-time PCR assays for detection of Mycoplasma pneumoniae. J. Clin. Microbiol. 52, 4078–4081. doi: 10.1128/JCM.02048-14
Dumke, R., Schurwanz, N., Lenz, M., Schuppler, M., Lück, C., and Jacobs, E. (2007). Sensitive detection of Mycoplasma pneumoniae in human respiratory tract samples by optimized real-time PCR approach. J. Clin. Microbiol. 45, 2726–2730. doi: 10.1128/JCM.00321-07
Dumke, R., Strubel, A., Cyncynatus, C., Nuyttens, H., Herrmann, R., Lück, C., et al. (2012). Optimized serodiagnosis of Mycoplasma pneumoniae infections. Diagn. Microbiol. Infect. Dis. 73, 200–203. doi: 10.1016/j.diagmicrobio.2012.02.014
Edin, A., Granholm, S., Koskiniemi, S., Allard, A., Sjöstedt, A., and Johansson, A. (2015). Development and laboratory evaluation of a real-time PCR assay for detecting viruses and bacteria of relevance for community-acquired pneumonia. J. Mol. Diagn. 17, 315–324. doi: 10.1016/j.jmoldx.2015.01.005
García-González, R., Costa-García, A., and Fernández-Abedul, M. T. (2015). Enzymatic amplification-free nucleic acid hybridisation sensing on nanostructured thick-film electrodes by using covalently attached methylene blue. Talanta 142, 11–19. doi: 10.1016/j.talanta.2015.03.028
Gotoh, K., Nishimura, N., Ohshima, Y., Arakawa, Y., Hosono, H., Yamamoto, Y., et al. (2012). Detection of Mycoplasma pneumoniae by loop-mediated isothermal amplification (LAMP) assay and serology in pediatric community-acquired pneumonia. J. Infect. Chemother. 18, 662–667. doi: 10.1007/s10156-012-0388-5
Henderson, K. C., Benitez, A. J., Ratliff, A. E., Crabb, D. M., Sheppard, E. S., Winchell, J. M., et al. (2015). Specificity and strain-typing capabilities of nanorod array-surface enhanced raman spectroscopy for Mycoplasma pneumoniae detection. PLoS ONE 10:e0131831. doi: 10.1371/journal.pone.0131831
Henderson, K. C., Sheppard, E. S., Rivera-Betancourt, O. E., Choi, J. Y., Dluhy, R. A., Thurman, K. A., et al. (2014). The multivariate detection limit for Mycoplasma pneumoniae as determined by nanorod array-surface enhanced Raman spectroscopy and comparison with limit of detection by qPCR. Analyst 139, 6426–6434. doi: 10.1039/C4AN01141D
Hirama, T., Minezaki, S., Yamaguchi, T., Kishi, E., Kodama, K., Egashira, H., et al. (2014). HIRA-TAN: a real-time PCR-based system for the rapid identification of causative agents in pneumonia. Respir. Med. 108, 395–404. doi: 10.1016/j.rmed.2013.11.018
Holter, J. C., Müller, F., Bjørang, O., Samdal, H. H., Marthinsen, J. B., Jenum, P. A., et al. (2015). Etiology of community-acquired pneumonia and diagnostic yields of microbiological methods: a 3-year prospective study in Norway. BMC Infect. Dis. 15:64. doi: 10.1186/s12879-015-0803-5
Ieven, M., and Loens, K. (2013). Should serology be abolished in favor of PCR for the diagnosis of Mycoplasma pneumoniae infections? Curr. Pediatr. Rev. 9, 304–313. doi: 10.2174/157339630904131223110501
Ieven, M., Ursi, D., Van Bever, H., Quint, W., Niesters, H. G., and Goossens, H. (1996). Detection of Mycoplasma pneumoniae by two polymerase chain reactions and role of M. pneumoniae in acute respiratory tract infections in pediatric patients. J. Infect. Dis. 173, 1445–1452. doi: 10.1093/infdis/173.6.1445
Jain, S., Williams, D. J., Arnold, S. R., Ampofo, K., Bramley, A. M., Reed, C., et al. (2015). Community-acquired pneumonia requiring hospitalization among U.S. children. N. Engl. J. Med. 372, 835–845. doi: 10.1056/NEJMoa1405870
Ji, M., Lee, N. S., Oh, J. M., Jo, J. Y., Choi, E. H., Yoo, S. J., et al. (2014). Single-nucleotide polymorphism PCR for the detection of Mycoplasma pneumoniae and determination of macrolide resistance in respiratory samples. J. Microbiol. Methods 102, 32–36. doi: 10.1016/j.mimet.2014.04.009
Jiang, W., Yan, Y., Ji, W., Wang, Y., and Chen, Z. (2014). Clinical significance of different bacterial load of Mycoplasma pneumoniae in patients with Mycoplasma pneumoniae pneumonia. Braz. J. Infect. Dis. 18, 124–128. doi: 10.1016/j.bjid.2013.06.004
Kakuya, F., Kinebuchi, T., Fujiyasu, H., Tanaka, R., and Kano, H. (2014). Genetic point-of-care diagnosis of Mycoplasma pneumoniae infection using LAMP assay. Pediatr. Int. 56, 547–552. doi: 10.1111/ped.12327
Lambert, H. P. (1964). Eaton agent and other non-bacterial pneumonias. Public Health 78, 335–341. doi: 10.1016/S0033-3506(64)80050-0
Li, W., Liu, Y., Zhao, Y., Tao, R., Li, Y., and Shang, S. (2015). Rapid diagnosis of Mycoplasma pneumoniae in children with pneumonia by an immuno-chromatographic antigen assay. Sci. Rep. 5:15539. doi: 10.1038/srep15539
Liu, Y., Ye, X., Zhang, H., Wu, Z., and Xu, X. (2014). Rapid detection of Mycoplasma pneumoniae and its macrolide-resistance mutation by Cycleave PCR. Diagn. Microbiol. Infect. Dis. 78, 333–337. doi: 10.1016/j.diagmicrobio.2013.12.002
Loens, K., Goossens, H., and Ieven, M. (2010a). Acute respiratory infection due to Mycoplasma pneumoniae: current status of diagnostic methods. Eur. J. Clin. Microbiol. Infect. Dis. 29, 1055–1069. doi: 10.1007/s10096-010-0975-2
Loens, K., Mackay, W. G., Scott, C., Goossens, H., Wallace, P., and Ieven, M. (2010b). A multicenter pilot external quality assessment programme to assess the quality of molecular detection of Chlamydophila pneumoniae and Mycoplasma pneumoniae. J. Microbiol. Methods 82, 131–135. doi: 10.1016/j.mimet.2010.05.006
Loens, K., Ursi, D., Goossens, H., and Ieven, M. (2003). Molecular diagnosis of Mycoplasma pneumoniae respiratory tract infections. J. Clin. Microbiol. 41, 4915–4923. doi: 10.1128/JCM.41.11.4915-4923.2003
Loens, K., Van Heirstraeten, L., Malhotra-Kumar, S., Goossens, H., and Ieven, M. (2009). Optimal sampling sites and methods for detection of pathogens possibly causing community-acquired lower respiratory tract infections. J. Clin. Microbiol. 47, 21–31. doi: 10.1128/JCM.02037-08
Loens, K., van Loon, A. M., Coenjaerts, F., van Aarle, Y., Goossens, H., Wallace, P., et al. (2012). Performance of different mono- and multiplex nucleic acid amplification tests on a multipathogen external quality assessment panel. J. Clin. Microbiol. 50, 977–987. doi: 10.1128/JCM.00200-11
Medjo, B., Atanaskovic-Markovic, M., Radic, S., Nikolic, D., Lukac, M., and Djukic, S. (2014). Mycoplasma pneumoniae as a causative agent of community-acquired pneumonia in children: clinical features and laboratory diagnosis. Ital. J. Pediatr. 40:104. doi: 10.1186/s13052-014-0104-4
Miyashita, N., Kawai, Y., Tanaka, T., Akaike, H., Teranishi, H., Wakabayashi, T., et al. (2015). Diagnostic sensitivity of a rapid antigen test for the detection of Mycoplasma pneumoniae: comparison with real-time PCR. J. Infect. Chemother. 21, 473–475. doi: 10.1016/j.jiac.2015.02.007
Nir-Paz, R., Michael-Gayego, A., Ron, M., and Block, C. (2006). Evaluation of eight commercial tests for Mycoplasma pneumoniae antibodies in the absence of acute infection. Clin. Microbiol. Infect. 12, 685–688. doi: 10.1111/j.1469-0691.2006.01469.x
Nübling, C. M., Baylis, S. A., Hanschmann, K. M., Montag-Lessing, T., Chudy, M., Kress, J., et al. (2015). World health organization international standard to harmonize assays for detection of mycoplasma DNA. Appl. Environ. Microbiol. 81, 5694–5702. doi: 10.1128/AEM.01150-15
Nummi, M., Mannonen, L., and Puolakkainen, M. (2015). Development of a multiplex real-time PCR assay for detection of Mycoplasma pneumoniae, Chlamydia pneumoniae and mutations associated with macrolide resistance in Mycoplasma pneumoniae from respiratory clinical specimens. Springerplus 4:684. doi: 10.1186/s40064-015-1457-x
Pillet, S., Lardeux, M., Dina, J., Grattard, F., Verhoeven, P., Le Goff, J., et al. (2013). Comparative evaluation of six commercialized multiplex PCR kits for the diagnosis of respiratory infections. PLoS ONE 8:e72174. doi: 10.1371/journal.pone.0072174
Poritz, M. A., Blaschke, A. J., Byington, C. L., Meyers, L., Nilsson, K., Jones, D. E., et al. (2011). FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS ONE 6:e26047. doi: 10.1371/journal.pone.0026047
Puppe, W., Weigl, J., Gröndahl, B., Knuf, M., Rockahr, S., von, B. P., et al. (2013). Validation of a multiplex reverse transcriptase PCR ELISA for the detection of 19 respiratory tract pathogens. Infection 41, 77–91. doi: 10.1007/s15010-012-0298-6
Ratliff, A. E., Duffy, L. B., and Waites, K. B. (2014). Comparison of the illumigene Mycoplasma DNA amplification assay and culture for detection of Mycoplasma pneumoniae. J. Clin. Microbiol. 52, 1060–1063. doi: 10.1128/JCM.02913-13
Schmitt, B. H., Sloan, L. M., and Patel, R. (2013). Real-time PCR detection of Mycoplasma pneumoniae in respiratory specimens. Diagn. Microbiol. Infect. Dis. 77, 202–205. doi: 10.1016/j.diagmicrobio.2013.07.016
She, R. C., Thurber, A., Hymas, W. C., Stevenson, J., Langer, J., Litwin, C. M., et al. (2010). Limited utility of culture for Mycoplasma pneumoniae and Chlamydophila pneumoniae for diagnosis of respiratory tract infections. J. Clin. Microbiol. 48, 3380–3382. doi: 10.1128/JCM.00321-10
Shen, H., Zhu, B., Wang, S., Mo, H., Wang, J., Li, J., et al. (2015). Association of targeted multiplex PCR with resequencing microarray for the detection of multiple respiratory pathogens. Front. Microbiol. 6:532. doi: 10.3389/fmicb.2015.00532
Simões, E. A., Patel, C., Sung, W. K., Lee, C. W., Loh, K. H., Lucero, M., et al. (2013). Pathogen chip for respiratory tract infections. J. Clin. Microbiol. 51, 945–953. doi: 10.1128/JCM.02317-12
Spuesens, E. B., Fraaij, P. L., Visser, E. G., Hoogenboezem, T., Hop, W. C., van Adrichem, L. N., et al. (2013). Carriage of Mycoplasma pneumoniae in the upper respiratory tract of symptomatic and asymptomatic children: an observational study. PLoS Med. 10:e1001444. doi: 10.1371/journal.pmed.1001444
Talkington, D. F., Shott, S., Fallon, M. T., Schwartz, S. B., and Thacker, W. L. (2004). Analysis of eight commercial enzyme immunoassay tests for detection of antibodies to Mycoplasma pneumoniae in human serum. Clin. Diagn. Lab Immunol. 11, 862–867. doi: 10.1128/cdli.11.5.862-867.2004
Waites, K. B., Balish, M. F., and Atkinson, T. P. (2008). New insights into the pathogenesis and detection of Mycoplasma pneumoniae infections. Future Microbiol. 3, 635–648. doi: 10.2217/17460913.3.6.635
Waites, K. B., and Talkington, D. F. (2004). Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. 17, 697–728. doi: 10.1128/CMR.17.4.697-728.2004
Weinberg, G. A., Schnabel, K. C., Erdman, D. D., Prill, M. M., Iwane, M. K., Shelley, L. M., et al. (2013). Field evaluation of TaqMan Array Card (TAC) for the simultaneous detection of multiple respiratory viruses in children with acute respiratory infection. J. Clin. Virol. 57, 254–260. doi: 10.1016/j.jcv.2013.03.016
Zhang, L., Zong, Z. Y., Liu, Y. B., Ye, H., and Lv, X. J. (2011). PCR versus serology for diagnosing Mycoplasma pneumoniae infection: a systematic review & meta-analysis. Indian J. Med. Res. 134, 270–280.
Zhao, F., Cao, B., He, L. H., Yin, Y. D., Tao, X. X., Song, S. F., et al. (2012). Evaluation of a new real-time PCR assay for detection of Mycoplasma pneumoniae in clinical specimens. Biomed. Environ. Sci. 25, 77–81. doi: 10.3967/0895-3988.2012.01.011
Keywords: Mycoplasma pneumoniae, serology, nucleic acid amplification test, technological developments
Citation: Loens K and Ieven M (2016) Mycoplasma pneumoniae: Current Knowledge on Nucleic Acid Amplification Techniques and Serological Diagnostics. Front. Microbiol. 7:448. doi: 10.3389/fmicb.2016.00448
Received: 29 January 2016; Accepted: 18 March 2016;
Published: 31 March 2016.
Edited by:
Cécile Bébéar, University of Bordeaux—Institut National de la Recherche Agronomique, FranceReviewed by:
Enno Jacobs, Technische Universität Dresden, GermanyOlivia Peuchant, Université de Bordeaux, France
Copyright © 2016 Loens and Ieven. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Margareta Ieven, Z3JlZXQuaWV2ZW5AdXphLmJl
 Katherine Loens
Katherine Loens Margareta Ieven*
Margareta Ieven*