- 1Intensive Care Unit, Beijing Haidian Hospital, Haidian Section of Peking University Third Hospital, Beijing, China
- 2Department of Gastroenterology and Hepatology, Chinese PLA General Hospital, Beijing, China
- 3Department of Respiratory Medicine, Beijing Tsinghua Changgung Hospital, Medical Center, Tsinghua University, Beijing, China
- 4Nanlou Respiratory, Diseases Department, Chinese PLA General Hospital, Beijing, China
Background: Hospital-acquired pneumonia (HAP) due to Achromobacter has become a substantial concern in recent years. However, HAP due to Achromobacter in the elderly is rare.
Methods: A retrospective analysis was performed on 15 elderly patients with HAP due to Achromobacter spp., in which the sequence types (STs), integrons, biofilm production and antibiotic resistance of the Achromobacter spp. were examined.
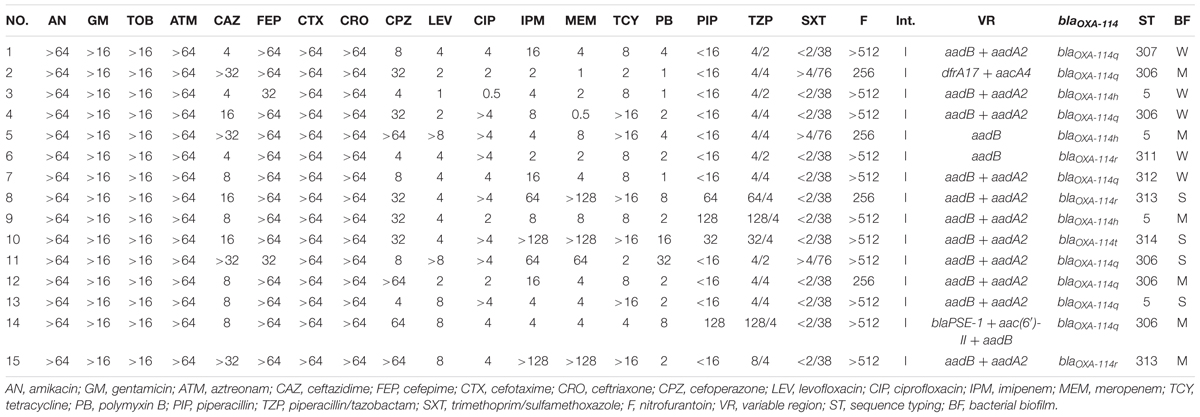
Results: The mean age of the 15 elderly patients was 88.8 ± 5.4 years. All patients had at least three underlying diseases and catheters. Clinical outcomes improved in 10 of the 15 patients after antibiotic and/or mechanical ventilation treatment, but three patients had chronic infections lasting more than 1 year. The mortality rate was 33.3% (5/15). All strains were resistant to aminoglycosides, aztreonam, nitrofurantoin, and third- and fourth-generation cephalosporins (except ceftazidime and cefoperazone). Six new STs were detected. The most frequent ST was ST306. ST5 was identified in two separate buildings of the hospital. ST313 showed higher MIC in cephalosporins, quinolones and carbapenems, which should be more closely considered in clinical practice. All strains produced biofilm and had integron I and blaOXA-114-like. The main type was blaOXA-114q. The variable region of integron I was different among strains, and the resistance gene of the aminoglycosides was most commonly inserted in integron I. Additionally, blaPSE-1 was first reported in this isolate.
Conclusion: Achromobacter spp. infection often occurs in severely ill elders with underlying diseases. The variable region of integrons differs, suggesting that Achromobacter spp. is a reservoir of various resistance genes.
Introduction
Achromobacter is a ubiquitous, non-fermenting, gram-negative bacterium that lives in soil and aquatic environments. In recent years, many studies have shown that this opportunistic pathogen can lead to various infections, such as sepsis, bacteremia, urinary infection, endocarditis, and peritonitis, especially in immunocompromised individuals (Aisenberg et al., 2004). Moreover, this organism can colonize various medical devices, such as disinfectant-soaked unwoven cleaning cloth (Oie et al., 2012), intravascular pressure transducer (Gahrn-Hansen et al., 1988), and chlorhexidine (Vu-Thien et al., 1998).
To distinguish different Achromobacter strains, various methods have been applied. Similar to OXA-50-like enzymes from P. aeruginosa or OXA-51/69 from A. baumannii, the blaOXA-114-like gene is a known characteristic of Achromobacter (Doi et al., 2008; Turton et al., 2011). Additionally, multilocus sequence typing (MLST) is one of the best methods for use in epidemiological studies in the world. There are more than 300 STs in the MLST database distributed across the world. However, due to the low incidence of Achromobacter infections, no clone complex has been reported.
Elderly patients often have underlying diseases and frequently suffer from chronic infections, which may result in continuous exposure to various types of antibiotics. This exposure may contribute to antibiotic resistance. Based on previous studies, integrons and the inborn RND-Type Multidrug Efflux Pump (Bador et al., 2011) are the main reasons for antibiotic resistance. Various integrons (intI/intII) have been detected (Traglia et al., 2012). Additionally, extended-Spectrum β-Lactamases (ESBL) and metallo-b-lactamase (MBL) production by Achromobacter spp. has been detected in the last decade. In Europe, blaV EB and blaTMB inserted in integrons were found in Achromobacter spp. (Neuwirth et al., 2006; El Salabi et al., 2012). Various subtypes of blaVIM have been detected in integron I in Asia and Europe (Riccio et al., 2001; Shin et al., 2005; Sofianou et al., 2005). Infections, pandemic or sporadic, are often related to the location of resistance genes (chromosome/plasmid). Thus, the insertion of a new blaIMP-type gene in plasmids in A. xylosoxidans has attracted wide attention because it may spread easily (Yamamoto et al., 2012). Moreover, the inherent blaOXA-114-like genes may result in a higher level of OXA-114-mediated-lactam resistance because of the insertion sequence elements (Doi et al., 2008). Therefore, Achromobacter spp. is an emerging pathogen and is becoming a reservoir for horizontal genetic transfer elements involved in spreading antibiotic resistance (Traglia et al., 2012).
Hospital-acquired pneumonia (HAP) due to Achromobacter spp. has emerged as a substantial concern in recent years. Pulmonary involvement has been frequently reported in cases with underlying disease, especially in cystic fibrosis (CF) (Dunne and Maisch, 1995; Ferroni et al., 2002). In CF patients, A. xylosoxidans isolated from sputum increased gradually (Zemel et al., 2000; Liu et al., 2002). However, CF is a rare disease in China. In non-CF patients, several cases of pneumonia have been reported in patients with underlying malignancy (Aisenberg et al., 2004), those with IgM deficiency (Dworzack et al., 1978) and those on mechanical ventilation (Chandrasekar et al., 1986). Additionally, approximately half of these non-CF patients originated from the community, which may be related to the soil and aquatic environments (Swenson and Sadikot, 2015). However, HAP due to Achromobacter spp. in the elderly is rare.
The features of this organism in the elderly need to be urgently studied. Therefore, this study aimed to understand the clinical characteristics, epidemiology, biofilm production and integrons of Achromobacter spp. in elderly patients with HAP.
Materials and Methods
Patients
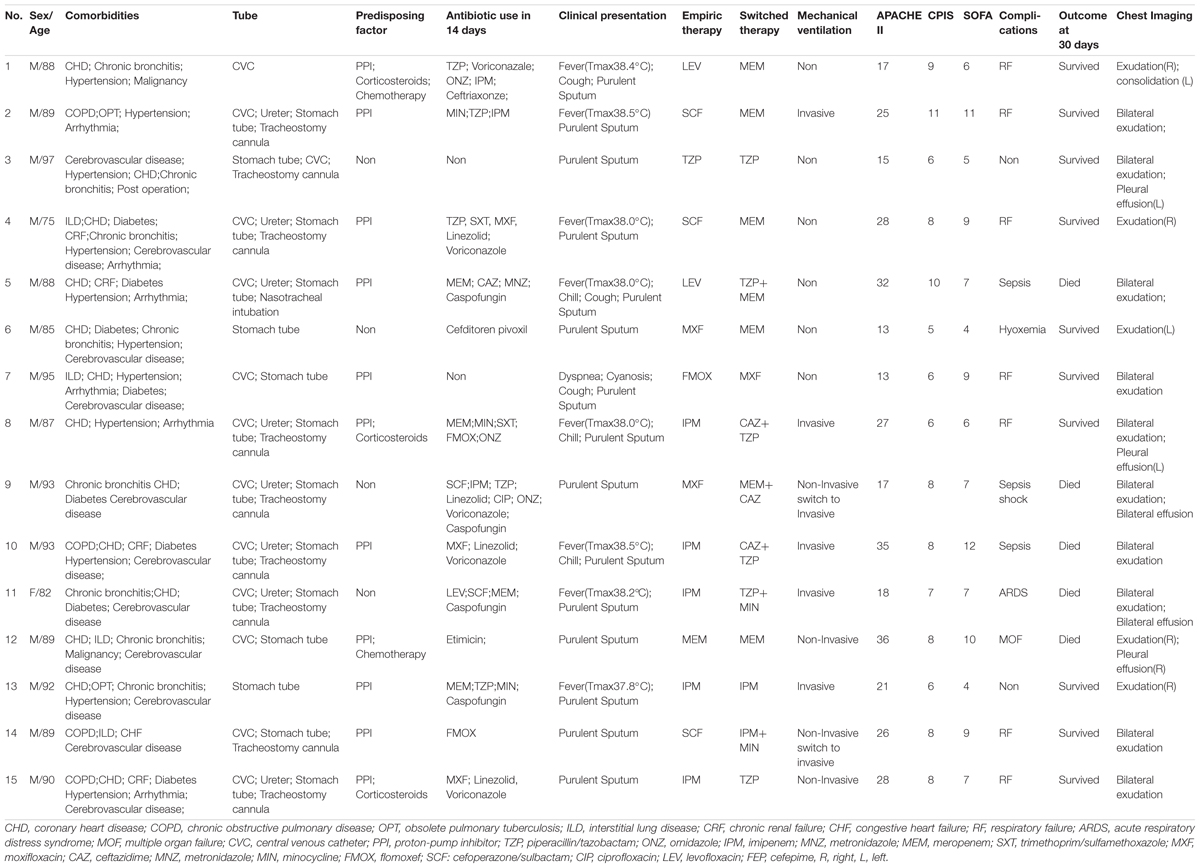
Fifteen patients with HAP due to Achromobacter spp. treated in a geriatric ward in China from September 2008 to May 2012 were enrolled in this study. The diagnostic criteria of HAP were as described previously (Rotstein et al., 2007). Patient demographics and clinical characteristics (underlying disease, invasive manipulation, therapy, and mortality) were collected. The Acute Physiology and Chronic Health Evaluation II (APACHE II), Sequential Organ Failure Assessment (SOFA) score and Clinical Pulmonary Infection Score (CPIS) were evaluated within the first 24 h. Chronic infection was defined as a positive sputum culture on at least three occasions over a 6-month period, as previously suggested (Trancassini et al., 2014).
Microbial Identification and blaOXA-114-like Analysis
Achromobacter spp. was cultured from sputum or aspirations more than twice and was the dominant species in the culture. All the strains were identified by the API 20 NE system and the Vitek II system as described previously (Otto-Karg et al., 2009; Tena et al., 2014). Species identification was confirmed by 16S ribosomal RNA gene sequencing (Mellmann et al., 2003). To further classify this organism, a specific PCR based on the detection of blaOXA-114-like was performed (Turton et al., 2011).
Multilocus Sequence Typing
The primers and reaction conditions of seven housekeeping genes (nusA, rpoB, eno, gltB, lepA, nuoL, nrdA) were designed by searching Pubmlst1. Allelic profiling and ST determination were also performed through the above website. Additionally, phylogenetic analysis of housekeeping genes and eBURST analysis were performed. The concatenation of the seven housekeeping genes (total of 2249 bp) of Achromobacter spp. was performed. A dendrogram was constructed from the concatenated sequences by the neighbor-joining method (MEGA 6.05). The eBURST analysis was conducted using eBURST V3.
Biofilm Production Assay
All isolates were individually cultured overnight in M-H medium (OXOID). A monoclonal sample from the M-H medium was mixed with LB medium to prepare a bacterial suspension of 0.5 units. Bacterial suspensions (200 μl) were inoculated in a 96-well plate followed by 48 h of incubation at 37°C in a bacterial incubator (German He Liman). Plankton bacteria were removed by washing three times with distilled water every 30 s. The wells were dyed at room temperature for 5 min and then incubated with 200 μl of 1% Crystal Violet dye for 20 min. They were then washed twice with distilled water and dried at 37°C for 30 min. Two hundred microliters of 95% ethanol was added into each well, and the optical density (OD) at 570 nm was measured in a microtiter auto reader (Thermo Fisher Scientific, US) (Trancassini et al., 2014; Zhang et al., 2016). ATCC Escherichia coli DH5a (Ac) was used as a negative control. The established criteria (four levels of biofilm-forming ability) were the following: (1) N: no biofilm producer (-), A ≤ Ac; (2) W: weakly positive (++), Ac < A ≤ 2Ac; (3) M: moderately positive (++), 2Ac < A ≤ 4Ac; and (4) S: strongly positive (+++), A > 4Ac (Stepanović et al., 2007).
Susceptibility Testing
Antimicrobial susceptibility testing was conducted using the microbroth dilution method as described previously (Yamamoto et al., 2012). The etest was employed for polymyxin B in M-H medium (OXOID) (Table 1). The results were interpreted using the 2015 Clinical and Laboratory Standards Institute (CLSI) guidelines.
BlaOXA-114-like Analyses
The dendrogram analysis of blaOXA-114-like was performed using MEGA (version 6.05). The alignment used for the tree calculation and various amino acid sequences of OXA enzymes were performed with Clustal X (version 1.83). To determine whether different subtypes of blaOXA-114 relate to different antibacterial spectra, antibacterial spectra were compared among these subtypes.
PCR
Total DNA was prepared and used as a template for PCR reactions. PCR screening for integrase (intI1, intI2, and intI3) genes was performed as described previously (Yamamoto et al., 2012). The primers used in this study are shown in Supplementary Table S1 (Woodford et al., 2006; Poirel et al., 2011; El Salabi et al., 2012; Poirel et al., 2012).
The primers designed for various regions were Pf: GCATCCAAGCAGCAAG and Pr: AAGCAGACTTGACCTGA. PCR was performed using the Pyrobest DNA Polymerase according to the manufacturer’s instructions (Takara). PCR products were resolved on 1.0% agarose gels, stained with ethidium bromide, and photographed with UV illumination. The PCR products were then sequenced after purifying the DNA by using the Gel Extraction Kit according to the manufacturer’s directions (Axygen). The products were sequenced by Major Biological Medicine Technology Co Ltd in Shanghai.
Nucleotide Sequence Accession Numbers
The nucleotide sequences of the various regions in the integrons reported in this study were submitted to the NCBI2 and assigned the accession numbers KT454532, KT719385, KT719386, and KT719387 (Figure 1).
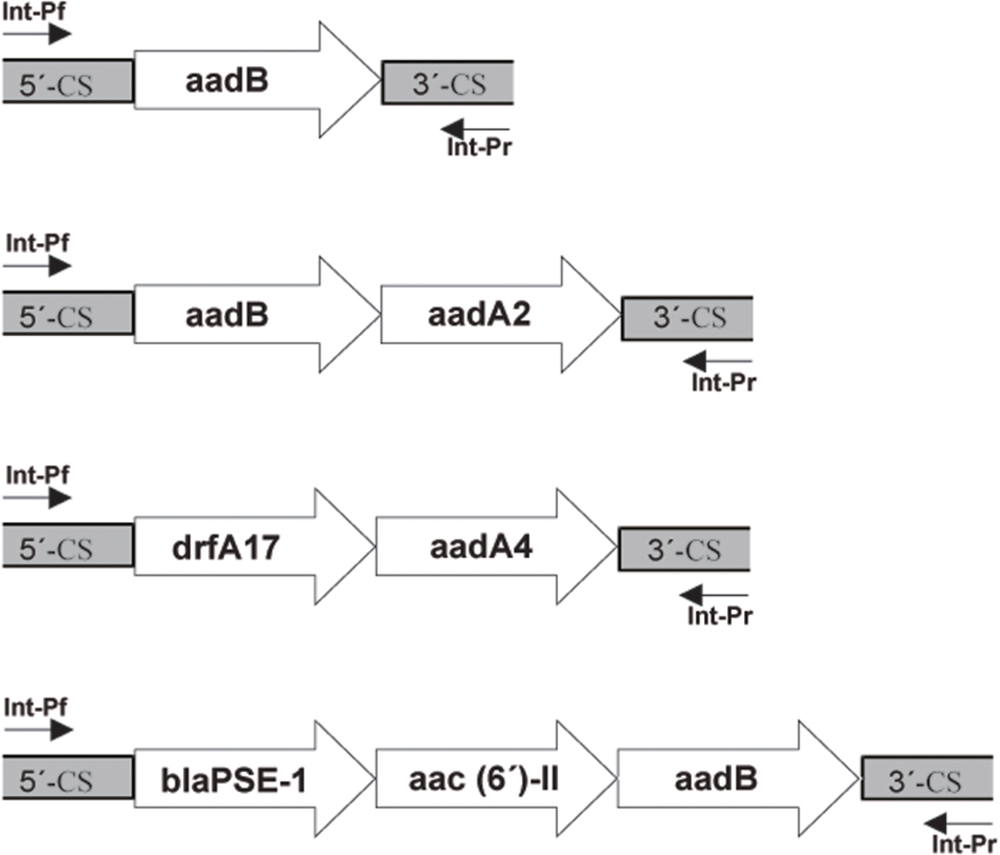
FIGURE 1. Schematic representation of the class 1 integron gene cassettes in Achromobacter spp. isolates.
Results
Clinical Characteristics
These infections first occurred in 2008, and 2, 3, 5, 1, and 3 infections happened per year from 2008 to 2012, respectively. Patient characteristics are summarized in Table 2 and Supplementary Table S2. In total, 14 males and 1 female were included in this study. The mean age was 88.8 ± 5.4 years. All the patients had more than 3 diseases, and 10 of them had 3 or more catheters. The primary underlying diseases were coronary heart disease (CHD) and chronic pulmonary disease. Eleven patients received proton pump inhibitors (PPI), three patients received corticosteroids and two received chemotherapy. Almost all these patients were treated with various antimicrobial therapies within the previous 14 days. Chest imaging of all patients showed patchy exudation. Pleural effusion was found in 33.3% (5/15) of cases, and one case showed consolidation. The mean APACHE II, SOFA and CPIS scores were 23.4 ± 7.7, 7.4 ± 2.7, and 7.6 ± 1.64, respectively. Twelve patients were given a different antibiotic regimen after the antimicrobial susceptibility test. Nine patients were treated by mechanical ventilation. Three patients had chronic infections lasting more than 1 year. Of those with infections, 10 were alive, and 5 (33.3%) died 30 days after infection. The fatal cases were more likely to have sepsis (n = 3).
Diversity of blaOXA-114-like
All Achromobacter spp. strains had positive results for the amplification of blaOXA-114-like (Figure 2). To further study the sequence diversity of blaOXA-114-like in Achromobacter spp., amino acid sequence comparison of the blaOXA-114-like gene was conducted and an evolutionary tree were constructed (Figure 3). The most frequent type was blaOXA-114q, which accounted for 53.3% (8/15) of isolates. Other genes included blaOXA-114r (three cases), blaOXA-114h (three cases) and blaOXA-114t (one case). There were amino acid differences among all of the isolates.
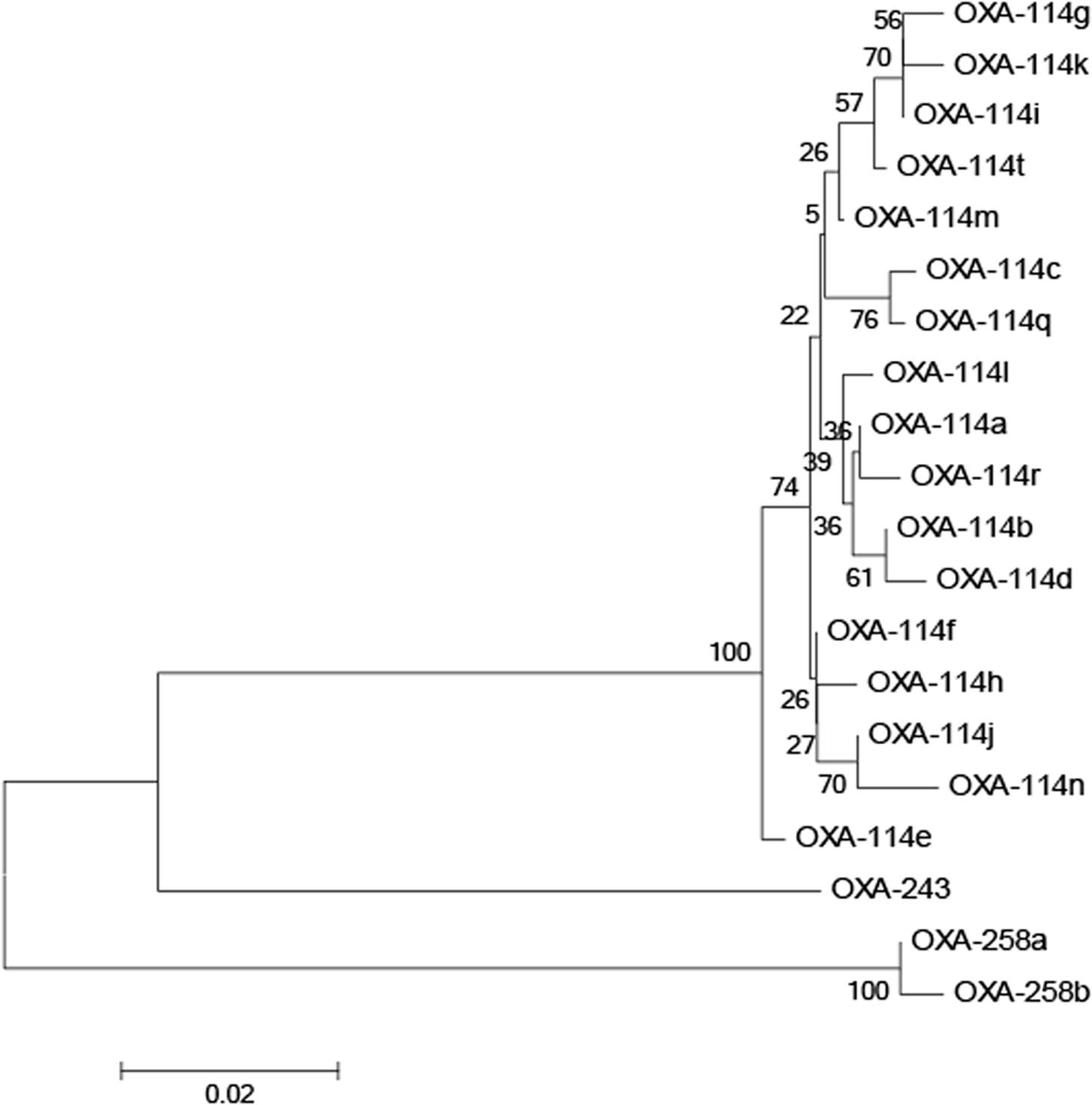
FIGURE 3. Dendrogram analysis of OXA enzymes in Achromobacter spp. isolates. The tree was inferred for various blaOXA-114 types, blaOXA-243, and blaOXA-253 using the neighbor-joining method.
MLST and BF
Based on MLST, four cases belonged to ST5, and these cases were distributed in two separate buildings of the hospital. Six new STs were detected (ST306, 307, 311, 312, 313, 314). Five cases belonged to ST306, and 2 belonged to ST313 (Figure 4). Phylogenetic analysis of housekeeping genes showed that there was a less obvious relationship between ST5 and ST314 (Figure 5), which were found in cases from the general medicine department in a different building from where the other STs were isolated. Additionally, all STs available in the database were analyzed via eBURST. The results show that ST313 belongs to the ST1 CC (clone complex), and HAP caused by this organism may occur sporadically (Figure 6).
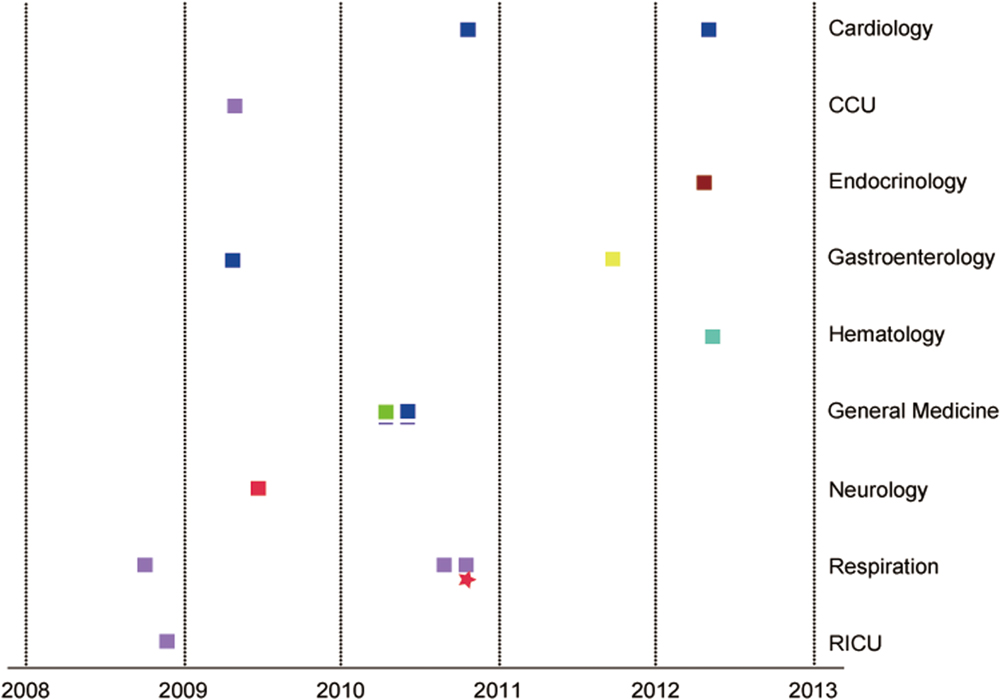
FIGURE 4. The distribution of the 15 patients by admission department and year.  : ST311
: ST311  :ST307
:ST307  :ST312
:ST312  :ST5
:ST5  :ST314
:ST314  :ST313
:ST313  :ST313
:ST313  :ST306. CCU, cardiac intensive care unit; RICU, respiratory intensive care unit. General Medicine (the blue underline) is in another building. The same sequence type is shown in the same color square. The square represents male gender. The star represents female gender.
:ST306. CCU, cardiac intensive care unit; RICU, respiratory intensive care unit. General Medicine (the blue underline) is in another building. The same sequence type is shown in the same color square. The square represents male gender. The star represents female gender.
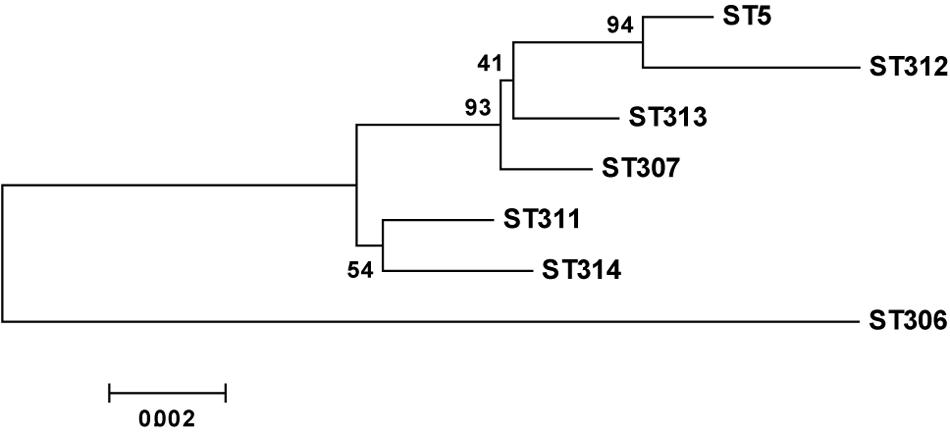
FIGURE 5. Neighbor-joining dendrogram of concatenated sequences of seven housekeeping genes from the MLST database.
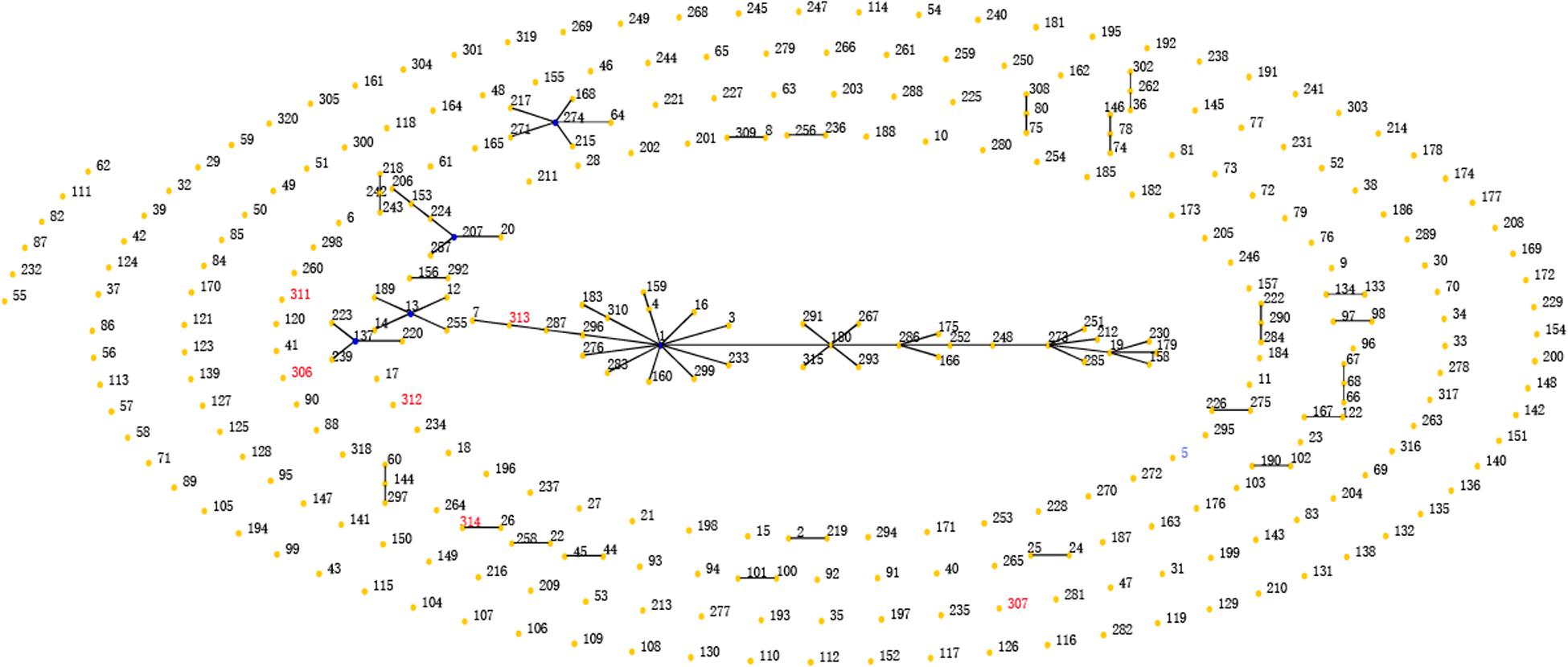
FIGURE 6. eBRUST analysis of all STs of Achromobacter. Circles represent different STs; blue represents a primary founder, yellow represents a subgroup founder, red denotes STs found in the present study dataset only.
All strains were able to produce a biofilm on 96-well plates (Table 1). Nearly half of the strains were moderate biofilm producers, four strains were strong producers, and five strains were weak producers.
Antibiotic Resistance and Integron Variable Region
All strains were resistant to aminoglycosides, aztreonam, nitrofurantoin, and third- and fourth-generation cephalosporins (except ceftazidime and cefoperazone). Approximately half of these strains were resistant to ciprofloxacin, tetracycline and imipenem, but the sensitivity to piperacillin/tazobactam and meropenem was close to 70%.
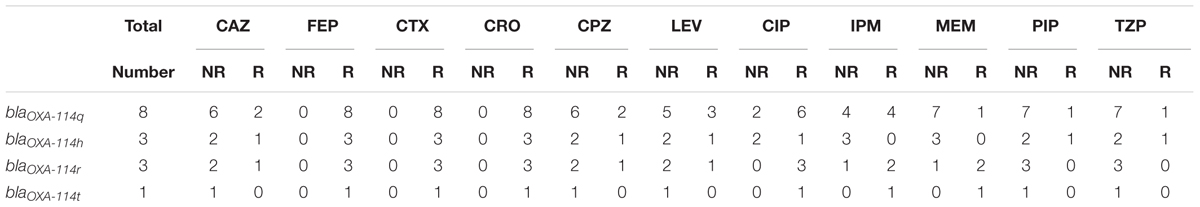
All the blaOXA-114-like type strains showed high resistance to cefepime, cefotaxime and ceftriaxone and high sensitivity to piperacillin and piperacillin/tazobactam. It is interesting that both blaOXA-114t and blaOXA-114r strains are resistant to ciprofloxacin and show higher resistance to carbapenems than other subtypes (Table 3, Figure 7). However, due to the small sample, a large sample analysis should be conducted to further examine this finding.
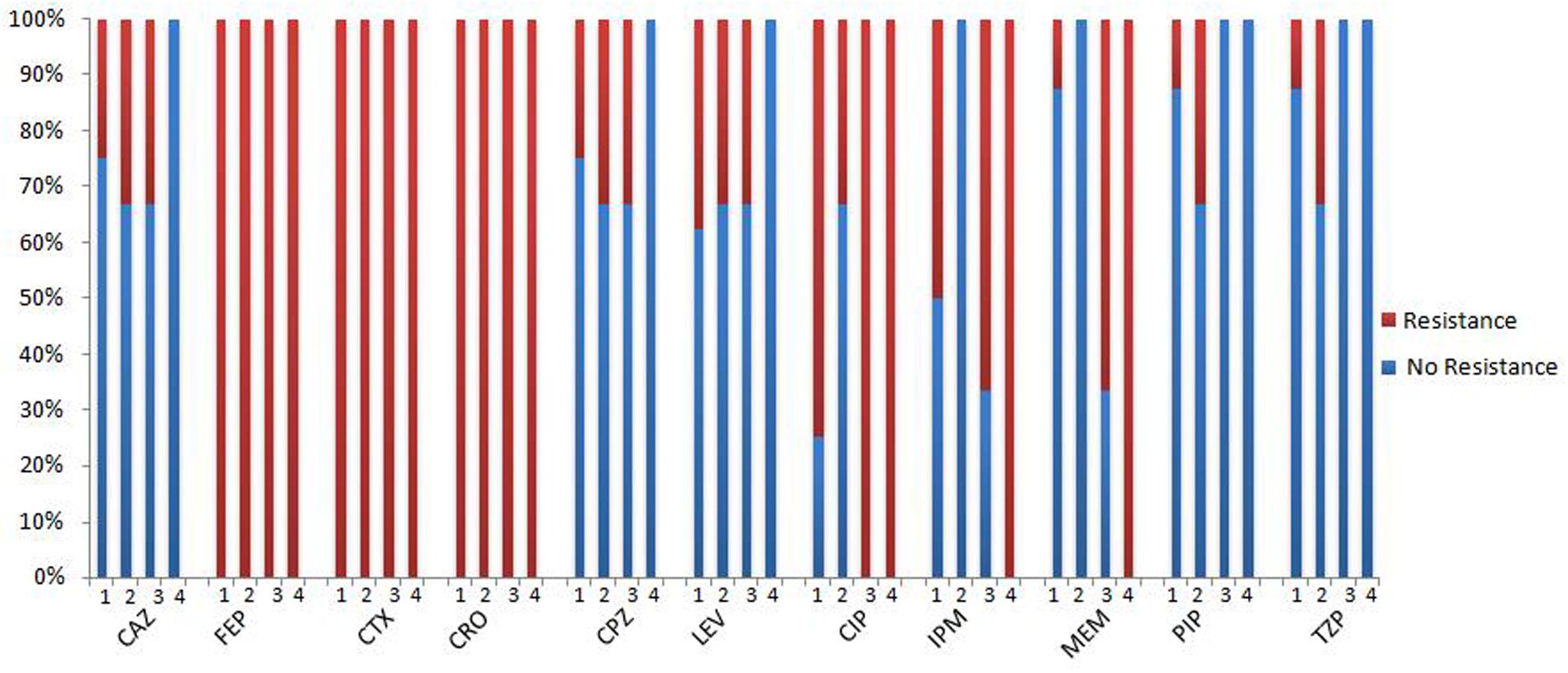
FIGURE 7. The antibacterial spectrum of blaOXA-114-like -containing strains. 1: blaOXA-114q, 2: blaOXA-114q, 3: blaOXA-114q, 4: blaOXA-114q; CAZ, ceftazidime; FEP, cefepime; CTX, cefotaxime; CRO, ceftriaxone; CPZ, cefoperazone; LEV, levofloxacin; CIP, ciprofloxacin; IPM, imipenem; MEM, meropenem; PIP, piperacillin; TZP, piperacillin/tazobactam.
We examined three types of integrons (intI, intII, intIII), and all the strains had intI, but neither intII nor intIII were found. In the variable region in intI, aadB + aadA2 were detected in 10 strains, aadB was inserted in two strains, dfrA17+ aacA4 in one isolate, and blaPSE-1+aac(6′)-II + aadB in one isolate (Supplementary Figure S1).
Discussion
Achromobacter spp. have been frequently detected by the cystic pulmonary fibrosis (CF) center in recent years (Lambiase et al., 2011; Pereira et al., 2011; Amoureux et al., 2013b; Llorca Otero et al., 2016). A study showed that colonization mostly occurred in elderly patients who had more pronounced lung damage and poorer lung function (De Baets et al., 2007). Chronic pulmonary disease and respiratory tract infections often occur in elderly patients. In our study, elderly patients had more than three diseases, particularly CHD, chronic pulmonary disease and cerebrovascular disease. Additionally, 10 had 3 or more indwelling catheters. These catheters may contribute to the higher APACHE II and SOFA scores. The mortality rate was 33.3% (5/15), which is higher than that in other studies (Gómez-Cerezo et al., 2003; Aisenberg et al., 2004). Piperacillin/tazobactam and meropenem showed a strong ability to treat the infection, consistent with the in vitro susceptibility tests. In our study, nine patients (60%) received mechanical ventilation. This organism can colonize humidifiers, which is an issue that needs to be addressed when a patient receives mechanical ventilation (Tena et al., 2005).
More than 300 types of Achromobacter spp. have been recorded in the MLST database. ST5 was first cultured from a CF patient in USA. In our hospital, four cases were infected by this type from 2009 to 2012 in two buildings. These patients did not appear to be clustered in space or time. Moreover, the MIC of carbapenems for ST5 was lower. From 2008 to 2010, ST306 was detected in the Respiration and CCU. Interestingly, the same ST showed various antibiotic-resistance spectra, indicating different resistance mechanisms. The other STs (ST307, ST311, ST312, ST313, and ST314) did not appear to be clustered in space or time. Additionally, the location and timing were not consistent with the six non-clustered cases. However, no strain was detected from doctors’ hands or from medical equipment. The source of their infection could not be determined. Two strains (ST313) showed higher MIC for cephalosporins, quinolones and carbapenems, which should be considered in clinical practice.
It has been proposed that blaOXA-114-like is an inherent gene for Achromobacter xylosoxidans (Turton et al., 2011). Additionally, the diversity of blaOXA-114-like has been reported (Amoureux et al., 2013a). BlaOXA-114q, blaOXA-114r, blaOXA-114h and blaOXA-114t were also detected in our study, which confirmed the diversity of this gene. We have listed the similarities and differences in amino acids among these genes in our isolates. Further studies are needed to understand the function of these differences.
In our study, the strains isolated from patients who were supported by mechanical ventilation had stronger biofilm production. Additionally, the strong biofilm producers (No. 8, 10, 11) showed higher MIC of carbapenems. These characteristics may be shared by other Gram-negative biofilm producers.
Various regions of integrons can carry various resistance genes. In Argentina, class 1 integrons and class 2 integrons were detected in an Achromobacter spp., including various antibiotic-resistant genes (Traglia et al., 2012). In the present study, only class 1 integrons were detected, and all strains were resistant to aminoglycosides, consistent with the three types of genes (aadB, aadA2 and aacA4) detected in the integron.
The overuse of antibiotics may be a reason for the increasing opportunistic infections with resistant microorganisms (Tan et al., 2002). In vitro, these isolates showed high sensitivity to trimethoprim/sulfamethoxazole, polymyxin B, meropenem and piperacillin/tazobactam. In clinical practice, due to the hepatic and renal toxicity of trimethoprim/sulfamethoxazole and polymyxin B, meropenem and piperacillin/tazobactam are frequently used in therapy. Additionally, in our study, nine patients survived after meropenem and piperacillin/tazobactam therapy. However, four isolates showed high resistance to imipenem in vitro. In a previous study, the blaVIM located in integron I was reported in both Asia and Europe (Riccio et al., 2001; Shin et al., 2005; Sofianou et al., 2005). Additionally, a new type of blaIMP inserted in plasmids was detected in A. xylosoxidans, which could easily trigger a pandemic of carbapenem-resistant isolates (Yamamoto et al., 2012). Unfortunately, metallo-b-lactamase (MBL) gene cassettes involved in carbapenem resistance were not detected in the integrons. Other mechanisms may account for the carbapenem resistance observed in this study. Thus, whole genome sequencing may be needed for further study.
Many studies showed that dfrA was frequently detected in integrons (Yu et al., 2003, 2004). In our study, dfrA17 was found in integron I, which was once detected in patients with urinary tract infection from another Gram-negative organism in Korea. However, the clinical specimens of our patients were sputum samples. The spread of dfrA17 may be related to the plasmid (Yu et al., 2003). The blaPSE-1 gene was first reported in Achromobacter spp. This gene can induce carbenicillin-hydrolyzing beta-lactamases aimed at carboxypenicillins, ureidopenicillins and cefsulodin. These enzymes belong to molecular class A and functional group 2c (Bush et al., 1995), which can be frequently detected in Gram-negative organisms. These results indicate that this organism may be a reservoir of horizontal genetic transfer elements for spreading antibiotic resistance (Collis and Hall, 1995).
This study has several limitations. First, it is a small retrospective study conducted over 5 years. Therefore, the source of infection could not be determined, and our results cannot be generalized to other populations. Second, these cases of Achromobacter spp. in China are not routinely reported. Third, the mechanism of resistance to carbapenems was unclear. Whole genome sequencing may be needed in further studies.
Author Contributions
XF: study design, data collection, data analysis, writing of the manuscript; CL: study design, data collection and analysis, writing of the manuscript. Chao L: study design, data collection and analysis, writing of the manuscript; FP: data collection and analysis, writing of the manuscript; JG: data analysis, writing of the manuscript; YJ: data analysis, writing of the manuscript; LQ, YJ, and WY: data collection; All authors have read and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported by the National Basic Research Program of China (973 program) No. 2014CB744400.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00621
Footnotes
References
Aisenberg, G., Rolston, K. V., and Safdar, A. (2004). Bacteremia caused by Achromobacter and Alcaligenes species in 46 patients with cancer (1989-2003). Cancer 101, 2134–2140. doi: 10.1002/cncr.20604
Amoureux, L., Bador, J., Fardeheb, S., Mabille, C., Couchot, C., Massip, C., et al. (2013a). Detection of Achromobacter xylosoxidans in hospital, domestic, and outdoor environmental samples and comparison with human clinical isolates. Appl. Environ. Microbiol. 79, 7142–7149. doi: 10.1128/AEM.02293-13
Amoureux, L., Bador, J., Siebor, E., Taillefumier, N., Fanton, A., and Neuwirth, C. (2013b). Epidemiology and resistance of Achromobacter xylosoxidans from cystic fibrosis patients in Dijon, Burgundy: first French data. J. Cyst. Fibros. 12, 170–176. doi: 10.1016/j.jcf.2012.08.005
Bador, J., Amoureux, L., Duez, J. M., Drabowicz, A., Siebor, E., and Llanes, C. (2011). First description of an RND-type multidrug efflux pump in Achromobacter xylosoxidans, AxyABM. Antimicrob. Agents Chemother. 55, 4912–4914. doi: 10.1128/AAC.00341-11
Bush, K., Jacoby, G. A., and Medeiros, A. A. (1995). A functional classification scheme for b-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39, 1211–1233. doi: 10.1128/AAC.39.6.1211
Chandrasekar, P. H., Arathoon, E., and Levine, D. P. (1986). Infections due to Achromobacter xylosoxidans. Case report and review of the literature. Infection 14, 279–282. doi: 10.1007/BF01643962
Collis, C. M., and Hall, R. M. (1995). Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39, 155–162. doi: 10.1128/AAC.39.1.155
De Baets, F., Schelstraete, P., Van Daele, S., Haerynck, F., and Vaneechoutte, M. (2007). Achromobacter xylosoxidans in cystic fibrosis: prevalence and clinical relevance. J. Cyst. Fibros. 6, 75–78. doi: 10.1016/j.jcf.2006.05.011
Doi, Y., Poirel, L., Paterson, D. L., and Nordmann, P. (2008). Characterization of a naturally occurring class D beta-lactamase from Achromobacter xylosoxidans. Antimicrob. Agents Chemother. 52, 1952–1956. doi: 10.1128/AAC.01463-07
Dunne, W. M. Jr., and Maisch, S. (1995). Epidemiological investigation of infections due to Alcaligenes species in children and patients with cystic fibrosis: use of repetitive-element sequence polymerase chain reaction. Clin. Infect. Dis. 20, 836–841. doi: 10.1093/clinids/20.4.836
Dworzack, D. L., Murray, C. M., Hodges, G. R., and Barnes, W. G. (1978). Community-acquired bacteremic Achromobacter xylosoxidans type IIIa pneumonia in a patient with idiopathic IgM deficiency. Am. J. Clin. Pathol. 70, 712–717. doi: 10.1093/ajcp/70.4.712
El Salabi, A., Borra, P. S., Toleman, M. A., Samuelsen, O., and Walsh, T. R. (2012). Genetic and biochemical characterization of a novel metallo-beta-lactamase, TMB-1, from an Achromobacter xylosoxidans strain isolated in Tripoli, Libya. Antimicrob. Agents Chemother. 56, 2241–2245. doi: 10.1128/AAC.05640-11
Ferroni, A., Sermet-Gaudelus, I., Abachin, E., Quesne, G., Lenoir, G., Berche, P., et al. (2002). Use of 16S rRNA gene sequencing for identification of nonfermenting gram negative bacilli recovered from patients attending a single cystic fibrosis center. J. Clin. Microbiol. 40, 3793–3797. doi: 10.1128/JCM.40.10.3793-3797.2002
Gahrn-Hansen, B., Alstrup, P., Dessau, R., Fuursted, K., Knudsen, A., Olsen, H., et al. (1988). Outbreak of infection with Achromobacter xylosoxidans from contaminated intravascular pressure transducers. J. Hosp. Infect. 12, 1–6. doi: 10.1016/0195-6701(88)90115-6
Gómez-Cerezo, J., Suárez, I., Ríos, J. J., Peña, P., García de Miguel, M. J., de José, M., et al. (2003). Achromobacter xylosoxidans bacteremia: a 10-year analysis of 54 cases. Antimicrob. Agents Chemother. 39, 1211–1233.
Lambiase, A., Catania, M. R., Del Pezzo, M., Rossano, F., Terlizzi, V., Sepe, A., et al. (2011). Achromobacter xylosoxidans respiratory tract infections in cystic fibrosis patients. Eur. J. Clin. Microbiol. Infect. Dis. 30, 973–980. doi: 10.1007/s10096-011-1182-5
Liu, L., Coenye, T., Burns, J. L., Whitby, P. W., Stull, T. L., and Lipuma, J. J. (2002). Ribosomal DNA-directed PCR for identification of Achromobacter (Alcaligenes) xylosoxidans recovered from sputum samples from cystic fibrosis patients. J. Clin. Microbiol. 40, 1210–1213. doi: 10.1128/JCM.40.4.1210-1213.2002
Llorca Otero, L., Girón Moreno, R., Buendía Moreno, B., Valenzuela, C., Guiu Martínez, A., and Alarcón Cavero, T. (2016). Achromobacter xylosoxidans infection in an adult cystic fibrosis unit in Madrid. Enferm. Infect. Microbiol. Clin. 34, 184–187. doi: 10.1016/j.eimc.2015.05.006
Mellmann, A., Cloud, J. L., Andrees, S., Blackwood, K., Carroll, K. C., Kabani, A., et al. (2003). Evaluation of RIDOM, MicroSeq, and GenBank services in the molecular identification of Nocardia species. Int. J. Med. Microbiol. 293, 359–370. doi: 10.1078/1438-4221-00271
Neuwirth, C., Freby, C., Ogier-Desserrey, A., Perez-Martin, S., Houzel, A., Péchinot, A., et al. (2006). VEB-1 in Achromobacter xylosoxidans from Cystic Fibrosis Patient, France. Emerg. Infect. Dis. 12, 1737–1739. doi: 10.3201/eid1211.060143
Oie, S., Arakawa, J., Furukawa, H., Matsumoto, S., Matsuda, N., and Wakamatsu, H. (2012). Microbial contamination of a disinfectant-soaked unwoven cleaning cloth. J. Hosp. Infect. 82, 61–63. doi: 10.1016/j.jhin.2012.06.010
Otto-Karg, I., Jand, S., Müller, T., Stirzel, B., Frosch, M., Hebestreit, H., et al. (2009). Validation of Vitek 2 nonfermenting gram-negative cards and Vitek 2 version 4.02 software for identification and antimicrobial susceptibility testing of nonfermenting gram-negative rods from patients with cystic fibrosis. J. Clin. Microbiol. 47, 3283–3288. doi: 10.1128/JCM.00505-09
Pereira, R. H., Carvalho-Assef, A. P., Albano, R. M., Folescu, T. W., Jones, M. C., Leão, R. S., et al. (2011). Achromobacter xylosoxidans: characterization of strains in Brazilian cystic fibrosis patients. J. Clin. Microbiol. 49, 3649–3651. doi: 10.1128/JCM.05283-11
Poirel, L., Potron, A., and Nordmann, P. (2012). OXA-48-like carbapenemases: the phantom menace. J. Antimicrob. Chemother. 67, 1597–1606. doi: 10.1093/jac/dks121
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Riccio, M. L., Pallecchi, L., Fontana, R., and Rossolini, G. M. (2001). In70 of plasmid pAX22, a bla(VIM-1)-containing integron carrying a new aminoglycoside phosphotransferase genecassette. Antimicrob. Agents Chemother. 45, 1249–1253. doi: 10.1128/AAC.45.4.1249-1253.2001
Rotstein, C., Evans, G., Born, A., Grossman, R., Light, R. B., Magder, S., et al. (2007). Clinical practice guidelines for hospital– Clinical practice guidelines for hospital-acquired pneumonia and ventilator associated pneumonia in adults. Can. J. Infect. Dis. Med. Microbiol. 19, 19–53.
Shin, K. S., Han, K., Lee, J., Hong, S. B., Son, B. R., and Youn, S. J. (2005). Imipenem-resistant Achromobacter xylosoxidans carrying blaVIM-2-containing class 1 integrant. Diagn. Microbiol. Infect. Dis. 53, 215–220. doi: 10.1016/j.diagmicrobio.2005.06.018
Sofianou, D., Markogiannakis, A., Metzidie, E., Pournaras, S., and Tsakris, A. (2005). VIM-2 metallolactamase inAchromobacter xylosoxidans in Europe. Eur. J. Clin. Microbiol. Infect. Dis. 24, 854–855. doi: 10.1007/s10096-005-0054-2
Stepanović, S., Vuković, D., Hola, V., Di Bonaventura, G., Djukić, S., Cirković, I., et al. (2007). Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 115, 891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x
Swenson, C. E., and Sadikot, R. T. (2015). Achromobacter respiratory infections. Ann. Am. Thorac. Soc. 12, 252–258. doi: 10.1513/AnnalsATS.201406-288FR
Tan, K., Conway, S. P., Brownlee, K. G., Etherington, C., and Peckham, D. G. (2002). Alcaligenes infection in cystic fibrosis. Pediatr. Pulmonol. 34, 101–104. doi: 10.1002/ppul.10143
Tena, D., Carranza, R., Barberá, J. R., Valdezate, S., Garrancho, J. M., Arranz, M., et al. (2005). Outbreak of long-term intravascular catheter-related bacteremia due to Achromobacter xylosoxidans subspecies xylosoxidans in a hemodialysis unit. Eur. J. Clin. Microbiol. Infect. Dis. 24, 727–732. doi: 10.1007/s10096-005-0028-4
Tena, D., Martínez, N. M., Losa, C., and Solís, S. (2014). Skin and soft tissue infection caused by Achromobacter xylosoxidans: report of 14 cases. Scand. J. Infect. Dis. 46, 130–135. doi: 10.3109/00365548.2013.857043
Traglia, G. M., Almuzara, M., Merkier, A. K., Adams, C., Galanternik, L., Vay, C., et al. (2012). Achromobacter xylosoxidans: an emerging pathogen carrying different elements involved in horizontal genetic transfer. Curr. Microbiol. 65, 673–678. doi: 10.1007/s00284-012-0213-5
Trancassini, M., Iebba, V., Citerà, N., Tuccio, V., Magni, A., Varesi, P., et al. (2014). Outbreak of Achromobacter xylosoxidans inan Italian Cystic fibrosis center: genome variability, biof ilm production, antibiotic resistance, and motility in isolated strains. Front. Microbiol. 5:138. doi: 10.3389/fmicb.2014.00138
Turton, J. F., Mustafa, N., Shah, J., Hampton, C. V., Pike, R., and Kenna, D. T. (2011). Identification of Achromobacter xylosoxidans by detection of the blaOXA-114-like gene intrinsic in this species. Diagn. Microbiol. Infect. Dis. 70, 408–411. doi: 10.1016/j.diagmicrobio.2011.02.007
Vu-Thien, H., Darbord, J. C., Moissenet, D., Dulot, C., Dufourcq, J. B., Marsol, P., et al. (1998). Investigation of an outbreak of wound infections due to Alcaligenes xylosoxidans transmitted by chlorhexidine in a burns unit. Eur. J. Clin. Microbiol. Infect. Dis. 17, 724–726. doi: 10.1007/s100960050168
Woodford, N., Ellington, M. J., Coelho, J. M., Turton, J. F., Ward, M. E., Brown, S., et al. (2006). Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27, 351–353. doi: 10.1016/j.ijantimicag.2006.01.004
Yamamoto, M., Nagao, M., Hotta, G., Matsumura, Y., Matsushima, A., Ito, Y., et al. (2012). Molecular characterization of IMP-type metallo-β-lactamases among multidrug-resistant Achromobacter xylosoxidans. Antimicrob. Agents Chemother. 67, 2110–2113. doi: 10.1093/jac/dks179
Yu, H. S., Lee, J. C., Kang, H. Y., Jeong, Y. S., Lee, E. Y., Choi, C. H., et al. (2004). Prevalence of dfr genes associated with integrons and dissemination of dfrA17 among urinary isolates of Escherichia coli in Korea. J. Antimicrob. Chemother. 53, 445–450. doi: 10.1093/jac/dkh097
Yu, H. S., Lee, J. C., Kang, H. Y., Ro, D. W., Chung, J. Y., Jeong, Y. S., et al. (2003). Changes in gene cassettes of class I integrons among Escherichia coli isolates from urine specimens collecte in Kroea during the lsat two decades. J. Clin. Microbiol. 41, 5429–5433. doi: 10.1128/JCM.41.12.5429-5433.2003
Zemel, B. S., Jawad, A. F., Fitzsimmons, S., and Stallings, V. A. (2000). Longitudinal relationship among growth, nutritional status, and pulmonary function in children with cystic fibrosis: analysis of the Cystic Fibrosis Foundation National CF Patient Registry. J. Pediatr. 137, 374–380. doi: 10.1067/mpd.2000.107891
Keywords: hospital-acquired pneumonia, Achromobacter, biofilm, elderly, integron
Citation: Liu C, Pan F, Guo J, Yan W, Jin Y, Liu C, Qin L and Fang X (2016) Hospital Acquired Pneumonia Due to Achromobacter spp. in a Geriatric Ward in China: Clinical Characteristic, Genome Variability, Biofilm Production, Antibiotic Resistance and Integron in Isolated Strains. Front. Microbiol. 7:621. doi: 10.3389/fmicb.2016.00621
Received: 05 February 2016; Accepted: 15 April 2016;
Published: 09 May 2016.
Edited by:
John W. A. Rossen, University of Groningen, NetherlandsReviewed by:
Kai Zhou, University Medical Center Groningen, NetherlandsYunlong Li, Wadsworth Center, USA
Copyright © 2016 Liu, Pan, Guo, Yan, Jin, Liu, Qin and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangqun Fang, ZmFuZ3hpYW5ncXVuMzAxckAxNjMuY29t; Long Qin, cWlubG9uZzYxMkAxNjMuY29t
†These authors are co-first authors.
 Chao Liu1†
Chao Liu1† Xiangqun Fang
Xiangqun Fang