- Shanghai Key Laboratory of Veterinary Biotechnology, Key Laboratory of Urban Agriculture (South), Ministry of Agriculture, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai, China
Staphylococcus aureus (S. aureus) is a Gram-positive pathogen causing a variety of infections in humans and animals. Extensive use of antibiotics has led to the emergence of methicillin-resistant S. aureus (MRSA). As an alternative antibacterial agent against drug-resistant S. aureus, a lytic phage, designated SLPW, was isolated from fecal sewage in a pig farm. The SLPW was morphologically classified under Podoviridae and contains a double-stranded DNA genome. The genome of SLPW was 17,861 bp (29.35% G+C) containing 20 open reading frames and lacked regions encoding lysogeny-related integrase gene and cI repressor gene. Phage SLPW showed a broad host range and high efficiency of plating against various types of S. aureus. One-step growth curve showed a short latency period (10 min) and a long lytic period (120 min). Phage SLPW remained stable under a wide range of temperatures or pH and was almost unaffected in chloroform or ultraviolet light. Further, it efficiently lysed MRSA strains in vitro and in vivo. Intraperitoneal phage administration at 1 h post-infection cured the mice and reduced the bacterial expression of inflammatory cytokines in mice. Specifically, the phage SLPW displayed a wide antibacterial spectrum. It was therapeutically effective against intra-abdominal infection in mice harboring different multilocus sequence typing (MLST) types of S. aureus strains. Therefore, phage SLPW is a potential therapeutic agent against MRSA infections.
Introduction
Staphylococcus aureus (S. aureus) is the most virulent pathogen causing various diseases, including skin abscesses, pneumonia, endocarditis, and osteomyelitis, in humans and animals (Lowy, 1998; Plata et al., 2009). The two major sources of infection include community and hospital (Engemann et al., 2003). The bacterial strains are resistant to many antibiotics, and especially to methicillin and vancomycin. The emergence and prevalence of methicillin-resistant S. aureus (MRSA) and vancomycin-resistant S. aureus (VRSA) underscores the need for development of effective therapeutic alternatives (Sasidharan et al., 2011; Gardete and Tomasz, 2014).
In the last 15 years, there has been a marked increase in the number of identified Staphylococcus phages, with tremendous progress in therapeutic interventions targeting Staphylococcus, especially S. aureus (Hsieh et al., 2011). Phages are the most common organisms on the planet and represent great diversity in host range. S. aureus phages target pathogens in diseases, such as bacteremia, eye infections, and S. aureus-associated lung infections (Wills et al., 2005; Kazmierczak et al., 2015). Compared with traditional antibiotics, bacteriophages are cost-effective without serious side effects, and are virulent especially against drug-resistant bacteria (Borysowski et al., 2011; Kazmierczak et al., 2015). Further, phages generally recognize specific receptors on bacterial cell membrane, without affecting human, or animal cells. Therefore, the side effects in eukaryotic hosts are minimal (Sulakvelidze et al., 2001). Studies involving S. aureus phages show effective and comprehensive antimicrobial activity in vitro and in vivo (Capparelli et al., 2007; Gutierrez et al., 2015).
In this study, we isolated a lytic phage, SLPW, from fecal sewage in a pig farm. We report the wide host range, adequate stability and strong bacteriolytic activity of this phage. Specifically, the phage SLPW was safe and effective against MRSA infection in mice.
Materials and Methods
Ethics Statement
Animal experiments were carried out according to the animal welfare standards approved by the Ethical Committee for Animal Experiments of Shanghai Jiao Tong University, China. All animal experiments complied with the guidelines of the Animal Welfare Council of China.
Bacterial Strains and Culture Conditions
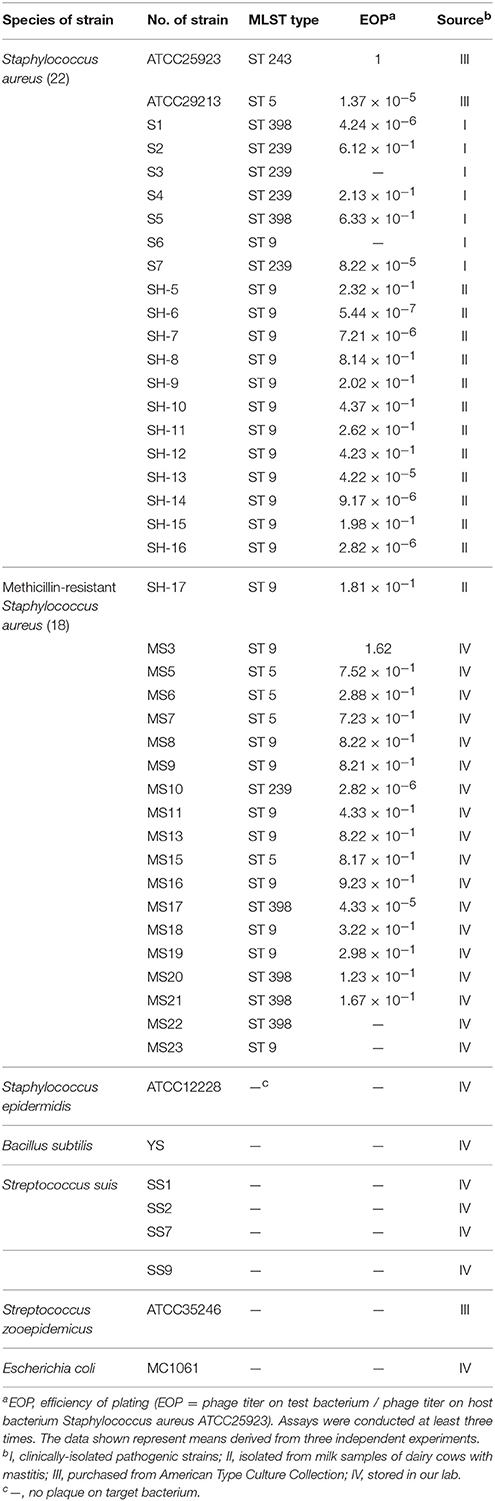
In this study, 38 S. aureus strains (18 MRSA strains, 7 clinically isolated pathogenic strains and 13 strains isolated from milk samples of dairy cows with mastitis) and 8 other strains (Staphylococcus epidermidis ATCC12228, Bacillus subtilis YS, S. zooepidemicus ATCC35246, 4 Streptococcus suis and Escherichia coli MC1061) were used (Table 1). Two reference strains of S. aureus ATCC 25923 and ATCC 29213 from the American Type Culture Collection (ATCC) were also used. All the strains were grown in Todd–Hewitt broth (THB) and brain heart infusion (BHI) or agar medium supplemented with 2% (vol/vol) fetal bovine serum (Gibco, Invitrogen Corp., Carlsbad, CA) at 37°C.
Phage Isolation, Purification, and Host Range Determination
The method of Matsushiro et al. was adopted for the isolation of S. aureus phages with some modifications (Matsushiro and Okubo, 1972). Seventy-four samples, including 44 dust swabs and 30 fecal samples were suspended in SM buffer [NaCl 5.8 g/L, MgSO4 · 7H2O 2 g/L, 1 M Tris·HCl (pH7.5) 50 ml/L, and 2% gelatin 5 ml/L] and centrifuged at 5000 × g for 20 min at 4°C. The supernatants were filtered through 0.22-μm pore membranes and evaluated for the presence of lytic phages using different S. aureus isolates on BHI plates. After overnight incubation, bacterial plaque formation suggested the presence of lytic phage, which was purified after three rounds of single-plaque isolation.
For purification, a single-phage plaque was precipitated in the presence of 10% (wt/vol) polyethylene glycol (PEG) 8000 and 1 M NaCl at 4°C for at least 1 h. The precipitate was collected by centrifugation at 10,000 × g for 10 min at 4°C and suspended in SM buffer. After the addition of 0.5 g/mL CsCl, the mixture was layered on top of CsCl step gradients (densities of 1.15, 1.45, 1.50, and 1.70 g/mL) in Ultra-Clear centrifugation tubes and centrifuged at 28,000 × g for 2 h at 4°C, and dialyzed in sodium chloride–magnesium sulfate buffer [100 mM NaCl, 10 mM MgSO4 · 7H2O, and 50 mM Tris·HCl (pH 7.5)]. Phages were stored at 4°C for further experiments.
The host range of the phage was defined by the double-layered agar method described by Adams (1959). The SLPW phage was inoculated with all the 47 strains listed in Table 1 and then monitored for plaque formation.
Transmission Electron Microscopy (TEM) of Phage Particles
The purified phage sample was loaded onto a copper grid for 7 min followed by negative staining with 2% (vol/vol) uranyl acetate (pH 6.7) and drying. The phage morphology was observed using a FEI TEM Tecnai G2 Spirit Biotwin (FEI, Hillsboro, US) at an accelerating voltage of 80 kV.
Restriction Enzyme Digestion of Phage Genomic DNA
Purified phage genomic DNA was prepared as described previously by Son et al. (2010). For the identification of nucleic acid type, purified phage genomic DNA was subjected to nuclease treatment using DNase I (20 U/μL), RNase A (5 U/μL), and Mung bean nuclease (20 U/μL) at 37°C for 1 h. Restriction site analysis of the phage was conducted by digesting purified phage genomic DNA with 10 U Xho I, EcoR I, Hind III, Ned I, and Not I for 1 h at 37°C. Products of digested phage nucleic acid were separated by 0.8% (wt/vol) agarose gel electrophoresis.
Genome Sequencing and Annotation
Shotgun sequencing was used for Phage SLPW whole genome analysis. Sequence alignments were carried out using the Accelrys DS Gene software package of Accelrys Inc. (USA). Putative open reading frames were suggested using the algorithms of the software packages Accelrys Gene v2.5 (Accelrys Inc.) and ORF Finder (NCBI). Identity values were calculated using different BLAST algorithms (http://www.ncbi.nlm.nih.gov/BLAST/) at the NCBI homepage. The sequence of phage SLPW has been submitted to NCBI (GenBank accession number: KU992911).
Assay of Optimal Multiplicity of Infection (MOI)
Overnight cultures of S. aureus ATCC 25923 strain were diluted 1:100 in fresh BHI and incubated at 37°C with shaking until early logarithmic growth phase (optical density at 600 nm, 0.4–0.6), diluted 1:10, and mixed with phages at different MOIs. After 3.5 h incubation at 37°C, the mixture was centrifuged at 5000 × g for 20 min at 4°C and the supernatants were filtered through 0.22-μmporesize membranes. The phage titer in the supernatant was immediately determined using a double-layer agar plate method. This assay was performed at least in triplicate.
One-Step Growth
For determination of one-step growth of phage SLPW, we used S. aureus ATCC 25923 as the host strain because it displayed the largest clearance zone in a spot test and was lysed rapidly to yield a clear lysate in liquid culture. One-step growth experiments were performed using a modified method described previously (Pajunen et al., 2000). Briefly, SLPW phage was added at a MOI of 0.1 to the cells of S. aureus and allowed to adsorb for 15 min at 37°C. The mixture was then centrifuged at 10,000 × g for 1 min. After the supernatants were removed, the pellets containing the phage-infected bacterial cells were suspended in fresh BHI and incubated with shaking at 180 rpm and 37°C. Partial samples were obtained at 10 min intervals and the titrations from the aliquots were immediately determined using the double-layer agar plate method. This assay was performed at least in triplicate.
Phage Stability
Phage stability was determined at different temperatures (25, 37, 45, 50, 55, 60, 65, and 70°C), using an aliquot of phage SLPW obtained after 1 h. The titers of the phage lysate were assayed using a double-layer agar plate method. The phage stability at different pH-values was tested by determining the titers after dilution of the phage lysates (1:100) in SM buffer and stored at 37°C for 3 h. To analyze the chemical stability, the phage SLPW was treated with chloroform (5, 25, 50, or 75%, vol/vol) for 6, 12, 18, and 24 h at 4°C. In addition, the phage SLPW was also exposed to ultraviolet light treatment for 10, 20, 30, 40, 50, and 60 min, and titrated immediately using a double-layer agar plate method.
Efficiency of Plating
Phage SLPW was screened against S. aureus strains using the efficiency of plating method (EOP = phage titer on test bacterium/phage titer on host bacterium) to determine the effectiveness against a variety of target bacteria. Ten-fold serial dilutions of phage suspensions (100 μL) were mixed with 100 μL of the target or host bacterium (grown overnight at 37°C) and incubated for 5 min at room temperature (25°C) and plated as double layers on THB (Viscardi et al., 2008).
Phage Bacteriolytic Activity In vitro
Overnight cultures of S. aureus culture were diluted 1:100 in fresh THB liquid medium incubated at 37°C with shaking at 180 rpm until an early-exponential host bacterial culture (optical density at 600 nm, 0.4–0.6) was reached. Phage SLPW was added at MOI of 0.01, 1, and 100, and an identical S. aureus culture with the same volume phage diluent was used as the control. The mixture was then grown at 37°C with shaking at 180 rpm. The phage bacteriolytic activity was assessed by monitoring the cell absorbance of the culture solution (OD600) at 30-min intervals for up to 4 h, and this assay was performed in triplicate.
Phage Protection Studies
Female BALB/c mice (6 weeks of age) were purchased from the Experimental Animal Center, Shanghai Jiao Tong University. Overnight cultures of S. aureus were diluted 1:100 in fresh THB liquid medium incubated at 37°C with shaking at 180 rpm to an early-exponential host bacterial culture (optical density at 600 nm, 0.4–0.6). Cells were pelleted and washed twice with phosphate-buffered saline (PBS). The mice were infected with a dose of 1 × 109 CFU in 0.2 mL of the S. aureus strain. The bacterial cells were injected unilaterally into the abdominal cavities of mice, and 0.2 mL of the purified phage samples (1 × 109 PFU) were injected into the other side immediately, at 60 and 120 min after a bacterial challenge. The controls included uninfected mice administered with 0.2 mL of phage in SM buffer. The mouse survival rate was recorded daily for 7 days. The CFU or PFU organ burden in spleen, lung, and blood was determined by sacrificing groups of six mice at 6, 12, and 24 h after phage SPLW administration. Each sample was homogenized in 1 mL PBS and serially diluted in PBS. CFU were evaluated by plating each dilution on THB agar plates. The PFU were evaluated by plating each dilution on the double-layer agar plate.
Cytokine Assays
To evaluate the antimicrobial effects of phage, 0.2 mL of phage in SM buffer (1 × 109 PFU) was administered intraperitoneally at 1 h after infection with S. aureus (1 × 109 CFU). SM buffer alone was administered to uninfected mice serving as control groups. Spleen tissues were removed from mice 6, 12, and 24 h after injection with phage SLPW. Tissues were homogenized in 1 mL of lysis buffer (Qiagen, West Sussex, UK), followed by centrifugation at 2000 × g for 10 min. The supernatants were sterilized with a millipore filter (0.45-μm pore size).
Total RNA was isolated from the supernatants using an AllPrep RNA microkit (Qiagen). The cDNA synthesis was performed using the PrimeScript RT reagent kit (TaKaRa, Dalian, China) according to the manufacturer's instructions. The mRNA levels were measured using two-step relative qRT-PCR. The β-actin housekeeping gene was amplified as an internal control. The sequences of the primers for tumor necrosis factor alpha (TNF-α), interleukin-6 (IL-6), interleukin-1β (IL-1β), and β-actin are listed in Table 2. Gene expression was normalized to the expression of the housekeeping gene β-actin. Real-time PCR was performed using a SYBR Premix Ex Taq kit (TaKaRa) and CFX ConnectTM RT-PCR system (BIO-RAD, Hercules, USA). The comparative cycle threshold (2−ΔΔCT) method was used to analyze the mRNA levels.
Statistical Analyses
Experimental data points were plotted using GraphPad Prism 5 (GraphPad Software, Inc., San Diego, CA). Data were expressed as mean values ± standard errors of the means (SEM). The phage protection analyses were performed using the non-parametric Mann-Whitney U-test. A P < 0.05 was considered significant.
Results and Discussion
Phage Isolation and Host Range Determination
In this study, we isolated a lytic S. aureus phage designated as SLPW, from fecal sewage in a pig farm of Shanghai (China) in 2013. Using S. aureus ATCC25923 as the host strain, the phage plaques measuring 1–2 mm in diameter were obtained. The phage SLPW had a strong ability to produce plaques on S. aureus strains. Among the 40 S. aureus strains, 36 (90%) isolates were lysed by SLPW (Table 1). Furthermore, the SLPW phage showed strong lytic activity against the majority of MRSA strains (16 of 18 strains), suggesting a potential therapeutic role in MRSA infection. However, no plaque production was observed in Staphylococcus epidermidis, Bacillus subtilis, S. zooepidemicus, Streptococcus suis, and E. coli strains investigated (Table 1).
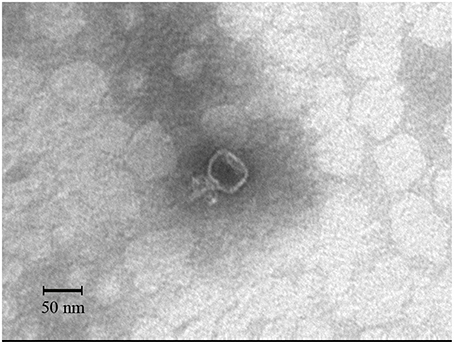
The morphology of the isolated phage SLPW was determined. Electron microscopy showed that the SLPW particle had an isometric head of 49.5 ± 1.5 nm and a short, non-contractile tail measuring 19.5 ± 1.5 nm long (Figure 1). Thus, it was morphologically similar to phages of the family Podoviridae according to the classification of International Committee on Taxonomy of Viruses (ICTV; Adams et al., 2014).
Phage Nucleic Acid Type and Genome Description
The purified phage genomic DNA was subjected to digestion by different nucleases. The results showed that the genome of phage SLPW was completely digested by DNase I but not by RNase A or Mung bean nuclease (Figure 2), suggesting that phage SLPW was a double-stranded DNA. Purified phage SLPW genomic DNA could be digested with several restriction endonucleases including Xho I, EcoR I, Hind III, and Ned I (Figure 3).
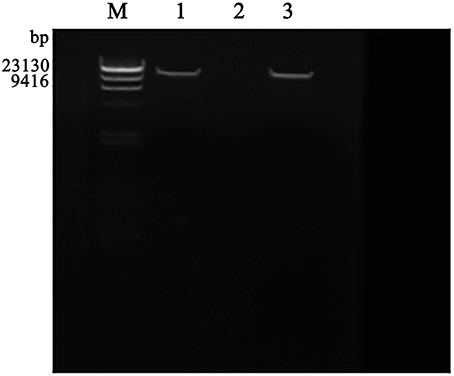
Figure 2. Agarose gel electrophoresis of phage SLPW genome digested with nuclease. Lane M: λ-Hind III digest DNA Marker, Lane 1–3: phage SLPW genome digested with RNaseA, DNase I, and Mung Bean Nuclease, respectively.
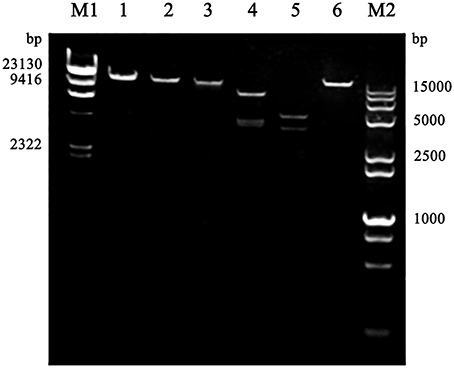
Figure 3. Restriction digestion patterns of phage SLPW DNA. Lane M1: λ-Hind III digested DNA Marker, Lane 1: phage SLPW genome, Line 2–6: phage SLPW genome digested with Xho I, EcoR I, Hind III, Ned I, and Not I, respectively, Line M2: DL 15000+2000 marker.
The complete nucleotide sequence of phage SLPW was determined. The SLPW genome comprises 17,861 bp with an average G+C content of 29.35%, which is similar to that of the lytic Staphylococcus phages S13', PSa3, and 66 (Table S1). As shown in Table S2, 20 open reading frames (ORFs) were defined as potential genes of SLPW. Genes involved in packaging, head, tail, lysis, and DNA replication showed high homology with other phages listed in Figure S1 and Table S2 (Kwan et al., 2005). However, a few unknown proteins of SLPW including Gp4, Gp6, and Gp20 showed a lower degree of similarity than those of other Podoviridae Staphylococcus phages (Table S2), which may lead to functional discrepancy. The genes of SLPW encoding lysin (Gp14) and holin (Gp10) showed high similarity with Staphylococcus lytic phages listed in Table S2, which also showed a broad host range and strong lytic ability against S. aureus (Kraushaar et al., 2013). Comparison of the genome structure with Staphylococcus prophages suggested that SLPW was a lytic phage, which lacked specific integration-related and cI repressor genes, devoid of lysogenic characteristics (Kwan et al., 2005; Hoshiba et al., 2010; Biswas et al., 2014).
Determination of Optimal Multiplicity of Infection (MOI) and One-Step Growth Curve
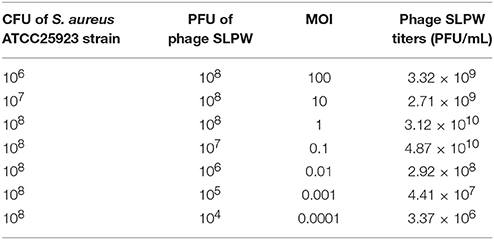
The results showed that the optimal MOI of phage SLPW was 0.1, which was the highest titer attained by the phage lysates (4.87 × 1010 PFU/mL; Table 3). Based on the optimal MOI, we established a one-step growth curve. Short latency period (10 min) and large burst size (estimated at 95.3 PFU per infected cell; Figure 4) suggest lytic nature of the SLPW phage and a higher lytic activity than the previously published lytic S. aureus phages (Han et al., 2013; Li and Zhang, 2014).
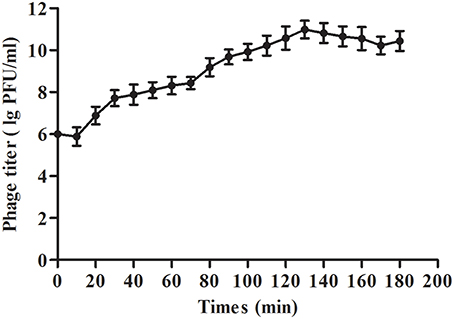
Figure 4. One-step growth curve of phage SLPW in S. aureus. Phage SLPW was co-incubated with S. aureus ATCC25923 strain cultured at an MOI of 0.1 for 15 min at 37°C. The mixture was centrifuged to remove non-absorbed phage. The re-suspended pellets were incubated at 37°C and sampled at 10 min intervals over a period of 3 h. Phage titer was measured. Results are shown as means ± SEM from triplicate experiments. The latent period was 10 min: interval between the absorption and the beginning of the initial burst. The burst size was estimated at 95.3 PFU per infected cell, which was the ratio of the final count of liberated phage particles to the initial count of infected bacterial cells.
Phage Stability
The potential clinical role of phage SLPW was evaluated by determining their physical and chemical stabilities. The thermal stability of the phages was investigated at different temperatures. We found that the activity of phage SLPW was stable at temperatures up to 45°C. Higher temperatures resulted in progressive inactivation. Phage SLPW was completely inactivated when heated to 65°C (Figure 5A). The pH stability was studied in SM buffers at a pH range of 2–12. Phage SLPW showed a relatively high survival rate (more than 80%) at a pH ranging from 6 to 10. Beyond these values, the activity decreased dramatically (Figure 5B). Further, the viability of phage SLPW was almost unaffected in the presence of 5, 25, 50, and 75% chloroform as shown in Figure 5C. Ultraviolet irradiation assay showed that about 90% of phage SLPW survived UV light (30 w, 30 cm wave-length) treatment ranging from 10 to 60 min (Figure 5D).
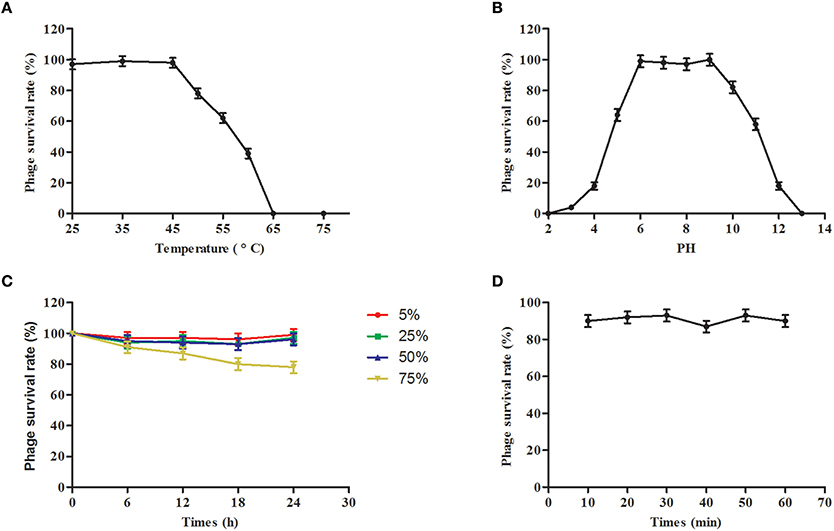
Figure 5. Stability tests of phage SLPW. (A) Thermostability: Phage SLPW was incubated at various temperatures as indicated. Samples were collected after 1 h; (B) pH stability: Phage SLPW was incubated under different pH conditions for 3 h; (C) Chloroform stability: Phage SLPW was treated with chloroform (5, 25, 50, or 75%, vol/vol) for 6, 12, 18, and 24 h; (D) Ultraviolet light stability: Phage SLPW was exposed to UV light for 10, 20, 30, 40, 50, and 60 min. The overall results were expressed as survival rates, and were titrated immediately using double-layer agar plate method. Results are shown as means ± SEM from triplicate experiments.
Studies suggested a probable relationship between phage structure and survival under adverse environmental conditions (Lasobras et al., 1997). Ackermann et al indicated that tailed phages remain comparatively steady in adverse conditions (Ackermann et al., 2004). Under harsh conditions, such as strong ultraviolet light, and large temperature fluctuations, phages belonging to Myoviridae protect themselves from extremely dry environment via intercellular location in pseudo-lysogens or biofilms created by bacterial hosts (Jonczyk et al., 2011). Phages from Podoviridae family may be extremely resistant to dry environment and survive large temperature fluctuations (Prigent et al., 2005). Our studies suggest that SLPW, which belongs to Podoviridae, showed a broad range of thermal and pH stability and strong resistance to chloroform and ultraviolet light treatment. Based on the above studies, tailed phages generally show great ability to adapt to adverse conditions, contributing to the development of phagotherapy.
Bacteriolytic Activity In vitro
The phage SLPW bacteriolytic activity was tested in an early-exponential phase culture of S. aureus ATCC25923 and MS3 strains. The growth of these strains steadily declined at an MOI 1 and was completely inhibited at MOI 100 directly after phage administration (Figure 6). However, when the culture was administered using phage SLPW at MOI 0.01, the absorbance (OD600) continued to increase during the incubation (Figure 6). The results suggested that SLPW was highly effective against S. aureus in vitro and an MOI 1 was used for therapeutic study in vivo.
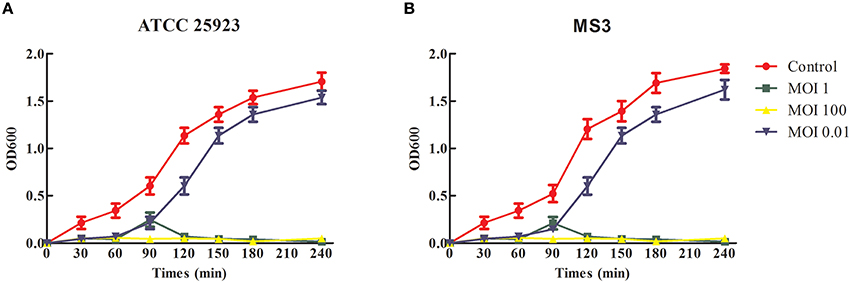
Figure 6. Bacteriolytic activity of SLPW against S. aureus in vitro. Early exponential cultures of S. aureus (A) ATCC25923 and (B) MS3 strains were co-cultured with SLPW phage at MOIs of 0.01, 1, and 100, respectively. S. aureus cultured with a similar volume of phage diluent was used as a control. Results are shown as means ± SEM from triplicate experiments.
Phage Therapeutic Study
Although studies indicate successful outcomes with topical phage treatment of human and animal infections involving S. aureus, few studies focused on the treatment of acute and lethal infection (Chhibber et al., 2013; Pincus et al., 2015). To investigate the virulence of MRSA strain MS3 in BALB/c mice, the mortality of mice was recorded daily, and followed over a period of 7 days after infection. Injection with 1 × 109 and 1 × 107 bacterial cells resulted in 100 and 0% death, respectively (data not shown). These doses were therefore used in subsequent experiments.
To evaluate the therapeutic potential of phage SLPW in vivo, assays were performed using BALB/c mice after infection with high concentrations of S. aureus (1 × 109 CFU/mouse). The results showed that mice treated with phage immediately (0 h) or at 1 h post-infection showed significantly higher survival rates than the control groups (infected mice treated with SM buffer) after 7 days. The survival rates following phage therapy at 0, 1, and 2 h were about 80, 80, and 50% after 7 days, respectively (Figure 7). Recent studies have shown that immediate phage treatment provided better protection than delayed administration against bacterial infection in mice (Watanabe et al., 2007; Hsieh et al., 2011). However, our study indicated that the survival rates between immediate and delayed therapy (1 h post-infection) by phage SLPW were similar (Figure 7). This finding confirmed that phage SLPW showed higher efficiency and sensitivity against S. aureus in vivo. In addition, phage SLPW exhibited satisfactory therapeutic effect against different sequence types of S. aureus following multilocus sequence typing (MLST; Figure 8).
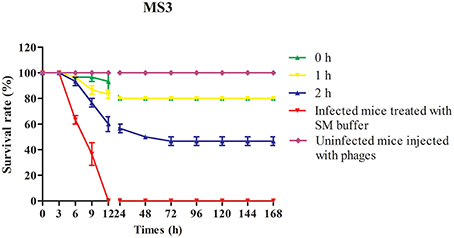
Figure 7. Survival curves of mice infected with S. aureus MS3 strain and treated with phage SLPW. Phage (MOIs of 1) was administered intraperitoneally into mice at 0, 1, or 2 h post-infection and the survival rates were recorded. Data shown are representative of three independent experiments using 10 mice per group, and displayed as mean ± SEM.
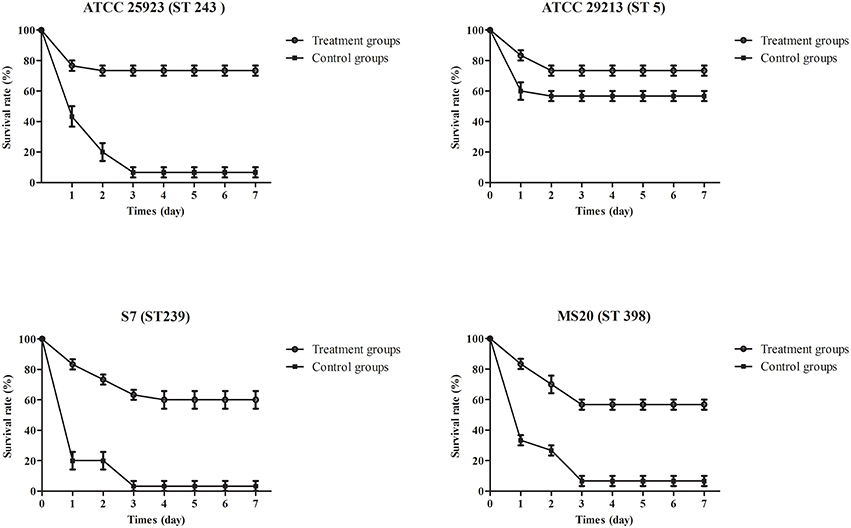
Figure 8. Survival curves of mice infected with different S. aureus strains and treated with phage SLPW. Phage (MOIs of 1) was injected intraperitoneally into mice at 1 h post-infection and the survival rates were recorded. Infected mice treated with SM buffer served as control. Data shown are representative of three independent experiments using 10 mice per group, and displayed as mean ± SEM.
Treatment efficacy was evaluated by examining bacterial colonization in the organs and blood of mice after phage therapy (1 × 107 PFU/mouse) following 1 h of infection with a non-lethal dosage of S. aureus (1 × 107 CFU/mouse). Mice infected with S. aureus strains showed relatively high pathogen density in organs (Takemura-Uchiyama et al., 2014; Li et al., 2015). However, the mice treated with SLPW showed significantly lower S. aureus levels in the spleen and lung than the control groups at every time point (Figure 9) suggesting that SLPW was therapeutically effective against systemic infection caused by S. aureus. However, the S. aureus concentrations in the blood of the phage-administered groups were slightly lower than in the control groups (Figure 9). The results also showed that phage titers in the blood and organs at 24 h after therapy were significantly increased by nearly four orders of magnitude in the spleen and two orders of magnitude in the blood and lung compared with those in the uninfected control groups (Figure 10). Based on these results, we confirmed that SLPW contributed to resistance to S. aureus and enhanced mouse survival. Unfortunately, the therapeutic effect of SLPW in uninfected control mice was temporary (Figure 10) and phage therapy before infection with S. aureus showed no effect on survival (data not shown).
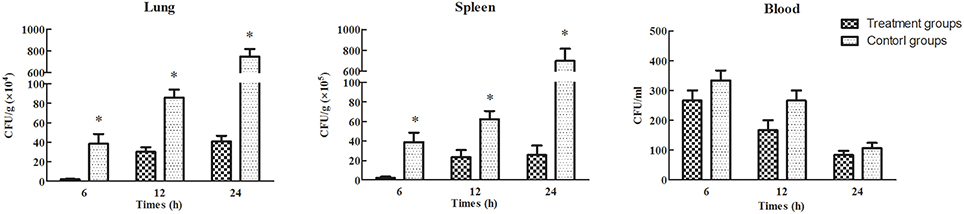
Figure 9. S. aureus concentrations in the blood and organs of mice treated with phage SLPW at 1 h post-infection. Six mice were sacrificed from each mouse group at different time points. SM buffer was administered to infected mice, which served as control. Results are shown as means ± SEM from triplicate experiments. The Mann-Whitney U-test was used to compare the phage concentration data. Significant differences (P < 0.01) are indicated by asterisks.
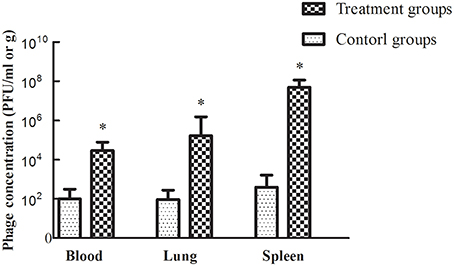
Figure 10. Figure 10 Phage SLPW concentrations in the blood and organs of mice treated with phage SLPW at 1 h post-infection. Six mice were sacrificed from each group at 24 h after the experiment. Phage was injected into uninfected mice, which served as controls. Results are shown as means ± SEM from triplicate experiments. The Mann-Whitney U-test was used to compare the phage concentration data. Significant differences (P < 0.01) are indicated by asterisks.
The strength and efficiency of the host immune response depends on the level of pro-inflammatory cytokines including IL-1β, IL-6, and TNF-α, which are indicators of the severity of infection (Wang et al., 2014; Lee et al., 2015). The results showed that phage therapy of uninfected mice (medium-treated groups) did not alter the cytokines levels (Figure 11). Therefore, phage treatment was considered safe in mice and cytokine experiments showed no bias. The IL-1β, IL-6, and TNF-α mRNA levels were significantly lower in the phage-administered mice (treatment groups) than in untreated mice (infected groups) at every time point (Figure 11). The results suggested that phage treatment successfully attenuated inflammation caused by S. aureus in mice.
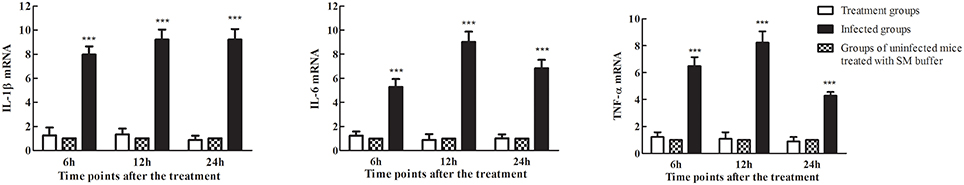
Figure 11. Levels of the pro-inflammatory cytokines IL-1β, IL-6, and TNF-α in the spleens of mice treated with phage SLPW at 1 h after infection. Six mice were sacrificed from each mouse group at different time points. SM buffer was administered to uninfected control mice. The values of pro-inflammatory cytokines in the control groups were normalized to 1.0. Levels of IL-1β, IL-6, and TNF-α mRNA were normalized to β-actin mRNA levels and were expressed as n-fold increases with respect to the control. Results are shown as means ± SEM from triplicate experiments. The Mann-Whitney U-test was used to compare the cytokine levels between infected groups and treatment groups. Significant differences (P < 0.001) are shown by asterisks.
The adverse effects associated with bactericidal agents include the release of large amounts of pathogen-associated molecular patterns recognized by Toll-like receptors and induction of pro-inflammatory cytokines in mammals (Ginsburg, 2002; Horner and Raz, 2003). However, our studies showed that treatment of uninfected mice with phage SLPW showed no significant differences in the expression of pro-inflammatory cytokines (IL-1β, IL-6, and TNF-α) during 24 h (Figure 11). In addition, phage therapy of uninfected mice resulted in similar survival as mice injected with normal saline after 7 days (data not shown). Our findings suggest that therapy using phage SLPW was safe, although phage residues in tissues may influence normal microflora in the human body (Endersen et al., 2014). Therefore, active therapy using phage SLPW is expected to be effective in the treatment of severe systemic infection caused by S. aureus.
Conclusion
In conclusion, our study investigates a lytic phage SLPW, which exhibits a wide host range, strong lytic activity and relative stability under various conditions. Rodent studies demonstrate a protective role of the phage SLPW in mice against MRSA infection, suggesting a potential antimicrobial role. Controlled clinical studies are needed to investigate the findings in animal studies.
Author Contributions
JS, YY, ZW, and PZ designed experiments; ZW and PZ carried out experiments; ZW, WJ, QF and HW analyzed experimental results; ZW and JS wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by National Natural Science Foundation of China (31172381, 31372500, and 31272580), the Special Fund for Public Welfare Industry of Chinese Ministry of Agriculture (201303041), grants from the Basic Research Programs of Science and Technology Commission Foundation of Shanghai (12JC1404700) and the Key Project of Shanghai Municipal Agricultural Commission (2014-3-1).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00934
References
Ackermann, H. W., Tremblay, D., and Moineau, S. (2004). Long-term bateriophage preservation. WFCC Newslett. 38, 35–40.
Adams, M. J., Lefkowitz, E. J., King, A. M. Q., and Carstens, E. B. (2014). Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2014). Arch. Virol. 159, 2831–2841. doi: 10.1007/s00705-014-2114-3
Biswas, A., Mandal, S., and Sau, S. (2014). The N-terminal domain of the repressor of Staphylococcus aureus phage Phi11 possesses an unusual dimerization ability and DNA binding affinity. PLoS ONE 9:e95012. doi: 10.1371/journal.pone.0095012
Borysowski, J., Lobocka, M., Miedzybrodzki, R., Weber-Dabrowska, B., and Gorski, A. (2011). Potential of bacteriophages and their lysins in the treatment of MRSA current status and future perspectives. Biodrugs 25, 347–355. doi: 10.2165/11595610-000000000-00000
Capparelli, R., Parlato, M., Borriello, G., Salvatore, P., and Iannelli, D. (2007). Experimental phage therapy against Staphylococcus aureus in mice. Antimicrob. Agents Chemother. 51, 2765–2773. doi: 10.1128/AAC.01513-06
Chhibber, S., Kaur, T., and Kaur, S. (2013). Co-therapy using lytic bacteriophage and linezolid: effective treatment in eliminating methicillin resistant Staphylococcus aureus (MRSA) from diabetic foot infections. PLoS ONE 8:e56022. doi: 10.1371/journal.pone.0056022
Endersen, L., O'Mahony, J., Hill, C., Ross, R. P., McAuliffe, O., and Coffey, A. (2014). Phage therapy in the food industry. Annu. Rev. Food Sci. Technol. 5, 327–349. doi: 10.1146/annurev-food-030713-092415
Engemann, J. J., Carmeli, Y., Cosgrove, S. E., Fowler, V. G., Bronstein, M. Z., Trivette, S. L., et al. (2003). Adverse clinical and economic outcomes attributable to methicillin resistance among patients with Staphylococcus aureus surgical site infection. Clin. Infect. Dis. 36, 592–598. doi: 10.1086/367653
Gardete, S., and Tomasz, A. (2014). Mechanisms of vancomycin resistance in Staphylococcus aureus. J. Clin. Invest. 124, 2836–2840. doi: 10.1172/JCI68834
Ginsburg, I. (2002). Role of lipoteichoic acid in infection and inflammation. Lancet Infect. Dis. 2, 171–179. doi: 10.1016/S1473-3099(02)00226-8
Gutierrez, D., Vandenheuvel, D., Martinez, B., Rodriguez, A., Lavigne, R., and Garcia, P. (2015). Two phages, phiIPLA-RODI and phiIPLA-C1C, lyse mono- and dual-species Staphylococcal biofilms. Appl. Environ. Microbiol. 81, 3336–3348. doi: 10.1128/AEM.03560-14
Han, J. E., Kim, J. H., Hwang, S. Y., Choresca, C. H. Jr., Shin, S. P., Jun, J. W., et al. (2013). Isolation and characterization of a Myoviridae bacteriophage against Staphylococcus aureus isolated from dairy cows with mastitis. Res. Vet. Sci. 95, 758–763. doi: 10.1016/j.rvsc.2013.06.001
Horner, A. A., and Raz, E. (2003). Do microbes influence the pathogenesis of allergic diseases? building the case for Toll-like receptor ligands. Curr. Opin. Immunol. 15, 614–619. doi: 10.1016/j.coi.2003.09.021
Hoshiba, H., Uchiyama, J., Kato, S., Ujihara, T., Muraoka, A., Daibata, M., et al. (2010). Isolation and characterization of a novel Staphylococcus aureus bacteriophage, phiMR25, and its therapeutic potential. Arch. Virol. 155, 545–552. doi: 10.1007/s00705-010-0623-2
Hsieh, S. E., Lo, H. H., Chen, S. T., Lee, M. C., and Tseng, Y. H. (2011). Wide host range and strong lytic activity of Staphylococcus aureus lytic phage Stau2. Appl. Environ. Microbiol. 77, 756–761. doi: 10.1128/AEM.01848-10
Jonczyk, E., Klak, M., Miedzybrodzki, R., and Gorski, A. (2011). The influence of external factors on bacteriophages-review. Folia Microbiol. (Praha). 56, 191–200. doi: 10.1007/s12223-011-0039-8
Kazmierczak, Z., Gorski, A., and Dabrowska, K. (2015). Facing antibiotic resistance: Staphylococcus aureus phages as a medical tool. Viruses 7:1667. doi: 10.3390/V7041667
Kraushaar, B., Thanh, M. D., Hammerl, J. A., Reetz, J., Fetsch, A., and Hertwig, S. (2013). Isolation and characterization of phages with lytic activity against methicillin-resistant Staphylococcus aureus strains belonging to clonal complex 398. Arch. Virol. 158, 2341–2350. doi: 10.1007/s00705-013-1707-6
Kwan, T., Liu, J., DuBow, M., Gros, P., and Pelletier, J. (2005). The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc. Natl. Acad. Sci. U.S.A. 102, 5174–5179. doi: 10.1073/pnas.0501140102
Lasobras, J., Muniesa, M., Frias, J., Lucena, F., and Jofre, J. (1997). Relationship between the morphology of bacteriophages and their persistence in the environment. Water Sci. Technol. 35, 129–132. doi: 10.1016/S0273-1223(97)00247-3
Lee, J. K., Luchian, T., and Park, Y. (2015). Effect of regular exercise on inflammation induced by drug-resistant Staphylococcus aureus 3089 in ICR mice. Sci. Rep. 5:16364. doi: 10.1038/srep16364
Li, L., and Zhang, Z. (2014). Isolation and characterization of a virulent bacteriophage SPW specific for Staphylococcus aureus isolated from bovine mastitis of lactating dairy cattle. Mol. Biol. Rep. 41, 5829–5838. doi: 10.1007/s11033-014-3457-2
Li, Z. P., Li, J., Qu, D., Hou, Z., Yang, X. H., Zhang, Z. D., et al. (2015). Synthesis and pharmacological evaluations of 4-hydroxycoumarin derivatives as a new class of anti-Staphylococcus aureus agents. J. Pharm. Pharmacol. 67, 573–582. doi: 10.1111/jphp.12343
Lowy, F. D. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532. doi: 10.1056/NEJM199808203390806
Matsushiro, A., and Okubo, S. (1972). [New method of phage isolation]. Tanpakushitsu Kakusan Koso 17(Suppl.), 292–298.
Pajunen, M., Kiljunen, S., and Skurnik, M. (2000). Bacteriophage phiYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 182, 5114–5120. doi: 10.1128/JB.182.18.5114-5120.2000
Pincus, N. B., Reckhow, J. D., Saleem, D., Jammeh, M. L., Datta, S. K., and Myles, I. A. (2015). Strain specific phage treatment for Staphylococcus aureus infection is influenced by host immunity and site of infection. PLoS ONE 10:e0124280. doi: 10.1371/journal.pone.0124280
Plata, K., Rosato, A. E., and Wegrzyn, G. (2009). Staphylococcus aureus as an infectious agent: overview of biochemistry and molecular genetics of its pathogenicity. Acta Biochim. Pol. 56, 597–612.
Prigent, M., Leroy, M., Confalonieri, F., Dutertre, M., and DuBow, M. S. (2005). A diversity of bacteriophage forms and genomes can be isolated from the surface sands of the Sahara Desert. Extremophiles 9, 289–296. doi: 10.1007/s00792-005-0444-5
Sasidharan, S., Prema, B., and Yoga, L. L. (2011). Antimicrobial drug resistance of Staphylococcus aureus in dairy products. Asian Pac. J. Trop. Biomed. 1, 130–132. doi: 10.1016/S2221-1691(11)60010-5
Son, J. S., Kim, E. B., Lee, S. J., Jun, S. Y., Yoon, S. J., Kang, S. H., et al. (2010). Characterization of Staphylococcus aureus derived from bovine mastitis and isolation of two lytic bacteriophages. J. Gen. Appl. Microbiol. 56, 347–353. doi: 10.2323/jgam.56.347
Sulakvelidze, A., Alavidze, Z., and Morris, J. G. Jr. (2001). Bacteriophage therapy. Antimicrob. Agents Chemother. 45, 649–659. doi: 10.1128/AAC.45.3.649-659.2001
Takemura-Uchiyama, I., Uchiyama, J., Osanai, M., Morimoto, N., Asagiri, T., Ujihara, T., et al. (2014). Experimental phage therapy against lethal lung-derived septicemia caused by Staphylococcus aureus in mice. Microbes Infect. 16, 512–517. doi: 10.1016/j.micinf.2014.02.011
Viscardi, M., Perugini, A. G., Auriemma, C., Capuano, F., Morabito, S., Kim, K. P., et al. (2008). Isolation and characterisation of two novel coliphages with high potential to control antibiotic-resistant pathogenic Escherichia coli (EHEC and EPEC). Int. J. Antimicrob. Agents 31, 152–157. doi: 10.1016/j.ijantimicag.2007.09.007
Wang, Z., Guo, C., Xu, Y., Liu, G., Lu, C., and Liu, Y. (2014). Two novel functions of hyaluronidase from Streptococcus agalactiae are enhanced intracellular survival and inhibition of proinflammatory cytokine expression. Infect. Immun. 82, 2615–2625. doi: 10.1128/IAI.00022-14
Watanabe, R., Matsumoto, T., Sano, G., Ishii, Y., Tateda, K., Sumiyama, Y., et al. (2007). Efficacy of bacteriophage therapy against gut-derived sepsis caused by Pseudomonas aeruginosa in mice. Antimicrob. Agents Chemother. 51, 446–452. doi: 10.1128/AAC.00635-06
Keywords: Staphylococcus aureus, MRSA, phage SLPW, infection, therapy
Citation: Wang Z, Zheng P, Ji W, Fu Q, Wang H, Yan Y and Sun J (2016) SLPW: A Virulent Bacteriophage Targeting Methicillin-Resistant Staphylococcus aureus In vitro and In vivo. Front. Microbiol. 7:934. doi: 10.3389/fmicb.2016.00934
Received: 22 February 2016; Accepted: 31 May 2016;
Published: 15 June 2016.
Edited by:
Pilar García, Consejo Superior de Investigaciones Científicas, SpainReviewed by:
Peter Mullany, University College London, UKDinesh Sriramulu, Shres Consultancy (Life Sciences), India
Maia Merabishvili, Queen Astrid Military Hospital, Belgium
Copyright © 2016 Wang, Zheng, Ji, Fu, Wang, Yan and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaxian Yan, eWFueWF4aWFuQHNqdHUuZWR1LmNu;
Jianhe Sun, c3VuamhlQHNqdHUuZWR1LmNu
 Zhaofei Wang
Zhaofei Wang Jianhe Sun
Jianhe Sun