- 1School of Life Sciences, Jawaharlal Nehru University, New Delhi, India
- 2School of Environmental Sciences, Jawaharlal Nehru University, New Delhi, India
Understanding the mechanism of photosynthate transfer at symbiotic interface by fungal monosaccharide transporter is of substantial importance. The carbohydrate uptake at the apoplast by the fungus is facilitated by PiHXT5 hexose transporter in root endophytic fungus Piriformospora indica. The putative PiHXT5 belongs to MFS superfamily with 12 predicted transmembrane helices. It possess sugar transporter PFAM motif (PF0083) and MFS superfamily domain (PS50850). It contains the signature tags related to glucose transporter GLUT1 of human erythrocyte. PiHXT5 is regulated in response to mutualism as well as glucose concentration. We have functionally characterized PiHXT5 by complementation of hxt-null mutant of Saccharomyces cerevisiae EBY.VW4000. It is involved in transport of multiple sugars ranging from D-glucose, D-fructose, D-xylose, D-mannose, D-galactose with decreasing affinity. The uncoupling experiments indicate that it functions as H+/glucose co-transporter. Further, pH dependence analysis suggests that it functions maximum between pH 5 and 6. The expression of PiHXT5 is dependent on glucose concentration and was found to be expressed at low glucose levels (1 mM) which indicate its role as a high affinity glucose transporter. Our study on this sugar transporter will help in better understanding of carbon metabolism and flow in this agro-friendly fungus.
Introduction
Mycorrrhiza represents 400–100 million year of plant–fungus co-evolution due to nutritional dependency on each other. It has been credited with the most predominant association (70–90% of terrestrial plant species) in nature (Smith and Read, 2010). In comparison to the direct uptake of phosphate (Pi) by plant root, mycorrhiza dependent pathway is proved to be most effective pathway to scavenge soil Pi and to deliver it to the root cortical cells. Most of the mycorrhizal fungi, belongs to the phylum Glomeromycota and Basidiomycota, are soil inhabitants and utilize up to 30% of photosynthates in exchange of nutrient supplied from soil. Piriformospora indica is an root endophytic fungus that belongs to order sabacinale (Varma et al., 1999; Weiss et al., 2011). P. indica has several beneficiary effects on their host plants which include nutrient transport like phosphate, increase in biomass and grain yield and provide resistance to host plant against various abiotic and biotic stresses (Waller et al., 2005; Baltruschat et al., 2008; Sherameti et al., 2008; Kumar et al., 2009; Oelmüller et al., 2009; Vadassery et al., 2009; Sun et al., 2010; Yadav et al., 2010; Jogawat et al., 2013; Trivedi et al., 2013). As the arbuscular mycorrhizal fungi (AMF) cannot be cultured axenically, therefore their use in sustainable agriculture for crop improvement was not so successful. P. indica has an advantage over AMF as it has a broad-host spectrum as compared to AMF (Peskan-Berghofer et al., 2004) and can be propagated axenically. Its stable transformation system has been developed, so it can be easily manipulated genetically to understand different biological mechanisms and can be used as a model to study plant–fungal interaction (Yadav et al., 2010). P. indica grows progressively inter- and intra-cellularly and forms coiled structure similar to arbuscules of AMF (Gianinazzi-Pearson, 1996; Schäfer and Kogel, 2009). It creates a microsymbiont that involves the infolding of the periarbuscular membrane of the host plant around the fungal wall, by forming an apoplastic space enclosed by symbiotic interface (Parniske, 2004; Harrison, 2005; Felle et al., 2009). Here, the bidirectional nutrient exchange involving phosphate and carbohydrate takes place (Shachar-Hill et al., 1995; Karandashov and Bucher, 2005). Symbiotic relation is so important that mycorrhizal plants can solely depend on phosphate acquisition by this route (Liu et al., 1998; Smith et al., 2003). Carbohydrate is the price paid by plant in exchange of phosphate supplied (Bonfante and Anca, 2009; Plett and Martin, 2011). It has been proposed that at root-fungus symbiotic interface, first the passive diffusion of carbohydrates and Pi occurs via plant and fungal plasma membrane respectively, followed by uptake of these nutrients by active transport driven by H+-ATPase(s) (Smith et al., 2001). However, the mechanism related to the carbohydrate and Pi transportation across symbiotic interface is still unknown. In case of mycorrhizal fungi, very less information is available on carbohydrate transporters active during symbiosis. A high-affinity glucose transporter AmMST1 was characterized from ectomycorrhizal fungi Amanita muscaria (Smith et al., 2001), another high-affinity hexose transporter TbHXT1 was characterized from Tuber borchii Vittadini (Polidori et al., 2007). In Laccaria bicolor genome, 15 putative hexose transporter genes are identified. Three of the above genes (LbMST1.2, LbMST1.3, and LbMST3.1) have been characterized as high-affinity glucose transporters (Lopez-Pedrosa et al., 2006). Only two reports are available related to the hexose transporters from AMF which are involved in the symbiosis like GpMST1 from symbiotic fungus Geosiphon pyriformis and MST2 from Glomus intraradices (Schubler et al., 2006; Helber et al., 2011), both of these proteins are high affinity glucose transporters with a broad substrate specificity for different monosaccharides.
Recent genome sequencing of P. indica genome has revealed presence of 19 putative hexose transporter genes (Zuccaro et al., 2011). The presence of high number of putative hexose transporter genes suggest that there are different type of hexose uptake system to function under different conditions. Till date, no report is available on hexose transporters in mutualistic endophyte P. indica. The characterization of hexose transporter genes are important in order to understand the mechanism of carbohydrate uptake during symbiosis. For the first time, we have characterized a monosaccharide transporter PiHXT5 (accession number CCA71201.1) from P. indica. PiHXT5 was found upregulated during colonization of P. indica with maize plant as compare to axenically grown fungus. Functional studies suggest that PiHXT5 encode for a high-affinity hexose transporter and its activity depends on proton gradient and pH.
Materials and Methods
Fungal, Plant Materials, and Growth Conditions
Piriformospora indica (Verma et al., 1998) was used throughout the study. Zea mays (var. pro33) was used for plant–fungus interaction (Yadav et al., 2010). Escherichia coli DH5-α was used for cloning purposes (Inoue et al., 1990). Saccharomyces cerevisiae hexose uptake mutant EBYVW.4000 (MATa _hxt1-17 _gal2 _stl1 _agt1 _mph2_mph3 leu2-3, 112 ura3-52 trp1-289 his3-_1 MAL2-8c SUC2; Wieczorke et al., 1999) was used for functional characterization and kinetic studies of PiHXT5. P. indica culture was maintained in modified Aspergillus minimal medium (Hill and Kafer, 2001) or in modified MN medium (Bécard and Fortin, 1988). The yeast strain EBY.VW4000 unless otherwise mentioned, was maintained on YPD medium supplemented with 2% maltose instead of glucose. Plasmid p112A1NE transformed E. coli DH5-α was maintained on LB agar plates containing ampicillin (Sigma). Z. mays seeds were washed with Tween-20 detergent and later surface sterilized with 75% ethanol for 2 min followed by treatment with 0.75% NaClO for 5 min and after washing six times with sterile distilled water, seeds were treated at 60°C in sterile water for 5 min (Kumar et al., 2009). Seeds were kept on water agar plates for germination (0.8% Bacto Agar; Difco, Detroit, MI, USA) at 25°C in the dark (Gamborg and Phillips, 2013).
Bioinformatics Analysis
For identification of hexose transporters in P. indica genome, blastX algorithm1 was used. For modeling PiHXT5 transmembrane (TM) domains, TM helix segments were predicted with various computer programs; e.g., SOSUI, HMMTOP, TMHMM, and TMpred (Von Heijne, 1994; Hirokawa et al., 1998; Krogh et al., 2001; Tusnady and Simon, 2001). A consensus of 12 TM helices emerged and the boundary of each predicted helix was identified based on empirical rules derived from known membrane protein structures. Sequence alignments were done with ClustalW and BLOSUM62 with a gap penalty of 10 for insertion and 5 for extension (Henikoff and Henikoff, 1992; Thompson et al., 1994). Multiple Sequence Alignment were performed to search the conserved signature tags of the glucose transporters derived from Homo sapiens (GLUT1, GLUT2, GLUT3) and E. coli (XylE; Iancu et al., 2013; Deng et al., 2014; Wisedchaisri et al., 2014).
Expression Analysis of PiHXT5, PiHXT8, and PiHXT9 during Colonization
Expression of P. indica PiHXT5 (GenBank ID: CAFZ01000110.1), PiHXT8 (GenBank ID: CAFZ01000044.1), and PiHXT9 (GenBank ID: CAFZ01000021.1) during colonization with maize plants were compared with axenically grown P. indica. For this purpose, P. indica was grown in Aspergillus minimal media in 111 mM (or 2% w/v) glucose and 10 mM phosphate (Hill and Kafer, 2001) for 7 days and then subsequently transferred to MN media (Bécard and Fortin, 1988) with 111 mM glucose and low phosphate (10 μM KH2PO4) and grown for 15 days. For colonization, radicals germinated seedlings were first submerged into macerated P. indica and incubated for 4 days on water agar and finally transferred to MN with low phosphate (10 μM) and allowed to grow for 15 days, whereas in case of control plants autoclaved double distilled water was used. The MN medium was changed weekly. At 15 days of colonization, maize roots were harvested, and RNA was isolated from these roots. The plants were maintained in an environmentally controlled green house at 30 ± 2°C and 16 h light/8 h dark and relative humidity 60–70%, with light intensity of 1000 Lux. To check colonization, root segments (1.0 cm approximately) were heated on 10% KOH for 15 min, treated with 1 N HCl and were stained with 0.05% trypan blue overnight at RT(or stained at 60°C for 1 h) and mounted on lactophenol (Kumar et al., 2009). Observation was done under light microscope (Leica Microscope. Type 020-518.500, Germany). Percent colonization was calculated for the inoculated plants using the following formula (Mcgonigle et al., 1990).
Further, to study the effect of different glucose concentration on expression pattern of PiHXT5, P. indica was grown in KF medium at 10 mM Phosphate and 111 mM glucose for 7 days and subsequently transferred to MN media (Bécard and Fortin, 1988) containing 10 mM phosphate and different glucose concentrations ranging from very low to high glucose concentration (1 and 500 mM) and grown for 5 days at 110 rpm and 30 ± 2°C temperature. After 5 days of growth, P. indica culture was harvested and RNA was isolated. For quantitative real time PCR (qRT-PCR) analysis the expression of PiHXT5 in all these conditions was compared with P. indica grown in optimum glucose concentration (111 mM glucose).
Isolation of RNA, cDNA Synthesis, and qRT-PCR
Total RNA was isolated from P. indica colonized maize plant roots and from axenically grown fungus. Samples were harvested and washed with sterile water and frozen in liquid nitrogen. These tissue samples were then crushed in liquid nitrogen and RNA was isolated using TRIzol reagent according to the protocol provided by the manufacturer (Invitrogen, USA). The concentration of RNA was quantified by Nanodrop-2000 (Thermoscientific) spectrophotometer and RNA integrity was checked on denaturing formaldehyde agarose gel with 1.2% agarose by fractionating the 10 μg of total RNA. For cDNA synthesis 1.5 μg of total RNA was treated with DNase1 (Thermoscientific) and cDNA was synthesized using Thermoscientific First strand synthesis Kit. Real time PCR was performed with gene specific primer pairs (Table 1) and SYBR Green I (TakaRa) using ABI 7500 Real-Time PCR System (Applied Biosystems) according to manufacturer’s instructions. The cycling conditions were as follows: denaturation was done at 94°C for 5 min for one cycle. Subsequently, for 40 cycles denaturation was done at 95°C for 15 s, annealing was done at 57°C for 30 s and extension was done at 68°C for 30 s. P. indica translational elongation factor gene (PiTef) was used as a reference.
Isolation of PiHXT5 Coding DNA Sequence (CDS)
PiHXT5 CDS was amplified with gene specific primer pair (Table 1) using Pfu polymerase (Thermoscientific). The cycling conditions were as follows: 94°C for 3 min (one cycle), 94°C for 40 s, 59°C for 30 s, 72°C for 3 min (35 cycles) and 72°C for 5 min (one cycle). Amplified PCR product was cloned into pGEM-T Easy vector (Promega). The pGEMT-PiHXT5 construct was transformed in E. coli DH5-α cell and transformants were confirmed by colony PCR with gene specific primers (Table 1). Plasmid was isolated from the positive clones and confirmed by restriction digestion using Pst1 and BamH1. Further cloning was confirmed by sequencing.
Rapid Amplification of cDNA Ends (RACE)
The full-length cDNA sequences of PiHXT5 were obtained after rapid amplification of cDNA ends (RACE) using the protocols of the SMART (Clontech) kits. The 5′ RACE library was prepared by the incorporation of a “smart oligo” at the 5′-end of the reverse-transcribed cDNA. 5′-end of the PiHXT5 gene was amplified from 5′ RACE library using a gene-specific oligonucleotide (GSP1). Further amplicon specificity is enhanced in secondary PCR using the same gene-specific oligonucleotide (GSP1). The GSP1 primer is located at 312–331 bp downstream of the ATG codon. For 3′ RACE library, the 3′-end of the PiHXT5 was amplified from 3′ RACE library using gene specific oligonucleotide (GSP1′ and GSP2′). The GSP1′ and GSP2′ primers are located at 1288–1308 bp and 1463–1483 downstream of the ATG codon. Primers were designed such that of 331 bp along with 5′ UTR were amplified. Similarly, amplicon of 312 and 137 bp with 3′ UTR in primary and nested PCR, were obtained respectively. Amplicons were analyzed by 1.5% agarose gel electrophoresis and purified using the GeneJet gel extraction kit (Thermo scientific, Lithuania, EU). Purified 5′ RACE and 3′ RACE fragment was cloned into the T-A cloning vector pGEM-T Easy and 10 random colonies of each were sequenced.
CLUSTALW2 and Phylogenetic Analysis
Phylogenetic relationship study was done including deduced amino acid sequence of PiHXT5 from P. indica and the sugar transporters sequences from other fungi, plants and human. Sequences were obtained from GenBank databases. Multiple sequence alignment was done using CLUSTALW22 and MULTIALIN algorithum (Corpet, 1988). Phylogenetic studies were done using neighbor-joining method by MEGA 5 software. Bootstrap step was conducted using 1000 replicates to support the inferred clades (Felsenstein, 1985). The tree is drawn to scale having branch lengths in the same units as those of the evolutionary distances used to deduce the phylogenetic tree. Poisson correction method (Zuckerkandl and Pauling, 1965) was used to compute evolutionary distances which represent units of the number of amino acid substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). We have used 54 amino acid sequences for the analysis purpose and a total of 296 positions were used for phylogenetic analysis. The sequence of the sugar transporters ST1 from Medicago truncatula and Z. mays were used as outgroups.
Complementation and Growth Assay
For complementation, S. cerevisiae hexose transporter deficient strain EBY.VW4000 was used. The pGEM-T-PiHXT5 construct digested with Pst1 and BamH1 and the PiHXT5 fragment obtained was sub-cloned into p112A1NE vector (Leggewie et al., 1997) which was previously treated with same enzymes. The ligation was done using T4 DNA ligase (Thermoscientific). E. coli DH5-α cells were transformed with p112A1NE-PiHXT5 construct and recombinant clones were confirmed by colony PCR with PiHXT5 specific primers. Further cloning was confirmed by digestion of plasmid with Pst1 and BamH1 and confirmed by sequencing. EBY.VW4000 mutant was transformed with p112A1NE-PiHXT5 construct by LiCl-PEG method (Gietz and Schiestl, 1991; Riesmeier et al., 1992). Procedures and medium used for growth and selection of transformants were similar as described (Bun-Ya et al., 1991). The transformants were first selected on their ability to grow on YNB medium deficient in tryptophan (Boles and Hollenberg, 1997) containing 1% maltose as sole carbon source and later the single colonies of these transformants were transferred to the same medium with 2% glucose. The growth of PiHXT5 containing yeast was compared with EBY.VW4000 containing only p112A1NE vector. The recombinant clones which were able to grow on glucose were selected for further experiments.
14C-Glucose Transport Assay and Biochemical Kinetics Characterization
For kinetic study, yeast cells were grown to an OD600 of 0.5–0.7 in YNB medium without tryptophan and with 2% maltose. The cells were harvested by centrifugation at 4000 rpm for 5 min. Further, the harvested cells were washed two times with buffer A (50 mM potassium phosphate, 0.6 M sorbitol, pH 6.5) and re-suspended in the same buffer to a final concentration of 109 cell/ml. 14C-glucose uptake was done as described previously (Polidori et al., 2007; Wahl et al., 2010). To start sugar uptake 100 μl of yeast cell suspension was mixed with 100 μl of D-glucose (5, 25, 100, 500, 1, and 10 mM non-radioactive D-glucose plus 5 μM 14C-glucose, specific activity 0.2 μCi/μL). After 30, 60, 120, and 240 s, 50 μl aliquots from each reaction mix were transferred to 4 ml of ice-cold buffer A, to block sugar uptake. Cells were filtered onto a GF/C grade filter paper (Whatman Glass microfiber filters), and washed twice with 10 ml of ice-cold buffer. Then the filters were transferred into scintillation vials containing 5 ml of scintillation cocktail ‘O’ (CDH) and the radioactivity was measured with scintillation counter (Liquid Scintillation Analyzer TRI-CARB 2100TR; Packard). Amount of the glucose retained on the filters was determined by using the specific activity of the substrate (14C-glucose) solutions which are used during the experiment. The competition experiments were performed in the presence of different unlabeled substrates D-glucose, D-fructose, D-mannose, D-galactose, D-xylose. In the competition experiments, yeast strain EBYVW.4000 expressing PiHXT5 was incubated (1 min at 30°C) with 25 μM 14C-glucose in the presence of five-fold excess of different sugars. The uptake rate of 14C-glucose is reported in percentage of control (without competitor), and 100% uptake corresponds to 225 pmol.min-1.108 cells-1. All uptake experiments were repeated with independent samples at least three times. Kinetic studies of PiHXT5 were measured by uptake rate as a function of glucose concentration. The kinetic parameters were determined using the Michaelis–Menten kinetics plot, and all the data were analyzed by non-linear regression using the Enz-Fitter program. The standard errors were evaluated by the same program. Sugar uptake inhibition experiments were performed using the uncoupling compound 2,4-dinitrophenol (DNP) and H+-ATPase inhibitor sodium vanadate at a final concentration of 100 μM, respectively, in the presence of 25 μM 14C-glucose. pH dependence of PiHXT5 for glucose transportation was analyzed by adjusting the potassium phosphate buffer at different pH values ranging from 2 to 8 and then 14C-glucose uptake was measured as described above, at pH 2–8.
Results
Identification of Sugar Transporters from P. indica Genome
Blastx search of first P. indica (Genus ID: 65672) genome sequencing project3 using already characterized fungal hexose transporters such as GiMST2 from G. intraradices, GpMST1 from G. pyriformis, ScHXT1 from S. cerevisiae and TbHXT1 from Tuber borchii as query, a total of 19 putative sugar transporter sequences were identified in P. indica genome. Three of these putative sugar transporters with accession number CCA70422.1, CCA68995.1, and CCA67897.1 were showing highest homology with query proteins. The predicted protein size of these putative 19 proteins was 568–487 amino acids. The putative function for two of these proteins with accession number CCA69504.1 and CCA70422.1 was assigned as 4-hydroxy benzoate transporter and quinate transporter, respectively in pedant database. For one other putative hexose transporter with accession number CCA68994.1, only a partial sequence of 67 amino acids is given in database and was found to be a hypothetical protein. Based on these data three sequences were rejected. These 16 putative sugar transporter sequences were further analyzed by SACS HMMTOP program for their membrane topology, we found that nine out of 16 protein sequences were showing typical MFS transporter membrane arrangement with 12 transmembrane helices, with a long intracellular loop between helix 6 and 7 and cytosolic N and C-terminal. Finally nine sequences were aligned using Multialin and ClustalW2 with hGLUT1, hGLUT2, hGLUT3, and XylE to find conserved regions in P. indica hexose transporter family. The hGLUT1 and XylE crystal structures were available that explained its functional motifs. Multiple sequence alignment shows presence of many conserved residues in these proteins. They all contain two conserved GR-[KR] motifs, which are present in MFS transporters and the PETKG sequence, which is highly conserved among sugar transporters. P. indica putative sugar transporters show 17–60% homology with other sugar transporters. Invariant and highly conserved sugar porter family signature motifs [GR]-[P]-[PESPR]-[V-GR]-[LFP]- [PETKG] are found in PiHXT5, PiHXT8, and PiHXT9 similar to hGLUT1-3 and XylE (Figure 1). The structural and functional significance of these motifs has been examined previously. The PiHXT5, PiHXT8, and PiHXT9 were found to exhibit high sequence similarity to hGLUT1-3 (Figure 1). This strongly suggests that these transporters are the member of the sugar porter family.
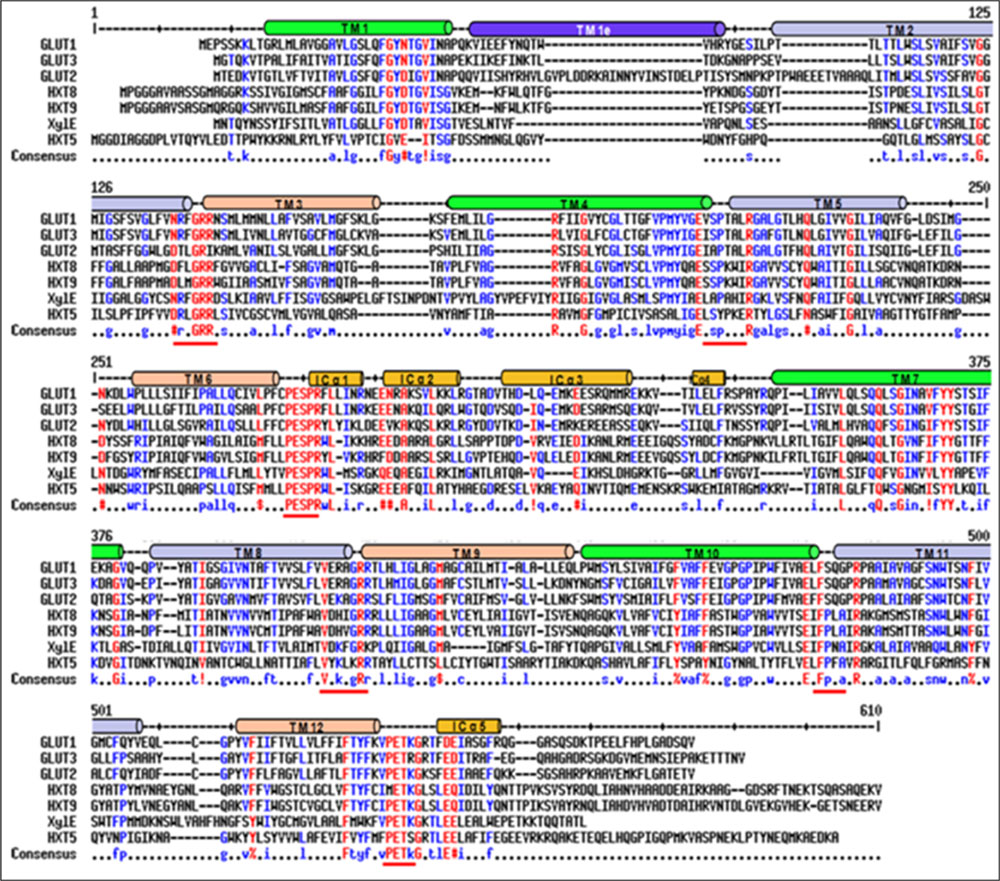
FIGURE 1. Sequence alignment of Piriformospora indica PiHXT5, PiHXT8, and PiHXT9 with hGLUT1-3 and XylE. Secondary structural elements of GLUT1 are indicated above the sequence alignment. Invariant and highly conserved amino acids are red and blue, respectively. The conserved sugar porter family signature motifs are underscored with red lines.
PiHXT5, a Fungal Carbohydrate Transporter Expression is Induced during Colonization
Recently, plant–fungal symbiotic stage is reflected by induced expression pattern of specific AMF carbohydrate transporter as well as plant mycorrhiza specific phosphate transporter. It was shown during symbiosis, Glomus sp. monosaccharide transporter MST2 as well as plant phosphate transporter PT4 of Solanum tuberosum (St PT4) and Medicago truncatula (Mt PT4) gets induced as the marker of symbiotic stage. Interestingly, Glomus sp. monosaccharide transporter MST2, whose expression is undetected at spore stage, follows the expression pattern of PT4 during colonization. Similarly we inquired that, which sugar transporter of P. indica is induced during the colonizing stage, that may help the fungus to acquire carbohydrate from host plant. It was observed that P. indica colonization in plant root is a time-dependent process. Further we found 65% colonization at day 15 which was characterized by the presence of intracellular pear-shaped chlamydospores (Figure 2). To identify the hexose transporters that are regulated in response to mutualistic interaction with maize plant, qRT-PCR analysis was done. Out of nine sugar transporters, three (PiHXT5, PiHXT8, and PiHXT9) found to be upregulated during the colonization stage as compared to non-colonizing or axenically grown fungus. Further, as compared to PiHXT8 and PiHXT9, PiHXT5 expression was found to be highly elevated during colonization with host plant (Figure 3A). PiHXT5 was found 64-fold up-regulated after 15 days of colonization as compared to non-colonized P. indica. However as compared to PiHXT5, both PiHXT8 and PiHXT9 expression were found to be non-significant (Figure 3A). Therefore, only PiHXT5 was selected for further study.
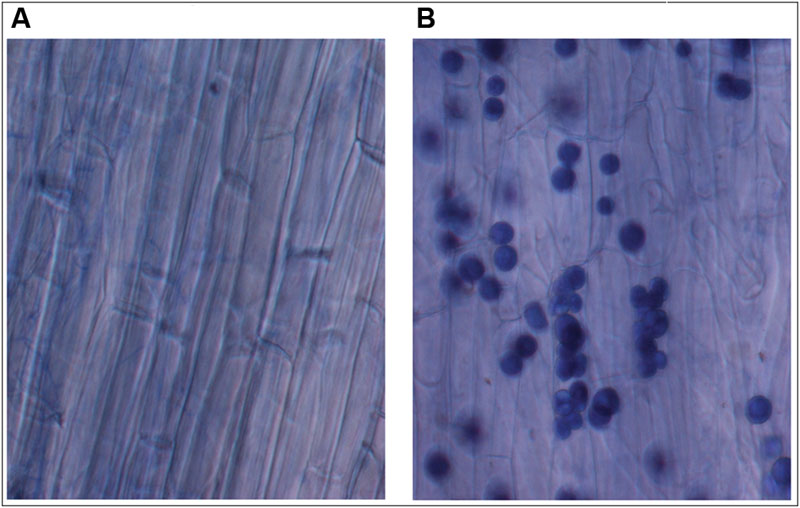
FIGURE 2. Trypan blue staining of maize plant roots to show the colonization of maize roots by P. indica. (A) Control maize roots without P. indica. (B) Maize root cortical cells showing intracellular chlamydospores of P. indica after 15 days of colonization.
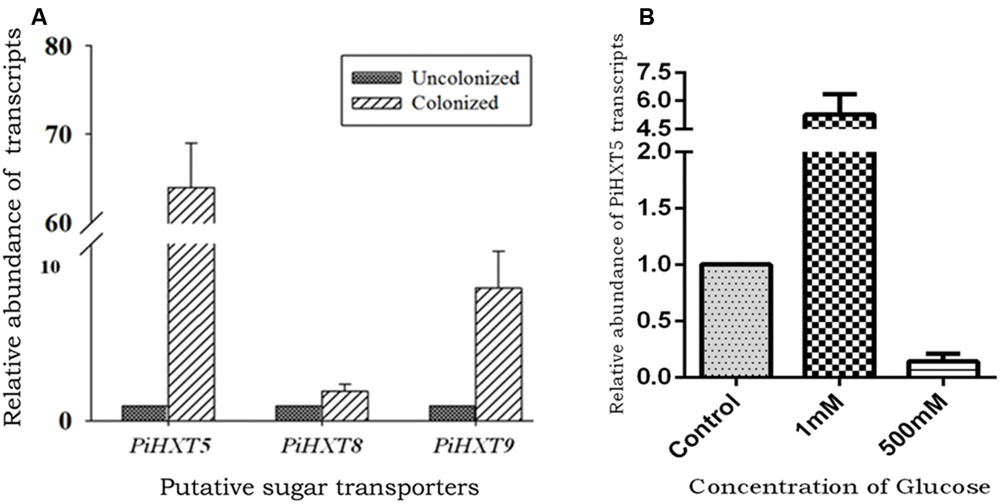
FIGURE 3. Expression analysis of PiHXT5 by qRT-PCR. (A) Effect of colonization of P. indica with maize plant on PiHXT5, PiHXT8, and PiHXT9 expression. qRT-PCR was performed to quantify relative expression level of PiHXT5 in axenically grown P. indica and in colonized state of P. indica after 15 days of colonization at low phosphate (10 μM) in MN media. (B) Effect of different D-glucose concentrations on expression of PiHXT5. P. indica was grown in MN with different D-glucose concentrations (1 and 500 mM) for 5 days and qRT-PCR was performed to quantify PiHXT5 relative expression level in different glucose concentrations, the expression was compared with control where P. indica was grown in 111 mM glucose. qRT-PCR was performed using gene specific primer and SYBR green I. The comparative Ct method was applied to analyze the data. For experimental samples, targeted quantity was determined and divided by the target quantity of the calibrator (PiTef). Thus, the calibrator becomes the 1X sample, and all other quantities are expressed as an n-fold difference relative to the PiTef.
Glucose Deficiency Triggers the Expression of PiHXT5
We also investigated the effect of glucose on the expression of the PiHXT5. The normal glucose concentration in routine culture was established as 111 mM (or 2% w/v). Therefore, we compared the expression of PiHXT5 at different glucose concentration, i.e., 1 and 500 mM with respect to 111 mM. Interestingly, very low glucose concentration (1 mM) was found to induce (five-fold) the expression of PiHXT5 in axenic culture as compared to optimum glucose level (111 mM). However, PiHXT5 was found to be suppressed (10-fold) at higher glucose concentration (500 mM) as compared to optimum glucose level (111 mM). This expression pattern of PiHXT5 suggests its high-affinity nature (Figure 3B).
Isolation of CDS, 5′ and 3′ UTR
The PiHXT5 CDS was amplified, cloned and sequenced from cDNA of P. indica. The gene was found to be 1.599 Kb in length (Figure 4). The sequencing of cloned PiHXT5 CDS shows the presence of 21 extra nucleotide (888–908 downstream of ATG) as compared to derived CDS sequence of predicted hexose transporter (GenBank CAFZ01000110.1) from P. indica genome sequence. PiHXT5 encodes a polypeptide of 532 as compared to initially predicted 525 amino acids having a relative molecular mass of 59.69 kDa.
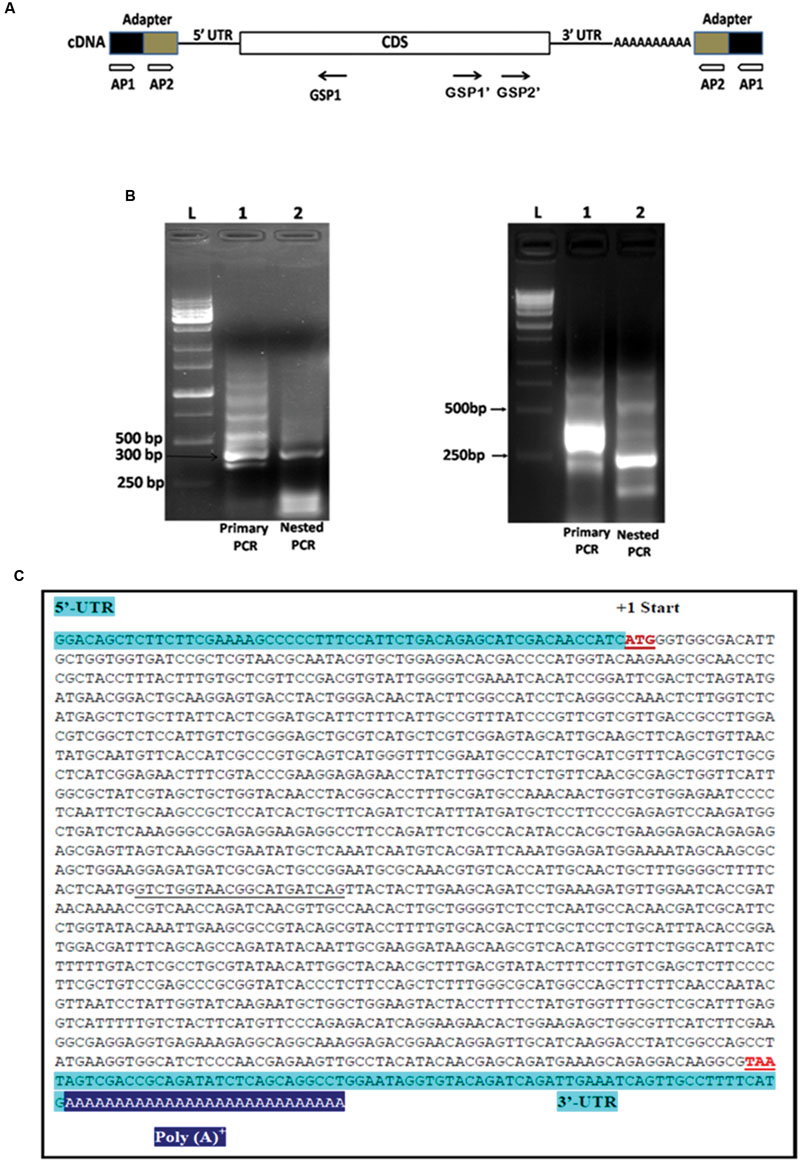
FIGURE 4. Rapid amplification of cDNA ends (RACE) analysis for the full length gene of PiHXT5. (A) Schematic presentation of 5′ and 3′ RACE amplification. Two RACE assays were conducted by using two adapter primers for both of 5′ RACE and 3′ RACE (AP1 and AP2) and PiHXT5 GSP1 (nucleotides 312–331 bp) for 5′ RACE and GSP1′ (nucleotides 1288–1308 bp) and GSP2′ (nucleotides 1463–1483 bp) for 3′ RACE. In the first 5′ RACE primary PCR were set with AP1-GSP1 primer pairs (each of lanes 1 in figure B) and nested PCR with AP2-GSP1 (each of lanes 2 in figure B). In 3′ RACE the primary PCR and nested PCR were conducted with different set of primers AP1-GSP1′ and AP2-GSP2′, respectively. (B) Analysis of the primary and nested PCR products on agarose gel, The 1 Kb DNA size marker are shown in extreme left lane marked by letter L. (C) Full length sequence of PiHXT5 gene of P. indica. Sequences of the PiHXT5 5′ UTR and 3′ UTR are shown in shaded letters. The first start codon (methionine) is numbered +1. The sequencing of cloned PiHXT5 CDS shows the presence of 21 extra nucleotide (underlined) as compared to available CDS sequence of predicted hexose transporter (GenBank CCA71201.1).
To isolate 5′ UTR, 5′ RACE analysis was performed by using reverse PiHXT5 gene specific primers GSP1 (Figure 4B). Primary PCR was performed with outer primer AP1 and reverse primer GSP1 by using total RNA from P. indica which shows a multiple bands on the agarose gel (Figure 4B). The primary PCR products were used as template for nested PCR by using AP2 and GSP1. Single prominent product was obtained from nested PCR (Figure 4B). It can be predicted from this result that single transcriptional start site is present for PiHXT5 gene. The 5′ RACE product was cloned into T-A cloning vector pGEM-T Easy. The sequencing of 10 different randomly selected clones shows that they have the same sequence. The first base of this product was therefore assigned position -57, and it is located 57 bp upstream of PiHXT5 start codon (Figure 4C). Similarly, 3′ RACE analysis was performed by successive primary and nested PCR using AP1-GSP1′ and AP2-GSP2′ primers, respectively. The 3′ UTR product is 73 bp long excluding the poly A-tail downstream of PiHXT5 stop codon.
Homology and Phylogenetic Analysis
Blastx analysis showed 57–75% identity with many putative fungal hexose transporters. It has 23% identity with other hexose transporters such as GiMST2 (ADM21463.1), GpMST1 (CAJ77495.1), TbHXT1 (AAY26391), and ScHXT1 (AAA34700.1; Table 2). Phylogenetic analysis suggests that PiHXT5 cluster together with uncharacterized fungal hexose transporters from Auricularia delicate (EJD41559.1), Trametes versicolor (EIW61310.1), Coprinopsis cinerea okayama (XP001839222.1), Paxillus involutus (AAT91304.1), and Aspergillus flavus (XP002374068.1). These branches include closely related hexose transporters from basiodiomycota or ascomycota fungi (Figure 5). The putative membrane topology of PiHXT5 was deduced by HMMTOP program which shows 12 transmembrane helices with cytosolic N and C-terminal, and a long intracellular loop between helix 6 and 7. PROSITE shows that PiHXT5 belongs to MFS, we have identified two G-R-[KR] motifs identified in PiHXT5 polypeptide, which are typical of these transporter proteins. The first one is located between 2 and 3 transmembrane domain and the second between transmembrane domains 8 and 9. Moreover, sugar transporter PFAM motif (PF0083) was also present in PiHXT5 polypeptide at position 45–480 and MFS superfamily domain (PS50850) is present at position 36–471 (Figure 6, Table 3).
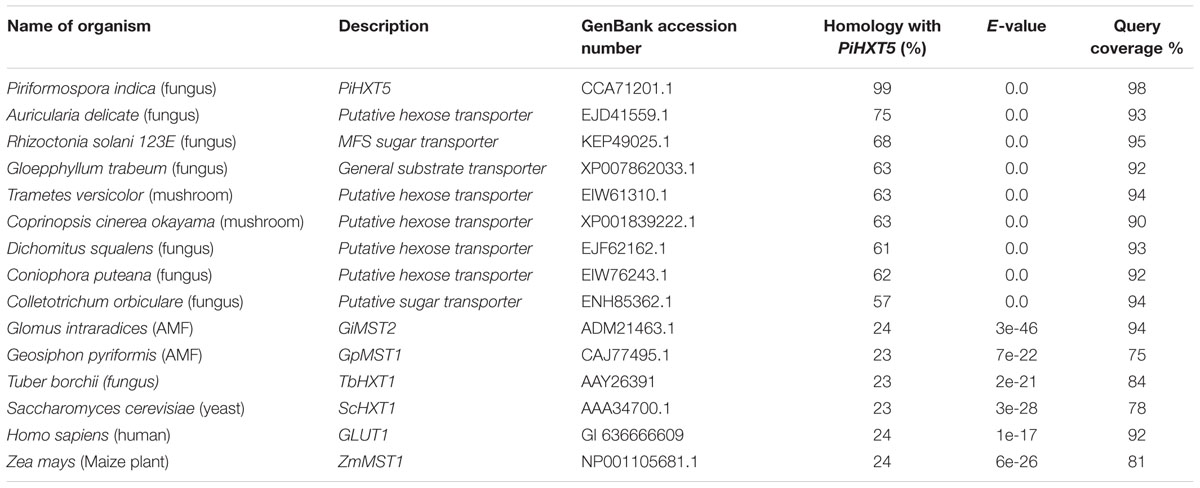
TABLE 2. Percentage homology between PiHXT5 and hexose transporters from fungi, bacteria, plant, and animal.

FIGURE 5. Phylogeny of deduced protein sequence of PiHXT5. Phylogenetic tree of fungal monosaccharide transporter protein sequences. The dendrogram was generated by Mega 5 software using the neighbor-joining method for the construction of the phylogeny (Saitou and Nei, 1987). Bootstrap test were performed using 1000 replicates. The branch lengths are proportional to the phylogenetic distance. The sequence of the sugar transporters ST1 from Zea mays and Medicago truncatula were used as outgroups. P. indica PiHXT5 cluster together with uncharacterized fungal hexose transporters from basidiomycota fungi.
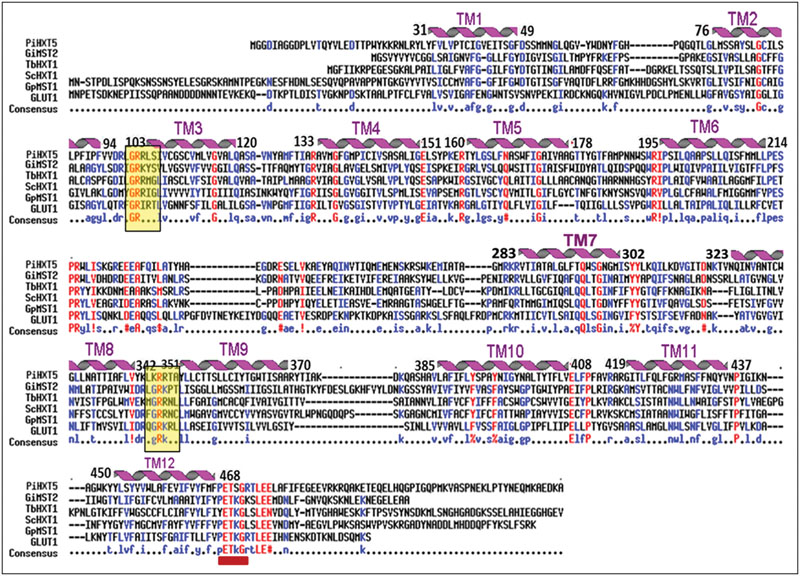
FIGURE 6. Alignment of the predicted amino acid sequence of PiHXT5 with Glomus intraradices (GiMST2), Tuber borchii (TbHXT1), Saccaharomyces cerevisiae (ScHXT1) and Geosiphon pyriformis (GpMST1) and Homo sapiens (hGLUT1) by using MULTIALIN. The degree of sequence conservation at each position amino acids is shown in red, and low consensus amino acids are shown in blue. Membrane-spanning domains (TM) of PiHXT5 as predicted by HMMTOP are shown as helices over the corresponding amino acid sequences indicated by numerals (TM1–TM12). Prediction of functional motifs in PiHXT5 polypeptide was performed with PROSITE data base (available on-line). The PETKG sequence, which is highly conserved among sugar transporters, is indicated with red bar. Two GR-[KR] motifs typical of the transporters belonging to the MFS superfamily are highlighted by yellow color (Consensus symbols: “!” is any sequence of IV, “$” is any sequence of LM, “%” is any sequence of FY, and “#” is any sequence of NDQEBZ.).
Complementation Assay
In order to functionally characterize PiHXT5 and to investigate its kinetic properties and substrate specificity, it was cloned in yeast expression vector p112A1NE under the control of ADH1 promoter. This construct p112A1NE-PiHXT5 was transformed into S. cerevisiae strain EBY.VW4000. Because EBY.VW4000 is devoid of any hexose transporter, its growth was found retarded on glucose, as compare to EBY.VW4000 complemented with PiHXT5 or its parental strain harboring only p112A1NE plasmid vector. Growth of EBY.VW4000 transformed with p112A1NE-PiHXT5 construct or empty expression vector and untransformed EBY.VW4000 was compared in YNB medium containing all the amino acids except tryptophan and 2% D-glucose as carbon source at OD650 for 2 h at 30°C. The expression of PiHXT5 cDNA in mutant yeast enabled the mutant to grow significantly as compare to mutant harboring only p112A1NE vector and untransformed EBY.VW4000.
PiHXT5 is a Broad Substrate Range Monosaccharide Transporter
To characterize PiHXT5 biochemically, we expressed PiHXT5 in glucose transport deficient mutant of S. cerevisiae EBY.WV4000. Glucose uptake by EBY.VW4000 harboring PiHXT5 was further confirmed by measuring uptake of 14C-glucose. EBY.VW4000 expressing PiHXT5 accumulated 14C-glucose at a rate significantly higher than the background rate shown by the control EBY.VW4000 cells. To investigate the substrate specificity, competition experiment was performed, where glucose uptake was challenged with five-fold molar excess of competitive sugar. The competition assay was performed in presence of D-glucose, D-fructose, D-mannose, D-galactose, and D-xylose. Using 14C-labeled sugars, we showed that PiHXT5 is able to transport Glc, Fru, Xyl, Man, and Gal with decreasing affinity in that order (Figure 7A).
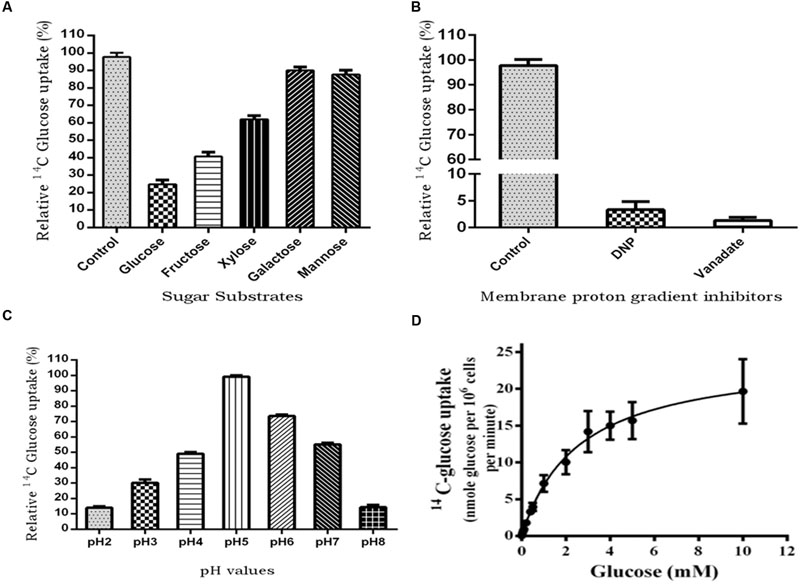
FIGURE 7. 14C-glucose uptake in PiHXT5-expressing yeast strain EBY.VW4000. (A) Substrate competition on the respective sugars; PiHXT5 is able to transport D-glucose (highly), D-fructose, and D-xylose (moderate). It was weakly able to transport D-galactose and D-mannose. (B) Incubation of PiHXT5 expressing yeast cells in presence of protonophore (DNP) and plasma membrane H+-ATPase inhibitor (vanadate) for 5 min strongly inhibits glucose uptake. (C) The optimum pH for glucose uptake by PiHXT5 is pH5 and highest uptake is found between pH 5 and 6. The readings are relative to negative control (14C-glucose transport in strain EBY.VW4000 transformed with vector only; D) Michaelis–Menten kinetics of glucose uptake rates (pH 6.5) indicates a Km of 2.56 ± 0.18 mM and Vmax of 24.5 ± 0.69 nmol/min/106 cells. Error bars represent SD; n = 3.
The 14C-glucose uptake rate of PiHXT5 was found significantly lower in presence of protonophore DNP and plasma membrane H+-ATPase inhibitor sodium vanadate as compared to control (absence of these chemicals; Figure 7B). The PiHXT5 lose its transport property in the presence of protonophore, which indicates its secondary nature of H+ co-transport. To obtain the optimum pH value for function of PiHXT5, EBY.VW4000 expressing PiHXT5 was subjected to 14C-glucose transport at different pH value ranging from pH 2 to 8. We found that glucose transport activity of PiHXT5 is pH dependent. The optimum value for glucose transport by PiHXT5 is between pH 5 and 6 (Figure 7C). Affinity studies show that the apparent Km of PiHXT5 is 2.56 ± 0.18 mM. The Km is closest to the highest affinity monosachharide transporter reported so far. The glucose uptakes by cells expressing PiHXT5 follow Michaelis–Menten kinetics with an apparent Km of 2.56 ± 0.18 mM (Vmax 24.5 ± 0.69 nmol/min/106 cells), which suggests it is a high-affinity transporter (Figure 7D).
Discussion
The carbon flow from the photoautotrophic plants to fungal heterotrophs plays the key role in the formation of symbiotic relationship (Ho and Trappe, 1973). The mechanism of transportation of photosynthate from the host plant cortical cells to the fungal tissue involves three major steps (1) Release of sucrose into apoplastic space by gradient diffusion (2) Degradation of sucrose by cell wall invertase into simpler molecules like fructose and glucose (3) Subsequent, transport across fungal membrane into the intra-radical hyphae (Gianinazzi-Pearson, 1996; Hodge et al., 2010). The mechanism of carbohydrate transported across symbiotic interface is still unexplored. Monosaccharide transporters plays critical role in transportation of nutrients across plasma membrane (Lalonde et al., 2004). It would be of utmost importance to identify and characterize the fungal transporters involved in uptake of carbohydrate from the symbiotic interface for the survival and maintenance of the fungal partner. In this study, we have analyzed putative hexose transporters from genome of mutualistic endophyte P. indica, that confers nutrient enrichment (e.g., N, P, K, S, Mg, Co, Cu, Zn, etc.) in plant under deprived conditions (Varma et al., 1999; Yadav et al., 2010). There are 19 putative hexose transporters in P. indica genome, which shows 15–91% homology with each other. Similar number of hexose transporters were reported in Laccaria bicolor (López et al., 2008), S. cerevisiae (Ozcan et al., 1998; Fan et al., 2002), Candida albicans (Fan et al., 2002). The presence of GR-[KR] motifs conserved in MFS transporters and the PETKG sequence, signature tag of sugar transporters (Pao et al., 1998; Sun et al., 2012; Deng et al., 2014) in PiHXT5 assign it to sugar porter family.
We reported the role of AMF like fungus P. indica hexose transporter during symbiosis, having the ability to transport glucose as well as plant cell wall derived sugars. Till date, only report available related to the hexose transporters from AMF which are involved in the plant symbiosis is MST2 from G. intraradices (Helber et al., 2011).
Our study reveals the role of symbiotic signaling in the regulation of fungal Hexose transporter and subsequent carbohydrate supply to P. indica when it colonizes the host plant. Using the benefit of axenic culture of P. indica on synthetic media, we are able to differentiate the expression pattern of fungal carbohydrate transporters in presence and absence of symbiotic signals (Harrison, 1999, 2005). Thus, it deciphers the effect of mutualism on the expression pattern of hexose transporters. In our study PiHXT5 is 64-fold upregulated during colonization as compared to axenic state. Our data supports the previous study, in which MST2 from G. intraradices have shown to be induced during symbiosis (Helber et al., 2011). Further, we have a novel finding that the hexose transporters are regulated by extracellular glucose concentration. Low glucose concentration induced the expression of PiHXT5. This may be explained by the fact that rate of photosynthesis is regulated by many environmental factors which affects the carbohydrate supply to the sink tissues like root cortical cells. Subsequently, there may be decrease in concentration of sugar in apoplast (Sonnewald et al., 1997), therefore to maintain the constant C flux, fungus adapts by inducing hexose transporter expression. Similar reports were present on glucose transporters in S. cerevisiae (Carlson, 1999), but not in mutualistic fungus. Present work will open some novel questions like how the mycorrhizal glucose transfer mechanism works in apoplast? Is photosynthetic activity of the plant determines the transcriptome of the symbiotic fungus? Is symbiotic signaling and glucose signaling is co-related? These questions needs warrant investigation.
Further, the PiHXT5 also shows a broad range of substrate specificity. The result is in agreement with previous study which showed the versatility of MST2 as a sugar transporter. It transports multiple substrates such as xylose, glucuronic, galacturonic acid, mannose, and galactose efficiently, beside glucose (Helber et al., 2011). Similarly, the sugar transporter from G. pyriformis have shown that mannose was more efficiently transported than glucose (Schubler et al., 2006). This result suggests like MST2, PiHXT5 might transport not only glucose but also cell wall constituting monosaccharides. Previous cytological and biochemical studies have shown the presence of non-assembled primary plant cell wall components, such as β-1,4-glucans, xyloglucans, non-sterified polygalacturonans, and arabinogalactan proteins at the plant–fungal interface in AM symbiosis (Perotto et al., 1995; Gianinazzi-Pearson, 1996). This implies that during C starvation, the fungus can feed on “amorphous cell wall components” mainly glucose, mannose, galactose, xylose, and arabinose (York et al., 1986). It has been reported that the AMF is able to feed on cell wall components (Ho and Trappe, 1973; Hodge et al., 2010). Other studies have shown systemic activation of a plant xyloglucan endotransglucosylase/hydrolyase gene (XTH) in mycorrhizal roots (Maldonado-Mendoza et al., 2005), xyloglucanase capacity of AMF (Rejon-Palomares et al., 1996) which supports the above theory. Similar observations were reported from GpMST1 from cyanobacterial symbiotic fungus G. pyriformis and MST2 from G. intraradices (Redecker and Raab, 2006; Helber et al., 2011), both of these proteins are high affinity glucose transporters with a broad substrate specificity for different monosaccharides. This adaptation of acquiring C from cell wall components suits well for biotrophic modus of life.
PiHXT5 protein is expressed in heterologous system leading to functional complementation. Since we did not get the spot assay, phenotypic complementation was not observed, similar results were obtained by Helber et al. (2011). We found that PiHXT5 is a high- affinity glucose transporter as it has Km value 2.5 mM and Vmax of 24 nmol/min/106 cells. Other high-affinity glucose transporters of fungus and plants found to have similar Km and Vmax, e.g., fungal GpMST1 has Km of ∼1.2 mM and Uromyces fabae HXT1 has Km 0.36 mM (Voegele et al., 2001) and TbHXT1 has Km 38 μM and Glomus MST2 has Km 33 μM for glucose thus support our findings. Most highly expressed sugar transporter like STP1, STP4, STP7, and STP13 from Arabidopsis thaliana (Büttner, 2010) reported to have Km 15–74 μM. The high affinity nature of PiHXT5 is consistence with data from qRT-PCR as its expression was higher at low glucose levels. Similar reports are presents showing how eukaryotic cells sense availability of glucose, and through glucose sensors (Snf3 and Rgt2) generate a signal for altering the expression of hexose transporters in S. cerevisiae (Ozcan and Johnston, 1999). Radioactive sugar uptake experiments using 14C-glucose shows that yeast cells expressing PiHXT5 were able to transport glucose. Substrate specificity experiments using 14C-glucose with five-fold molar excess of D-fructose, D-xylose, D-galactose, and D-mannose shows that PiHXT5 has highest specificity for D-glucose, although it can also transport D-fructose and D-xylose, which are components of plant cell wall. pH dependence studies of PiHXT5 shows that the maximum transport activity of radioactive glucose was found between pH 5 and 6. The glucose transport activity of PiHXT5 dependent on membrane proton gradient and also on H+-ATPases as glucose uptake was found blocked in presence of protonophore DNP and plasma membrane H+-ATPase inhibitor sodium vanadate, similar results were also found in case of MST2 from G. intraradices and GpMST1 from G. pyriformis, HXT1 from U. fabae and also in case of three hexose transporters HXT B, C, and E from A. nidulans, where glucose transport was found sensitive to protonophore CCCP (Carbonyl Cyanide 3-ChloroPhenylhydrazone), suggesting that they function as energy dependent H+/glucose symporters (Requena et al., 2003). We suggest that PiHXT5 uptake monosachharides using the H+ gradient generated by this enzyme. The present study provides functional and biochemical characterization of PiHXT5 from P. indica. Our study will open new vistas to understand the glucose transport and metabolism in this symbiotically important fungus. The characterization of first hexose transporter from this fungus will also help in understanding the nutrient exchange mechanism between plants and P. indica.
Author Contributions
MR, SR, MK, and VD have performed the experiments. MK, SR, AJ, and MD have designed the experiments. Chemicals were provided by AJ and MD. Project was supervised by AJ and MD. MS is written by AJ and MD.
Funding
AJ is thankful to Department of Science and Technology (DST)-SERB Government of India for providing grant (EMR/2015/000169). AJ and MD are thankful to Jawaharlal Nehru University for providing DST-PURSE-II and UGC-Resource NET-working grant.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
MR and SR thankful to University Grant Commission (UGC) and Council for Scientific and Industrial Research (CSIR), Government of India respectively for providing fellowship. MK and VD are thankful to Indian Council of Medical Research (ICMR) and CSIR respectively for providing fellowship. We are very thankful to Prof. Eckhard Boles, Institut für Molekulare Biowissenschaften Goethe-Universität Frankfurt Germany for providing the EBY.VW4000 strain.
Footnotes
References
Baltruschat, H., Fodor, J., Harrach, B. D., Niemczyk, E., Barna, B., Gullner, G., et al. (2008). Salt tolerance of barley induced by the root endophyte Piriformospora indica is associated with a strong increase in antioxidants. New Phytol. 180, 501–510. doi: 10.1111/j.1469-8137.2008.02583.x
Bécard, G., and Fortin, J. (1988). Early events of vesicular–arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol. 108, 211–218. doi: 10.1111/j.1469-8137.1988.tb03698.x
Boles, E., and Hollenberg, C. P. (1997). The molecular genetics of hexose transport in yeasts. FEMS Microbiol. Rev. 21, 85–111. doi: 10.1111/j.1574-6976.1997.tb00346.x
Bonfante, P., and Anca, I.-A. (2009). Plants, mycorrhizal fungi, and bacteria: a network of interactions. Ann. Rev. Microbiol. 63, 363–383. doi: 10.1146/annurev.micro.091208.073504
Bun-Ya, M., Nishimura, M., Harashima, S., and Oshima, Y. (1991). The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol. Cell. Biol. 11, 3229–3238. doi: 10.1128/MCB.11.6.3229
Büttner, M. (2010). The Arabidopsis sugar transporter (AtSTP) family: an update. Plant Biol. 12, 35–41. doi: 10.1111/j.1438-8677.2010.00383.x
Carlson, M. (1999). Glucose repression in yeast. Curr. Opin. Microbiol. 2, 202–207. doi: 10.1016/S1369-5274(99)80035-6
Corpet, F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890. doi: 10.1093/nar/16.22.10881
Deng, D., Xu, C., Sun, P. C., Wu, J. P., Yan, C. Y., Hu, M. X., et al. (2014). Crystal structure of the human glucose transporter GLUT1. Nature 510, 121–125. doi: 10.1038/nature13306
Fan, J., Chaturvedi, V., and Shen, S.-H. (2002). Identification and phylogenetic analysis of a glucose transporter gene family from the human pathogenic yeast Candida albicans. J. Mol. Evol. 55, 336–346. doi: 10.1007/s00239-002-2330-4
Felle, H. H., Waller, F., Molitor, A., and Kogel, K.-H. (2009). The mycorrhiza fungus Piriformospora indica induces fast root-surface pH signaling and primes systemic alkalinization of the leaf apoplast upon powdery mildew infection. Mol. Plant Microbe. Interact. 22, 1179–1185. doi: 10.1094/MPMI-22-9-1179
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Gamborg, O. L., and Phillips, G. (2013). Plant Cell, Tissue and Organ Culture: Fundamental Methods. Berlin: Springer Science & Business Media.
Gianinazzi-Pearson, V. (1996). Plant cell responses to arbuscular mycorrhizal fungi: getting to the roots of the symbiosis. Plant Cell 8, 1871–1883. doi: 10.1105/tpc.8.10.1871
Gietz, R. D., and Schiestl, R. H. (1991). Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast 7, 253–263. doi: 10.1002/yea.320070307
Harrison, M. J. (1999). Molecular and cellular aspects of the arbuscular mycorrhizal symbiosis. Ann. Rev. Plant. Physiol. Plant. Hol. Biol. 50, 361–389. doi: 10.1146/annurev.arplant.50.1.361
Harrison, M. J. (2005). Signaling in the arbuscular mycorrhizal symbiosis. Annu. Rev. Microbiol. 59, 19–42. doi: 10.1146/annurev.micro.58.030603.123749
Helber, N., Wippel, K., Sauer, N., Schaarschmidt, S., Hause, B., and Requena, N. (2011). A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp is crucial for the symbiotic relationship with plants. Plant Cell 23, 3812–3823. doi: 10.1105/tpc.111.089813
Henikoff, S., and Henikoff, J. G. (1992). Amino-Acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. U.S.A. 89, 10915–10919. doi: 10.1073/pnas.89.22.10915
Hill, T., and Kafer, E. (2001). Improved protocols for Aspergillus minimal medium: trace element and minimal medium salt stock solutions. Fungal Genet. Newslett. 48, 20–21.
Hirokawa, T., Boon-Chieng, S., and Mitaku, S. (1998). SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378–379. doi: 10.1093/bioinformatics/14.4.378
Ho, I., and Trappe, J. (1973). Translocation of 14C from Festuca plants to their endomycorrhizal fungi. Nature 244, 30–31.
Hodge, A., Helgason, T., and Fitter, A. (2010). Nutritional ecology of arbuscular mycorrhizal fungi. Fungal Ecol. 3, 267–273. doi: 10.1016/j.funeco.2010.02.002
Iancu, C. V., Zamoon, J., Woo, S. B., Aleshin, A., and Choe, J. Y. (2013). Crystal structure of a glucose/H+ symporter and its mechanism of action. Proc. Natl. Acad. Sci. U.S.A 110, 17862–17867. doi: 10.1073/pnas.1311485110
Inoue, H., Nojima, H., and Okayama, H. (1990). High-Efficiency Transformation of Escherichia coli with Plasmids. Gene 96, 23–28. doi: 10.1016/0378-1119(90)90336-P
Jogawat, A., Saha, S., Bakshi, M., Dayaman, V., Kumar, M., Dua, M., et al. (2013). Piriformospora indica rescues growth diminution of rice seedlings during high salt stress. Plant Signal. Behav. 8:e26891. doi: 10.4161/psb.26891
Karandashov, V., and Bucher, M. (2005). Symbiotic phosphate transport in arbuscular mycorrhizas. Trends Plant Sci. 10, 22–29. doi: 10.1016/j.tplants.2004.12.003
Krogh, A., Larsson, B., Von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kumar, M., Yadav, V., Tuteja, N., and Johri, A. K. (2009). Antioxidant enzyme activities in maize plants colonized with Piriformospora indica. Microbiology 155, 780–790. doi: 10.1099/mic.0.019869-0
Lalonde, S., Wipf, D., and Frommer, W. B. (2004). Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Annu. Rev. Plant. Biol. 55, 341–372. doi: 10.1146/annurev.arplant.55.031903.141758
Leggewie, G., Willmitzer, L., and Riesmeier, J. W. (1997). Two cDNAs from potato are able to complement a phosphate uptake-deficient yeast mutant: identification of phosphate transporters from higher plants. Plant Cell 9, 381–392. doi: 10.1105/tpc.9.3.381
Liu, C., Muchhal, U. S., Uthappa, M., Kononowicz, A. K., and Raghothama, K. G. (1998). Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus. Plant Physiol. 116, 91–99. doi: 10.1104/pp.116.1.91
López, M. F., Dietz, S., Grunze, N., Bloschies, J., Weiß, M., and Nehls, U. (2008). The sugar porter gene family of Laccaria bicolor: function in ectomycorrhizal symbiosis and soil-growing hyphae. New Phytol. 180, 365–378. doi: 10.1111/j.1469-8137.2008.02539.x
Lopez-Pedrosa, A., Gonzalez-Guerrero, M., Valderas, A., Azcon-Aguilar, C., and Ferrol, N. (2006). GintAMT1 encodes a functional high-affinity ammonium transporter that is expressed in the extraradical mycelium of Glomus intraradices. Fungal Genet. Biol. 43, 102–110. doi: 10.1016/j.fgb.2005.10.005
Maldonado-Mendoza, I. E., Dewbre, G. R., Blaylock, L., and Harrison, M. J. (2005). Expression of a xyloglucan endotransglucosylase/hydrolase gene, Mt-XTH1, from Medicago truncatula is induced systemically in mycorrhizal roots. Gene 345, 191–197. doi: 10.1016/j.gene.2004.10.028
Mcgonigle, T. P., Miller, M. H., Evans, D. G., Fairchild, G. L., and Swan, J. A. (1990). A new method which gives an objective-measure of colonization of roots by vesicular arbuscular mycorrhizal fungi. New Phytol. 115, 495–501. doi: 10.1111/j.1469-8137.1990.tb00476.x
Oelmüller, R., Sherameti, I., Tripathi, S., and Varma, A. (2009). Piriformospora indica, a cultivable root endophyte with multiple biotechnological applications. Symbiosis 49, 1–17. doi: 10.1007/s13199-009-0009-y
Ozcan, S., Dover, J., and Johnston, M. (1998). Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 17, 2566–2573. doi: 10.1093/emboj/17.9.2566
Ozcan, S., and Johnston, M. (1999). Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 63, 554–569.
Pao, S. S., Paulsen, I. T., and Saier, M. H. (1998). Major facilitator superfamily. Microbio. Mol. Biol. Rev. 62, 1–34.
Parniske, M. (2004). Molecular genetics of the arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 7, 414–421. doi: 10.1016/j.pbi.2004.05.011
Perotto, S., Peretto, R., Faccio, A., Schubert, A., Bonfante, P., and Varma, A. (1995). Ericoid mycorrhizal fungi: cellular and molecular bases of their interactions with the host plant. Can. J. Bot. 73, 557–568. doi: 10.1139/b95-296
Peskan-Berghofer, T., Shahollari, B., Giong, P. H., Hehl, S., Markert, C., Blanke, V., et al. (2004). Association of Piriformospora indica with Arabidopsis thaliana roots represents a novel system to study beneficial plant-microbe interactions and involves early plant protein modifications in the endoplasmic reticulum and at the plasma membrane. Physiol. Plant. 122, 465–477. doi: 10.1111/j.1399-3054.2004.00424.x
Plett, J. M., and Martin, F. (2011). Blurred boundaries: lifestyle lessons from ectomycorrhizal fungal genomes. Trends Genet. 27, 14–22. doi: 10.1016/j.tig.2010.10.005
Polidori, E., Ceccaroli, P., Saltarelli, R., Guescini, M., Menotta, M., Agostini, D., et al. (2007). Hexose uptake in the plant symbiotic ascomycete Tuber borchii Vittadini: biochemical features and expression pattern of the transporter TBHXT1. Fungal Genet. Biol. 44, 187–198. doi: 10.1016/j.fgb.2006.08.001
Redecker, D., and Raab, P. (2006). Phylogeny of the Glomeromycota (arbuscular mycorrhizal fungi): recent developments and new gene markers. Mycologia 98, 885–895. doi: 10.3852/mycologia.98.6.885
Rejon-Palomares, A., Garcia-Garrido, J., Ocampo, J., and Garcia-Romera, I. (1996). Presence of xyloglucan-hydrolyzing glucanases (xyloglucanases) in arbuscular mycorrhizal symbiosis. Symbiosis 21, 249–261.
Requena, N., Breuninger, M., Franken, P., and Ocón, A. (2003). Symbiotic status, phosphate, and sucrose regulate the expression of two plasma membrane H+-ATPase genes from the mycorrhizal fungus Glomus mosseae. Plant Physiol. 132, 1540–1549. doi: 10.1104/pp.102.019042
Riesmeier, J. W., Willmitzer, L., and Frommer, W. B. (1992). Isolation and characterization of a sucrose carrier cdna from spinach by functional expression in yeast. EMBO J. 11, 4705–4713.
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Schäfer, P., and Kogel, K.-H. (2009). “The sebacinoid fungus Piriformospora indica: an orchid mycorrhiza which may increase host plant reproduction and fitness,” in Plant Relationships. The Mycota, Vol. 5, eds H. B. Deising and K. Esser (Heidelberg: Springer), 99–112.
Schubler, A., Martin, H., Cohen, D., Fitz, M., and Wipf, D. (2006). Characterization of a carbohydrate transporter from symbiotic glomeromycotan fungi. Nature 444, 933–936. doi: 10.1038/nature05364
Shachar-Hill, Y., Pfeffer, P. E., Douds, D., Osman, S. F., Doner, L. W., and Ratcliffe, R. G. (1995). Partitioning of intermediary carbon metabolism in vesicular-arbuscular mycorrhizal leek. Plant Physiol. 108, 7–15.
Sherameti, I., Venus, Y., Drzewiecki, C., Tripathi, S., Dan, V. M., Nitz, I., et al. (2008). PYK10, a beta-glucosidase located in the endoplasmatic reticulum, is crucial for the beneficial interaction between Arabidopsis thaliana and the endophytic fungus Piriformospora indica. Plant J. 54, 428–439. doi: 10.1111/j.1365-313X.2008.03424.x
Smith, S. E., Dickson, S., and Smith, F. A. (2001). Nutrient transfer in arbuscular mycorrhizas: how are fungal and plant processes integrated? Funct. Plant Biol. 28, 685–696. doi: 10.1071/PP01152
Smith, S. E., Smith, F. A., and Jakobsen, I. (2003). Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiol. 133, 16–20. doi: 10.1104/pp.103.024380
Sonnewald, U., Westergaard, N., and Schousboe, A. (1997). Glutamate transport and metabolism in astrocytes. Glia 21, 56–63. doi: 10.1002/(SICI)1098-1136(199709)21:1<56::AID-GLIA6>3.0.CO;2-#
Sun, C., Johnson, J. M., Cai, D., Sherameti, I., Oelmüller, R., and Lou, B. (2010). Piriformospora indica confers drought tolerance in Chinese cabbage leaves by stimulating antioxidant enzymes, the expression of drought-related genes and the plastid-localized CAS protein. J. Plant Physiol. 167, 1009–1017. doi: 10.1016/j.jplph.2010.02.013
Sun, L., Zeng, X., Yan, C., Sun, X., Gong, X., Rao, Y., et al. (2012). Crystal structure of a bacterial homologue of glucose transporters GLUT1-4. Nature 490, 361–366. doi: 10.1038/nature11524
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). Clustal-W - improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Trivedi, D. K., Bhatt, H., Pal, R. K., Tuteja, R., Garg, B., Johri, A. K., et al. (2013). Structure of RNA-interacting cyclophilin A-like protein from Piriformospora indica that provides salinity-stress tolerance in plants. Sci. Rep. 3:3001. doi: 10.1038/srep03001
Tusnady, G. E., and Simon, I. (2001). The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849–850. doi: 10.1093/bioinformatics/17.9.849
Vadassery, J., Ranf, S., Drzewiecki, C., Mithöfer, A., Mazars, C., Scheel, D., et al. (2009). A cell wall extract from the endophytic fungus Piriformospora indica promotes growth of Arabidopsis seedlings and induces intracellular calcium elevation in roots. Plant J. 59, 193–206. doi: 10.1111/j.1365-313X.2009.03867.x
Varma, A., Verma, S., Sahay, N., Bütehorn, B., and Franken, P. (1999). Piriformospora indica, a cultivable plant-growth-promoting root endophyte. Appl. Environ. Microbiol. 65, 2741–2744.
Verma, S., Varma, A., Rexer, K.-H., Hassel, A., Kost, G., Sarbhoy, A., et al. (1998). Piriformospora indica, gen. et sp. nov., a new root-colonizing fungus. Mycologia 60, 896–903. doi: 10.2307/3761331
Voegele, R. T., Struck, C., Hahn, M., and Mendgen, K. (2001). The role of haustoria in sugar supply during infection of broad bean by the rust fungus Uromyces fabae. Proc. Natl. Acad. Sci. U.S.A. 98, 8133–8138. doi: 10.1073/pnas.131186798
Von Heijne, G. (1994). Membrane proteins: from sequence to structure. Annu. Rev. Biophys. Biomol. Struct. 23, 167–192. doi: 10.1146/annurev.bb.23.060194.001123
Wahl, R., Wippel, K., Goos, S., Kämper, J., and Sauer, N. (2010). A novel high-affinity sucrose transporter is required for virulence of the plant pathogen Ustilago maydis. PLoS Biol. 8:e1000303. doi: 10.1371/journal.pbio.1000303
Waller, F., Achatz, B., Baltruschat, H., Fodor, J., Becker, K., Fischer, M., et al. (2005). The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc. Natl. Acad. Sci. U.S.A 102, 13386–13391. doi: 10.1073/pnas.0504423102
Weiss, M., Sykorova, Z., Garnica, S., Riess, K., Martos, F., Krause, C., et al. (2011). Sebacinales everywhere: previously overlooked ubiquitous fungal endophytes. PLoS ONE 6:e16793. doi: 10.1371/journal.pone.0016793
Wieczorke, R., Krampe, S., Weierstall, T., Freidel, K., Hollenberg, C. P., and Boles, E. (1999). Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 464, 123–128. doi: 10.1016/S0014-5793(99)01698-1
Wisedchaisri, G., Park, M. S., Iadanza, M. G., Zheng, H. J., and Gonen, T. (2014). Proton-coupled sugar transport in the prototypical major facilitator superfamily protein XylE. Nat. Commun. 5:4521. doi: 10.1038/ncomms5521
Yadav, V., Kumar, M., Deep, D. K., Kumar, H., Sharma, R., Tripathi, T., et al. (2010). A phosphate transporter from the root endophytic fungus Piriformospora indica plays a role in phosphate transport to the host plant. J. Biol. Chem. 285, 26532–26544. doi: 10.1074/jbc.M110.111021
York, W. S., Darvill, A. G., Mcneil, M., Stevenson, T. T., and Albersheim, P. (1986). Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 118, 3–40. doi: 10.1016/0076-6879(86)18062-1
Zuccaro, A., Lahrmann, U., Guldener, U., Langen, G., Pfiffi, S., Biedenkopf, D., et al. (2011). endophytic life strategies decoded by genome and transcriptome analyses of the mutualistic root symbiont Piriformospora indica. PLoS Pathog. 7:e1002290. doi: 10.1371/journal.ppat.1002290
Keywords: root-endophyte, Piriformospora indica, MFS superfamily, hexose transporter, glucose uptake
Citation: Rani M, Raj S, Dayaman V, Kumar M, Dua M and Johri AK (2016) Functional Characterization of a Hexose Transporter from Root Endophyte Piriformospora indica. Front. Microbiol. 7:1083. doi: 10.3389/fmicb.2016.01083
Received: 20 April 2016; Accepted: 28 June 2016;
Published: 22 July 2016.
Edited by:
Pankaj Kumar Arora, M. J. P. Rohilkhand University, IndiaReviewed by:
Jay Prakash Verma, Banaras Hindu University, IndiaAshutosh Kumar, Indian Institute of Technology Delhi, India
Copyright © 2016 Rani, Raj, Dayaman, Kumar, Dua and Johri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Atul K. Johri, YWtqb2hyaTE0QHlhaG9vLmNvbQ== Meenakshi Dua, bWVlbmFrc2hpNzJAaG90bWFpbC5jb20=
†These authors have contributed equally to this work.
 Mamta Rani
Mamta Rani Sumit Raj
Sumit Raj Vikram Dayaman
Vikram Dayaman Manoj Kumar
Manoj Kumar Meenakshi Dua2*
Meenakshi Dua2* Atul K. Johri
Atul K. Johri