- 1Division of Bacteriology, National Institute of Cholera and Enteric Diseases, Kolkata, India
- 2Maharishi Valmiki Infectious Diseases Hospital, Delhi, India
- 3Infectious Diseases and Immunology Division, Council of Scientific and Industrial Research-Indian Institute of Chemical Biology, Kolkata, India
- 4Collaborative Research Center of Okayama University for Infectious Diseases in India, National Institute of Cholera and Enteric Diseases, Kolkata, India
- 5Center for Human Microbial Ecology, Translational Health Science and Technology Institute, Faridabad, India
Incidence of epidemic Vibrio cholerae serogroup O139 has declined in cholera endemic countries. However, sporadic cholera caused by V. cholerae O139 with notable genetic changes is still reported from many regions. In the present study, 42 V. cholerae O139 strains isolated from 2001 to 2006 in Delhi, India, were retrospectively analyzed to understand their phenotype and molecular characteristics. The majority of isolates were resistant to ampicillin, furazolidone and nalidixic acid. Though the integrative conjugative element was detected in all the O139 isolates, the 2004–2006 isolates remained susceptible to co-trimoxazole, chloramphenicol, and streptomycin. Cholera toxin genotype 1 was present in the majority of the O139 isolates while few had type 3 or a novel type 4. In the cholera toxin encoding gene (ctx) restriction fragment length polymorphism, the majority of the isolates harbored three copies of CTX element, of which one was truncated. In this study, the ctx was detected for the first time in the small chromosome of V. cholerae O139 and one isolate harbored 5 copies of CTX element, of which 3 were truncated. The ribotype BII pattern was found in most of the O139 isolates. Three V. cholerae O139 isolated in 2001 had a new ribotype BVIII. Pulsed-field gel electrophoresis analysis revealed clonal variation in 2001 isolates compared to the 2004–2006 isolates. Molecular changes in V. cholerae O139 have to be closely monitored as this information may help in understanding the changing genetic features of this pathogen in relation to the epidemiology of cholera.
Introduction
The aquatic bacterium Vibrio cholerae is the causative agent of cholera or cholera-like diarrhea in humans. Of the 206 serogroups identified in this species (Yamai et al., 1997), the serogroups O1 and O139 are responsible for global cholera epidemics. V. cholerae serogroup O1 is further divided into two biotypes, classical and El Tor and each has two distinct serotypes, Inaba and Ogawa. The classical biotype was associated with cholera in first six pandemics (Sack et al., 2004). The current 7th cholera pandemic is represented by V. cholerae O1 El Tor biotype, which became dominant from 1961 and gradually replaced the classical biotype from the global cholera scenario. V. cholerae O139 serogroup emerged in 1992 by replacing the El Tor biotype in the Indian subcontinent and spread to more than 14 countries in the following years (Nair et al., 1994a; Siddique et al., 1996; Ramamurthy et al., 2003). Emergence of V. cholerae O139 serogroup was thought to be the beginning of the 8th cholera pandemic considering the rapid spread of the pathogen (Nair et al., 1994b). However, after causing large cholera epidemics in 1993, the serogroup O139 disappeared abruptly from the endemic scenario ensuing resurgence of V. cholerae O1 El Tor biotype in cholera endemic regions (Sharma et al., 1997). Until late 1999, there has been periodic shift between El Tor and O139 in India and Bangladesh (Basu et al., 2000; Faruque et al., 2003a). In 2008, the incidence of V. cholerae O139 in China was 32% among cholera cases (WHO, 2009) and continued until 2012 (Zhang et al., 2014).
In V. cholerae O139, changes in the antimicrobial susceptibility patterns and arrangement of genetic elements, especially the organization of ribosomal RNA operons, location, and arrangement of cholera toxin prophages (CTXΦ) were reported during its emergence on several occasions (Sharma et al., 1997; Faruque et al., 2003a; Nandi et al., 2003; Chatterjee et al., 2007; Ghosh et al., 2008). Initial genetic analysis showed that emergence of V. cholerae O139 may be due to the insertion of a novel 35-kb wbf gene that encodes O139-somatic (O) antigen in a V. cholerae serogroup O22 strain or due to the loss of a 22-kb wbe region in a V. cholerae O1 that encodes the O1 antigen (Yamasaki et al., 1999). The whole genome sequence analysis by Chun et al. (2009) confirmed the above finding, i.e., substitution of the gene cluster coding for the O139 antigen took place by horizontal gene transfer but not the deletion.
Based on the amino acid changes, the B-subunits of CT have been designated into several CT-genotypes or ctxB alleles (Safa et al., 2008; Raychoudhuri et al., 2009). CT genotyping (ctxB allele) can be made using Mismatch amplification mutation assay (MAMA) PCR (Morita et al., 2008). CT genotype 1 is reported in strains of the classical biotype worldwide and in US Gulf Coast, genotype 2 is found in El Tor biotype strains from Australia, and genotype 3 is prevalent in El Tor biotype from the 7th pandemic and the Latin American epidemic strains (Olsvik et al., 1993). V. cholerae O1 El Tor isolates that produces classical CT is a newly emerged trait, which is said to be associated with the severity of the illness (Siddique et al., 2010) with a large number of cholera outbreaks (Nair et al., 2006; Safa et al., 2008; Raychoudhuri et al., 2009). CT encoding genes of O1 and O139 serogroups is carried by a filamentous CTXΦ, which is known to use the toxin-coregulated pili (TCP) as its receptor (Waldor and Mekalanos, 1996). V. cholerae O139 harboring CTXclassΦ and CTXcalcΦ has been described based on the difference in the sequence of rstR that encodes for the repressor protein of the CTXΦ (Faruque et al., 2003a; Bhattacharya et al., 2006; Raychoudhuri et al., 2010).
This study was undertaken to understand the phenotype and genetic changes of V. cholerae O139 isolated from sporadic hospitalized cholera cases in Delhi during 2001–2006. The outcome of this study may be useful to comprehend the epidemiology of V. cholerae O139.
Materials and Methods
Bacterial Strains
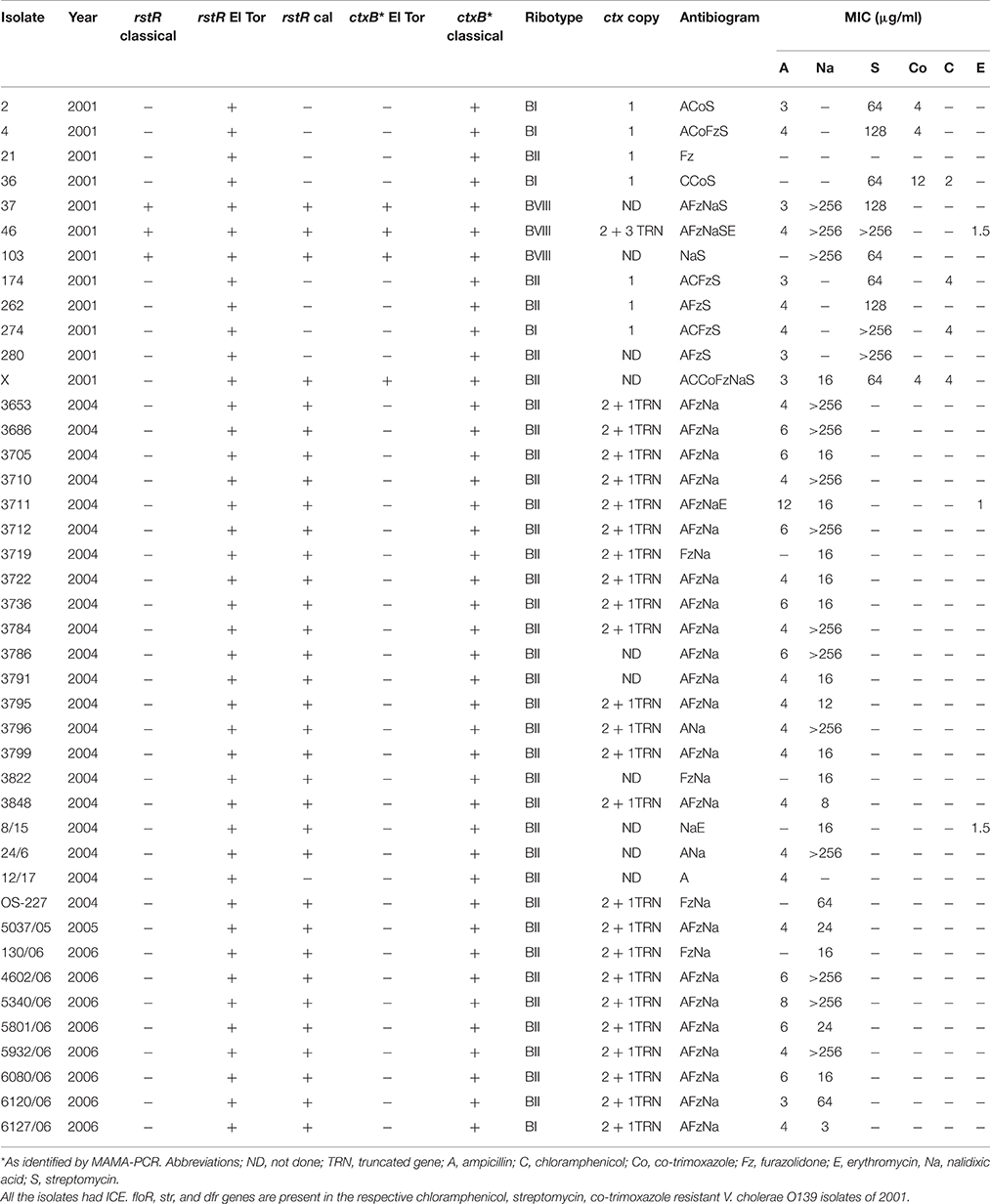
V. cholerae O139 was isolated from cholera patients admitted at the Maharishi Valmiki Infectious Diseases Hospital, Delhi. Between 2001 and 2006, 42 isolates individually isolated strains were included in this study (Table 1). V. cholerae O1 569B (classical biotype), N16961 (El Tor biotype), and SG-24 (serogroup O139) were used as reference strains. In the pulsed-field gel electrophoresis (PFGE), Salmonella enterica serotype Braenderup strain H9812 was used as the molecular size standard.
Bacteriology and Serotyping
V. cholerae isolates were grown on thiosulphate-citrate-bile salt-sucrose (TCBS) agar (Eiken, Tokyo, Japan) at 37°C for 16–18 h. Typical sucrose fermenting yellow colonies was further streaked on Luria agar (LA, Difco, Detroit, MD, USA) and subsequently used in the rapid biochemical identification (Nair et al., 1987). Presumptively identified V. cholerae isolates were further confirmed by oxidase test and confirmed serologically by slide agglutination test using O1 and O139 monoclonal antibodies prepared at the National Institute of Cholera and Enteric Diseases, Kolkata, India (Garg et al., 1994; Ramamurthy et al., 1995).
Antimicrobial Susceptibility
Antimicrobial susceptibility testing was performed using commercially available disks (Difco) following the Clinical and Laboratory Standard Institute guidelines (CLSI, 2014). The concentration of antibiotics in the disc was as follows: ampicillin (10 μg), chloramphenicol (30 μg), co-trimoxazole (sulfamethoxazole/trimethoprim, 1.25/23.45 μg), ciprofloxacin (5 μg), furazolidone (100 μg), norfloxacin (10 μg), gentamycin (10 μg), nalidixic acid (30 μg), neomycin (30 μg), streptomycin (10 μg), tetracycline (30 μg), and erythromycin (15 μg). Except for furazolidone, the minimal inhibitory concentrations (MICs) of antibiotics (ampicillin, chloramphenicol, erythromycin, nalidixic acid, streptomycin, sulfamethoxazole/trimethoprim) were determined by E-test (AB bioMérieux, Solna, Sweden).
Extraction of Chromosomal DNA
Modified method of Murray and Thompson (1980) was used for V. cholerae genomic DNA extraction.
Polymerase Chain Reaction (PCR) Assay
Multiplex PCRs were used for the detection of rfb genes encoding the somatic antigen of O139/O1, CT encoding gene (ctxA), and biotypes based on the allelic difference in the tcpA gene (Keasler and Hall, 1993; Hoshino et al., 1998). Simplex PCR assays with specific primers were made for the detection of rstR alleles (Bhattacharya et al., 2006). MAMA-PCR was made to detect the presence of ctxB alleles (CT genotypes) as described previously (Morita et al., 2008). Location of CTX prophage in chromosome II was confirmed by PCR using published methods (Maiti et al., 2006). To confirm the presence of integrative conjugative element (ICE) that carries the SXT element, two sets of primers were used in this study. Primers 10SF13 (5′-TTGTGGTGGAAA GAGGGTG-3′), SXT-13 (5′-CCAACAAAGAAC AGTTTGACTC-3′), and ORF-16 (5′-CATCTACCA CTTCATAGGCAGG-3′), YND-2 (5′-CAGCTTAAC TCACCAAGGAC-3′) were designed using conserved right and left terminal ends of the ICE, respectively. In addition, floR, str, and dfr genes encoding chloramphenicol, streptomycin, co-trimoxazole resistance was identified using published methods (Hochhut et al., 2001). In these PCRs, V. cholerae 569B, N16961, and SG-24 were used as reference strains. PCR assays were performed using an automated thermocycler (Gene Amp PCR system 9700, Applied Biosystems, Foster City, CA).
DNA Sequencing
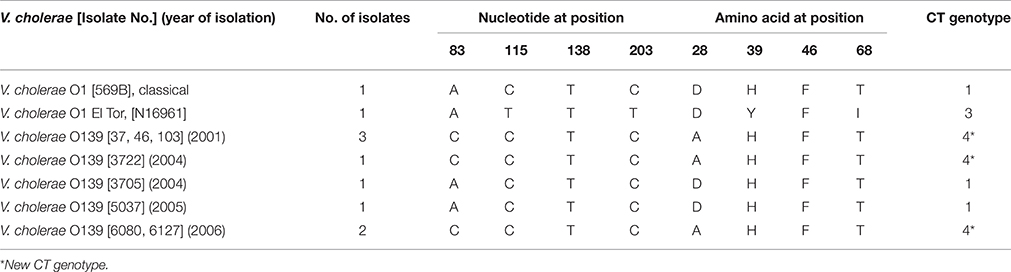
The 460 bp region of ctxB gene was amplified by PCR from eight representative isolates of V. cholerae O139 covering all the years (Olsvik et al., 1993). The amplified product was purified using a PCR purification kit (Qiagen, Hilden, Germany) and used directly as a template for nucleotide sequencing. Both the strands of DNA were sequenced with BigDye terminator cycle sequencing kit using an automated sequencer ABI 3700 (Applied Biosystems). The nucleotide and amino acid sequences were compared with the sequences available in the GenBank. The nucleotide sequence data generated with five representative isolates of V. cholerae O139 were submitted to the GenBank with accession numbers from GQ892075 to GQ892079.
Ribotyping
A 7.5-kb BamH1 (Fermentas, Waltham, MA, USA) fragment of plasmid pKK3535 containing the 16S and 23S rRNA genes of Escherichia coli was used as a rRNA probe (Brosius et al., 1981). Standard V. cholerae ribotyping was followed in this study (Faruque et al., 2000). Instead of radioisotope, we used chemiluminescent dye (Gene Images Alkaphos direct labeling and detection system, Amersham Biosciences, UK) in the DNA hybridization analysis.
ctxA RFLP
Restriction enzymes HindIII, PstI, and BglII (Fermentas) were used for the digestion of V. cholerae O139 chromosomal DNA and immobilized on nylon membranes (Amersham International). The CT encoding gene (ctxA) probe was a 540-bp XbaI-ClaI (Fermentas) fragment cloned into the plasmid pKTN901 using EcoR1 linkers (Kaper et al., 1988). The 267-bp cep probe was derived from EcoR1 (Fermentas) digested pSC01 plasmid.
Pulsed-Field Gel Electrophoresis (PFGE)
PFGE of V. cholerae O139 was performed as described previously for V. cholerae O1 (Cooper et al., 2006). PFGE profiles were analyzed using the BioNumerics version 4.0 software (Applied Maths, Sint Martens Latem, Belgium). The tagged image file formats were normalized by using the universal S. enterica serotype Braenderup (H9812) size standard on each gel against the reference in the database. In the dendrogram analysis, the PFGE profiles were matched using the Dice coefficient and unweighted pair group method using arithmetic averages (UPGMA). Clustering of PFGE profiles was made using 1.5% band position tolerance window and 1.5% optimization.
Results
Identification
Conventional serology and multiplex PCRs employed in this study confirmed all the isolates as V. cholerae O139.
Antimicrobial Susceptibility
In the antimicrobial susceptibility testing by disc diffusion assay, more than 60% of the V. cholerae O139 isolates were resistant to ampicillin, furazolidone, and nalidixic acid displaying the antibiogram as AFzNa (Table 1). The susceptibility pattern of V. cholerae O139 isolated during 2001 differed from the rest of the study period by displaying resistance to chloramphenicol, co-trimoxazole, and streptomycin. During the same year, 66% of the isolates were susceptible to nalidixic acid. However, in the subsequent years (2004–2006), all most all the isolates were resistant to ampicillin, furazolidone, and nalidixic acid (Table 1). For neomycin, 23 isolates showed reduced susceptibility and 19 remained susceptible (data not shown). The MIC values varied considerably for ampicillin (4–12 μg/ml), co-trimoxazole (4–12 μg/ml), nalidixic acid (3 to >256 μg/ml), and streptomycin (64 to >256 μg/ml). MIC for chloramphenicol (2–4 μg/ml) and erythromycin (1–1.5 μg/ml) remained low (Table 1).
Analysis of Virulence Loci, ICE and Antimicrobial Resistance Encoding Genes
The O139 isolates uniformly harbored ctxA with an El Tor allele of tcpA. In the MAMA-PCR, all the isolates were identified as CT genotype 1. In addition, four isolates (37, 46, 103, and X) collected in 2001 exhibited CT genotype 3 (Table 1). The amplified ctxB gene from eight isolates was directly sequenced. The deduced amino acid sequence analysis identified heterogeneity in the B subunit of CT. Some of the 2004 and 2005 isolates had aspartic acid (D), histidine (H), phenylalanine (F), and threonine (T) at positions 28, 39, 46, and 68, respectively in the CtxB, which is similar to the CT genotype 1 of the V. cholerae O1 classical 569B strain (Table 2). However, the isolates representing 2001, 2004, and 2006 had amino acids alanine (A), H, F, T at positions 28, 39, 46, 68, respectively, which has been classified as CT genotype 4. This genotype was described in our previous report as genotype 5 with V. cholerae O139 isolates from Bangladesh (Bhuiyan et al., 2009). Subsequently, this was corrected in our publication in 2010 (Raychoudhuri et al., 2010).
About 80% of the isolates possessed more than one allele of rstR, one being the El Tor type (rstRET) and the other with rstRcalc type. Interestingly, three 2001 isolates (37, 46, and 103) carried all the three rstR alleles, i.e., rstRCl, rstRET, and rstRCalc. These isolates belonged to a new ribotype (Table 1). ICE was present in all the V. cholerae O139 isolates as confirmed by two sets of primers. V. cholerae O139 isolated in 2001 that were resistant to chloramphenicol, streptomycin and co-trimoxazole respectively harbored floR, str, and dfr genes.
ctxA RFLP
Twenty four V. cholerae O139 isolated during 2004–2006 displayed two tandemly arranged copies of intact CTX prophages with cep, orfU, ace, zot, and ctxAB as a 23 Kb fragment (Figure 1A). These CTX prophages were closely bordered with a 5 Kb truncated prophage (without ctxAB) as detected by the cep probe (Table 1, Figure 1A). Seven V. cholerae O139 isolated in 2001 had a single copy of CTX prophage as detected by 8 Kb ctx/cep probes (Table 1, Figure 1B). One isolate harbored two entire copies of CTX prophages as detected by ctx probe along with 3 truncated phages that were detected as three 5 Kb fragments by cep probe (Table 1, Figure 1C). Mapping could not be accomplished for 10 isolates with the applied strategy in this study.
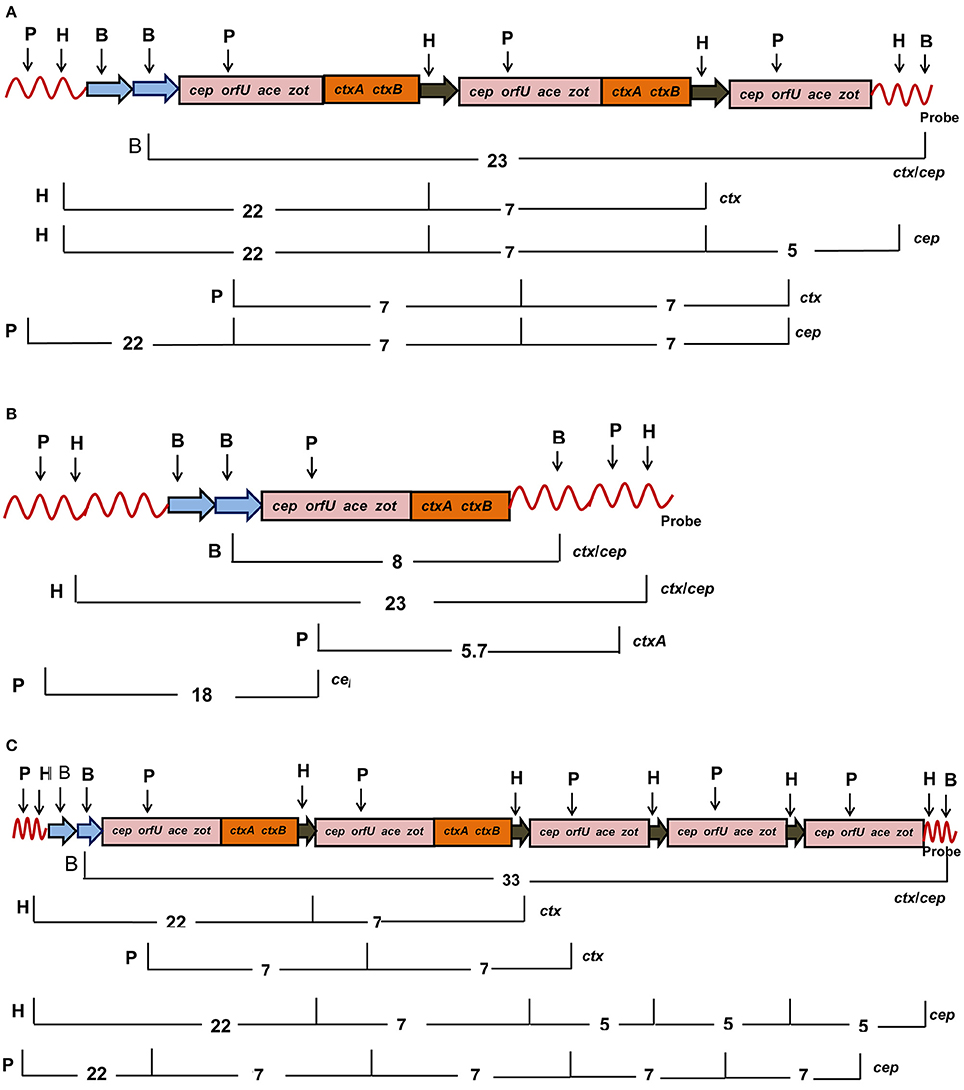
Figure 1. Mapping of CTX genetic element consisting of cep, orfU, ace, zot, and ctxAB genes in V. cholerae O139 after digestion of chromosomal DNA with HindIII (H), PstI (P), BglII (B), and probed with ctxA. (A) Eighteen V. cholerae O139 isolates had two intact copies (pink and brown) and one truncated CTX elements (pink). (B) Six V. cholerae O139 isolates harbored single intact copy of the CTX element (pink and brown). (C) The isolate 46 had two intact copies (pink and brown) and three copies of the truncated (pink) CTX elements. Restriction sites are abbreviated as B, H, and P for BglII, HindIII, and PstI, respectively. List of probes used for hybridization was given at the right hand side. The number denotes length of the DNA fragments in Kb after digestion with the marked restriction enzymes on the left hand side.
Chromosomal Location of CTX Prophages
Three of the 2001 isolates (37, 46, and 103) carried CTX prophages on both the chromosomes, which were confirmed by PCR with specific primers for chromosome I and II of V. cholerae (Maiti et al., 2006). In the rest of the V. cholerae O139 isolates, the CTX prophages remained in chromosome 1. To our knowledge, this is the first report indicating the presence of CTX prophages on chromosome II in V. cholerae O139.
Ribotyping
V. cholerae O139 isolates exhibited three different ribotypes (Table 1, Figure 2). Ribotype BII was predominant in 34 isolates, while 5 isolates exhibited BI ribotype. All the isolates of 2004–2005 exhibited ribiotype BII pattern (Table 1). Ribotype patterns of 2001 isolates had mixture of BI (with 4 isolates) and BII (with 5 isolates). Interestingly, three isolates (37, 46, and 103) identified in 2001 exhibited a new ribotype pattern (Table 1, Figure 2). These three isolates had an extra DNA band around the 2-Kb region (Figure 2). This could be the new ribotype BVIII of V. cholerae O139.
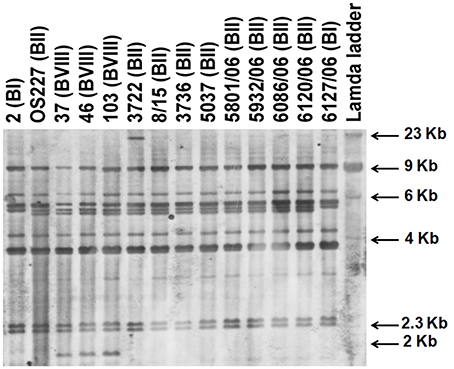
Figure 2. Ribotyping of V. cholerae O139 isolates after digestion of chromosomal DNA with BglI. Letter in parentheses against each isolate indicate the ribotype number.
Pulsed-Field Gel Electrophoresis (PFGE)
Among the 9 2001 isolates, 8 different PFGE profiles were identified demonstrating the diversity of their genomes (Figure 3, cluster A). However, 3 isolates of 2001 belongs to ribotype BVIII were closely related in the PFGE. V. cholerae O139 isolated during 2004–2006 had similar PFGE profiles (Figure 3, cluster B), but diverged from the other isolates of 2001. A consistent correlation existed in both ribotyping and PFGE methods as most of the isolates having BII ribotype pattern were placed in clusters B. In addition, the dendrogram displayed subtypes among V. cholerae O139 isolates with the BII and BVIII ribotypes at about 97% similarity level (Figure 3).
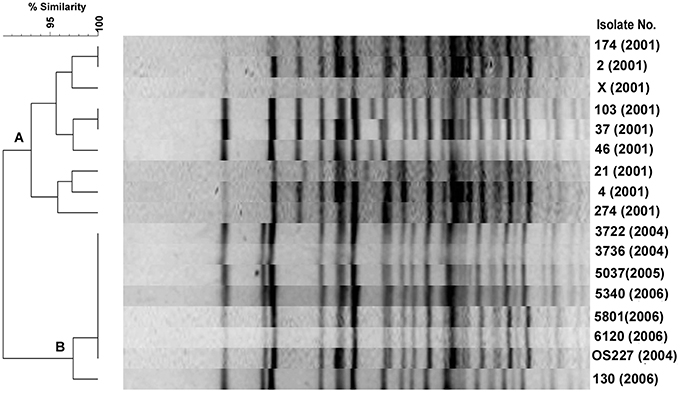
Figure 3. PFGE of V. cholerae O139 isolates after digestion of chromosomal DNA with NotI. The TIFF image of the gel was analyzed using Bionumeric software to generate the dendrogram. Percent similarity was shown at the left hand side.
Discussion
One of the phenotypic markers used in the epidemiology of cholera is the antimicrobial susceptibility patterns. In this study, V. cholerae O139 isolates were resistant to ampicillin, furazolidone, and nalidixic acid, a trend observed in majority of the V. cholerae O1 serotype Inaba isolated during 2004–2005 from different parts of India (Dutta et al., 2006). The O139 isolates identified in 1992 were resistant to chloramphenicol, co-trimoxazole, and streptomycin (Mukhopadhyay et al., 1998). The reemerged V. cholerae O139 during 1996–1997 in India and Bangladesh showed susceptibility toward co-trimoxazole (Mitra et al., 1998; Faruque et al., 2003a).
In V. cholerae O1 and O139, mobile ICE that carried antimicrobial resistance genes in the variable region expressed resistance to chloramphenicol, co-trimoxazole, and streptomycin (Hochhut et al., 2001). In this study, ICE was detected in all the O139 isolates. However, only some of the 2001 isolates were resistant to chloramphenicol, streptomycin, and co-trimoxazole and harbored floR, str, and dfr. These resistance-encoding genes were not present in other isolates in the ICE variable region. Early studies conducted during the emergence of V. cholerae O139 in India showed a trend of resistance to neomycin (Mukhopadhyay et al., 1998). In this study, the O139 isolates were either susceptible or showed reduced susceptibility to neomycin. As seen in previous reports, all the V. cholerae O139 isolates remained susceptible to norfloxacin, tetracycline, and ciprofloxacin, which are used in the treatment of cholera (Basu et al., 2000).
The CT genotype of V. cholerae O1 El Tor isolates from many countries has changed from CT genotype 3 to 1 (Safa et al., 2008; Raychoudhuri et al., 2009) and such changes were detected in strains associated with large cholera outbreaks in India and Bangladesh (Kumar et al., 2009; Nguyen et al., 2009; Taneja et al., 2009). CT genotype 4 has closest homology to CT genotype 1 with a difference of only single nucleotide (nucleotide cytosine instead of adenine) at position 83 (Raychoudhuri et al., 2010). Overall, our finding matches with the observation made in V. cholerae O139 isolated during 1998, 2000, and 2002 from Bangladesh and Kolkata, respectively (Bhuiyan et al., 2009; Raychoudhuri et al., 2010). Compared to El Tor, the hybrid isolates with CT genotype 4 have caused larger cholera outbreaks with more severe clinical symptoms (Kumar et al., 2009; Nguyen et al., 2009; Taneja et al., 2009; Siddique et al., 2010).
Epidemiologically, the CTXΦ appear to be very important as they show the genetic changes among V. cholerae O1/O139 that emerged during different periods (Faruque et al., 2000; Qu et al., 2003). In the ctxA RFLP analysis, three prophages were encountered in different years. The unusual genetic features of the three 2001 isolates of V. cholerae O139 includes identification of the new ribotype BVIII pattern, the presence of three rstR allele types, CTX prophages of the classical type, and integration of CTX prophage in both the chromosomes. Epidemiologically, the new ribotypes of V. cholerae O1/O139 has been identified along with changes in the CTX prophage or rstR allele (Faruque et al., 1997). Considering several genetic events in the past, it has been inferred that the V. cholerae O139 may have multiple origins with different progenitors (Faruque et al., 2003b; Garg et al., 2003; Qu et al., 2003).
Genesis of V. cholerae O1 El Tor from the classical biotype, the emergence of the serogroup O139, and existence of El Tor that produces classical CT suggests that the V. cholerae is in a continuous state of adaptability, resulting in generation of new serogroups and/or new variants of the same serogroup. Our results suggest that the genome of V. cholerae O139 is dynamic and has undergone several changes since its emergence in 1992. Continuous surveillance and proper monitoring of V. cholerae O139 are however needed to detect subtle genetic changes in the genomes and its implications in its epidemiology, pathogenesis and persistence. Future studies should focus on epigenetic studies to find answers to the question as to why the O139 serogroup has disappeared from cholera endemic regions despite several genetic changes.
Author Contributions
RG, NS, KH, GC, and GP isolated and identified the pathogens, performed phenotypic characterization and all the genetic analysis. RB, AM, SS, GN, and TR conceived the idea analyzed the data and wrote the manuscript. All authors were involved in the compilation of the report and approved the final version.
Funding
This work supported in part by the Japan Agency for Medical Research and Development, Ministry of Education, Culture, Sports, Science and Technology, Japan, Council of Scientific and Industrial Research, and Indian Council of Medical Research, New Delhi, India.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Basu, A., Garg, P., Datta, S., Chakraborty, S., Bhattacharya, T., Khan, A., et al. (2000). Vibrio cholerae O139 in Calcutta, 1992-1998: incidence, antibiograms, and genotypes. Emerg. Infect. Dis. 6, 139–147. doi: 10.3201/eid0602.000206
Bhattacharya, T., Chatterjee, S., Maiti, D., Bhadra, R. K., Takeda, Y., Nair, G. B., et al. (2006). Molecular analysis of the rstR and orfU genes of the CTX prophages integrated in the small chromosomes of environmental Vibrio cholerae non-O1, non-O139 strains. Environ. Microbiol. 8, 526–634. doi: 10.1111/j.1462-2920.2005.00932.x
Bhuiyan, N. A., Nusrin, S., Alam, M., Morita, M., Watanabe, H., Ramamurthy, T., et al. (2009). Changing genotypes of cholera toxin (CT) of Vibrio cholerae O139 in Bangladesh and description of three new CT genotypes. FEMS Immunol. Med. Microbiol. 57, 136–141. doi: 10.1111/j.1574-695X.2009.00590.x
Brosius, J., Ullrich, A., Raker, M. A., Gray, A., Dull, T. J., Gutell, R. R., et al. (1981). Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid 6, 112–118. doi: 10.1016/0147-619X(81)90058-5
Chatterjee, S., Ghosh, K., Raychoudhuri, A., Pan, A., Bhattacharya, M. K., Mukhopadhyay, A. K., et al. (2007). Phenotypic and genotypic traits and epidemiological implication of Vibrio cholerae O1 and O139 strains in India during 2003. J. Med. Microbiol. 56, 824–832. doi: 10.1099/jmm.0.46982-0
Chun, J., Grim, C. J., Hasan, N. A., Lee, J. H., Choi, S. Y., Haley, B. J., et al. (2009). Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 106, 15442–15447. doi: 10.1073/pnas.0907787106
CLSI (2014). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fourth Informational Supplement. CLSI Document M100-S24. Wayne, PA: Clinical and Laboratory Standards Institute.
Cooper, K. L., Luey, C. K., Bird, M., Terajima, J., Nair, G. B., Kam, K. M., et al. (2006). Development and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping of Vibrio cholerae. Foodborne Pathog. Dis. 3, 51–58. doi: 10.1089/fpd.2006.3.51
Dutta, B., Ghosh, R., Sharma, N. C., Pazhani, G. P., Taneja, N., Raychowdhuri, A., et al. (2006). Spread of cholera with newer clones of Vibrio cholerae O1 El Tor, serotype Inaba, in India. J. Clin. Microbiol. 44, 3391–3393. doi: 10.1128/JCM.00632-06
Faruque, S. M., Ahmed, K. M., Siddique, A. K., Zaman, K., Alim, A. R., and Albert, M. J. (1997). Molecular analysis of toxigenic Vibrio cholerae O139 Bengal strains isolated in Bangladesh between 1993 and 1996: evidence for emergence of a new clone of the Bengal vibrios. J. Clin. Microbiol. 35, 2299–2306.
Faruque, S. M., Chowdhury, N., Kamruzzaman, M., Ahmad, Q. S., Faruque, A. S., Salam, M. A., et al. (2003a). Reemergence of epidemic Vibrio cholerae O139, Bangladesh. Emerg. Infect. Dis. 9, 1116–1122. doi: 10.3201/eid0909.020443
Faruque, S. M., Sack, D. A., Sack, R. B., Colwell, R. R., Takeda, Y., and Nair, G. B. (2003b). Emergence and evolution of Vibrio cholerae O139. Proc. Natl. Acad. Sci. U.S.A. 100, 1304–1309. doi: 10.1073/pnas.0337468100
Faruque, S. M., Saha, M. N., Asadulghani, Bag, P. K., Bhadra, R. K., Bhattacharya, S. K., et al. (2000). Genomic diversity among Vibrio cholerae O139 strains isolated in Bangladesh and India between 1992 and 1998. FEMS Microbiol. Lett. 184, 279–284. doi: 10.1111/j.1574-6968.2000.tb09027.x
Garg, P., Aydanian, A., Smith, D. J., Glenn, M. Jr., Nair, G. B., and Stine, O. C. (2003). Molecular epidemiology of O139 Vibrio cholerae: mutation, lateral gene transfer, and founder flush. Emer. Infect. Dis. 9, 810–814. doi: 10.3201/eid0907.020760
Garg, S., Ramamurthy, T., Mukhopadhyay, A. K., Deb, B. C., Nair, G. B., Shimada, T., et al. (1994). Production and cross-reactivity patterns of a panel of high affinity monoclonal antibodies to Vibrio cholerae O139 Bengal. FEMS Immunol. Med. Microbiol. 8, 293–298. doi: 10.1111/j.1574-695X.1994.tb00455.x
Ghosh, R., Nair, G. B., Tang, L., Morris, J. G., Sharma, N. C., Ballal, M., et al. (2008). Epidemiological study of Vibrio cholerae using variable number of tandem repeats. FEMS Microbiol. Lett. 288, 196–201. doi: 10.1111/j.1574-6968.2008.01352.x
Hochhut, B., Lotfi, Y., Mazel, D., Faruque, S. M., Woodgate, R., and Waldor, M. K. (2001). Molecular analysis of the antibiotic resistance gene clusters in the Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45, 2991–3000. doi: 10.1128/AAC.45.11.2991-3000.2001
Hoshino, K., Yamasaki, S., Mukhopadhyay, A. K., Chakraborty, S., Basu, A., Bhattacharya, S. K., et al. (1998). Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol. Med. Microbiol. 20, 201–207. doi: 10.1111/j.1574-695X.1998.tb01128.x
Kaper, J. B., Morris, J. G. Jr., and Nishibuchi, M. (1988). “DNA probes for pathogenic Vibrio species,” in DNA Probes for Infectious Diseases, ed F. C. Tenover (Boca Raton, FL: CRC Press), 65–77.
Keasler, S. P., and Hall, R. H. (1993). Detecting and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet 341, 1661. doi: 10.1016/0140-6736(93)90792-F
Kumar, P., Jain, M., Goel, A. K., Bhadauria, S., Sharma, S. K., Kamboj, D. V., et al. (2009). A large cholera outbreak due to a new cholera toxin variant of the Vibrio cholerae O1 El Tor biotype in Orissa, Eastern India. J. Med. Microbiol. 58, 234–238. doi: 10.1099/jmm.0.002089-0
Maiti, D., Das, B., Saha, A., Nandy, R. K., Nair, G. B., and Bhadra, R. K. (2006). Genetic organization of pre-CTX and CTX prophages in the genome of an environmental Vibrio cholerae non-O1, non-O139 strain. Microbiology 152, 3633–3641. doi: 10.1099/mic.0.2006/000117-0
Mitra, R., Basu, A., Dutta, D., Nair, G. B., and Takeda, Y. (1998). Resurgence of Vibrio cholerae O139 Bengal with altered antibiogram in Calcutta, India. Lancet 348, 1181. doi: 10.1016/S0140-6736(05)65326-3
Morita, M., Ohnishi, M., Arakawa, E., Bhuiyan, N. A., Nusrin, S., Alam, M., et al. (2008). Development and validation of a mismatch amplification mutation PCR assay to monitor the dissemination of an emerging variant of Vibrio cholerae O1 biotype El Tor. Microbiol. Immunol. 52, 314–317. doi: 10.1111/j.1348-0421.2008.00041.x
Mukhopadhyay, A. K., Basu, A., Garg, P., Bag, P. K., Ghosh, A., Bhattacharya, S. K., et al. (1998). Molecular epidemiology of reemergent Vibrio cholerae O139 Bengal in India. J. Clin. Microbiol. 36, 2149–2152.
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. doi: 10.1093/nar/8.19.4321
Nair, G. B., Bhattacharya, S. K., and Deb, B. C. (1994b). Vibrio cholerae O139 Bengal: the eighth pandemic strain of cholera. Indian J. Public Health 38, 33–36.
Nair, G. B., Misra, S., Bhadra, R. K., and Pal, S. C. (1987). Evaluation of the multitest medium for rapid presumptive identification of Vibrio cholerae from environmental sources. Appl. Environ. Microbiol. 53, 1203–1205.
Nair, G. B., Qadri, F., Holmgren, J., Svennerholm, A. M., Safa, A., Bhuiyan, N. A., et al. (2006). Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J. Clin. Microbiol. 44, 4211–4213. doi: 10.1128/JCM.01304-06
Nair, G. B., Ramamurthy, T., Bhattacharya, S. K., Mukhopadhyay, A. K., Garg, S., Bhattacharya, M. K., et al. (1994a). Spread of Vibrio cholerae O139 Bengal in India. J. Infect. Dis. 169, 1029–1034. doi: 10.1093/infdis/169.5.1029
Nandi, S., Maiti, D., Saha, A., and Bhadra, R. K. (2003). Genesis of variants of Vibrio cholerae O1 biotype El tor: role of the CTXÐď array and its position in the genome. Microbiology 149, 89–97. doi: 10.1099/mic.0.25599-0
Nguyen, B. M., Lee, J. H., Cuong, N. T., Cho, S. Y., Hien, N. T., Anh, D. D., et al. (2009). Cholera outbreaks caused by an altered Vibrio cholerae O1 El Tor biotype strain producing classical cholera toxin B in Vietnam in 2007 to 2008. J. Clin. Microbiol. 47, 1568–1571. doi: 10.1128/JCM.02040-08
Olsvik, O., Wahlberg, J., Petterson, B., Uhlén, M., Popovic, T., Wachsmuth, I. K., et al. (1993). Use of automated sequencing of polymerase chain reaction-generated amplicons to identify three types of cholera toxin subunit B in Vibrio cholerae O1 strains. J. Clin. Microbiol. 31, 22–25.
Qu, M., Xu, J., Ding, Y., Wang, R., Liu, P., Kan, B., et al. (2003). Molecular epidemiology of Vibrio cholerae O139 in China: polymorphism of ribotypes and CTX elements. J. Clin. Microbiol. 41, 2306–2310. doi: 10.1128/JCM.41.6.2306-2310.2003
Ramamurthy, T., Garg, S., and Nair, G. B. (1995). Monoclonal antibodies against Ogawa-specific and Ogawa-Inaba common antigenic determinants of Vibrio cholerae & their diagnostic utility. Indian J. Med. Res. 101, 10–12.
Ramamurthy, T., Yamasaki, S., Takeda, Y., and Nair, G. B. (2003). Vibrio cholerae O139 Bengal: odyssey of a fortuitous variant. Microbes Infect. 5, 329–344. doi: 10.1016/S1286-4579(03)00035-2
Raychoudhuri, A., Mukherjee, P., Ramamurthy, T., Nandy, R. K., Takeda, Y., Nair, G. B., et al. (2010). rstR alleles of Vibrio cholerae O139 strains isolated from Kolkata over a decade. FEMS Microbiol. Lett. 303, 107–115. doi: 10.1111/j.1574-6968.2009.01856.x
Raychoudhuri, A., Patra, T., Ghosh, K., Ramamurthy, T., Nandy, R. K., Takeda, Y., et al. (2009). Classical ctxB in Vibrio cholerae O1, Kolkata, India. Emerg. Infect. Dis. 15, 131–132. doi: 10.3201/eid1501.080543
Sack, D. A., Sack, R. B., Nair, G. B., and Siddique, A. K. (2004). Cholera. Lancet 363, 223–233. doi: 10.1016/S0140-6736(03)15328-7
Safa, A., Sultana, J., Dac Cam, P., Mwansa, J. C., and Kong, R. Y. (2008). Vibrio cholerae O1 hybrid El Tor strains, Asia and Africa. Emerg. Infect. Dis. 14, 987–988. doi: 10.3201/eid1406.080129
Sharma, C., Maiti, S., Mukhopadhyay, A. K., Basu, A., Basu, I., Nair, G. B., et al. (1997). Unique organization of the CTX genetic element in Vibrio cholerae O139 strains which reemerged in Calcutta, India, in September 1996. J. Clin. Microbiol. 35, 3348–3350.
Siddique, A. K., Akram, K., Zaman, K., Mutsuddy, P., Eusof, A., and Sack, R. B. (1996). Vibrio cholerae O139: how great is the threat of a pandemic? Trop. Med. Int. Health 1, 393–398. doi: 10.1046/j.1365-3156.1996.d01-54.x
Siddique, A. K., Nair, G. B., Alam, M., Sack, D. A., Huq, A., Nizam, A., et al. (2010). El Tor cholera with severe disease: a new threat to Asia and beyond. Epidemiol. Infect. 138, 347–352. doi: 10.1017/S0950268809990550
Taneja, N., Mishra, A., Sangar, G., Singh, G., and Sharma, M. (2009). Outbreaks caused by new variants of Vibrio cholerae O1 El Tor, India. Emerg. Infect. Dis. 15, 352–354. doi: 10.3201/eid1502.080943
Waldor, M. K., and Mekalanos, J. J. (1996). Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272, 1910–1914. doi: 10.1126/science.272.5270.1910
Yamai, S., Okitsu, T., Shimada, T., and Katsube, Y. (1997). Distribution of serogroups of Vibrio cholerae non-O1 non-O139 with specific reference to their ability to produce cholera toxin, and addition of novel serogroups. Kansenshogaku Zasshi 71, 1037–1045. doi: 10.11150/kansenshogakuzasshi1970.71.1037
Yamasaki, S., Shimizu, T., Hoshino, K., Ho, S. T., Shimada, T., Nair, G. B., et al. (1999). The genes responsible for O-antigen synthesis of Vibrio cholerae O139 are closely related to those of Vibrio cholerae O22. Gene 237, 321–332. doi: 10.1016/S0378-1119(99)00344-3
Keywords: V. cholerae O139, ribotypes, CT genotype, CTX prophage, PFGE
Citation: Ghosh R, Sharma NC, Halder K, Bhadra RK, Chowdhury G, Pazhani GP, Shinoda S, Mukhopadhyay AK, Nair GB and Ramamurthy T (2016) Phenotypic and Genetic Heterogeneity in Vibrio cholerae O139 Isolated from Cholera Cases in Delhi, India during 2001–2006. Front. Microbiol. 7:1250. doi: 10.3389/fmicb.2016.01250
Received: 01 October 2015; Accepted: 27 July 2016;
Published: 09 August 2016.
Edited by:
Andres M. Perez, University of Minnesota, USAReviewed by:
Paras Jain, Albert Einstein College of Medicine, USADaniela Ceccarelli, Wageningen University and Research Centre, Netherlands
Christopher John Grim, United States Food and Drug Administration, USA
Copyright © 2016 Ghosh, Sharma, Halder, Bhadra, Chowdhury, Pazhani, Shinoda, Mukhopadhyay, Nair and Ramamurthy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thadavarayan Ramamurthy, dHJhbXVAdGhzdGkucmVzLmlu
 Raikamal Ghosh
Raikamal Ghosh Naresh C. Sharma2
Naresh C. Sharma2 Rupak K. Bhadra
Rupak K. Bhadra Goutam Chowdhury
Goutam Chowdhury Gururaja P. Pazhani
Gururaja P. Pazhani Thadavarayan Ramamurthy
Thadavarayan Ramamurthy