- 1Centre of Biological Engineering, University of Minho, Braga, Portugal
- 2Laboratory of Microbiology, Wageningen University, Wageningen, Netherlands
An anaerobic thermophilic strain (strain PCO) was isolated from a syngas-converting enrichment culture. Syngas components cannot be used by strain PCO, but the new strain is very tolerant to carbon monoxide (pCO = 1.7 × 105 Pa, 100% CO). 16S rRNA gene analysis and DNA-DNA hybridization revealed that strain PCO is a strain of Thermoanaerobacter thermohydrosulfuricus. The physiology of strain PCO and other Thermoanaerobacter species was compared, focusing on their tolerance to carbon monoxide. T. thermohydrosulfuricus, T. brockii subsp. finnii, T. pseudethanolicus, and T. wiegelii were exposed to increased CO concentrations in the headspace, while growth, glucose consumption and product formation were monitored. Remarkably, glucose conversion rates by Thermoanaerobacter species were not affected by CO. All the tested strains fermented glucose to mainly lactate, ethanol, acetate, and hydrogen, but final product concentrations differed. In the presence of CO, ethanol production was generally less affected, but H2 production decreased with increasing CO partial pressure. This study highlights the CO resistance of Thermoanaerobacter species.
Introduction
Thermophiles thrive at austere and unusual conditions and their evolutionary significance and biotechnological potential have triggered microbiological research over the last decades (Turner et al., 2007; Wagner and Wiegel, 2008; Yoneda et al., 2015). There is a continuous biotechnological interest in highly thermostable enzymes, which make this kind of organisms very attractive. Thermophilic bacteria of the class Clostridia, such as members of the genera Clostridium, Thermoanaerobacter, Thermoanaerobacterium, and Caldicellulosiruptor, are currently used as biocatalysts for the production of biofuels or other chemicals of interest (Hemme et al., 2010; Carere et al., 2012). Specifically members of the Thermoanaerobacter genus are utilized to produce ethanol and hydrogen (H2) from a variety of saccharides (Jessen and Orlygsson, 2012). Within thermophiles, an organism from Thermoanaerobacter genus—T. ethanolicus—is one of the most well-studied ethanol-producing bacteria (Wiegel and Ljungdahl, 1981; Lacis and Lawford, 1991). A less common substrate, carbon monoxide (CO), is used by T. thermohydrosulfuricus subsp. carboxydovorans and T. kivui. T. thermohydrosulfuricus subsp. carboxydovorans can grow with CO as sole electron donor (25% in the headspace), producing H2 and CO2 (Balk et al., 2009). T. thermohydrosulfuricus shares 99% similarity of the 16S rRNA gene sequence and over 70% DNA-DNA hybridization with T. thermohydrosulfuricus subsp. carboxydovorans, but only the latter one can use CO. Growth of the homoacetogenic T. kivui with CO diluted with CO2/N2 or CO2/H2 was described by Kevbrina et al. (1996). Recently, Weghoff and Müller (2016) reported the ability of T. kivui to grow on only CO (100% in the headspace), producing acetate and hydrogen. Carboxydotrophic metabolism in Thermoanaerobacter species is normally not assessed, and it is not known if they can endure CO or even adapt to grow on CO, as recently reported for T. kivui (Weghoff and Müller, 2016). In this work we isolated Thermoanaerobacter thermohydrosulfuricus strain PCO from a thermophilic syngas-converting enrichment, but this strain appears unable to oxidize CO. The main objectives of this work were (1) to characterize and determine the CO tolerance of Thermoanaerobacter thermohydrosulfuricus strain PCO, and (2) to compare the effect of CO on growth, glucose consumption and product formation of strain PCO and of four close relative species from the Thermoanaerobacter genus.
Materials and Methods
Enrichments and Isolation
Suspended sludge from a thermophilic anaerobic municipal solid waste digester (Barcelona, Spain) was used as inoculum for starting up syngas-converting enrichments. Microbial cultures were enriched with synthetic syngas (mixture of 60% CO, 10% CO2, and 30% H2, total pressure 1.7 × 105 Pa) as sole carbon and energy source (Alves et al., 2013). Isolation of strain PCO was done using soft agar (1.5%, w/v) incubations and liquid medium serial dilutions, with 20 mM pyruvate as sole substrate. Sodium pyruvate was added to the medium from a 1M filter-sterilized stock solution. A phosphate-buffered mineral medium was used, containing (per liter): Na2HPO4, 1.63 g; NaH2PO4, 1.02 g; resazurin, 0.5 g; NH4Cl, 0.3 g; CaCl2·2H2O, 0.11 g; MgCl2·6H2O, 0.10 g; NaCl, 0.3 g; 1 mL of acid and alkaline trace element stock each, and 0.2 ml of vitamin stock. Trace elements and vitamins were prepared as described previously (Stams et al., 1993). Before inoculation, medium was reduced with sodium sulfide (0.8 mM final concentration). Bottles were incubated in the dark at 55°C while shaken at 100 rpm (liquid cultures) or standing (soft-agar cultures). Colonies were picked from soft-agar incubations, inoculated in fresh liquid medium containing pyruvate (20 mM). Cultures were further purified by subsequent serial dilutions alternating with soft-agar colony picking. Purity of the culture was checked by microscopic examination after growth with different substrates (Olympus CX41, Tokyo, Japan). Direct sequencing of the 16S rRNA gene and denaturing gradient gel electrophoresis (DGGE) were also applied to check the genetic purity of the culture.
DNA Isolation, PCR and DGGE
Genomic DNA from strain PCO was extracted using the FastDNA SPIN kit for soil (MP Biomedicals, Solon, OH), according to the manufacturer's instructions. The 16S rRNA gene was directly amplified from genomic DNA by PCR, using the primer set 027F/1492R (Nübel et al., 1996) and the following PCR program: pre-denaturation, 2 min at 95°C; 30 cycles of denaturation, 30 s at 95°C, annealing, 40 s at 52°C, and elongation, 90 s at 72°C; and post-elongation, 5 min, at 72°C. For DGGE analysis, the 16S rRNA gene was partially amplified from genomic DNA with primer set U968GC-f/L1401-r (Lane, 1991; Muyzer et al., 1993). The thermocycling program used for PCR-DGGE amplification was: pre-denaturation, 5 min at 95°C; 35 cycles of denaturation, 30 s at 95°C, annealing, 40 s at 56°C, and elongation, 90 s at 72°C; and post-elongation, 5 min at 72°C. DGGE was performed using a DCode system (Bio-Rad, Hercules, CA). Gels contained 8% (wt/vol) polyacrylamide (37.5:1 acrylamide/bis-acrylamide) and a linear denaturing gradient of 30–60%, with 100% of denaturant corresponding to 7 M urea and 40% (vol/vol) formamide. Electrophoresis was performed for 16 h at 85 V and 60°C in a 0.5x Tris-Acetate–EDTA buffer. DGGE gels were stained with silver nitrate (Sanguinetti et al., 1994).
Sequencing and Phylogenetic Analysis
PCR products obtained from 16S rRNA gene amplification were purified using the PCR Clean Up kit NucleoSpin Extract II (Macherey-Nagel, Düren, Germany) and sequenced directly at Eurofins MWG Operon (Ebersberg, Germany). Partial sequences were assembled using the alignment editor BioEdit v7.0.9 software package (Hall, 1999). Similarity searches for the 16S rRNA gene sequence derived from strain PCO were performed using the NCBI BLAST search program within the GenBank database (Altschul et al., 1990). The 16S rRNA gene sequence of Thermoanaerobacter strain PCO is available in the DDBL/EMBL/GenBank databases under the accession number HF586422.
Characterization of Strain PCO and Cultivation of Thermoanaerobacter Strains
Unless otherwise stated, all the physiological tests of strain PCO and its close relatives (T. thermohydrosulfuricus, T. brockii subsp. finnii, T. pseudethanolicus, and T. wiegelii) were performed using a bicarbonate-buffered mineral salt medium (Stams et al., 1993). Type strains of Thermoanaerobacter thermohydrosulfuricus (DSM 527T), T. brockii subsp. finnii (DSM 3389T), T. pseudethanolicus (DSM 2355T), and T. wiegelii (DSM 10319T) were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ; German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany). Growth of strain PCO was tested with the following substrates (at a concentration of 20 mM unless indicated otherwise): acetate, arabinose, cellobiose, cellulose (5 g L−1), ethanol, formate, fructose, galactose, glucose, glycerol, glycine, lactate, lactose, maltose, mannitol, mannose, methanol, pectin (5 g L−1), propionate, pyruvate, raffinose, ribose, sorbitol, starch (5 g L−1), sucrose, trehalose, xylan (5 g L−1), xylose, yeast extract (5 g L−1), CO (from 20 to 100% CO, 1.7 × 105 Pa), and H2/CO2 (80/20%, 1.7 × 105 Pa). Utilization of different electron acceptors (elemental sulfur, AQDS, sulfate, sulfite, thiosulfate, nitrate, and nitrite) by strain PCO was done using pyruvate (20 mM) as electron donor, while glucose (20 mM) was for tests with T. thermohydrosulfuricus, T. brockii subsp. finnii, T. pseudethanolicus, and T. wiegelii. Pyruvate (20 mM) was used to test the optimum growth temperature (range 20–85°C) and pH (range 5.7–8.0) of strain PCO. All the assays were done in duplicate. Cell growth was determined by measuring optical density at 600 nm with a spectrophotometer (U-1500 Hitachi, Tokyo, Japan). Cell morphology of strain PCO was examined by phase contrast microscopy (Leica DM 2000, Wetzlar, Germany). Cells from active cultures of strain PCO were stained using standard Gram staining techniques. The DNA–DNA hybridization analysis and the G+C content of the DNA were determined by the identification service of the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ; German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany).
Carbon Monoxide Tolerance Tests
Strain PCO, Thermoanaerobacter thermohydrosulfuricus, T. brockii subsp. finnii, T. pseudethanolicus, and T. wiegelii were tested for CO tolerance. All cultures were incubated with 0, 50, and 100% CO (pCO/P, where “pCO” is the CO partial pressure and “P” the total gas pressure). Additionally, strain PCO was incubated with 25 and 75% CO. Initial total pressure was 1.7 × 105 Pa in all the assays; N2 was used to pressurize the headspace for CO percentages lower than 100%. The tests were performed using an anaerobic phosphate-buffered mineral salt medium and glucose (20 mM) was used as carbon and energy source. Bottles were incubated in the dark, at 55°C and shaken at 100 rpm. All the assays were done in duplicate. Growth of the strains was determined by measuring optical density increase at 600 nm with a spectrophotometer (U-1500 Hitachi, Tokyo, Japan). The statistical significance of the differences detected in glucose conversion rates and end products production was evaluated using single factor analysis of variances (ANOVA).
Analytical Methods
Soluble substrates and intermediates (sugars, organic acids, and alcohols) were measured using a HPLC Thermo Electron equipment with a Shodex SH1821 column and equipped with a RI detector. The mobile phase used was sulfuric acid (0.01 N) at a flow rate of 0.6 mL min−1. Column temperature was set at 60°C. Inorganic anions were analyzed by chromatography using a HPLC Dionex system, equipped with an Ionpac AS22 column, and ED40 electrochemical detector. Column temperature and pressure varied between 35–40°C and 130 × 105–160 × 105 Pa. Gaseous compounds (CO, CO2, H2) were analyzed by gas chromatography on a GC-2014 Shimadzu with a thermal conductivity detector. CO2 was analyzed with a CP Poraplot Q column (25 m length, 0.53 mm internal diameter; film thickness, 20 μm). Helium was used as carrier gas at a flow rate of 15 mL min−1, and the temperatures in the injector, column, and detector were 60, 33, and 130°C. CO and H2 were analyzed with a Molsieve 13X column (2 m length, 3 mm internal diameter). Argon was used as carrier gas at a flow rate of 50 mL min−1, and temperatures in the injector, column, and detector were 80, 100, and 130°C.
Results and Discussion
Physiological Characterization of Strain PCO and Comparison with Closely Related Thermoanaerobacter Species
Strain PCO was isolated from the thermophilic syngas-converting enrichment described by Alves et al. (2013). Isolation was performed using pyruvate as sole carbon and energy source. Strain PCO has a G+C content of the DNA of 34.5 mol % and shares 98% identity with the 16S rRNA gene of Thermoanaerobacter thermohydrosulfuricus (the 16S rRNA gene sequence of strain PCO is available in the DDBL/EMBL/GenBank databases under the accession number HF586422). DNA-DNA hybridization between the two strains was 100%, confirming that strain PCO is a strain of Thermoanaerobacter thermohydrosulfuricus. Strain PCO was not a predominant microorganism in the syngas enriched culture, as shown by the molecular characterization (Alves et al., 2013). However, its presence indicates that it can grow on metabolic byproducts and/or dead cells at high CO concentrations. Strain PCO formed terminal round endospores, which is a characteristic of Thermoanaerobacter species (Wiegel and Ljungdahl, 1981; Lee et al., 1993, 2007; Kim et al., 2001; Balk et al., 2009; Shaw et al., 2010). Cells of strain PCO are straight rods and normally occur singly (Figure 1). Strain PCO had an optimum growth temperature of 70°C; no growth was detected below 37°C or above 75°C. The optimum pH for growth was between 6.5 and 7.5. Strain PCO is a very versatile organism that can utilize a range of different substrates, such as: arabinose, cellobiose, cellulose, fructose, galactose, glucose, lactose, maltose, mannitol, mannose, pectin, pyruvate, raffinose, ribose, sorbitol, starch, sucrose, trehalose, xylan, xylose, and yeast extract. No growth occurred with acetate, ethanol, formate, glycerol, glycine, lactate, methanol, propionate, CO (from 20 to 100% CO, total pressure 1.7 × 105 Pa), and H2/CO2 (80/20%, total pressure 1.7 × 105 Pa). The main products detected and quantified from glucose fermentation by strain PCO were lactate, ethanol, acetate, and H2 (Figure 2) which are typically formed from glucose by most of the Thermoanaerobacter species (Wiegel and Ljungdahl, 1981; Lee et al., 1993; Kim et al., 2001; Lee et al., 2007; Balk et al., 2009; Shaw et al., 2010). Strain PCO is able to reduce elemental sulfur and AQDS, but sulfate, sulfite, thiosulfate, nitrate, and nitrite could not serve as electron acceptors. The comparison between the morphological, biochemical and physiological characteristics of strain PCO and its close relatives is presented in Table 1. All of them can use thiosulfate as electron acceptor, but strain PCO cannot. Even though strain PCO and T. thermohydrosulfuricus are the same species, strain PCO can be differentiated because of its ability to grow and ferment cellulose and reduce AQDS.
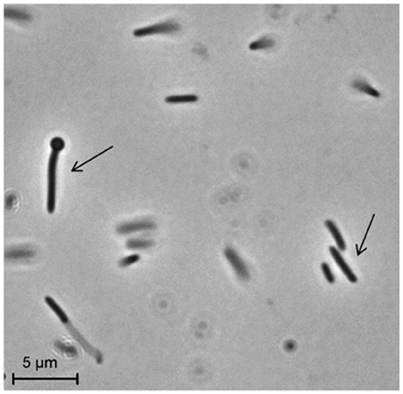
Figure 1. Phase-contrast micrographs showing morphology of cells of strain PCO. The arrows indicate vegetative and sporulating cells. Bar, 5 μm.
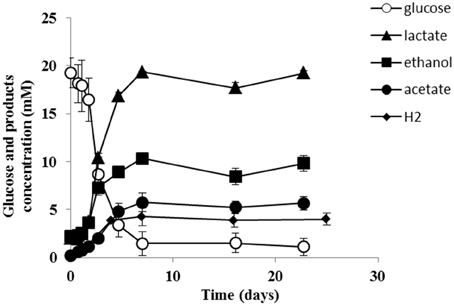
Figure 2. Glucose conversion and products formation by strain PCO over time. The results represent the average of duplicate experiments.
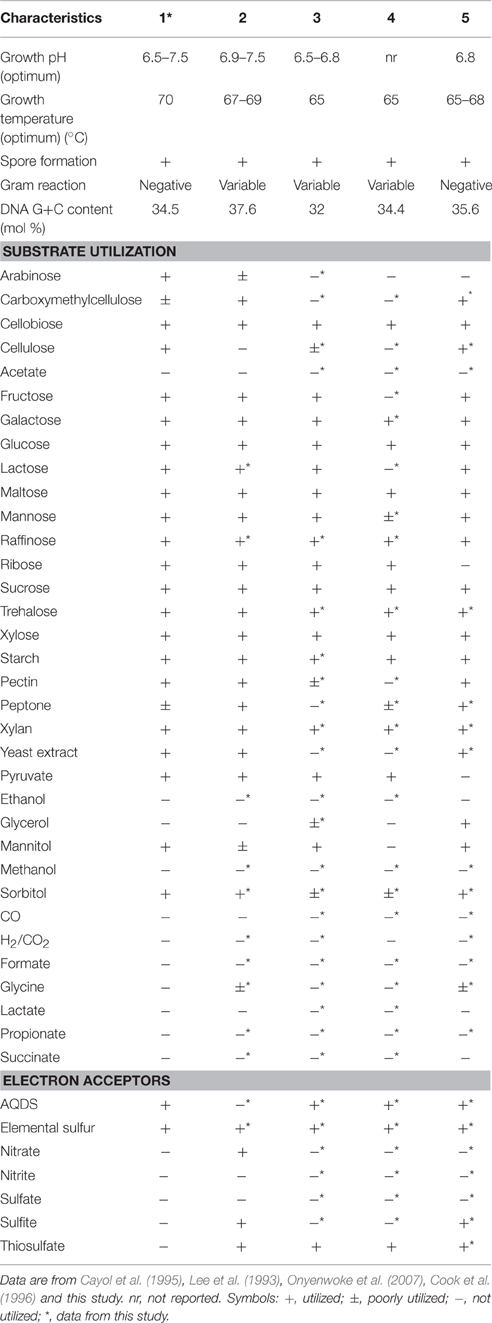
Table 1. Physiological and biochemical characteristics of (1) strain PCO and its phylogenetic related species: (2) Thermoanaerobacter thermohydrosulfuricus (DSM 527T), (3) T. brockii subsp. finnii (DSM 3389T), (4) T. pseudethanolicus (DSM 2355T), and (5) T. wiegelii (DSM 10319T).
Carbon Monoxide Tolerance of Strain PCO and Related Thermoanaerobacter sp.
When strain PCO was cultured with 20 mM of glucose and subjected to different CO concentrations in the headspace (0, 25, 50, 75, and 100%, total pressure 1.7 × 105 Pa) no significant differences were found in glucose consumption rates (Table 2). Carbon monoxide concentrations of 75% or lower did not affect substantially product formation from glucose (Figures 3A, 4A), except for H2. Hydrogen production by strain PCO decreased significantly (P = 0.0001) from 3.94 ± 0.55 to 1.52 ± 0.13 mM when cultures were incubated with 0 and 25% CO in the headspace, respectively (Figure 3A). When testing different CO partial pressures, from 25 to 100%, a decrease in H2 production is observed, with H2 production of < 0.02 mmol L−1 at 100% CO. The other end products from glucose conversion were only affected when strain PCO was exposed to 100% CO. There was a significant decrease on the final production of lactate (P = 0.01) and ethanol (P = 0.004) only when comparing cultures grown with 75 and 100% CO (Figure 4A). These results could be explained due to the fact that this strain had been isolated from a syngas-converting culture, that was in contact with high CO concentrations for over 1 year (Alves et al., 2013), which might have increased the tolerance of strain PCO to CO. There are only two Thermoanaerobacter species able to use CO: T. thermohydrosulfuricus subsp. carboxydovorans and T. kivui (Balk et al., 2009; Weghoff and Müller, 2016); for other Thermoanaerobacter species the ability to convert CO was never reported. The ability to utilize CO as carbon and energy source and/or the ability to tolerate CO by T. thermohydrosulfuricus, T. brockii subsp. finnii, T. pseudethanolicus, and T. wiegelii were tested in this study. None of the tested Thermoanaerobacter species could utilize CO (Table 1), but all species could grow and completely convert glucose in the presence of 0, 50, or 100% of CO in the headspace (Table 3, Figures 3, 4). This suggests that CO tolerance is a characteristic present among Thermoanaerobacter genus, and not only a property of strain PCO. Carbon recovery at the end of the incubations was nearly 100% for all the growth tests (Table 3). All the tested strains produced the same identified and quantified end products from glucose fermentation, i.e., hydrogen, lactate, acetate, and ethanol, as expected from previous reports (Kim et al., 2001; Lee et al., 2007; Balk et al., 2009; Shaw et al., 2010). Nevertheless, final product concentrations (Figures 3, 4) varied for the different tested strains. Ethanol production was in general less affected by the presence of different CO concentrations. A significant decrease in ethanol production was only observed in the presence of 100% CO and just for two of the five microorganisms tested: strain PCO (P = 0.004) and T. wiegelii (P = 0.002). H2 production by all the tested strains decreased significantly, even in the presence of low CO concentrations. In the presence of 100% CO, H2 production by the Thermoanaerobacter species tested was decreased by 75–95%. Acetate concentration decreased with increasing CO percentage (reduction between 25 and 50%); the exception was strain PCO for which the final acetate concentration was significantly higher in the presence of high CO percentage [acetate final concentration in cultures with 0 and 100% CO were 5.4 ± 0.6 and 8.3 ± 0.5 mM, respectively (P = 0.0003)]. These results show the metabolic changes in versatile Thermoanaerobacter species upon addition of CO, resulting in a general decrease in H2 production. Although none of the tested Thermoanaerobacter strains could convert CO, all were able to withstand CO. The lower H2 production suggests that CO is inhibiting the hydrogenases of Thermoanaerobacter species. Hydrogenases catalyze the oxidation of hydrogen or the reduction of protons. From recent genomic studies, it was confirmed that [Fe-Fe]-hydrogenases, responsible for hydrogen production, are present and well-conserved in all of the Thermoanaerobacter species. However, another class of hydrogenases, [Ni-Fe]-hydrogenases, is present only in T. thermohydrosulfuricus and T. wiegelii (Verbeke et al., 2013; Bhattacharya et al., 2015). [Fe-Fe] or iron-only hydrogenases are known to be more sensitive to CO than [Ni-Fe]-hydrogenases (Diender et al., 2015), which corroborate the results obtained regarding to the effect of CO on hydrogen production from glucose conversion by Thermoanaerobacter species. Hydrogenases were shown to be specifically inhibited by carbon monoxide, since CO binds at the active site of the enzyme (Purec et al., 1962; Gutiérrez-Sánchez et al., 2010; Baffert et al., 2011; Matsumoto et al., 2011; Bertsch and Müller, 2015). Genomic analysis of T. kivui revealed the presence of genes encoding for carbon monoxide dehydrogenases (CODH) and Ech-hydrogenases complexes (which are responsible for hydrogen production by carboxydotrophic organisms; Hess et al., 2014), although its ability to convert CO was only reported very recently after adaption to increasing concentrations of CO (Weghoff and Müller, 2016). Proper adaption to CO may be the key for achieving CO conversion by Thermoanaerobacter species which contain the necessary genomic machinery. Therefore, the high tolerance to CO and the potential of some microorganisms for CO utilization, make the members of Thermoanaerobacter genus important for the biotechnological use of syngas/industrial CO-rich gases. Thermophilic microorganisms including members of Thermoanaerobacter genus are interesting catalysts for production of biofuels (Carere et al., 2012; Verbeke et al., 2013; Hess et al., 2014; Bhattacharya et al., 2015; Sant'Anna et al., 2015). From this perspective the present study is important as CO is a way to steer the formation of fermentation products. Further research is needed to get a better insight into how at a molecular level carbon monoxide affects product formation in Thermoanaerobacter species.
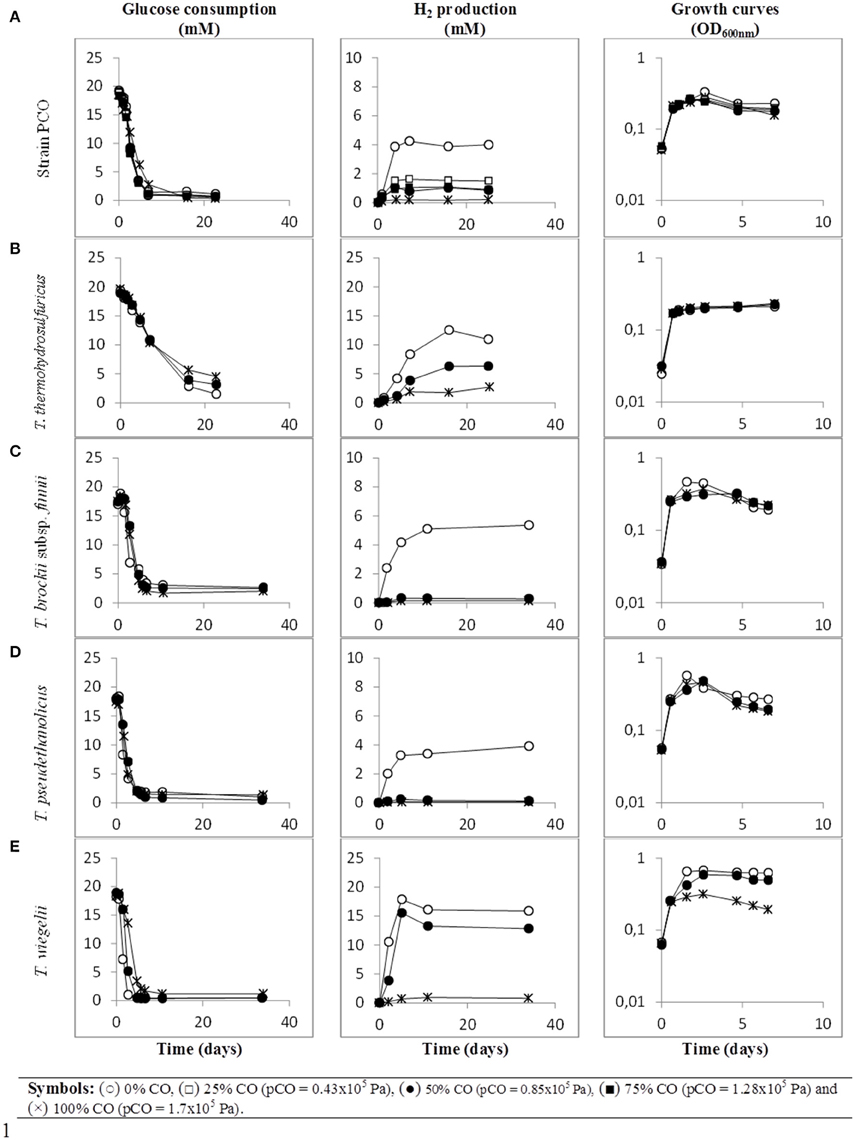
Figure 3. Glucose conversion, production of H2 from glucose and growth curves over time by (A) strain PCO, (B) Thermoanaerobacter thermohydrosulfuricus (DSM 527T), (C) T. brockii subsp. finnii (DSM 3389T), (D) T. pseudethanolicus (DSM 2355T), and (E) T. wiegelii (DSM 10319T), in incubations with different CO concentration in the gas phase (0, 25, 50, 75, or 100% CO). Plotted are the average data of duplicate experiments. The values of optical density were plotted vs. time on a logarithmic scale.
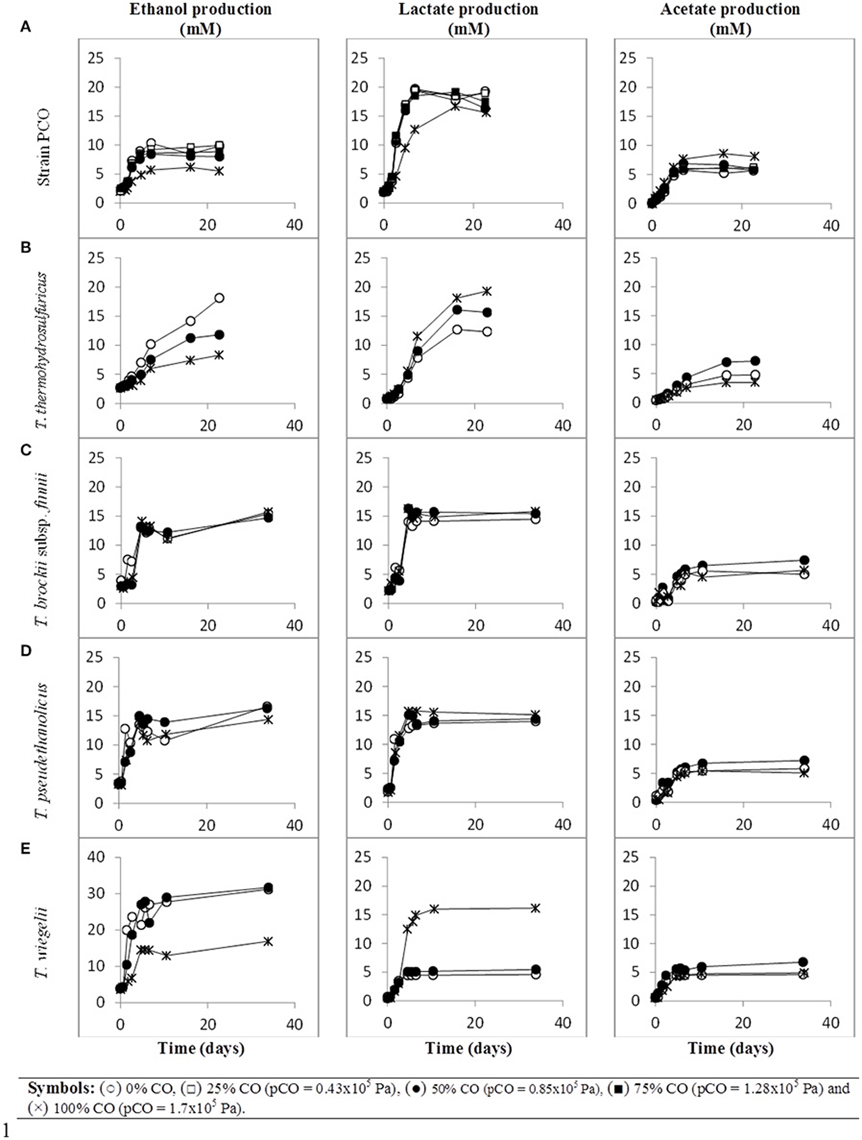
Figure 4. Production of ethanol and organic acids (lactate and acetate) from glucose, over time, by (A) strain PCO, (B) Thermoanaerobacter thermohydrosulfuricus (DSM 527T), (C) T. brockii subsp. finnii (DSM 3389T), (D) T. pseudethanolicus (DSM 2355T), and (E) T. wiegelii (DSM 10319T), in incubations with different CO concentration in the gas phase (0, 25, 50, 75, or 100% CO). Plotted are the average data of duplicate experiments.
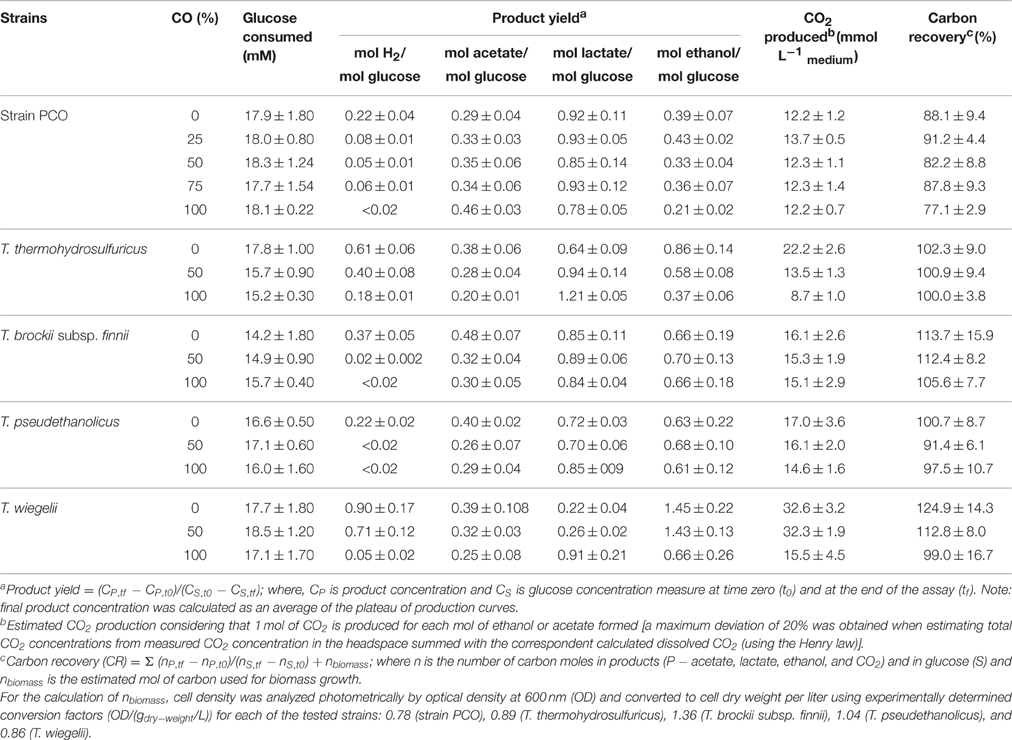
Table 3. Effect of CO partial pressure on glucose conversion and carbon recovery (%) by strain PCO, Thermoanaerobacter thermohydrosulfuricus (DSM 527T), T. brockii subsp. finnii (DSM 3389T), T. pseudethanolicus (DSM 2355T), and T. wiegelii (DSM 10319T).
Author Contributions
JA planned and performed the experiments, data interpretation and wrote the manuscript. MA assisted in the design of the study, participated in data interpretation as well as revisions of the final manuscript. CP assisted in the design of the study, participated in data interpretation as well as revisions of the final manuscript. AS assisted in the design of the study, participated in data interpretation as well as revisions of the final manuscript. DS conceived the study, participated in the planning and coordination of the study, and revised the manuscript. All authors read and gave approval for publication of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the Portuguese Foundation for Science and Technology (FCT) under the scope of the strategic funding of UID/BIO/04469/2013 unit and COMPETE 2020 (POCI-01-0145-FEDER-006684). The financial support from FCT and European Social Fund (POPH-QREN) through post-doc grant SFRH/BPD/104837/2014 given to JA is gratefully acknowledged. AS and DS are supported by an advanced ERC grant (project 323009) and a Gravitation grant (project 024.002.002) of the Netherlands Ministry of Education, Culture and Science and the Netherlands Science Foundation (NWO).
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Alves, J. I., Stams, A. J., Plugge, C. M., Alves, M. M., and Sousa, D. Z. (2013). Enrichment of anaerobic syngas-converting bacteria from thermophilic bioreactor sludge. FEMS Microbiol. Ecol. 86, 590–597. doi: 10.1111/1574-6941.12185
Baffert, C., Bertini, L., Lautier, T., Greco, C., Sybirna, K., Ezanno, P., et al. (2011). CO disrupts the reduced H-cluster of FeFe hydrogenase. A combined DFT and protein film voltammetry study. J. Am. Chem. Soc. 133, 2096–2099. doi: 10.1021/ja110627b
Balk, M., Heilig, H. G., van Eekert, M. H., Stams, A. J. M., Rijpstra, I. C., Sinninghe-Damste, J. S., et al. (2009). Isolation and characterization of a new CO-utilizing strain, Thermoanaerobacter thermohydrosulfuricus subsp. carboxydovorans, isolated from a geothermal spring in Turkey. Extremophiles 13, 885–894. doi: 10.1007/s00792-009-0276-9
Bertsch, J., and Müller, V. (2015). Bioenergetic constraints for conversion of syngas to biofuels in acetogenic bacteria. Biotechnol. Biofuels 8:210. doi: 10.1186/s13068-015-0393-x
Bhattacharya, P., Barnebey, A., Zemla, M., Goodwin, L., Auer, M., and Yannone, S. M. (2015). Complete genome sequence of the chromate-reducing bacterium Thermoanaerobacter thermohydrosulfuricus strain BSB-33. Stand. Genomic Sci. 10:74. doi: 10.1186/s40793-015-0028-7
Carere, C. R., Rydzak, T., Verbeke, T. J., Cicek, N., Levin, D. B., and Sparling, R. (2012). Linking genome content to biofuel production yields: a meta-analysis of major catabolic pathways among select H2 and ethanol-producing bacteria. BMC Microbiol. 12:295. doi: 10.1186/1471-2180-12-295
Cayol, J. L., Ollivier, B., Patel, B. K., Ravot, G., Magot, M., Ageron, E., et al. (1995). Description of Thermoanaerobacter brockii subsp. lactiethylicus subsp. nov., isolated from a deep subsurface French oil well, a proposal to reclassify Thermoanaerobacter finnii as Thermoanaerobacter brockii subsp. finnii comb. nov., and an emended description of Thermoanaerobacter brockii. Int. J. Syst. Bacteriol. 45, 783–789. doi: 10.1099/00207713-45-4-783
Cook, G. M., Rainey, F. A., Patel, B. K., and Morgan, H. W. (1996). Characterization of a new obligately anaerobic thermophile, Thermoanaerobacter wiegelii sp. nov. Int. J. Syst. Bacteriol. 46, 123–127. doi: 10.1099/00207713-46-1-123
Diender, M., Stams, A. J. M., and Sousa, D. Z. (2015). Pathways and bioenergetics of anaerobic carbon monoxide fermentation. Front. Microbiol. 6:1275. doi: 10.3389/fmicb.2015.01275
Gutiérrez-Sánchez, C., Rudiger, O., Fernandez, V. M., De Lacey, A. L., Marques, M., and Pereira, I. A. (2010). Interaction of the active site of the Ni-Fe-Se hydrogenase from Desulfovibrio vulgaris Hildenborough with carbon monoxide and oxygen inhibitors. J. Biol. Inorg. Chem. 15, 1285–1292. doi: 10.1007/s00775-010-0686-2
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hemme, C. L., Mouttaki, H., Lee, Y. J., Zhang, G., Goodwin, L., Lucas, S., et al. (2010). Sequencing of multiple clostridial genomes related to biomass conversion and biofuel production. J. Bacteriol. 192, 6494–6496. doi: 10.1128/JB.01064-10
Hess, V., Poehlein, A., Weghoff, M. C., Daniel, R., and Müller, V. (2014). A genome-guided analysis of energy conservation in the thermophilic, cytochrome-free acetogenic bacterium Thermoanaerobacter kivui. BMC Genomics 15:1139. doi: 10.1186/1471-2164-15-1139
Jessen, J. E., and Orlygsson, J. (2012). Production of ethanol from sugars and lignocellulosic biomass by Thermoanaerobacter J1 isolated from a hot spring in Iceland. J. Biomed. Biotechnol. 2012, 186982. doi: 10.1155/2012/186982
Kevbrina, M. V., Ryabokon, A. M., and Pusheva, M. A. (1996). Acetate formation from CO-containing gas mixtures by free and immobilized cells of the Thermophilic homoacetogenic bacterium Thermoanaerobacter kivui. Microbiology 65, 753–757.
Kim, B. C., Grote, R., Lee, D. W., Antranikian, G., and Pyun, Y. R. (2001). Thermoanaerobacter yonseiensis sp. nov., a novel extremely thermophilic, xylose-utilizing bacterium that grows at up to 85 degrees C. Int. J. Syst. Evol. Microbiol. 51, 1539–1548. doi: 10.1099/00207713-51-4-1539
Lacis, L. S., and Lawford, H. G. (1991). Thermoanaerobacter ethanolicus growth and product yield from elevated levels of xylose or glucose in continuous cultures. Appl. Environ. Microbiol. 57, 579–585.
Lane, D. J. (1991). Nucleic Acid Techniques in Bacterial Systematics. Chichester: John Wiley & Sons.
Lee, Y.-E., Jain, M. K., Lee, C., Lowe, S. E., and Zeikus, J. G. (1993). Taxonomic distinction of saccharolytic thermophilic anaerobes: description of Thermoanaerobacterium xylanolyticum gen. nov., sp. nov., and Thermoanaerobacterium saccharolyticum gen. nov., sp. nov.; Reclassification of Thermoanaerobium brockii, Clostridium thermosulfurogenes, and Clostridium thermohydrosulfuricum E100-69 as Thermoanaerobacter brockii comb. nov., Thermoanaerobacterium thermosulfurigenes comb. nov., and Thermoanaerobacter thermohydrosulfuricus comb. nov., respectively; and transfer of Clostridium thermohydrosulfuricum 39E to Thermoanaerobacter ethanolicus. Int. J. Syst. Bacteriol. 43, 41–51. doi: 10.1099/00207713-43-1-41
Lee, Y. J., Dashti, M., Prange, A., Rainey, F. A., Rohde, M., Whitman, W. B., et al. (2007). Thermoanaerobacter sulfurigignens sp. nov., an anaerobic thermophilic bacterium that reduces 1 M thiosulfate to elemental sulfur and tolerates 90 mM sulfite. Int. J. Syst. Evol. Microbiol. 57, 1429–1434. doi: 10.1099/ijs.0.64748-0
Matsumoto, T., Kabe, R., Nonaka, K., Ando, T., Yoon, K. S., Nakai, H., et al. (2011). Model study of CO inhibition of [NiFe]hydrogenase. Inorg. Chem. 50, 8902–8906. doi: 10.1021/ic200965t
Muyzer, G., de Waal, E. C., and Uitterlinden, A. G. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700.
Nübel, U., Engelen, B., Felske, A., Snaidr, J., Wieshuber, A., Amann, R. I., et al. (1996). Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178, 5636–5643.
Onyenwoke, R. U., Kevbrin, V. V., Lysenko, A. M., and Wiegel, J. (2007). Thermoanaerobacter pseudethanolicus sp. nov., a thermophilic heterotrophic anaerobe from Yellowstone National Park. Int. J. Syst. Evol. Microbiol. 57, 2191–2193. doi: 10.1099/ijs.0.65051-0
Purec, L., Krasna, A. I., and Rittenberg, D. (1962). The inhibition of hydrogenase by carbon monoxide and the reversal of this inhibition by light. Biochemistry 1, 270–275. doi: 10.1021/bi00908a013
Sanguinetti, C. J., Dias Neto, E., and Simpson, A. J. (1994). Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques 17, 914–921.
Sant'Anna, F. H., Lebedinsky, A. V., Sokolova, T. G., Robb, F. T., and Gonzalez, J. M. (2015). Analysis of three genomes within the thermophilic bacterial species Caldanaerobacter subterraneus with a focus on carbon monoxide dehydrogenase evolution and hydrolase diversity. BMC Genomics 16:757. doi: 10.1186/s12864-015-1955-9
Shaw, A. J., Hogsett, D. A., and Lynd, L. R. (2010). Natural competence in Thermoanaerobacter and Thermoanaerobacterium species. Appl. Environ. Microbiol. 76, 4713–4719. doi: 10.1128/AEM.00402-10
Stams, A. J. M., van Dijk, J. B., Dijkema, C., and Plugge, C. M. (1993). Growth of syntrophic propionate-oxidizing bacteria with fumarate in the absence of methanogenic bacteria. Appl. Environ. Microbiol. 59, 1114–1119.
Turner, P., Mamo, G., and Karlsson, E. N. (2007). Potential and utilization of thermophiles and thermostable enzymes in biorefining. Microb. Cell Fact. 6:9. doi: 10.1186/1475-2859-6-9
Verbeke, T. J., Zhang, X., Henrissat, B., Spicer, V., Rydzak, T., Krokhin, O. V., et al. (2013). Genomic evaluation of Thermoanaerobacter spp. for the construction of designer co-cultures to improve lignocellulosic biofuel production. PLoS ONE 8:e59362. doi: 10.1371/journal.pone.0059362
Wagner, I. D., and Wiegel, J. (2008). Diversity of thermophilic anaerobes. Ann. N. Y. Acad. Sci. 1125, 1–43. doi: 10.1196/annals.1419.029
Weghoff, M. C., and Müller, V. (2016). CO metabolism in the thermophilic acetogen Thermoanaerobacter kivui. Appl. Environ. Microbiol. 82, 2312–2319. doi: 10.1128/AEM.00122-16
Wiegel, J., and Ljungdahl, L. G. (1981). Thermoanaerobacter ethanolicus gen. nov., spec. nov., a new extreme thermophilic anaerobic bacterium. Arch. Microbiol. 128, 343–348. doi: 10.1007/BF00405910
Keywords: syngas, carbon monoxide, Thermoanaerobacter, sugar fermentation, ethanol, hydrogen
Citation: Alves JI, Alves MM, Plugge CM, Stams AJM and Sousa DZ (2016) Comparative Analysis of Carbon Monoxide Tolerance among Thermoanaerobacter Species. Front. Microbiol. 7:1330. doi: 10.3389/fmicb.2016.01330
Received: 31 May 2016; Accepted: 11 August 2016;
Published: 29 August 2016.
Edited by:
Eric Altermann, AgResearch, New ZealandReviewed by:
Jose M. Bruno-Barcena, North Carolina State University, USAChristopher L. Hemme, University of Rhode Island, USA
Copyright © 2016 Alves, Alves, Plugge, Stams and Sousa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joana I. Alves, am9hbmEuYWx2ZXNAZGViLnVtaW5oby5wdA==
 Joana I. Alves
Joana I. Alves M. Madalena Alves1
M. Madalena Alves1 Caroline M. Plugge
Caroline M. Plugge Alfons J. M. Stams
Alfons J. M. Stams Diana Z. Sousa
Diana Z. Sousa