- 1Division of Genetics and Molecular Biology, Institute of Biological Sciences, Faculty of Science, University of Malaya, Kuala Lumpur, Malaysia
- 2Novel Bacteria and Drug Discovery Research Group, School of Pharmacy, Monash University Malaysia, Bandar Sunway, Malaysia
- 3UKM Medical Molecular Biology Institute, UKM Medical Centre, Universiti Kebangsaan Malaysia, Kuala Lumpur, Malaysia
- 4Center of Health Outcomes Research and Therapeutic Safety, School of Pharmaceutical Sciences, University of Phayao, Phayao, Thailand
Introduction
Vibrio parahaemolyticus is a Gram-negative bacterium that naturally occurs in marine associated aquatic environments (Letchumanan et al., 2014; Malcolm et al., 2015). This bacterium causes highest number of seafood-associated gastroenteritis in many countries including United States and Asian countries (Scallan et al., 2011; Newton et al., 2012). V. parahaemolyticus is often been isolated from aquatic environments such as seawater and marine sediment, as well as from vertebrate and invertebrate seafood products (Shen et al., 2009). The most likely route of infection in humans is reported to be associated with consumption of raw or improperly cooked seafood (Daniels et al., 2000; Jun et al., 2014; Hazen et al., 2015; Raghunath, 2015; Law et al., 2015).
Recently, V. parahaemolyticus has been demonstrated to be a major source of infection in the aquaculture industry (Letchumanan et al., 2014; Soto-Rodriguez et al., 2015; Tey et al., 2015). Aquaculture farmers rely on a wide range of antibiotics to prevent (prophylactic use) and treat (therapeutic use) bacterial infections in fish and invertebrates (Cabello et al., 2013). The extensive use of antibiotics and other chemotherapeutics in aquaculture has led to the emergence of multidrug resistant strains in the biosphere (Letchumanan et al., 2015a, 2016; Rao and Lalitha, 2015). Multidrug resistant V. parahaemolyticus strains have been isolated and detected from shrimp in Thailand (Yano et al., 2014), Malaysia (Al-Othrubi et al., 2011; Sani et al., 2013; Letchumanan et al., 2015b,c) and China (Peng et al., 2010; Xu et al., 2014). Resistance toward clinically used antibiotics will eventually hamper the treatment of bacterial infections in humans and potentially increase the fatality rate (Daniels et al., 2000). Therefore, monitoring Vibrio species in aquaculture surroundings is crucial for both human health and the aquaculture industry.
In our previous study, we have isolated environmental V. parahaemolyticus strains from two types of Malaysian shrimp, Penaeus indicus and Solenocera subnuda. We detected the thermostable direct hemolysin (tdh) and thermostable direct related hemolysin (trh) virulence genes through a PCR based assay and studied the antibiotic resistance profile of all the isolated strains (Letchumanan et al., 2015c). V. parahaemolyticus VP152 was isolated from Penaeus indicus (Banana prawn) and originated from a supermarket sample. This strain did not possess both the tdh and trh virulence genes, which are responsible for causing diseases in humans and marine animals. Despite the fact that V. parahaemolyticus VP152 strain does not have tdh and trh virulence genes properties, the strain cannot be ignored in light of the fact that it exhibits multidrug resistance profiles toward 11/14 antibiotics tested. Based on the antibiotic susceptibility phenotype, the strain exhibited multiple-antibiotic resistance toward ampicillin, oxytetracycline, nalidixic acid, ampicillin/sulbactam, tetracycline, third generation cephalos porins (cefotaxime and ceftazidime), aminoglycosides (amikacin, kanamycin, and gentamicin) and trimethoprim/sulfameth oxazole (Letchumanan et al., 2015c).
This is a worrying situation as the antibiotic resistant profiles shown by V. parahaemolyticus VP152 include the recommended antimicrobial agents used in treatment of Vibrio spp. infections, including third generation cephalosporin, fluoroquinolones, aminoglycosides, tetracycline, gentamicin, trimethoprim/sulfamethoxazole (Daniels and Shafaie, 2000; Shaw et al., 2014). Therefore, the whole genome sequence of V. parahaemolyticus VP152 was studied with respect to the multidrug resistance profiles to gain a better understanding of the antibiotic resistant patterns. The availability of this genome sequence of V. parahaemolyticus VP152 will aid as a basis for further in-depth analysis of the antibiotic resistance profile of environmental V. parahaemolyticus.
Materials and Methods
Genome Sequencing and Assembly
Genomic DNA of VP152 strain was extracted using MasterpureTM DNA purification kit (Epicenter, Illumina Inc, Madison, WI, USA) and subjected to RNase (Qiagen, USA) treatment (Ser et al., 2015). The DNA quality was quantified using NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA), and a Qubit version 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA). Illumina sequencing library of genomic DNA was prepared using NexteraTM DNA Sample Preparation kit (Illumina, San Diego, CA, USA) and library quality was validated by a Bioanalyzer 2100 high sensitivity DNA kit (Agilent Technologies, Palo Alto, CA, USA) prior to sequencing. The genome of VP152 strain was sequenced on MiSeq platform with MiSeq Reagent Kit 2 (2 × 250 bp; Illumina Inc, San Diego, CA, USA). The trimmed sequences were de novo assembled with CLC Genomic Workbench version 5.1 (CLC Bio, Denmark).
Genome Annotation
Gene prediction was carried out using Prodigal 2.6, while rRNA and tRNA were analyzed using RNAmmer and tRNAscan SE version 1.21 (Lowe and Eddy, 1997; Lagesen et al., 2007; Hyatt et al., 2010). Gene prediction and annotation were performed using Rapid Annotation Search Tool (RAST; Aziz et al., 2008). Antibiotic resistance genes were analyzed using antibiotic resistance genes-ANNOTation (ARG-ANNOT; Gupta et al., 2014).
Results
Genome Characteristics
The genome of V. parahaemolyticus VP152 consists of 4,982,021 bp with mean genome coverage of 183.46-fold and with an average G+C content of 53.4% (Table 1). A total of 4809 genes was predicted of which 4638 were identified as protein coding genes. There are 91 RNA genes consisting of 11 rRNAs and 80 tRNAs.
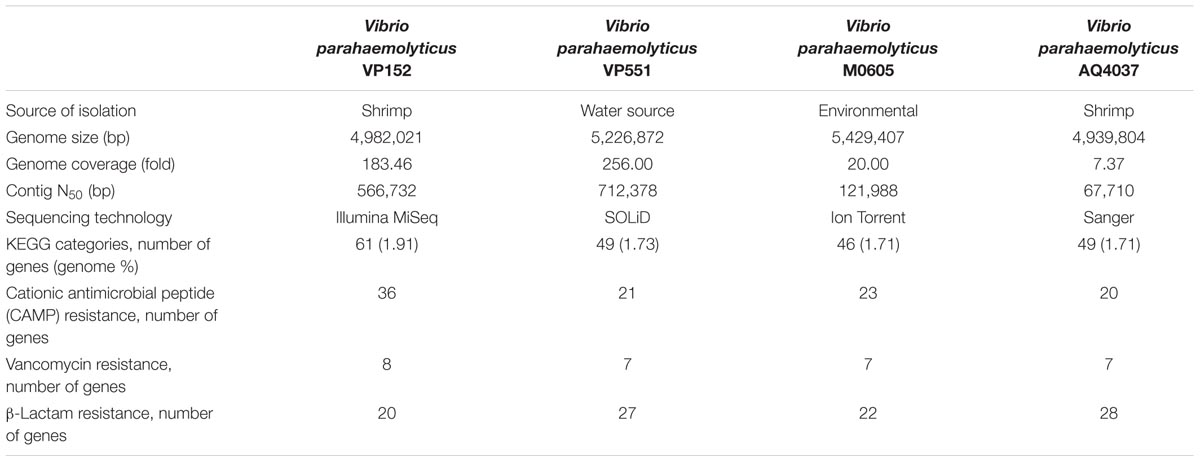
TABLE 1. Comparison of genome sequence of Vibrio parahaemolyticus VP152 with other genome sequences.
Virulence and Antimicrobial Resistance Genes
The analysis obtained from RAST server revealed 573 subsystems (Figure 1). The annotated genome has 97 genes responsible for resistance to antibiotic and toxic compounds including seven genes for mdtABCD multidrug resistance cluster, 19 genes for multidrug resistance efflux pumps, four genes for β-lactamase and two genes aminoglycoside adenylyltransferases. The genome sequence of V. parahaemolyticus VP152 was compared with three environmental V. parahaemolyticus strains, in order to delineate the similarities between the four strains. The genome size of V. parahaemolyticus VP152 was similar to strains of V. parahaemolyticus and contained several antibiotic resistance genes as shown in Table 1. Also, further comparison of hemolysin genes present in V. parahaemolyticus VP152 and the selected strains revealed no significant differences.
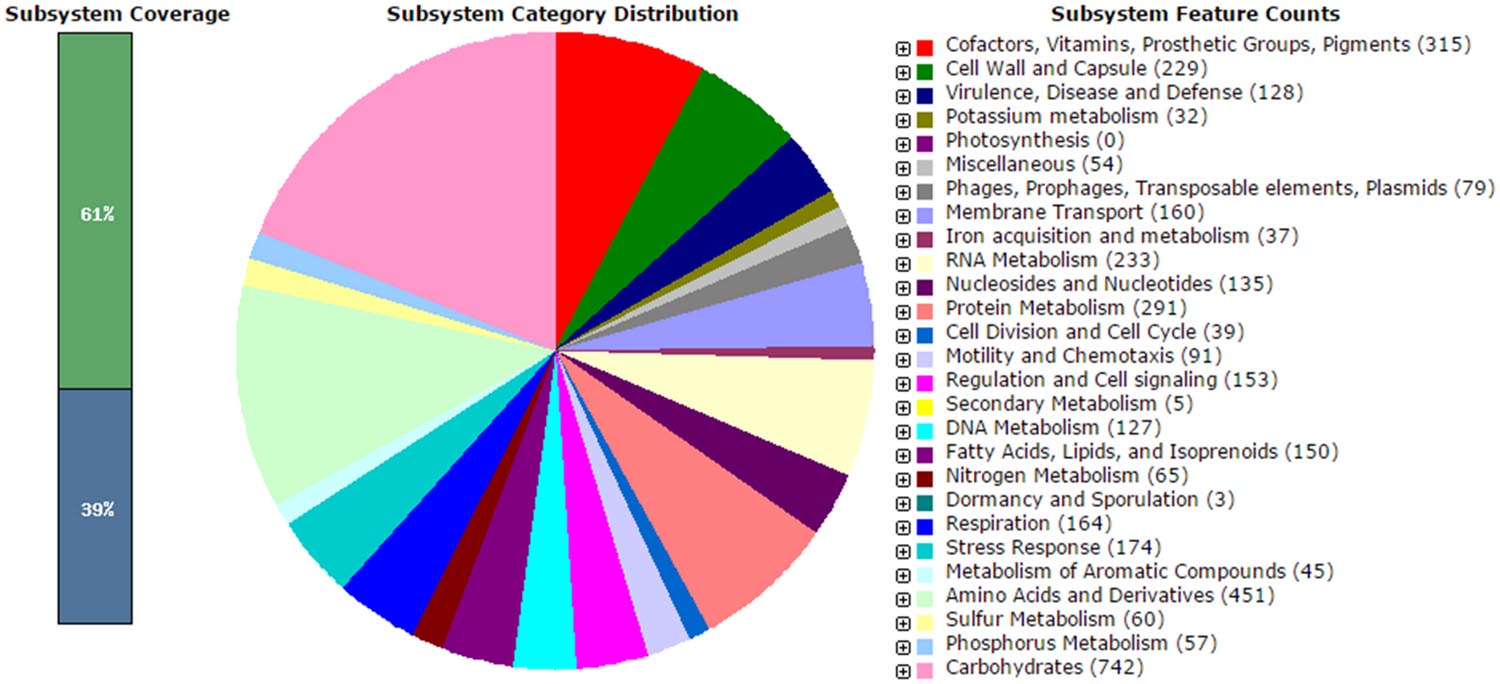
FIGURE 1. Subsystem category distribution of Vibrio parahaemolyticus VP152 (based on RAST annotation server).
The genome analysis on ARG-ANNOT noted the presences of tetracycline resistant gene, Tet and Tet-2 gene within the genome. The presence of these genes is closely related to the phenotypic resistance shown by the strain toward oxytetracycline and tetracycline. Furthermore, β-lactam resistance-related gene, bla gene of VP152 exhibited 99% similarities when compared to other V. parahaemolyticus strain and Vibrio species. The phenotypic resistance shown by V. parahaemolyticus VP152 toward ampicillin, ampicillin/sulbactam, cefotaxime and ceftazidime is closely related to the gene coding β-lactamase in the genome. The gene coding aminoglycosides adenylyltransferase of V. parahaemolyticus VP152 confers resistance phenotype observed toward amikacin, kanamycin, and gentamicin. Based on the annotation tools and detailed analysis of V. parahaemolyticus VP152 genome using PlasmidFinder, the genome of V. parahaemolyticus VP152 did not recover any plasmid sequence. Even though these genes were commonly found in plasmids, some of the Vibrio species including V. coralliilyticus and V. alginolyticus carry these genes in their chromosomes (Costa et al., 2015). Therefore, the resistant genes observed in V. parahaemolyticus VP152 are chromosome mediated.
The multidrug resistance profile seen in the phenotype and genes of V. parahaemolyticus VP152 genome illustrates how extensive antibiotics have been utilized in the aquaculture industry. The resistance phenotype observed in this strain could be triggered by the extensive use of permitted antibiotics in the Asian aquaculture industry namely oxytetracycline, tetracycline, quinolone, sulphonamides, and trimethoprim (Rico et al., 2012; Yano et al., 2014). The resistance toward third generation cephalosporins seen in V. parahaemolyticus VP152 would further hamper the treatment of Vibrio species infection in future. This situation is cause for concern, and warrants more stringent surveillance in the use of antibiotics, as well as the resultant antibiotic resistance in clinically important bacterial species. In summary, the whole genome sequence of V. parahaemolyticus VP152 will be useful in future studies to determine antimicrobial resistance and virulence attributes as well as mechanisms that enhance its environmental or host fitness.
Nucleotide Sequence Accession Numbers
This genome sequence data of VP152 strain sequenced under this study has been deposited in DDBJ/EMBL/GenBank under Accession No. LCUL00000000. The version described in this paper is the first version, LCUL01000000. The genome sequences data are available in FASTA, annotated GenBank flat file, graphical and ASN.1 formats.
Author Contributions
The experiments, data analysis and manuscript writing were performed by VL and H-LS, while W-ST, N-SA, B-HG, K-GC, and L-HL provided vital guidance, technical support, and proofreading for the work. The research project was founded by L-HL.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by University of Malaya for High Impact Research Grant (UM-MOHE HIR Nature Microbiome Grant No. H-50001-A000027) awarded to K-GC., MOSTI eScience Fund (06-02-10-SF0300) and External Industry Grants from Biotek Abadi Sdn Bhd (vote no. GBA-808138 & GBA-808813) awarded to L-HL.
References
Al-Othrubi, S. M., Alfizah, H., Son, R., Humin, N., and Rahaman, J. (2011). Rapid detection and E-test antimicrobial susceptibility testing of Vibrio parahaemolyticus isolated from seafood and environmental sources in Malaysia. Saudi Medic. J. 32, 400–406.
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Cabello, F. C., Godfrey, H. P., Tomova, A., Ivanova, L., Dölz, H., Millanao, A., et al. (2013). Antimicrobial use in aquaculture re-examined: its relevance to antimicrobial resistance and to animal and human health. Appl. Eviron. Microbiol. 15, 1917–1942. doi: 10.1111/1462-2920.12134
Costa, R. A., Araujo, R. L., Souza, O. V., and Vieira, H. S. F. (2015). Antibiotic-resistant Vibrios in farmed shrimp. BioMed. Res. Int. 2015, 1–5. doi: 10.1155/2015/602078
Daniels, N., and Shafaie, A. (2000). A review of pathogenic Vibrio infections for clinicians. J. Infect. Medic. 17, 665–685.
Daniels, N. A., MacKinnon, L., Bishop, R., Altekruse, S., Ray, B., Hammond, R. M., et al. (2000). Vibrio parahaemolyticus infections in the United States, 1973–1998. J. Infect. Dis. 181, 1661–1666. doi: 10.1086/315459
Gupta, S. K., Padmanabhan, B. R., Diene, S. M., Rojas, R. L., Kemf, M., Landraud, L., et al. (2014). ARG-ANNOT, a new bioinformatics tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemo. 58, 212–220. doi: 10.1128/AAC.01310-13
Hazen, T. H., Lafon, P. C., Garrett, N. M., Lowe, T. M., Silberger, D. J., Rowe, L. A., et al. (2015). Insights into the environmental reservoir of pathogenic Vibrio parahaemolyticus using comparative genomics. Front. Microbiol. 6:204. doi: 10.3389/fmicb.2015.00204
Hyatt, D., Chen, G. L., Locascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Jun, J. W., Kim, H. J., Yun, S. K., Chai, J. Y., and Park, S. C. (2014). Eating oysters without risk of vibriosis: application of a bacteriophage against Vibrio parahaemolyticus in oysters. Int. J. Food Microbiol. 188, 31–35. doi: 10.1016/j.ijfoodmicro.2014.07.007
Lagesen, K., Hallin, P., Rodland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Law, J. W.-F., Ab Mutalib, N. S., Chan, K.-G., and Lee, L.-H. (2015). Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front. Microbiol. 5:770. doi: 10.3389/fmicb.2014.00770
Letchumanan, V., Chan, K.-G., and Lee, L.-H. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence and advance molecular identification techniques. Front. Microbiol. 5:705. doi: 10.3389/fmicb.2014.00705
Letchumanan, V., Chan, K.-G., and Lee, L.-H. (2015a). An insight of traditional plasmid curing in Vibrio species. Front. Microbiol. 6:735. doi: 10.3389/fmicb.2015.00735
Letchumanan, V., Chan, K.-G., Pusparajah, P., Saokaew, S., Duangjai, A., Goh, B.-H., et al. (2016). Insights into bacteriophage application in controlling Vibrio species. Front. Microbiol. 7:1114. doi: 10.3389/fmicb.2016.01114
Letchumanan, V., Pusparajah, P., Loh, T. H. T., Yin, W.-F., Lee, L.-H., and Chan, K.-G. (2015b). Occurrence and antibiotic resistance of Vibrio parahaemolyticus from shellfish in Selangor, Malaysia. Front. Microbiol. 6:1417. doi: 10.3389/fmicb.2015.01417
Letchumanan, V., Yin, W.-F., Lee, L.-H., and Chan, K.-G. (2015c). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 6:33. doi: 10.3389/fmicb.2015.00033
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.0955
Malcolm, T. T. H., Cheah, Y. K., Radzi, C. W. J. W. M., Kasim, F. A., Kantilal, H. K., John, T. Y. H., et al. (2015). Detection and quantification of pathogenic Vibrio parahaemolyticus in shellfish by using multiplex PCR and loop-mediated isothermal amplification assay. Food Cont. 47, 664–671. doi: 10.1016/j.foodcont.2014.08.010
Newton, A., Kendall, M., Vugia, D. J., Henao, O. L., and Mahon, B. E. (2012). Increasing rates of vibriosis in the United States, 1996-2010: review of surveillance data from 2 systems. Clin. Infect. Dis. 54, 391–395. doi: 10.1093/cid/cis243
Peng, F. M., Jiang, D. Y., Ruan, H. H., Liu, H. Q., and Zhou, L. P. (2010). Pathogenic investigation on a food poisoning induced by Vibrio parahaemolyticus. Prev. Med. Trib. 16, 746–747.
Raghunath, P. (2015). Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front. Microbiol. 5:805. doi: 10.3389/fmicb.2014.00805
Rao, B. M., and Lalitha, K. V. (2015). Bacteriophage for aquaculture: are they beneficial or inimical. Aquaculture 437, 146–154. doi: 10.1016/j.aquaculture.2014.11.039
Rico, A., Satapornvanit, K., Haque, M. M., Min, J., Nguyen, P. T., Telfer, T., et al. (2012). Use of chemicals and biological products in Asian aquaculture and their potential environmental risks: a critical review. Rev. Aqua. 4, 75–93. doi: 10.1111/j.1753-5131.2012.01062.x
Sani, N. A., Ariyawansa, S., Babji, A. S., and Hashim, J. K. (2013). The risk assessment of Vibrio parahaemolyticus in cooked black tiger shrimps (Penaeus monodon) in Malaysia. Food Cont. 31, 546–552. doi: 10.1016/j.foodcont.2012.10.018
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., and Roy, S. L. (2011). Foodborne illness acquired in the United States e major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1707.110572
Ser, H. L., Tan, W. S., Cheng, H. J., Yin, W. F., Chan, K. G., and Lee, L. H. (2015). Draft genome of amylolytic actinobacterium, Sinomonas humi MUSC 117T isolated from intertidal soil. Mar. Genomics 24, 209–210. doi: 10.1016/j.margen.2015.05.012
Shaw, K. S., Goldstein, R. E. R., He, X., Jacobs, J. M., Crump, B. C., and Sapkota, A. R. (2014). Antimicrobial susceptibility of Vibrio vulnificus and Vibrio parahaemolyticus recovered from recreational and commercial areas of Chesapeake Bay and Maryland Coastal Bays. PLoS ONE 9:89616. doi: 10.1371/journal.pone.0089616
Shen, X., Cai, Y., Liu, C., Liu, W., Hui, Y., and Su, Y.-C. (2009). Effect of temperature on uptake and survival of Vibrio parahaemolyticus in oysters (Crasssostrea plicatula). Int. J. Food. Microbiol. 136, 129–132. doi: 10.1016/j.ijfoodmicro.2009.09.012
Soto-Rodriguez, S. A., Gomez-Gil, B., Lozano-Olvera, R., BetancourtLozano, M., and Morales-Covarrubias, M. S. (2015). Field and experimental evidence of Vibrio parahaemolyticus as the causative agent of acute hepatopancreatic necrosis disease of cultured shrimp (Litopenaeus vannamei) in northwestern Mexico. Appl. Environ. Microbiol. 81, 1689–1699. doi: 10.1128/AEM.03610-14
Tey, Y. H., Jong, K. J., Fen, S. Y., and Wong, H. C. (2015). Occurrence of Vibrio parahaemolyticus, Vibrio cholerae, and Vibrio vulnificus in the aquacultural environments of Taiwan. J. Food Prot. 78, 969–976. doi: 10.4315/0362-028X.JFP-14-405
Xu, X., Wu, Q., Zhang, J., Cheng, J., Zhang, S., and Wu, K. (2014). Prevalence, pathogenicity, and serotypes of Vibrio parahaemolyticus in shrimp from Chinese retail markets. Food Cont. 46, 81–85. doi: 10.1016/j.foodcont.2014.04.042
Keywords: Vibrio parahaemolyticus, seafood, Penaeus indicus, antibiotic resistance, genome
Citation: Letchumanan V, Ser H-L, Tan W-S, Ab Mutalib N-S, Goh B-H, Chan K-G and Lee L-H (2016) Genome Sequence of Vibrio parahaemolyticus VP152 Strain Isolated from Penaeus indicus in Malaysia. Front. Microbiol. 7:1410. doi: 10.3389/fmicb.2016.01410
Received: 19 July 2016; Accepted: 25 August 2016;
Published: 07 September 2016.
Edited by:
Jennifer Ronholm, Health Canada, CanadaReviewed by:
Hongxia Wang, University of Alabama at Birmingham, USAJessica L. Jones, United Stated Food and Drug Administration, USA
Copyright © 2016 Letchumanan, Ser, Tan, Ab Mutalib, Goh, Chan and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Learn-Han Lee, bGVlLmxlYXJuLmhhbkBtb25hc2guZWR1 bGVlbGVhcm5oYW5AeWFob28uY29t Kok-Gan Chan, a29rZ2FuQHVtLmVkdS5teQ==
 Vengadesh Letchumanan
Vengadesh Letchumanan Hooi-Leng Ser
Hooi-Leng Ser Wen-Si Tan
Wen-Si Tan Nurul-Syakima Ab Mutalib
Nurul-Syakima Ab Mutalib Bey-Hing Goh
Bey-Hing Goh Kok-Gan Chan
Kok-Gan Chan Learn-Han Lee
Learn-Han Lee