- School of Public Health, Guangdong Pharmaceutical University, Guangzhou, China
While some studies have defined Staphylococcus aureus based on its clonal complex and resistance pattern, few have explored the relations between the genetic lineages and antibiotic resistance patterns and immune evasion cluster (IEC) genes. Our aim was to investigate the potential relationship between phenotypic and molecular characteristics so as to reveal livestock-associated S. aureus in humans. The study participants were interviewed, and they provided two nasal swabs for S. aureus analysis. All S. aureus and methicillin-resistant S. aureus (MRSA) were tested for antibiotic susceptibility, multilocus sequence type and IEC genes. Of the 1162 participants, 9.3% carried S. aureus, including MRSA (1.4%) and multidrug-resistant S. aureus (MDRSA, 2.8%). The predominant multidrug-resistant pattern among MDRSA isolates was non-susceptibility to erythromycin, clindamycin and tetracycline. The most common S. aureus genotypes were ST7, ST6, ST188, and ST59, and the predominant MRSA genotype was ST7. Notably, the livestock-associated S. aureus isolates (IEC-negative CC9, IEC-negative tetracycline-resistant CC398, and IEC-negative tetracycline-resistant CC5) were found in people with no occupational livestock contact. These findings reveal a potential relationship between S. aureus CCs and IEC genes and antibiotic resistance patterns in defining livestock-associated S. aureus in humans and support growing concern about the potential livestock-to-human transmission of livestock-associated S. aureus by non-occupational livestock contact.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is an important cause of nosocomial infections worldwide (Woodford and Livermore, 2009). The prevalence of MRSA in hospitals is approximately 50–80% in some Asian countries, such as Korea, Taiwan, Hong Kong, Thailand, and Vietnam (Song et al., 2011). Despite the high prevalence of healthcare-associated MRSA (HA-MRSA) in Asia, only a few epidemic clones have been identified; sequence type (ST) 239 has been reported in China, India, Thailand and Taiwan, and ST5 has been identified in Japan and South Korea (Ko et al., 2005). Recent reports have indicated that the epidemiology of MRSA is undergoing a major change following the emergence of community-associated MRSA (CA-MRSA) strains in patients without any risk factors or previous health-care contact (David and Daum, 2010; DeLeo et al., 2010). CA-MRSA outbreaks have been reported worldwide, and successful clones are usually associated with specific geographical locations (Deurenberg and Stobberingh, 2008; Chuang and Huang, 2013). Clones with multilocus ST59 and ST30 account for most of the CA-MRSA cases commonly reported in Asia (Chuang and Huang, 2013). The epidemiologic history of MRSA has been reshaped since the emergence of livestock-associated MRSA (LA-MRSA) in humans. Note that the predominant LA-MRSA lineage in Europe and North America is ST398, while in Asia, ST9 isolates are most prevalent (Chuang and Huang, 2015).
The reports of CA-MRSA have raised concerns about the prevalence and risk factors of MRSA in community populations. The prevalence of CA-MRSA varies substantially worldwide, ranging from less than 1% to more than 50% in different countries (Deurenberg and Stobberingh, 2008; Chuang and Huang, 2013). While some researchers have defined S. aureus (including MRSA) based on the clonal complex (CC) and resistance pattern, few have incorporated recent evidence suggesting that absence of the immune evasion cluster (IEC) genes may be a useful indicator of S. aureus livestock adaptation (Sung et al., 2008; Verkaik et al., 2011; Price et al., 2012; Rinsky et al., 2013). The IEC gene region at the 3′ end of βC-φs encodes up to four different immune evasion molecules [highly specific antiopsonic molecule (scn), chemotaxis and phagocyte activation (chp), inhibiting human α-defensins (sak), and staphylococcal enterotoxin (sea/sep)]; these mobile genetic elements move around in the population (van Wamel et al., 2006). The scn gene has been detected at a relatively high frequency among S. aureus isolates obtained from humans (90–100%) compared to those obtained from livestock (2–35%) (Sung et al., 2008; Verkaik et al., 2011; Price et al., 2012; Rinsky et al., 2013). A recent study in Germany indicated that IEC was absent from MRSA CC398 in pigs and pig farmers; however, IEC was found in MRSA CC398 from veterinary personnel (Cuny et al., 2015). Notably, there are few published studies of IEC genes among S. aureus isolates obtained from Asian populations, and it remains unclear whether there is a potential relation between the genetic lineages (ST types) and antibiotic resistance patterns and IEC genes.
In the present study, we aimed to examine the prevalence of carriage and antimicrobial susceptibility and the molecular characteristics (ST types and IEC genes) of MRSA and multidrug-resistant S. aureus (MDRSA) among healthy adults in Guangdong, China. We also investigated the potential relationship between the genetic lineages (ST types) of S. aureus isolates and antibiotic resistance patterns and IEC genes to reveal the potential risk of livestock-to-human transmission of livestock-associated S. aureus by non-occupational livestock contact.
Materials and Methods
Ethics Statement
This study was approved by the Ethics Committee of Guangdong Pharmaceutical University, and it was performed in accordance with the approved guidelines. All participants provided signed informed consent.
Population and Questionnaire
This cross-sectional study was conducted in Guangdong province, China, between November 2013 and November 2014. A multistage sample design was used to obtain a representative sample. First, four cities were randomly chosen from 21 cities in Guangdong province. Second, in each city, we selected two factories to achieve a sample size of 300 non-farm workers (i.e., workers from hardware factories or biscuit factories) with no occupational livestock contact. Finally, all workers in the selected venues were sampled to participate in this study. The following eligibility criteria were applied: (1) an age of 15 years or older, (2) an ability to speak and understand Chinese, (3) no current employment at a health care facility, (4) no acute infectious diseases in the last 7 days, and (5) no antibiotic use in the last 7 days. After obtaining informed consent, a face-to-face questionnaire was administered to collect demographic data and the influencing factors for S. aureus (including MRSA) carriage, such as sex, age (years), pet contact (e.g., dogs and cats) in homes in the last year, physical examination in a medical facility (including clinics, hospitals, community health station, and nursing homes) in the last month, antibiotic use in the last month and smoking (defined as “has smoked over 100 cigarettes in their lifetime and has smoked in the past month”). Note that none of the participants reported any occupational animal contact.
Sample Collection and Culture Methods
Two nasal swabs were taken from each participant (sterile cotton-wool swabs were used in both anterior nares), and the swabs were stored in 5 ml of enrichment broth containing 1% tryptone, 7.5% NaCl, 1% mannitol and 0.25% yeast extract at 4°C during transportation and incubated at 35 ± 1°C for 24 h. To isolate S. aureus, a loopful of the broth was plated on mannitol salt agar and incubated at 35 ± 1°C for 24–48 h. Per plate, one representative colony of each different suspected morphology was selected and purified on 5% sheep blood agar plates, and gram staining and catalase and coagulase tests were performed. S. aureus isolates were identified based on the above-mentioned positive tests.
Antimicrobial Susceptibility Testing
All S. aureus isolates were tested for their susceptibility to 12 antimicrobials [i.e., penicillin (10 units), cefoxitin (30 μg), erythromycin (15 μg), clindamycin (2 μg), trimethoprim-sulfamethoxazole(25 μg), linezolid (30 μg), tetracycline (30 μg), rifampin (5 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), gentamicin (10 μg), and quinupristin-dalfopristin (15 μg)] using the Kirby-Bauer disk diffusion method, according to the Clinical and Laboratory Standards Institute guidelines 2013 (CLSI, 2013). S. aureus strain ATCC 25923 was used for the quality control. S. aureus isolates were classified as MDRSA if they were MRSA or non-susceptible to ≥3 classes of antibiotics (Magiorakos et al., 2012). S. aureus isolates resistant to cefoxitin were identified as suspect MRSA, according to CLSI (2013).
Molecular Characterization
All cefoxitin-resistant isolates were also tested through PCR for carriage of the mecA gene using the previously described primers (Zhang et al., 2004). The presence of IEC genes (scn, chp, sak, sea, and sep) was tested through PCR for all S. aureus isolates (van Wamel et al., 2006). Multilocus sequence typing (MLST) for all S. aureus is a nucleotide sequence-based approach for seven housekeeping genes. Alleles and sequence types were determined using the MLST database, and singletons or members of a clonal complex were determined using the eBURST algorithm (accessible at http://eburst.mlst.net).
Statistical Analysis
All data were entered in duplicate into an EpiData (version 3.1) database (the EpiData Association, Odense Denmark). The relationships between potential influencing factors and S. aureus and MRSA carriage were examined using Pearson's chi-squared test and multiple logistic regression models. We also conducted separate analyses to examine the phenotypic and molecular characteristics of S. aureus isolates between methicillin-sensitive S. aureus (MSSA) and MRSA isolates using Pearson's chi-squared test. A two-sided p-value for statistical significance was defined as p ≤ 0.05. All statistical analyses were performed using STATA version 14.0 (StataCorp LP, College Station, Texas, USA).
Results
Participant Characteristics
The participant characteristics are provided in Table 1. A total of 1162 individuals were interviewed and participated in this survey. Approximately half of the participants were women (520/1162, 44.8%). The mean age (± standard deviation) was 36.7 ± 9.4 years, and the participant ages ranged from 15 to 60 years. Of the 1162 participants, 108 (9.3%) carried S. aureus, including 16 (1.4%) MRSA isolates. The overall prevalence of MDRSA carriage was 2.8% (32/1162).
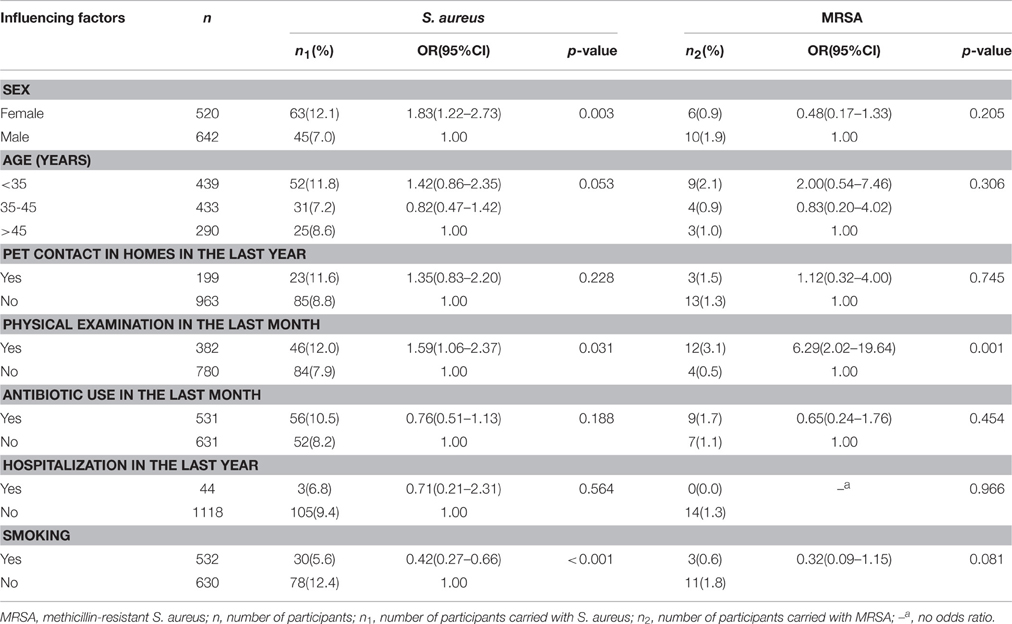
Table 1. Influencing factors of nasal carriage of S. aureus and MRSA among the 1162 study participants.
Influencing Factors Associated with S. aureus and MRSA carriage
The prevalence of S. aureus carriage was higher in females than in males (12.1% vs. 7.0%, OR = 1.83, 95% CI = 1.22–2.73). The prevalence of S. aureus carriage was lower in smokers than in no-smokers (5.6% vs. 12.4%; OR = 0.42, 95% CI = 0.27–0.66). Compared to individuals with no physical examinations, individuals with physical examinations experienced a significantly higher risk of S. aureus carriage (7.9% vs. 12.0%, OR = 1.59, 95% CI = 1.06–2.37) and MRSA carriage (0.5% vs. 3.1%; OR = 6.29, 95% CI = 2.02–19.64) (Table 1). In the multiple logistic regression model of sex, physical examination and smoking, there were significant differences in S. aureus carriage in terms of smoking status (adjusted OR = 0.45, 95% CI = 0.24–0.87) and physical examination (adjusted OR = 1.52, 95% CI = 1.01–2.31), but there was no significant difference in S. aureus carriage according to sex (adjusted OR = 1.06, 95% CI = 0.58–1.93).
Antimicrobial Susceptibility
The antimicrobial susceptibility among the 108 S. aureus isolates was based on CLSI (2013), with 92 (85.2%) classified as MSSA and 16 (14.8%) classified as MRSA (Table 2). Penicillin resistance was the most common phenotype observed in the MSSA (89.1%) and MRSA (100%) isolates. Compared with the MSSA isolates, the MRSA isolates had a higher risk of resistance to erythromycin (28.3% for MSSA vs. 62.5% for MRSA, p = 0.011), clindamycin (35.9% for MSSA vs. 68.8% for MRSA, p = 0.025), rifampin (3.3% for MSSA vs. 18.8% for MRSA, p = 0.041), and ciprofloxacin (3.3% for MSSA vs. 25.0% for MRSA, p = 0.009).
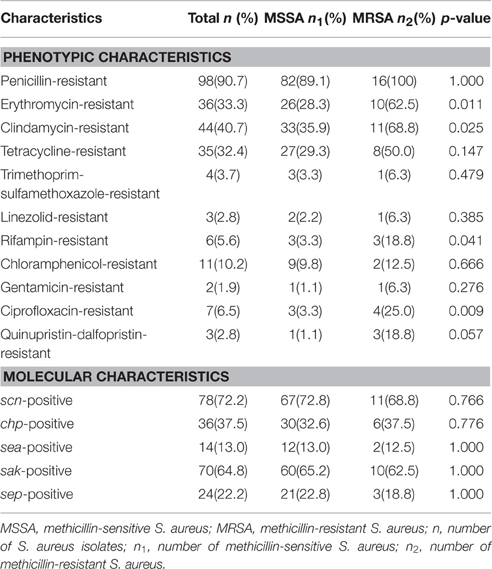
Table 2. Phenotypic and molecular characteristics of MSSA and MRSA carried by the study participants in Guangdong, China.
The proportion of MDRSA among all S. aureus isolates was 29.6% (32/108), including 17.4% (16/92) among the MSSA isolates. Among the 16 multidrug-resistant MSSA isolates, the most common pattern of multiple resistance was non-susceptibility to erythromycin, clindamycin, and tetracycline (68.8%, 11/16). Among the 16 MRSA isolates, the most common pattern of multiple resistance was non-susceptibility to cefoxitin, erythromycin, and clindamycin (62.5%, 10/16; Table 3).
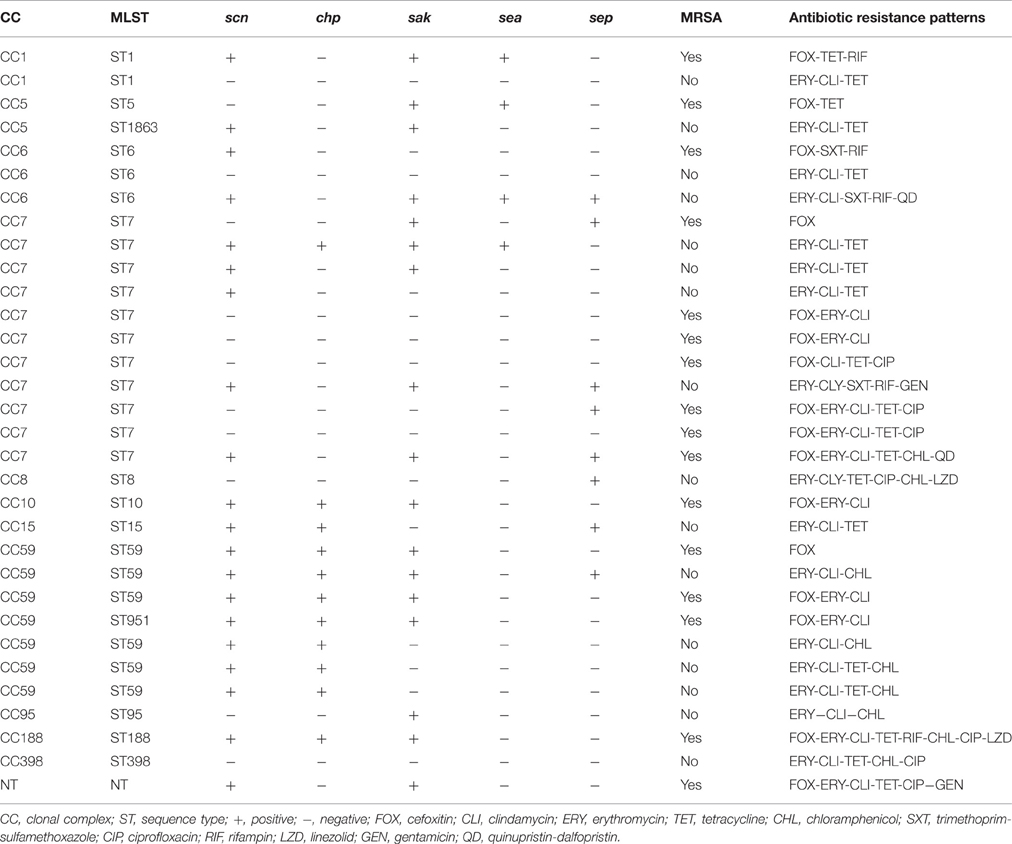
Table 3. Antibiotic susceptibility patterns of multidrug-resistant S. aureus isolates carried by the study participants in Guangdong, China.
MLST Typing and IEC Genes
Twenty unique STs belonging to 18 CCs were identified from 108 S. aureus (Figure 1), except for 5 untypeable isolates. ST7 (24/108, 22.2%), ST6 (16/108, 14.8%), ST188 (15/108, 13.8%), and ST59 (10/108, 9.3%) were the most prevalent STs. Note that ST1, ST5, ST6, ST7, ST10, ST59, ST188, and ST951 were found in both MRSA and MSSA isolates.
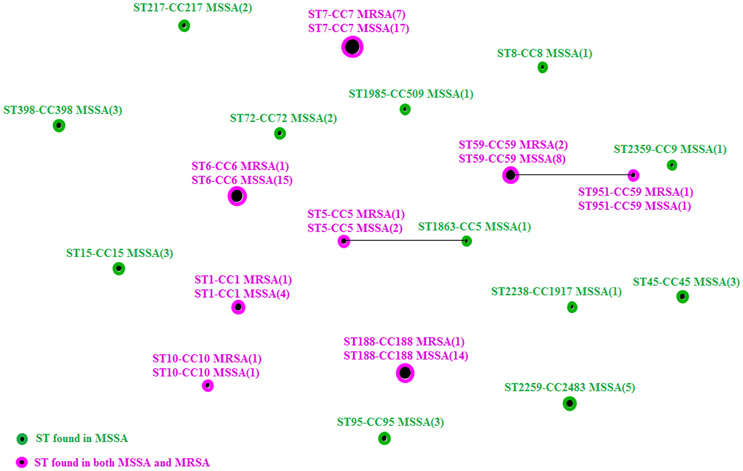
Figure 1. A diagram produced using the eBURST algorithm with the stringent (default) group definition based on the MLST data from this study, comparing the relationship between MSSA and MRSA isolates. (MSSA, methicillin-sensitive S. aureus; MRSA, methicillin-resistant S. aureus).
In terms of the IEC genes, scn was present in 78 (72.2%) of the S. aureus isolates, sak was in 70 (64.8%), chp was present in 36 (37.5%), sep was present in 24 (22.2%), and sea was present in 14 (13.0%) (Table 2). Note that the proportions of IEC genes were similar in the MSSA and MRSA isolates (Table 2). Notably, the IEC(sea and sep)-negative MSSA CC9 (ST2359) was found in a pet owner, and the IEC(scn, chp, sak, sea, and sep)-negative tetracycline-resistant MDRSA ST398 and the IEC-negative (scn, chp, and sep) tetracycline-resistant MRSA ST5 were found in participants without pet contact (Table 3).
Discussion
The results of the present study showed that 9.3% participants carried S. aureus, including 1.4% MRSA and 2.8% MDRSA, with the predominant multidrug-resistant pattern being non-susceptibility to erythromycin, clindamycin and tetracycline. The most common S. aureus genotypes were ST7, ST6, ST188, and ST59, with the predominant MRSA being ST7. Note that we found livestock-associated S. aureus isolates (IEC-negative CC9, IEC-negative tetracycline-resistant CC398, and IEC-negative tetracycline-resistant CC5) in participants with no occupational livestock contact, indicating a potential livestock-to-human transmission of livestock-associated S. aureus by non-occupational livestock contact.
S. aureus and MRSA nasal carriage in healthy adult population have been previously suggested to be approximately 10.7–37.1% and 0.2–8.6% (Gorwitz et al., 2008; Hamdan-Partida et al., 2010; Best et al., 2011; den Heijer et al., 2013; Wang et al., 2013; Yan et al., 2015), respectively, which is similar to our results (9.3% for S. aureus and 1.4% for MRSA). As reported in previous studies, the prevalences of S. aureus and MRSA were 10.7 and 1.9%, respectively, in Southeastern China (Wang et al., 2013) and 16.5 and 0.33%, respectively, in Northern China (Yan et al., 2015). Imparity results among healthy people in other countries have been recently reported. The carriage rates of S. aureus and MRSA were 30.4 and 1.2% in the United States (Gorwitz et al., 2008), 37.1 and 8.6% in Mexico (Hamdan-Partida et al., 2010), 18 and 0.2% in New Zealand (Best et al., 2011), and 21.6 and 0.3% in Europe (den Heijer et al., 2013), respectively. The prevalence of S. aureus in China is lower than that in American-European countries. There may be several reasons for this difference. In addition to geological and environmental factors, the prevalence of S. aureus can be influenced by study design (cross-sectional study or follow-up study), sampling sites (only anterior nares or multiple sites), screening and isolating methods for S. aureus (conventional biochemical methods or PCR test), and cultivation (enrichment or no enrichment).
Our study extends the previous knowledge in this area by identifying several predictors of S. aureus (including MRSA) carriage, which may help to inform clinical decision making when treating community-acquired infections. The finding that health care exposure (such as hospitalization, outpatient visit, surgery, and contact with healthcare workers) is significantly associated with MRSA carriage has been widely reported in the literature (Fritz et al., 2011; Rodriguez et al., 2014). Our study also indicated that physical examination was significantly associated with MRSA carriage. In addition, our study provides important evidence that smoking might be a protective factor against the nasal colonization of S. aureus, which aligns with previous studies (Wang et al., 2009; Olsen et al., 2012; Andersen et al., 2013). It might be that smoking creates a microenvironment in the nose that protects against the growth of S. aureus. A recent study conducted in northern Germany with a non-hospitalized adult population reported that males were more likely to carry S. aureus (Mehraj et al., 2014), indicating gender specific risk factors, which are not yet well understood but also align with observations from the USA, Norway and Denmark (Graham et al., 2006; Skråmm et al., 2011; Andersen et al., 2013). However, our study found no significant sex differences in S. aureus carriage after adjusting for smoking and physical examination, which is consistent with reports from Colombia (Rodriguez et al., 2014). Possible reasons for these findings include host genetics or human innate immune factors, hand-hygiene practice, smoking status, or vitamin D levels (Anderson et al., 2008; Johannessen et al., 2012; Olsen et al., 2012).
Infections caused by multidrug-resistant bacteria are associated with worse health outcomes and higher expenditures (Cardoso et al., 2012). However, few studies have examined the prevalence of MDRSA in healthy people. A recent study in North Carolina reported a 8.0% prevalence of MDRSA carriage among hog slaughter/processing plant workers compared with a 6.5% prevalence among household members, and the higher multidrug-resistant rate among workers may be associated with the use of multiple antimicrobials in hog feed (Neyra et al., 2014). In our study, the prevalence of multidrug-resistant S. aureus among health people was 2.8%, which was higher than a previous report of household members of livestock operation workers in America (0%) (Rinsky et al., 2013). Note that in our study, the most common pattern of multiple resistance among the participants with MSSA was non-susceptibility to erythromycin, clindamycin and tetracycline, and the most common pattern of multiple resistance among the participants with MRSA was non-susceptibility to cefoxitin, erythromycin and clindamycin, which aligned with a recent study of nine European countries (den Heijer et al., 2013). Erythromycin resistance mediates inducible macrolides, lincosamides and streptogramin B (MLSB), and phenotype erythromycin, together with the erm genes, encodes methylation of 23S rRNA binding, which is commonly shared by clindamycin drug classes (Weisblum et al., 1971; Schwarz et al., 2002). In collaboration, erythromycin resistance and clindamycin resistance are mutually intensified, which causes MRDSA isolates to appear. Therefore, future studies must direct more attention to exploring the specific discrepancy of multiple resistance pattern and resistant genes among S. aureus species.
In our study, the most common S. aureus genotypes were ST7, ST6, ST188, and ST59, with ST7 as the predominant MRSA. Previously seldom noted in Chinese isolates, ST7 was the major clone among S. aureus isolates associated with skin and soft tissue infections (Yu et al., 2015). Notably, in a study of invasive community-acquired S. aureus infections in Chinese children, more than half of the ST7 isolates were associated with multi-site infections (Qiao et al., 2014). ST59 is a common genetic background among CA-MRSA isolates in Asian-Pacific populations, including children in Taiwan (Wang et al., 2004), outpatients in Japan (Yamaguchi et al., 2015), and skin and soft tissue infection patients in Australia (Coombs et al., 2010), but it has also been recently observed in healthy Spanish carriers (Argudin et al., 2013) and homeless youths in San Francisco (Pan et al., 2005).
It is well known that S. aureus CC398 and CC9 isolates colonize livestock and can spread to humans. Recent studies of S. aureus CC398 indicated that the best genetic marker for human-associated S. aureus (including MRSA) CC398 was positive for φ3 bacteriophage carrying IEC genes chp and scn, while the best genetic marker for livestock-associated S. aureus CC398 was positive for tet(M) (McCarthy et al., 2011, 2012). In addition, a follow-up study of American industrial hog operation workers reported that scn-negative S. aureus primarily belonged to CC398 and CC9 and 82% of scn-negative S. aureus isolates were tetracycline-resistant, but no scn-positive S. aureus demonstrated tetracycline resistance (Nadimpalli et al., 2015). In our study, the livestock-associated IEC-negative tetracycline-susceptible MSSA CC9 (ST2359) was found in a participant with pet contact, and the livestock-associated IEC-negative tetracycline-resistant MDRSA ST398 was found in a participant without pet contact. Therefore, our findings support growing concern about the potential livestock-to-human transmission of livestock-associated S. aureus by non-occupational livestock contact.
Several studies have indicated that S. aureus ST5 isolates are relatively common in the swine industry in America and European countries (Frana et al., 2013; Hau et al., 2015; Linhares et al., 2015). In contrast to ST398 and ST9, the presence of ST5 in swine has raised additional public health concerns because ST5 is among the most prevalent lineages causing clinical infections in humans (Hau et al., 2015). The prominence of ST5 in clinical disease is believed to result from the acquisition of bacteriophages containing virulence and IEC genes. Previous studies have revealed that none of the swine isolates contained IEC genes (scn and sak), while these genes were present in 90% of the isolates from humans with no swine contact (Hau et al., 2015). Additionally, MRSA ST5 isolates of chicken origin were tetracycline resistant and had no IEC genes (sea, sak, scn, and chp) (Monecke et al., 2013). Similarly, the livestock-associated IEC-negative (scn, chp, and sep) tetracycline-resistant MRSA ST5 was found in a participant with no pet contact in our study, and some possible modes of exposure, including contact with air or waste releases from livestock farms, contact with contaminated meats and animals, and person-to-person contact, could not be ruled out.
This study contributed to the literature by exploring the potential relationship between genetic lineages (ST types) of S. aureus isolates and antibiotic resistance patterns and IEC genes. However, some limitations should be considered when interpreting our results. First, this study was conducted at only one time point (using a cross-sectional design), thus, we could not understand whether the carriage of MRSA among healthy people was transient or persistent. At the same time, we can only describe associations between influencing factors and MRSA carriage, not a causal conclusion. The results from this study must be confirmed and improved in a longitudinal study. Second, only the anterior nares were used as a sampling site, which may underestimate the true prevalence of S. aureus and MRSA. However, these measures have been widely used in previous similar studies in other countries, and the results were consistent with previous studies, as noted above. In addition, in this large population study, to simplify the collection of microbial samples and to compare the results with other similar studies, we chose to limit the data collection to nasal swabs. Third, a novel mecA homolog denominated mecC has been found in S. aureus lineages typically associated with cattle (i.e., CC130, CC1943, and C425). Although screening MRSA using cefoxitin allows the identification of mecC-MRSA, further studies are need to identify the mecC gene among S. aureus being phenotypic MRSA but negative for mecA.
In conclusion, this study provides insight into the phenotypic and genotypic determinants of S. aureus (including MRSA and MDRSA) isolates from healthy adults, and finds a potential relationship between S. aureus CCs and IEC genes and antibiotic resistance patterns in defining livestock-associated S. aureus in humans. The livestock-associated S. aureus isolates were found in participants with no occupational livestock contact; therefore, these findings support growing concern about the potential livestock-to-human transmission of livestock-associated S. aureus by non-occupational livestock contact.
Author Contributions
YF and XW carried out the samplings, the molecular genetic studies and drafted the manuscript. YF, XW, and LL participated to the samplings and performed the experiments. ZY participated in the design of the study. XY and SC coordinated the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81602901) and the Innovation and Strong School Project of Guangdong Pharmaceutical University (No. 2014KQNCX138). The funders played no role in the study design or data collection, analysis, and interpretation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Andersen, P. S., Larsen, L. A., Fowler, V. J., Stegger, M., Skov, R. L., and Christensen, K. (2013). Risk factors for Staphylococcus aureus nasal colonization in Danish middle-aged and elderly twins. Eur. J. Clin. Microbiol. Infect. Dis. 32, 1321–1326. doi: 10.1007/s10096-013-1882-0
Anderson, M. E., Lefebvre, S. L., and Weese, J. S. (2008). Evaluation of prevalence and risk factors for methicillin-resistant Staphylococcus aureus colonization in veterinary personnel attending an international equine veterinary conference. Vet. Microbiol. 129, 410–417. doi: 10.1016/j.vetmic.2007.11.031
Argudin, M. A., Argumosa, V., Mendoza, M. C., Guerra, B., and Rodicio, M. R. (2013). Population structure and exotoxin gene content of methicillin-susceptible Staphylococcus aureus from Spanish healthy carriers. Microb. Pathog. 54, 26–33. doi: 10.1016/j.micpath.2012.09.001
Best, N., Fraser, J. D., Rainey, P. B., Roberts, S. A., Thomas, M. G., and Ritchie, S. R. (2011). Nasal carriage of Staphylococcus aureus in healthy Aucklanders. N.Z. Med. J. 124, 31–39.
Cardoso, T., Ribeiro, O., Aragao, I. C., Costa-Pereira, A., and Sarmento, A. E. (2012). Additional risk factors for infection by multidrug-resistant pathogens in healthcare-associated infection: a large cohort study. BMC Infect. Dis. 12:375. doi: 10.1186/1471-2334-12-375
Chuang, Y. Y., and Huang, Y. C. (2013). Molecular epidemiology of community-associated methicillin-resistant Staphylococcus aureus in Asia. Lancet Infect. Dis. 13, 698–708. doi: 10.1016/S1473-3099(13)70136-1
Chuang, Y. Y., and Huang, Y. C. (2015). Livestock-associated methicillin-resistant Staphylococcus aureus in Asia: an emerging issue? Int. J. Antimicrob. Agents 45, 334–340. doi: 10.1016/j.ijantimicag.2014.12.007
Clinical Laboratory Standards Institute (2013). Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Third Informational Supplement. CLSI document M100-S23. Wayne, PA: CLSI.
Coombs, G. W., Monecke, S., Ehricht, R., Slickers, P., Pearson, J. C., Tan, H. L., et al. (2010). Differentiation of clonal complex 59 community-associated methicillin-resistant Staphylococcus aureus in Western Australia. Antimicrob. Agents Chemother. 54, 1914–1921. doi: 10.1128/AAC.01287-09
Cuny, C., Abdelbary, M., Layer, F., Werner, G., and Witte, W. (2015). Prevalence of the immune evasion gene cluster in Staphylococcus aureus CC398. Vet. Microbiol. 177, 219–223. doi: 10.1016/j.vetmic.2015.02.031
David, M. Z., and Daum, R. S. (2010). Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 23, 616–687. doi: 10.1128/CMR.00081-09
DeLeo, F. R., Otto, M., Kreiswirth, B. N., and Chambers, H. F. (2010). Community-associated methicillin-resistant Staphylococcus aureus. Lancet 375, 1557–1568. doi: 10.1016/S0140-6736(09)61999-1
den Heijer, C. D., van Bijnen, E. M., Paget, W. J., Pringle, M., Goossens, H., Bruggeman, C. A., et al. (2013). Prevalence and resistance of commensal Staphylococcus aureus, including methicillin-resistant S aureus, in nine European countries: a cross-sectional study. Lancet Infect. Dis. 13, 409–415. doi: 10.1016/S1473-3099(13)70036-7
Deurenberg, R. H., and Stobberingh, E. E. (2008). The evolution of Staphylococcus aureus. Infect. Genet. Evol. 8, 747–763. doi: 10.1016/j.meegid.2008.07.007
Frana, T. S., Beahm, A. R., Hanson, B. M., Kinyon, J. M., Layman, L. L., Karriker, L. A., et al. (2013). Isolation and characterization of methicillin-resistant Staphylococcus aureus from pork farms and visiting veterinary students. PLoS ONE 8:e53738. doi: 10.1371/journal.pone.0053738
Fritz, S. A., Krauss, M. J., Epplin, E. K., Burnham, C. A., Garbutt, J., Dunne, W. M., et al. (2011). The natural history of contemporary Staphylococcus aureus nasal colonization in community children. Pediatr. Infect. Dis. J. 30, 349–351. doi: 10.1097/INF.0b013e3181fe075e
Gorwitz, R. J., Kruszon-Moran, D., McAllister, S. K., McQuillan, G., McDougal, L. K., Fosheim, G. E., et al. (2008). Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001-2004. J. Infect. Dis. 197, 1226–1234. doi: 10.1086/533494
Graham, P. R., Lin, S. X., and Larson, E. L. (2006). A U.S. population-based survey of Staphylococcus aureus colonization. Ann. Intern. Med. 144, 318–325. doi: 10.7326/0003-4819-144-5-200603070-00006
Hamdan-Partida, A., Sainz-Espuñes, T., and Bustos-Martínez, J. (2010). Characterization and persistence of Staphylococcus aureus strains isolated from the anterior nares and throats of healthy carriers in a Mexican community. J. Clin. Microbiol. 48, 1701–1705. doi: 10.1128/JCM.01929-09
Hau, S. J., Sun, J., Davies, P. R., Frana, T. S., and Nicholson, T. L. (2015). Comparative prevalence of immune evasion complex genes associated with beta-hemolysin converting bacteriophages in MRSA ST5 isolates from Swine, Swine facilities, humans with Swine contact, and humans with no Swine contact. PLoS ONE 10:e142832. doi: 10.1371/journal.pone.0142832
Johannessen, M., Sollid, J. E., and Hanssen, A. M. (2012). Host- and microbe determinants that may influence the success of S. aureus colonization. Front. Cell. Infect. Microbiol. 2:56. doi: 10.3389/fcimb.2012.00056
Ko, K. S., Lee, J. Y., Suh, J. Y., Oh, W. S., Peck, K. R., Lee, N. Y., et al. (2005). Distribution of major genotypes among methicillin-resistant Staphylococcus aureus clones in Asian countries. J. Clin. Microbiol. 43, 421–426. doi: 10.1128/JCM.43.1.421-426.2005
Linhares, L. L., Yang, M., Sreevatsan, S., Munoz-Zanzi, C. A., Torremorell, M., and Davies, P. R. (2015). The effect of anatomic site and age on detection of Staphylococcus aureus in pigs. J. Vet. Diagn. Invest. 27, 55–60. doi: 10.1177/1040638714559598
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
McCarthy, A. J., van Wamel, W., Vandendriessche, S., Larsen, J., Denis, O., Garcia-Graells, C., et al. (2012). Staphylococcus aureus CC398 clade associated with human-to-human transmission. Appl. Environ. Microbiol. 78, 8845–8848. doi: 10.1128/AEM.02398-12
McCarthy, A. J., Witney, A. A., Gould, K. A., Moodley, A., Guardabassi, L., Voss, A., et al. (2011). The distribution of mobile genetic elements (MGEs) in MRSA CC398 is associated with both host and country. Genome Biol. Evol. 3, 1164–1174. doi: 10.1093/gbe/evr092
Mehraj, J., Akmatov, M. K., Strömpl, J., Gatzemeier, A., Layer, F., Werner, G., et al. (2014). Methicillin-sensitive and methicillin-resistant Staphylococcus aureus nasal carriage in a random sample of non-hospitalized adult population in northern Germany. PLoS ONE 9:e107937. doi: 10.1371/journal.pone.0107937
Monecke, S., Ruppelt, A., Wendlandt, S., Schwarz, S., Slickers, P., Ehricht, R., et al. (2013). Genotyping of Staphylococcus aureus isolates from diseased poultry. Vet. Microbiol. 162, 806–812. doi: 10.1016/j.vetmic.2012.10.018
Nadimpalli, M., Rinsky, J. L., Wing, S., Hall, D., Stewart, J., Larsen, J., et al. (2015). Persistence of livestock-associated antibiotic-resistant Staphylococcus aureus among industrial hog operation workers in North Carolina over 14 days. Occup. Environ. Med. 72, 90–99. doi: 10.1136/oemed-2014-102095
Neyra, R. C., Frisancho, J. A., Rinsky, J. L., Resnick, C., Carroll, K. C., Rule, A. M., et al. (2014). Multidrug-resistant and methicillin-resistant Staphylococcus aureus (MRSA) in hog slaughter and processing plant workers and their community in North Carolina (USA). Environ. Health Perspect. 122, 471–477. doi: 10.1289/ehp.1306741
Olsen, K., Falch, B. M., Danielsen, K., Johannessen, M., Ericson, S. J., Thune, I., et al. (2012). Staphylococcus aureus nasal carriage is associated with serum 25-hydroxyvitamin D levels, gender and smoking status. The tromso staph and skin study. Eur. J. Clin. Microbiol. Infect. Dis. 31, 465–473. doi: 10.1007/s10096-011-1331-x
Pan, E. S., Diep, B. A., Charlebois, E. D., Auerswald, C., Carleton, H. A., Sensabaugh, G. F., et al. (2005). Population dynamics of nasal strains of methicillin-resistant Staphylococcus aureus–and their relation to community-associated disease activity. J. Infect. Dis. 192, 811–818. doi: 10.1086/432072
Price, L. B., Stegger, M., Hasman, H., Aziz, M., Larsen, J., Andersen, P. S., et al. (2012). Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. MBio 3:e00305–11. doi: 10.1128/mBio.00305-11
Qiao, Y., Ning, X., Chen, Q., Zhao, R., Song, W., Zheng, Y., et al. (2014). Clinical and molecular characteristics of invasive community-acquired Staphylococcus aureus infections in Chinese children. BMC Infect. Dis. 14:582. doi: 10.1186/s12879-014-0582-4
Rinsky, J. L., Nadimpalli, M., Wing, S., Hall, D., Baron, D., Price, L. B., et al. (2013). Livestock-associated methicillin and multidrug resistant Staphylococcus aureus is present among industrial, not antibiotic-free livestock operation workers in North Carolina. PLoS ONE 8:e67641. doi: 10.1371/journal.pone.0067641
Rodríguez, E. A., Correa, M. M., Ospina, S., Atehortúa, S. L., and Jiménez, J. N. (2014). Differences in epidemiological and molecular characteristics of nasal colonization with Staphylococcus aureus (MSSA-MRSA) in children from a university hospital and day care centers. PLoS ONE 9:e101417. doi: 10.1371/journal.pone.0101417
Schwarz, S., Kehrenberg, C., and Ojo, K. K. (2002). Staphylococcus sciuri gene erm(33), encoding inducible resistance to macrolides, lincosamides, and streptogramin B antibiotics, is a product of recombination between erm(C) and erm(A). Antimicrob. Agents Chemother. 46, 3621–3623. doi: 10.1128/AAC.46.11.3621-3623.2002
Skråmm, I., Moen, A. E., and Bukholm, G. (2011). Nasal carriage of Staphylococcus aureus: frequency and molecular diversity in a randomly sampled Norwegian community population. APMIS 119, 522–528. doi: 10.1111/j.1600-0463.2011.02758.x
Song, J. H., Hsueh, P. R., Chung, D. R., Ko, K. S., Kang, C. I., Peck, K. R., et al. (2011). Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: an ANSORP study. J. Antimicrob. Chemother. 66, 1061–1069. doi: 10.1093/jac/dkr024
Sung, J. M., Lloyd, D. H., and Lindsay, J. A. (2008). Staphylococcus aureus host specificity: comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology 154, 1949–1959. doi: 10.1099/mic.0.2007/015289-0
van Wamel, W. J., Rooijakkers, S. H., Ruyken, M., van Kessel, K. P., and van Strijp, J. A. (2006). The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 188, 1310–1315. doi: 10.1128/JB.188.4.1310-1315.2006
Verkaik, N. J., Benard, M., Boelens, H. A., de Vogel, C. P., Nouwen, J. L., Verbrugh, H. A., et al. (2011). Immune evasion cluster-positive bacteriophages are highly prevalent among human Staphylococcus aureus strains, but they are not essential in the first stages of nasal colonization. Clin. Microbiol. Infect. 17, 343–348. doi: 10.1111/j.1469-0691.2010.03227.x
Wang, C. C., Lo, W. T., Chu, M. L., and Siu, L. K. (2004). Epidemiological typing of community-acquired methicillin-resistant Staphylococcus aureus isolates from children in Taiwan. Clin. Infect. Dis. 39, 481–487. doi: 10.1086/422642
Wang, J. T., Liao, C. H., Fang, C. T., Chie, W. C., Lai, M. S., Lauderdale, T. L., et al. (2009). Prevalence of and risk factors for colonization by methicillin-resistant Staphylococcus aureus among adults in community settings in Taiwan. J. Clin. Microbiol. 47, 2957–2963. doi: 10.1128/JCM.00853-09
Wang, T., Yang, Y., Chen, Y., Mei, Y., Gu, B., Pan, S., et al. (2013). Resistance analysis of the major opportunistic pathogens in community healthy adults in Nanjing, China. Chin. J. Clin. 7, 171–175.
Weisblum, B., Siddhikol, C., Lai, C. J., and Demohn, V. (1971). Erythromycin-inducible resistance in Staphylococcus aureus: requirements for induction. J. Bacteriol. 106, 835–847.
Woodford, N., and Livermore, D. M. (2009). Infections caused by Gram-positive bacteria: a review of the global challenge. J. Infect. 59(Suppl. 1), S4–S16. doi: 10.1016/s0163-4453(09)60003-7
Yamaguchi, T., Okamura, S., Miura, Y., Koyama, S., Yanagisawa, H., and Matsumoto, T. (2015). Molecular characterization of community-associated methicillin-resistant Staphylococcus aureus isolated from skin and pus samples of outpatients in Japan. Microb. Drug Resist. 21, 441–447. doi: 10.1089/mdr.2014.0153
Yan, X., Song, Y., Yu, X., Tao, X., Yan, J., Luo, F., et al. (2015). Factors associated with Staphylococcus aureus nasal carriage among healthy people in Northern China. Clin. Microbiol. Infect. 21, 157–162. doi: 10.1016/j.cmi.2014.08.023
Yu, F., Liu, Y., Lv, J., Qi, X., Lu, C., Ding, Y., et al. (2015). Antimicrobial susceptibility, virulence determinant carriage and molecular characteristics of Staphylococcus aureus isolates associated with skin and soft tissue infections. Braz. J. Infect. Dis. 19, 614–622. doi: 10.1016/j.bjid.2015.08.006
Zhang, K., Sparling, J., Chow, B. L., Elsayed, S., Hussain, Z., Church, D. L., et al. (2004). New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 42, 4947–4955. doi: 10.1128/JCM.42.11.4947-4955.2004
Keywords: community-acquired, livestock-associated, methicillin-resistant S. aureus, multidrug-resistant S. aureus
Citation: Fan Y, Wang X, Li L, Yao Z, Chen S and Ye X (2016) Potential Relationship between Phenotypic and Molecular Characteristics in Revealing Livestock-Associated Staphylococcus aureus in Chinese Humans without Occupational Livestock Contact. Front. Microbiol. 7:1517. doi: 10.3389/fmicb.2016.01517
Received: 04 August 2016; Accepted: 09 September 2016;
Published: 27 September 2016.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaCopyright © 2016 Fan, Wang, Li, Yao, Chen and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sidong Chen, Y2hlbnNpZG9uZzFAMTI2LmNvbQ==
Xiaohua Ye, c21hbGx0b21hdG9AMTYzLmNvbQ==
†These authors have contributed equally to this work.
 Yanping Fan†
Yanping Fan† Xiaolin Wang
Xiaolin Wang Ling Li
Ling Li Xiaohua Ye
Xiaohua Ye