- 1Department of Entomology, International Crops Research Institute for the Semi-Arid Tropics, Patancheru, India
- 2Department of Biochemistry, Gulbarga University, Kalaburagi, India
Bacillus thuringiensis toxin proteins are deployed in transgenic plants for pest management. The present studies were aimed at characterization of gut bacterial proteases involved in activation of inactive Cry1Ac protoxin (pro-Cry1Ac) to active toxin in Helicoverpa armigera. Bacterial strains were isolated from H. armigera midgut andscreened for their proteolytic activation toward pro-Cry1Ac. Among 12 gut bacterial isolates seven isolates showed proteolytic activity, and proteases from three isolates (IVS1, IVS2, and IVS3) were found to be involved in the proteolytic conversion of pro-Cry1Ac into active toxin. The proteases from IVS1, IVS2, and IVS3 isolates were purified to 11.90-, 15.50-, and 17.20-fold, respectively. The optimum pH and temperature for gut bacterial protease activity was 8.0 and 40°C. Maximum inhibition of total proteolytic activity was exerted by phenylmethane sulfonyl fluoride followed by EDTA. Fluorescence zymography revealed that proteases from IVS1, IVS2, and IVS3 were chymotrypsin-like and showing protease band at ~15, 65, and 15 kDa, respectively. Active Cry1Ac formed from processing pro-Cry1Ac by gut bacterial proteases exhibited toxicity toward H. armigera. The gut bacterial isolates IVS1, IVS2, and IVS3 showed homology with B. thuringiensis (CP003763.1), Vibrio fischeri (CP000020.2), and Escherichia coli (CP011342.1), respectively. Proteases produced by midgut bacteria are involved in proteolytic processing of B. thuringiensis protoxin and play a major role in inducing pathogenicity of B. thuringiensis toxins in H. armigera.
Introduction
Cotton bollworm, Helicoverpa armigera (Hubner) (Lepidoptera: Noctuidae) is polyphagous pest. It is distributed all over the world, and feeds on more than 250 plant species, including the agriculturally important crops cotton, tomato, sunflower, corn, pigeonpea, chickpea, and vegetable and field crops (Sharma, 2005). Overdependence on insecticide use has not only resulted in development of insect resistance to insecticides, but also leaves harmful residues on food products. Therefore, plants expressing toxin genes produced from the bacteria Bacillus thuringiensis have been deployed on a large scale for controlling insect pests (Sharma et al., 2004; James, 2013).
Bacillus thuringiensis is an aerobic, gram-positive, spore-forming bacterium that produces delta-endotoxins (Schnepf et al., 1998; De Maagd et al., 2001). The B. thuringiensis formulation sprays have been widely used as a biopesticide for controlling the insect pests (Sanahuja et al., 2011). After ingestion by the larvae, the B. thuringiensis protoxin undergo proteolysis in midgut under the alkaline environment, the active toxin formed will binds to receptors on the midgut epithelium, and then active toxin inserted into the membrane, creates membrane pores and eventually results in cell lysis leading to death of insect (Grochulski et al., 1995; Aronson and Shai, 2001). Pest insects had gained resistance to B. thuringiensis toxins and the cause of resistance may be due to lack of major gut protease involved in protoxin cleavage (Oppert et al., 1997), variations in the expression pattern of midgut proteases (Keller et al., 1996; Karumbaiah et al., 2007), improper processing of protoxin by proteases (Li et al., 2004; Rajagopal et al., 2009), and reduced binding of the active toxin to the receptors on midgut epithelium (Wang et al., 2007; Nair et al., 2013). Resistance to B. thuringiensis toxins is also associated with changes in expression of receptors, glycosylphosphatidyl-inositol (GPI) anchored alkaline phosphatases (ALPs) (Fuentes et al., 2011), GPI anchored aminopeptidases-N (APN) (Tiewsiri and Wang, 2011), cadherin (CAD) (Fabrick et al., 2014), and ABC transporter loci (Baxter et al., 2011).
Some insects had gained resistance to B. thuringiensis in the absence of midgut bacteria. Midgut bacteria are required for B. thuringiensis pathogenicity, and there is an obligate association between B. thuringiensis subsp. kurstaki HD-1 and midgut microbiota in some insect species [e.g., Lymantria dispar (L.), Pieris rapae (L.), and Vanessa cardui (L.)] (Broderick et al., 2009). Elimination of gut bacteria by oral administration of antibiotics reduced the B. thuringiensis insecticidal activity in gypsy moth (Broderick et al., 2006) and in H. armigera (Paramasiva et al., 2014; Visweshwar et al., 2015), however, the B. thuringiensis toxicity is restored by inoculation of a gut-associated strain of Enterobacter in gypsy moth (Broderick et al., 2006). There are reports that midgut bacteria are not necessary for B. thuringiensis insecticidal activity in pink bollworm, Pectinophora gossypiella (Saunders) (Broderick et al., 2009) and Plutella xylostella (L.) (Raymond et al., 2009).
In earlier studies (Visweshwar et al., 2015), we found that the insecticidal activity of B. thuringiensis toxins was reduced in H. armigera larvae eliminated with gut bacteria using antibiotic cocktail. The present studies were therefore carried out to know whether the proteases produced from H. armigera gut bacteria are involved in proteolytic processing of pro-Cry1Ac to active toxin and also to understand the role of gut bacterial proteases in inducing Cry1Ac toxicity in H. armigera.
Materials and Methods
Insect Culture
The H. armigera larvae were reared on chickpea based artificial diet under controlled laboratory conditions [Temperature at 26 ± 1°C, 60–70% relative humidity, and photoperiod of 16:8 h (L:D)] (Chitti Babu et al., 2014) in the insect rearing laboratory at the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Patancheru, Telangana, India.
Isolation of Gut Bacteria form H. armigera Larvae
The neonates were reared on artificial diet till they attain fourth-instar. The healthy early fourth-instar larvae were kept for starvation for 3 h, then surface sterilized with distilled water fallowed by 70% ethanol. Larvae were transferred to Petri plates containing paraffin, immobilized with surgical pins, covered with sterile water, and dissected using surgical blade. All the dissecting instruments were autoclaved before use. Only midgut portions were collected in sterile 0.1 M phosphate buffer, pH 7.0, homogenized and serially diluted aseptically. Different dilutions (10-3, 10-5, and 10-7) were inoculated on half strength tryptic soya-agar (TSA) media by spread plate method. The media was incubated at 30°C for 48 h. Total 10 larvae were used for isolation of midgut and all the steps were performed under sterile conditions in laminar air flow hood.
Determination of Proteolytic Activity in the Bacterial Culture
To determine the bacterial proteolytic activity, single bacterial colony on TSA media was selected and inoculated on skim milk agar media (0.5% skim milk and 1.5% agar). The inoculated media was incubated at 30°C for 24 h. The bacteria with proteolytic activity formed a clear zone around the inoculum. Protease producing bacterial colonies were picked for further characterization.
Production of Proteases from Bacterial Culture
The midgut bacteria showing proteolytic activity were selected and screened for the protease production on Glucose-Yeast-Peptone (GYP) media (Kumar et al., 2002) [glucose (10 g/l), yeast extract (5 g/l), peptone (5 g/l), MgSO4 7H2O (0.2 g/l), K2HPO4 (1 g/l), pH 9.5]. GYP broth was inoculated with 10% (v/v) of 24 h old seed culture prepared in nutrient broth and the culture was incubated at 30°C for 48 h. The culture was centrifuged at 10,000 rpm for 15 min, pellet was discarded and supernatant used as protease source.
Gut Bacterial Protease Activity Assay
Total protease activity in gut bacteria was determined by using azocasein (Sigma-Aldrich, India) as a substrate (Vinod et al., 2010; Visweshwar et al., 2015). The culture extract (50 μl) was mixed with the 500 μl substrate (1% azocasein in 0.1 M glycine-NaOH buffer, pH 8.0) and incubated for 50 min. Then 200 μl of 5% trichloroaceticacid (TCA) was added and centrifuged at 10000 rpm for 10 min at 25°C. To the supernatant, equal volume of 1 N NaOH was added and absorbance recorded at 450 nm. Units for total protease activity (UA) were calculated by using the equation units activity (UA) = ABS450 nm/[time (min) x volume of enzyme (ml)].
Purification of Gut Bacterial Proteases
The midgut bacterial culture supernatant was partially purified by salt precipitation. Ammonium sulfate was dissolved slowly in culture supernatant to achieve 75% saturation. After 16 h, the precipitate was recovered by centrifugation at 10,000 × g for 20 min. The precipitate was dissolved in 10 mM Tris-HCl buffer (pH 8.0), and dialyzed overnight against the same buffer. The dialyzed enzyme preparation was applied on diethylaminoethyl-cellulose (DEAE-cellulose) (Sigma-Aldrich, India) column (50 cm × 2 cm) pre-equilibrated with 10 mM Tris-HCl (pH 8.0) and un-adsorbed protein fractions were eluted with the same buffer. The enzyme was eluted with a gradient of 0.1–1 M NaCl in the same buffer at a flow rate of 1 ml/min. In each eluted fraction, total protease activity was determined using azocasein as substrate. The active fractions containing protease activity were pooled, concentrated by Amicon® Pro Affinity Concentrator (Millipore, USA) and then loaded on Sephadex G-75 (Sigma-Aldrich, India) column (90 cm × 2 cm), previously equilibrated with 0.2 M phosphate buffer (pH 7.5) and developed at a flow rate of 0.5 ml/min. Five milliliter fractions were eluted with the same buffer and fractions containing protease activity were pooled, concentrated to 2 ml by Amicon® Pro Affinity Concentrator. The protein concentration was quantified using bovine serum albumin (BSA) as standard protein (Lowry et al., 1951).
Bacterial Proteases in Protoxin–Toxin Conversion
Pro-Cry1Ac was isolated by fermenting B. thuringiensis subsp. kurstaki 4D4 strain (supplied by Daniel R. Zeigler, Bacillus Genetic Stock Center, Ohio State University, Columbus, OH, USA) at 30°C in Luria-Bertaini (LB) medium (Shao et al., 1998). After 48 h, the fermented medium was collected and kept at 4°C until cells lysis. The spore and crystal mixture were precipitated by centrifugation at 10,000 g × 10 min, the resultant pellet was washed three times with 1 M NaCl to remove the endogenous proteases, followed with distilled water. Delta-endotoxins were selectively dissolved in 2% β-mercaptoethanol-NaOH buffer, pH 10.7, and centrifuged at 10,000 g × 20 min under 4°C. Pro-Cry1Ac was dissolved in the supernatant. Acetic acid (2 mM) was added to the supernatant to adjust pH 4.4, and then centrifuged at 10,000 g for 30 min. The precipitate of pro-Cry1Ac was collected, dialyzed against water, lyophilized, and stored at -20°C.
Gut bacterial proteases were incubated with pro-Cry1Ac for 1 h at room temperature and the samples were electrophoresed on sodium dodecyl polyacrylamide gel electrophoresis (SDS-PAGE) using 10% (w/v) running gel (Laemmli, 1970) under reducing conditions. The same samples were subjected to ELISA using B. thuringiensis-Cry1Ac ELISA kit (Agdia®, India) for quantification of active Cry1Ac. For immunodetection, the proteins were separated by SDS-PAGE as described above and electroblotted onto a nitrocellulose membrane in electrophoretic transfer buffer (48 mM Tris base, 39 mM glycine, 20% methanol and 1.3 mM SDS, pH 9.2). The membrane was blocked in PBST (155 mM NaCl, 1.1 mM KH2PO4, 3.0 mM K2HPO4 7H2O, pH 7.4, 0.25% Tween-20) plus 5% skimmed milk for 1 h at room temperature and then incubated with primary antibody in PBST overnight at 4°C. The primary antibody used for Western blot analyses was monoclonal mouse anti-Cry1Ac antibody [1:1000 (v/v); RDI, USA]. The membranes were then washed three times with PBST, followed by incubation with horseradish peroxidase-conjugated secondary antibody [1:4000 (v/v); Agdia®, India] in PBST. The immunoreactivity was detected by incubating the membrane in PBS containing 0.06% (w/v) diaminobenzidine, 0.018% NiCl2 (w/v), and 0.3% H2O2 (v/v).
Effect of pH and Temperature on the Activity of Gut Bacterial Proteases
Effect of pH on gut bacterial proteases was determined by incubating the proteases with substrate azocasein at varying pH values ranging from 2.0 to 12.0, with intervals of 2 units using 100 mM aconitate buffer for pH 2.0–5.0, 100 mM sodium phosphate buffer for pH 6.0–7.0 and 100 mM glycine-NaOH for pH 8.0–12.0. The influence of temperature on gut bacterial protease was studied by incubating the proteases with the substrate azocasein at different temperatures ranging from 20 to 70°C, with intervals of 10°C for 10 min under the standard assay conditions. The total proteolytic activity was determined as described above.
Effect of Inhibitors on Protease Activity
For inhibitory assays, a suitable amount of inhibitor (5 mM) was pre-incubated with H. armigera gut bacterial protease extract (50 μl) for 30 min at room temperature. After incubation, total protease activity was determined by using azocasein as a substrate. Chymotrypsin activity was estimated by using substrate N-succinyl-alanine-alanine-proline-phenylalanine-p-nitroanilide (SAAPFpNA) (Sigma-Aldrich, India) (Li et al., 2004; Visweshwar et al., 2015). One unit of enzyme activity was defined as the amount of enzyme catalyzing the hydrolysis of 1 μmol substrate per minute at 30°C. Protease inhibitors used were TPCK (N-tosyl-L-phenylalanine chloromethyl ketone), E64 [N-(N-(L-3-trans-corboxirane-2-carbonyl)-L-leucyl) agmatine], leupeptin, aprotonin, EDTA (ethylene diamine tetra acetic acid), and PMSF (phenylmethane sulfonyl fluoride). Residual protease activity in presence of inhibitors was presented as relative to activity in control (activity in the absence of protease inhibitor). All the protease inhibitors were purchased from Roche Diagnostics, Mumbai, India.
Fluorescence Zymogram Analysis of Midgut Bacterial Proteases
The gut bacterial proteases were subjected to 10% SDS-PAGE under non-reducing conditions. After electrophoresis, the gel was treated with 2.5% triton X-100 for 10 min to remove the SDS. The gel was incubated in trypsin assay buffer [50 mM Tris-HCl, pH 8.0, 10 mM CaCl2, 0.005% Triton X-100 and 50 μM N-tert-butoxycarbonyl-glutamine-alanine-arginine-7-amido-4-methyl coumarin (Boc-Gln-Ala-Arg-MCA)] (Sigma-Aldrich, India) or chymotrypsin assay buffer [50 mM Tris-HCl, pH 8.0, 10 mM CaCl2, 0.005% Triton X-100 and 50 μM N-succinyl-alanine-alanine-proline-phenylalanine-7-amido-4-methyl coumarin (Suc-Ala-Ala-Pro-Phe-MCA)] (Sigma-Aldrich, India) at 37°C for 30 min. The gel was then washed in distilled water and observed for fluorescent bands of trypsin and chymotrypsin activity in Gel Documentation System under UV-B transillumination (Bio-Rad Laboratories, Hercules, CA, USA) (385 nm = AMC group excitation wavelength) (Yasothornsrikul and Hook, 2000). In one lane, standard molecular weight markers were loaded to determine the molecular mass of the purified gut bacterial proteases. After electrophoresis, the lane was sliced and stained with Coomassie brilliant blue (R-250) and then de-stained.
Preparation of Active Cry1Ac Toxin
The purified crystal δ-endotoxin was solubilized in 0.1 M carbonate/dithiothreitol buffer containing 1 M sodium chloride and incubated with proteases (5:1 v/v) from midgut bacteria at 30°C for 60 min. Pro-Cry1Ac digestion was stopped with 1 mM PMSF and the samples were centrifuged at 16,000 g for 10 min. The toxin-containing supernatant was harvested and protein concentration was determined by the method of Lowry et al. (1951).
Elimination of Gut Bacteria from H. armigera
To eliminate the gut bacteria, eggs laid by H. armigera adults were surface sterilized by immersing in distilled water followed by 2% sodium hypochlorite and then allowed to hatch. Stock solutions of antibiotic cocktail were prepared using antibiotics, gentamycin, neomycin, chloramphenicol, ampicillin, streptomycin, rifampicin, and penicillin (Sigma-Aldrich, India) (stock: 0, 0.2, 0.4, 0.6, 0.8, 1.0 mg/ml each in distilled water). The neonates were released on sterilized artificial diet treated with 100 μl of antibiotic cocktail (from each stock solution) per gram of diet and the larvae were reared until they reach fourth-instar. The early fourth-instar larva was surface sterilized, midguts (10 no.) were removed aseptically and homogenized in sterile distilled water. The gut homogenates were serially diluted and spread on nutrient agar (NA) media and incubated at 30°C for 24 h. The microbial counts were converted into colony forming units per mg of whole gut. There were three replications for each treatment and 10 larvae in each replication in a completely randomized design (CRD).
Insect Bioassay
For insect bioassay, pro-Cry1Ac and Cry1Ac toxin produced after proteolytic processing of pro-Cry1Ac by three gut bacterial (IVS1, IVS2, and IVS3) proteases were used. Fifty microliters of pro-Cry1Ac and active Cry1Ac toxin (0.1, 0.3, 0.6, 1.2, 2.4, 4.8, and 9.6 μg) samples were coated uniformly to the surface (2 cm2) of an artificial diet (antibiotic cocktail untreated) and also to artificial diet pre-treated with 100 μl of antibiotic cocktail per gram diet (antibiotic cocktail stock: 1 mg/ml of each antibiotic) and allowed to dry. Each early fourth-instar larva reared on artificial diet alone and antibiotic cocktail treated diet was placed onto the surface of diet treated with toxin and diet treated with antibiotic cocktail plus toxin, respectively. The larvae were reared under optimal conditions (see Insect Culture). Insect mortality was scored after 5 days. There were three replications for each treatment and 10 larvae in each replication in a CRD. The LC50 values (effective concentration to kill 50% of the H. armigera larvae) were calculated using probit analysis (LC50 software program, version 1.5 developed by EPA; Finney, 1971).
To test whether midgut bacteria were contributing to Cry1Ac induced mortality directly, the early fourth-instar larvae pre-treated with antibiotic cocktail were transferred to diet treated with both pro-Cry1Ac and gut bacterial culture. There were three replications for each treatment and 10 larvae in each replication in a CRD. The mortality data was recorded after 5 days.
MALDI-TOF Mass Spectrometry
The selected protease band from IVS1, IVS2, and IVS3 isolates were excised from the Coomassie Blue stained gel, and washed thrice with water for 10 min, and stored in milli Q water. MALDI time-of-flight mass spectrometry (TOFMS) analysis of proteases were performed using a MALDI-tandem time-of-flight (TOF/TOF) mass spectrometer (Bruker Autoflex III Smartbeam; Bruker Daltonics, Bremen, Germany). Protein identification was done by database searches (PMF and MS/MS) using MASCOT program1 employing Biotools software (Bruker Daltonics) (Himabindu et al., 2013). The similarity search for mass values was done with existing digests and sequence information from NCBInr and Swiss Prot database. The other search parameters were: fixed modification of carbamidomethyl (C), variable modification of oxidation (M), enzyme trypsin, peptide charge of 1+ and monoisotropic. According to the MASCOT probability analysis (P < 0.05), only significant hits were accepted for protein identification. The multiple alignment was made using Clustal Omega software.
Characterization of Gut Protease Producing Bacterial Strains
The gut bacterial strain, involved in conversion of protoxin–toxin was characterized by biochemical and molecular methods. For biochemical characterization of the proteolytic strain, tests such as nitrate reduction, citrate utilization, H2S production, catalase, gelatin liquification, starch hydrolysis, and urease test were performed as per standard protocols (Hemaraj et al., 2013). Results were analyzed as per Bergey’s Manual of Systematic Bacteriology (Holding and Shewan, 1974).
Molecular characterization was carried out using 16S rRNA gene sequence analysis. DNA was isolated from a single bacterial colony using BiopureTM kits (Bioaxis DNA Research Centre, Hyderabad, India) for bacterial genomic DNA isolation. 16S rRNA gene was amplified using 16S universal primers (forward, AGAGTTTGATCCTGGCTCAG; reverse, ACGGCTACCTTGTTACGACTT). Forward and reverse sequencing reaction of PCR amplicon was carried out on ABI PRISM® 377 Genetic Analyzer to obtain the sequence. The sequences were deposited in the GenBank. Related sequences were obtained from the GenBank database (National Center for Biotechnology information) using BLAST. A phylogenetic tree was constructed by the neighbor-joining method using the MEGA 7.0 software (Kumar et al., 2016).
Results
Purification and Activity of Proteases Isolated from Midgut Bacteria of H. armigera
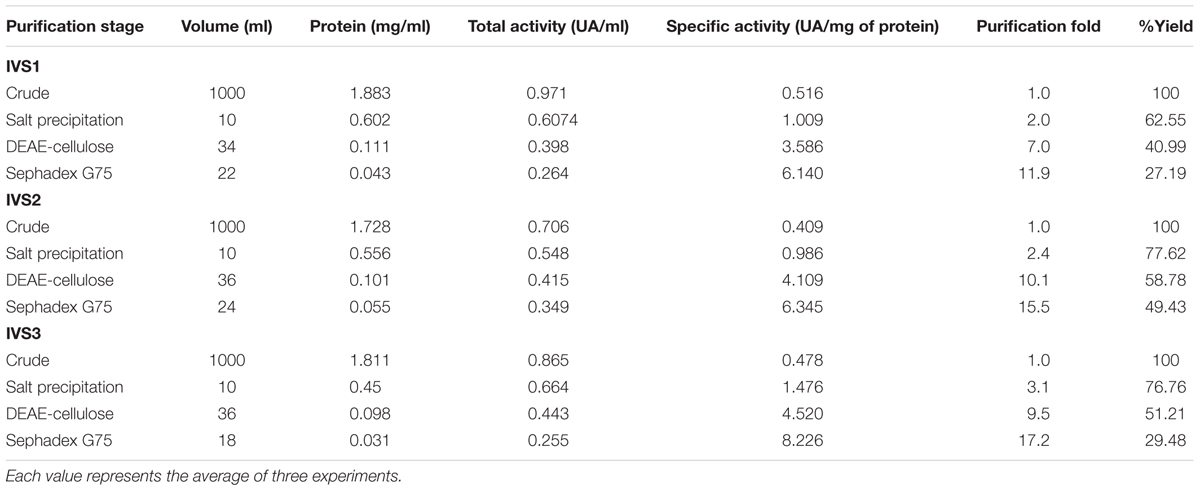
Twelve dominant microbial colonies were isolated from the midgut of H. armigera. Amongst these, only seven gut bacterial isolates exhibited protease activity, as indicated by formation of a clear zone around the inoculum on skim milk agar media, and these isolates were named as IVS-1 to 7 (Figure 1). The midgut bacterial proteases were salt precipitated and were further purified by ion-exchange and gel permeation chromatography (Table 1). Proteases from IVS1 isolate were purified to 27.19% yield with 11.90-fold increase in activity, and the specific activity was 6.14 UA/mg of protein. Proteases from IVS2 isolate were purified to 49.43% yield with 15.50-fold increase in activity. Specific activity of protease was 6.34 UA/mg of protein. Proteases from IVS3 isolate were purified to 29.48% yield with 17.20-fold increment in activity, and the specific activity was 8.22 UA/mg of protein (data has been provided for only three isolates of gut bacteria which were able to convert pro-Cry1Ac to active toxin).
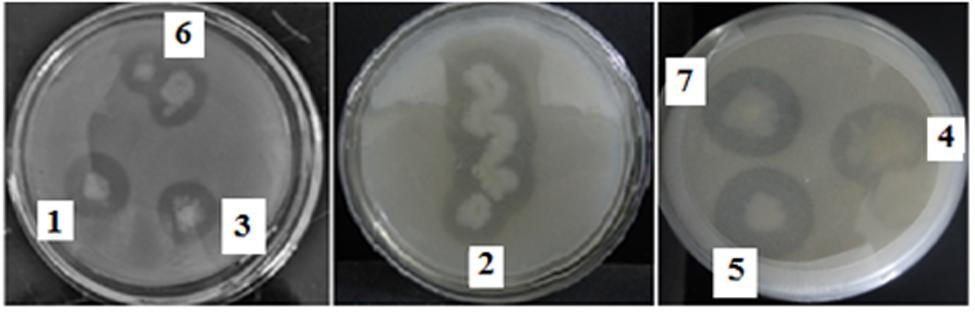
FIGURE 1. Screening of Helicoverpa armigera midgut bacterial isolates for protease activity. Gut bacterial isolates were inoculated on agar plate containing skimmed milk casein and incubated for 24 h at 30°C. The clear zone around the inoculum indicated the hydrolysis of skimmed milk casein by the proteases secreted by gut bacteria.
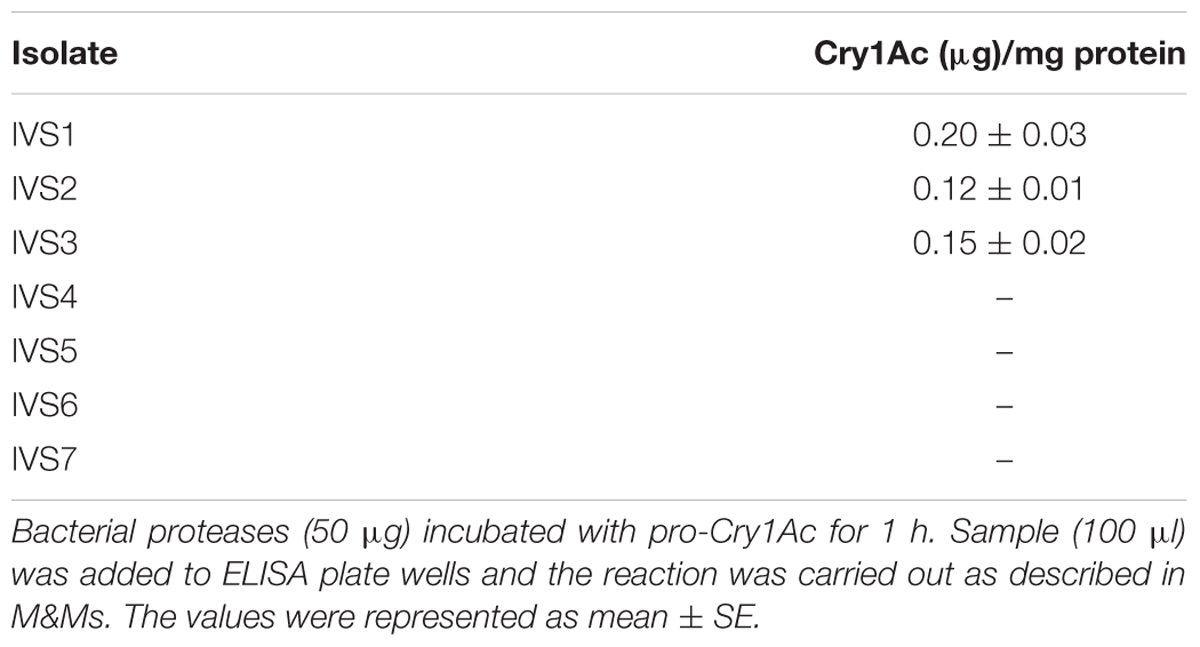
Purified proteases from seven protease producing gut bacterial isolates were incubated with pro-Cry1Ac for 1 h at room temperature. Proteases from only three isolates (IVS1, IVS2, and IVS3) were involved in proteolytic processing of pro-Cry1Ac which showed a band corresponding to active Cry1Ac toxin at 65 kDa on SDS-PAGE (Figure 2A), while remaining four isolates (IVS4, IVS5, IVS6, and IVS7) did not show any band correspond to 65 kDa. This was also confirmed by detecting the proteolytic samples for the presence of active toxin using ELISA (Table 2) and Western blot (Figure 2B).
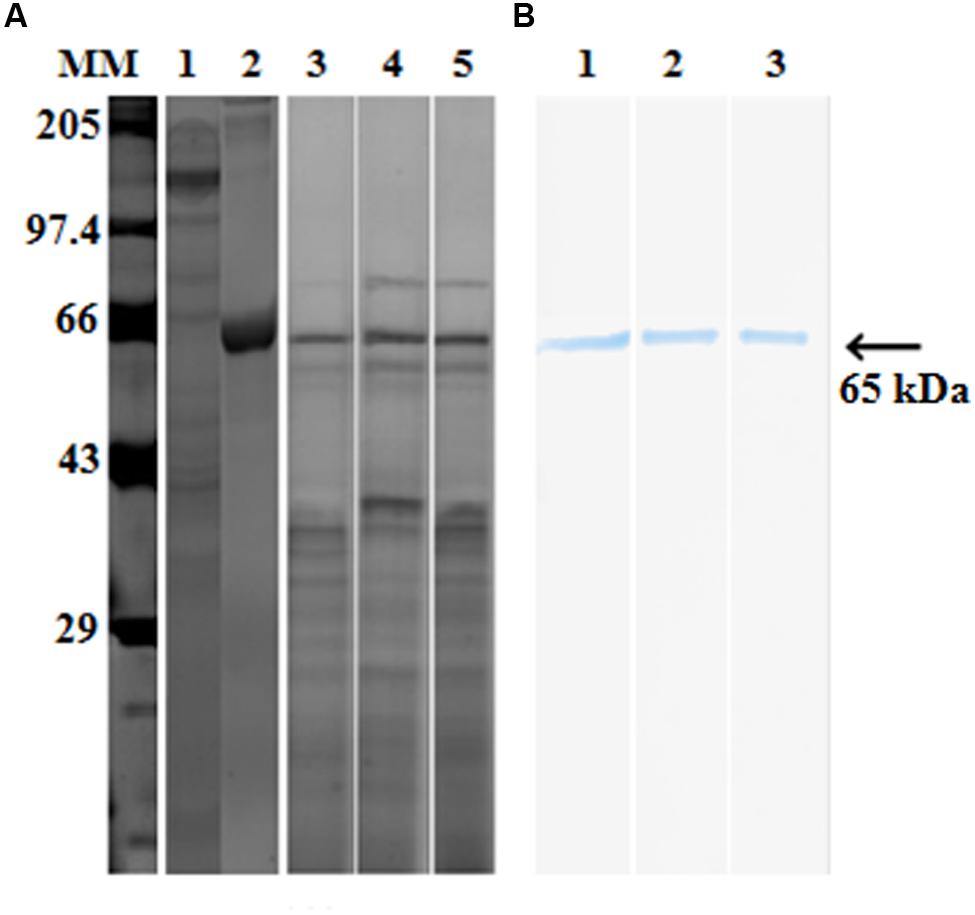
FIGURE 2. Proteolytic processing of pro-Cry1Ac to active toxin by midgut bacterial proteases. Proteases (30 μg) from midgut bacteria were incubated with pro-Cry1Ac (10 μg) for 1 h. Active Cry1Ac (band at 65 kDa) was detected on (A) SDS-PAGE [molecular markers (MMs), lane 1- pro-Cry1Ac, lane 2- active Cry1Ac, lane 3 – IVS1, lane 4 – IVS2 and lane 5 – IVS3] and (B) Western blot (lane 1- IVS1, lane 2- IVS2, and lane 3- IVS3).
Proteolytic gut bacterial isolates IVS1, IVS2, and IVS3 were characterized by biochemical and molecular methods. Microscopic observations showed that all the three bacterial isolates were rod shaped. Biochemical tests confirmed that all the three isolates showed negative results for H2S production, urease test, and citrate utilization test (Table 3). Based on the biochemical tests and 16S rRNA gene sequence analysis the three isolates, IVS1, IVS2, and IVS3 were identified as B. thuringiensis HD-789 (CP003763.1; 99% similarity), Vibrio fischeri ES114 (CP000020.2; 95% similarity) and Escherichia coli GM-4792 (CP011342.1; 99% similarity), respectively. The sequences have been deposited in GenBank under the accession numbers KT714122 (IVS1), KT714123 (IVS2), and KT714124 (IVS3) (The phylogenetic tree for IVS1, IVS2, and IVS3 isolates was provided as Supplementary Figure S1).

TABLE 3. Morphological and biochemical characterization of proteolytic bacterial isolates from Helicoverpa armigera.
Effect of pH and Temperature on Gut Bacterial Protease Activity
The H. armigera gut bacterial protease activity was observed over the pH range 2.0–12.0, with maximum activity in alkaline environment [pH 8.0 for proteases from IVS1 (61.8 UA/mg of protein), IVS2 (39.87 UA/mg of protein), and IVS3 (50.08 UA/mg of protein)] (Figure 3). These proteases had a relatively low activity in the acidic pH range from 2.0 to 6.0. After pre-incubating at different temperatures, i.e., 20, 30, 40, 50, 60, and 70°C at pH 8 for 10 min, the H. armigera gut bacterial proteases were found to be stable at a temperature range from 30 to 50°C, with optimum activity at 40°C for proteases from IVS1 (62.37 UA/mg of protein), IVS2 (43.21 UA/mg of protein), and IVS3 (51.99 UA/mg of protein) (Figure 4). A rapid decrease in enzyme activity was observed above 50°C.
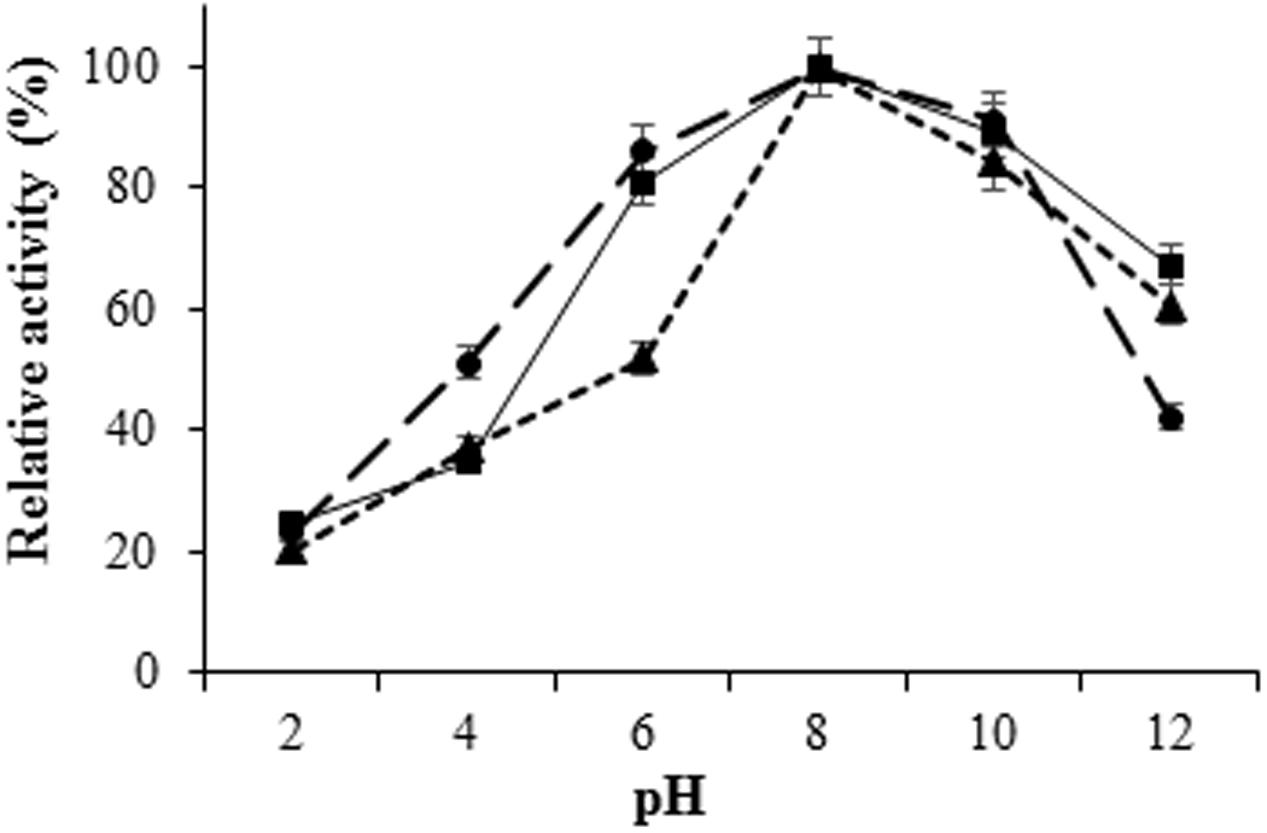
FIGURE 3. Effect of pH on activity of proteases from gut bacteria in H. armigera. The midgut bacterial proteases were incubated with azocasein at different pH ranging from 2.0 to 12.0 and the total protease activity was measured. Total protease activity in (■) IVS1, (▲) IVS2, and (●) IVS3 isolates was determined. Each value represents the average of three experiments.
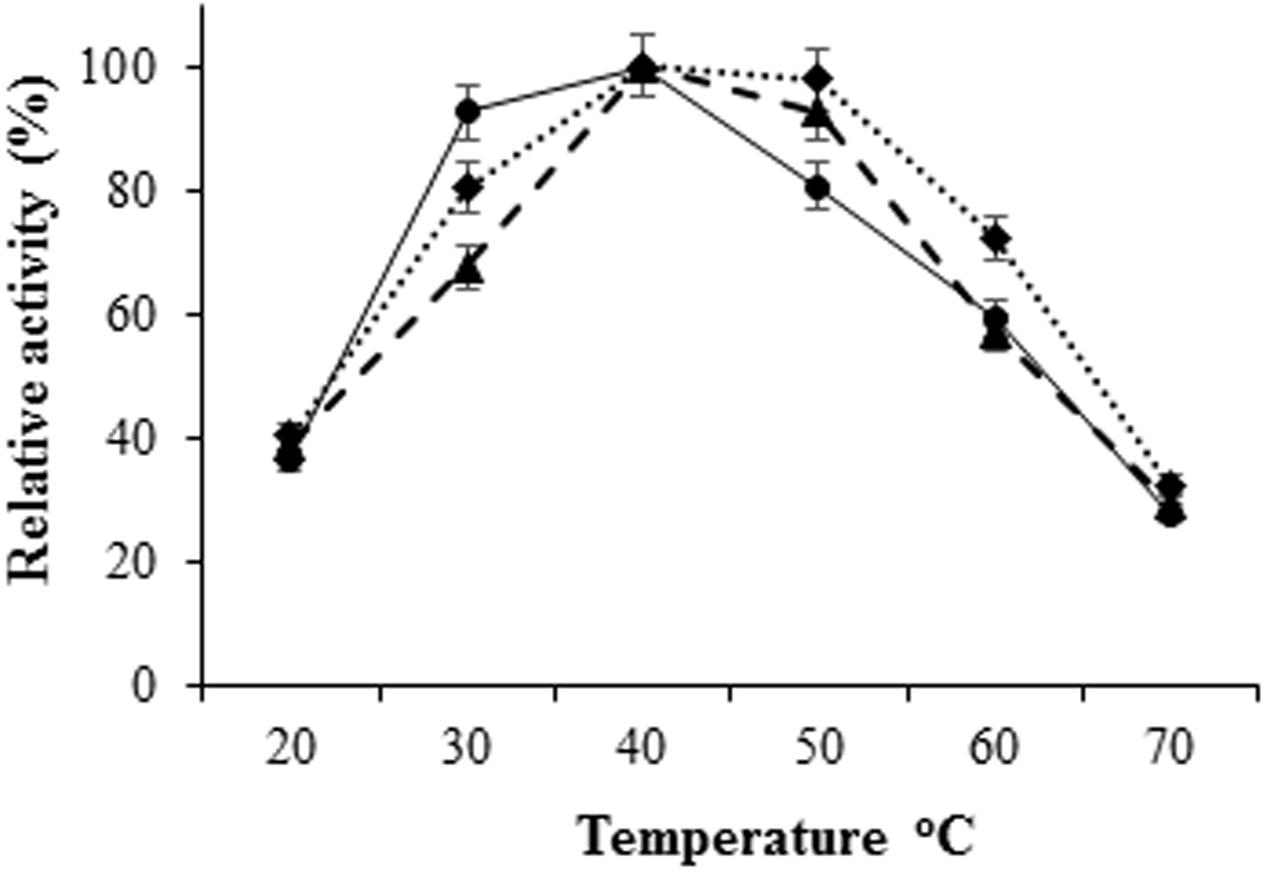
FIGURE 4. Effect of temperature on activity of proteases from gut bacteria in H. armigera. The bacterial proteases were incubated with azocasein dissolved in 0.1 M glycine-NaOH buffer at different temperatures ranging from 20 to 70°C. Total proteolytic activity was measured at 450 nm. Effect of temperature on proteases from (◆) IVS1, (●) IVS2, and (▲) IVS3 isolates was determined. Each value represents the average of three experiments.
Effect of Protease Inhibitors on Gut Bacterial Protease Activity
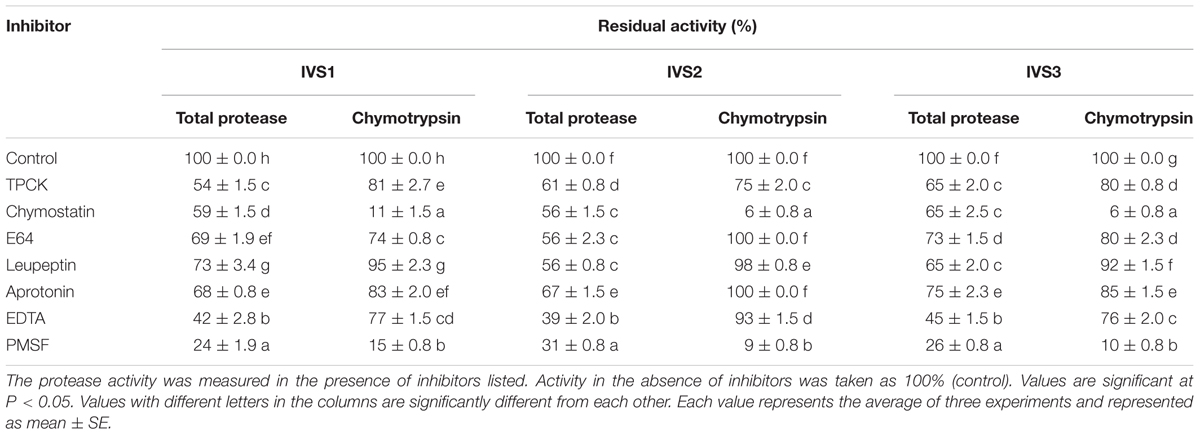
Phenylmethane sulfonyl fluoride exerted maximum inhibition of H. armigera gut bacterial total proteolytic activity, followed by EDTA (Table 4). PMSF inhibited 76, 69, and 74% of the total proteolytic activity in IVS1, IVS2, and IVS3 isolates, respectively. EDTA inhibited 58, 61, and 55% of the total proteolytic activity in IVS1, IVS2, and IVS3 isolates, respectively. Less than 50% inhibition of total proteolytic activity was seen in case of chymostatin (IVS1-41, IVS2-44, and IVS3-35%), E64 (IVS1-31, IVS2-44, and IVS3-27%), leupeptin (IVS1-27, IVS2-44, and IVS3-35%), and aprotonin (IVS1-32, IVS2-33, and IVS3-25%). Chymostatin and PMSF inhibited 88 and 85% of chymotrypsin activity in IVS1, 94 and 91% of chymotrypsin activity in IVS2 and 94 and 90% of chymotrypsin activity in IVS3.
Fluorescence Zymography of Bacterial Proteases
Fluorescent zymogram analysis performed for the detection of trypsin- and chymotrypsin-like proteases in gut bacterial isolates IVS1, IVS2, and IVS3. No band was observed on trypsin fluorescent zymogram, whereas, a single chymotrypsin-like band with molecular mass of ~15 kDa each in IVS1, and IVS3, and of ~65 kDa in IVS2 was visualized due to the liberated AMC (7-amino-4-methyl-coumarin) of the substrate, peptide-MCA, under UV transillumination (Figure 5).
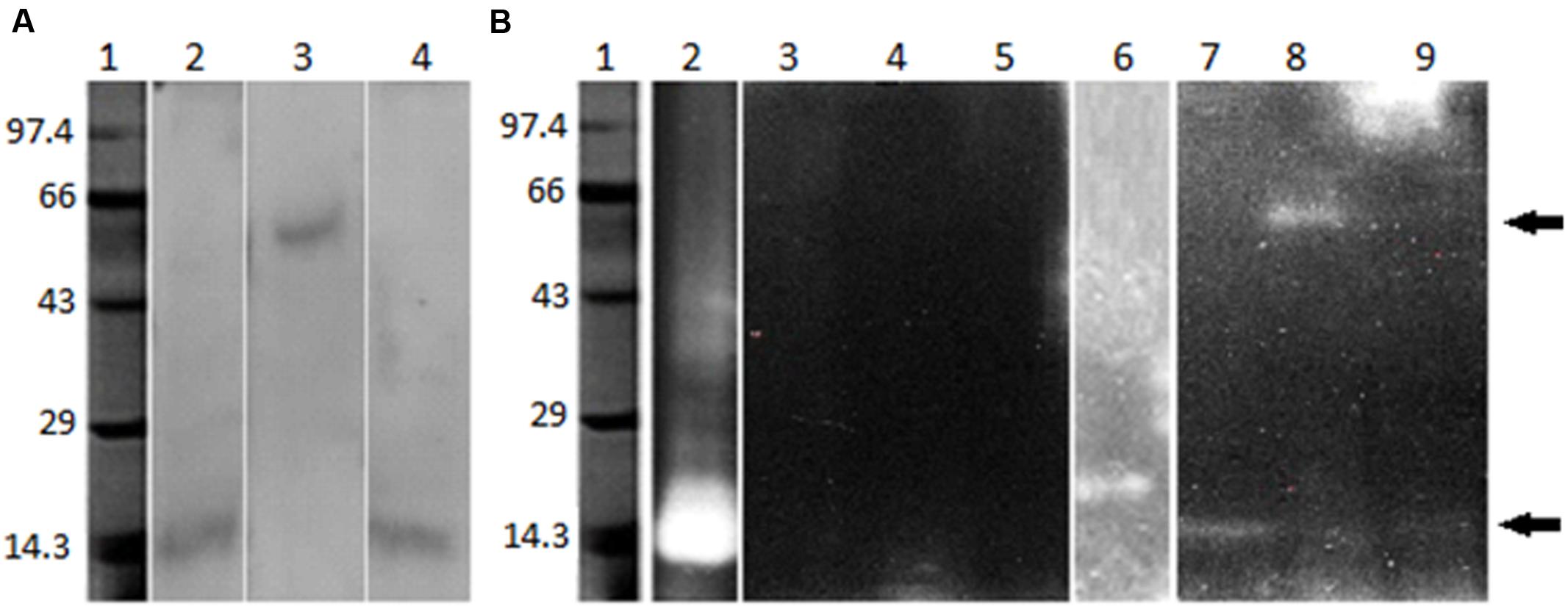
FIGURE 5. Visualization of midgut bacterial proteases. Sephadex G-75 column eluted bacterial protease (5 μg) were electrophoresed on 10% SDS-PAGE. After electrophoresis the gel was incubated in the trypsin or chymotrypsin assay buffer as described in text. Bacterial protease bands were visualized on (A) SDS-PAGE (Lane 1 – Molecular weight markers, lane 2 – proteases from IVS1, lane 3 – IVS2, lane 4 – IVS3). (B) Fluorescence zymography for the detection of trypsin- and chymotrypsin-like activity in the bacterial isolates (lane 1 – molecular weight markers, lane 2 – bovine trypsin, lane 3 – proteases from IVS1, lane 4 – IVS2, lane 5 – IVS3, lane 6 – bovine chymotrypsin, lane 7 – IVS1, lane 8 – IVS2 and lane 9 – IVS3. Lane 2 to 5 are for identification of trypsin proteases and 6 to 9 are for chymotrypsin proteases).
Insect Bioassay
Antibiotic cocktail was used to eliminate gut bacteria from H. armigera. As the concentration of antibiotic cocktail increased, the microbial colony count was decreased on nutrient agar culture plates. Among the five different concentrations tested, antibiotic cocktail at 80 and 100 μg/g diet inhibited all the gut bacteria (Supplementary Figure S2), and 100 μg of antibiotic cocktail per gram diet was further used to eliminate the gut bacteria from H. armigera larvae. Pro-Cry1Ac was treated with proteases from three gut bacterial isolates (IVS1, IVS2, and IVS3) and the active Cry1Ac toxin formed was used for insect bioassay. Significant increase in mortality rate was observed in larvae reared on pro-Cry1Ac treated diet [R2 = 84.96%; (LC50 = 5.38 μg, 95% confidence limits: lower = 2.72 and upper = 23.85)], and pro-Cry1Ac plus antibiotic cocktail treated diet [R2 = 83.70%; (LC50 = 9.85 μg, 95% confidence limits: lower = 4.34 and upper = 34.72)] (Figure 6A). Significant increase in mortality rate was observed in larvae reared on active Cry1Ac treated diet [R2 = 82.60%; (LC50 = 3.15 μg, 95% confidence limits: lower = 1.49 and upper = 12.86)], and active Cry1Ac plus antibiotic cocktail treated diet [R2 = 80.95%; (LC50 = 6.11 μg, 95% confidence limits: lower = 2.81 and upper = 44.43)] (Figure 6B).
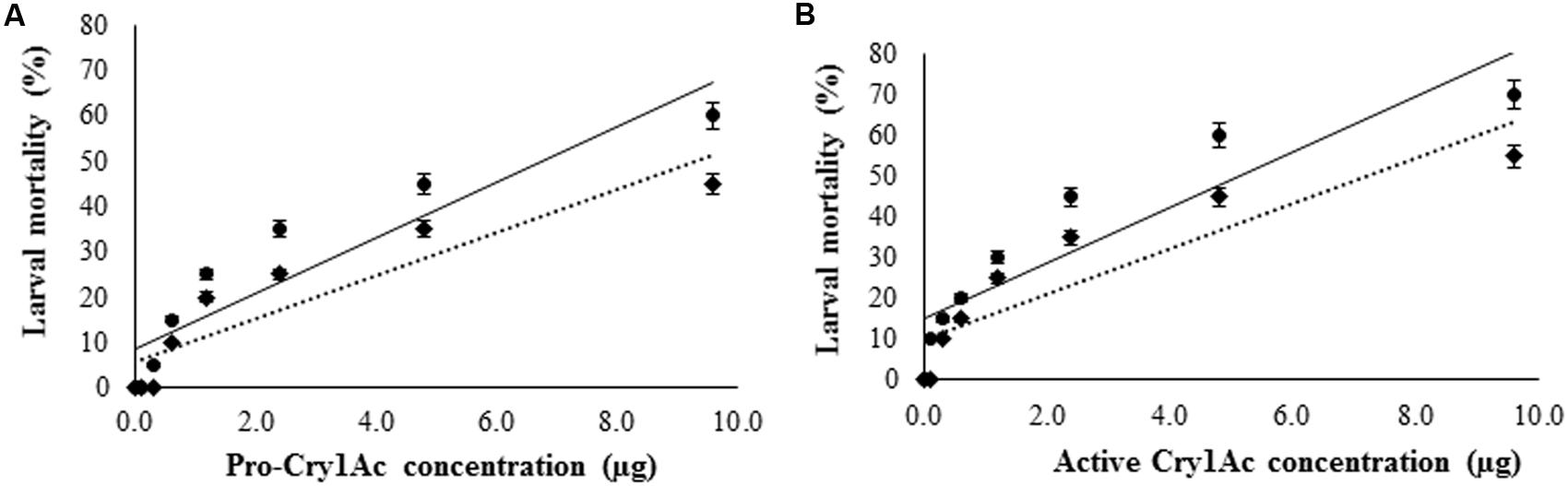
FIGURE 6. Effect of Cry1Ac on larval survival. The early fourth-instar larvae were used for bioassay. The larvae were reared on (A) pro-Cry1Ac and (B) active Cry1Ac (formed after the processing of pro-Cry1Ac by gut bacterial proteases) toxin treated artificial diet. (●) mortality percent in larvae reared on toxin treated artificial diet and (◆) mortality percent in larvae reared on artificial diet treated with both toxin and antibiotic cocktail. To eliminate gut bacteria the larvae were reared on diet treated with 100 μl of antibiotic cocktail (stock: 1 mg/ml of each antibiotic) per gram diet till fourth-instar, and then reared on antibiotic cocktail containing diet treated with toxin. There were three replications for each treatment and 10 larvae in each replication in a completely randomized design (CRD). Each value represents the average of three experiments.
The mortality rate triggered by Cry1Ac toxin was 100% in larvae reared on artificial diet which was reduced to 25% in the larvae reared on diet treated with antibiotic cocktail (Figure 7). However, in the larvae pre-treated with antibiotic cocktail when fed on diet treated with gut bacterial culture, the mortality rate triggered by Cry1Ac toxin was nearly that of larvae reared on artificial diet alone.
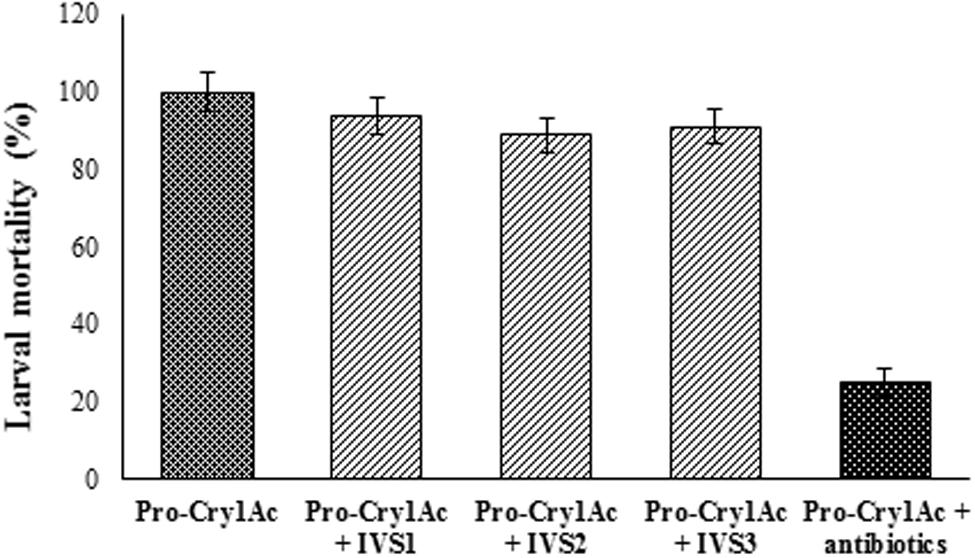
FIGURE 7. Cry1Ac toxicity in antibiotic cocktail fed H. armigera larvae. Larvae were reared until the fourth-instar on sterile artificial diet amended with 100 μl of antibiotic cocktail (Stock: 1 mg/ml of each antibiotic) per gram of diet. The antibiotic pre-treated early fourth-instar larvae were transferred to artificial diet treated with gut bacterial culture and pro-Cry1Ac (15 μg/ml of diet) and reared for 5 days. Data was recorded for Cry1Ac toxin mortality in larvae reared on artificial diet, antibiotic cocktail treated diet and gut bacteria treated diet. Equal amount of culture from IVS1, IVS2, and IVS3 isolates was added to the artificial diet. There were three replications for each treatment and 10 larvae in each replication in a CRD. Each value represents the average of three experiments.
Peptide Sequence Analysis
The highest peak in peptide sequence from IVS1 proteases [1374.81 Da (GLNVHPMHIGGSR)] was showing similarity with metal-dependent hydrolases from Bacillus sp., IVS2 proteases [1959.09 Da (IAVGGDSAGGNLAAVLSLMAR)] with alpha/beta hydrolase Afipia massiliensis, and IVS3 proteases [858.52 Da (RQVSASPPAT)] with serine protease pepA from Mycobacterium tuberculosis. The peptide sequences were subjected to protein-protein BLAST (BLAST-P) to obtain the nearest homologous neighbors, and putative conserved domains (The results for peptide sequence analysis for IVS1, IVS2, and IVS3 isolates were provided as Supplementary Figures S3–S5, respectively).
Discussion
Bacillus thuringiensis toxins have been deployed on a large-scale in transgenic crops for pest management (Sharma et al., 2004; James, 2013). Large-scale cultivation of transgenic crops will exercise a strong selection pressure on the target insect pests, and continuous exposure of insect pests to B. thuringiensis toxins in transgenic plants will lead to development of insect resistance to transgenic crops (Carriere et al., 2010; Huang et al., 2011). The gut bacteria perform numerous functions including digestion, nutrient concentration, nitrogen fixation, and detoxification of defensive compounds (Broderick et al., 2003; Dillon and Dillon, 2004). Gut bacteria produce a wide variety of extra-cellular enzymes, including proteases. Midgut bacteria influence the insecticidal activity of B. thuringiensis to insect pests (Broderick et al., 2006, 2009; Paramasiva et al., 2014; Visweshwar et al., 2015). The midgut bacteria of insect host have either synergistic (Raymond et al., 2009) or protective (Broderick et al., 2006, 2009; Paramasiva et al., 2014; Visweshwar et al., 2015) effect, but the underlying mechanisms are unclear. In the present study we have isolated three isolates of protease producing gut bacteria and characterized their proteases which are involved in protoxin to toxin conversion.
Twelve dominant gut bacterial colonies were isolated from midgut of H. armigera, and only seven bacterial isolates exhibited proteolytic activity. Proteases from the gut bacterial isolates were purified by implying ammonium sulfate precipitation, DEAE-cellulose and Sephadex G-75 columns. Active fractions containing protease activity were pooled and incubated with the pro-Cry1Ac (130 kDa) and the liberated active toxin (65 kDa) was detected using SDS-PAGE, ELISA and Western blotting (Figure 2). Among the seven protease producing bacterial strains, proteases from only three strains (IVS1, IVS2, and IVS3) were able to convert inactive pro-Cry1Ac to active Cry1Ac toxin. The results suggest that the proteases produced by the midgut bacteria were involved in proteolytic processing of pro-Cry1Ac into active Cry1Ac, and thereby influencing the toxicity of B. thuringiensis to H. armigera. The Cry11Aa1 protoxin is hydrolyzed by proteases produced from bacteria (Ibarra and Federici, 1986). The B. thuringiensis subsp. kurstaki mediates endogenous proteolytic processing of 132 kDa protoxin to an active 66 kDa toxin only under denaturing/reducing conditions (Kumar and Venkateswerlu, 1997). The nucleotide homology search of IVS1, IVS2, and IVS3 isolates showed similarity with B. thuringiensis, V. fischeri, and E. coli, respectively. IVS2 and IVS3 isolates belong to the phylum Proteobacteria, while IVS1 isolate belongs to Firmicutes.
The optimum pH and temperature for the gut bacterial protease activity were 8 and 40°C, respectively (Figures 3 and 4), indicating that the proteases were alkaline in nature. Although some activity was detected at acidic pH, the maximum activity was observed at a slightly alkaline environment (pH 8.0). Gut bacterial proteases are characterized by their high activity at alkaline pH (8.0–12.0), with optimal temperature between 50 and 70°C (Al-Shehri et al., 2004). Among the seven protease inhibitors tested, the maximum inhibition of total proteolytic activity was exerted by serine protease inhibitor, PMSF followed by EDTA, indicates that the proteases were of serine type. Maximum inhibition of the protease activities was observed with chymostatin indicates that proteases were of chymotrypsin type. Fluorescence zymography of H. armigera gut bacterial proteases showed that the purified bacterial proteases were chymotrypsin-like, and their molecular weights were around 15 kDa for protease from IVS1 and IVS3, and 65 kDa from IVS2 isolate (Figure 5). In general, molecular mass of bacterial proteases ranges between 15 and 45 kDa (Kumar and Takagi, 1999; Gupta et al., 2002; Olivieri et al., 2002; Niu et al., 2006; Disney et al., 2008).
With the increased concentration of pro-Cry1Ac and active Cry1Ac the mortality rate was increased in larvae reared on artificial diet and diet treated with antibiotic cocktail. However, the reduced mortality rate was observed in the larvae reared on artificial diet treated with antibiotic cocktail compared to the larvae reared on artificial diet only (Figure 6). The observations are further supported from the earlier made observations that B. thuringiensis does not kill the larvae of the gypsy moth in the absence of indigenous midgut bacteria (Broderick et al., 2006). Elimination of the gut microbial community by oral administration of antibiotics abolished the insecticidal activity of B. thuringiensis in Lymantria dispar (L.), Pieris rapae (L.), and Vanessa cardui (L.) (Broderick et al., 2009) and in H. armigera (Paramasiva et al., 2014; Visweshwar et al., 2015). In antibiotic fed H. armigera larvae, after feeding protease producing gut bacterial culture (IVS1, IVS2, and IVS3) the mortality rate was triggered nearly that of control larvae (Larvae un-treated with antibiotics; Figure 7). Reduction in gypsy moth larval mortality was accompanied by reduced populations of Enterococcus spp. and Enterobacter spp. from the midguts of larvae (Broderick et al., 2006). After re-establishment of Enterobacter spp., the insecticidal activity was restored (Broderick et al., 2006). The mortality data was similar for active Cry1Ac produced from processing of pro-Cry1Ac by proteases purified from gut bacteria IVS1, IVS2, and IVS3.
Peptide sequence analysis from IVS1 and IVS2 showed similarity with hydrolases, and IVS3 with serine protease pepA. Serine proteases are involved in the activation of Cry protoxins through proteolytic removal of peptide fragments (Broehan et al., 2008). Several Bacillus species produce proteases (Beg and Gupta, 2003; Banik and Prakash, 2004; Gerze et al., 2005). Protease activity associated with whole cells of V. fischeri strain ES114 is a product of putative cell membrane-associated aminopeptidase (PepN; Fidopiastis et al., 2012). E. coli is one of the most important pathogens, and it contains numerous proteases, including metallo-, serine- and aspartic-proteases capable of catalyzing casein and some oxidized proteins (Rozkov and Enfors, 2004).
In summary, the gut bacterial proteases isolated from H. armigera were involved in proteolytic cleavage of pro-Cry1Ac to active toxin, indicates that proteases produced by the H. armigera gut bacteria influenced the proteolytic processing of protoxin to toxin, which results in insect mortality. Any changes in relative abundance and diversity of gut bacteria in insect pests will have a major bearing on development of insect resistance to B. thuringiensis-transgenic crops. Therefore, there is a need for understanding the role of midgut microbes in evolution of insect resistance to transgenic crops to develop strategies for deployment of transgenic crops for sustainable crop production.
Author Contributions
HS, SK, AM, and VR designed the experiments. VR performed the experiments, analyzed the results and wrote the paper. HS, SK, and AM corrected the paper. All the authors approve the final version for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the staff, Insect Rearing Laboratory, Entomology, for providing the insects throughout the study. Mr. VR would like to thank University Grants Commission (UGC), New Delhi, India for the financial assistance in the mode of BSR fellowship in science for meritorious students. We thank to Dr. Arunasree, Assistant Professor, School of Life Sciences, Hyderabad Central University, Hyderabad, India, for helping in proteases sequence analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01567
Footnotes
References
Al-Shehri, L., Abdul-Rahman, M., and Yassar, S. (2004). Production and some properties of protease produced by Bacillus licheniformis isolated from Tihametaseer, Saudi Arabia. Pak. J. Biol. Sci. 7, 1631–1635. doi: 10.3923/pjbs.2004.1631.1635
Aronson, A. I., and Shai, Y. (2001). Why Bacillus thuringiensis insecticidal toxins are so effective: unique features of their mode of action. FEMS Microbiol. Lett. 195, 1–8. doi: 10.1111/j.1574-6968.2001.tb10489.x
Banik, R. M., and Prakash, M. (2004). Laundry detergent compatibility of the alkaline protease from Bacillus cereus. Microbiol. Res. 159, 135–140. doi: 10.1016/j.micres.2004.01.002
Baxter, S. W., Badenes-Perez, F. R., Morrison, A., Vogel, H., Crickmore, N., Kain, W., et al. (2011). Parallel evolution of Bacillus thuringiensis toxin resistance in Lepidoptera. Genetics 189, 675–679. doi: 10.1534/genetics.111.130971
Beg, Q. K., and Gupta, R. (2003). Purification and characterization of an oxidation stable, thiol-dependent serine alkaline protease from Bacillus mojavensis. Enzyme Microbial. Technol. 32, 294–304. doi: 10.1016/S0141-0229(02)00293-4
Broderick, N. A., Goodman, R. M., Handelsman, J., and Raffa, K. F. (2003). Effect of host diet and insect source on synergy of gypsy moth (Lepidoptera: Lymantriidae) mortality to Bacillus thuringiensis subsp kurstaki by zwittermicin A. Environ. Entomol. 32, 387–391. doi: 10.1603/0046-225X-32.2.387
Broderick, N. A., Raffa, K. F., and Handelsman, J. (2006). Midgut bacteria required for Bacillus thuringiensis insecticidal activity. Proc. Natl. Acad. Sci. U.S.A. 103, 15196–15199. doi: 10.1073/pnas.0604865103
Broderick, N. A., Robinson, C. J., McMahon, M. D., Holt, J., Handelsman, J., and Raffa, K. F. (2009). Contributions of gut bacteria to Bacillus thuringiensis-induced mortality vary across a range of Lepidoptera. BMC Biol. 7:11. doi: 10.1186/1741-7007-7-11
Broehan, G., Kemper, M., Driemeier, D., Vogelpohl, I., and Merzendorfer, H. (2008). Cloning and expression analysis of midgut chymotrypsin-like proteinases in the tobacco hornworm. J. Insect. Physiol. 54, 1243–1252. doi: 10.1016/j.jinsphys.2008.06.007
Carriere, Y., Crowder, D., and Tabashnik, B. E. (2010). Evolutionary ecology of adaptation to Bt crops. Evol. Appl. 3, 561–573. doi: 10.1111/j.1752-4571.2010.00129.x
Chitti Babu, G., Sharma, H. C., Madhumati, T., Raghavaiah, G., Murthy, K. V. M. K., and Rao, V. S. (2014). A semi-synthetic chickpea flour based diet for long-term maintenance of laboratory culture of Helicoverpa armigera. Ind. J. Entomol. 76, 336–340.
De Maagd, R. A., Bravo, A., and Crickmore, N. (2001). How Bacillus thuringiensis has evolved specific toxins to colonize the insect world. Trends Genet. 17, 193–199. doi: 10.1016/S0168-9525(01)02237-5
Dillon, R. J., and Dillon, V. M. (2004). The gut bacteria of insects: nonpathogenic interactions. Annu. Rev. Entomol. 49, 71–92. doi: 10.1146/annurev.ento.49.061802.123416
Disney, D. R., Vilela, D. M., Silvestre, M. P. C., and Schwan, R. F. (2008). Alkaline protease from Bacillus sp. isolated from coffee bean grown on cheese whey. World J. Microbiol. Biotechnol. 24, 2027–2034. doi: 10.1007/s11274-008-9706-6
Fabrick, J. A., Ponnuraj, J., Sing, A., Tanwar, R. K., Unnithan, G. C., Yelich, A. J., et al. (2014). Alternative splicing and highly variable cadherin transcripts associated with field-evolved resistance of pink bollworm to Bt cotton in India. PLoS ONE 9:e97900. doi: 10.1371/journal.pone.0097900
Fidopiastis, P. M., Rader, B. A., Gerling, D. G., Gutierrez, N. A., Watkins, K. H., Frey, M. W., et al. (2012). Characterization of a Vibrio fischeri aminopeptidase and evidence for its influence on an early stage of squid colonization. J. Bacteriol. 194, 3995–4002. doi: 10.1128/JB.00108-12
Fuentes, J. L., Karumbaiah, L., Jakka, S. R. K., Ning, C., Liu, C., Wu, K., et al. (2011). Reduced levels of membrane-bound alkaline phosphatases are common to Lepidopteran strains resistant to cry toxins from Bacillus thuringiensis. PLoS ONE 6:e17606. doi: 10.1371/journal.pone.0017606
Gerze, A., Omay, D., and Guvenilir, Y. (2005). Partial purification and characterization of protease enzyme from Bacillus subtilis and Bacillus megatherium. Appl. Biochem. Biotechnol. 12, 335–345. doi: 10.1385/ABAB:121:1-3:0335
Grochulski, P., Masson, L., Borisova, S., Pusztai-Carey, M., Schwartz, J. L., Brousseau, R., et al. (1995). Cry1A(a) insecticidal toxin: crystal structure and channel formation. J. Mol. Biol. 54, 447–464. doi: 10.1006/jmbi.1995.0630
Gupta, R., Beg, Q. K., Khan, S., and Chauhan, B. (2002). An overview on fermentation, downstream processing and properties of microbial alkaline proteases. Appl. Microbiol. Biotechnol. 60, 381–395. doi: 10.1007/s00253-002-1142-1
Hemaraj, V., Diksha, S., and Avneet, G. (2013). A review on commonly used biochemical test for bacteria. Innov. J. Life Sci. 1, 1–7.
Himabindu, V. K., Kumar, R., Rameshwar, S., and Sreelakshmi, Y. (2013). Chromoplast-specific carotenoid-associated protein appears to be important for enhanced accumulation of carotenoids in hp1 tomato fruits. Plant Physiol. 161, 2085–2101. doi: 10.1104/pp.112.212191
Holding, A. J., and Shewan, J. M. (1974). “Genera of uncertain affiliation,” in Bergey’s Manual of Determinative Bacteriology, 8th Edn, eds R. E. Buchanan and N. E. Gibbons (Baltimore: William and Wilkins).
Huang, F., Andow, D. A., and Buschman, L. L. (2011). Success of the high-dose/refuge resistance management strategy after 15 years of Bt crop use in North America. Entomol. Exp. Appl. 140, 1–16. doi: 10.1111/j.1570-7458.2011.01138.x
Ibarra, J. E., and Federici, B. A. (1986). Isolation of a relatively nontoxic 65-kilodalton protein inclusion from the parasporal body of Bacillus thuringiensis subsp. israelensis. J. Bacteriol. 165, 527–533.
James, C. (2013). Global Status of Commercialized Biotech/GM Crops. ISAAA Briefs 46. Ithaca, NY: ISAAA.
Karumbaiah, L., Oppert, B., Jurat-Fuentes, J. L., and Adang, M. J. (2007). Analysis of midgut proteinases from Bacillus thuringiensis-susceptible and -resistant Heliothis virescens (Lepidoptera: Noctuidae). Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 146, 139–146. doi: 10.1016/j.cbpb.2006.10.104
Keller, M., Sneh, B., Strizhov, N., Prudovsky, E., Regev, A., Koncz, C., et al. (1996). Digestion of δ-endotoxin by gut proteases may explain reduced sensitivity of advanced instar larvae of Spodoptera littoralis to CryIC. Insect Biochem. Mol. Biol. 26, 365–373. doi: 10.1016/0965-1748(95)00102-6
Kumar, C. G., and Takagi, H. (1999). Microbial alkaline proteases: from a bioindustrial viewpoint. Biotechnol. Adv. 17, 561–594. doi: 10.1016/S0734-9750(99)00027-0
Kumar, H. D., Gajju, H., and Bhalla, T. C. (2002). Production of a thermostable protease by Bacillus sp. APR-4. J. Microbiol. Biotechnol. Environ. Sci. 4, 533–540.
Kumar, N. S., and Venkateswerlu, G. (1997). Involvement of an endogenous metalloprotease in the activation of protoxin in Bacillus thuringiensis subsp. kurstaki. Biochem. Mol. Biol. Int. 42, 901–908.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Laemmli, U. K. (1970). Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Li, H., Oppert, B., Higgins, R. A., Huang, F., Zhu, K. Y., and Buschman, L. L. (2004). Comparative analysis of proteinase activities of Bacillus thuringiensis-resistant and -susceptible Ostrinia nubilalis (Lepidoptera: Crambidae). Insect Biochem. Mol. Biol. 34, 753–762. doi: 10.1016/j.ibmb.2004.03.010
Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951). Protein measurement with the folin phenol reagent. J. Biol. Chem. 193, 265–275.
Nair, R., Kalia, V., Aggarwal, K. K., and Gujar, G. T. (2013). Variation in the cadherin gene sequence of Cry1Ac susceptible and resistant Helicoverpa armigera (Lepidoptera: Noctuidae) and the identification of mutant alleles in resistant strains. Curr. Sci. 104, 215–223.
Niu, Q., Huang, X., Zhang, L., Li, Y., Li, J., Yang, J., et al. (2006). A neutral protease from Bacillus nematocida, another potential virulence factor in the infection against nematodes. Arch. Microbiol. 185, 439–448. doi: 10.1007/s00203-006-0112-x
Olivieri, F., Zanetti, M. E., Oliva, C. R., Covarrubias, A. A., and Casalongue, C. A. (2002). Characterization of an extracellular serine protease of Fusarium eumartii and its action on pathogenesis related proteins. Eur. J. Plant. Pathol. 108, 63–72. doi: 10.1023/A:1013920929965
Oppert, B., Kramer, K. J., Beeman, R. W., Johnson, D., and McGaughey, W. H. (1997). Proteinase-mediated insect resistance to Bacillus thuringiensis toxins. J. Biol. Chem. 272, 23473–23476. doi: 10.1074/jbc.272.38.23473
Paramasiva, I., Sharma, H. C., and Krishnayya, P. V. (2014). Antibiotics influence the toxicity of the delta endotoxins of Bacillus thuringiensis towards the cotton bollworm, Helicoverpa armigera. BMC Microbiol. 14:200. doi: 10.1186/1471-2180-14-200
Rajagopal, R., Arora, N., Sivakumar, S., Rao, N. G. V., Nimbalkar, S. A., and Bhatnagar, R. K. (2009). Resistance of Helicoverpa armigera to Cry1Ac toxin from Bacillus thuringiensis is due to improper processing of the protoxin. Biochem. J. 419, 309–316. doi: 10.1042/BJ20081152
Raymond, B., Johnston, P. R., Wright, D. J., Ellis, R. J., Crickmore, N., and Bonsall, M. B. (2009). A mid-gut microbiota is not required for the pathogenicity of Bacillus thuringiensis to diamondback moth larvae. Environ. Microbiol. 11, 2556–2563. doi: 10.1111/j.1462-2920.2009.01980.x
Rozkov, A., and Enfors, S. O. (2004). Analysis and control of proteolysis of recombinant proteins in Escherichia coli. Adv. Biochem. Eng. Biotechnol. 89, 163–195.
Sanahuja, G., Banakar, R., Twyman, R. M., Capell, T., and Christou, P. (2011). Bacillus thuringiensis: a century of research, development and commercial applications. Plant Biotechnol. J. 9, 283–300. doi: 10.1111/j.1467-7652.2011.00595.x
Schnepf, E., Crickmore, N., and VanRie, J. (1998). Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62, 775–806.
Shao, Z., Cui, Y., Liu, X., Yi, H., Ji, J., and Yu, Z. (1998). Processing of δ-endotoxin of Bacillus thuringiensis subsp. kurstaki HD-1 in Heliothis armigera midgut juice and the effects of protease inhibitors. J. Invertbr. Pathol. 72, 73–81. doi: 10.1006/jipa.1998.4757
Sharma, H. C. (2005). Heliothis/Helicoverpa Management Emerging Trends and Strategies for Future Research. New Delhi: Oxford and IBH Publishing Co.
Sharma, H. C., Sharma, K. K., and Crouch, J. H. (2004). Genetic transformation of crops for insect resistance: potential and limitations. Crit. Rev. Plant Sci. 23, 47–72. doi: 10.1080/07352680490273400
Tiewsiri, K., and Wang, P. (2011). Differential alteration of two aminopeptidase N associated with resistance to Bacillus thuringiensis toxin Cry1Ac in cabbage looper. Proc. Natl. Acad. Sci. U.S.A. 108, 14037–14042. doi: 10.1073/pnas.1102555108
Vinod, D. P., Sharma, H. C., and Kochole, M. S. (2010). In vivo inhibition of Helicoverpa armigera gut pro-proteinase activation by non-host plant protease inhibitors. J. Insect Physiol. 56, 1315–1324. doi: 10.1016/j.jinsphys.2010.04.003
Visweshwar, R., Sharma, H. C., Akbar, S. M. D., and Sreeramulu, K. (2015). Elimination of gut microbes with antibiotics confers resistance to Bacillus thuringiensis toxin proteins in Helicoverpa armigera (Hubner). Appl. Biochem. Biotechnol. 177, 1621–1637. doi: 10.1007/s12010-015-1841-6
Wang, P., Zhao, J. Z., Rodrigo-Simon, A., Kain, W., Janmaat, A. F., Shleton, A. M., et al. (2007). Mechanism of resistance to Bacillus thuringiensis toxin Cry1Ac in greenhouse population of cabbage looper, Trichopusia ni. Appl. Environ. Microbiol. 73, 1199–1207. doi: 10.1128/AEM.01834-06
Keywords: Helicoverpa armigera, midgut bacteria, proteases, Cry1Ac proteins, transgenics
Citation: Regode V, Kuruba S, Mohammad AS and Sharma HC (2016) Isolation and Characterization of Gut Bacterial Proteases Involved in Inducing Pathogenicity of Bacillus thuringiensis Toxin in Cotton Bollworm, Helicoverpa armigera. Front. Microbiol. 7:1567. doi: 10.3389/fmicb.2016.01567
Received: 12 July 2016; Accepted: 20 September 2016;
Published: 06 October 2016.
Edited by:
Marc Bramkamp, Ludwig Maximilian University of Munich, GermanyReviewed by:
David Rivers, Loyola University Maryland, USAEitan Ben-Dov, Achva Academic College, Israel
Copyright © 2016 Regode, Kuruba, Mohammad and Sharma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hari C. Sharma, aC5zaGFybWFAY2dpYXIub3Jn
 Visweshwar Regode1,2
Visweshwar Regode1,2 Hari C. Sharma
Hari C. Sharma