Introduction
Hyperthermophilic archaea have been isolated from high-temperature environments such as geothermally heated soils, sulfur-rich hot springs, and submarine volcanic habitats; optimal growth of these organisms occurs above 80°C (Stetter, 1999, 2006, 2013). The genus Vulcanisaeta belongs to the family Thermoproteaceae, order Thermoproteales, phylum Crenarchaeota, and was first proposed by Itoh et al. (2002). It currently includes 3 validly named species, that is, Vulcanisaeta distributa (Itoh et al., 2002), V. souniana (Itoh et al., 2002), and V. thermophila (Yim et al., 2015), as per the List of Prokaryotic Names with Standing in Nomenclature database (Parte, 2014). Members of the genus Vulcanisaeta are rod-shaped, anaerobic, hyperthermophilic, and acidophilic (Itoh et al., 2002). To date, 15 genomes, including two complete genomes, V. distributa and “Vulcanisaeta moutnovskia” (Mavromatis et al., 2010; Gumerov et al., 2011), have been reported for the genus Vulcanisaeta, as per the NCBI genome database (http://www.ncbi.nlm.nih.gov/genome/).
Hyperthermophilic enzymes are stable and active at high temperatures of >70°C (Vieille et al., 1996). These enzymes can be studied using model systems to elucidate enzyme mechanisms and evolution of proteins stable at high temperatures and to determine the higher temperature limit for enzyme stability (Vieille and Zeikus, 2001). In a previous study, V. thermophila CBA1501T (= ATCC BAA-2415T = JCM 17228T) was isolated from solfataric soil in the Republic of the Philippines (Yim et al., 2015). It was found to grow at 75–90°C, pH 4.0–6.0, and 0–1.0% (w/v) NaCl, with optimal growth at 85°C, pH 5.0, and 0% (w/v) NaCl. Here, a genome sequence of V. thermophila CBA1501T has been reported and information of hyperthermophilic enzymes of high biotechnological value has been provided.
Materials and Methods
Culture Conditions and DNA Extraction
In a previous study, we isolated V. thermophila CBA1501T from the solfataric soil of the Mayon volcano in the Republic of the Philippines (Yim et al., 2015) and cultivated it on modified JCM medium no. 236 (M236) (containing per liter salt base solution: 2.94 g trisodium citrate dihydrate, 0.5 g yeast extract, 10.0 ml trace vitamins, 1.0 mg resazurin, 0.5 g Na2S·9H2O, and 20 mM thiosulfate). For DNA extraction, the strain was enriched at 80°C in M236 medium, using a serum bottle. Its genomic DNA was extracted using the G-spin total DNA extraction kit (iNtRON Biotechnology, Korea) and QuickGene DNA tissue kit S (Kurabo, Japan).
Genome Sequencing, Assembly, and Annotation
The genome sequences of V. thermophila CBA1501T were sequenced at a read length of 300 bp using the Illumina MiSeq system, with paired-end library [insert size, 634–1101 bp (average 852 bp), computed by CLC Genomics Workbench 7.5.1 (CLC bio, Denmark)] constructed using the Nextera DNA Library Prep kit (illumina, USA), according to the manufacturer's instructions (Moon et al., 2015). A total of 6,939,438 reads (with 688-fold coverage) were assembled using CLC Genomics Workbench 7.5.1 with default parameters as follows: masking mode, no masking; mismatch cost, 2; insertion cost, 3; deletion cost, 3; length cost, 3; length fraction, 0.5; similarity fraction, 0.8; global alignment, no; auto-detect paired distances, yes; non-specific match handling map, randomly. To identify ribosomal RNA and transfer RNA, RNAmmer 1.2 (Lagesen et al., 2007) and tRNAscan v. 1.3.1 (Lowe and Eddy, 1997), respectively, were used. Protein coding sequences (CDSs) identification was performed using PRODIGAL v. 2.6.2 (Hyatt et al., 2012), and functional annotation was performed using EggNOG v. 4.1 (Powell et al., 2014), SEED subsystems (Overbeek et al., 2014), Swiss-Prot (UniProt, 2015), and KEGG (Kanehisa et al., 2016) databases with the USEARCH v. 8.0.1517 program (Edgar, 2010).
Phylogenetic Analysis
Similarities based on 16S rRNA gene sequences were calculated using EzBioCloud (http://www.ezbiocloud.net). Phylogenetic tree based on 16S rRNA gene sequences was constructed using MEGA5 (Tamura et al., 2011) with the neighbor-joining (Saitou and Nei, 1987), maximum-parsimony (Kluge and Farris, 1969), and maximum-likelihood (Felsenstein, 1981) methods, based on 1000 randomly generated trees.
Comparative Genomic Analysis
For comparative analysis, reference genome sequences of closely related strains of the genus Vulcanisaeta were selected using the NCBI genome database (http://www.ncbi.nlm.nih.gov/genome/): V. distributa JCM 11215 (BBCT00000000), V. distributa DSM 14429 (CP002100), V. distributa JCM 11217 (BBBJ00000000), V. souniana JCM 11219 (BBBK00000000), V. moutnovskia 768-28 (CP002529), and Vulcanisaeta sp. strains CIS_19 (LOCG00000000), JCM 14467 (BBDM00000000), JCM 16159 (BBDN00000000), and JCM 161 (BBDO00000000). To determine the similarity between genome sequences, orthologous average nucleotide identity (OrthoANI) values of CBA1501T and related strains in the genus Vulcanisaeta were calculated using the Orthologous Average Nucleotide Identity Tool (Lee et al., 2015), and a phylogenetic tree based on OrthoANI values was obtained using the EzBioCloud Comparative Genomics Database (EzCgDb; Chunlab; http://cg.ezbiocloud.net/). Annotated genomes of CBA1501T and other related strains were subjected to homology search using the UBLAST program (Ward and Moreno-Hagelsieb, 2014) for pan-genome analysis. Then, pan-genome orthologous groups (POGs) were constructed using EzCgDb.
Results
General Genomic Features of V. thermophila CBA1501T
The draft genome sequence of V. thermophila CBA1501T was 2,022,594 bp in length, with a G+C content of 49.1 mol % in 10 contigs. The largest contig was 791,731 bp long, and the N50 value was 634,758 bp. The genome was found to contain 2170 CDSs, one 16S-23S-5S rRNA gene operon, and 41 tRNA genes. Genomic features are shown in Figure 1. On the basis of information from the EggNOG v. 4.1 database, 1927 genes were categorized into Clusters of Orthologous Groups of proteins (COGs) functional groups. The most abundant COG category was “Function unknown” (S; 729 genes), followed by “Energy production and conversion” (C; 178 genes), “Amino acid transport and metabolism” (E; 168 genes), “Translation, ribosomal structure and biogenesis” (J; 159 genes), “Carbohydrate transport and metabolism” (G; 96 genes), and “Coenzyme transport and metabolism” (H; 87 genes). Among the SEED subsystem categories, “Carbohydrates” (181 genes), “Amino Acids and Derivatives” (171 genes), “Protein Metabolism” (142 genes) and “Cofactors, Vitamins, Prosthetic Groups, Pigments” (116 genes) were the most dominant categories (>10% of a total of 1,094 matched SEED subsystem categories).
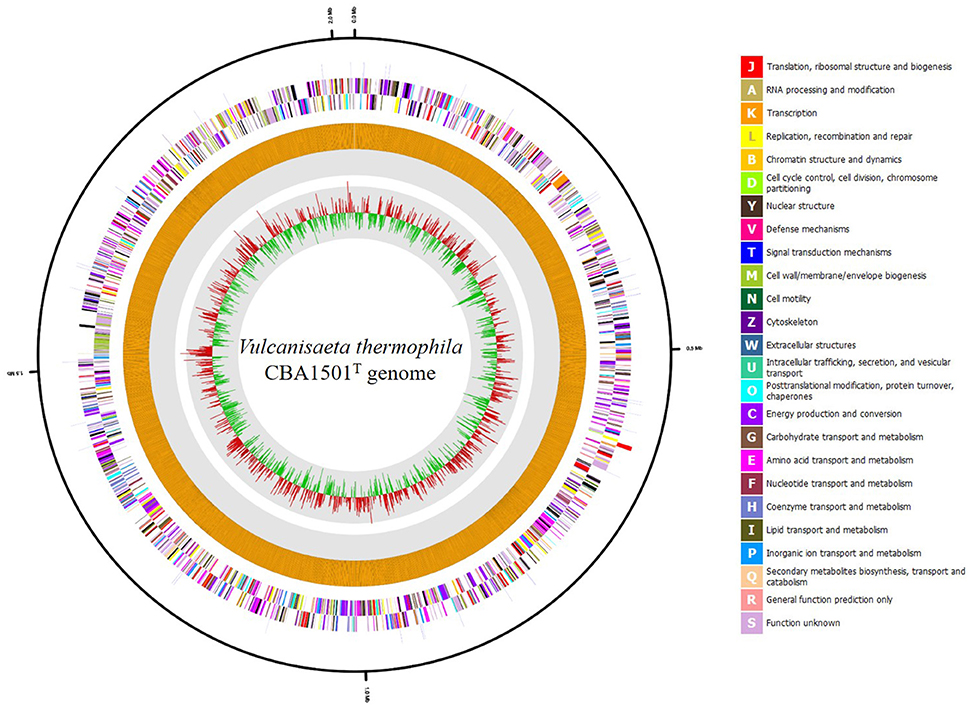
Figure 1. Graphic circular map of the Vulcanisaeta thermophila CBA1501T genome. Outer circle shows genes on the sense and antisense strands (colored according to COG categories), and RNA genes (red, tRNA; blue, rRNA) are shown from the outside of the circle to the center. Inner circles show the GC skew, with yellow and blue indicating positive and negative values, respectively; the GC content is indicated in red and green. This genome map was visualized using CLgenomics 1.52 (Chun Lab Inc.).
Phylogenetic Analysis
V. thermophila CBA1501T had the greatest 16S rRNA gene sequence similarity with the following (in this order): V. distributa DSM 14429T (98.6%), Stygiolobus azoricus DSM 6296T (98.6%), V. souniana IC-059T (97.5%), Caldivirga maquilingensis IC-167T (94.6%), Pyrobaculum ferrireducens 1860T (93.8%), Pyrobaculum islandicum DSM 4184T (93.6%), Thermoproteus uzoniensis 768-20 (93.6%), Pyrobaculum organotrophum JCM 9190T (93.5%), and Thermoproteus thermophilus CBA1502T (93.2%). The phylogenetic analysis indicated that the strain CBA1501T clustered with species of the genus Vulcanisaeta (Supplementary Figure 1A).
Comparative Genomics Data
V. thermophila CBA1501T had lesser than 73% orthoANI values with all of the related strains in the genus Vulcanisaeta (Supplementary Table 1). In the orthoANI values-based dendrogram, the strain CBA1501T was located as an outgroup to the other related strains in Vulcanisaeta (Supplementary Figure 1B). These results indicate that V. thermophila CBA1501T is evolutionarily distinct from other related strains. The pan-genome analysis showed that 10 genomes in the genus Vulcanisaeta have the core genome, comprised of 979 POGs. In contrast, only the genome of strain CBA1501T had 211 POGs as a singleton. Among these singletons, various enzymes, including arylformamidase, shikimate kinase, formyl-CoA transferase, xanthine dehydrogenase, hydrogensulfite reductase, and amidase, were detected.
In conclusion, the information provided here is useful as the genome of V. thermophila CBA1501T will provide insights into the metabolism of hyperthermophilic archaea and aid in identifying opportunities for biotechnological applications of novel hyperthermophilic enzymes.
Data Access
The genome sequences of V. thermophila CBA1501T (=ATCC BAA-2415T = JCM 17228T) were deposited in the DDBJ under the accession numbers BCLI01000001-BCLI01000010 (http://www.ncbi.nlm.nih.gov/Traces/wgs/BCLI01). The annotated data of V. thermophila CBA1501T based on SEED subsystems is accessible on SEED viewer v. 2.0 by logging in with the guest account (Genome ID 6666666.192913, username: guest, password: guest) at the web address: http://rast.nmpdr.org/seedviewer.cgi?page=Organism&organism=6666666.192913.
Author Contributions
SWR designed and coordinated all the experiments. KJY performed cultivation, DNA extraction and purification. JYK, HSS, YBK, D-GL, JK, and K-SO performed the sequencing, genome assembly, gene prediction, gene annotation and comparative genomic analysis. JYK, KJY, and SWR wrote manuscript. All authors have read the manuscript and approved.
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (2015R1D1A1A09061039), project fund from the Center for Analytical Research of Disaster Science of the Korea Basic Science Institute (C36703).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01639
References
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376.
Gumerov, V. M., Mardanov, A. V., Beletsky, A. V., Prokofeva, M. I., Bonch-Osmolovskaya, E. A., Ravin, N. V., et al. (2011). Complete genome sequence of “Vulcanisaeta moutnovskia” strain 768-28, a novel member of the hyperthermophilic crenarchaeal genus Vulcanisaeta. J. Bacteriol. 193, 2355–2356. doi: 10.1128/JB.00237-11
Hyatt, D., LoCascio, P. F., Hauser, L. J., and Uberbacher, E. C. (2012). Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics 28, 2223–2230. doi: 10.1093/bioinformatics/bts429
Itoh, T., Suzuki, K., and Nakase, T. (2002). Vulcanisaeta distributa gen. nov., sp. nov., and Vulcanisaeta souniana sp. nov., novel hyperthermophilic, rod-shaped crenarchaeotes isolated from hot springs in Japan. Int. J. Syst. Evol. Microbiol. 52(Pt 4), 1097–1104. doi: 10.1099/00207713-52-4-1097
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462. doi: 10.1093/nar/gkv1070
Kluge, A. G., and Farris, F. S. (1969). Quantitative phyletics and the evolution of anurans. Syst. Zool. 18, 1–32.
Lagesen, K., Hallin, P., Rødland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Lee, I., Kim, Y. O., Park, S. C., and Chun, J. (2015). OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964.
Mavromatis, K., Sikorski, J., Pabst, E., Teshima, H., Lapidus, A., Lucas, S., et al. (2010). Complete genome sequence of Vulcanisaeta distributa type strain (IC-017). Stand. Genomic Sci. 3, 117–125. doi: 10.4056/sigs.1113067
Moon, J. S., Choi, H. S., Shin, S. Y., Noh, S. J., Jeon, C. O., and Han, N. S. (2015). Genome sequence analysis of potential probiotic strain Leuconostoc lactis EFEL005 isolated from kimchi. J. Microbiol. 53, 337–342. doi: 10.1007/s12275-015-5090-8
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Parte, A. C. (2014). LPSN–list of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 42, D613–D616. doi: 10.1093/nar/gkt1111
Powell, S., Forslund, K., Szklarczyk, D., Trachana, K., Roth, A., Huerta-Cepas, J., et al. (2014). eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res. 42, D231–D239. doi: 10.1093/nar/gkt1253
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Stetter, K. O. (1999). Extremophiles and their adaptation to hot environments. FEBS Lett. 452, 22–25.
Stetter, K. O. (2006). History of discovery of the first hyperthermophiles. Extremophiles 10, 357–362. doi: 10.1007/s00792-006-0012-7
Stetter, K. O. (2013). A brief history of the discovery of hyperthermophilic life. Biochem. Soc. Trans. 41, 416–420. doi: 10.1042/BST20120284
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
UniProt, C. (2015). UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212. doi: 10.1093/nar/gku989
Vieille, C., Burdette, D. S., and Zeikus, J. G. (1996). Thermozymes. Biotechnol. Annu. Rev. 2, 1–83.
Vieille, C. Zeikus, G. J. (2001). Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65, 1–43. doi: 10.1128/MMBR.65.1.1-43.2001
Ward, N., and Moreno-Hagelsieb, G. (2014). Quickly finding orthologs as reciprocal best hits with BLAT, LAST, and UBLAST: how much do we miss? PLoS ONE 9:e101850. doi: 10.1371/journal.pone.0101850
Keywords: Vulcanisaeta thermophila, genome sequence, archaea, hyperthermophile, hyperthermophilic enzyme
Citation: Kim JY, Yim KJ, Song HS, Kim YB, Lee D-G, Kwon J, Oh K-S and Roh SW (2016) Genomic Analysis of Vulcanisaeta thermophila Type Strain CBA1501T Isolated from Solfataric Soil. Front. Microbiol. 7:1639. doi: 10.3389/fmicb.2016.01639
Received: 16 August 2016; Accepted: 30 September 2016;
Published: 19 October 2016.
Edited by:
Frank T. Robb, University of Maryland, Blatimore, USAReviewed by:
David L. Bernick, University of California, Santa Cruz, USANikolai Ravin, Research Center for Biotechnology Russian Academy of Sciences, Russia
Lydia Kreuter, University of Maryland, Baltimore, USA
Copyright © 2016 Kim, Yim, Song, Kim, Lee, Kwon, Oh and Roh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seong Woon Roh, c2VvbmcxOEBnbWFpbC5jb20=
 Joon Yong Kim
Joon Yong Kim Kyung June Yim
Kyung June Yim Dong-Gi Lee
Dong-Gi Lee Seong Woon Roh
Seong Woon Roh