- 1Department of Microbiology and Plant Pathology, Forestry and Agriculture Biotechnology Institute, University of Pretoria, Pretoria, South Africa
- 2Natural Products Research Institute, College of Pharmacy, Seoul National University, Seoul, Republic of Korea
- 3Department of Chemistry, Hankuk University of Foreign Studies, Yongin, Republic of Korea
- 4Department of Genetics, Forestry and Agriculture Biotechnology Institute, University of Pretoria, Pretoria, South Africa
Common saprophytic fungi are seldom present in Protea infructescences, which is strange given the abundance of mainly dead plant tissue in this moist protected environment. We hypothesized that the absence of common saprophytic fungi in Protea infructescences could be due to a special symbiosis where the presence of microbes producing antifungal compounds protect the infructescence. Using a culture based survey, employing selective media and in vitro antifungal assays, we isolated antibiotic producing actinomycetes from infructescences of Protea repens and P. neriifolia from two geographically separated areas. Isolates were grouped into three different morphological groups and appeared to be common in the Protea spp. examined in this study. The three groups were supported in 16S rRNA and multi-locus gene trees and were identified as potentially novel Streptomyces spp. All of the groups had antifungal activity in vitro. Streptomyces sp. Group 1 had inhibitory activity against all tested fungi and the active compound produced by this species was identified as fungichromin. Streptomyces spp. Groups 2 and 3 had lower inhibition against all tested fungi, while Group 3 showed limited inhibition against Candida albicans and Sporothrix isolates. The active compound for Group 2 was also identified as fungichromin even though its production level was much lower than Group 1. The antifungal activity of Group 3 was linked to actiphenol. The observed antifungal activity of the isolated actinomycetes could contribute to protection of the plant material against common saprophytic fungi, as fungichromin was also detected in extracts of the infructescence. The results of this study suggest that the antifungal Streptomyces spp. could play an important role in defining the microbial population associated with Protea infructescences.
Introduction
Species of the genus Protea (Proteaceae) are shrubs and small trees distributed throughout sub-Saharan Africa (Rourke, 1980). A total of 114 species are known of which the greatest diversity occurs in the Cape Floristic Region (Rourke, 1980). The flowers of Protea spp. are arranged into inflorescences (Rourke, 1980) and the mature seeds are stored for long periods of time in these structures before they are released when environmental conditions are suitable for seed germination (Bond, 1985). Mature, closed inflorescences containing seeds are referred to as infructescences. The infructescences of many species remain closed until the time of dispersal and consist of abundant dead plant tissue in a moist, warm, and often insect-colonized environment.
A remarkable feature of the closed infructescences of Protea spp. is that very few fungi are found colonizing them at any given time, despite the fact that they provide the perfect conditions in which saprotrophic fungi would typically thrive (Marais and Wingfield, 1994; Zhou and Hyde, 2001; Roets et al., 2013). One would expect to find common saprophytes such as species of Trichoderma, Penicillium and the many other fungi frequently encountered under similar conditions in nature, but this is not the case. A number of studies have shown that before onset of senescence this enclosure is mainly occupied by a specialized group of ascomycete fungi, often referred to as the ophiostomatoid fungi, belonging to either the Ophiostomatales or Microascales (Marais and Wingfield, 1994; Marais et al., 1998; Wingfield et al., 1999; Roets et al., 2008, 2010, 2013). The ophiostomatoid associates of Protea infructescences from Southern Africa mainly reside in the genera Sporothrix and Knoxdaviesia (= Gondwanamyces) (Marais and Wingfield, 1994; Marais et al., 1998; Wingfield et al., 1999; Roets et al., 2008, 2010, 2013; de Beer et al., 2013a,b, 2016).
Our hypothesis to explain the absence of saprophytes in the infructescences is that antibiotic producing bacteria may suppress their growth in these environments. The aim of this study was to consider whether antibiotic producing bacteria occur in the infructescences of Protea repens and P. neriifolia. The antifungal activity of such isolates against common saprophytes and the ophiostomatoid associates that are typically present were also determined.
Materials and Methods
Isolation of Bacterial Strains
P. repens infructescences (Figure 1) were collected at Pringle Bay and at the J.S. Marais Botanical gardens near Stellenbosch, and P. neriifolia infructescences from Pringle Bay and Franschhoek, in the Western Cape Province of South Africa. The infructescences were aseptically opened and parts from the individual flowers, outer involucral bracts, inner involucral bracts, and involucral receptacle were removed. Tissue samples from the different floral parts were suspended in a solution of 6% yeast extract and 0.05% SDS (Sodium Dodecyl Sulphate) and incubated at 40°C for 20 min (Hayakawa and Nonomura, 1989) followed by dilution to 10−6 to remove the antimicrobial detergent effect of SDS (Hayakawa and Nonomura, 1989). These suspensions were then plated onto three types of artificial media: YEME as described by Cafaro and Currie (2005), glycerol-asparagine agar (Pridham and Lyons, 1961) and starch casein agar (Kuster and Williams, 1964).
Plates were incubated at 28°C for approximately 14 days, whilst being continuously monitored. Colonies provisionally identified as actinomycetes based on morphology were obtained in pure culture by repeated isolation on YEME (Cafaro and Currie, 2005). Isolates were then grouped based on similar colony morphology. Representatives were selected from each group for further study. Care was taken to include isolates from each host species and location sampled.
DNA Extraction and Sequencing
Genomic DNA was isolated by adding mycelium from each single pure colony into Prepman Ultra™ solution (Applied Biosystems). For identification of isolates, the 16S ribosomal RNA gene region was selected for sequencing. For further phylogenetic placement of isolates, the trpB, rpoB, and gyrB gene regions were amplified. The 16S rRNA gene region was amplified using primers pA and pH designed by Edwards et al. (1989). The methods from Guo et al. (2008) were used for the trpB and rpoB PCR reactions and those from Rong et al. (2009) for the gyrB gene. All reactions were performed using SuperTherm taq polymerase (Southern Cross Biotechnology, Cape Town, South Africa) on an Applied Biosystems Veriti Thermal Cyler. The 16S rRNA (Edwards et al., 1989), gyrB (Rong et al., 2009), rpoB and trpB (Guo et al., 2008) amplicons were sequenced using the methods previously described. Purified PCR products were sequenced using the ABI BigDye system (Applied Biosystems) and purified using sodium acetate precipitation. Sequences were analyzed on an ABI 3130 sequence analyzer (Applied Biosystems).
Phylogenetic Analysis
Isolates obtained were initially identified by comparing their 16S rRNA sequences to all type strain sequences available in the RDP database using the Seqmatch program hosted by the Ribosomal Database Project II (RDP-II) (Maidak et al., 2001). The closest matching sequences of validly described species were downloaded and included in a single data set with our sequences. Mycobacterium tuberculosis was used as an outgroup.
The sequences for the 16S rRNA, gyrB, rpoB, and trpB (Supplementary Table S1) were concatenated along with those from selected reference strains using SequenceMatrix software (Vaidya et al., 2010). Sequence ends were trimmed using Bioedit (Hall, 1999) and the sequences aligned using MAFFT (Katoh et al., 2005). The best-fitting nucleotide substitution model was determined using JModeltest version 0.1 (Posada, 2008) and selected using the Akaike Information Criteria (Posada and Buckley, 2004). A maximum-likelihood phylogeny was produced with PhyML version 3.0 (Guindon et al., 2009) using the TIM3 nucleotide substitution model for 16S rRNA and the TVM+I+G nucleotide substitution model for the concatenated data set (Guindon et al., 2009; Darriba et al., 2012). Tree confidence was measured using a bootstrap search with 1000 replicates (Felsenstein, 1985). The tree was visualized using Mega 4 (Tamura et al., 2007).
In vitro Antifungal Assays of Streptomyces spp. Strains from Protea Infructescences
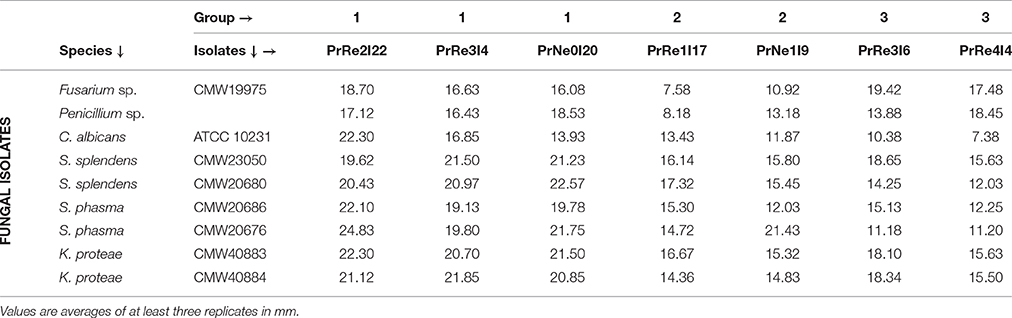
Antifungal bioassays were performed using the method described by Cafaro and Currie (2005). Bacterial isolates used in bioassays were randomly selected, with at least two isolates chosen from each group (Table 1). The fungal isolates selected were representative of fungi occurring in infructescences of Protea spp. and ubiquitous saprophytic fungi. Knoxdaviesia proteae and Sporothrix splendens are the most common fungi in the infructescences of P. repens, and S. phasma is common in P. neriifolia (Roets et al., 2009). Fusarium and Penicillium spp. are ubiquitous saprophytic fungi and have occasionally been reported from the infructescences of Protea spp. (Lee et al., 2005; Visagie et al., 2014). Candida albicans is sensitive to many antifungal compounds and is a common isolate used in antifungal assays which could serve as a control for antifungal activities observed. All fungi were retrieved from the CMW culture collections (Forestry and Agriculture Biotechnology Institute, University of Pretoria, South Africa; Table 2), except for C. albicans (retrieved from the ATCC) and the Penicillium sp., which was isolated from a P. repens infructescence (data not shown). The clear zones between fungal growth and putative antibiotic producing bacterial isolate were measured. This is referred to as the zone of inhibition and is used as an indication of the potency of antibiotic and the level of susceptibility of the test organism (Cafaro and Currie, 2005).
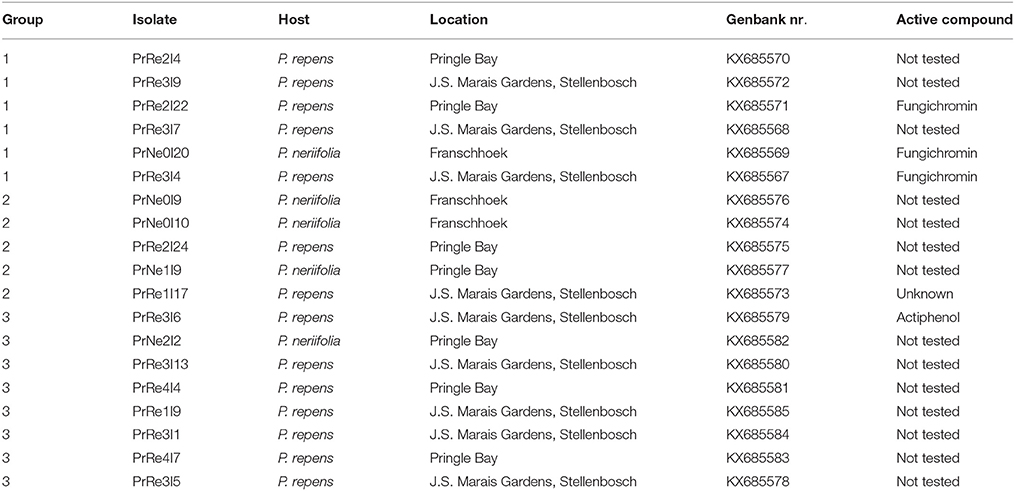
Table 1. Isolates of Streptomyces obtained during the present study, their respective sources, and groupings based on culture morphology and 16S rRNA gene sequences and potential antifungal compounds.
Profiling Secondary Metabolite Production of Streptomyces spp.
All the Streptomyces strains were cultivated in 200 mL YEME (4 g yeast extract, 10 g malt extract, and 4 g glucose in 1 L distilled water) medium in 500 mL Erlenmeyer flasks. The profiles of secondary metabolites of the bacterial strains were analyzed by LC/MS every 2 days. When the cultures were monitored, 10 mL aliquot of each culture was extracted with 10 mL of ethyl acetate (EtOAc). The extracts were concentrated in vacuo and resuspended in 1 mL of methanol (MeOH). After filtering the methanol-soluble extracts, 10 μL from each concentrated extract in MeOH was injected to LC/MS (Agilent 1200 Series HPLC coupled with Agilent 6130 single quad mass spectrometer). Gradient solvent conditions were consistently used (0~20 min: 10~100% CH3CN in H2O, 20~25 min: 100% CH3CN/flow rate: 0.7 mL/min, 0.1% formic acid in CH3CN and H2O). A diode array UV detector scanned from 200 to 700 nm. The mass spectrometric analyses were performed on both positive and negative ion modes. The acquired LC/MS profiles were analyzed with Agilent Chemstation 4.3 software.
Identification of Major Secondary Metabolites from Streptomyces spp. Strains
The UV spectra of the major common secondary metabolites detected in the LC/MS chemical profiling were compared with the house-built UV spectral library interlinked with Agilent Chemstation 4.3 software. For further analysis, the major secondary metabolite of Streptomyces Group 1 was produced by large-scale (18 L) cultivation of a representative strain (PrRe2I22). The whole culture was extracted with 27 L ethyl acetate to yield 5 g of dry material. The extract was fractionated by a reversed-phase (C18) chromatographic column (20, 40, 60, 80, and 100% MeOH in water). The major metabolite eluted in the 60 and 80% MeOH fractions. These fractions were subjected to further purification by HPLC (Kromasil 100-5-C18 250 × 10 mm, flow rate 2 mL/min, UV 360 nm detection, with 60% aqueous MeOH, 0.1% formic acid) to isolate the major compound. The major compound eluted at 35 min to yield 15 mg as yellow soild. The structure of the major metabolite of the strain PrRe2I22 was identified by NMR spectroscopic analysis including 1H, 13C, COSY (correlation spectroscopy), HSQC (heteronuclear single quantum correlation), and HMBC (heteronuclear multiple bond correlation) experiments as well as literature comparison (Noguchi et al., 1988). The major metabolite of Group 2 was compared with that of Group 1 because their spectroscopic and chromatographic properties were identical. The major compound produced by Group 3 was identified by the UV and MS data acquired by LC/MS analysis and 1H and 13C NNR spectra.
In vitro Antifungal Assay of Fungichromin Produced by Streptomyces spp. Strains
Colonies of the fungal strains were inoculated onto the center of 90 mm petri dishes containing YEME agar and incubated at 28°C. After 4 days, four 6 mm diameter sterile paper disks were placed on each inoculated agar petri dish. Sterile paper disks were imbued with three concentrations (10, 20, 50 μL) of fungichromin (10 mg/mL) dissolved in DMSO, and for the negative control 10 μL DMSO was used. Every petri dish was monitored for 28 days.
Detection of Fungichromin Directly in an Infructescence Specimen
An infructescence specimen was dissected into the involucral bracts and florets. Then each part was soaked in MeOH and organic compounds were extracted with MeOH for 3 h. The MeOH extracts were analyzed by LC/MS using the following gradient conditions; 0~20 min: 10~100% CH3CN in H2O, 20~25 min: 100% CH3CN/flow rate: 0.7 mL/min, 0.1% formic acid in CH3CN and H2O. A diode array UV detector scanned from 200 to 700 nm. The mass spectrometric analyses were performed on both positive and negative ion modes. Matrix-assisted laser desorption/ionization (MALDI) MS was also performed against pure fungichromin and the infructescence specimen. For MALDI MS of pure fungichromin, 0.2 mg/mL fungichromin in MeOH (0.5 μL) was first spotted and dried on the 384-well MALDI target plate (ASTA Inc., Suwon, Korea) followed by 0.5 μL of the matrix solution. The matrix solution was 10 mg/mL 2, 5-dihydroxybenzoic acid (DHB) in acetonitrile/water (50:50 v/v). For direct analysis of infructescence specimen, dissected florets were first attached onto the MALDI target plate by using a conductive double-sided tape and gently dried under moderate vacuum (~50 Torr). After drying, the DHB matrix solution was sprayed on a floret surface by using an airbrush. MALDI mass spectra were recorded in the positive reflectron mode by using an ABI 4800 Plus MALDI-TOF/TOF analyzer (Applied Biosystems, Foster City, CA, USA).
Results
Isolations of Bacterial Strains
A total of 83 putative actinomycetes were obtained. All of these isolates could be classified in one of three morphological groups. Isolates residing in Streptomyces sp. Group 1 had soft black colonies, produced gray spores and a yellow substrate pigment was observed in surrounding growth medium. Members belonging to Streptomyces sp. Group 2 produced yellow translucent colonies with gray spores. A red substrate pigment was produced by all isolates in this group. Streptomyces sp. Group 3 had white colonies and produced light gray spores while a red substrate pigment was observed. From the total number of isolates, 20 representing the different morphological groups were selected (Table 1) for further detailed identification.
Isolates of Streptomyces sp. Group 1 were from P. repens in Pringle Bay and Stellenbosch, and P. neriifolia in Franschhoek. Streptomyces sp. Group 2 isolates were from both hosts at all locations sampled, while Streptomyces sp. Group 3 isolates were from P. neriifolia in Pringle Bay and P. repens in Pringle Bay and Stellenbosch (Table 1).
DNA Sequencing and Phylogenetic Analysis
The aligned 16S rRNA data set consisting of sequences obtained in the present study and those of type strains from RDP database consisted of approximately 650 characters from the species-specific α-region of the 16S rRNA gene sequence (Kataoka et al., 1997). Maximum likelihood analysis confirmed that isolates having a similar morphology also grouped together based on 16S rRNA sequences (Figure 2). Furthermore, all the isolates collected in this study grouped among species of Streptomyces.
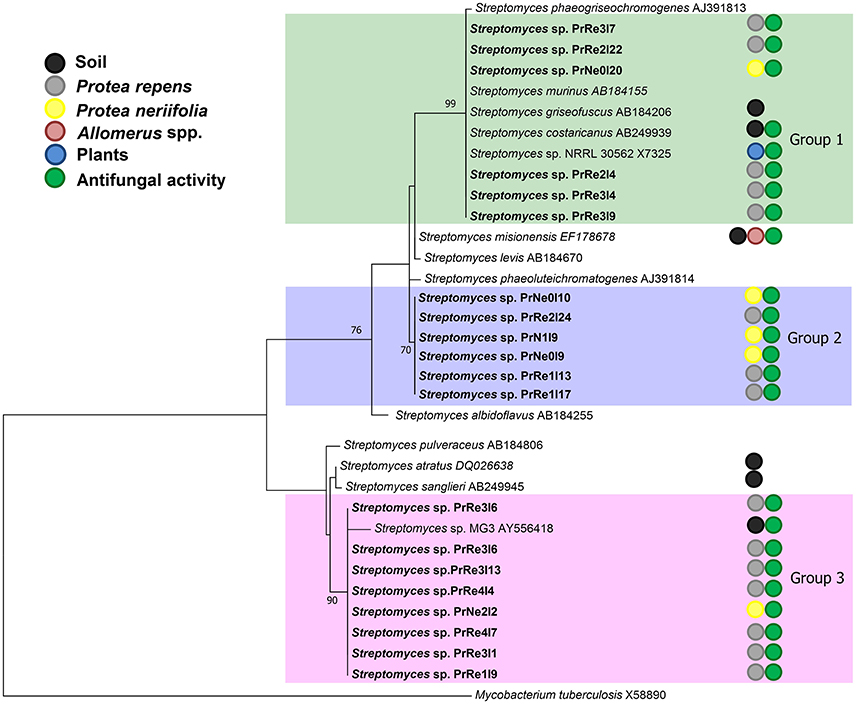
Figure 2. A ML tree exhibiting the phylogenetic relationships based on 16S rRNA gene sequence of Streptomyces species obtained during the present study. Isolate numbers from this study are printed in bold type. Groups of isolates are indicated by shaded areas.
According to the 16S rRNA maximum likelihood phylogeny, isolates belonging to Streptomyces sp. Group 1 formed a well-supported, distinct clade with the type strains of S. griseofuscus, S. murinus, S. phaegriseochromogenes, S. costaricanus, and an undescribed Streptomyces sp. NRRL 30562 from snakevine in Australia (Castillo et al., 2002, 2006, Figure 2). The ML phylogenetic tree based on the four combined genes was able to separate our isolates in this group into two clades; one which included isolates from P. repens (PrRe2I4, PrRe2I22, PrRe3I4) with 90% bootstrap support, while PrNe0I20 from P. neriifolia formed a sister clade and grouped with the type strain of S. murinus with 77% bootstrap support (Figure 3). These two clades formed a larger group which was separated (93% bootstrap support) from the S. costaricanus, S. phaeogriseochromogenes, and S. griseofuscus (Figure 3). Isolates from this group could belong to S. murinus or the proposed species S. padanus (Xiong et al., 2012).
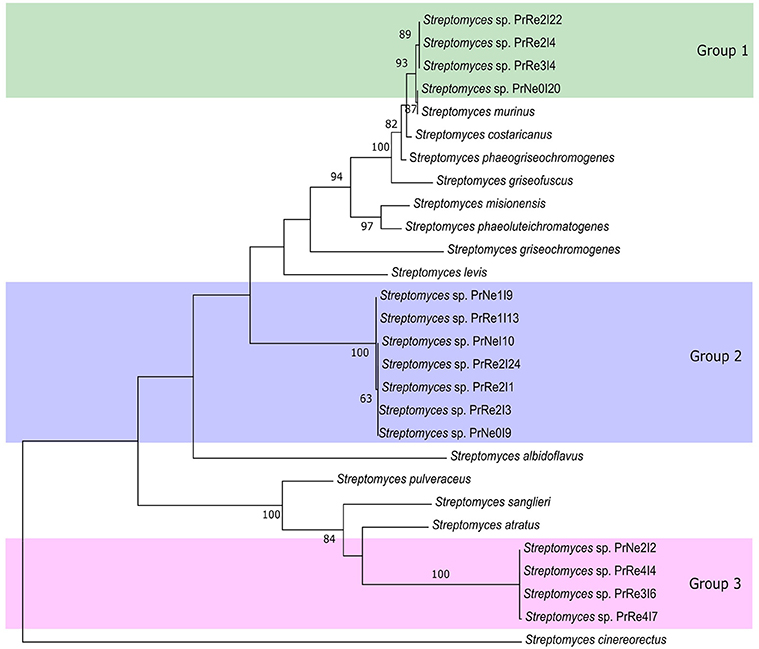
Figure 3. Maximum likelihood tree showing phylogenetic placement of our isolates based on concatenated 16S rRNA-trpB-rpoB-gyrB alignments. Only bootstrap values above 65% are indicated. Shaded areas indicate groups of isolates.
In the 16S rRNA phylogenetic tree, isolates in Streptomyces sp. Group 2 formed its own clade which did not include any type strain sequences (Figure 2). The closest type strains were those from S. misionensis, S. levis, and S. phaeoluteichromatogenes (Figure 2). In the MLSA phylogenetic tree, these isolates also clustered separately with considerable distance to all type strains and with 100% bootstrap support and thus most likely represents an undescribed species. Interestingly, this clade formed a subgroup containing isolates from both P. repens and P. neriifolia.
Members of Streptomyces sp. Group 3 formed a distinct cluster with 93% bootstrap support with an unidentified strain (MG3) from soil in Germany (Hoster et al., 2005). According to our MLSA, these isolates remained in a distinct cluster, with 100% bootstrap support and were separated from the type strains of S. atratus and S. sanglieri (Figure 3).
In vitro Antifungal Assays
Seven bacterial isolates were included in the assays and these represented the three different groups identified from Protea infructescences (Table 2, Figure 4). Isolates residing in Streptomyces Group 1 generally had strong inhibitory activities against all tested fungi and the antifungal activity against Sporothrix spp. and K. proteae was generally higher than against saprophytic test fungi (Table 2, Figure 4).
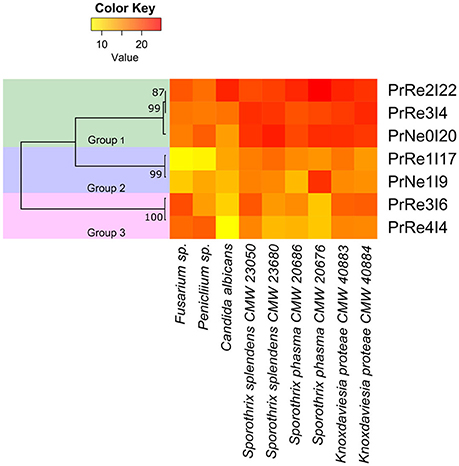
Figure 4. Heatmap illustrating in vitro bioassay results. Values represented from 0 to 22.8 on a continuous scale from yellow to red. Phylogenetic relationships between bacterial isolates are presented to the left of the heatmap.
Isolates belonging to Streptomyces Group 2 was able to inhibit the growth of all tested fungi, although this was generally limited and much lower than observed for members of Group 1 and only limited inhibition of the two saprophyte test fungi was recorded (Table 2, Figure 4). Streptomyces sp. Group 3 had noticeable inhibition against Penicillium and Fusarium isolates, similar to the inhibition observed for Group 1. Interestingly, isolates from this group had lower levels of inhibition against the Sporothrix spp. tested fungi, while K. proteae was generally inhibited to levels similar to Penicillium and Fusarium. The isolates from Group 3 also had much lower inhibitory activity against C. albicans.
Identification of Major Secondary Metabolites Produced by Streptomyces spp. Strains and Antifungal Activity
To identify secondary metabolites from the Streptomyces spp. strains responsible for the growth inhibition of fungi, we cultivated Groups 1 to 3 Streptomyces spp. strains in liquid culture medium. Based on the LC/MS (liquid chromatography and mass spectrometry) chemical profiling of the EtOAc extracts of the cultures, a common major secondary metabolite in Group 1 and 2 strains was distinctively detected. This compound displayed a typical UV spectral feature (λmax at 325, 340, and 362 nm), indicating the existence of five conjugated double bonds, and the low resolution molecular ion [M+Na]+ at m/z 693. Comparison of the UV spectrum of the compound with our house-built UV library interlinked with the LC/MS software clearly indicated that this metabolite was similar to pentamycin, which bears a pentaene moiety. For definite structure identification, a large culture (18 L) of the Streptomyces sp. strain (PrRe2I22) was prepared. Subsequently, we analyzed 1H (Supplementary Figure S1), 13C (Supplementary Figure S2), and two-dimensional NMR spectra of the compound including 1H-1H correlation (COSY; Supplementary Figure S3), heteronuclear single quantum coherence (HSQC; Supplementary Figure S4), and heteronuclear multiple bond correlation (HMBC; Supplementary Figure S5). The spectroscopic analysis and the comparison of the spectroscopic data with the literature (Noguchi et al., 1988) unequivocally identified the compound of Groups 1 and 2 as fungichromin, which is also called pentamycin, a polyene polyketide secondary metabolite as we expected (Figure 5). Therefore, the major secondary metabolite of Groups 1 and 2 Streptomyces strains was elucidated as fungichromin. We confirmed that fungichromin is responsible for the antifungal activity observed using an antifungal assay (Figure 5). Petri dishes were observed daily for 28 days (Figure 5). Fungichromin inhibited the growth of the fungi in a dose-dependent manner (Figure 5).
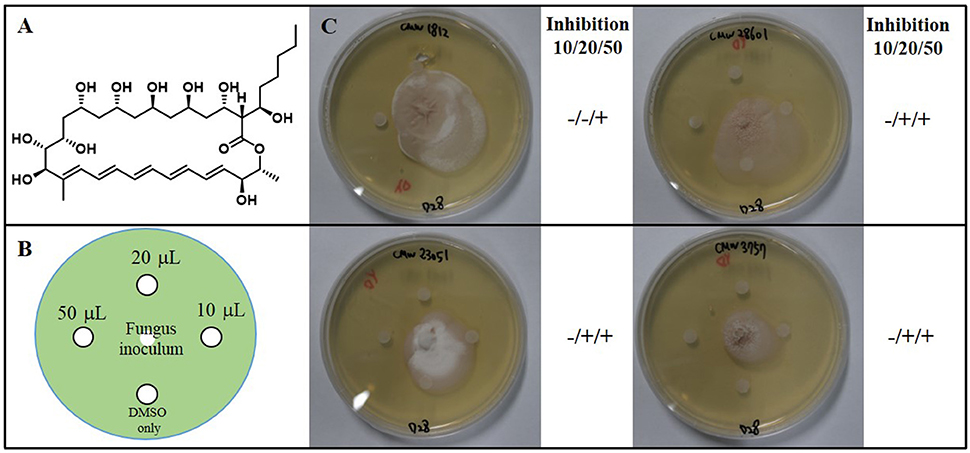
Figure 5. (A) The chemical structure of fungichromin. (B) Petri plate antifungal activity assay, showing the placement of three concentrations (10 μL, 20 μL, 50 μL) of fungichromin (10 mg/mL) dissolved in DMSO, the control (DMSO only, 10 μL), and the placement of the fungal inoculum. (C) Representative image examples of fungichromin activity against Sporothrix africana CMW 1812, S. variecibatus CMW 23051, S. protea-sedis CMW 28601, Knoxdaviesia proteae CMW 3757, 28 days after inoculation.
A common major secondary metabolite in Group 3 strains was also detected. This compound showed a distinct UV spectral feature (λmax at 220, 260, and 350 nm) and the low resolution molecular ion [M+H]+ at m/z 276. Compared with our house-built UV library, the molecular ion detected in MS, and 1H and 13C NMR spectra (Supplementary Figures S6, S7), this metabolite was identified as actiphenol (Highet and Prelog, 1959; Sun et al., 2014), which belongs to the cycloheximide class (Figure 6).
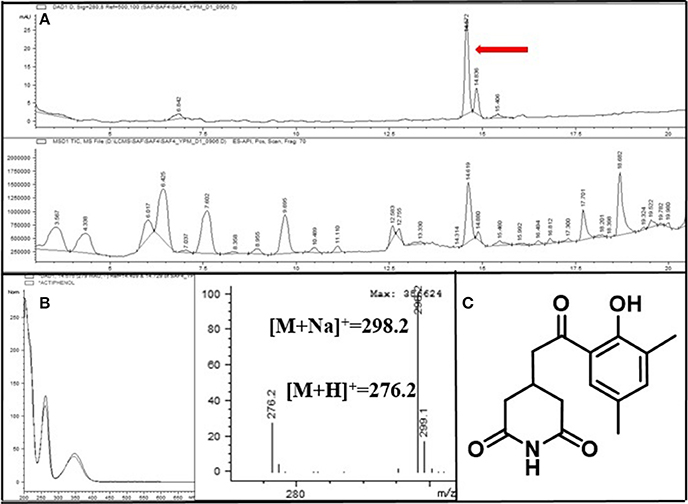
Figure 6. (A) LC/MS chemical analysis of the liquid culture of the Streptomyces strain PrRe3I6. UV chromatogram at 280 nm (upper), and mass chromatogram (lower). (B) UV and mass spectra of the major secondary metabolite, actiphenol. (C) The chemical structure of actiphenol.
Detection of Fungichromin in Plant Material
The LC/MS analysis of the MeOH extraction of the infructescence specimen clearly detected fungichromin in the involucral bracts and florets. The exact ion [M+Na]+ at m/z 693 was observed at the identical retention time to that of pure fungichromin (Figure 7). We also attempted to detect fungichromin directly from the infructescence specimen by MALDI MS. Figure 8 shows MALDI mass spectra of pure fungichromin and the floret part of the infructescence specimen. For the pure fungichromin, sodium and potassium adduct ions of fungichromin ([M + Na]+ and [M + K]+) were clearly detected at m/z 693.50 and 709.48, respectively (Figure 8A). In addition to these ions, another intense signal was observed at m/z 1363.50 and this signal was found to be sodium adduct ions of fungichromin dimer ([2M + Na]+) by MALDI tandem MS (MS/MS). After confirming that fungichromin is readily detected by MALDI MS, we performed direct analysis of the infructescence specimen by MALDI MS. Figure 8B shows a chemical fingerprint obtained directly from this specimen. From the floret part of the infructescence specimen, both sodium adduct ions of fungichromin and its dimer were detected by MALDI MS. Although the signal corresponding to [M + Na]+ at m/z 693.50 was crowded by other metabolite ion signals, the signal corresponding to [2M + Na]+ at m/z 1363.50 was distinctly visible (Figure 8B).
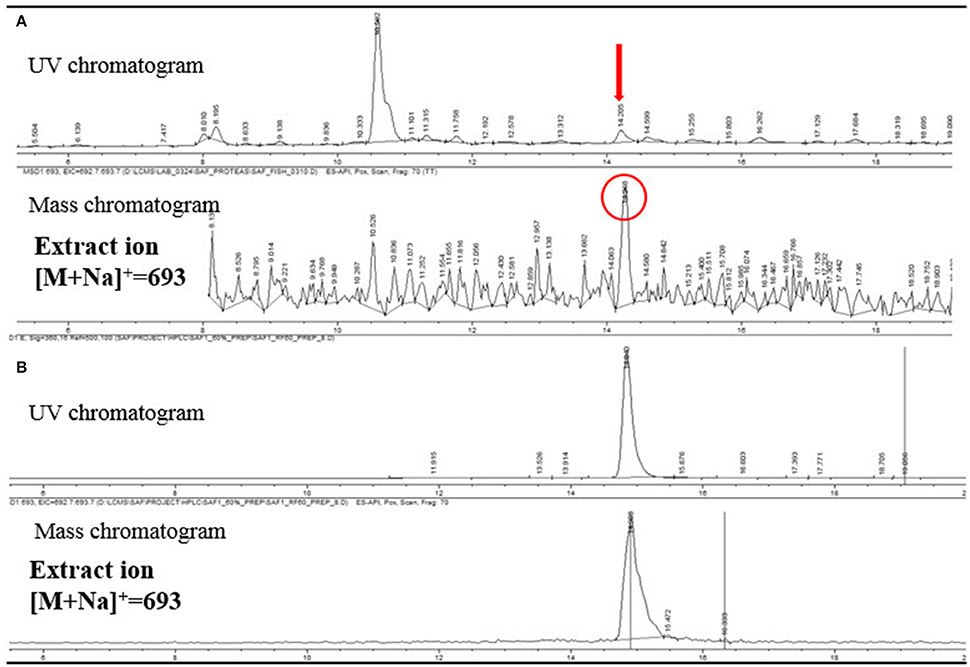
Figure 7. (A) LC/MS analysis of the extract of a Protea infructescence specimen. UV chromatogram (upper) and mass chromatogram (lower). (B) LC/MS analysis of pure fungichromin (UV detection at 360 nm).
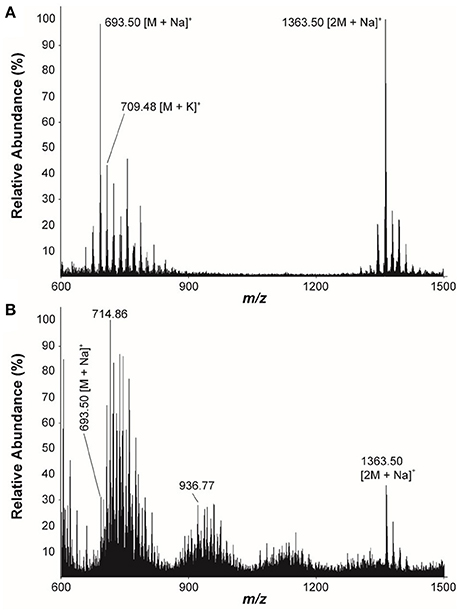
Figure 8. Positive ion-mode MALDI mass spectra of (A) The pure fungichromin and (B) The infructescence specimen (floret part).
Discussion
This study represents the first discovery of antibiotic producing actinomycete bacteria in the infructescences of Protea spp. In all, three main groups of actinomycetes were identified and they were all species of Streptomyces. Bioassays showed that some of these bacteria were antagonistic to saprotrophic fungi found on the outside of Protea infructescences. Results of this study suggest that the presence of actinomycetes that produce antifungal compounds might contribute to the absence of saprotrophic fungi in the nutrient rich, moist and warm environment found in Protea infructescences (Roets et al., 2012).
Isolates from Streptomyces spp. Group 1 had identical 16S rRNA sequences to Streptomyces sp. NRRL 30562 (Castillo et al., 2002), an endophyte from Kennedia nigriscans and identified as S. padanus, a species that has not yet been validly described. The type strain most closely resembling our isolates was S. murinus (Frommer, 1959). Strains identified as S. padanus, isolated from soil (Xiong et al., 2012) has also been reported from mountain laurel (Streptomyces sp. AOK-30) (Ericaceae) (Nishimura et al., 2002), which could be reinfected to become more resistant to fungal pathogens (Nishimura et al., 2002). Streptomyces padanus generally produces the antibiotics actinomycin (Kurosawa et al., 2006) and fungichromin (Shih et al., 2003; Xiong et al., 2012). Fungichromin was also identified as the active compound associated with this group of isolates. Production of this compound under natural conditions was also demonstrated as it could be detected in the infructescence material.
Our MLSA showed that Streptomyces sp. Group 2 is an undescribed species and most closely resemble the type strains of S. phaeoluteichromatogenes, S. misionensis, and S. levis. The origin of S. phaeoluteichromatogenes and S. levis is not known, but S. misionensis was originally isolated from soil in Argentina. Strains identified as S. misionensis have been used as biocontrol agents against Fusarium pathogens of Lilium spp. (Chung et al., 2011). Streptomyces misionensis has also been detected on ants (Allomerus spp.) and was found to antagonize fungal pathogens of the ant colonies (Seipke et al., 2012).
Streptomyces sp. Group 3 was shown to be an undescribed species, most similar to S. atratus and S. sanglieri, both species of which have been reported from soil (Manfio et al., 2003). Streptomyces sanglieri produces the antibiotic lactonamycin Z that has antibacterial and antitumor activity (Zhang et al., 2008). Streptomyces sp. MG3, the closest matching sequence from Genbank, was isolated from soil in Germany and had strong antifungal activity due to the production of a chitinase (Hoster et al., 2005).
All three groups of isolates were found on both P. repens and P. neriifolia infructescences, and it is clear that actinomycetes are commonly present in the infructescences of Protea plants. Our results are concordant with other studies that detected antibiotic producing Streptomyces spp. in plants, with one from a member of the family Proteaceae. Isolates residing in Streptomyces sp. Group 1 are very similar to those found in K. nigriscans in Australia, which produce munumbicin A, B, C, and D, active against many pathogenic bacteria and fungi (Castillo et al., 2002, 2006). Grevillea pteridifolia, a member of the Proteaceae from Australia, is a host to Streptomyces spp. that produce the range of antibiotics known as the kakadumycins (Castillo et al., 2003). Monstera sp. harbors Streptomyces endophytes producing the coronamycin antibiotics (Ezra et al., 2004). All of these are thought to provide some protection to the host plant (Strobel and Daisy, 2003).
Most of the species related to the isolates obtained in this study originate predominately from soil, where Streptomyces spp. most commonly occur (Kampfer, 2006). Members of the mite family Edbakerellidae commonly occur in soil, and several new species of this family have recently been discovered in Protea infructescences (Theron et al., 2012). Streptomyces spp. have spores that are well-adapted to arthropod dispersal (Ruddick and Williams, 1972; Chater, 2006). Furthermore, it has been shown that S. griseus spores adhere to the exoskeletons of mites, effectively facilitating its spread (Goodfellow and Williams, 1983) and it is reasonable to hypothesize that these mites play a role in maintaining Streptomyces spp. in Protea infructescences.
Streptomycetes are able to digest several complex carbohydrates and nitrogenous wastes including a multitude of nutrients inaccessible to many other organisms (Kaltenpoth, 2009). They also utilize chitin from fungal debris or insect exoskeletons, lignocellulose, and several other plant associated polymers (Crawford, 1978; Kampfer, 2006). Due to the potential of Streptomyces to grow in very diverse environments and the ease with which they are dispersed by arthropods (Ruddick and Williams, 1972; Goodfellow and Williams, 1983), they could potentially play an important role in the interaction between insects, mites, fungi, and Protea spp.
All three groups of Streptomyces spp. isolated in this study had noticeable antifungal activity. Members from Streptomyces spp. Group 1 had the strongest antifungal activity against all tested fungi. Fungichromin, a polyene antifungal compound, is known to inhibit a broad spectrum of fungi (Dixon and Walsh, 1996). Streptomyces spp. Groups 2, of which the major secondary metabolite is also fungichromin, had stronger inhibitory activity against the ophiostomatoid fungi than against the non-ophiostomatoid saprophytes, possibly due to their slower growth and an optimal level of fungichromin production. Members of Streptomyces spp. Group 3 produced actiphenol, a compound belonging to the antifungal cycloheximide class (Highet and Prelog, 1959). This group allowed for better growth of Sporothrix spp. and C. albicans, known to be tolerant of cycloheximide (Yamada-Okabe and Yamada-Okabe, 2002; Roets et al., 2008). No possible benefit to K. proteae could be deduced from in vitro bioassay results, but it is clear that common saprotrophic fungi are sensitive to Streptomyces spp. from Protea plants.
There are two potential benefits provided to the plant by actinomycete bacteria. Protea infructescences provide an enclosed environment with the ideal requirements for fungal growth. They are, therefore, at some risk from invasion by degrading saprophytes and pathogens. Furthermore, once seeds have spread and come into contact with soil, they will come into contact with more potentially harmful microbes. Streptomyces spp. have the ability to provide continued protection as has been shown in maize where the levels of seed pathogenic fungi have been reduced through treatment with a mixture of Streptomyces strains (Bressan, 2003). Thus, Streptomyces spp. most likely provide various advantages to Protea infructescences and these would have provided a positive force to maintain the relationship between them.
Future studies should determine the mechanism of inoculation of Streptomyces spp. in Protea spp., including their ability to be spread and grazed upon by arthropods inhabiting Protea infructescences. Possible mutualistic interactions need to be investigated, which can be achieved by wider sampling across geographic regions and over time, and should include the effect of more variables such as temperature, rainfall, soil type, and associated animal diversity. Functional metagenomics is an emerging field of study that may also be applied to this niche. Knowledge of the fungal and bacterial secondary metabolite gene clusters present in this niche may provide very important answers toward understanding the complex interactions between Protea spp. and the diversity of microorganisms and animals associated with them.
Author Contributions
MW, ZdB, BS, ZH, and SV conceptualized the study. ZH did isolations and assays in the laboratory. ZH, ZdB, and SV did phylogenetic and assay data analyses. DO designed chemical analysis and interpreted chemical data. KM performed LC/MS and NMR analysis and identified antifungal compounds. MB purified and identified actiphenol by NMR spectroscopic data. SC contributed to mass spectrometric analysis. ZH, SV, KM, DO, ZdB, SC, BS, and MW wrote and edited the text. ZH and KM contributed equally to this work.
Funding
We acknowledge financial support from the Department of Science and Technology (DST)-National Research Foundation (NRF) Centre of Excellence in Tree Health Biotechnology (CTHB), the National Research Foundation (NRF), and the University of Pretoria, South Africa. This work was also supported by a National Research Foundation of Korea Grant funded by the Korean Government (Ministry of ICT and Future Planning; No. 2014R1A2A1A11053477).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Francois Roets (University of Stellenbosch) for assistance with field collections.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01657/full#supplementary-material
References
Bond, W. J. (1985). Canopy-stored seed reserves (serotiny) in cape proteaceae. S. Afr. J. Bot. 51, 181–185. doi: 10.1016/S0254-6299(16)31669-6
Bressan, W. (2003). Biological control of maize seed pathogenic fungi by use of actinomycetes. Biocontrol 48, 233–240. doi: 10.1023/A:1022673226324
Cafaro, M. J., and Currie, C. R. (2005). Phylogenetic analysis of mutualistic filamentous bacteria associated with fungus-growing ants. Can. J. Microbiol. 51, 441–446. doi: 10.1139/w05-023
Castillo, U. F., Strobel, G. A., Ford, E. J., Hess, W. M., Porter, H., Jensen, J. B., et al. (2002). Munumbicins, wide-spectrum antibiotics produced by Streptomyces NRRL 30562, endophytic on Kennedia nigriscans. Microbiology 148, 2675–2685. doi: 10.1099/00221287-148-9-2675
Castillo, U. F., Strobel, G. A., Mullenberg, K., Condron, M. M., Teplow, D. B., Fogliano, V., et al. (2006). Munumbicins E-4 an E-5: novel broad spectrum antibiotics from Streptomyces NRRL 3052. FEMS Microbiol. Lett. 255, 296–300. doi: 10.1111/j.1574-6968.2005.00080.x
Castillo, U., Harper, J. K., Strobel, G. A., Sears, J., Alesi, K., Ford, E., et al. (2003). Kakadumycins, novel antibiotics from Streptomyces sp. NRRL 30566, an endophyte of Grevillea pteridifolia. FEMS Microbiol. Lett. 224, 183–190. doi: 10.1016/S0378-1097(03)00426-9
Chater, K. F. (2006). Streptomyces inside-out: new perspective on the bacteria that provide us with antibiotics. Philos. Trans. R. Soc. B 361, 761–768. doi: 10.1098/rstb.2005.1758
Chung, W. C., Wu, R. S., Hsu, C. P., Huang, H. C., and Huang, J. W. (2011). Application of antagonistic rhizobacteria for control of Fusarium seedling blight and basal rot of lily. Australas. Plant Pathol. 40, 269–276. doi: 10.1007/s13313-011-0040-3
Crawford, D. L. (1978). Lignocellulose decomposition by selected Streptomyces strains. Appl. Environ. Microbiol. 35, 1041–1045.
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772. doi: 10.1038/nmeth.2109
de Beer, Z. W., Duong, T. A., and Wingfield, M. J. (2016). The divorce of Sporothrix and Ophiostoma: solution to a problematic relationship. Stud. Mycol. 83, 165–191. doi: 10.1016/j.simyco.2016.07.001
de Beer, Z. W., Seifert, K. A., and Wingfield, M. J. (2013a). “A nomenclator for ophiostomatoid genera and species in the ophiostomatales and microascales,” in The Ophiostomatoid Fungi: Expanding Frontiers, eds K. A. Seifert, Z. W. de Beer and M. J. Wingfield (Utrecht: CBS), 245–322.
de Beer, Z. W., Seifert, K. A., and Wingfield, M. J. (2013b). “The ophiostomatoid fungi: their dual position in the Sordariomycetes,” in The Ophiostomatoid Fungi: Expanding Frontiers, eds K. A. Seifert, Z. W. de Beer and M. J. Wingfield (Utrecht: CBS), 1–19.
Dixon, D. M., and Walsh, T. J. (1996). “Antifungal agents,” in Chapter 76: Medical Microbiology, 4th Edn., ed S. Baron (Galveston, TX: University of Texas Medical Branch at Galveston).
Edwards, U., Rogall, T., Blöcker, H., Ernde, M., and Böttger, E. (1989). Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17, 7843–7853. doi: 10.1093/nar/17.19.7843
Ezra, D., Castillo, U. F., and Strobel, G. A. (2004). Coronamycins, peptide antibiotics produced by a verticillate Streptomyces sp. (MSU-2110) endophyte on Monstera sp. Microbiology 150, 785–793. doi: 10.1099/mic.0.26645-0
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Frommer, W. (1959). Zur Systematik der Actinomycin bildenden Streptomyceten. Arch. Mikrobiol. 32, 187–206. doi: 10.1007/BF00425228
Goodfellow, M., and Williams, S. T. (1983). Ecology of actinomycetes. Annu. Rev. Microbiol. 37, 189–216. doi: 10.1146/annurev.mi.37.100183.001201
Guindon, S., Delsuc, F., Dufayard, J. F., and Gascuel, O. (2009). Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 537, 113–137. doi: 10.1007/978-1-59745-251-9_6
Guo, Y., Zheng, W., Rong, X., and Huang, Y. (2008). A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: use of multilocus sequence analysis for streptomycete systematics. Int. J. Syst. Evol. Micrbiol. 58, 149–159. doi: 10.1099/ijs.0.65224-0
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hayakawa, M., and Nonomura, H. (1989). A new method for the intensive isolation of actinomycetes from soil. Actinomycetologica 3, 95–104. doi: 10.3209/saj.3_95
Highet, R. J., and Prelog, V. (1959). Stoffwechselprodukte von Actinomyceten. XVIII. Actiphenol. Helv. Chim. Acta 42, 1523–1526. doi: 10.1002/hlca.19590420514
Hoster, F., Schmitz, J. E., and Daniel, R. (2005). Enrichment of chitinolytic microorganisms: isolation and characterization of a chitinase exhibiting antifungal activity against phytopathogenic fungi from a novel Streptomyces strain. Appl. Microbiol. Biotechnol. 66, 434–442. doi: 10.1007/s00253-004-1664-9
Kaltenpoth, M. (2009). Actinobacteria as mutualists: general healthcare for insects? Trends Microbiol. 17, 529–535. doi: 10.1016/j.tim.2009.09.006
Kampfer, P. (2006). “The family streptomycetaceae, Part I: taxonomy,” in The Prokaryotes: A Handbook on the Biology of Bacteria, eds M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (Berlin: Springer), 538–604.
Kataoka, M., Ueda, K., Kudo, T., Seki, T., and Yoshida, T. (1997). Application of the variable region in 16S rDNA to create and index for rapid species identification in the genus Streptomyces. FEMS Microbiol. Lett. 151, 249–255. doi: 10.1111/j.1574-6968.1997.tb12578.x
Katoh, K., Kuma, K., Toh, H., and Miyata, T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518. doi: 10.1093/nar/gki198
Kurosawa, K., Bui, V. P., VanEssendelft, J. L., Willis, L. B., Lessard, P. A., Ghiviriga, T., et al. (2006). Characterization of Streptomyces MITKK-103, a newly isolated actinomycin X2-producer. Appl. Microbiol. Biotechnol. 72, 145–154. doi: 10.1007/s00253-005-0240-2
Kuster, E., and Williams, S. T. (1964). Selection of media for isolation of Streptomycetes. Nature 202, 298–299. doi: 10.1038/202928a0
Lee, S., Roets, F., and Crous, P. W. (2005). Protea infructescences represent a unique fungal niche. Fungal Divers. 19, 69–78. Available online at: http://www.fungaldiversity.org/fdp/sfdp/19-4.pdf
Maidak, B. L., Cole, J. R., Lilburn, T. G., Parker, C. T. Jr., Saxman, P. R., Farris, R. J., et al. (2001). The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29, 173–174. doi: 10.1093/nar/29.1.173
Manfio, G. P., Atalan, E., Zakrzewska-Czerwinska, J., Mordarski, M., Rodríguez, C., Collins, M. D., et al. (2003). Classification of novel soil streptomycetes as Streptomyces aureus sp. nov., Streptomyces laceyi sp. nov. and Streptomyces sanglieri sp. nov. Antonie van Leeuwenhoek 83, 245–255. doi: 10.1023/A:1023332427794
Marais, G. J., and Wingfield, M. J. (1994). Fungi associated with infructescences of Protea species in South Africa including a new species of Ophiostoma. Mycol. Res. 98, 369–374. doi: 10.1016/S0953-7562(09)81191-X
Marais, G. J., Wingfield, M. J., Viljoen, C. D., and Wingfield, B. D. (1998). A new Ophiostomatoid genus from Protea infructescences. Mycologia 90, 136–141. doi: 10.2307/3761023
Nishimura, T., Meguro, A., Hasegawa, S., Nakagawa, Y., Shimizu, M., and Kunoh, H. (2002). An endophytic actinomycete, Streptomyces sp. AOK-30, isolated from mountain laurel and its antifungal activity. J. Gen. Plant Pathol. 68, 390–397. doi: 10.1007/PL00013109
Noguchi, H., Harrison, P. H., Arai, K., Nakashima, T. T., Trimble, L. A., and Vederas, J. C. (1988). Biosynthesis and full NMR assignment of fungichromin, a polyene antibiotic from Streptomyces cellulose. J. Am. Chem. Soc. 110, 2938–2945. doi: 10.1021/ja00217a041
Posada, D. (2008). jModeltest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. doi: 10.1093/molbev/msn083
Posada, D., and Buckley, T. R. (2004). Model selection and model averaging in phylogenetics: advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst. Biol. 53, 793–808. doi: 10.1080/10635150490522304
Pridham, T. G., and Lyons, A. J. (1961). Streptomyces albus (Rossi Doria) waksman et henrici: taxonomic study of strains labeled Streptomyces albus. J. Bacteriol. 81, 431–441.
Roets, F., de Beer, Z. W., Wingfield, M. J., Crous, P. W., and Dreyer, L. L. (2008). Ophiostoma gemellus and Sporothrix variecibatus from mites infesting Protea infructescences in South Africa. Mycologia 100, 496–510. doi: 10.3852/07-181R
Roets, F., Theron, N., Wingfield, M. J., and Dreyer, L. L. (2012). Biotic and abiotic constraints that facilitate host exclusivity of Gondwanamyces and Ophiostoma on Protea. Fungal Biol. 116, 49–61. doi: 10.1016/j.funbio.2011.09.008
Roets, F., Wingfield, B. D., de Beer, Z. W., Wingfield, M. J., and Dreyer, L. L. (2010). Two new Ophiostoma species from Protea caffra in Zambia. Persoonia 24, 18–28. doi: 10.3767/003158510X490392
Roets, F., Wingfield, M. J., Crous, P. W., and Dreyer, L. L. (2009). Fungal radiation in the cape floristic region: an analysis based on Gondwanamyces and Ophiostoma. Mol. Phylogenet. Evol. 51, 111–119. doi: 10.1016/j.ympev.2008.05.041
Roets, F., Wingfield, M. J., Crous, P. W., and Dreyer, L. L. (2013). “Taxonomy and ecology of ophiostomatoid fungi associated with Protea infructescences,” in The Ophiostomatoid Fungi, eds K. A. Seifert, Z. W. de Beer, and M. J. Wingfield (Utrecht: CBS), 177–187.
Rong, X., Guo, Y., and Huang, Y. (2009). Proposal to reclassify the Streptomyces albidoflavus clade on the basis of multilocus sequence analysis and DNA-DNA hybridization, and taxonomic elucidation of Streptomyces griseus subsp. solvifaciens. Syst. Appl. Microbiol. 32, 314–322. doi: 10.1016/j.syapm.2009.05.003
Ruddick, S. M., and Williams, S. T. (1972). Studies on the ecology of actinomycetes in soil V. Some factors influencing the dispersal and adsorption of spores in soil. Soil Biol. Biochem. 4, 93–100. doi: 10.1016/0038-0717(72)90046-6
Seipke, R. F., Barke, J., Ruiz-Gonzalez, M. X., Orivel, J., Yu, D. W., and Hutchings, M. I. (2012). Fungus-growing Allomerus ants are associated with antibiotic-producing actinobacteria. Antonie van Leeuwenhoek 101, 443–447. doi: 10.1007/s10482-011-9621-y
Shih, H. D., Liu, Y. C., Hsu, F. L., Mulabagal, V., Dodda, R., and Huang, J. W. (2003). Fungichromin: a substance from Streptomyces padanus with inhibitory effects on Rhizoctonia solani. J. Agric. Food Chem. 51, 95–99. doi: 10.1021/jf025879b
Strobel, G., and Daisy, B. (2003). Bioprospecting for microbial endophytes and their natural products. Microbiol. Mol. Biol. Rev. 67, 491–502. doi: 10.1128/MMBR.67.4.491-502.2003
Sun, D., Sun, W., Yu, Y., Li, Z., Deng, Z., and Lin, S. (2014). A new glutarimide derivative from marine sponge-derived Streptomyces anulatus S71. Nat. Prod. Res. 28, 1602–1606. doi: 10.1080/14786419.2014.928877
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Theron, N., Roets, F., Dreyer, L. L., Esler, K. J., and Ueckermann, E. A. (2012). A new genus and eight new species of tydeoidea (Acari: Trombidiformes) from Protea species in South Africa. Int. J. Acarol. 38, 257–273. doi: 10.1080/01647954.2011.619576
Vaidya, G., Lohman, D. J., and Meier, R. (2010). SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Visagie, C. M., Seifert, K. A., Houbraken, J., Samson, R. A., and Jacobs, K. (2014). Diversity of Penicillium section Citrina within the fynbos biome of South Africa, including a new species from a Protea repens infructescence. Mycologia 106, 537–552. doi: 10.3852/13-256
Wingfield, B. D., Viljoen, C. D., and Wingfield, M. J. (1999). Phylogenetic relationships of ophiostomatoid fungi associated with Protea infructescences in South Africa. Mycol. Res. 103, 1616–1620. doi: 10.1017/S0953756299008990
Xiong, Z. Q., Zhang, Z. P., Li, J. H., Wei, S. J., and Tu, G. Q. (2012). Characterization of Streptomyces padanus JAU4234, a producer of actinomycin X2, fungichromin, and a new polyene macrolide antibiotic. Appl. Environ. Microbiol. 78, 589–592. doi: 10.1128/AEM.06561-11
Yamada-Okabe, T., and Yamada-Okabe, H. (2002). Characterization of the CaNAG3, CaNAG4, and CaNAG6 genes of the pathogenic fungus Candida albicans: possible involvement of these genes in the susceptibilities of cytotoxic agents. FEMS Microbiol. Lett. 212, 15–21. doi: 10.1111/j.1574-6968.2002.tb11238.x
Zhang, X., Alemany, L. B., Fiedler, H. P., Goodfellow, M., and Parry, R. J. (2008). Biosynthetic investigations of lactonamycin and lactonamycin Z: cloning of the biosynthetic gene clusters and discovery of an unusual starter unit. Antimicrob. Agents Chemother. 52, 574–585. doi: 10.1128/AAC.00717-07
Keywords: Sporothrix, Candida albicans, antifungal, Protea, infructescence, fungichromin, actiphenol, Streptomyces
Citation: Human ZR, Moon K, Bae M, de Beer ZW, Cha S, Wingfield MJ, Slippers B, Oh D-C and Venter SN (2016) Antifungal Streptomyces spp. Associated with the Infructescences of Protea spp. in South Africa. Front. Microbiol. 7:1657. doi: 10.3389/fmicb.2016.01657
Received: 14 August 2016; Accepted: 05 October 2016;
Published: 02 November 2016.
Edited by:
Michael Thomas-Poulsen, University of Copenhagen, DenmarkReviewed by:
Johannes F. Imhoff, GEOMAR Helmholtz Centre for Ocean Research Kiel (HZ), GermanyKi Hyun Kim, Sungkyunkwan University, Republic of Korea
Copyright © 2016 Human, Moon, Bae, de Beer, Cha, Wingfield, Slippers, Oh and Venter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong-Chan Oh, ZG9uZ2NoYW5vaEBzbnUuYWMua3I=
Stephanus N. Venter, ZmFudXMudmVudGVyQHVwLmFjLnph
†These authors have contributed equally to this work.
 Zander R. Human
Zander R. Human Kyuho Moon
Kyuho Moon Munhyung Bae2
Munhyung Bae2 Dong-Chan Oh
Dong-Chan Oh Stephanus N. Venter
Stephanus N. Venter