- 1Department of Respiratory Diseases, Chinese PLA General Hospital, Beijing, China
- 2Department of Infection Management and Disease Control, Chinese PLA General Hospital, Beijing, China
- 3Testing Center of HMI, Chinese PLA General Hospital, Beijing, China
- 4Department of Clinical Pharmacology, Chinese PLA General Hospital, Beijing, China
Stenotrophomonas maltophilia is a common nosocomial pathogen that causes high morbidity and mortality. Because of its inherent extended antibiotic resistance, therapeutic options for S. maltophilia are limited, and sulfamethoxazole/trimethoprim (SXT) is the only first-line antimicrobial recommended. However, with the spread of dihydropteroate synthase (sul1 and sul2) genes, global emergence of SXT resistance has been reported. There is an urgent need to develop a rapid and sensitive but cost-efficient method to monitor the dissemination of sul genes. In this study, we developed loop-mediated isothermal amplification (LAMP) assays for sul1 and sul2 using real-time turbidity and hydroxy naphthol blue coloration methods. The assays could quickly detect sul genes with high sensitivity and specificity. The LAMP detection limit was 0.74 pg/reaction of extracted genomic DNA for sul1 and 2.6 pg/reaction for sul2, which were both 10-fold more sensitive than the corresponding traditional PCR assays. Additionally, the LAMP assays could positively amplify DNA from sul1-producing strains, but not from the negative controls. We then used the LAMP assays to investigate the dissemination of sul genes among S. maltophilia isolates from patients in three hospitals in Beijing, China. Among 450 non-duplicated samples collected during 2012–2014, 56 (12.4%) strains were SXT-resistant. All these SXT-resistant strains were positive for sul genes, with 35 (62.5%) carrying sul1, 17 (30.4%) carrying sul2, and 4 (7.1%) carrying both sul1 and sul2, which indicated that sul genes were the predominant resistance mechanism. Of 394 SXT-susceptible strains, 16 were also sul-positive. To provide epidemiological data for the appropriate choice of antimicrobials for treatment of sul-positive S. maltophilia, we further tested the susceptibility to 18 antimicrobials. Among these, sul-positive strains showed the highest susceptibility to tetracycline derivatives, especially minocycline (MIC50/MIC90, 0.5/4; susceptibility rate, 95.4%). Ticarcillin-clavulanate and new fluoroquinolones (moxifloxacin and levofloxacin) also showed some in vitro activity. Apart from these three kinds of antimicrobials, other agents showed poor activity against sul-positive strains.
Introduction
Stenotrophomonas maltophilia is the third most common non-fermentative gram-negative bacillus isolated from hospitals and has emerged as an important opportunistic pathogen (Looney et al., 2009). It is associated with a broad range of serious human infections, including pneumonia, bloodstream infections, and urinary tract infections, especially in immunocompromised patients (Looney et al., 2009; Chang et al., 2015). An increasing incidence and considerable morbidity have been reported in recent decades (Falagas et al., 2009; Looney et al., 2009). Since S. maltophilia shows a high level of intrinsic resistance to most commonly used antimicrobial agents, including most cephalosporins, carbapenems, aminoglycosides, and quinolones, its treatment is extremely difficult. Sulfamethoxazole/trimethoprim (SXT) is the only first-line antimicrobial recommended for treatment because of its good antibacterial activity in vitro and low incidence of resistance (Abbott et al., 2011; Wu et al., 2012; Wei et al., 2015). Although other mechanisms, such as dfrA and SmeDEF genes, might influence the minimal inhibitory concentration (MIC), the sul genes have generally been recognized as the predominant resistance mechanism (Toleman et al., 2007; Hu et al., 2011). The sul genes are usually located on mobile genetic elements, and can mobilize horizontally and vertically, assisted by the spread of class 1 integrons and insertion sequence common region (ISCR) elements. The global emergence of SXT resistance mediated by the dissemination of sul genes has been reported (Toleman et al., 2007). There is an urgent need to develop a rapid, simple, and cost-effective assay for the epidemiological investigation and surveillance of the dissemination of sul genes, which could provide epidemiological data to guide the establishment of infection control measures and the choice of initial antimicrobials. Loop-mediated isothermal amplification (LAMP) is a novel DNA amplification method. It was first developed in the year 2000 and can efficiently and specifically amplify DNA under isothermic conditions within 60 min using Bst DNA polymerase (Notomi et al., 2000; Dong et al., 2015a). The only requirement to perform LAMP assays is having access to a constant-temperature apparatus. Moreover, by adding hydroxy naphthol blue (HNB), the results can be evaluated by the naked eye, thus avoiding the process of agarose gel electrophoresis that is required for traditional PCR. LAMP assays have been used for the rapid diagnosis of infectious diseases and the surveillance of epidemics of bacteria, viruses, and parasites (Sun et al., 2010; Wang et al., 2011; Soo et al., 2013). Recently, the technique has also been used for the surveillance of genes encoding various carbapenemase enzymes (Qi et al., 2012; Nakano et al., 2015; Yamamoto et al., 2015; Kim et al., 2016), but there have been no reports on the use of LAMP for the detection of SXT-resistance genes.
The objectives of this study were to establish a rapid, simple, and cost-effective assay for the detection of sul genes in S. maltophilia and to investigate the dissemination and antimicrobial susceptibility of S. maltophilia carrying sul genes. These data are essential to guide the establishment of infection control measures and the choice of an appropriate initial antibacterial agent. First, we developed LAMP assays for the detection of sul1 and sul2 and confirmed their specificity and sensitivity using two different detection methods, namely sample turbidity and HNB coloration. We then used the LAMP assays to study the dissemination of sul genes in three hospitals in Beijing, China during 2012–2014. Finally, the susceptibility of the sul-positive S. maltophilia strains to 18 commonly used antimicrobial agents was investigated.
Materials and Methods
Bacterial Isolation and Identification
In this study, during 2012–2014, 450 non-duplicated clinical S. maltophilia isolates were collected from hospitalized patients in the Chinese PLA General Hospital, Peking Union Medical College Hospital, and Air Force General Hospital in Beijing, China. Bacterial species identification was performed using a Vitek® II bacterial identification system (bioMérieux, Marcy-l’Étoile, France) and further confirmed by species-specific PCR (Whitby et al., 2000). Three sul1 (X2, P121, and K106) and sul2 (X133, P57, and K14) positive control DNAs were used in the establishment of the LAMP assays and were confirmed to carry sul genes by PCR and sequencing of the PCR products. S. maltophilia ATCC 13637, K279a, and P129 were used as negative controls for estimating the specificity of the LAMP assays. Whole genome sequencing of these strains has been performed and they were confirmed to carry genes encoding multidrug-efflux pumps, β-lactamases, and aminoglycoside-modifying enzymes, but they did not carry sul genes (Crossman et al., 2008; Davenport et al., 2014).
The research protocol was approved by the Ethics Committee of Chinese PLA General Hospital, Peking Union Medical College Hospital, and Air Force General Hospital, and written informed consent was obtained from patients. Our research protocol did not affect patients’ health, safety, or privacy.
PCR Detection of sul Genes
Whole genomic DNA (including plasmid DNA) was extracted using the protocol described by a previous study (Yang et al., 2014). The bacterial strains were cultured in brain–heart infusion broth at 37°C. Genomic DNA was extracted using a Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, USA) according to the manufacturer’s recommended protocol. The extracted DNA was used as a template in the LAMP and PCR reactions.
To compare the sensitivity of LAMP assays and traditional PCR, sul1 detection was performed using the PCR primers shown in Table 1. The target products were 250 bp for sul1 and 276 bp for sul2. The PCR cycling parameters were: initial PCR activation, 95°C for 5 min; amplification, 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s; final extension step, 72°C for 10 min. A 25-μl reaction volume was used, and the products were separated by 1% (w/v) agarose (Amresco, Solon, OH, USA) gel electrophoresis and stained with ethidium bromide. Images were documented with a Gel DocTM EQ imaging system (Bio-Rad, Hercules, CA, USA).
LAMP Reaction
The sequences CP011493 (80815–81654) and CP009257 (2540172–2540987) obtained from the NCBI GenBank database were used for the design of sul1-specific and sul2-specific PCR and LAMP primers, respectively. The primers were designed and analyzed by PrimerExplorer software1. The sequences and locations of primers for sul1 and sul2 are both shown in Table 1.
The LAMP reactions were performed in a final volume of 25 μl containing 12.5 μl reaction mixture, 1 μl Bst DNA polymerase, and 2 μl template using the Loopamp DNA Amplification Kit (Loopamp DNA Amplification Kit; Eiken Chemical, Co., Ltd, Tochigi, Japan) for real-time turbidimetry or with the further addition of 1 μl HNB solution (Eiken Chemical, Co., Ltd) for visual detection. Primers were used at the concentrations of 1.6 μM for the forward internal primer and backward internal primer, 0.8 μM for the loop forward primer and loop backward primer, and 0.2 μM for the forward outer primer and backward outer primer. The reactions were performed in the reaction tubes (Eiken Chemical, Co., Ltd) for 60 min at 65°C.
Two different methods, namely sample turbidity and HNB coloration, were used to detect the LAMP products. Real-time changes in turbidity were monitored by spectrophotometric analysis by recording the optical density (650 nm) every 6 s with a Loopamp Realtime Turbidimeter (LA-320c; Eiken Chemical, Co., Ltd). For direct visual detection, 1 μl of HNB detection reagent was added to the reaction. The amount of DNA template used in the LAMP reaction mixture was equal to that used in PCR reactions. Each experiment was performed at least three times.
Antimicrobial Susceptibility Testing
Eighteen clinically used antibacterial agents were tested in our study, as follows: SXT, ceftazidime, minocycline, levofloxacin, chloramphenicol, ticarcillin/clavulanate, tigecycline, doxycycline, moxifloxacin, cefepime, cefoperazone/sulbactam, meropenem, amikacin, colistin, fosfomycin, aztreonam, azithromycin, and rifampicin. Tigecycline was obtained from Wyeth Pharmaceutical (Wyeth Pharmaceutical, Philadelphia, PA, USA); moxifloxacin was obtained from Bayer Healthcare (Bayer Pharma, Wuppertal, AG, Germany); Colistin was purchased from Sigma-Aldrich (St. Louis, MO, USA); other antimicrobial agents were purchased from the National Institute for the Control of Pharmaceutical and Biological Products (NICPBP, Beijing, China). The antimicrobial powders were used to prepare stock solutions, as described by the Clinical and Laboratory Standards Institute (CLSI, 2015). MICs of the sul-positive strains were determined for each isolate by the Mueller–Hinton agar (Difco, Franklin Lakes, NJ, USA) macrodilution method. Pseudomonas aeruginosa ATCC 27853, Staphylococcus aureus ATCC 29213, and Escherichia coli ATCC 25922 were used as quality control strains to ensure the accuracy and reliability of the assays. Interpretive breakpoints for susceptibility are available only for SXT, ceftazidime, minocycline, levofloxacin, chloramphenicol, and ticarcillin/clavulanate (CLSI, 2015). Other antimicrobial agents with no published breakpoint criteria for S. maltophilia were interpreted with reference to those for Acinetobacter spp. or Enterobacteriaceae published by CLSI (2015) (Table 3).
Results
Establishment of LAMP Assays for the Detection of sul1 and sul2
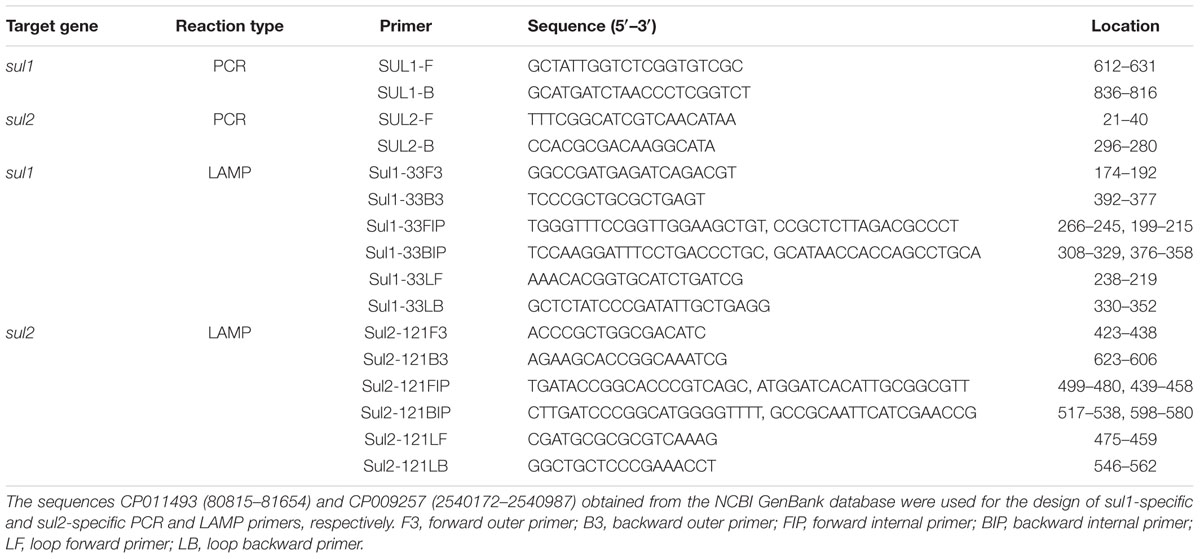
The sul1-specific and sul2-specific LAMP primers were designed and analyzed using PrimerExplorer software (Table 1). To compare the detection limit of the LAMP method with that of the conventional PCR method, genomic DNA was extracted from S. maltophilia X2 and serially diluted 10 times from 37.0 ng/μL to 3.7 fg/μL. As shown in Figure 1, the detection limits of the real-time turbidity and HNB methods were 0.74 pg/reaction of extracted whole genomic DNA, 10-fold more sensitive than that of the PCR assay (7.4 pg/reaction) (Figure 1). Similarly, the detection limit of LAMP specific to sul2 was 2.6 pg/reaction, which is 10 times more sensitive than that obtained for the conventional PCR method (Figure 2).
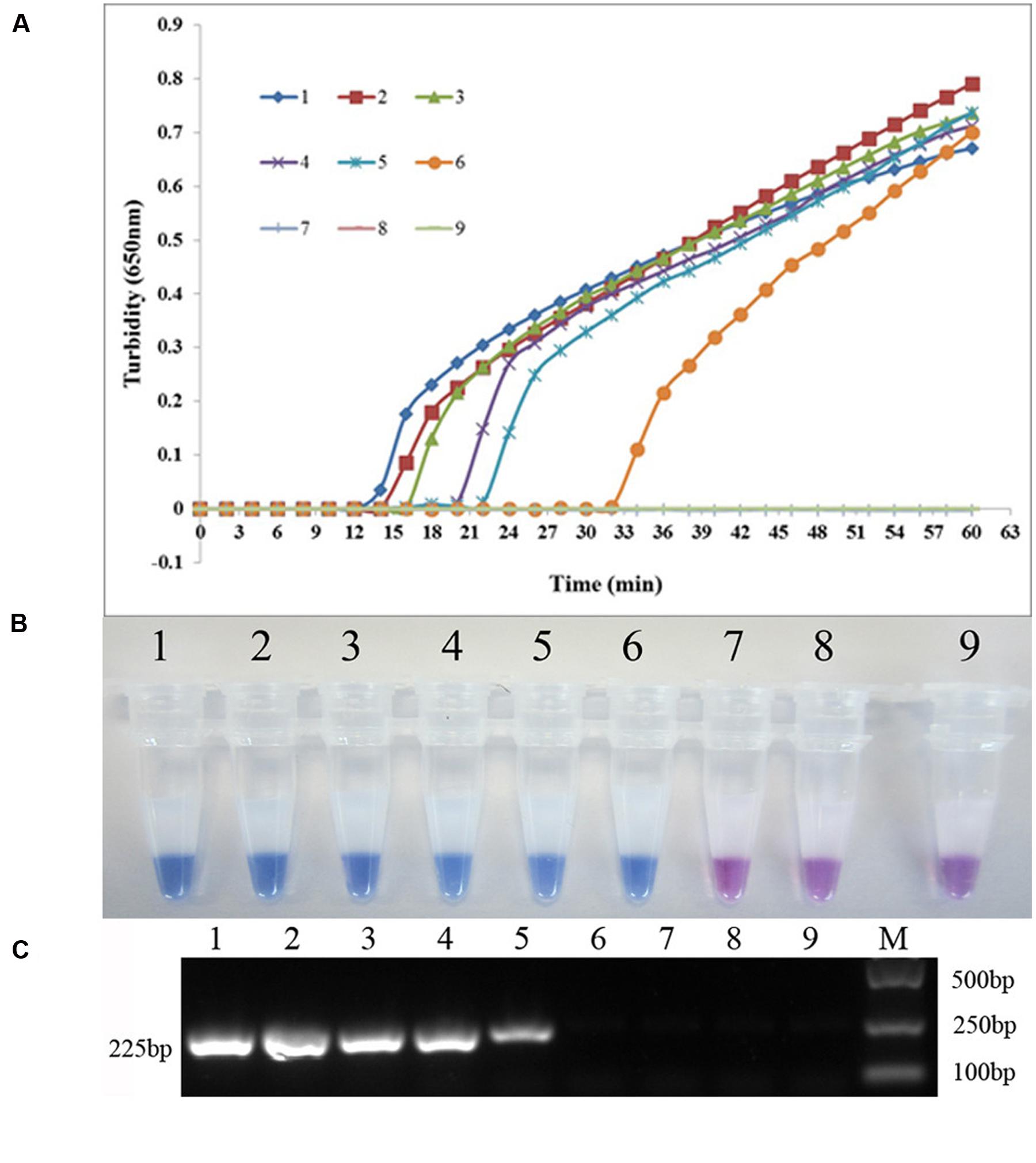
FIGURE 1. Comparison of the sensitivities of LAMP and conventional PCR assays for detecting sul1 in Stenotrophomonas maltophilia. (A) Turbidity was monitored every 6 s using a Loopamp Realtime Turbidimeter with detection at 650 nm. The reaction was performed at 65°C. (B) The results were visualized by adding hydroxy naphthol blue (HNB) to the reaction mixture before the reaction. (C) PCR products were separated by 1% (w/v) agarose gel electrophoresis and stained with ethidium bromide. (1) 74.0 ng; (2) 7.4 ng; (3) 0.74 ng; (4) 74.0 pg; (5) 7.4 pg; (6) 0.74 pg; (7) 74.0 fg; (8) 7.4 fg; (9) blank control (double-distilled water).
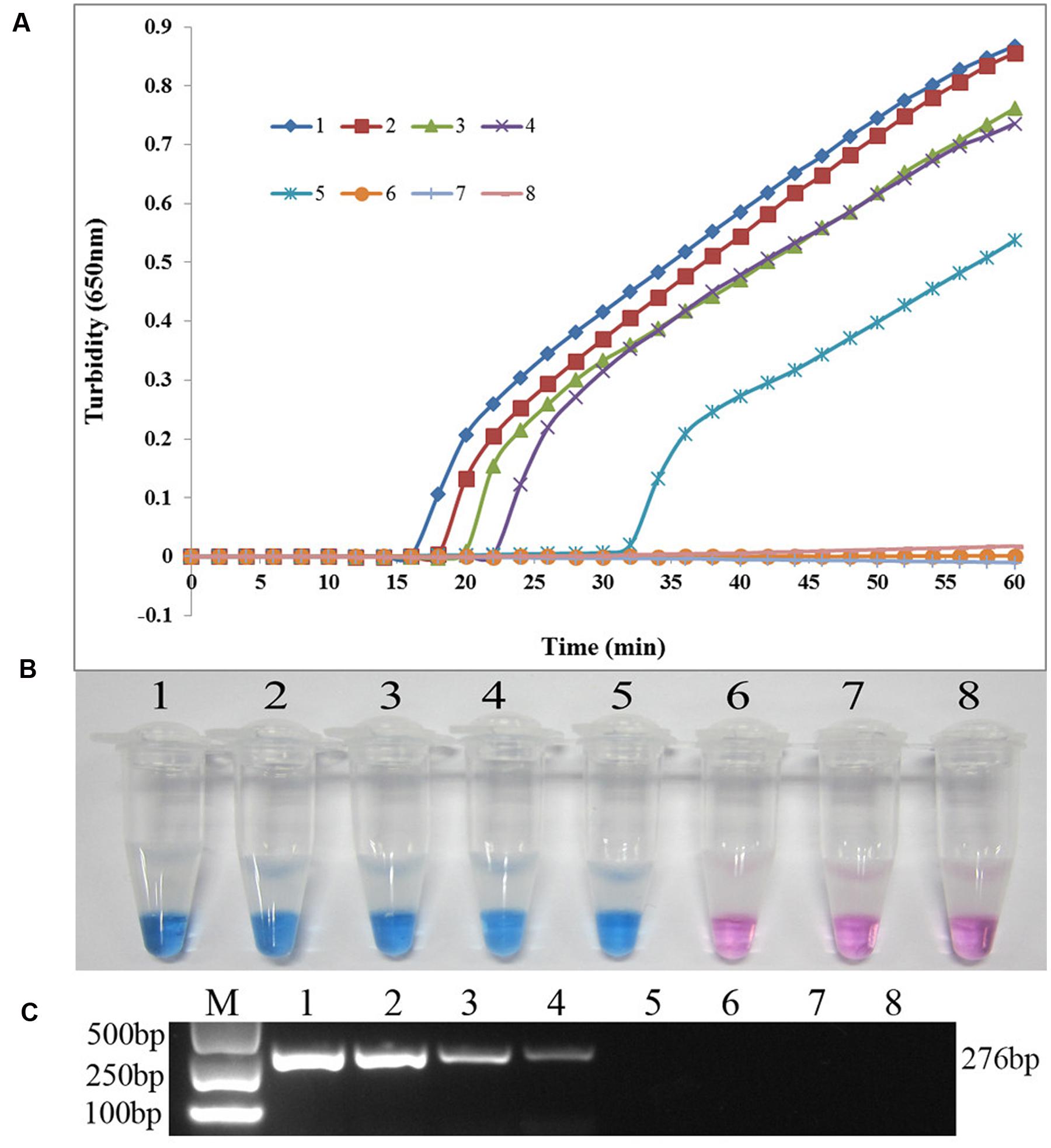
FIGURE 2. Comparison of the sensitivities of LAMP and conventional PCR assays for detecting sul2 in S. maltophilia. (A) Turbidity was monitored every 6 s using a Loopamp Realtime Turbidimeter with detection at 650 nm. The reaction was performed at 65°C. (B) The results were visualized by adding HNB to the reaction mixture before the reaction. (C) PCR products were separated by 1% (w/v) agarose gel electrophoresis and stained with ethidium bromide. (1) 26.0 ng; (2) 2.6 ng; (3) 0.26 ng; (4) 26.0 pg; (5) 2.6 pg; (6) 0.26 pg; (7) 26.0 fg; (8) blank control (double-distilled water).
The specificity of the LAMP assay for detecting sul1 was evaluated using X2, P121, and K106 carrying sul1 as positive controls, S. maltophilia ATCC 13637, K279a, and P129 as negative controls, and distilled water as blank control. As shown in Figure 3, both methods positively amplified DNA from sul1-producing strains, but not from the negative and blank controls. The results showed that the assay was highly specific for detecting sul1. The same results were observed for the developed sul2 LAMP assay (Figure 4).
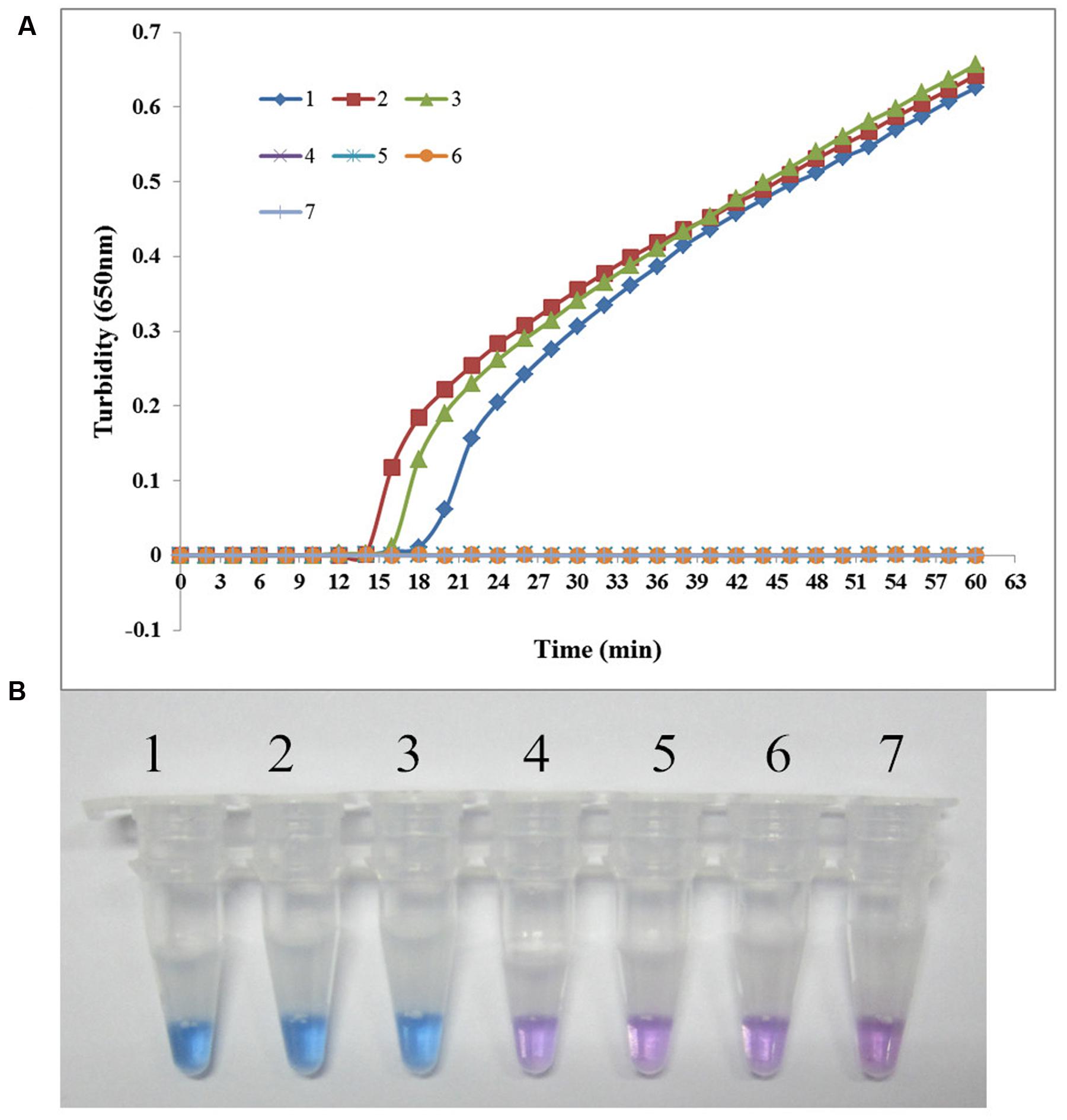
FIGURE 3. Specificity of the LAMP reaction for detecting sul1 in S. maltophilia. (A) Turbidity was monitored every 6 s using a Loopamp Realtime Turbidimeter with detection at 650 nm. The reaction was performed at 65°C. (B) The results were visualized by adding HNB to the reaction mixture before the reaction. (1) X2 (positive control for sul1 from Peking Union Medical College Hospital); (2) P121 (positive control for sul1 from Chinese PLA General Hospital); (3) K106 (positive control for sul1 from Chinese PLA General Hospital); (4) S. maltophilia K279a; (5) S. maltophilia ATCC 13637; (6) P129 (negative control for sul from Chinese PLA General Hospital, confirmed by genome sequencing); (7) double-distilled water.
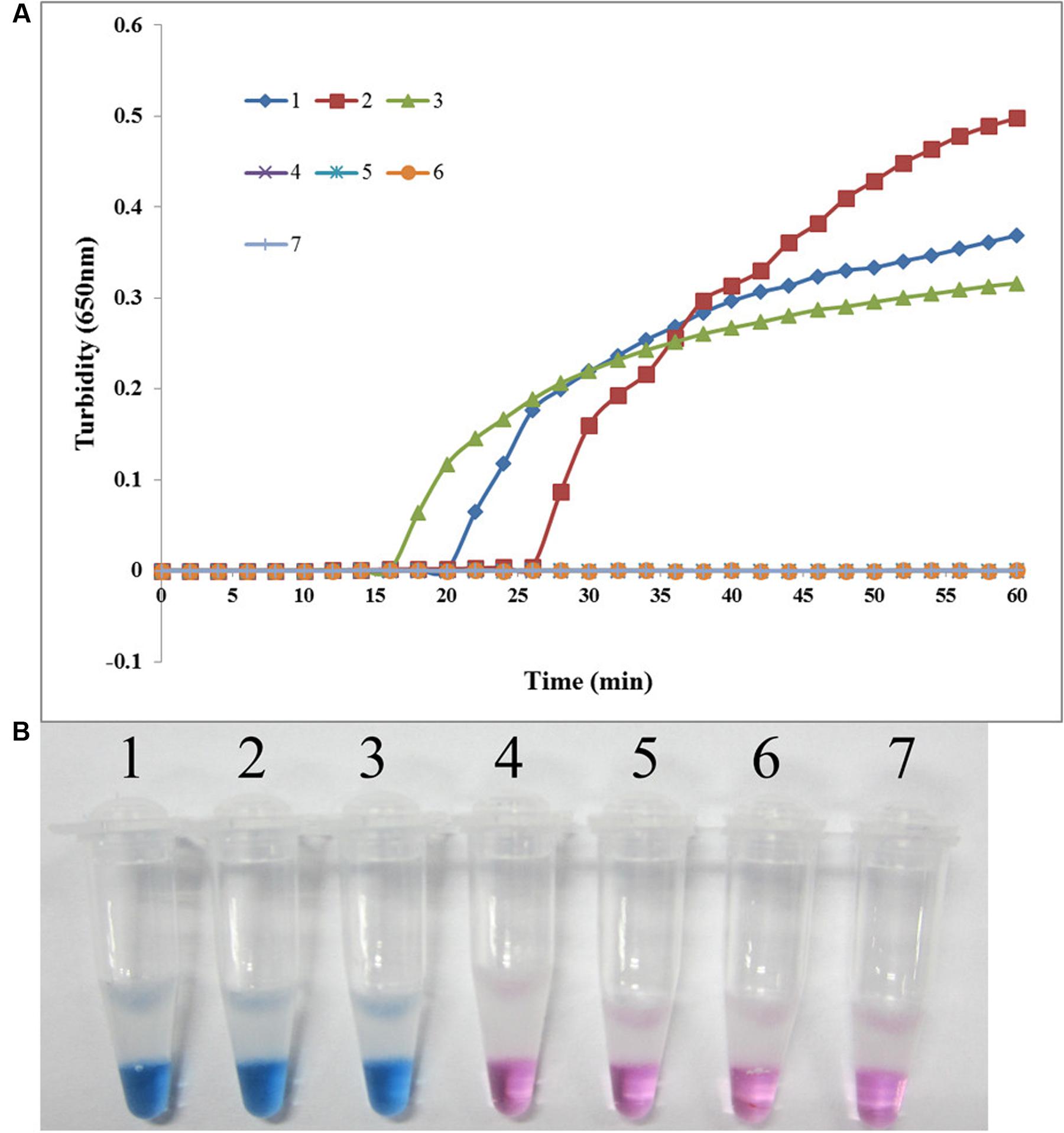
FIGURE 4. Specificity of the LAMP reaction for detecting sul2 in S. maltophilia. (A) Turbidity was monitored every 6 s using a Loopamp Realtime Turbidimeter with detection at 650 nm. The reaction was performed at 65°C. (B) The results were visualized by adding HNB to the reaction mixture before the reaction. (1) X133 (positive control for sul2 from Peking Union Medical College Hospital); (2) P57 (positive control for sul2 from Chinese PLA General Hospital); (3) K14 (positive control for sul2 from Chinese PLA General Hospital); (4) S. maltophilia K279a; (5) S. maltophilia ATCC 13637; (6) P129 (negative control for sul from Chinese PLA General Hospital, confirmed by genome sequencing); (7) double-distilled water.
Dissemination of sul Genes in S. maltophilia
During 2012–2014, 450 non-duplicated S. maltophilia strains were collected in our study, and 56 (12.4%) strains were resistant to SXT. As shown in Table 2, all these 56 SXT-resistant strains were tested positive for sul genes, among which 35 (62.5) strains carried sul1, 17 (30.4%) strains carried sul2, and 4 (7.1%) strains carried both sul1 and sul2. Among 394 SXT-susceptible strains, 16 (4.1%) carried sul genes.
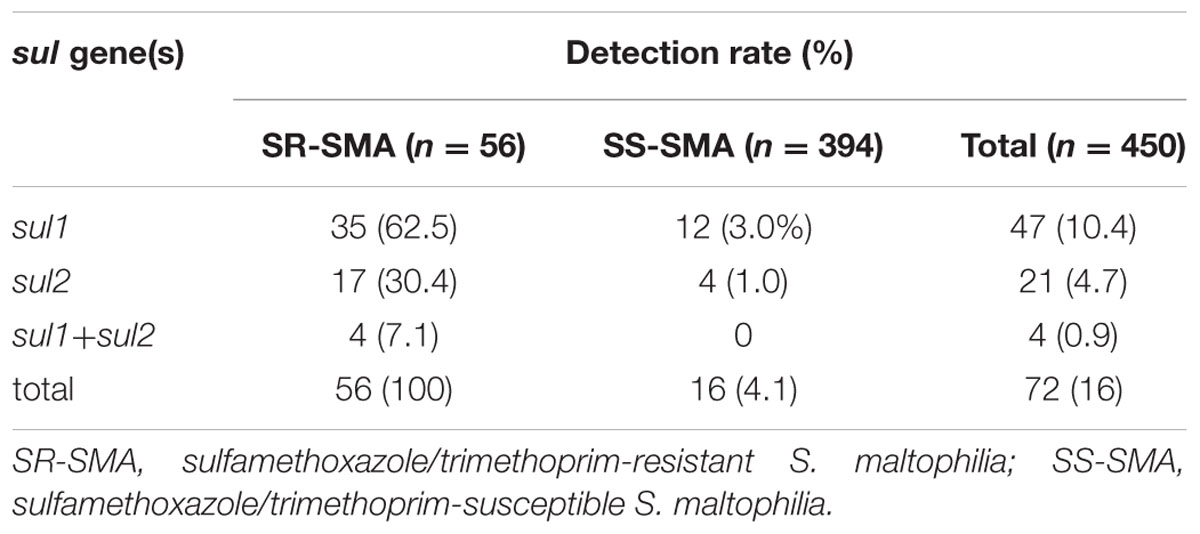
TABLE 2. Detection rates of sul genes in Stenotrophomonas maltophilia isolated from China during 2012–2014.
Antimicrobial Susceptibility of sul-Positive S. maltophilia
Among 18 clinically used antimicrobials, the susceptibility rates of S. maltophilia were 95.4% for minocycline, 68.1% for doxycycline and tigecycline, 50.0% and 59.7% for levofloxacin and moxifloxacin, respectively, and 69.4% for ticarcillin-clavulanate. The susceptibility rates for 12 other antimicrobial agents were less than 50%, while for the agents meropenem, amikacin, colistin, ceftazidime, chloramphenicol, aztreonam, sulfamethoxazole/trimethoprim, azithromycin, and rifampicin, S. maltophilia had resistance rates exceeding 70% (Table 3).
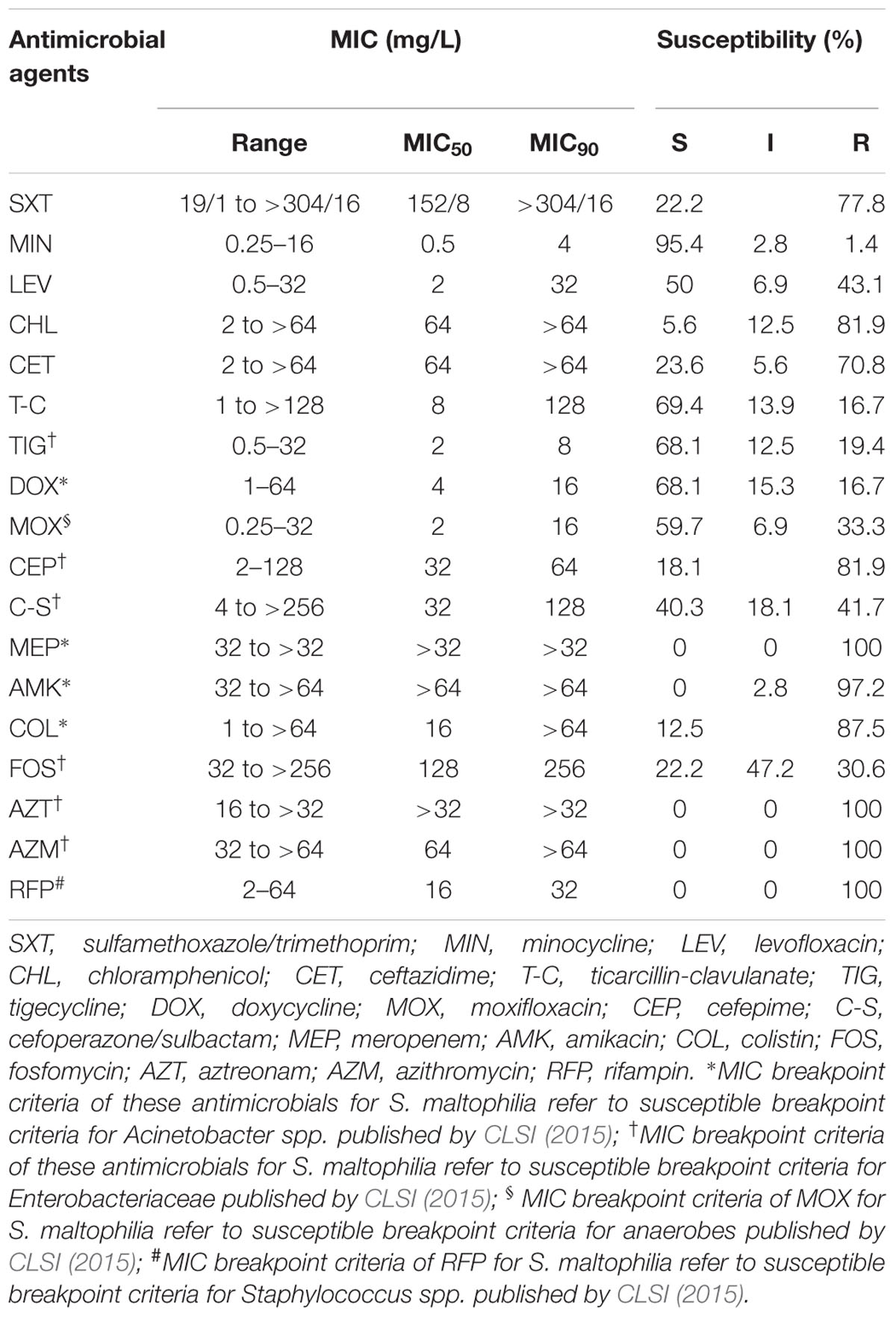
TABLE 3. In vitro susceptibility of 72 sul-positive S. maltophilia strains to 18 clinically used antimicrobials.
Discussion
Stenotrophomonas maltophilia is an opportunistic pathogen, mainly causing pneumonia and bloodstream infections in immunocompromised patients, with high morbidity and mortality (Falagas et al., 2009; Looney et al., 2009; Chang et al., 2015). Because of its intrinsic and acquired resistance to most commonly clinical used antibiotics, its treatment is extremely difficult, and SXT is the only first-line drug recommended (Looney et al., 2009; Chang et al., 2015), with an overall susceptibility rate of higher than 90% (Abbott et al., 2011; Wu et al., 2012; Chang et al., 2015; Wei et al., 2015). Resistance to SXT in S. maltophilia will lead to more treatment failure and higher mortality, and the global emergence of an increasing tendency toward SXT resistance mediated by the dissemination of sul genes has been reported. Surveillance of the dissemination of sul genes by rapid and cost-efficient LAMP assays could contribute to the control of SXT resistance and the reduction of S. maltophilia infection incidence. In the current study, we designed primers specific for both sul1 and sul2 for use in LAMP assays to monitor the prevalence of sul genes among clinical isolates of S. maltophilia. The LAMP detection method has several advantages over traditional PCR as a tool for use in epidemiological surveys. First, LAMP is generally less time- and labor-intensive, and it proceeds under an isothermal condition within 60 min (Notomi et al., 2000; Dong et al., 2015a; Li et al., 2015). Second, LAMP is more cost-efficient than conventional PCR, since it only requires access to a constant-temperature apparatus, such as a common laboratory water bath. Third, LAMP is more sensitive than traditional PCR. The LAMP detection limit was 0.74 pg/reaction for sul1 and 2.6 pg/reaction for sul2, which were both 10-fold more sensitive than the corresponding traditional PCR assays. Despite having a higher sensitivity than traditional PCR, the LAMP assays remained specific, and the negative controls used for estimating the specificity were all negative in the LAMP assays. Moreover, after adding HNB, the reaction results could be evaluated by the naked eye, which not only avoids the process of agarose gel electrophoresis that is required with conventional PCR methods, but also reduces the chance of cross-contamination (Liu et al., 2012; Solanki et al., 2013; Yang et al., 2014; Dong et al., 2015b).
Using these LAMP assays, we surveyed the prevalence of sul genes among clinical isolates of S. maltophilia in three hospitals in Beijing. We isolated 56 SXT-resistant strains and found that they all carried sul genes. The results indicated that the carriage of sul1 and/or sul2 might be the predominant mechanism of SXT resistance in Beijing, China, as has been reported from other domestic and international surveys (Chang et al., 2007; Toleman et al., 2007; Hu et al., 2011; Chung et al., 2015; Kaur et al., 2015). We also detected both sul1 and sul2 in some SXT-susceptible S. maltophilia strains. Recent studies also have reported the detection of sul1 and sul2 in SXT-susceptible S. maltophilia isolates (Chang et al., 2007; Hu et al., 2016). Therefore, the phenotypic breakpoints are not reliable for genotypic identification. In addition, SXT resistance breakpoints are established from a clinical efficacy perspective that also considers the dosage, pharmacokinetics/pharmacodynamics, and clinical trials. SXT treatment might fail for these strains, since resistance could emerge during treatment through the up-regulation of sul genes expression.
There are limited antimicrobial options for infection due to S. maltophilia because of its extensive resistance to most antibiotics. The occurrence of acquired resistance to SXT mediated by the dissemination of sul genes increases the difficulty of selecting an appropriate choice for treatment. To provide epidemiological data for the choice of appropriate antimicrobial agents for the treatment of sul-positive S. maltophilia, we further tested their susceptibility to 18 clinically used antimicrobials. In previous studies, ceftazidime and ticarcillin/clavulanate were reported as the most effective β-lactam drugs against S. maltophilia (Chang et al., 2015). However, only 23% of sul-positive strains remained susceptible to ceftazidime in our study, and ceftazidime seemed not to be a good choice due to exhibiting poor in vitro activity. Nearly 70% of sul-positive S. maltophilia strains remained susceptible to ticarcillin/clavulanate, and ticarcillin/clavulanate seemed to be a good choice for the treatment of sul-positive strains. However, the recommendation for the use of ticarcillin-clavulanate should be made with caution, since the L1/L2 β-lactamases carried by S. maltophilia have strong activity and together can inactivate nearly all β-lactam antibiotics.
New fluoroquinolones, such as moxifloxacin and levofloxacin, have been reported to show good in vitro activity and could be alternative therapeutic options against S. maltophilia (Valdezate et al., 1999; Nicodemo and Paez, 2007; Looney et al., 2009). Fluoroquinolones are now popular alternatives because of their lesser side effects compared to those of SXT. A clinical comparison of SXT and levofloxacin showed that their overall microbiological cure and clinical success rates showed no statistically significant difference, but the rate of adverse events was significantly lower in the levofloxacin group than in the SXT group (Cho et al., 2014; Wang et al., 2014). However, in our study, nearly 50% of sul-positive S. maltophilia strains were non-susceptible to moxifloxacin and levofloxacin. Moreover, it has been reported that the overuse of fluoroquinolones worldwide has resulted in a higher resistance rate among many pathogenic bacteria, including S. maltophilia (Chang et al., 2014; Pien et al., 2015). Therefore, it would be prudent to use them as initial empirical antibiotics for sul-positive strains.
Tetracycline derivatives, such as tigecycline, minocycline, and doxycycline, have been reported to be the most active agents against S. maltophilia other than SXT, even in patients with cystic fibrosis. Global surveillance studies have reported that minocycline was found to be significantly more active than other tetracyclines, and the susceptibility rate of S. maltophilia to minocycline exceeded 97% across all geographic regions (Gales et al., 2008; Chen et al., 2012; Milne and Gould, 2012; Wu et al., 2012; Chung et al., 2013; Castanheira et al., 2014; Sader et al., 2014). The results of our study agreed with the aforementioned observations, and the tetracycline derivatives we tested, including minocycline, doxycycline, and tigecycline, all showed good in vitro activity against clinical isolates of sul-negative S. maltophilia. Minocycline showed a particularly high in vitro activity, with S. maltophilia showing a susceptibility rate of 95% against this drug. Tetracycline derivatives might be an appropriate therapeutic option for these sul-positive drugs, based on their excellent in vitro activities. Ten other agents, including colistin, aztreonam, meropenem, amikacin, azithromycin, fosfomycin, and rifampicin, showed little activity against sul-positive S. maltophilia strains, and they should not be recommended for use as monotherapy to treat these strains. Epidemiological data might contribute to guiding the choice of an appropriate initial antimicrobial for the treatment of sul-positive S. maltophilia and reducing the frequency of treatment failures. However, choosing an initial antimicrobial agent by referring to epidemiological data alone is not always adequate, and making more precise treatment choices based on antimicrobial susceptibility testing is encouraged.
Conclusion
We have developed rapid, simple, and cost-efficient LAMP assays for the detection of the SXT resistance genes sul1 and sul2 with high sensitivity and specificity. These assays could contribute to the epidemiological investigation and surveillance of the dissemination of sul genes, which could provide epidemiological data to guide the establishment of infection control measures and the choice of initial antimicrobials. Using these methods, 450 non-duplicated S. maltophilia strains isolated from three hospitals in Beijing, China were screened, and 72 (16.0%) of these strains tested positive for sul genes. These sul-positive strains showed poor susceptibility to most clinically used antimicrobials apart from tetracycline derivatives, ticarcillin-clavulanate, moxifloxacin, and levofloxacin. The developed LAMP assays provide a useful tool for the surveillance of sul genes among clinical S. maltophilia strains, since these assays are more sensitive, faster, and more cost-efficient than the conventional PCR method.
Author Contributions
YoL and YuL conceived and designed experiments. JZ, YX, WL, WN, CW, and RW executed experiments. JZ, YX, and WN performed and wrote the manuscript. CW and RW helped to edit the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 30872155), Mega-projects of China (No. BWS13J027, AWS11L009, 11BJZ01), and the Special Grant for the Prevention and Control of Infectious Diseases of China (No. 2013ZX10004 805-003, 2013ZX10004 217-002).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We are grateful to Peking Union Medical College Hospital and Air Force General Hospital for kindly providing S. maltophilia strains and helpful information.
Footnotes
References
Abbott, I. J., Slavin, M. A., Turnidge, J. D., Thursky, K. A., and Worth, L. J. (2011). Stenotrophomonas maltophilia: emerging disease patterns and challenges for treatment. Expert Rev. Anti Infect. Ther. 9, 471–488. doi: 10.1586/eri.11.24
Castanheira, M., Mendes, R. E., and Jones, R. N. (2014). Update on Acinetobacter species: mechanisms of antimicrobial resistance and contemporary in vitro activity of minocycline and other treatment options. Clin. Infect. Dis. 59(Suppl.6), S367–S373. doi: 10.1093/cid/ciu706
Chang, L. L., Lin, H. H., Chang, C. Y., and Lu, P. L. (2007). Increased incidence of class 1 integrons in sulfamethoxazole/trimethoprim-resistant clinical isolates of Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 59, 1038–1039. doi: 10.1093/jac/dkm034
Chang, Y. T., Lin, C. Y., Chen, Y. H., and Hsueh, P. R. (2015). Update on infections caused by Stenotrophomonas maltophilia with particular attention to resistance mechanisms and therapeutic options. Front. Microbiol. 6:893. doi: 10.3389/fmicb.2015.00893
Chang, Y. T., Lin, C. Y., Lu, P. L., Lai, C. C., Chen, T. C., Chen, C. Y., et al. (2014). Stenotrophomonas maltophilia bloodstream infection: comparison between community-onset and hospital-acquired infections. J. Microbiol. Immunol. Infect. 47, 28–35. doi: 10.1016/j.jmii.2012.08.014
Chen, Y. H., Lu, P. L., Huang, C. H., Liao, C. H., Lu, C. T., Chuang, Y. C., et al. (2012). Trends in the susceptibility of clinically important resistant bacteria to tigecycline: results from the tigecycline in vitro surveillance in taiwan study, 2006 to 2010. Antimicrob. Agents Chemother. 56, 1452–1457. doi: 10.1128/AAC.06053-11
Cho, S. Y., Kang, C.-I., Kim, J., Ha, Y. E., Chung, D. R., Lee, N. Y., et al. (2014). Can levofloxacin be a useful alternative to trimethoprim-sulfamethoxazole for treating Stenotrophomonas maltophilia bacteremia? Antimicrob. Agents Chemother. 58, 581–583. doi: 10.1128/AAC.01682-13
Chung, H. S., Hong, S. G., Kim, Y. R., Shin, K. S., Whang, D. H., Ahn, J., et al. (2013). Antimicrobial susceptibility of stenotrophomonas maltophilia isolates from Korea, and the activity of antimicrobial combinations against the isolates. J. Korean Med. Sci. 28, 62–66. doi: 10.3346/jkms.2013.28.1.62
Chung, H. S., Kim, K., Hong, S. S., Hong, S. G., Lee, K., and Chong, Y. (2015). The sul1 gene in Stenotrophomonas maltophilia with high-level resistance to sulfamethoxazole/trimethoprim. Ann. Lab. Med. 35, 246–249. doi: 10.3343/alm.2015.35.2.246
CLSI (2015). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement (M100-S25). Wayne, PA: Clinical and Laboratory Standards Institute.
Crossman, L. C., Gould, V. C., Dow, J. M., Vernikos, G. S., Okazaki, A., Sebaihia, M., et al. (2008). The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 9, R74. doi: 10.1186/gb-2008-9-4-r74
Davenport, K. W., Daligault, H. E., Minogue, T. D., Broomall, S. M., Bruce, D. C., Chain, P. S., et al. (2014). Complete Genome Sequence of Stenotrophomonas maltophilia Type Strain 810-2 (ATCC 13637). Genome Announc. 2, e974–e914. doi: 10.1128/genomeA.00974-14
Dong, D., Liu, W., Li, H., Wang, Y., Li, X., Zou, D., et al. (2015a). Survey and rapid detection of Klebsiella pneumoniae in clinical samples targeting the rcsA gene in Beijing, China. Front. Microbiol. 6:519. doi: 10.3389/fmicb.2015.00519
Dong, D., Zou, D., Liu, H., Yang, Z., Huang, S., Liu, N., et al. (2015b). Rapid detection of Pseudomonas aeruginosa targeting the toxA gene in intensive care unit patients from Beijing, China. Front. Microbiol. 6:1100. doi: 10.3389/fmicb.2015.01100
Falagas, M. E., Kastoris, A. C., Vouloumanou, E. K., Rafailidis, P. I., Kapaskelis, A. M., and Dimopoulos, G. (2009). Attributable mortality of Stenotrophomonas maltophilia infections: a systematic review of the literature. Future Microbiol. 4, 1103–1109. doi: 10.2217/fmb.09.84
Gales, A. C., Sader, H. S., and Fritsche, T. R. (2008). Tigecycline activity tested against 11808 bacterial pathogens recently collected from US medical centers. Diagn. Microbiol. Infect. Dis. 60, 421–427. doi: 10.1016/j.diagmicrobio.2007.10.017
Hu, L. F., Chang, X., Ye, Y., Wang, Z. X., Shao, Y. B., Shi, W., et al. (2011). Stenotrophomonas maltophilia resistance to sulfamethoxazole/trimethoprim mediated by acquisition of sul and dfrA genes in a plasmid-mediated class 1 integron. Int. J. Antimicrob. Agents 37, 230–234. doi: 10.1016/j.ijantimicag.2010.10.025
Hu, L. F., Xu, X. H., Yang, H. F., Ye, Y., and Li, J. B. (2016). Role of sul2 gene linked to transposase in resistance to sulfamethoxazole/trimethoprim among Stenotrophomonas maltophilia isolates. Ann. Lab. Med. 36, 73–75. doi: 10.3343/alm.2016.36.1.73
Kaur, P., Gautam, V., and Tewari, R. (2015). Distribution of class 1 integrons, sul1 and sul2 genes among clinical isolates of Stenotrophomonas maltophilia from a tertiary care hospital in North India. Microb. Drug Resist. 21, 380–385. doi: 10.1089/mdr.2014.0176
Kim, H. J., Kim, H. S., Lee, J. M., Yoon, S. S., and Yong, D. (2016). Rapid detection of Pseudomonas aeruginosa and Acinetobacter baumannii harboring bla(VIM-2), bla(IMP-1) and bla(OXA-23) genes by using loop-mediated isothermal amplification methods. Ann. Lab. Med. 36, 15–22. doi: 10.3343/alm.2016.36.1.15
Li, P., Niu, W., Li, H., Lei, H., Liu, W., Zhao, X., et al. (2015). Rapid detection of Acinetobacter baumannii and molecular epidemiology of carbapenem-resistant A. baumannii in two comprehensive hospitals of Beijing, China. Front. Microbiol. 6:997. doi: 10.3389/fmicb.2015.00997
Liu, W., Zou, D., Li, Y., Wang, X., He, X., Wei, X., et al. (2012). Sensitive and rapid detection of the new Delhi metallo-beta-lactamase gene by loop-mediated isothermal amplification. J. Clin. Microbiol. 50, 1580–1585. doi: 10.1128/JCM.06647-11
Looney, W. J., Narita, M., and Muhlemann, K. (2009). Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect. Dis. 9, 312–323. doi: 10.1016/s1473-3099(09)70083-0
Milne, K. E., and Gould, I. M. (2012). Combination antimicrobial susceptibility testing of multidrug-resistant Stenotrophomonas maltophilia from cystic fibrosis patients. Antimicrob. Agents Chemother. 56, 4071–4077. doi: 10.1128/AAC.00072-12
Nakano, R., Nakano, A., Ishii, Y., Ubagai, T., Kikuchi-Ueda, T., Kikuchi, H., et al. (2015). Rapid detection of the Klebsiella pneumoniae carbapenemase (KPC) gene by loop-mediated isothermal amplification (LAMP). J. Infect. Chemother. 21, 202–206. doi: 10.1016/j.jiac.2014.11.010
Nicodemo, A. C., and Paez, J. I. (2007). Antimicrobial therapy for Stenotrophomonas maltophilia infections. Eur. J. Clin. Microbiol. Infect. Dis. 26, 229–237. doi: 10.1007/s10096-007-0279-3
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28, E63. doi: 10.1093/nar/28.12.e63
Pien, C. J., Kuo, H. Y., Chang, S. W., Chen, P. R., Yeh, H. W., Liu, C. C., et al. (2015). Risk factors for levofloxacin resistance in Stenotrophomonas maltophilia from respiratory tract in a regional hospital. J. Microbiol. Immunol. Infect. 48, 291–295. doi: 10.1016/j.jmii.2013.09.005
Qi, J., Du, Y., Zhu, X., Bai, H., Luo, Y., and Liu, Y. (2012). A loop-mediated isothermal amplification method for rapid detection of NDM-1 gene. Microb. Drug Resist. 18, 359–363. doi: 10.1089/mdr.2011.0220
Sader, H. S., Farrell, D. J., Flamm, R. K., and Jones, R. N. (2014). Variation in potency and spectrum of tigecycline activity against bacterial strains from U.S. medical centers since its approval for clinical use (2006 to 2012). Antimicrob. Agents Chemother. 58, 2274–2280. doi: 10.1128/AAC.02684-13
Solanki, R., Vanjari, L., Ede, N., Gungi, A., Soory, A., and Vemu, L. (2013). Evaluation of LAMP assay using phenotypic tests and conventional PCR for detection of blaNDM-1 and blaKPC genes among carbapenem-resistant clinical Gram-negative isolates. J. Med. Microbiol. 62, 1540–1544. doi: 10.1099/jmm.0.059907-0
Soo, P. C., Tseng, C. C., Ling, S. R., Liou, M. L., Liu, C. C., Chao, H. J., et al. (2013). Rapid and sensitive detection of Acinetobacter baumannii using loop-mediated isothermal amplification. J. Microbiol. Methods 92, 197–200. doi: 10.1016/j.mimet.2012.11.020
Sun, J., Najafzadeh, M. J., Vicente, V., Xi, L., and de Hoog, G. S. (2010). Rapid detection of pathogenic fungi using loop-mediated isothermal amplification, exemplified by Fonsecaea agents of chromoblastomycosis. J. Microbiol. Methods 80, 19–24. doi: 10.1016/j.mimet.2009.10.002
Toleman, M. A., Bennett, P. M., Bennett, D. M., Jones, R. N., and Walsh, T. R. (2007). Global emergence of sulfamethoxazole/trimethoprim resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg. Infect. Dis. 13, 559–565. doi: 10.3201/eid1304.061378
Valdezate, S., Vindel, A., Baquero, F., and Canton, R. (1999). Comparative in vitro activity of quinolones against Stenotrophomonas maltophilia. Eur. J. Clin. Microbiol. Infect. Dis. 18, 908–911. doi: 10.1007/s100960050430
Wang, B., Wang, Y., Tian, Z. J., An, T. Q., Peng, J. M., and Tong, G. Z. (2011). Development of a reverse transcription loop-mediated isothermal amplification assay for detection of Porcine teschovirus. J. Vet. Diagn. Invest. 23, 516–518. doi: 10.1177/1040638711403427
Wang, Y. L., Scipione, M. R., Dubrovskaya, Y., and Papadopoulos, J. (2014). Monotherapy with fluoroquinolone or trimethoprim-sulfamethoxazole for treatment of Stenotrophomonas maltophilia infections. Antimicrob. Agents Chemother. 58, 176–182. doi: 10.1128/AAC.01324-13
Wei, C., Ni, W., Cai, X., and Cui, J. (2015). A Monte Carlo pharmacokinetic/pharmacodynamic simulation to evaluate the efficacy of minocycline, tigecycline, moxifloxacin, and levofloxacin in the treatment of hospital-acquired pneumonia caused by Stenotrophomonas maltophilia. Infect. Dis. (Lond). 47, 846–851. doi: 10.3109/23744235.2015.1064542
Whitby, P. W., Carter, K. B., Burns, J. L., Royall, J. A., LiPuma, J. J., and Stull, T. L. (2000). Identification and detection of Stenotrophomonas maltophilia by rRNA-directed PCR. J. Clin. Microbiol. 38, 4305–4309.
Wu, H., Wang, J. T., Shiau, Y. R., Wang, H. Y., Lauderdale, T. L., and Chang, S. C. (2012). A multicenter surveillance of antimicrobial resistance on Stenotrophomonas maltophilia in Taiwan. J. Microbiol. Immunol. Infect. 45, 120–126. doi: 10.1016/j.jmii.2011.09.028
Yamamoto, N., Hamaguchi, S., Akeda, Y., Santanirand, P., Kerdsin, A., Seki, M., et al. (2015). Clinical specimen-direct LAMP: a useful tool for the surveillance of blaOXA-23-positive carbapenem-resistant Acinetobacter baumannii. PLoS ONE 10:e0133204. doi: 10.1371/journal.pone.0133204
Keywords: loop-mediated isothermal amplification, LAMP, Stenotrophomonas maltophilia, dihydropteroate synthase, sul gene, sulfamethoxazole, minocycline
Citation: Zhao J, Xing Y, Liu W, Ni W, Wei C, Wang R, Liu Y and Liu Y (2016) Surveillance of Dihydropteroate Synthase Genes in Stenotrophomonas maltophilia by LAMP: Implications for Infection Control and Initial Therapy. Front. Microbiol. 7:1723. doi: 10.3389/fmicb.2016.01723
Received: 11 April 2016; Accepted: 14 October 2016;
Published: 26 October 2016.
Edited by:
Bart Devreese, Ghent University, BelgiumReviewed by:
Xian-Zhi Li, Health Canada, CanadaBai Changqing, 307th Hospital of PLA, China
Liuyu Huang, Academy of Military Medical Sciences, China
Changyun Ye, Chinese Center for Disease Control and Prevention, China
Copyright © 2016 Zhao, Xing, Liu, Ni, Wei, Wang, Liu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youning Liu, bGl1eW91bmluZ3BsYTMwMUAxNjMuY29t Yunxi Liu, eXVueGlsaXU2NTExQDE2My5jb20=
†These authors have contributed equally to this work.
 Jin Zhao1†
Jin Zhao1† Rui Wang
Rui Wang Youning Liu
Youning Liu