- 1State Key Laboratory of Agro-Biotechnology, China Agricultural University, Beijing, China
- 2Key Laboratory of Pollinating Insect Biology, Ministry of Agriculture, Institute of Apicultural Research, Chinese Academy of Agricultural Sciences, Beijing, China
Co-infection of none-coding satellite RNAs (sat-RNAs) usually inhibits replication and attenuates disease symptoms of helper viruses. However, we find that the sat-RNA of Beet black scorch virus (BBSV) and low temperature (18°C) additively enhance the systemic infection of BBSV in Nicotiana benthamiana. Northern blotting hybridization revealed a relatively reduced accumulation of BBSV-derived small interfering RNAs (siRNAs) in presence of sat-RNA as compared to that of BBSV alone. Cloning and sequencing of total small RNAs showed that more than 50% of the total small RNAs sequenced from BBSV-infected plants were BBSV-siRNAs, whereas the abundance of sat-siRNAs were higher than BBSV-siRNAs in the sat-RNA co-infected plants, indicating that the sat-RNA occupies most of the silencing components and possibly relieves the RNA silencing-mediated defense against BBSV. Interestingly, the 5′ termini of siRNAs derived from BBSV and sat-RNA were dominated by Uridines (U) and Adenines (A), respectively. Besides, the infection of BBSV alone and with sat-RNA induce down-regulation of miR168 and miR403, respectively, which leads to high accumulation of their targets, Argonaute 1 (AGO1) and AGO2. Our work reveals the profiles of siRNAs of BBSV and sat-RNA and provides an additional clue to investigate the complicated interaction between the helper virus and sat-RNA.
Introduction
The satellite RNAs (sat-RNAs) of plant viruses rely on the helper virus for replication and encapsidation but share little or no sequence similarity with the helper virus genome (Murant and Mayo, 1982; Hu et al., 2009). Based on genome size, the sat-RNAs are divided into two classes: large sat-RNAs encoding a nonstructural protein and small sat-RNAs without coding capacity (Simon et al., 2004). Sat-RNAs usually inhibit the replication of helper viruses and may alter the disease symptoms of the helper viruses depending on the interaction among host factors, helper viruses, and sat-RNAs (Collmer and Howell, 1992; Roossinck et al., 1992; Hull, 2002). Despite the fact that there are sat-RNAs that reduce the disease symptoms without influencing the accumulation of helper viruses, most sat-RNAs reduce both disease symptoms and titer of the helper virus, probably by competing with the helper virus for common replication factors (Roossinck et al., 1992; Hull, 2002). However, there are exceptions in which sat-RNAs enhance the disease symptoms during co-infections. For example, a few isolates of Cucumber mosaic virus (CMV) sat-RNAs can exacerbate the disease symptoms in tobacco and tomato hosts because of a specific sequence carried by the sat-RNAs (review in Roossinck et al., 1992; Simon et al., 2004). Similarly, sat-RNA of Groundnut rosette virus (GRV) is largely responsible for the groundnut rosette symptom and the transmission between natural hosts of GRV (Murant et al., 1988; Robinson et al., 1999). Interestingly, unlike the sat-RNAs of CMV and GRV, the sat-RNA C of Turnip crinkle virus (TCV) often enhances the visible symptoms of TCV in a host-dependent manner (Li, 1990). Since sat-RNA C is capable of interfering the encapsidation of TCV genomic RNAs thereby to boost the accumulation of free coat proteins, which are RNA silencing suppressor (Wang and Simon, 1999; Thomas et al., 2003; Zhang and Simon, 2003), these observations suggest that some sat-RNAs enhance helper virus disease symptoms by directly or indirectly suppressing RNA silencing. So far, to our knowledge, there is no report to document an enhancement of helper virus titers during co-infection with sat-RNAs (Hull, 2002; Simon et al., 2004).
In the past few years, the progressive understanding of RNA silencing results in more information about the interaction between plants and the pathogens. The RNA silencing in plants has been found to have numerous functions in the course of plant growth (Baulcombe, 2004; Meister and Tuschl, 2004; Chapman and Carrington, 2007), among which one of the important function is to provide an adaptive immune system counteracting pathogens, including viruses, bacteria, and so on (Ding et al., 2004; Voinnet, 2005; Ding and Voinnet, 2007). In Arabidopsis, the cascade of DCL2/3/4, AGO1/2/3/5/7/10, and RDR1/6 have been shown to be involved in antiviral RNA silencing pathways (review in Huang et al., 2016). Accordingly, the viruses have evolved diverse viral suppressors of RNA silencing (VSRs) to inhibit distinct steps in the silencing pathway (Li and Ding, 2006; Dı́az-Pendón and Ding, 2008). At present, about 35 individual VSR families have been reported in plant viruses (Ding and Voinnet, 2007). The co-evolution processes of silencing and suppressing reveal complex interaction between virus and host plant in the long history of co-existence (Ding and Voinnet, 2007). Some subviral pathogens, such as viroids and satellites, are also influenced by the pressure of RNA silencing and evolved effective secondary structures to avoid or minimize the small RNA-mediated silencing (Wang et al., 2004). The secondary structure of Potato spindle-tube viroid (PSTVd) was also found to induce silencing but could be resistant to RISC-mediated cleavage (Itaya et al., 2007). Recent studies demonstrate that sat-RNAs-derived siRNAs can directly silence host genes, which is responsible for sat-RNA-induced disease symptom (Shimura et al., 2011; Smith et al., 2011).
Beet black scorch virus (BBSV) was firstly reported in northern China in the late 1980s and lately identified as a new species of genus Betanecrovirus (Cao et al., 2002; Yuan et al., 2006; King et al., 2011). BBSV induced the symptom of black scorched leaves and necrotic fibrous roots in the sugar beet plants in late spring, causing severe yield loss in the plantation areas. Two isolates from the provinces of Ningxia and Xinjiang have been reported in China, designated as BBSV-N and BBSV-X respectively (Cao et al., 2002; Xi et al., 2006), which exhibit 99.45% similarity in nucleotide sequence. In addition, BBSV-Co was reported in Colorado of Unite States (Weiland et al., 2006, 2007), which shared 93% similarity with BBSV-N, and different BBSV isolates were identified in Iran and Spain (Koenig and Valizadeh, 2008; González-Vázquez et al., 2009). During the serial propagation of BBSV, a gain-of-function mutant harboring a single nucleotide substitution at nucleotide (nt) 3477 in the 3′UTR induce higher infectivity than wild-type BBSV in N. benthamiana (Xu et al., 2012). In addition to the viral genome RNA, another 615 nt single stranded RNA has been identified as a satellite RNA in the isolate of BBSV-X (Guo et al., 2005). During the replication of sat-RNA of BBSV, various forms such as monomers, dimers, and tetramers are accumulated, and the dimer form plays an intermediate role in replication (Guo et al., 2005).
In this study, we first showed that the satellite RNA enhance the pathogenesis and accumulation of BBSV in N. benthamiana plants under at or below room temperature. Further analyses including cloning and sequencing of siRNAs derived from BBSV and its sat-RNA, suggest that sat-RNA may alleviate RNAi mediated antiviral silencing to enhance the systemic infection of BBSV by acting as surrogacy of the helper virus.
Materials and Methods
Plant Materials and Virus Inoculation
N. benthamiana plants were grown in a growth chamber with a 16-h-light/8-h-dark cycle at 25°C. Three leaves of N. benthamiana, typically at the four-leaf stage in 1-month-old, were used for inoculation (Xu et al., 2012). A BBSV variant, BBSV-m294 (abbreviated as Bm, GenBank accession no. JN635330.1) that caused severe symptom in N. benthamiana and obtained after a passage of propagation (Xu et al., 2012). The sat-RNA (GenBank accession no. NC_006460.1) of BBSV were used for inoculation by the method reported previously (Xu et al., 2012). After inoculation, plants were grown at 18 and 25°C conditions. Three systemic leaves were harvested at 12 dpi for northern blot analysis and sequencing of small RNA.
RNA Isolation and Northern Blot Analysis
Total RNA was extracted using TRIzol® Reagent (Invitrogen, USA). For detection of viral genomic RNA and mRNA of BBSV and its sat-RNA, 2 μg total RNA extracted from mock or virus-infected plants was used for hybridization using indicated gene-specific 32P-radiolabled cDNA probes corresponding to the 3′UTR fragment of BBSV or the full-length sat-RNA, respectively as described (Xu et al., 2012). For small RNA gel blots, 10 μg total RNA were separated on 17% denaturing polyacrylamide gel (PAGE) and transferred to nylon membranes (GE Healthcare, UK). DNA oligonucleotides corresponding to the sequences of BBSV (nt 155–176, nt 769–788, nt 823–842, nt 1259–1278, nt 1762–1781, nt 2020–2039, nt 2266–2287, and nt 3115–3134) or sat-RNA (nt 132–152, nt 395–416 and nt 506–527) were synthesized respectively. The mixtures of antisense oligonucleotides corresponding to BBSV and sat-RNA were labeled with [γ-32P] ATP as probes used for hybridization at 40°C for 16–20 h in PerfectHyb plus buffer (Sigma-Aldrich). The membranes were washed in 2 × SSC (0.3 M NaCl and 0.03 M sodium citrate) containing 0.2% SDS for 30 min and then twice with 1 × SSC containing 0.1% SDS for 20 min both at 50°C.
Small RNA Library Sequencing and Analysis
The small RNA libraries were generated following the manufacturer's protocol (Illumina, California, USA). Briefly, separated by electrophoresis, RNA fractions with sizes between 18–30 nt corresponding to the small RNA population were purified, and cloned using NEBNext® Multiplex Small RNA Library Prep Set for Illumina kit. The final products were quantified on the Agilent DNA 1000 chip and sequencing was performed by use of an Illumina Hiseq 2500-SE50 (Illumina, California, USA). The Illumina sequencing reads were first trimmed to remove the adaptor sequence to get clean reads. The trimmed sequencing reads were then blasted to the Bm (JN635330.1) and sat-RNA (NC_006460.1) and the sequences with full matched were considered as Bm or sat-RNA small RNAs. The clean reads were also blasted in miRBase for miRNAs. The data of small RNA libraries was deposited in GenBank with accession number GSE80694.
Quantitative RT-PCR Analyses
To measure expression levels of miR168 and miR403, stem-loop quantitative RT-PCR technique was used as previously described (Varkonyi-Gasic and Hellens, 2011). The quantitative expression of DCLs, AGOs and RDR6 mRNAs were checked by real-time RT-PCR as described previously (Kotakis et al., 2010). Primers used for quantitative analysis above are listed (Table S1).
Results
Low Temperature or sat-RNA Co-infection Enhances the Infection by BBSV Variant m294
In our previous studies, a series of BBSV spontaneous variants were isolated from serial propagation of wild-type BBSV in Chenopodium amaranticolor and N. benthamiana. The typical variant BBSV-m294 (abbreviated as Bm, GenBank accession no. JN635330.1) causes more severe symptoms than wild-type BBSV at low temperature (18°C) (Xu et al., 2012). To determine the impact of environment temperature on Bm infection, N. benthamiana plants were mechanically inoculated with Bm and maintained at 18 or 25°C, respectively. At 12 dpi, the infected plants were photographed as shown in Figure 1. The Bm-infected plants induced typical yellow curling symptoms on systemic leaves at 18°C, whereas the Bm induced very few yellow chlorotic mottle spots in upper leaves at 25°C (Figure 1, middle panel).
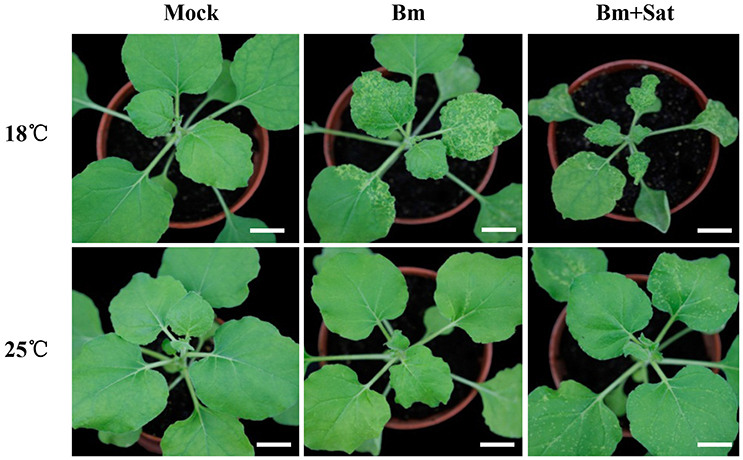
Figure 1. Co-infection of sat-RNA and low temperature additively improved the systemic infectivity of BBSV. Symptom development in N. benthamiana inoculated by buffer (Mock), Bm alone and co-infected with its sat-RNA at 18 and 25°C. The infected seedlings were photographed at 12 days after inoculation with purified Bm or sat-RNA-associated Bm at a concentration of 100 μg/mL. Bars = 2 cm.
BBSV infection is naturally associated with satellite RNAs (sat-RNAs), which depend on BBSV for replication and movement but share no sequence homology with the helper viral genome (Guo et al., 2005). To determine if the sat-RNA affects the pathogenicity of BBSV in different temperatures, we further inoculated N. benthamiana plants with Bm alone or with its sat-RNA at 18 or 25°C. In contrast with very few infection lesions by Bm alone, clearly visible disease symptom was observed in the systemically infected leaves infected by Bm and its sat-RNA at 25°C (Figure 1, bottom panel). Moreover, the viral symptom induced by Bm and its sat-RNA was further enhanced at 18°C than at 25°C (Figure 1, upper panel).
Collectively, both low temperature and co-infection with sat-RNA additively enhance the pathogenicity of BBSV.
Sat-RNA Co-infection Enhances Bm Accumulation but Reduces the Production of Bm-Derived siRNAs
In order to examine the accumulation of BBSV along with or without its sat-RNA in different temperature, we further carried out northern blot hybridizations to detect the genomic and subgenomic RNA of Bm, as well as sat-RNA. In consistence with symptom observations, the genomic and subgenomic RNA of Bm accumulated to significantly higher levels at 18°C than that of Bm at 25°C (Figure 2A, lane 2 and 5). In addition, the accumulation level of Bm genomic RNA was higher in sat-RNA co-infection samples than Bm alone at either low or room temperature (Figure 2A, upper panel). Thus, these results indicate that both low temperature and co-infection with its sat-RNA additively enhance Bm accumulation in N. benthamiana plants.
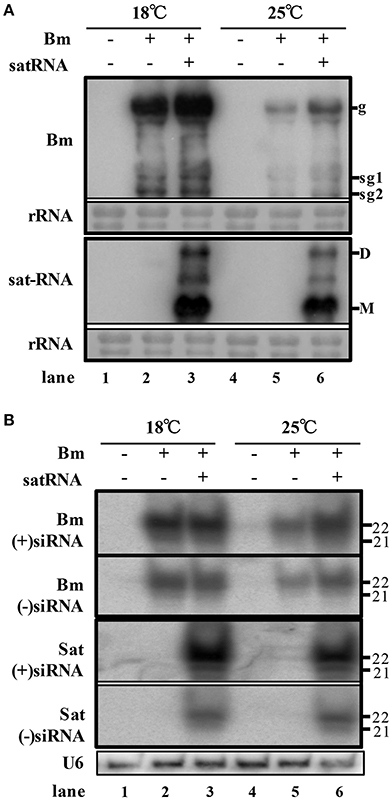
Figure 2. Low temperature and co-infection with its sat-RNA relatively relieve the silencing potency targeting to BBSV. (A) Accumulation of viral RNA from Bm (top panel) and sat-RNA (bottom panel) in systemically infected leaves. Genomic (g) and sub-genomic RNA1 and RNA2 (sg1 and sg2) were indicated on the right at the top panel. Sat-RNA monomer (M) and dimer (D) were indicated on the right at the bottom panel. Methylene blue-stained rRNA was used as a loading control. (B) northern blot analysis of Bm- and sat-RNA -derived siRNAs by probes hybridizing to sense and anti-sense genome regions. U6 were indicated as loading controls. The positions of 21 and 22 nt RNAs are indicated on the right.
To investigate the RNA silencing-mediated antiviral defense, the accumulation of the small interfering RNAs derived from the sat-RNA and its helper virus were analyzed through northern blot hybridizations. The Bm- and sat-RNA-derived siRNAs were readily detected in systemic leaves and all the viral siRNAs were mostly 22-nt in length followed by 21-nt (Figure 2B), which revealed that the replications of Bm and its sat-RNA strongly triggered the host RNA silencing. The accumulation level of BBSV-derived siRNAs was similar in the plants infected by Bm alone and co-infection with sat-RNA at 18°C (Figure 2B, compare lane 2 and 3), despite of the fact that BBSV genomic RNA accumulated to higher levels in the presence of sat-RNA than that of Bm alone (Figure 2A, compare lane 2 and 3). This finding indicated that the presence of sat-RNA relatively decreased the production of Bm-derived siRNAs in the co-infected plants at 18°C. Significantly, sat-siRNAs accumulated to very high levels in the co-infected leaves, regardless of temperature conditions (Figure 2B, sat-siRNAs lane 3 and 6). These results demonstrated that the high-level accumulation of the sat-RNA and its derived siRNAs may saturate the potency of antiviral silencing targeting Bm, which relieves the silencing targeting to Bm.
Sat-RNA Reduces the Production of Bm-Derived siRNAs by Saturating DCL2 and DCL4 Function during Co-infection
To characterize the population of the siRNAs derived from Bm and its sat-RNA, total small RNAs were cloned from the systemically infected leaves of N. benthamiana plants maintained at different temperatures. After trimming the linker sequences, the small RNA reads of 18- to 30-nt in length were further analyzed. More than 16 million small RNA reads, including endogenous small RNAs and virus-derived siRNAs, were obtained from each sample (Table 1). Notably, in the Bm-infected leaves, approximately 69.2% of total sequenced small RNAs were perfectly match or complementary to the genome of Bm (Table 1), indicating that Bm genomic and subgenomic RNAs served as the major substrates of the host Dicer enzyme(s) in N. benthamiana plants infected with Bm alone. In the BBSV/sat-RNA co-infected plants at 18°C, however, 29.6 and 43.2% of total small RNAs were mapped to Bm and its sat-RNA, respectively (Table 1). The similar results were obtained in the systemic leaves infected by Bm alone or with sat-RNA at 25°C (Table 1). These results are consistent with the above northern blot analysis and strongly indicate that sat-RNA is the predominant substrate of the host Dicer enzyme(s), leading to reduced production ratio of the helper viral siRNAs in the BBSV/sat-RNA co-infected plants.
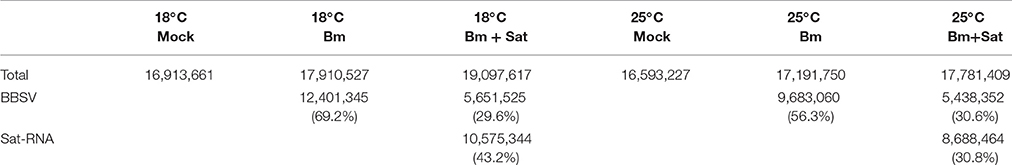
Table 1. The amount of small RNAs isolated from systemic leaves of N. benthamiana inoculated with buffer (Mock), Bm alone or co-inoculated with the sat-RNA at low (18°C) and room (25°C) temperature.
We further analyzed the polarity of virus-derived siRNAs and found different profiles of viral siRNAs derived from Bm and sat-RNA. Nearly equal amount of positive and negative stranded Bm-siRNAs accumulated in all small RNA samples (Figure 3A). However, in sat-RNA co-infected leaves, a clear prevalence for sense strand of sat-siRNAs was observed under both 18 and 25°C temperature conditions, representing 97.6 and 96.2% of the total sat-siRNAs, respectively (Figure 3B). The distinct polarity of Bm- and sat-siRNAs might due to their different replication processes, in which, positive-stranded RNA viruses usually use dsRNA as an intermediate template for genomic RNA synthesis (Kovalev et al., 2014), and most circular satellite RNAs utilize rolling cycle mechanism for its replication that produce abundant plus strands and few minus templates (Branch and Robertson, 1984; Bruening et al., 1991).
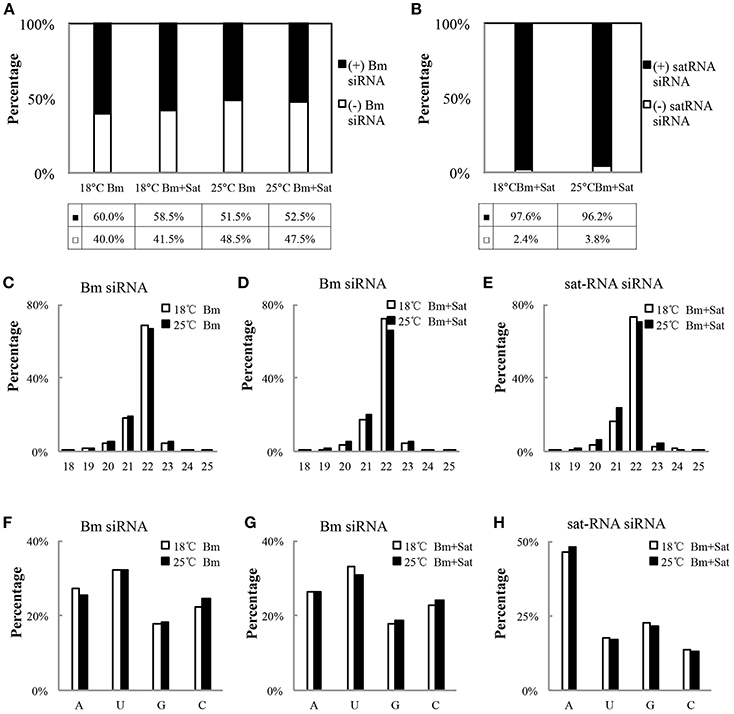
Figure 3. Profiles of the Bm- and sat-RNA -derived siRNAs. The total siRNAs were isolated from Bm-inoculated and sat-RNA co-inoculated systemic leaves grown at 18 and 25°C conditions at 12 dpi. The 18- to 30-nt siRNA from Bm- and sat-RNA were analyzed. Relative abundance of siRNA from the positive strand (black column) and the negative strand (white column) of BBSV genomic RNA (A) or sat-RNA (B). The relative percentages of (+) siRNA and (−) siRNA to total siRNAs are shown in the bottom. (C–E) showed the size distributions of Bm- and sat-RNA- derived siRNAs in the different treatment as indicated. (F–H) showed relative frequency of 5′-terminal nucleotide of Bm- and sat-RNA -derived siRNAs with respect to the amount of individual nucleotides in the Bm genome (F,G) or sat-RNA genome (H).
With regard to size distribution, both Bm-siRNAs and sat-siRNAs are dominated by 22-nt reads (65.3–73.6%), followed by 21-nt reads (16.3–28.9%) and other length reads (Figures 3C–E), which suggests that the N. benthamiana homologs of DCL2 and DCL4 are the predominant Dicers involved in the biogenesis of viral siRNA from both Bm and sat-RNA. The dominance of 22-nt siRNAs of BBSV and sat-RNA is consistent with Cymbidium ring spot virus (CymRSV)-derived vsRNAs in N. benthamiana plants (Donaire et al., 2009).
The previous studies have reported that the selective loading of small RNAs into specific AGOs is determined by the 5′-terminal nucleotides of siRNAs (Mi et al., 2008; Montgomery et al., 2008). To determine potential interactions between viral siRNAs and distinct AGO complexes, we analyzed the 5′ terminial of viral siRNAs derived from Bm- and sat-siRNAs (Figures 3F–H). Bm-siRNAs are dominated by uridines (5′U) with the ratio of 31.0–33.2%, and followed in order by adenines (A), cytidines (C), and guanines (G), which is consistent in the samples infected by Bm alone or with sat-RNA under different temperature (Figures 3F,G). In contrast, sat-RNA-derived siRNAs exhibited a clear predominance of A at 5′ end (46.2–48.0%) (Figure 3H). These trends were not affected by two temperature conditions. In contrast, there is no obvious preference of nucleotides in the composition of BBSV and sat-RNA genome (BBSV: A 24.8%, C 23.7%, G 25.8%, U 25.7%; sat-RNA: A 26.0%, C 22.0%, G 24.4%, U 27.6%). Considering different AGOs preferred different 5′-terminal first nucleotide (Mi et al., 2008; Montgomery et al., 2008), these results indicate an involvement of different AGOs, mainly AGO1, and AGO2, in the antiviral silencing targeting Bm and sat-RNA, respectively.
Sat-RNA-Derived siRNAs are Predominantly Processed from the Highly Structured Region of the sat RNA Genome
To further explore the frequencies of Bm-siRNAs and sat-siRNAs in the Bm and sat-RNA genomes, we mapped the positive- and negative-stranded viral siRNAs to the top and bottom of genomes of Bm and sat-RNA, respectively. Note that two different scales were used to accommodate the high abundance of siRNAs. The Bm-siRNAs were almost continuously but heterogeneously distributed along Bm genome and exhibited similar patterns with or without sat-RNA (Figure 4A). However, the sat-siRNAs exhibited several peak distribution features in all sat-RNA co-inoculation samples (Figure 4B). The most abundant sat-siRNAs were peaked in the positive strand of nt 396–417 (Figure 4B), where a highly structured region was predicted to be formed by using Mfold software (Figure S1). Notably, the distribution patterns of Bm- and sat-RNA-derived siRNAs remained unchanged in all the virus-inoculated samples at both 18 and 25°C, indicating that the profiles of Bm- and sat-siRNAs were not a result of sequencing biases (Figure 4, Figure S2).
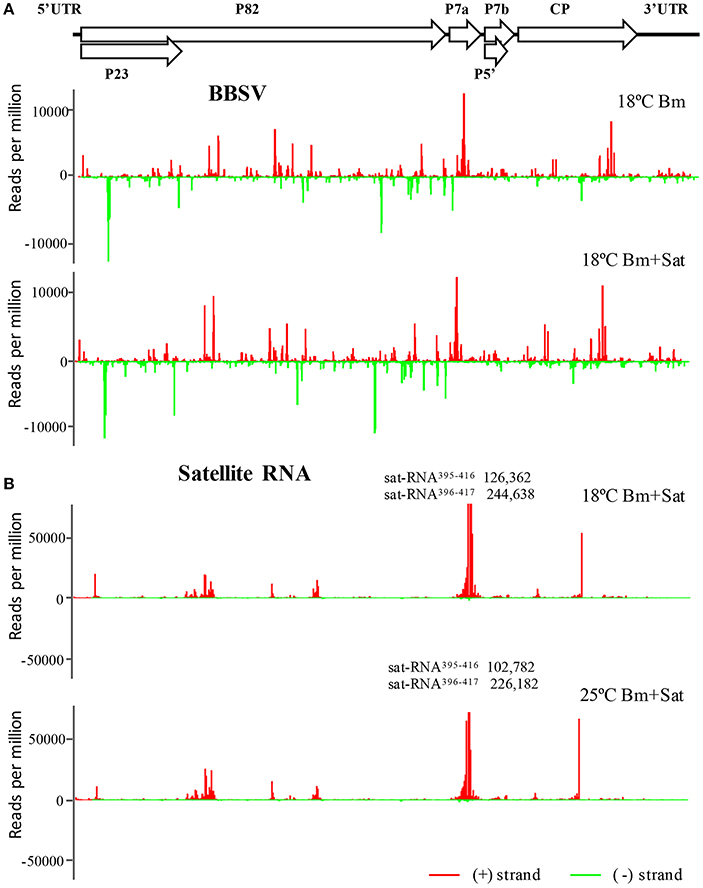
Figure 4. The abundant distribution of siRNAs on Bm and sat-RNA genomes. siRNAs derived from the viral genome of Bm (A) and sat-RNA (B) are shown in red above (positive strand) or green below (negative strand) the horizontal line. X axis represents the length of the genome, and Y axis the counts of the siRNAs per million of total viral-derived siRNAs. The Bm genome organization is shown schematically above the graphs with coding open reading frames indicated as open arrows. Note that the read counts of two (+) sat-siRNAs (sat-RNA395−416 and sat-RNA396−417) are out of scale and indicated (B).
Bm Infection or Co-infection with Bm Sat-RNA Perturbs the Expression of Antiviral Silencing Genes
We also analyzed miRNA expression from total small RNA reads. Virus infection induced down-regulation of miR164, miR166, miR167, miR168, and miR403, and up-regulation of miR172 and miR397, after inoculation of Bm alone or with sat-RNA at two temperatures (Figures 5A,D; Figure S3), indicating that a series of host miRNAs were affected by virus infection. Among these miRNAs, miR168, and miR403 target the mRNAs of AGO1 and AGO2, respectively, which are the main antiviral silencing components. Therefore, the expression levels of miR168 and miR403 were further confirmed by quantitative RT-PCR (Figures 5B,E). Interestingly, the down-regulation of miR168 level was more obvious in Bm samples than that in sat-RNA co-inoculation samples, which is consistent in 18 and 25°C (Figure 5B). Accordingly, AGO1, the target of miR168, was up-regulated and the mRNA level was higher in Bm samples than that with sat-RNA (Figure 5C), suggesting a main role of antiviral AGO1 in Bm inoculation leaves. However, miR403, which negatively regulates AGO2 mRNAs, exhibited down-regulation level to a larger extent in sat-RNA co-inoculation samples than that in Bm samples (Figure 5E), and its target AGO2 were up-regulated higher in sat-RNA co-inoculation leaves (Figure 5F), indicating AGO2 as a major antiviral component of RNA silencing in sat-RNA involving leaves. These results were consistent with the results of 5′ nucleotide bias analysis of viral siRNAs, in which predominant U in Bm-siRNAs were mediated by AGO1, whereas prominent A preference in sat-siRNAs were mainly AGO2 involved (Figures 2A,B).
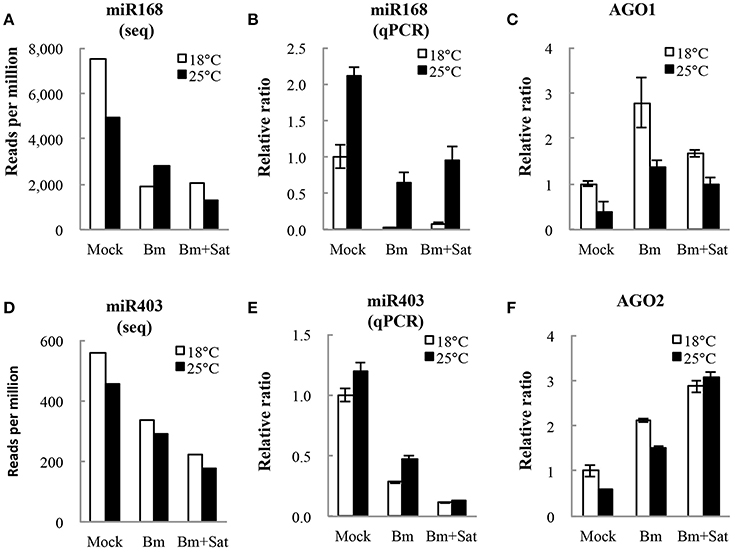
Figure 5. miR168 and miR403 expression levels and their targets AGO1 and AGO2 analysis. (A,D) showed the amount of miR168 and miR403 per million total reads in small RNA library. (B,E) showed quantitative RT-PCR validation of the relative expression levels of miR168 and miR403. (C,F) showed relative expression of AGO1 and AGO2 transcripts targeted by miR168 and miR403, respectively. Error bars represent mean standard error calculated from three biological replicates.
We also detected the accumulation of other RNA silencing components DCL2, DCL4, and RDR6 mRNA levels in N. benthamiana by quantitative real-time RT-PCR (Figure 6). Compared with mock, Bm alone or with sat-RNA consistently induced relatively high levels of DCL2 and DCL4 mRNA accumulation (Figures 6A,B), which is consistent with the sequencing data of dominant 22- and 21-nt length siRNAs. It is interesting that no significant changes in the accumulation of RDR6 mRNAs in sat-RNA co-inoculation samples compared with Bm alone (Figure 6B), perhaps due to its primary function of stably producing ta-siRNAs that is most important for plant growth. All these data suggest that the host plants exert different expression patterns to Bm and sat-RNA for antiviral silencing.
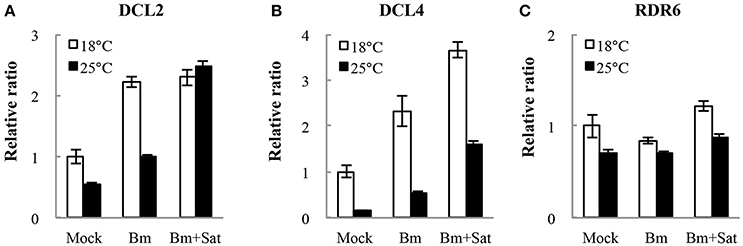
Figure 6. Quantitative RT-PCR analysis of the expression of RNA silencing-related N. benthamiana DCL2 (A), DCL4 (B), and RDR6 (C) mRNAs. Error bars represent mean standard error calculated from three biological replicates. The information and primer sequences used for amplification of DCLs and RDR6 were listed in Table S1.
Discussion
Sat-RNAs are viral parasites and depend on their helper viruses for replication, encapsidation and movement in the host plants. Sat-RNAs are usually involved in the interaction between their helper viruses and plant hosts by modulating the accumulation level of helper viruses and symptom induction. In this study, we found that the sat-RNA of BBSV facilitated the systemic infection of the helper virus in N. benthamiana. Our results also show that BBSV is temperature sensitive and the systemic infection is enhanced at lower temperature. Analysis of siRNAs derived from the sat-RNA and the helper virus by both northern blotting and cloning/sequencing revealed that virus infection triggers high levels of RNA silencing and that sat-RNA co-replication produces highly abundant sat-siRNAs to relieve the silencing pressure for helper virus. Meanwhile, virus infection induced high levels of expression of DCLs and AGOs of RNA silencing as main antiviral elements. Our findings reveal that the helper virus (Bm) is probably mainly targeted by AGO1-associated complex, and the sat-RNA is silenced by AGO2-associated complex. These results illustrate the sat-RNA could benefit the helper virus, especially when the helper virus is confronted with strong defense of host plants.
It is known that plant RNA-directed RNA polymerases (RDRs) were involved in antiviral silencing and that high temperature enhances antiviral silencing in N. benthamiana because RDR6 may be inactive at the low temperature (Xie et al., 2001; Szittya et al., 2003; Qu et al., 2005). In Arabidopsis and N. benthamiana, RDR6 has been shown playing an important role in the host antiviral defense (Dalmay et al., 2000; Mourrain et al., 2000; Schwach et al., 2005). Thus, our observation that milder symptom by BBSV in N. benthamiana at the higher temperature might be due to the improvement of RDR6 functions, but not of its mRNA levels shown in Figure 6C. The incomplete or aborted viral RNAs produced in the process of BBSV replication would be recognized as aberrant RNAs to be converted into dsRNAs de novo by RDR6 or other RDRs, strengthening the silencing to degrade the viral RNAs.
Although the replication of defective interfering RNA (DI RNAs), sat-RNAs and viroids produces abundant siRNAs, these sub-viral RNAs are resistant to RNA silencing, because highly structured RNAs may be poor targets for RNA cleavages by RNA-induced silencing complex (Wang et al., 2004; Du et al., 2007; Gomez and Pallas, 2007; Itaya et al., 2007). Unlike DI RNAs, however, sat-RNAs have no sequence homology with the helper virus so that the abundant siRNAs derived from the sat-RNA are not able to enhance BBSV silencing. Therefore, we propose that the efficient sat-RNA replication yields abundant substrates for host Dicers, which compete for the silencing factors and thereby facilitate infection and spread of the helper virus. It should be pointed out that sat-RNA modulation of helper virus silencing reported in this study is independent of the sequence homology between sat-RNA and the helper virus reported previously for TCV sat-RNA C, which is 3′ co-terminal with the helper viral RNAs (Zhang and Simon, 2003).
It is known that sat-RNAs compete for the replication machinery with the helper virus, which could enhance the survival of the host to the benefit of the helper virus (Hull, 2002). Our work suggests a new function for sat-RNAs in the antiviral silencing of the helper virus that is also beneficiary to the infection and spread of the helper virus. We propose that these properties of sat-RNAs play a key role in the emergence and evolution of sat-RNAs.
Author Contributions
XW and JY conceived and designed the experiments; JX and DeL performed the experiments; JX and XW analyzed the data and drafted the manuscript. YZ, YW, CH and DaL participated in experimental coordination and revision of the manuscript; XW and JY proofread and finalized the manuscript.
Funding
This work was supported by grants from National Science Foundation of China (31370176, 31500123, and 31322004).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Songyu Liu for analyzing the small RNA libraries of BBSV and its sat-RNA.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01771/full#supplementary-material
References
Branch, A. D., and Robertson, H. D. (1984). A replication cycle for viroids and other small infectious RNA's. Science 223, 450–455. doi: 10.1126/science.6197756
Bruening, G., Passmore, B. K., van Tol, H., Buzayan, J. M., and Feldstein, P. A. (1991). Replication of a plant virus satellite RNA: evidence favors transcription of circular templates of both polarities. Mol. Plant Microbe Interact. 4, 219–225. doi: 10.1094/MPMI-4-219
Cao, Y. H., Cai, Z. N., Ding, Q., Li, D. W., Han, C. G., Yu, J. L., et al. (2002). The complete nucleotide sequence of Beet black scorch virus (BBSV), a new member of the genus necrovirus. Arch. Virol. 147, 2431–2435. doi: 10.1007/s00705-002-0896-1
Chapman, E. J., and Carrington, J. C. (2007). Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 8, 884–896. doi: 10.1038/nrg2179
Collmer, C. W., and Howell, S. H. (1992). Role of satellite RNA in the expression of symptoms caused by plant viruses. Annu. Rev. Phytopathol. 30, 419–442. doi: 10.1146/annurev.py.30.090192.002223
Dalmay, T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D. C. (2000). An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101, 543–553. doi: 10.1016/S0092-8674(00)80864-8
Díaz-Pendón, J. A., and Ding, S. W. (2008). Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Phytopathol. 46, 303–326. doi: 10.1146/annurev.phyto.46.081407.104746
Ding, S. W., Li, H., Lu, R., Li, F., and Li, W. X. (2004). RNA silencing: a conserved antiviral immunity of plants and animals. Virus Res. 102, 109–115. doi: 10.1016/j.virusres.2004.01.021
Ding, S. W., and Voinnet, O. (2007). Antiviral immunity directed by small RNAs. Cell 130, 413–426. doi: 10.1016/j.cell.2007.07.039
Donaire, L., Wang, Y., Gonzalez-Ibeas, D., Mayer, K. F., Aranda, M. A., and Llave, C. (2009). Deep-sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology 392, 203–214. doi: 10.1016/j.virol.2009.07.005
Du, Q. S., Duan, C. G., Zhang, Z. H., Fang, Y. Y., Fang, R. X., Xie, Q., et al. (2007). DCL4 targets Cucumber mosaic virus satellite RNA at novel secondary structures. J. Virol. 81, 9142–9151. doi: 10.1128/JVI.02885-06
Gómez, G., and Pallás, V. (2007). Mature monomeric forms of Hop stunt viroid resist RNA silencing in transgenic plants. Plant J. 51, 1041–1049. doi: 10.1111/j.1365-313X.2007.03203.x
González-Vázquez, M., Ayala, J., García-Arenal, F., and Fraile, A. (2009). Occurrence of Beet black scorch virus infecting sugar beet in Europe. Plant Dis. 93, 21–24. doi: 10.1094/PDIS-93-1-0021
Guo, L. H., Cao, Y. H., Li, D. W., Niu, S. N., Cai, Z. N., Han, C. G., et al. (2005). Analysis of nucleotide sequences and multimeric forms of a novel satellite RNA associated with Beet black scorch virus. J. Virol. 79, 3664–3674. doi: 10.1128/JVI.79.6.3664-3674.2005
Hu, C. C., Hsu, Y. H., and Lin, N. S. (2009). Satellite RNAs and satellite viruses of plants. Viruses 1, 1325–1350. doi: 10.3390/v1031325
Huang, J., Yang, M., Lu, L., and Zhang, X. (2016). Diverse functions of small RNAs in different plant-pathogen communications. Front. Microbiol. 7:1552. doi: 10.3389/fmicb.2016.01552
Itaya, A., Zhong, X., Bundschuh, R., Qi, Y., Wang, Y., Takeda, R., et al. (2007). A structured viroid RNA serves as a substrate for dicer-like cleavage to produce biologically active small RNAs but is resistant to RNA-induced silencing complex-mediated degradation. J. Virol. 81, 2980–2994. doi: 10.1128/JVI.02339-06
King, A. M. Q., Adams, M. J., Carstens, E. B., and Lefkowitz, E. J. (2011). Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Elsevier Inc.
Koenig, R., and Valizadeh, J. (2008). Molecular and serological characterization of an Iranian isolate of Beet black scorch virus. Arch. Virol. 153, 1397–1400. doi: 10.1007/s00705-008-0121-y
Kotakis, C., Vrettos, N., Kotsis, D., Tsagris, M., Kotzabasis, K., and Kalantidis, K. (2010). Light intensity affects RNA silencing of a transgene in Nicotiana benthamiana plants. BMC Plant Bio. 10:220. doi: 10.1186/1471-2229-10-220
Kovalev, N., Pogany, J., and Nagy, P. D. (2014). Template role of double-stranded RNA in tombusvirus replication. J. Virol. 88, 5638–5651. doi: 10.1128/JVI.03842-13
Li, F., and Ding, S. W. (2006). Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 60, 503–531. doi: 10.1146/annurev.micro.60.080805.142205
Li, X. S. A. (1990). Symptom intensification on cruciferous hosts by the virulent satellite RNA of Turnip crinkle virus. Phytopathology 80, 238–242. doi: 10.1094/Phyto-80-238
Meister, G., and Tuschl, T. (2004). Mechanisms of gene silencing by double-stranded RNA. Nature 431, 343–349. doi: 10.1038/nature02873
Mi, S., Cai, T., Hu, Y., Chen, Y., Hodges, E., Ni, F., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5' terminal nucleotide. Cell 133, 116–127. doi: 10.1016/j.cell.2008.02.034
Montgomery, T. A., Howell, M. D., Cuperus, J. T., Li, D., Hansen, J. E., Alexander, A. L., et al. (2008). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133, 128–141. doi: 10.1016/j.cell.2008.02.033
Mourrain, P., Béclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J. B., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101, 533–542. doi: 10.1016/S0092-8674(00)80863-6
Murant, A. F., and Mayo, M. A. (1982). Satellites of plant viruses. Annu. Rev. Phytopathol. 20, 49–68. doi: 10.1146/annurev.py.20.090182.000405
Murant, A. F., Rajeshwari, R., Robinson, D. J., and Raschké, J. H. (1988). A satellite RNA of Groundnut rosette virus that is largely responsible for symptoms of groundnut rosette disease. J. Gen. Virol. 69, 1479–1486. doi: 10.1099/0022-1317-69-7-1479
Qu, F., Ye, X., Hou, G., Sato, S., Clemente, T. E., and Morris, T. J. (2005). RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J. Virol. 79, 15209–15217. doi: 10.1128/JVI.79.24.15209-15217.2005
Robinson, D. J., Ryabov, E. V., Raj, S. K., Roberts, I. M., and Taliansky, M. E. (1999). Satellite RNA is essential for encapsidation of groundnut rosette umbravirus RNA by groundnut rosette assistor luteovirus coat protein. Virology 254, 105–114. doi: 10.1006/viro.1998.9527
Roossinck, M. J., Sleat, D., and Palukaitis, P. (1992). Satellite RNAs of plant viruses: structures and biological effects. Microbiol. Mol. Biol. Rev. 56, 265–279.
Schwach, F., Vaistij, F. E., Jones, L., and Baulcombe, D. C. (2005). An RNA-dependent RNA polymerase prevents meristem invasion by Potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138, 1842–1852. doi: 10.1104/pp.105.063537
Shimura, H., Pantaleo, V., Ishihara, T., Myojo, N., Inaba, J.-I., Sueda, K., et al. (2011). A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog. 7:e1002021. doi: 10.1371/journal.ppat.1002021
Simon, A. E., Roossinck, M. J., and Havelda, Z. (2004). Plant virus satellite and defective interfering RNAs: new paradigms for a new century. Annu. Rev. Phytopathol. 42, 415–437. doi: 10.1146/annurev.phyto.42.040803.140402
Smith, N. A., Eamens, A. L., and Wang, M. B. (2011). Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog. 7:e1002022. doi: 10.1371/journal.ppat.1002022
Szittya, G., Silhavy, D., Molnár, A., Havelda, Z., Lovas, A., Lakatos, L., et al. (2003). Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J. 22, 633–640. doi: 10.1093/emboj/cdg74
Thomas, C. L., Leh, V., Lederer, C., and Maule, A. J. (2003). Turnip crinkle virus coat protein mediates suppression of RNA silencing in Nicotiana benthamiana. Virology 306, 33–41. doi: 10.1016/S0042-6822(02)00018-1
Varkonyi-Gasic, E., and Hellens, R. P. (2011). “Quantitative stem-loop RT-PCR for detection of microRNAs,” in RNAi and Plant Gene Function Analysis: Methods and Protocols, Vol. 744, eds H. Kodama and A. Komamine (New York, NY: Humana Press), 145–157.
Voinnet, O. (2005). Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6, 206–220. doi: 10.1038/nrg1555
Wang, J., and Simon, A. E. (1999). Symptom attenuation by a satellite RNA in vivo is dependent on reduced levels of virus coat protein. Virology 259, 234–245. doi: 10.1006/viro.1999.9781
Wang, M. B., Bian, X. Y., Wu, L. M., Liu, L. X., Smith, N. A., Isenegger, D., et al. (2004). On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. U.S.A. 101, 3275–3280. doi: 10.1073/pnas.0400104101
Weiland, J. J., Larson, R. L., Freeman, T. P., and Edwards, M. C. (2006). First report of Beet black scorch virus in the United States. Plant Dis. 90, 828. doi: 10.1094/PD-90-0828B
Weiland, J. J., Van Winkle, D., Edwards, M. C., Larson, R. L., Shelver, W. L., Freeman, T. P., et al. (2007). Characterization of a U.S. isolate of Beet black scorch virus. Phytopathology 97, 1245–1254. doi: 10.1094/PHYTO-97-10-1245
Xi, D. H., Cao, Y. H., Guo, L. H., Yuan, X. F., Cai, Z. N., Han, C. G., et al. (2006). Construction and pathogenicity of infectious cDNA clone of Beet black scorch virus xinjiang isolate. Bing Du Xue Bao 22, 309–313.
Xie, Z., Fan, B., Chen, C., and Chen, Z. (2001). An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proc. Natl. Acad. Sci. U.S.A. 98, 6516–6521. doi: 10.1073/pnas.111440998
Xu, J., Wang, X., Shi, L., Zhou, Y., Li, D., Han, C., et al. (2012). Two distinct sites are essential for virulent infection and support of variant satellite RNA replication in spontaneous Beet black scorch virus variants. J. Gen. Virol. 93, 2718–2728. doi: 10.1099/vir.0.045641-0
Yuan, X., Cao, Y., Xi, D., Guo, L., Han, C., Li, D., et al. (2006). Analysis of the subgenomic RNAs and the small open reading frames of Beet black scorch virus. J. Gen. Virol. 87, 3077–3086. doi: 10.1099/vir.0.81928-0
Keywords: beet black scorch virus, satellite RNA, RNA silencing, temperature, siRNAs
Citation: Xu J, Liu D, Zhang Y, Wang Y, Han C, Li D, Yu J-L and Wang X-B (2016) Improved Pathogenicity of a Beet Black Scorch Virus Variant by Low Temperature and Co-infection with Its Satellite RNA. Front. Microbiol. 7:1771. doi: 10.3389/fmicb.2016.01771
Received: 16 August 2016; Accepted: 21 October 2016;
Published: 04 November 2016.
Edited by:
Feng Li, Huazhong Agricultural University, ChinaReviewed by:
Jianguo Wu, Fujian Agriculture and Forestry University, ChinaFei Yan, ZheJiang Academy of Agricultural Sciences, China
Rui Lu, LSI, USA
Copyright © 2016 Xu, Liu, Zhang, Wang, Han, Li, Yu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xian-Bing Wang, d2FuZ3hpYW5iaW5nQGNhdS5lZHUuY24=
 Jin Xu
Jin Xu Deshui Liu1
Deshui Liu1 Yongliang Zhang
Yongliang Zhang Chenggui Han
Chenggui Han Dawei Li
Dawei Li Xian-Bing Wang
Xian-Bing Wang