- 1College of Veterinary Medicine, Nanjing Agricultural University, Nanjing, China
- 2Animal Quarantine Laboratory, Jiangsu Entry-Exit Inspection and Quarantine Bureau, Nanjing, China
- 3Shanghai Entry-Exit Inspection and Quarantine Bureau, Shanghai, China
- 4Division of Molecular Biology, Center for Food Safety and Applied Nutrition, United States Food and Drug Administration, Laurel, MD, USA
Foodborne outbreaks are a serious public health and food safety concern worldwide. There is a great demand for rapid, sensitive, specific, and accurate methods to detect microbial pathogens in foods. Conventional methods based on cultivation of pathogens have been the gold standard protocols; however, they take up to a week to complete. Molecular assays such as polymerase chain reaction (PCR), sequencing, microarray technologies have been widely used in detection of foodborne pathogens. Among molecular assays, PCR technology [conventional and real-time PCR (qPCR)] is most commonly used in the foodborne pathogen detection because of its high sensitivity and specificity. However, a major drawback of PCR is its inability to differentiate the DNA from dead and viable cells, and this is a critical factor for the food industry, regulatory agencies and the consumer. To remedy this shortcoming, researchers have used biological dyes such as ethidium monoazide and propidium monoazide (PMA) to pretreat samples before DNA extraction to intercalate the DNA of dead cells in food samples, and then proceed with regular DNA preparation and qPCR. By combining PMA treatment with qPCR (PMA-qPCR), scientists have applied this technology to detect viable cells of various bacterial pathogens in foods. The incorporation of PMA into PCR-based assays for viability detection of pathogens in foods has increased significantly in the last decade. On the other hand, some downsides with this approach have been noted, particularly to achieve complete suppression of signal of DNA from the dead cells present in some particular food matrix. Nowadays, there is a tendency of more and more researchers adapting this approach for viability detection; and a few commercial kits based on PMA are available in the market. As time goes on, more scientists apply this approach to a broader range of pathogen detections, this viability approach (PMA or other chemicals such as platinum compound) may eventually become a common methodology for the rapid, sensitive, and accurate detection of foodborne pathogens. In this review, we summarize the development in the field including progress and challenges and give our perspective in this area.
Introduction
Foodborne pathogens such as Escherichia coli O157:H7, Salmonella spp., Staphylococcus aureus, Listeria monocytogenes, Campylobacter spp., and Vibrio parahaemolyticus have been a public health concern and there is a growing demand for rapid, sensitive, and accurate methods to detect these pathogens (Scallan et al., 2011). According to the Centers for Disease Control and Prevention (CDC), foodborne pathogens are responsible for more than 48 million illnesses, 128,000 hospitalizations, and 3,000 deaths in the United States each year (Scallan et al., 2011). In 2013, there was a total of 5,196 foodborne outbreaks reported in the European Union, resulting in 43,183 infected humans, 5,946 hospitalizations, and 11 deaths (Da Silva Felicio et al., 2015). The global impact of foodborne illnesses is evidenced by its significant economic impact. The costs of foodborne illness extend from the direct medical costs associated with the illness to costs incurred by the industry through product recalls, loss of consumer confidence, and litigation. It has been estimated that the aggregated annual costs of foodborne illness in the United States exceed 77 million dollars (Scharff, 2012). Given the public health and economic impact of foodborne illness, it is important to study the distribution of foodborne microbes in food production chains and develop reliable and rapid methods for pathogen detection.
Traditional culture and microscopy methods for detection of viable cells can be tedious, labor-intensive and time-consuming. Some methods enable viability to be assessed by staining techniques, such as BacLight fluorescence microscopy or acridine orange, flow cytometry coupled with dyes, and physiological tests such as for cellular respiration but do not allow for detection of specific pathogen species (Diaper and Edwards, 1994; Caron et al., 1998; Keer and Birch, 2003). These culture-based methods give rise to several challenges such as the isolation and identification of specific pathogens among a plethora of background microflora, and the detection of pathogens that occur at low levels (Sidhu and Toze, 2009). Selective media are used to reduce growth of background microorganisms, but not without introducing potential biases (Nocker et al., 2007b). Enrichment can be used to detect low level of pathogens, however, this may enable reproduction of injured cells, and subsequently overestimate pathogen density (Sidhu and Toze, 2009). On the other hand, culture-based methods encounter another issue that some human pathogens such as Campylobacter jejuni, E. coli, Helicobacter pylori, Klebsiella pneumoniae, L. monocytogenes, Pseudomonas aeruginosa, Salmonella Typhimurium, Shigella dysenteriae, and Vibrio cholerae may enter a “viable but non-culturable” (VBNC) physiological state, in which they are living but cannot be grown outside of their natural habitat (Lowder et al., 2000; Oliver, 2005). Furthermore, culture-based methods are also time-consuming and tedious (Nocker et al., 2007b). Molecular assays such as polymerase chain reaction (PCR) assays are rapid, sensitive, however, they may overestimate viable cell numbers due to amplification of DNA from dead cells and extracellular DNA within samples (Rudi et al., 2005), and thus may lead to unnecessary product recalls and economic losses (Liu and Mustapha, 2014). Therefore, accurate detection of viable bacteria in foods is critical and necessary in assessing the risk for foodborne outbreaks because only live pathogens constitute the risk of foodborne outbreaks.
Currently, two techniques are available for viability detection of foodborne pathogens. The first one is based on the detection of mRNA by using reverse-transcriptase PCR (RT-PCR). It is based on that bacterial transcripts are sensitive to degradation by intra- and extracellular RNases; and mRNA levels should rapidly decline after cell death. Hence, mRNA would only be limited to the viable cells within the population. The development of RT-PCR assays to detect foodborne pathogens such as E. coli O157:H7 (Ju et al., 2016) utilized this premise. However, this approach is affected negatively by a few factors. First, it requires expression of the target gene(s), which may vary under stress conditions. Second, handling RNA is tedious and cumbersome due to its liability to contamination. Overall, the use of RT-PCR assay is more adapted for gene expression studies than as a detection means for foodborne pathogens (Barbau-Piednoir et al., 2014). Third, some reports have concluded that mRNA disappears quickly after cell death, while others suggest that transcripts can persist for extended lengths of time (Ju et al., 2016).
The other technique for viable cell detection is an approach that uses PCR method in conjunction with biological dyes, ethidium monoazide (EMA) and propidium monoazide (PMA, a derivative of ethidium bromide). The approach can specifically detect DNA from cells with intact cell/wall membranes and the viability discrimination is based on the characteristics of EMA and PMA. These biological dyes are positively charged molecules and thus are excluded by intact, negatively charged, bacterial cell-walls, but can enter bacteria with compromised cell-wall/membranes (Nocker and Camper, 2006).
The mechanism of action of EMA/PMA has not been fully elucidated yet, but it could be as the result of a combination of following factors, (i) when a EMA or PMA solution is added to a mixture of intact and membrane-compromised cells, the chemical can selectively enter only the compromised cells; (ii) once inside the cell, the dye intercalates into nucleic acids and the presence of an azide group allows for a cross-linking between the dye and the DNA after exposure to strong visible light; (iii) the light leads to the formation of a highly reactive nitrene radical, which can react with any organic molecule in its proximity including the bound DNA; (iv) this modification strongly inhibits the sequential DNA amplification in PCR; and (v) at the same time, when the cross-linking occurs, the light reacts with unbound excess dye with water molecules and the resulting hydroxylamine is no longer reactive, so the DNA from cells with intact membranes is not modified in the DNA extraction (Nocker et al., 2009). Therefore, by this mechanism, EMA or PMA can preferably intercalate DNA of the dead cells and thus prevent subsequent DNA amplification of dead cells by PCR as illustrated by Figure 1.
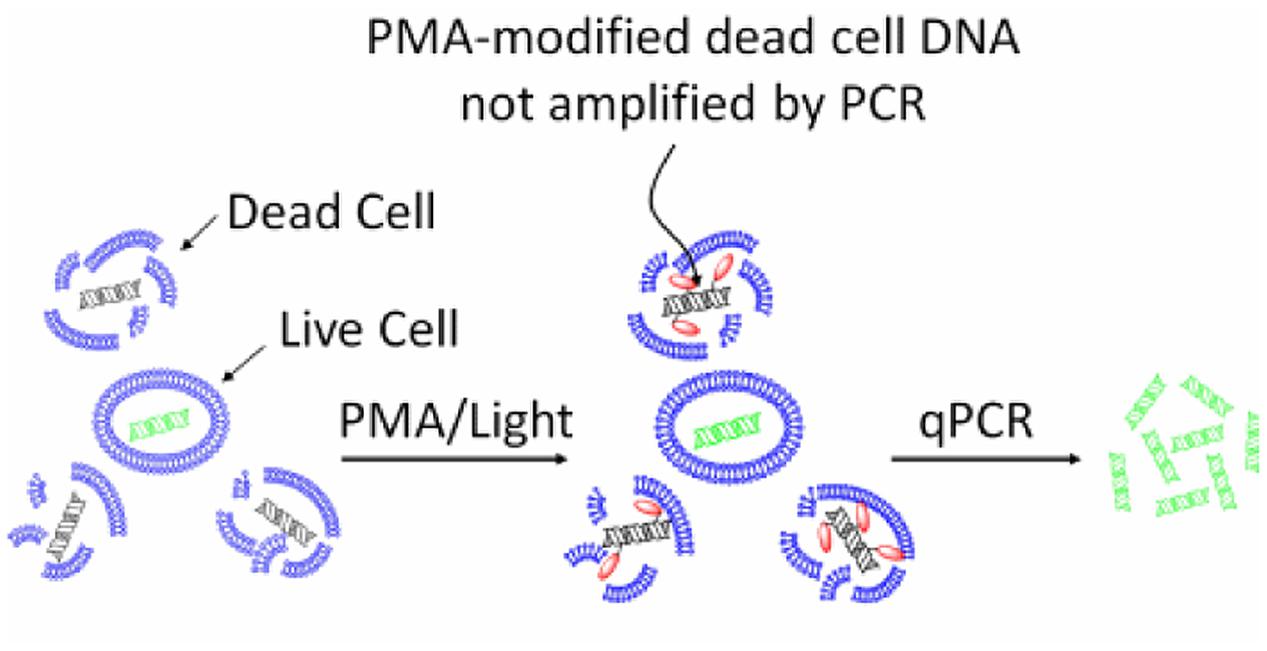
FIGURE 1. Principle of selective detection of viable cells using PMA dye (https://biotium.com/product/pmatm-dye-20mm-in-h2o/).
EMA/PMA-PCR assays have been applied to the detection of a variety of microbes, including bacterial cells and spores, fungi, viruses, and yeast in food and the environment (Agusti et al., 2010; Andorra et al., 2010; Josefsen et al., 2010; Fittipaldi et al., 2011; Li and Chen, 2012, 2013; Alonso et al., 2014; Law et al., 2015b). Recently, EMA/PMA-PCR assays have also been applied in bacterial studies on clinical samples, suggesting that this approach may have a potential alternative to diagnosis by microscopy and culture, and in clinical settings (Rogers et al., 2008; Miotto et al., 2012) or in new drug development (Caldas et al., 2014; Oliveira-Silva et al., 2015). In the article, we mainly focused on the application of this biological dye EMA/PMA in differentiation of viable cells of foodborne pathogens, E. coli O157:H7, Salmonella, S. aureus, L. monocytogenes, Campylobacter, and V. parahaemolyticus in foodborne pathogens and summarize the developments in this area.
Although EMA/PMA behaves nearly identically as intercalating stains, the two dyes differ in regard to their permeation through cell membranes. EMA, due to its chemical composition, is slightly more efficient in signal suppression than PMA, however, PMA is more effective than EMA in terms of live and dead discrimination. Numerous studies have used the EMA/PMA approach to detect viable cells of foodborne pathogens, and such work has yielded valuable data on the EMA/PMA efficiency and influence factors. The factors associated with efficiency of the viability detection includes the type and concentration of dye, concentration of organisms, type of food matrices, ratio between viable and dead cells, length of the PCR amplicon, physical condition of the sample, and light exposure conditions. In the study, we reviewed the development in application of the biological dyes, PMA/EMA, in differentiation of viable cells of foodborne pathogens including E. coli O157:H7, Salmonella, S. aureus, L. monocytogenes, Campylobacter, and V. parahaemolyticus; and we also discussed the challenges in using this approach and proposed strategies to remedy the drawbacks of this approach.
Application of EMA/PMA in Differentiation of Viable Cells of E. coli O157:H7 in Food
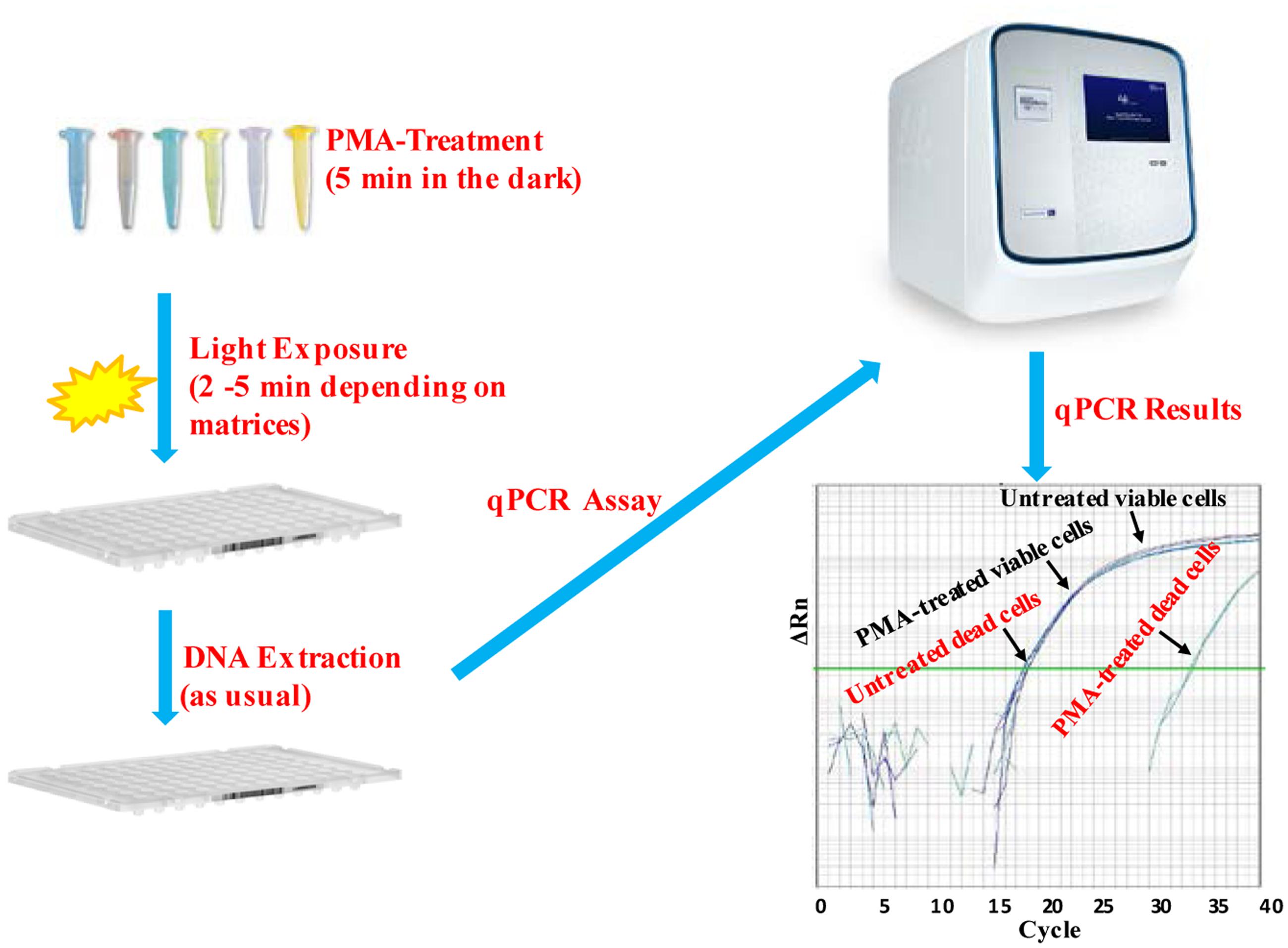
Escherichia coli O157:H7 is one of the most notorious foodborne pathogens, with an infectious dose of as low as a few hundred cells (Karmali, 2004). Beef, dairy products, juices, and fresh produce are foods that are often associated with E. coli O157:H7 outbreaks (Liu and Mustapha, 2014). Recently, numerous studies have applied EMA/PMA for differentiation of viable cells of E. coli O157:H7 in foods. Li and Chen (2012) selected ORF Z3276 as a unique detection target and applied PMA treatment to qPCR to accurately detect viable cells of E. coli O157:H7 in beef. They compared the different concentrations of PMA on the signal suppression of dead cells and found 50 μM PMA was the best concentration, which yielded strong signal suppression of dead cells and did not affect signal of viable cells (Table 1). In addition, the light exposure time was optimized at 2 min (Table 2). Subsequently, they optimized the PMA treatment conditions as the following: live/dead cell mixtures were added to PMA to a final concentration of 50 μM and incubated at room temperature in the dark for 5 min; the PMA-treated samples were exposed to a 650-W halogen light source, 20 cm from the samples for 2 min for the photo-induced cross-linking. After that the samples were subjected to regular DNA purification procedures and qPCR as illustrated in Figure 2. They demonstrated that this PMA-qPCR assay could detect 8 × 101 CFU/g mixed with 8 × 107 dead cells/g E. coli O157:H7 cells in spiked beef samples with an 8-h enrichment and that PMA treatment did not significantly affect the amplification of DNA from viable cells. In comparison, an EMA-qPCR assay could only detect at the 103 CFU/g E. coli O157:H7 cells in spiked beef samples after 8-h enrichment but failed to detect at 101 or 102 CFU/g E. coli O157:H7 cells in spiked beef samples with an 8-h enrichment (Wang et al., 2009). In comparison of the sensitivity of the two assays, Li and Chen (2012) attribute their enhanced sensitivity to three factors: first, the higher sensitivity of the PMA-qPCR can be attributed to the higher sensitivity of this qPCR assay itself; second, it may be due to the improved PMA treatment, as indicated by the smaller differences in CT values (0.5 CT value) between the PMA-treated and untreated viable cells; and third, PMA is more selective than EMA in inhibiting DNA amplification from dead cells. It is worth noting that the PMA-qPCR assay developed by Li and Chen (2012) has been well accepted by the scientific community and industry. Recently, a commercial kit has been developed based on Li and Chen’s findings by a U.S. company for detection of viable cells of E. coli O157:H71.
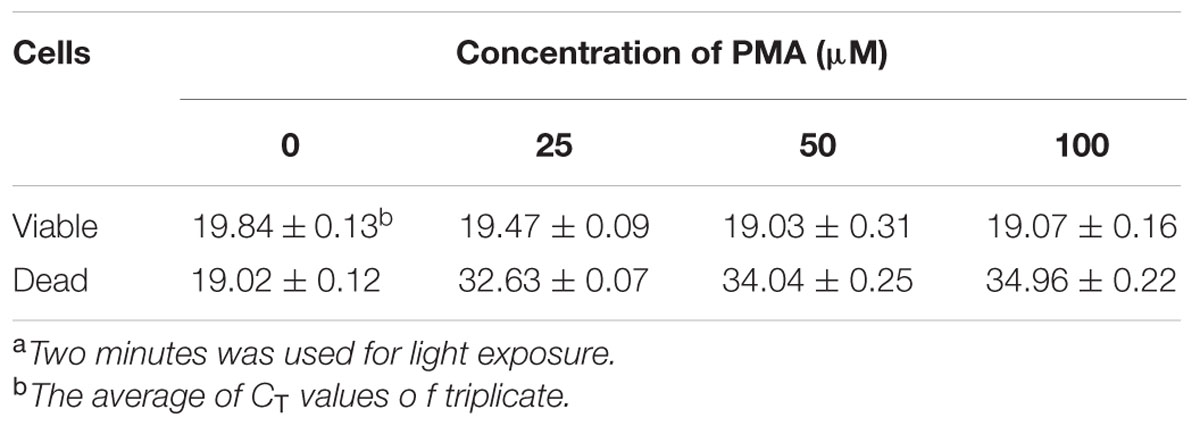
TABLE 1. Effect of different concentrations of PMA on signal suppression of dead cells of E. coli O157:H7 in PMA-qPCRa.
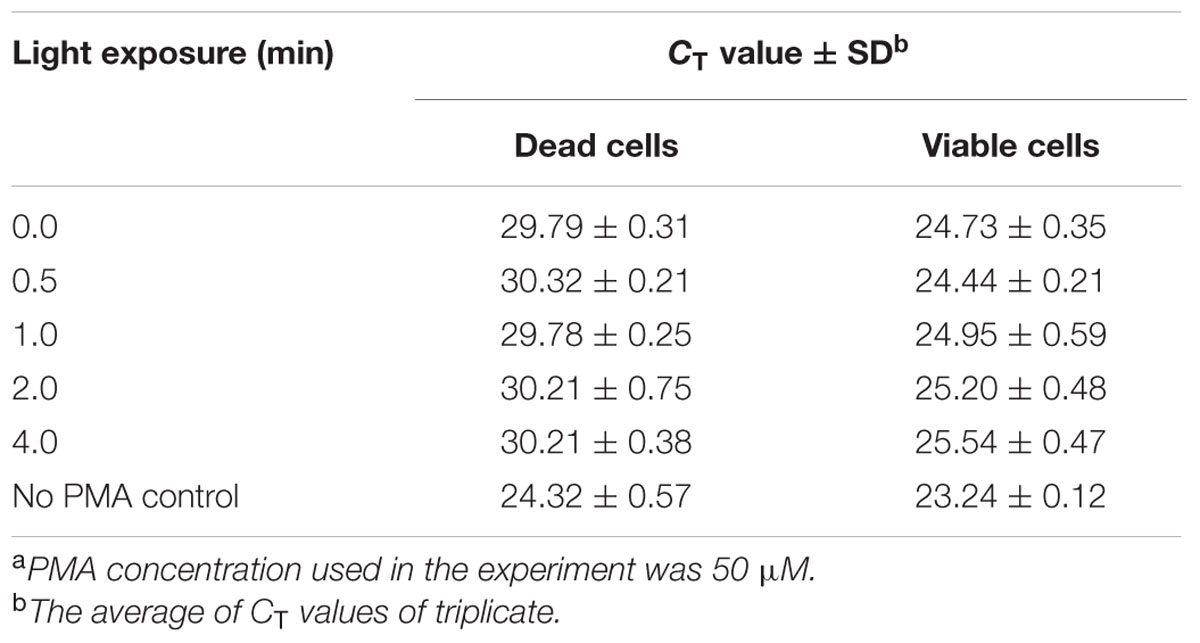
TABLE 2. Light exposure influence on PMA treatment of signal suppression of dead cells of E. coli O157:H7 in PMA-qPCRa.
Besides qPCR, researchers have also applied PMA/EMA to loop-mediated isothermal amplification (LAMP; Chen et al., 2011; Zhao et al., 2013). Chen et al. (2011) developed PMA-LAMP to detect Salmonella in produce and Zhao et al. (2013) combined PMA with LAMP to evaluate the inactivation effect on E. coli O157:H7 by slightly acidic electrolyzed water. Zhao et al. (2013) achieved detection limit of 1.6 × 102 CFU of E. coli O157:H7 per reaction by using rfbE gene as target gene and 3 μg/ml PMA as final concentration to treat the samples. Compared with PMA-qPCR, this PMA-LAMP assay had lower sensitivity, but it was more economical to run; it is particularly suitable for resource-limited labs to conduct large-scale detection.
Liu and Mustapha (2014) developed a PMA-qPCR assay for detection viable cells of E. coli O157:H7. They used 25 μM PMA to treat the cell mixtures with 10-min intensive light exposure. In the qPCR, the uidA gene and TaqMan were used. This PMA-qPCR assay could detect as low as 102 CFU/ml viable E. coli O157:H7 in pure culture and 105 CFU/g in ground beef in the presence of 106/g of dead cells. With an 8-h enrichment, 1 CFU/g viable E. coli O157:H7 in ground beef was detectable without interference from 106 dead cells/g (Liu and Mustapha, 2014). Additionally, other groups have also used PMA/EMA-qPCR to detect viable cells (Luo et al., 2010; Dinu and Bach, 2011; Elizaquivel et al., 2012; Liu et al., 2014). In contrast, in comparison of four different methods, culture, qPCR, RT-PCR, PMA-qPCR for quantitative detection of viable cells of E. coli O157:H7 in plant matrices, Ju et al. (2016) found that neither RT-PCR (with 2 log reduction) nor PMA-qPCR (with 3 log reduction) could efficiently suppress the DNA signals from dead cells.
EMA/PMA in Differentiation of Viable Cells of Salmonella in Food
Salmonella infections represent a considerable global burden with significant health and economic impacts. Salmonellosis is most often attributed to the consumption of contaminated foods such as poultry, beef, pork, eggs, milk, seafood, nut products, and fresh produce (Scallan et al., 2011). There is a need for the development of more sensitive, rapid, and inexpensive methods for detection of this pathogen in foods (Techathuvanan and D’Souza, 2012; Kokkinos et al., 2014). Recently, several studies applied EMA/PMA to PCR to detect viable cells of foodborne pathogens including Salmonella spp. in foods. However, it was noticed in some studies that sometimes the suppression of DNA signal of dead cells was incomplete. To address that issue, an efficient PMA-qPCR assay was developed by targeting a conserved region of the invA gene of Salmonella in conjunction with PMA treatment for detection of DNA from viable cells of Salmonella in food (Li and Chen, 2013). In that study, Li and Chen (2013) systematically compared efficiency of five different-sized amplicons ranging from 65 to 260 bp in signal suppression of dead cells in qPCR. The most efficient amplification was detected with Amplicon A (65 bp) with a CT value of 17.75, whereas the least amplification efficiency was found with Amplicon E (260 bp) with a CT value of 21.19. These data clearly demonstrated that the Amplicon D (130-bp) was the optimized amplicon in the PMA-qPCR assay. With the Amplicon D, the authors not only attained superior qPCR amplification efficiency with a CT value of 18.34, but also achieved high signal suppression of dead cells in qPCR with a CT value of 13.18 as shown in Table 3. Furthermore, this PMA-qPCR assay was capable of detecting live Salmonella cells in live/dead cell mixtures. In addition, the level of sensitivity achieved was 30 CFU/g live Salmonella cells from enriched spiked spinach samples as early as 4 h.
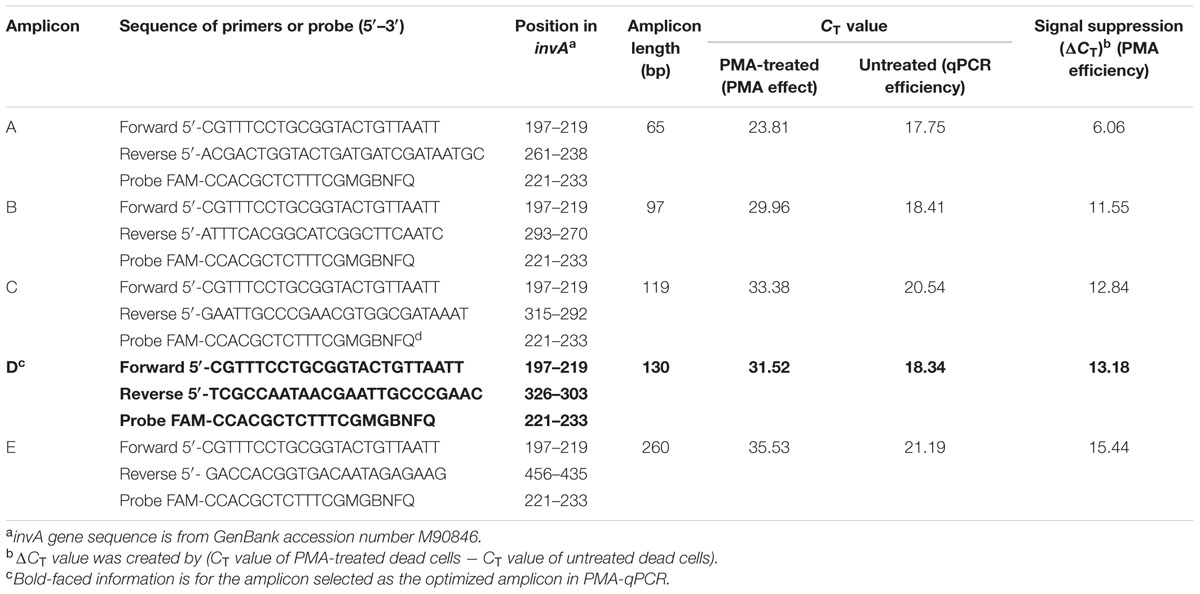
TABLE 3. Effect of amplicons of different length on signal suppression of Salmonella dead cells by PMA-qPCR.
It is worth noting that the optimal sized amplicon (130 bp) in the qPCR is quite different to that previously reported (Luo et al., 2010). In that work to detect viable cells of E. coli O157:H7, they found that PMA cannot efficiently exclude the DNA of the dead cells when the sizes of amplicons smaller than 190 bp were targeted by PCR. This is a significant size difference in designing amplicons for qPCR because the amplification efficiency of qPCR is affected by the sizes of amplicons and most of the qPCR instruments work best with amplicons under 150-bp. Therefore, selecting the right-sized amplicons for qPCR is a key factor to successfully detect viable foodborne pathogens (Li and Chen, 2013). Additionally, in the last few years, several more studies addressed the technical issues with the PMA-qPCR for detection of viable cells of Salmonella in food (Nocker et al., 2009; Liang et al., 2011; Banihashemi et al., 2012; Elizaquivel et al., 2012; Yang Y. et al., 2013; Barbau-Piednoir et al., 2014; Wu et al., 2015).
In order to improve the efficiency of PMA treatment, scientists have tried adding sarkosyl (0.2%) to the PMA solution and found that 0.2% sarkosyl increased PMA’s penetration to the dead cells with little effect on the viable cells (Wang et al., 2014; Li et al., 2015). Recently, a multiplex PMA-qPCR was developed to viable Legionella pneumophila, S. Typhimurium, and S. aureus in environmental waters (Li et al., 2015). The authors could detect S. Typhimurium and S. aureus with 3 CFU per reaction in their multiplex PMA-qPCR assay and further applied the assay to detect the multiple pathogens from rivers, canals, and tap water samples after simple water pretreatment. In the study, these authors optimized PMA treatment conditions as following: 30 μM PMA with 20 min of dark incubation and 10 min of light exposure. Scientists have also applied PMA approach to LAMP to detect viable cells of Salmonella in food. Chen et al. (2013) found PMA-LAMP was able detect viable cells in food with the detection limits comparable to that of PMA PMA-qPCR.
Besides PMA, EMA has been applied in viable cell detection of Salmonella in food. Wu et al. (2015) combined the EMA with LAMP to detect viable cells of Salmonella. They found that the concentration EMA is critical in effecting the DNA amplification of viable cells, i.e., if the EMA concentration used was <8.0 μg/ml, the DNA amplification of viable cell was not affected, otherwise, it would be significantly affected (Wu et al., 2015).
EMA/PMA in Differentiation of Viable Cells of Campylobacter in Food
Campylobacteriosis remains one of the most commonly reported bacterial foodborne disease in humans worldwide (Aase et al., 2000; Banihashemi et al., 2012). The incidence of campylobacteriosis has risen, with more than 200,000 confirmed cases in the European Union reported each year (Anonymous, 1996; D’Agostino et al., 2004). Campylobacter infections are clinically manifested by diarrhea, fever, and abdominal cramps, and, in certain cases, may be followed by long-term sequelae such as Guillain–Barré syndrome or reactive arthritis (Anonymous, 1996). Rapid pathogen detection is imperative for food manufacturers, public health agencies and clinicians alike. The sensitivity and specificity of detection of Campylobacter by PCR have been validated in the field. However, only viable Campylobacter cells can cause diseases and PCR or qPCR are not able to differentiate dead and viable cells.
To address this shortcoming of PCR, EMA/PMA has been combined with qPCR to detect viable cells of Campylobacter. Josefsen et al. (2010) developed a PMA-qPCR assay to detect the three major foodborne Campylobacter species (C. jejuni, Campylobacter coli, and Campylobacter lari) and found the PMA-qPCR quantification compared favorably with direct culture-based detection of Campylobacter in their study. The limit of detection of PMA-qPCR reached 102 CFU/g in the presence of dead cells. The specificity of the PMA-qPCR method was 100%, and it was shown to be more sensitive compared to the culture-based method. Eight chicken samples in that study were found to be Campylobacter positive by PMA-qPCR but not by culture (Josefsen et al., 2010). These PMA-QPCR results can be regarded as true positives due to the target-specific DNA probe-based PCR response according to ISO 20838 (Anonymous, 1996).
Traditional culture-based detection of Campylobacter bacteria, including enrichment, isolation, and confirmation, is a time-consuming procedure. Furthermore, bacterial cells may enter a VBNC state in which they may have the potential to cause human infection but are not detected by the culture method (Rollins and Colwell, 1986). Josefsen et al. (2010) developed a PMA-qPCR method to detect the infectious potential of the VBNC state. They found that the PMA-qPCR method was effective in assessing the risk of Campylobacter contamination including the infectious potential of the VBNC state cells in chicken carcass rinse. Banihashemi et al. (2012) also used PMA treatment and conventional PCR to tackle the issue of VBNC Campylobacter cells in food samples. Long amplicons were used in the PMA-PCR. The authors found when the length amplicon was <200 bp, the signal from DNA of the dead cells was not completely excluded, whereas, the length amplicon was >1.5k bp, the signal from DNA of the dead cells was completely excluded in the PMA-PCR assay (Banihashemi et al., 2012). Furthermore, the authors found that the signal of the DNA of the dead cells caused by UV-irradiation cannot be not excluded by PMA treatment (Banihashemi et al., 2012).
However, Seinige et al. (2014) gave a quite different view to the one that Flekna et al. (2007a) gave. Seinige et al. (2014) believed that EMA-qPCR was a suitable method for detection of viable Campylobacter from water samples, but the isolation technique and the type/quality of the water sample may impact the results. In general, most researchers preferred using PMA-qPCR than EMA-qPCR to detect viable cells Campylobacter.
EMA/PMA in Differentiation of Viable Cells of Vibrio parahaemolyticus in Food
Vibrio parahaemolyticus, a major foodborne pathogen known to cause gastroenteric infections, is often isolated from seawater, sediment, and a variety of seafood including oyster, clam, scallop, octopus, shrimp, crab, lobster, crawfish (Shen et al., 2009; Letchumanan et al., 2014). V. parahaemolyticus can cause diarrhea, vomiting, abdominal cramps, and, in rare cases, fever (Lin et al., 2005). Conventional culture-based techniques are laborious and time consuming. qPCR is rapid and sensitive, but its inability to discriminate between live and dead cells limits its applications. Therefore, EMA/PMA was used to combine qPCR assay to detect viable cells of V. parahaemolyticus. But so far only limited reports have been available reporting detection of viable cells of V. parahaemolyticus using EMA/PMA approach (Zhu et al., 2012; Fakruddin et al., 2013; Zhang et al., 2015a). Zhu et al. (2012) used PMA-qPCR to detect viable cells of V. parahaemolyticus from seafood. In comparison to culture-based methods, PMA-qPCR demonstrated advantage in detection of viable cells of V. parahaemolyticus. The authors used V. parahaemolyticus strains of different serotypes and 120 seafood samples to evaluate the sensitivity and specificity of the PMA-qPCR assay. They found that the sensitivity of the PMA-qPCR was 12 V. parahaemolyticus CFU per reaction for seafood samples, and the amount of DNA of pure culture samples was equivalent to 1.2 CFU per reaction. In addition, they found that 8 μM of PMA was the optimal concentration for PMA treatment of V. parahaemolyticus samples and PMA treatment became incomplete if the turbidity of the bacterial culture was over 10 Nephelometric Turbidity Unit (NTU) or OD600 nm greater than 0.8. Although the authors believed the PMA-qPCR was an effective tool for producing reliable quantitative data on viable V. parahaemolyticus in raw seafood (Zhu et al., 2012), there is more work needed to be done before the PMA-qPCR method can be widely used in detection of viable cells of V. parahaemolyticus in food.
EMA/PMA in Differentiation of Viable Cells of Staphylococcus aureus in Food
Staphylococcus aureus, a spherical and Gram-positive bacterium, is a major cause of skin, soft tissue, respiratory, bone, joint, and endovascular disorders (Lowy, 1998). It has also been recognized as a pathogen that causes outbreaks of food poisoning (Zhang et al., 2015b). S. aureus can contaminate a variety of foods such as salad, cheese, milk, fish, and meat (Alarcon et al., 2006; Vazquez-Sanchez et al., 2014). Routine detection of S. aureus in food is usually carried out by traditional methods based on the use of selective media (e.g., Baird–Parker agar) for direct enumeration or the recovery of isolates after enrichment in selective broth for 24–48 h at 37°C. Subsequently, the suspected colonies that are positive for DNase, and then coagulase production should be tested. This conventional method takes from 5 to 6 days and has low sensitivity and specificity (Alarcon et al., 2006). Hence, conventional method may underestimate the level of contamination (Zhang et al., 2015b).
In the last two decades, numerous PCR-based methods have been developed for the detection of foodborne pathogens to replace the time-consuming culture-based classical techniques (Johnson et al., 1991; Wilson et al., 1991; Hein et al., 2001, 2005; Verhoeven et al., 2012). The inability of PCR to differentiate between viable and dead cells is one of its major limitations (Nocker and Camper, 2006; Wang and Levin, 2006; Liu et al., 2012). To remedy this drawback of PCR, Kobayashi et al. (2009) combined the PMA treatment with qPCR to detect viable cells of S. aureus. They found that the PMA-qPCR assay inhibited the amplification of DNA from dead bacterial cells, and the qPCR results reflected the number of viable bacteria without being impacted by the presence of the dead bacteria. This approach of combining qPCR with PMA treatment has promise to limit false-positive PCR results when used to diagnose infections, but needs to be further validated in clinical samples (Kobayashi et al., 2009). Later on, Martinon et al. (2012a,b) also applied the PMA-qPCR approach to detect viable cells of S. aureus and other pathogens to assess the hygienic status of food contact surfaces within a commercial frozen meal factory. By comparison of plate counts, qPCR, PMA-qPCR, and Reagent D-qPCR, they found that the results from PMA-qPCR were slightly higher than those derived from plate counts. The authors believed that the PMA-qPCR results may reflect the real bacterial number in light of the presence of VBNC among bacterial populations (Martinon et al., 2012a). Zhang et al. (2015b) combined PMA with qPCR for selective detection of viable S. aureus in milk power and meat products and found that the PMA-qPCR assay was more specific and sensitive than conventional PCR, and the limit of detection was 3.0 × 102 CFU/g in spiked milk powder. The data indicated that the PMA treatment effectively eliminated the DNA amplification signals from dead cells but had little effect on viable cells (Zhang et al., 2015b).
EMA/PMA in Differentiation of Viable Cells of Listeria monocytogenes in Food
Listeria monocytogenes is one the most habitually investigated foodborne pathogens, whereas the L. monocytogenes outbreaks had the highest proportion of hospitalized cases as well as the highest proportion of deaths registered in the European Union (Anonymous, 2011; Law et al., 2015a). Flekna et al. (2007b) combined EMA with qPCR to detect viable cells of C. jejuni and L. monocytogenes. The authors tried to use different concentrations (1–100 μg/ml) EMA to treat viable and dead cells and concluded that EMA influences not only dead but also viable cells of C. jejuni and L. monocytogenes. Thus, EMA/real-time PCR is a poor indicator of cell viability (Flekna et al., 2007b). Scientists have compared EMA-qPCR with PMA-qPCR in detection of viable cells of L. monocytogenes (Pan and Breidt, 2007). In order to thoroughly evaluate the two dyes, the authors compared the influence of the environmental factors such as temperature. They found that the effect of EMA on viable cells correlated with the temperature used to treat the cells, whereas PMA did not show any effect on viable cells in regard to the temperature changes in the treatment. Furthermore, the authors found that the PMA-qPCR could be used for quantification of viable cells of L. monocytogenes in suspensions in which the ratio of dead cells to viable cells was no more than 104 and the concentration of live cells was no less than 103 CFU/ml. Compared with EMA, PMA was not found to penetrate live cells, as determined by the toxicity of the two dyes (Pan and Breidt, 2007). However, when other researchers applied the PMA-qPCR in detection of viable cells from food matrix, they had slightly different opinions on this assay. For example, Elizaquivel et al. (2012) developed a multiplex PMA-qPCR to detect viable cells of E. coli O157:H7, L. monocytogenes, and Salmonella from fresh-cut vegetables. The authors noted that when salad with high concentration of L. monocytogenes, PMA treatment cannot completely exclude the influence of the dead cells. Nevertheless, their data demonstrate that PMA-qPCR is a suitable technique for the detection and quantification of viable pathogens in fresh-cut vegetables at the levels normally found in vegetable samples (Elizaquivel et al., 2012).
Yang J. et al. (2013) have combined PMA with conventional PCR to detect viable cells of L. monocytogenes. The authors developed a multiplex PCR to simultaneously detect viable cells of S. Typhimurium, E. coli O157:H7, and L. monocytogenes in food products. In order to improve the sensitivity of the assay, magnetic nanobeads-based immunomagnetic separation was used to concentrate the target bacterial cells. Consequently, their results showed the detection limit of 8.4 × 103 CFU/g for L. monocytogenes in spiked food products (lettuce, tomato, and ground beef) (Yang J. et al., 2013). In a study comparing EMA with PMA in live-dead cell samples of four Gram-negative and four Gram-positive bacterial species, Nocker and Camper (2006) found PMA more impermeable to viable cells.
Which Dye Works Better for Viability Detection of Foodborne Pathogens?
Cawthorn and Witthuhn (2008) documented PMA more membrane-impermeant compared with EMA in a study for selective detection of viable Enterobacter sakazakii cells. It was concluded that EMA was less effective than PMA in selective amplification of DNA from viable cells and PMA was a useful alternative (van Frankenhuyzen et al., 2011). Besides viable cells of bacterial pathogens (Nocker and Camper, 2006, 2009; Nocker et al., 2007a; Pan and Breidt, 2007; Luo et al., 2010; Li and Chen, 2012, 2013), the PMA-qPCR has been applied to detect viable cells of fungi (Vesper et al., 2008), parasites (Brescia et al., 2009; Cancino-Faure et al., 2016), and viruses (Fittipaldi et al., 2010; Parshionikar et al., 2010; Kim and Ko, 2012; Sanchez et al., 2012; Coudray-Meunier et al., 2013; Fuster et al., 2016).
Limitations of the PMA Approach and the Remedies to Address the Issues
Like any other technologies or assays, PMA-qPCR has its limitations in detection of viable cells of foodborne pathogens in foods. PMA-PCR was first reported to effectively exclude the signal of dead bacteria (Nocker et al., 2006; Josefsen et al., 2010). Later, this approach was adapted by many scientists to detect viable cells of various foodborne pathogens (Cawthorn and Witthuhn, 2008; Josefsen et al., 2010; Liang et al., 2011; van Frankenhuyzen et al., 2011; Banihashemi et al., 2012; Li and Chen, 2012, 2013; Mamlouk et al., 2012; Soejima et al., 2012; Dinu and Bach, 2013; Singh et al., 2013; Yang Y. et al., 2013; Ditommaso et al., 2015; Li et al., 2015; Santiago et al., 2015; Cattani et al., 2016). The majority of the studies demonstrated that PMA-PCR effectively suppressed the signal of DNA from the dead cells. On the other hand, it was also found that PMA treatment does not always lead to complete removal of the qPCR signal of dead bacteria. Fittipaldi et al. (2010) summarized that incomplete suppression of the signal from dead will occur if (i) the amplicon size of the qPCR assay is short (Luo et al., 2010; Li and Chen, 2012; Schnetzinger et al., 2013); (ii) the target bacteria is at high concentration (Elizaquivel et al., 2012; Zhu et al., 2012; Li and Chen, 2013; Pacholewicz et al., 2013); (iii) the concentration of Mg2+ in the PCR reaction is not adapted (Nocker et al., 2006); or (iv) the fat content of food sample is high (Yang et al., 2011), and may also vary according to the “killing” treatment (Nocker et al., 2007a; Kobayashi et al., 2010; Yang et al., 2011; Liang and Keeley, 2012). Additionally, the turbidity of food samples may hamper light penetration and samples dilution is required for a thorough light exposure. Such dilution practically restricts the capacity of sample preparation and consequentially, it has to resort to extrapolation method to analyze the results, making it less accurate (van Frankenhuyzen et al., 2011).
With the downsides in using the PMA approach being recognized by scientists, the remedies have been proposed to address these issues. For example, “activity-labile compounds” as a possible alternative for PMA treatment was suggested (Nocker and Camper, 2009). Soejima et al. (2016) recommended to use platinum compounds. Platinum metals can be chelated by nucleic acid ligands in mammalian cells (Rosenberg et al., 1965, 1969; Lovejoy et al., 2008; Serrano et al., 2011). Platinum compounds do not depend on visible light to function and they are inexpensive. Using Pt compounds in viable detection can avoid the laborious procedures in PMA treatment (Cimino et al., 1991; Rudi et al., 2005; Nocker et al., 2006). More recently, Soejima et al. (2016) compared five platinum compounds with PMA in viable cell detection and indicated that this platinum-PCR method completely suppressed the signal of dead cells and enabled the specific detection of viable coliforms in milk at a concentration of 5–10 CFU/ml specified by EU/USA regulations after a 4-h process.
Conclusion and Perspective
Obviously, culture-based methods cannot match the challenges that the foodborne pathogens pose to the food safety and public health. There is a great demand for rapid, sensitive, specific, and accurate methodologies for pathogen detection in foods. Molecular assays such as PCR and LAMP methods have been demonstrated huge advantages in sensitivity, specificity, and speed. However, a major drawback of these assays is that their inability to differentiate viable and dead cells may overestimate risk of contamination of foodborne pathogens. To circumvent this shortcoming, recently, scientists have combined EMA/PMA to PCR or LAMP for accurate detection of viable cells of foodborne pathogens in foods. Numerous studies have used this approach for detection of viable cells of various foodborne pathogens, including E. coli O157:H7, Salmonella, S. aureus, V. parahaemolyticus, L. monocytogenes, and Campylobacter, and achieved various degree of success in surpassing the signals of DNA of the dead cells in the detection assay. In general, this PMA approach is the most practical means for detection of viable cells of foodborne pathogens.
As pathogen detection method, PCR, qPCR, or LAMP method each shows its advantages and shortcomings. PMA-qPCR is the most commonly used technique in foodborne detection because it is rapid, specific, sensitive, and quantitative. Although the sensitivity of PMA-LAMP is slightly lower than that of PMA-qPCR, it is simpler, more economic, and particularly suitable for the need of pathogen detection of resource-limited institutions.
PMA is more preferably used dye in viable cells detection of foodborne pathogens compared with EMA, which has been shown not only penetrates compromised membranes of dead cells but also penetrate membranes of viable cells, leading false negative results by numerous studies. While PMA-qPCR assays have been successfully applied to various foodborne pathogens, the drawbacks of this approach have been noted by researchers. For instance, the removal efficiency of DNA of dead cells is incomplete when small-sized amplicons (<130 bp) and/or high concentration of dead cells are used (Luo et al., 2010; Li and Chen, 2013); and food matrices used may influence the removal efficiency of DNA of dead cells (van Frankenhuyzen et al., 2011). To circumvent the shortcomings of this approach, several strategies can be taken in designing the PMA-qPCR assays: (i) a sensitive, specific and robust qPCR assay is the prerequisite for the development of a sound PMA-qPCR assay (Li and Chen, 2012); (ii) if the target sequence permits, select relatively large amplicon for the PMA-qPCR assay (>130 bp; Li and Chen, 2013); (iii) optimize the PMA treatment conditions such as the concentration of PMA and duration of PMA treatment and light exposure based on different organisms (Gram-positive and Gram-negative; Li and Chen, 2013); and (iv) enhance the PMA’s penetration by adding sodium lauroyl sarcosinate to the PMA solution (Wang et al., 2014; Li et al., 2015). Additionally, selecting alternative chemicals or compounds for PMA or EMA, such as “activity-labile compounds” (Nocker and Camper, 2009) or platinum compounds (Soejima et al., 2016) could be a promising strategy to further improve this viability technology for specific detection of viable pathogens in foods.
Author Contributions
BL conceived and contributed largely for the review, and DZ and FX contributed in writing, while all other authors contributed in organizing.
Funding
The work was done with the support from U.S. FDA and by the National Natural Science Foundation of China (31301460), the National “Youth Top-notch Talent” Support Program, the National Science and Technology Support Program of 2012BAK17B10, Jiangsu Collaborative Innovation Center of Meat Production and Processing, Quality and Safety Control and Jiangsu Province Science and Technology Support Program of BE20137334.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Aase, B., Sundheim, G., Langsrud, S., and Rorvik, L. M. (2000). Occurrence of and a possible mechanism for resistance to a quaternary ammonium compound in Listeria monocytogenes. Int. J. Food Microbiol. 62, 57–63. doi: 10.1016/S0168-1605(00)00357-3
Agusti, G., Codony, F., Fittipaldi, M., Adrados, B., and Morato, J. (2010). Viability determination of Helicobacter pylori using propidium monoazide quantitative PCR. Helicobacter 15, 473–476. doi: 10.1111/j.1523-5378.2010.00794.x
Alarcon, B., Vicedo, B., and Aznar, R. (2006). PCR-based procedures for detection and quantification of Staphylococcus aureus and their application in food. J. Appl. Microbiol. 100, 352–364. doi: 10.1111/j.1365-2672.2005.02768.x
Alonso, J. L., Amoros, I., and Guy, R. A. (2014). Quantification of viable Giardia cysts and Cryptosporidium oocysts in wastewater using propidium monoazide quantitative real-time PCR. Parasitol. Res. 113, 2671–2678. doi: 10.1007/s00436-014-3922-9
Andorra, I., Esteve-Zarzoso, B., Guillamon, J. M., and Mas, A. (2010). Determination of viable wine yeast using DNA binding dyes and quantitative PCR. Int. J. Food Microbiol. 144, 257–262. doi: 10.1016/j.ijfoodmicro.2010.10.003
Anonymous (1996). ISO 11290-1:Microbiology of food and Animal Feeding Stuffs – Horizontal Method for the Detection and Enumeration of Listeria monocytogenes – Part 1: Detection method. Geneva: ISO.
Anonymous (2011). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2009. EFSA J. 9:2090. doi: 10.2903/j.efsa.2011.2090
Banihashemi, A., Van Dyke, M. I., and Huck, P. M. (2012). Long-amplicon propidium monoazide-PCR enumeration assay to detect viable Campylobacter and Salmonella. J. Appl. Microbiol. 113, 863–873. doi: 10.1111/j.1365-2672.2012.05382.x
Barbau-Piednoir, E., Mahillon, J., Pillyser, J., Coucke, W., Roosens, N. H., and Botteldoorn, N. (2014). Evaluation of viability-qPCR detection system on viable and dead Salmonella serovar Enteritidis. J. Microbiol. Methods 103, 131–137. doi: 10.1016/j.mimet.2014.06.003
Brescia, C. C., Griffin, S. M., Ware, M. W., Varughese, E. A., Egorov, A. I., and Villegas, E. N. (2009). Cryptosporidium propidium monoazide-PCR, a molecular biology-based technique for genotyping of viable Cryptosporidium oocysts. Appl. Environ. Microbiol. 75, 6856–6863. doi: 10.1128/AEM.00540-09
Caldas, S., Caldas, I. S., Cecilio, A. B., Diniz, L. D., Talvani, A., Ribeiro, I., et al. (2014). Therapeutic responses to different anti-Trypanosoma cruzi drugs in experimental infection by benznidazole-resistant parasite stock. Parasitology 141, 1628–1637. doi: 10.1017/S0031182014000882
Cancino-Faure, B., Fisa, R., Alcover, M. M., Jimenez-Marco, T., and Riera, C. (2016). Detection and quantification of viable and nonviable trypanosoma cruzi parasites by a propidium monoazide real-time polymerase chain reaction assay. Am. J. Trop. Med. Hyg. 94, 1282–1289. doi: 10.4269/ajtmh.15-0693
Caron, G. N., Stephens, P., and Badley, R. A. (1998). Assessment of bacterial viability status by flow cytometry and single cell sorting. J. Appl. Microbiol. 84, 988–998. doi: 10.1046/j.1365-2672.1998.00436.x
Cattani, F., Barth, V. C. Jr., Nasario, J. S., Ferreira, C. A., and Oliveira, S. D. (2016). Detection and quantification of viable Bacillus cereus group species in milk by propidium monoazide quantitative real-time PCR. J. Dairy Sci. 99, 2617–2624. doi: 10.3168/jds.2015-10019
Cawthorn, D. M., and Witthuhn, R. C. (2008). Selective PCR detection of viable Enterobacter sakazakii cells utilizing propidium monoazide or ethidium bromide monoazide. J. Appl. Microbiol. 105, 1178–1185. doi: 10.1111/j.1365-2672.2008.03851.x
Chen, S., Li, X., Li, J., and Atwill, E. R. (2013). Rapid detection of Brucella spp. using loop-mediated isothermal amplification (LAMP). Methods Mol. Biol. 1039, 99–108. doi: 10.1007/978-1-62703-535-4_8
Chen, S., Wang, F., Beaulieu, J. C., Stein, R. E., and Ge, B. (2011). Rapid detection of viable salmonellae in produce by coupling propidium monoazide with loop-mediated isothermal amplification. Appl. Environ. Microbiol. 77, 4008–4016. doi: 10.1128/AEM.00354-11
Cimino, G. D., Metchette, K. C., Tessman, J. W., Hearst, J. E., and Isaacs, S. T. (1991). Post-PCR sterilization: a method to control carryover contamination for the polymerase chain reaction. Nucleic Acids Res. 19, 99–107. doi: 10.1093/nar/19.1.99
Coudray-Meunier, C., Fraisse, A., Martin-Latil, S., Guillier, L., and Perelle, S. (2013). Discrimination of infectious hepatitis A virus and rotavirus by combining dyes and surfactants with RT-qPCR. BMC Microbiol. 13:216. doi: 10.1186/1471-2180-13-216
Da Silva Felicio, M. T., Hald, T., Liebana, E., Allende, A., Hugas, M., Nguyen-The, C., et al. (2015). Risk ranking of pathogens in ready-to-eat unprocessed foods of non-animal origin (FoNAO) in the EU: initial evaluation using outbreak data (2007-2011). Int. J. Food Microbiol. 195, 9–19. doi: 10.1016/j.ijfoodmicro.2014.11.005
D’Agostino, M., Wagner, M., Vazquez-Boland, J. A., Kuchta, T., Karpiskova, R., Hoorfar, J., et al. (2004). A validated PCR-based method to detect Listeria monocytogenes using raw milk as a food model–towards an international standard. J. Food Prot. 67, 1646–1655.
Diaper, J. P., and Edwards, C. (1994). Survival of Staphylococcus aureus in lakewater monitored by flow cytometry. Microbiology 140(Pt 1), 35–42. doi: 10.1099/13500872-140-1-35
Dinu, L. D., and Bach, S. (2011). Induction of viable but nonculturable Escherichia coli O157:H7 in the phyllosphere of lettuce: a food safety risk factor. Appl. Environ. Microbiol. 77, 8295–8302. doi: 10.1128/AEM.05020-11
Dinu, L.-D., and Bach, S. (2013). Detection of viable but non-culturable Escherichia coli O157:H7 from vegetable samples using quantitative PCR with propidium monoazide and immunological assays. Food Control 31, 268–273. doi: 10.1016/j.foodcont.2012.10.020
Ditommaso, S., Giacomuzzi, M., Ricciardi, E., and Zotti, C. M. (2015). Viability-qPCR for detecting legionella: comparison of two assays based on different amplicon lengths. Mol. Cell. Probes 29, 237–243. doi: 10.1016/j.mcp.2015.05.011
Elizaquivel, P., Sanchez, G., and Aznar, R. (2012). Application of propidium monoazide quantitative PCR for selective detection of live Escherichia coli O157:H7 in vegetables after inactivation by essential oils. Int. J. Food Microbiol. 159, 115–121. doi: 10.1016/j.ijfoodmicro.2012.08.006
Fakruddin, M. D., Sultana, M., Ahmed, M. M., Chowdhury, A., and Choudhury, N. (2013). Multiplex PCR (polymerase chain reaction) assay for detection of E. coli O157:H7, Salmonella sp., Vibrio cholerae and Vibrio parahaemolyticus in spiked shrimps (Penaeus monodon). Pak. J. Biol. Sci. 16, 267–274. doi: 10.3923/pjbs.2013.267.274
Fittipaldi, M., Codony, F., Adrados, B., Camper, A. K., and Morato, J. (2011). Viable real-time PCR in environmental samples: can all data be interpreted directly? Microb. Ecol. 61, 7–12. doi: 10.1007/s00248-010-9719-1
Fittipaldi, M., Rodriguez, N. J., Codony, F., Adrados, B., Penuela, G. A., and Morato, J. (2010). Discrimination of infectious bacteriophage T4 virus by propidium monoazide real-time PCR. J. Virol. Methods 168, 228–232. doi: 10.1016/j.jviromet.2010.06.011
Flekna, G., Schneeweiss, W., Smulders, F. J., Wagner, M., and Hein, I. (2007a). Real-time PCR method with statistical analysis to compare the potential of DNA isolation methods to remove PCR inhibitors from samples for diagnostic PCR. Mol. Cell. Probes 21, 282–287. doi: 10.1016/j.mcp.2007.02.001
Flekna, G., Stefanic, P., Wagner, M., Smulders, F. J., Mozina, S. S., and Hein, I. (2007b). Insufficient differentiation of live and dead Campylobacter jejuni and Listeria monocytogenes cells by ethidium monoazide (EMA) compromises EMA/real-time PCR. Res. Microbiol. 158, 405–412. doi: 10.1016/j.resmic.2007.02.008
Fuster, N., Pinto, R. M., Fuentes, C., Beguiristain, N., Bosch, A., and Guix, S. (2016). Propidium monoazide RTqPCR assays for the assessment of hepatitis A inactivation and for a better estimation of the health risk of contaminated waters. Water Res. 101, 226–232. doi: 10.1016/j.watres.2016.05.086
Hein, I., Jorgensen, H. J., Loncarevic, S., and Wagner, M. (2005). Quantification of Staphylococcus aureus in unpasteurised bovine and caprine milk by real-time PCR. Res. Microbiol. 156, 554–563. doi: 10.1016/j.resmic.2005.01.003
Hein, I., Lehner, A., Rieck, P., Klein, K., Brandl, E., and Wagner, M. (2001). Comparison of different approaches to quantify Staphylococcus aureus cells by real-time quantitative PCR and application of this technique for examination of cheese. Appl. Environ. Microbiol. 67, 3122–3126. doi: 10.1128/AEM.67.7.3122-3126.2001
Johnson, W. M., Tyler, S. D., Ewan, E. P., Ashton, F. E., Pollard, D. R., and Rozee, K. R. (1991). Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin 1 in Staphylococcus aureus by the polymerase chain reaction. J. Clin. Microbiol. 29, 426–430.
Josefsen, M. H., Lofstrom, C., Hansen, T. B., Christensen, L. S., Olsen, J. E., and Hoorfar, J. (2010). Rapid quantification of viable Campylobacter bacteria on chicken carcasses, using real-time PCR and propidium monoazide treatment, as a tool for quantitative risk assessment. Appl. Environ. Microbiol. 76, 5097–5104. doi: 10.1128/AEM.00411-10
Ju, W., Moyne, A. L., and Marco, M. L. (2016). RNA-Based detection does not accurately enumerate living Escherichia coli O157:H7 cells on plants. Front. Microbiol. 7:223. doi: 10.3389/fmicb.2016.00223
Karmali, M. A. (2004). Infection by Shiga toxin-producing Escherichia coli: an overview. Mol. Biotechnol. 26, 117–122. doi: 10.1385/MB:26:2:117
Keer, J. T., and Birch, L. (2003). Molecular methods for the assessment of bacterial viability. J. Microbiol. Methods 53, 175–183. doi: 10.1016/S0167-7012(03)00025-3
Kim, S. Y., and Ko, G. (2012). Using propidium monoazide to distinguish between viable and nonviable bacteria, MS2 and murine norovirus. Lett. Appl. Microbiol. 55, 182–188. doi: 10.1111/j.1472-765X.2012.03276.x
Kobayashi, H., Oethinger, M., Tuohy, M. J., Hall, G. S., and Bauer, T. W. (2009). Improving clinical significance of PCR: use of propidium monoazide to distinguish viable from dead Staphylococcus aureus and Staphylococcus epidermidis. J. Orthop. Res. 27, 1243–1247. doi: 10.1002/jor.20872
Kobayashi, H., Oethinger, M., Tuohy, M. J., Hall, G. S., and Bauer, T. W. (2010). Distinction between intact and antibiotic-inactivated bacteria by real-time PCR after treatment with propidium monoazide. J. Orthop. Res. 28, 1245–1251. doi: 10.1002/jor.21108
Kokkinos, P. A., Ziros, P. G., Bellou, M., and Vantarakis, A. (2014). Loop-mediated isothermal amplification (LAMP) for the detection of salmonella in food. Food Anal. Methods 7, 512–526. doi: 10.1007/s12161-013-9748-8
Law, J. W.-F., Ab Mutalib, N.-S., Chan, K.-G., and Lee, L.-H. (2015a). An insight into the isolation, enumeration, and molecular detection of Listeria monocytogenes in food. Front. Microbiol. 6:1227. doi: 10.3389/fmicb.2015.01227
Law, J. W.-F., Ab Mutalib, N.-S., Chan, K.-G., and Lee, L.-H. (2015b). Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front. Microbiol. 5:770. doi: 10.3389/fmicb.2014.00770
Letchumanan, V., Chan, K.-G., and Lee, L.-H. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 5:705. doi: 10.3389/fmicb.2014.00705
Li, B., and Chen, J. Q. (2012). Real-time PCR methodology for selective detection of viable Escherichia coli O157:H7 cells by targeting Z3276 as a genetic marker. Appl. Environ. Microbiol. 78, 5297–5304. doi: 10.1128/AEM.00794-12
Li, B., and Chen, J. Q. (2013). Development of a sensitive and specific qPCR assay in conjunction with propidium monoazide for enhanced detection of live Salmonella spp. in food. BMC Microbiol. 13:273. doi: 10.1186/1471-2180-13-273
Li, H., Xin, H., and Li, S. F. (2015). Multiplex PMA-qPCR assay with internal amplification control for simultaneous detection of viable Legionella pneumophila, Salmonella typhimurium, and Staphylococcus aureus in environmental waters. Environ. Sci. Technol. 49, 14249–14256. doi: 10.1021/acs.est.5b03583
Liang, N., Dong, J., Luo, L., and Li, Y. (2011). Detection of viable Salmonella in lettuce by propidium monoazide real-time PCR. J. Food Sci. 76, M234–M237. doi: 10.1111/j.1750-3841.2011.02123.x
Liang, Z., and Keeley, A. (2012). Comparison of propidium monoazide-quantitative PCR and reverse transcription quantitative PCR for viability detection of fresh Cryptosporidium oocysts following disinfection and after long-term storage in water samples. Water Res. 46, 5941–5953. doi: 10.1016/j.watres.2012.08.014
Lin, Y. T., Kwon, Y. I., Labbe, R. G., and Shetty, K. (2005). Inhibition of Helicobacter pylori and associated urease by oregano and cranberry phytochemical synergies. Appl. Environ. Microbiol. 71, 8558–8564. doi: 10.1128/AEM.71.12.8558-8564.2005
Liu, H., Niu, Y. D., Li, J., Stanford, K., and Mcallister, T. A. (2014). Rapid and accurate detection of bacteriophage activity against Escherichia coli O157:H7 by propidium monoazide real-time PCR. Biomed Res. Int. 2014:319351.
Liu, Y., and Mustapha, A. (2014). Detection of viable Escherichia coli O157:H7 in ground beef by propidium monoazide real-time PCR. Int. J. Food Microbiol. 170, 48–54. doi: 10.1016/j.ijfoodmicro.2013.10.026
Liu, Y.-H., Wang, C.-H., Wu, J.-J., and Lee, G.-B. (2012). Rapid detection of live methicillin-resistant Staphylococcus aureus by using an integrated microfluidic system capable of ethidium monoazide pre-treatment and molecular diagnosis. Biomicrofluidics 6:34119. doi: 10.1063/1.4748358
Lovejoy, K. S., Todd, R. C., Zhang, S., Mccormick, M. S., D’aquino, J. A., Reardon, J. T., et al. (2008). cis-Diammine(pyridine)chloroplatinum(II), a monofunctional platinum(II) antitumor agent: uptake, structure, function, and prospects. Proc. Natl. Acad. Sci. U.S.A. 105, 8902–8907. doi: 10.1073/pnas.0803441105
Lowder, M., Unge, A., Maraha, N., Jansson, J. K., Swiggett, J., and Oliver, J. D. (2000). Effect of starvation and the viable-but-nonculturable state on green fluorescent protein (GFP) fluorescence in GFP-tagged Pseudomonas fluorescens A506. Appl. Environ. Microbiol. 66, 3160–3165. doi: 10.1128/AEM.66.8.3160-3165.2000
Lowy, F. D. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532. doi: 10.1056/NEJM199808203390806
Luo, J.-F., Lin, W.-T., and Guo, Y. (2010). Method to detect only viable cells in microbial ecology. Appl. Microbiol. Biotechnol. 86, 377–384. doi: 10.1007/s00253-009-2373-1
Mamlouk, K., Mace, S., Guilbaud, M., Jaffres, E., Ferchichi, M., Prevost, H., et al. (2012). Quantification of viable Brochothrix thermosphacta in cooked shrimp and salmon by real-time PCR. Food Microbiol. 30, 173–179. doi: 10.1016/j.fm.2011.09.012
Martinon, A., Cronin, U. P., Quealy, J., Stapleton, A., and Wilkinson, M. G. (2012a). Swab sample preparation and viable real-time PCR methodologies for the recovery of Escherichia coli, Staphylococcus aureus or Listeria monocytogenes from artificially contaminated food processing surfaces. Food Control 24, 86–94. doi: 10.1016/j.foodcont.2011.09.007
Martinon, A., Cronin, U. P., and Wilkinson, M. G. (2012b). Development of defined microbial population standards using fluorescence activated cell sorting for the absolute quantification of S. aureus using real-time PCR. Mol. Biotechnol. 50, 62–71. doi: 10.1007/s12033-011-9417-3
Miotto, P., Bigoni, S., Migliori, G. B., Matteelli, A., and Cirillo, D. M. (2012). Early tuberculosis treatment monitoring by Xpert(R) MTB/RIF. Eur. Respir. J. 39, 1269–1271. doi: 10.1183/09031936.00124711
Nocker, A., and Camper, A. K. (2006). Selective removal of DNA from dead cells of mixed bacterial communities by use of ethidium monoazide. Appl. Environ. Microbiol. 72, 1997–2004. doi: 10.1128/AEM.72.3.1997-2004.2006
Nocker, A., and Camper, A. K. (2009). Novel approaches toward preferential detection of viable cells using nucleic acid amplification techniques. FEMS Microbiol. Lett. 291, 137–142. doi: 10.1111/j.1574-6968.2008.01429.x
Nocker, A., Cheung, C. Y., and Camper, A. K. (2006). Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 67, 310–320. doi: 10.1016/j.mimet.2006.04.015
Nocker, A., Mazza, A., Masson, L., Camper, A. K., and Brousseau, R. (2009). Selective detection of live bacteria combining propidium monoazide sample treatment with microarray technology. J. Microbiol. Methods 76, 253–261. doi: 10.1016/j.mimet.2008.11.004
Nocker, A., Sossa-Fernandez, P., Burr, M. D., and Camper, A. K. (2007a). Use of propidium monoazide for live/dead distinction in microbial ecology. Appl. Environ. Microbiol. 73, 5111–5117. doi: 10.1128/AEM.02987-06
Nocker, A., Sossa, K. E., and Camper, A. K. (2007b). Molecular monitoring of disinfection efficacy using propidium monoazide in combination with quantitative PCR. J. Microbiol. Methods 70, 252–260. doi: 10.1016/j.mimet.2007.04.014
Oliveira-Silva, J. C., Machado-De-Assis, G. F., Oliveira, M. T., Paiva, N. C., Araujo, M. S., Carneiro, C. M., et al. (2015). Experimental benznidazole treatment of Trypanosoma cruzi II strains isolated from children of the jequitinhonha valley, minas gerais, Brazil, with Chagas disease. Mem. Inst. Oswaldo Cruz doi: 10.1590/0074-02760140260 [Epub ahead of print].
Pacholewicz, E., Swart, A., Lipman, L. J., Wagenaar, J. A., Havelaar, A. H., and Duim, B. (2013). Propidium monoazide does not fully inhibit the detection of dead Campylobacter on broiler chicken carcasses by qPCR. J. Microbiol. Methods 95, 32–38. doi: 10.1016/j.mimet.2013.06.003
Pan, Y., and Breidt, F. Jr. (2007). Enumeration of viable Listeria monocytogenes cells by real-time PCR with propidium monoazide and ethidium monoazide in the presence of dead cells. Appl. Environ. Microbiol. 73, 8028–8031. doi: 10.1128/AEM.01198-07
Parshionikar, S., Laseke, I., and Fout, G. S. (2010). Use of propidium monoazide in reverse transcriptase PCR to distinguish between infectious and noninfectious enteric viruses in water samples. Appl. Environ. Microbiol. 76, 4318–4326. doi: 10.1128/AEM.02800-09
Rogers, G. B., Stressmann, F. A., Koller, G., Daniels, T., Carroll, M. P., and Bruce, K. D. (2008). Assessing the diagnostic importance of nonviable bacterial cells in respiratory infections. Diagn. Microbiol. Infect. Dis. 62, 133–141. doi: 10.1016/j.diagmicrobio.2008.06.011
Rollins, D. M., and Colwell, R. R. (1986). Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. Appl. Environ. Microbiol. 52, 531–538.
Rosenberg, B., Vancamp, L., and Krigas, T. (1965). Inhibition of cell division in Escherichia coli by electrolysis products from a platinum electrode. Nature 205, 698–699. doi: 10.1038/205698a0
Rosenberg, B., Vancamp, L., Trosko, J. E., and Mansour, V. H. (1969). Platinum compounds: a new class of potent antitumour agents. Nature 222, 385–386. doi: 10.1038/222385a0
Rudi, K., Moen, B., Dromtorp, S. M., and Holck, A. L. (2005). Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl. Environ. Microbiol. 71, 1018–1024. doi: 10.1128/AEM.71.2.1018-1024.2005
Sanchez, G., Elizaquivel, P., and Aznar, R. (2012). Discrimination of infectious hepatitis A viruses by propidium monoazide real-time RT-PCR. Food Environ. Virol. 4, 21–25. doi: 10.1007/s12560-011-9074-5
Santiago, P., Moreno, Y., and Ferrus, M. A. (2015). Identification of viable Helicobacter pylori in drinking water supplies by cultural and molecular techniques. Helicobacter 20, 252–259. doi: 10.1111/hel.12205
Scallan, E., Griffin, P. M., Angulo, F. J., Tauxe, R. V., and Hoekstra, R. M. (2011). Foodborne illness acquired in the United States–unspecified agents. Emerg. Infect. Dis. 17, 16–22. doi: 10.3201/eid1707.110572
Scharff, R. L. (2012). Economic burden from health losses due to foodborne illness in the United States. J. Food Prot. 75, 123–131. doi: 10.4315/0362-028X.JFP-11-058
Schnetzinger, F., Pan, Y., and Nocker, A. (2013). Use of propidium monoazide and increased amplicon length reduce false-positive signals in quantitative PCR for bioburden analysis. Appl. Microbiol. Biotechnol. 97, 2153–2162. doi: 10.1007/s00253-013-4711-6
Seinige, D., Von Kockritz-Blickwede, M., Krischek, C., Klein, G., and Kehrenberg, C. (2014). Influencing factors and applicability of the viability EMA-qPCR for a detection and quantification of Campylobacter cells from water samples. PLoS ONE 9:e113812. doi: 10.1371/journal.pone.0113812
Serrano, F. A., Matsuo, A. L., Monteforte, P. T., Bechara, A., Smaili, S. S., Santana, D. P., et al. (2011). A cyclopalladated complex interacts with mitochondrial membrane thiol-groups and induces the apoptotic intrinsic pathway in murine and cisplatin-resistant human tumor cells. BMC Cancer 11:296. doi: 10.1186/1471-2407-11-296
Shen, X., Cai, Y., Liu, C., Liu, W., Hui, Y., and Su, Y. C. (2009). Effect of temperature on uptake and survival of Vibrio parahaemolyticus in oysters (Crassostrea plicatula). Int. J. Food Microbiol. 136, 129–132. doi: 10.1016/j.ijfoodmicro.2009.09.012
Sidhu, J. P., and Toze, S. G. (2009). Human pathogens and their indicators in biosolids: a literature review. Environ. Int. 35, 187–201. doi: 10.1016/j.envint.2008.07.006
Singh, G., Vajpayee, P., Bhatti, S., Ronnie, N., Shah, N., Mcclure, P., et al. (2013). Determination of viable Salmonellae from potable and source water through PMA assisted qPCR. Ecotoxicol. Environ. Saf. 93, 121–127. doi: 10.1016/j.ecoenv.2013.02.017
Soejima, T., Minami, J., and Iwatsuki, K. (2012). Rapid propidium monoazide PCR assay for the exclusive detection of viable Enterobacteriaceae cells in pasteurized milk. J. Dairy Sci. 95, 3634–3642. doi: 10.3168/jds.2012-5360
Soejima, T., Minami, J., Xiao, J. Z., and Abe, F. (2016). Innovative use of platinum compounds to selectively detect live microorganisms by polymerase chain reaction. Biotechnol. Bioeng. 113, 301–310. doi: 10.1002/bit.25711
Techathuvanan, C., and D’Souza, D. H. (2012). Reverse-transcriptase loop-mediated isothermal amplification as a rapid screening/monitoring tool for Salmonella enterica detection in liquid whole eggs. J. Food Sci. 77, M200–M205. doi: 10.1111/j.1750-3841.2011.02601.x
van Frankenhuyzen, J. K., Trevors, J. T., Lee, H., Flemming, C. A., and Habash, M. B. (2011). Molecular pathogen detection in biosolids with a focus on quantitative PCR using propidium monoazide for viable cell enumeration. J. Microbiol. Methods 87, 263–272. doi: 10.1016/j.mimet.2011.09.007
Vazquez-Sanchez, E. A., Rodriguez-Romero, M., Sanchez-Torres, L. E., Rodriguez-Martinez, S., Cancino-Diaz, J. C., Rodriguez-Cortes, O., et al. (2014). Peptidoglycan from Staphylococcus aureus has an anti-apoptotic effect in HaCaT keratinocytes mediated by the production of the cellular inhibitor of apoptosis protein-2. Microbiol. Immunol. 58, 87–95. doi: 10.1111/1348-0421.12126
Verhoeven, P. O., Grattard, F., Carricajo, A., Lucht, F., Cazorla, C., Garraud, O., et al. (2012). Quantification by real-time PCR assay of Staphylococcus aureus load: a useful tool for rapidly identifying persistent nasal carriers. J. Clin. Microbiol. 50, 2063–2065. doi: 10.1128/JCM.00157-12
Vesper, S., Mckinstry, C., Hartmann, C., Neace, M., Yoder, S., and Vesper, A. (2008). Quantifying fungal viability in air and water samples using quantitative PCR after treatment with propidium monoazide (PMA). J. Microbiol. Methods 72, 180–184. doi: 10.1016/j.mimet.2007.11.017
Wang, H., Gill, C. O., and Yang, X. (2014). Use of sodium lauroyl sarcosinate (sarkosyl) in viable real-time PCR for enumeration of Escherichia coli. J. Microbiol. Methods 98, 89–93. doi: 10.1016/j.mimet.2014.01.004
Wang, L., Li, Y., and Mustapha, A. (2009). Detection of viable Escherichia coli O157:H7 by ethidium monoazide real-time PCR. J. Appl. Microbiol. 107, 1719–1728. doi: 10.1111/j.1365-2672.2009.04358.x
Wang, S., and Levin, R. E. (2006). Discrimination of viable Vibrio vulnificus cells from dead cells in real-time PCR. J. Microbiol. Methods 64, 1–8. doi: 10.1016/j.mimet.2005.04.023
Wilson, I. G., Cooper, J. E., and Gilmour, A. (1991). Detection of enterotoxigenic Staphylococcus aureus in dried skimmed milk: use of the polymerase chain reaction for amplification and detection of staphylococcal enterotoxin genes entB and entC1 and the thermonuclease gene nuc. Appl. Environ. Microbiol. 57, 1793–1798.
Wu, G. P., Chen, S. H., and Levin, R. E. (2015). Application of ethidium bromide monoazide for quantification of viable and dead cells of Salmonella enterica by real-time loop-mediated isothermal amplification. J. Microbiol. Methods 117, 41–48. doi: 10.1016/j.mimet.2015.07.012
Yang, J., Lee, D., Afaisen, S., and Gadi, R. (2013). Inactivation by lemon juice of Escherichia coli O157:H7, Salmonella enteritidis, and Listeria monocytogenes in beef marinating for the ethnic food kelaguen. Int. J. Food Microbiol. 160, 353–359. doi: 10.1016/j.ijfoodmicro.2012.11.009
Yang, Y., Xu, F., Xu, H., Aguilar, Z. P., Niu, R., Yuan, Y., et al. (2013). Magnetic nano-beads based separation combined with propidium monoazide treatment and multiplex PCR assay for simultaneous detection of viable Salmonella Typhimurium, Escherichia coli O157:H7 and Listeria monocytogenes in food products. Food Microbiol. 34, 418–424. doi: 10.1016/j.fm.2013.01.004
Yang, X., Badoni, M., and Gill, C. O. (2011). Use of propidium monoazide and quantitative PCR for differentiation of viable Escherichia coli from E. coli killed by mild or pasteurizing heat treatments. Food Microbiol. 28, 1478–1482. doi: 10.1016/j.fm.2011.08.013
Zhang, Z., Liu, H., Lou, Y., Xiao, L., Liao, C., Malakar, P. K., et al. (2015a). Quantifying viable Vibrio parahaemolyticus and Listeria monocytogenes simultaneously in raw shrimp. Appl. Microbiol. Biotechnol. 99, 6451–6462. doi: 10.1007/s00253-015-6715-x
Zhang, Z., Liu, W., Xu, H., Aguilar, Z. P., Shah, N. P., and Wei, H. (2015b). Propidium monoazide combined with real-time PCR for selective detection of viable Staphylococcus aureus in milk powder and meat products. J. Dairy Sci. 98, 1625–1633. doi: 10.3168/jds.2014-8938
Zhao, X., Wang, J., Forghani, F., Park, J. H., Park, M. S., Seo, K. H., et al. (2013). Rapid detection of viable Escherichia coli O157 by coupling propidium monoazide with loop-mediated isothermal amplification. J. Microbiol. Biotechnol. 23, 1708–1716. doi: 10.4014/jmb.1306.06003
Keywords: viability detection, foodborne pathogens, propidium monoazide, ethidium monoazide, PMA-qPCR, outbreaks, false positive detection
Citation: Zeng D, Chen Z, Jiang Y, Xue F and Li B (2016) Advances and Challenges in Viability Detection of Foodborne Pathogens. Front. Microbiol. 7:1833. doi: 10.3389/fmicb.2016.01833
Received: 15 September 2016; Accepted: 01 November 2016;
Published: 22 November 2016.
Edited by:
Andrea Gomez-Zavaglia, Center for Research and Development in Food Cryotechnology (CIDCA, CONICET), ArgentinaReviewed by:
Séamus Fanning, University College Dublin, IrelandLearn-Han Lee, Monash University Malaysia Campus, Malaysia
Copyright © 2016 Zeng, Chen, Jiang, Xue and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baoguang Li, YmFvZ3VhbmcubGlAZmRhLmhocy5nb3Y= Feng Xue, ZmVuZ3h1ZTEyMTlAYWxpeXVuLmNvbQ==
 Dexin Zeng
Dexin Zeng Zi Chen1,2
Zi Chen1,2 Feng Xue
Feng Xue Baoguang Li
Baoguang Li