- 1Department of Microbiology, Faculty of Veterinary Medicine, Cairo University, Giza, Egypt
- 2Department of Poultry Diseases, Animal Health Research Institute, Giza, Egypt
- 3Department of Botany and Microbiology, College of Science, King Saud University, Riyadh, Saudi Arabia
- 4Department of Health Science, College of Applied Studies and Community Service, King Saud University, Riyadh, Saudi Arabia
- 5Department of Poultry Diseases, National Research Center, Giza, Egypt
- 6Department of Microbiology, Faculty of Veterinary Medicine, Sohag University, Sohag, Egypt
The use of antibiotics in farm management (growing crops and raising animals) has become a major area of concern. Its implications is the consequent emergence of antibiotic resistant bacteria (ARB) and accordingly their access into the human food chain with passage of antibiotic resistance genes (ARG) to the normal human intestinal microbiota and hence to other pathogenic bacteria causative human disease. Therefore, we pursued in this study to unravel the frequency and the quinolone resistance determining region, mecA and cfr genes of methicillin-susceptible Staphylococcus aureus (MSSA), methicillin-resistant S. aureus (MRSA), methicillin-resistant coagulase-negative staphylococci (MRCNS) and methicillin-susceptible coagulase-negative staphylococci (MSCNS) isolated from the retail trade of ready-to-eat raw chicken meat samples collected during 1 year and sold across the Great Cairo area. The 50 Staphylococcus isolated from retail raw chicken meat were analyzed for their antibiotic resistance phenotypic profile on 12 antibiotics (penicillin, oxacillin, methicillin, ampicillin-sulbactam, erythromycin, tetracycline, clindamycin, gentamicin, ciprofloxacin, chloramphenicol, sulfamethoxazole-trimethoprim, and vancomycin) and their endorsement of the quinolone resistance determining region, mecA and cfr genes. The isolation results revealed 50 isolates, CPS (14) and CNS (36), representing ten species (S. aureus, S. hyicus, S. epidermedius, S. lugdunensis, S. haemolyticus, S. hominus, S. schleiferi, S. cohnii, S. intermedius, and S. lentus). Twenty seven isolates were methicillin-resistant. Out of the characterized 50 staphylococcal isolates, three were MRSA but only 2/3 carried the mecA gene. The ARG that bestows resistance to quinolones, β-lactams, macrolides, lincosamides, and streptogramin B [MLS(B)] in MRSA and MR-CNS were perceived. According to the available literature, the present investigation was a unique endeavor into the identification of the quinolone-resistance-determining-regions, the identification of MRSA and MR-CNS from retail chicken meat in Egypt. In addition, these isolates might indicate the promulgation of methicillin, oxacillin and vancomycin resistance in the community and imply food safety hazards.
Introduction
Globally, health-conscious consumers have declined from consuming beef red meat which has been linked to heart disease, to choose leaner and easy digestible meats perceiving chicken, a white meat, to be a healthier option (Hathwar et al., 2012). Numerous potential vehicles of transmission of foodborne pathogens exist and commercial chicken meat has been identified as one of the most important food vehicles for these organisms, antimicrobial-resistant bacteria and antimicrobial resistance genes (Phillips et al., 2004; Aarestrup and Schwarz, 2006; Verraes et al., 2013). However, the bearing of foodborne pathogens in poultry meat and its by-products is a disturbing issue in the poultry industry due to its impact on public health and a challenge to the medical and veterinary officials worldwide (APUA, 2010a; Ruban and Fairoze, 2011). Epidemiologically, poultry meat is of paramount importance and still inculpated as prime source of human food poisoning (Kadariya et al., 2014). Although specific data on the burden of foodborne disease associated with Staphylococcus in poultry meat has been previously limited (Capita et al., 2002; Pesavento et al., 2007; Persoons et al., 2009), yet it has gained importance in the last couple of years (Bhargava et al., 2011; Hanson et al., 2011; Boost et al., 2013; He et al., 2013; Martins et al., 2013; Momtaz et al., 2013; Yurdakul et al., 2013; Islam et al., 2014; Khallaf et al., 2014; Xin et al., 2014; Abdalrahman et al., 2015; Owuna et al., 2015; Pinto et al., 2015; Bortolaia et al., 2016; Teramoto et al., 2016), it is considered to be significant to be a disturbing issue in the poultry industry due to its impact on public health and a challenge to the medical and veterinary officials worldwide (APUA, 2010a; Ruban and Fairoze, 2011).
The Genus Staphylococcus is very well-characterized consisting of 51 species and 27 sub-species (www.bacterio.net/staphylococcus.html). S. aureus is the most significant species within this genus, termed as coagulase-positive-Staphylococcus (CPS) by virtue of its versatility as a pathogen in humans and animals in addition to its being one of the causes of food intoxication (Jørgensen et al., 2005; Cunha, 2009). Other Staphylococcus species, collectively termed coagulase-negative-staphylococci (CNS), have gained importance as they have been implicated to be responsible for a variety of opportunistic infections in humans and animals (Vuong and Otto, 2002), their association with nosocomial infections in neonatal intensive care units and food poisoning in spite of the fact that, they are also not classical food poisoning bacteria (Cortes et al., 2013; Becker et al., 2014; Tong et al., 2015) as they are less pathogenic than S. aureus possessing a smaller array of virulence factors (Becker et al., 2014). Due to the ubiquity of many of the species within this group, their clinical significance has traditionally been dismissed, and when isolated from clinical specimens, the bacteria have merely been regarded as contaminants (Becker et al., 2014). This perception is, however, changing as many species have emerged as important causes of nosocomial infections, particularly in relation to foreign device-related infections and infections in immunocompromised patients (Ibrahem et al., 2009; Mathema et al., 2009). Almost half of all the CNS species that have been identified to date have been implicated in human infections (Lowy, 2013) and a PubMed search on CNS results in more than 15,000 references, reflecting the increasing medical impact of these bacteria (Becker et al., 2014).
The propensity for staphylococci to develop antimicrobial resistance is a cause for great concern in both human and veterinary medicine (Vanderhaeghen et al., 2010). Globally, antimicrobial resistance was highlightened as a priority issue (Davies and Davies, 2010; Acar and Moulin, 2012; WHO, 2012; World Economic Forum, 2013; One Health Commission, 2014) due to the increase in the public disquietness about the rampant promulgation of antimicrobial resistant bacteria (AMRB) causing a drop in the food supply as a result of treatment failure of the infected livestock, in addition to curtailment on international animal trade and human traveling. The connection between antibiotic use and antibiotic resistance profiles in chicken and human health has been reviewed by Marshall and Levy (2011) which also included methicillin-resistant S. aureus (MRSA) and which has been found in 12% of animal products—beef, veal, lamb, pork, and a variety of fowl—in Denmark (Normanno et al., 2007; de Boer et al., 2009). The European Food Safety Authority, the European Commission, and the European Centre for Disease Prevention, Control (ECDC), the U.S. National Antimicrobial Resistance Monitoring System (NARMS), a collaboration of the Food and Drug Administration (FDA), the Centers for Disease Control and Prevention (CDC), and the U.S. Department of Agriculture, routinely compile surveillance reports on antibiotic resistance in chicken from member countries which includes in some cases methicillin-susceptible S. aureus (MRSA). Significant levels of resistance are reported but the patterns vary considerably (CDDEP, 2015). The MRSA associated with animals and its relevance to human health has been highlighted by Pantosti (2012). Although, CNS is recognized as technologically/hygienically very important bacteria in food production and preservation (Hadžiosmanović et al., 2005; Šušković et al., 2010), yet its presence in food is beyond any doubt of public health significance due to the possible promulgation of AMRB and antimicrobial resistant genes (AMRG) (Zdolec et al., 2012a,b, 2013a,b; Dobranić et al., 2013; Chajęcka-Wierzchowska et al., 2015). In previous decades, a continuous loss of susceptibility toward most of the available antibiotics was recorded for CNS with a greater tendency to develop multidrug resistance (Taponen and Pyörälä, 2009) limiting present therapies posing a great threat to the health care system worldwide (Balaban and Rasooly, 2000; Anderson-Berry et al., 2011; deKraker et al., 2011; Jean-Baptiste et al., 2011; Marra et al., 2011). CNS of both animal and human origins are believed to serve as important reservoirs of antimicrobial resistance genes (Becker et al., 2014), which can transfer and integrate into the S. aureus genome leading to the emergence of new, potentially more resistant strains (Otto, 2012; Vitali et al., 2014). The mechanisms responsible for antimicrobial resistance in CNS are identical to those occurring in S. aureus (Livermore, 2000). Reports of methicillin-resistant strains among CNS are challenging due to the large proportion of methicillin-resistant strains and increasing numbers of isolates reinforcing the need to revise their importance to food safety (Bhargava and Zhang, 2014; Chah et al., 2014; Osman et al., 2016a,b). Therefore, screening of these elements is important for public health and despite the importance of such a screen, limited data are available for CNS at the species level among the chicken retail meat.
Consequently, our endeavor was to reveal the hypothetical implications that chicken meat may serve as a vector for the conveyance of AMRB and AMRG to humans. Therefore, we pursued in this study to probe the quinolone resistance determining region that has not been formerly analyzed, the mecA and cfr genes of methicillin-susceptible Staphylococcus aureus (MSSA), methicillin-resistant S. aureus (MRSA), methicillin-resistant coagulase-negative staphylococci (MRCNS) and methicillin-susceptible coagulase-negative staphylococci (MSCNS) isolated from the retail trade of ready-to-eat raw chicken meat sold in the retail market.
Materials and Methods
Sample Collection and Processing
Retail chicken (breasts and thighs) products (n = 100) were collected from 20 retail supermarkets and groceries in the Great Cairo Area during the year 2013. The purchased products at retail were placed in sterile plastic containment (zip-seal bags) then into ice chests with cold blocks to be immediately sent to the Department of Poultry Diseases, Animal Health Research, Institute, Dokki, laboratory for microbiological analyses. Upon arrival at the laboratory, the samples were transferred aseptically into stomacher bags with sterile tongs and weighed. Samples weighing 205 g were hand agitated with 250 ml of sterile 0.1% peptone broth. A 30 ml aliquot of sample wash was added to 30 ml of Baird Parker broth (2x concentration) with tellurite enrichment in a 250 ml sterile Erlenmeyer flask and incubated at 37°C for 18–24 h. After enrichment, 10 μl of broth was plated onto each of five antibiotic-supplemented Baird Parker agars with EY tellurite enrichment (BD) formulations and one un-supplemented formulation and incubated 18–24 h for growth. After enrichment, 10 μl was plated onto Baird Parker agar (BPA) with EY tellurite enrichment and incubated at 35°C 18–24 h for growth. Ten microliter were also plated onto CHROMagar MRSA plates and incubated at 35°C 24–48 h and examined for growth. Presumptive S. aureus (black colonies with clear halos on BPA) and presumptive MRSA (mauve colonies on CHROMagar) were initially screened using conventional methods: Gram staining, coagulase plasma test, hemolysis, catalase production, salt mannitol agar growth, Voges-Proskauer test, the maltose and trehalose fermentation test, susceptibility to polymyxin B test (300 lg, NewProv) and KOH test. Further confirmation was achieved by the API Staph kit. Individual colonies from all positive plates were streaked for isolated colonies twice before being frozen at −80°C in Brucella Broth (BD) containing 20% glycerol (Sigma). On the day of experimentation the isolates were subcultured onto Mueller-Hinton agar, stored at 27°C in tryptic soy broth and fetal calf serum containing 2% yeast extract and revitalized for phenotypic resistance testing and DNA preparation.
Antibiotic Susceptibility Test
Antibiotic resistance of the 50 isolated Staphylococcus spp. was examined using 12 commercial antibiotic discs and performed by the standard disc diffusion method (CLSI, 2013). S. aureus ATCC 25923 was plated as a control. After incubation at 37°C for 24 h the diameter of inhibition halos around the colonies was measured. The antibiotics used were: penicillin (10 μg), ampicillin-sulbactam (20 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), erythromycin (15 μg), tetracycline (30 μg), gentamicin (10 μg), clindamycin (2 μg), methicillin (5 μg), oxacillin (1 μg), vancomycin (30 μg), and sulfamethoxazole/trimethoprim (25 μg). In the present investigation, we followed the criteria and standardized international terminology for defining multidrug-resistant (MDR), extensively drug-resistant (XDR) and pandrug-resistant (PDR) in S. aureus that was created through a joint initiative by the European Centre for Disease Prevention and Control (ECDC) and the Centers for Disease Control and Prevention (CDC) (Magiorakos et al., 2012): MDR was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories (and one or more of these have to apply: an MRSA is always considered MDR by virtue of being an MRSA or/and non-susceptible to ≥1 agent in ≥3 antimicrobial categories), XDR was defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial categories (non-susceptible to ≥1 agent in all but ≤2 categories) and PDR was defined as non-susceptibility to all agents in all antimicrobial categories.
Extraction of Staphylococcal DNA and Amplification of the 16S rRNA Gene, Quinolone Resistance Determining Region, mecA and cfr Genes by PCR
Bacterial genomic DNA was made from all 50 isolates from 2 mL of bacterial suspension from an overnight culture in brain-heart infusion using a protocol previously described (Bakshi et al., 2005). The DNA pelletts were resuspended in an appropriate volume of TE solution (10 mM Tris-HCl, pH 8.0; 1 mM EDTA). Amplification was performed from the purified genomic DNA and a 0.2 mM concentration of each of the Staphylococcus-genus-specific primers (16S rRNA gene) was used. An internal control was integrated into the PCR-based assays to verify the efficiency of the amplifications and to ensure that significant PCR inhibition was absent. Genus-specific confirmation was carried out by PCR using the primers: F: AAC TCT GTT ATT AGG GAA GAA CA and R: CCA CCT TCC TCC GGT TTG TCA CC with annealing temperature: 68°C for 45 s to reveal an amplicon of bp 756 (Zhang et al., 2004). S. aureus ATCC 43300 was used as the positive control, while E. coli NCIMB 50034 was used as the negative control.
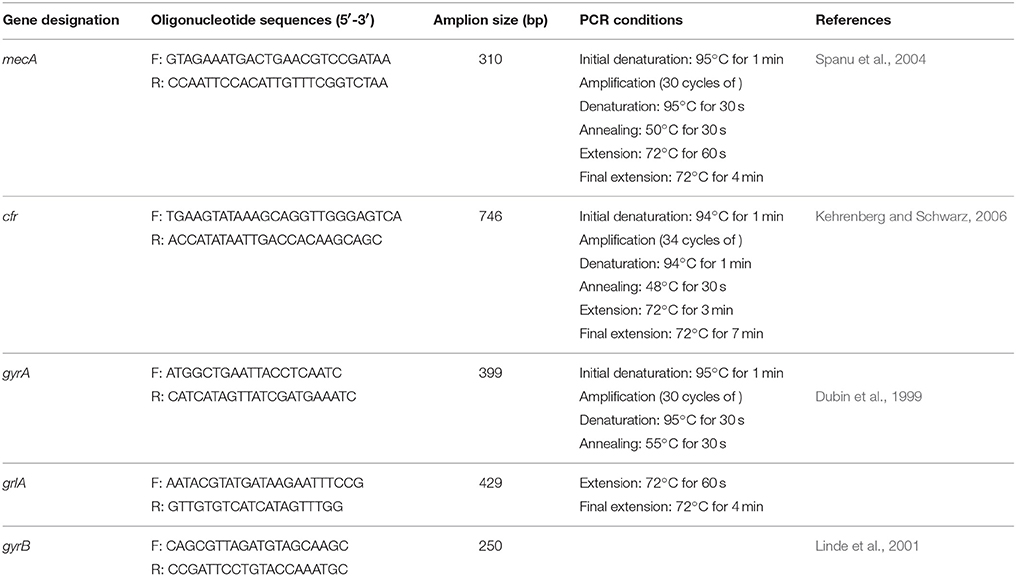
The confirmed staphylococci isolates were confirmed as MRSA using PCR to detect the presence of mecA (the gold-standard reference method for this analysis). Four additional antimicrobial resistance genes frequently reported in S. aureus conferring resistance in the quinolone-resistance-determining-regions (QRDRs) responsible for quinolone resistance (gyrA, gyrB, and grlA genes) and the cfr gene conferring resistance to several classes of antibiotics (oxazolidinones, phenicols, streptogramin compounds, lincosamidins, and pleuromutilins known as the PhLOPSA phenotype) were also amplified. Gene's targets, primers pair, nucleotide sequences, operative protocols, PCR amplification was performed as described previously (Table 1). Positive and negative controls were included with all PCR runs performed. All PCR assay runs incorporated a reagent control (without template DNA), positive and negative control (Streptococcus pyogenes ATCC 19615) processed in a fashion similar to that of the tested isolates. Three independent PCR amplifications were carried out with a GeneAmp PCR System 2400 (Perkin-Elmer, Weiterstadt, Germany). Two microliters of template DNA was added to 23 μL of distilled sterile water, and finally 25 μL of reaction mixture containing 10 mM Tris-HCl (pH8.3), 50 mM KCl, 2.5 mM MgCl2, 100 mM deoxynucleoside triphosphates, 1 mM of each primer and 3U of Taq polymerase were added. The amplicons were resolved by electrophoresis on 1.5% (w/v) agarose gel in TBE buffer, stained with ethidium bromide and photographed using a Polaroid Camera.
Results
The prevalence and diversity of the Staphylococcus species isolated from the chicken meat were found to be: out of the 100 chicken meat samples, 50 Staphylococcus species were isolated, which were further identified as, 14/50 CPS isolates differentiated into, S. aureus (n = 3), S. hyicus (n = 10), S. intermedius (n = 1); and 36/50 CNS isolates identified as S. epidermidis (n = 13), S. lugdunensis (n = 15), S. hemolyticus (n = 1), S. hominus (n = 3), S. lentus (n = 1), S. schleiferi (n = 2), and S. cohnii (n = 1).
Resistance Phenotypes of the Staphylococcus Isolates
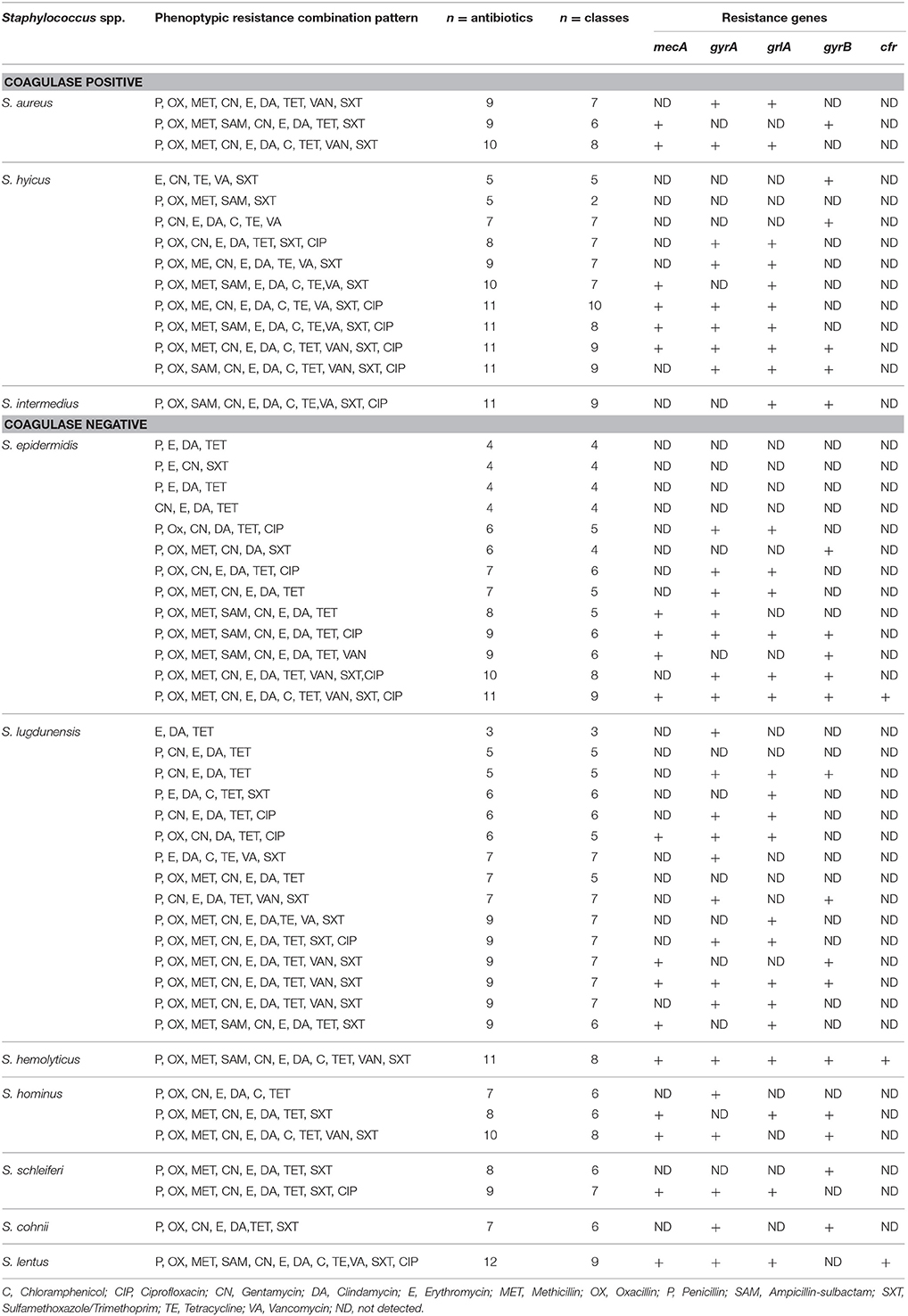
Table 2 shows the resistance phenotype of each of the 50 Staphylococcus isolates. Complete resistance to penicillin, gentamycin, clindamycin, oxacillin, and sulfamethoxazole/trimethoprim, with resistance to other important antimicrobials was also observed. The antimicrobial resistance profile of the 50 isolated Staphylococcus spp to different antibiotics was analyzed; none of the isolates were totally sensitive to the 12 tested antibiotics and 16/50 of the isolates were shown to be resistant to at least 3 antibiotics (S. lugdunensis) representing three classes. A small percentage of the isolates (less than 50%) demonstrated resistance to oxacillin, methicillin, erythromycin, chloramphenicol, ciprofloxacin, vancomycine, and tetracycline (Table 2). Also resistant to β-lactams, such as ampicillin (6/50), penicillin (22/50), methicillin (8/50), and oxacillin (13/50) was evident (Table 2). Forty seven out of the fifty isolates were resistant to penicillin and tetracycline.
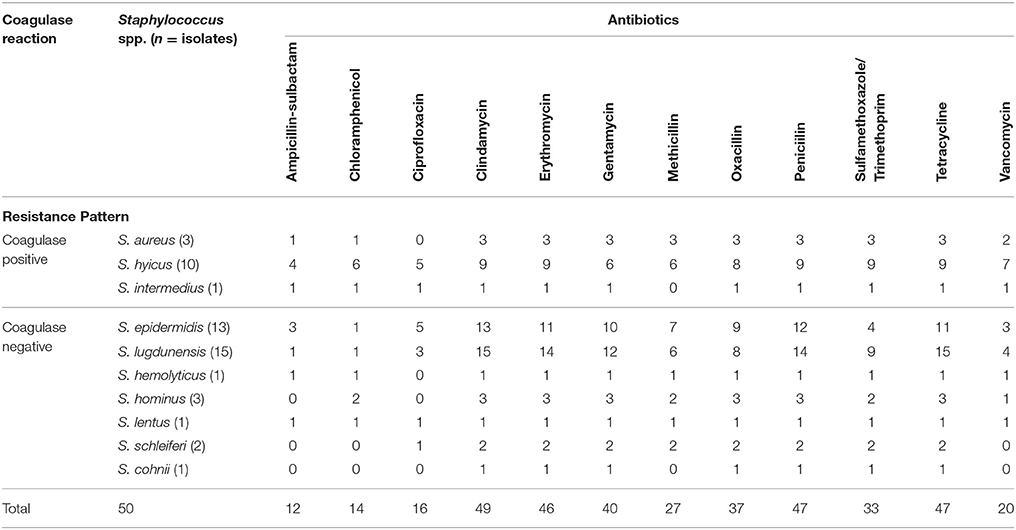
Table 2. Distribution of Staphylococcus spp. isolated from chicken meat samples according to their species diversity and multidrug resistance pattern.
The Distribution of Antibiotic Resistance Ratios of the Staphylococcus (S) Coagulase Positive (CP)-Negative (CN) That Are Resistant (MR) and Susceptible to Methicillin (MS)
Methicillin resistance in CPS and CNS was calculated to be 64.3 and 52.8%, respectively. The antimicrobial resistance patterns of the isolates are shown in Table 3. The resistance ratios to clindamycin, methicillin, oxacillin, penicillin, and sulfamethoxazole/trimethoprim in CPS and to clindamycin, gentamycin, methicillin, oxacillin and penicillin in CNS were found to be higher in the methicillin-resistant isolates compared to those that were susceptible to methicillin (p < 0.001). The lowest resistance ratio in the methicillin—resistant staphylococci was detected for ciprofloxacin and ampicillin-sulbactam in the CPS and to ampicillin-sulbactam, chloramphenicol and ciprofloxacin in the CNS. A comparison of the resistance patterns revealed that all isolates from chickens showed resistance from three to nine classes of antimicrobial agents (Table 4).
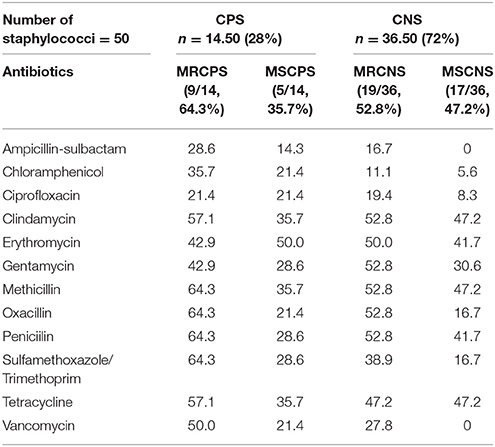
Table 3. The antibiotic resistance ratios of the Staphylococcus (S) Coagulase positive (CP)– negative (CN) that are resistant (MR) and susceptible to methicilin (MS) (%).
Eight isolates showed resistance to oxacillin, undetectable by mecA amplification. The three S. aureus isolates were resistant to 9–10 antimicrobials (Table 4). Two unique susceptibility profiles were identified among the S. aureus isolates, with many resistant to multiple clinically important antimicrobial classes (Table 4). In addition, it was noticed that 49/50 Staphylococcus isolates were MDR (non-susceptible to ≥1 agent in ≥3 antimicrobial classes) while 1/10 of the S. hyicus did not apply to this as it was only resistant to two classes of antibiotics although the number of antibiotics were five (P, OX, MET, SAM, and SXT). S. lentus was the only species to be PDR as it was resistant to the 12 antibiotics (P, OX, MET, SAM, CN, E, DA, C, TE, VA, SXT, and CIP) which represented nine classes and consequently, the remaining 48 Staphylococcus isolates were XDR as they were resistant to ≥1 agent in all but ≤2 classes (Table 4).
Antimicrobial Resistance Genes
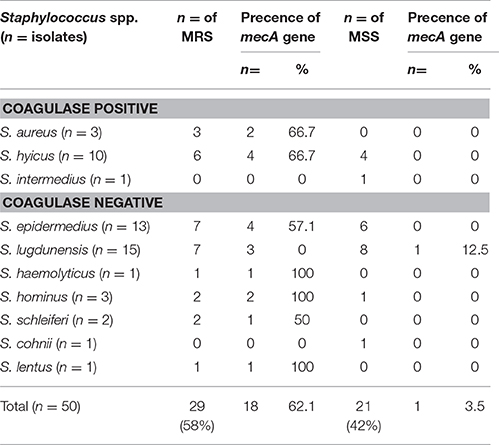
Of the 50 Staphylococcus isolates, two of the CPS species (2/3 S. aureus; 4/10 S. hyicus) were identified positive for mecA (6/50) while six out of the seven CNS species expressed the mecA gene (4/13 S. epidermedius; S. lugdunensis 4/15; S. haemolyticus 1/1; S. hominus 2/3; S. schleiferi 1/2 and S. lentus 1/1) (Table 4). With the exception of 1/8 S. lugdunensis isolates which was susceptible to oxacillin, mecA-positive, four Staphylococcus isolates (3/10 S. hyicus; 1/1 S. intermedius) were resistant to ciprofloxacin, gentamicin, erythromycin, methicillin, oxacillin, penicillin, sulfamethoxazole/trimethoprim, and vancomycin (Table 4). Forty seven out of the fifty isolates (47/50) were resistant to penicillin and tetracycline, while only one was resistant to chloramphenicol. Methicillin-resistant non-Staphylococcus aureus (MRNSA) isolates were resistant to nine of the 12 antimicrobials on the panel, including one to gentamicin.
Interestingly, two observations were recorded: (1) one of the three S. aureus did not carry the mecA gene and was not even phenotypically characterized as MRS (Table 5); and (2) the resistance gene mecA was present in the S. lugdunensis isolate (1/8) and which was phenotypically characterized as methicillin-susceptible non-Staphylococcus aureus (MSNSA) but at the same time phenotypically oxacillin resistant (Table 4). The resistance gene tested mecA was detected in 16/26 methicillin-resistant non-Staphylococcus aureus (MRNSA) isolates (Table 5), while cfr was not detected in any of the isolates (Table 4). Of the three MSSA isolates, two carried the gyrA gene and one carried the gyrB gene (Table 4). The 50 isolates of Staphylococcus species namely: aureus, hyicus, simulans, lugdunensis, hominus, hemolyticus, epidermidis, intermedius, and S. sciuri were non-cfr-carriers. Our susceptibility testing showed that, all non-cfr-carrying staphylococci (23/50) were resistant to the 12 antibiotics tested in the present with varying degrees. Of the 50 isolates, 40/50 isolates were gentamicin-resistant, 37/50 were oxacillin resistant while only 16/50 were ciprofloxacin-resistant. The results of antimicrobial susceptibility testing indicated that the non-cfr-carrying CNS exhibited also expanded resistance to antimicrobial agents other than those included in the cfr-mediated PhLOPSA phenotype (Table 4) [The Cfr rRNA Methyltransferase confers resistance to several classes of antibiotics (phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A; PhLOPSA phenotype)]. It was possible to conclude that 25/34 of the MR-NSA strains were potentially methicillin resistant. However, these results were evidently different from those obtained by PCR, in which the gene mecA was detected in only 16/47 of the CNSA isolates (Table 5).
Discussion
In previous investigations, the frequency of S. aureus in the chicken meat was found to be 33.3% in The Netherlands, 4% in Egypt, 22.7% in Switzerland and 65% in Japan (Schraft et al., 1992; Bakr et al., 2004; Kitai et al., 2005). The authors traced back the source of contamination to particular slaughterhouses in Switzerland while in Egypt poor hygienic and sanitary conditions in addition to the contaminated butchers and meat handlers during the process of slaughtering and evisceration of the bird (Kluytmans et al., 1995; Kitai et al., 2005) could be the main cause for the 50% prevalence recorded in the present investigation.
The coexistence of two subpopulations (one susceptible and the other resistant) within a culture of staphylococci could hinder an explicit detection of oxacillin/methicillin resistance (Bannerman, 2003). All cells in a culture may carry the genetic information for resistance, but only a small number may express the resistance in vitro. This phenomenon is termed heteroresistance and occurs in staphylococci resistant to penicillinase-stable penicillins, such as oxacillin. Cells expressing heteroresistance grow more slowly than the oxacillin-susceptible population and may be missed at temperatures above 35°C. This is why CLSI recommends incubating isolates being tested against oxacillin for a full 24 h before reading (CLSI, 2013). Oxacillin maintains its activity during storage better than methicillin and is more likely to detect heteroresistant strains. Previously, Petrelli et al. (2008) found a significant correlation between oxacillin resistance and resistance to erythromycin, clindamycin, gentamycin and ciprofloxacin, which is in queue with our findings; but on evaluating our results, penicillin and tetracycline resistance were also found to be significantly correlated with oxacillin resistance. It should be noted that, widespread veterinary usage of the fluoroquinolones, particularly in Europe, has been implicated (CVM, 2009; CVM/FDA, 2015) and that being a second-generation fluoroquinolone, is on the World Health Organization's List of Essential Medicines and the most important medications needed in a basic health system (WHO, 2015). Fearfully, this could lead to a rapid and sudden evolvement of resistance to ciprofloxacin and other fluoroquinolones, even during a course of treatment. This documentation is consistent with antecedent investigations and substantiates the allegations of MRSA causing healthcare-associated infections as being MDR to the traditionally prescribed drugs (erythromycin, clindamycin, fluoroquinolones, and tetracycline), while strains inducing community-associated infections are generally solely resistant to ß-lactam agents, erythromycin while feasibly fluoroquinolone resistant (Oliveira and de Lencastre, 2002).
Since 1996, MRSA strains with decreased susceptibility to vancomycin and strains fully resistant to vancomycin have been reported (CDC, 2013) a fact which should be taken into consideration in the Egyptian community as we detected a 20% resistance to vancomycin. Methicillin resistant Staphylococcus aureus (MRSA) and vancomycin resistant Staphylococcus aureus (VRSA) are some of the prominent pathogens that cause a wide variety of infections in humans and animals (Venkatesh et al., 2006; van Loo et al., 2007; Plata et al., 2009; Cuny et al., 2010; Gould, 2010; Grundmann et al., 2010). It should be emphasized that the present investigation highlights the importance of a sustainable surveillance program for any signs of vancomycin resistance emergence in Egypt concerning the medical and veterinary health condition in conjunction to typing and origin of the isolates. An increase in the cases of incompetent antibiotic treatment and mortality rates in avian disease (Han et al., 2013) is a common outcome of methicillin resistance, generating critical community health concern. Considering the limited options disease medication caused by MDR staphylococcal species, it is worrisome that CNS resistant to broad-spectrum antibiotics have been introduced in the community through the food chain. Regarding the CNS identified in this study, S. cohnii and S. lentus have been signified to yield enterotoxins (Podkowik et al., 2013).
Batista et al. (2013) have shown that oxacillin is more sensitive for predicting methicillin resistance, since 100% of isolates containing the mecA gene were phenotypically resistant to oxacillin, whereas only 13.3% were resistant to cefoxitin. Thus, the test using cefoxitin may not accurately reflect the phenotypic resistance of CNS, which has also been observed by other authors (Frigatto et al., 2005; Hung et al., 2011). To avoid misinterpretation, the detection of the mecA gene (the commonest gene that imparts oxacillin resistance in staphylococci) by nucleic acid amplification tests, such as the PCR, is currently considered the criterion standard for identification of MRSA strains (Zhang et al., 2004, 2008; Bagcigil et al., 2007; Zaraket et al., 2007), but this has not yet been established for CNS. The risk of transmission of resistance genes by horizontal interspecies transfer was clear and has been described by others (Bloemendaal et al., 2010; von Wintersdorff et al., 2016).
Foodborne pathogens, such as MRSA have become a global problem after being in North America, Europe and Asia. MRSA in chicken meat showed its highest prevalence from Germany (37.2%) (Feßler et al., 2011) followed by Netherlands (16.0%) (de Boer et al., 2009). On the other hand, low levels of MRSA in chicken meat were recorded in Japan (Kitai et al., 2005), Jordan (Quddoumi et al., 2006) to reach 0.3% in Korea (Lim et al., 2010) and 0% in Austrai (Zarfel et al., 2014). There are also reports of now the fourth phase has also been started which shows evidences that both animal and human MRSA has been detected on meat. Studies in Japan and Korea reported human MRSA from chicken meat (Lee, 2003; Kitai et al., 2005). Taiwanese study reported MRSA in chicken carcasses (Lin et al., 2009). MRSA have also been detected from the foods, such as meat products and raw chicken meat (Vanderhaeghen et al., 2010). Regardless of the ongoing endorsed methodizes for the MRSA susceptibility testing, Lee et al. (2004) demonstrated a variance in the phenotypic expression of resistance being affected by the cellular growth conditions and that several MRSA isolates which were phenotypically resistant to oxacillin, did not encode the mecA gene. MRSA can be found worldwide and Doyle et al. (2011) summarized the global survival results in raw meats, which indicated that the incidence of MRSA was infrequent in poultry meat when compared to the beef and pork meats and with variable prevalences between regions.
Antibiotic resistance patterns of individual pathogens to the drugs used to treat them vary considerably between and within countries. The discrepancies and uncomparable results between researchers, including us, was incriminated by Kluytmans et al. (1995), Chaibenjawong and Foster (2011), Doyle et al. (2011) and Omurtag et al. (2013) to be due to several criteria, such as: (i) sampling and culture methods differed among the studies; (ii) the condition of the meat when being sold is it packed or un-packed in the supermarkets or through the widely spread butcher shops in Great Cairo; (iii) MRSA carriers can contaminate meat during slaughter and processing; (iv) pathogen load on raw meat changes according to species; (v) heat treatment and cross-contamination change the pathogen titre on the meat; (vi) serving frequency and size, and demographic data; (vii) regional differences and social groups dissimilarities are conditions that must be taken into consideration; (viii) infected/colonized food handlers are favorable causes for meat contamination (ix) under conditions of temperature abuse, MSSA and MRSA cells could multiply on meat; (ix) the unjuridical use of antibiotics for animals; (x) different patterns of antibiotic use; (xi) distinct national disease burdens; (xii) disparities in access to first—and second-line treatments; and (xiii) the burden of coinfections.
Eventually, AMR drugs have become a global public health crisis implicating the importance of preventing contamination of food with MRSA, which could have trade consequences and exert coercion to those sovereignties with un-controlled antimicrobial practices to create and enforce proper danger administration strategies. Therefore, the use of antibiotics for animals and their documentation has become a must to aid in analyzing their use and consequent impact on public health to enact policy changes on the consumption of antibiotics (APUA, 2010b; WHO, 2013).
Conclusion
The occurrence of CNS in food should not be ignored nor their pathogenic potential considered as insignificant as a food-borne pathogen, rather safety measures should be taken to reduce or totally eliminate their occurrence in foods. The incidence of CNS may render food unsafe, as the clinical isolates have been reported to exude virulent traits (Fowoyo and Ogunbanwo, 2016). Our findings emphasize the need for improved hygiene practices during food processing and also during the distribution and consumption of the final food products to avoid the presence of CNS and CPS isolates in meat. The present study cynosures that the MRS has entered the food chain in Egypt through the raw chicken meat constituting a health hazard to consumers and should be given importance due to their zoonotic importance. This necessitates the urgent implementation of an energetic policy to ensure microbiological food safety to prevent the spread of MRS through the food chain by contamination of raw meat. In addition, these isolates might indicate the promulgation of methicillin, oxacillin, and vancomycin resistance in the community and imply food safety hazards.
Author Contributions
KO conceived, designed the experiments, analyzed the data and wrote the paper; KA, IM, AH, ZAG, and UA contributed their scientific advice during the work and MS revision; JB, AO, and AA performed the experiments, supplied and contributed reagents/materials/analysis tools.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for supporting the work through the research group project No.: RGP-176.
References
Aarestrup, F. M., and Schwarz, S. (2006). “Antimicrobial resistance in staphylococci and streptococci of animal origin,” in Antimicrobial Resistance in Bacteria of Animal Origin, ed F. M. Aarestrup (Washington, DC: ASM Press), 187–212.
Abdalrahman, L. S., Stanley, A., Wells, H., and Fakhr, M. K. (2015). Isolation, Virulence, and Antimicrobial Resistance of Methicillin-Resistant Staphylococcus aureus (MRSA) and Methicillin Sensitive Staphylococcus aureus (MSSA) Strains from Oklahoma Retail Poultry Meats. Int. J. Environ. Res. Public Health 12, 6148–6161. doi: 10.3390/ijerph120606148
Acar, J. F., and Moulin, G. (2012). Antimicrobial resistance: a complex issue. Rev. Sci. Tech. 31, 23–31. doi: 10.20506/rst.31.1.2098
Anderson-Berry, A., Brinton, B., Lyden, E., and Faix, R. G. (2011). Risk factors associated with development of persistent coagulase negative staphylococci bacteremia in the neonate and associated short-term and discharge morbidities. Neonatology 99, 23–31. doi: 10.1159/000292567
APUA (2010a). Policy Brief and Recommendations #4 Misuse of Antibiotics in Food Animal Production Antibiotic Misuse in Food Animals – Time for Change. Boston, MA: Alliance for the Prudent Use of Antibiotics.
APUA (2010b). Antibiotics in Food Animal Production: A Forty Year Debate. Vol. 28. Boston, MA: Alliance for the prudent use of antibiotics.
Bagcigil, F. A., Moodley, A., Baptiste, K. E., Jensen, V. F., and Guardabassi, L. (2007). Occurrence, species distribution, antimicrobial resistance and clonality of methicillin-and erythromycin-resistant staphylococci in the nasal cavity of domestic animals. Vet. Microbiol. 121, 307–315. doi: 10.1016/j.vetmic.2006.12.007
Bakr, W. M., Fawzi, M., and Hashish, M. H. (2004). Detection of coagulase positive staphylococci in meat products sold in Alexandria using two different media. J. Egypt Public Health Assoc. 79, 31–42.
Bakshi, C. S., Shah, D. H., Verma, R., Singh, R. K., and Malik, M. (2005). Rapid differentiation of Mycobacterium bovis and Mycobacterium tuberculosis based on a 12.7-kb fragment by a single tube multiplex-PCR. Vet. Microbiol. 109, 211–216. doi: 10.1016/j.vetmic.2005.05.015
Balaban, N., and Rasooly, A. (2000). Staphylococcal enterotoxins. Int. J. Food Microbiol. 61, 1–10. doi: 10.1016/S0168-1605(00)00377-9
Bannerman, T. L. (2003). “Staphylococcus, Micrococcus and other catalase-positive cocci that grow aerobically,” in Manual of Clinical Microbiology, 8th Edn., eds P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (Washington, DC: ASM Press), 384–404.
Batista, J. E., Ferreira, E. L., Nascimento, D. C., Ventura, R. F., de Oliveira, W. L., Leal, N. C., et al. (2013). Antimicrobial resistance and detection of the mecA gene besides enterotoxin-encoding genes among coagulase-negative Staphylococci isolated from clam meat of Anomalocardia brasiliana. Foodborne Pathog. Dis. 10, 1044–1049. doi: 10.1089/fpd.2013.1576
Becker, K., Heilmann, C., and Peters, G. (2014). Coagulase-negative staphylococci. Clin. Microbiol. Rev. 27, 870–926. doi: 10.1128/CMR.00109-13
Bhargava, K, Wang, X, Donabedian, S, Zervos, M, da Rocha, L, Zhang, Y. (2011). Methicillin-resistant Staphylococcus aureus in retail meat, Detroit, Michigan, USA [letter]. Emerg. Infect. Dis. 17, 1135–1137. doi: 10.3201/eid/1706.101905
Bhargava, K., and Zhang, Y. (2014). Characterization of methicillin-resistant coagulase-negative staphylococci (MRCoNS) in retail meat. Food Microbiol. 42, 56–60. doi: 10.1016/j.fm.2014.02.019
Bloemendaal, A. L., Brouwer, E. C., and Fluit, A. C (2010). Methicillin resistance transfer from Staphylocccus epidermidis to methicillin-susceptible Staphylococcus aureus in a patient during antibiotic therapy. PLoS ONE 5:e11841. doi: 10.1371/journal.pone.0011841
Boost, M. V., Wong, A., Ho, J., and O'Donoghue, M. (2013). Isolation of Methicillin-Resistant Staphylococcus aureus (MRSA) from retail meats in Hong Kong. Foodborne Pathog. Dis. 10, 705–710. doi: 10.1089/fpd.2012.1415
Bortolaia, V., Espinosa-Gongora, C., and Guardabassi, L. (2016). Human health risks associated with antimicrobial-resistant enterococci and Staphylococcus aureus on poultry meat. Clin. Microbiol. Infect. 22, 130–140. doi: 10.1016/j.cmi.2015.12.003
Capita, R., Alonso-Calleja, C., Garcı'a-Ferna'ndez, M. C., and Moreno, B. (2002). Characterization of Staphylococcus aureus isolated from poultry meat in Spain. Poult. Sci. 81, 414–421. doi: 10.1093/ps/81.3.414
CDC (2013). Methicillin-resistant Staphylococcus aureus (MRSA) Infections. Atlanta, GA: Centers for Disease Control and Prevention.
CDDEP (2015). Center for Disease Dynamics, Economics & Policy. 2015. State of the World's Antibiotics. Washington, DC: CDC.
Chah, K. F., Gómez-Sanz, E., Nwanta, J. A., Asadu, B., Agbo, I. C., Lozano, C., et al. (2014). Methicillin-resistant coagulase-negative staphylococci from healthy dogs in Nsukka, Nigeria. Braz. J. Microbiol. 45, 215–220. doi: 10.1590/S1517-83822014005000034
Chaibenjawong, P., and Foster, S. J. (2011). Desiccation tolerance in Staphylococcus aureus. Archiv. Microbiol. 193, 125–135. doi: 10.1007/s00203-010-0653-x
Chajęcka-Wierzchowska, W., Zadernowska, A., Nalepa, B., Sierpi_nska, M., and Łaniewska-Trokenheim, L. (2015). Coagulase-negative staphylococci (CoNS) isolated from ready-to-eat food of animal origin–phenotypic and genotypic antibiotic resistance. Food Microbiol. 46, 222–226. doi: 10.1016/j.fm.2014.08.001
CLSI (2013). Performance Standards for Antimicrobial Susceptibility Testing. CLSI Approved Standard M100-S23. Wayne, PA: Clinical and Laboratory Standards Institute.
Cortes, J. A., Leal, A. L., Montañez, A. M., Buitrago, G., Castillo, J. S., Guzman, L., et al. (2013). Frequency of microorganisms isolated in patients with bacteremia in intensive care units in Colombia and their resistance profiles. Braz. J. Infect. Dis. 17, 346–352. doi: 10.1016/j.bjid.2012.10.022
Cunha, M. (2009). “Staphylococcus aureus and bovine mastitis: a public health problem,” in Cross Infections: Types, Causes and Prevention, eds J. Dong and X. Liang (New York, NY: Nova Science Publishers), 117–128.
Cuny, C., Friedrich, A., Kozytska, S., Layer, F., Nübel, U., Ohlsen, K., et al. (2010). Emergence of methicillin-resistant Staphylococcus aureus (MRSA) in different animal species. Int. J. Med. Microbiol. 300, 109–117. doi: 10.1016/j.ijmm.2009.11.002
CVM/FDA (2015). Center for Veterinary Medicine/ Center for Veterinary Medicine. Update on Antimicrobial Resistance Activities at the FDA. U.S. FDA Office of International Programs 5th Biannual Educational Forum.
Davies, J., and Davies, D. (2010). Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 74, 417–433. doi: 10.1128/MMBR.00016-10
de Boer, E., Zwartkruis-Nahuis, J. T., Wit, B., Huijsdens, X. W., de Neeling, A. J., Bosch, T., et al. (2009). Prevalence of methicillin-resistant Staphylococcus aureus in meat. Int. J. Food Microbiol. 134, 52–56. doi: 10.1016/j.ijfoodmicro.2008.12.00710.1016/j.ijfoodmicro.2008.12.007
Dobranić, V., Zdolec, N., Račić, I., Vujnović, A., Zdelar-Tuk, M., Filipović, I., et al. (2013). Determination of enterotoxin genes in coagulase-negative staphylo-cocci from autochthonous Croatian fermented sausages. Vet. Arh. 83, 145–152.
Doyle, M. E., Hartmann, F. A., Lee, A. C., and Wong, R. I. (2011). Food Safety Review: White Paper on Sources of Methicillin-Resistant Staphylococcus aureus (MRSA) and Other Methicillin-Resistant Staphylococci: Implications for Our Food Supply? Available online at: http://fri.wisc.edu/docs/pdf/FRI_Brief_MRSA_Feb2011.pdf
Dubin, D. T., Fitzgibbon, J. E., Nahvi, M. D., and John, J. F. (1999). Topoisomerase sequences of coagulase-negative staphylococcal isolates resistant to ciprofloxacin or trovafloxacin. Antimicrob. Agents Chemother. 43, 1631–1637.
Feßler, A. T., Kadlec, K., Hassel, M., Hauschild, T., Eidam, C., Ehricht, R., et al. (2011). Characterization of methicillin-resistant Staphylococcus aureus isolates from food and food products of poultry origin in germany. Appl. Environ. Microbiol. 77, 7151–7157. doi: 10.1128/AEM.00561-11
Fowoyo, P. T., and Ogunbanwo, S. T. (2016). Virulence and toxigenicity of coagulase-negative Staphylococci in Nigerian traditional fermented foods. Can. J. Microbiol. 62, 572–578. doi: 10.1139/cjm-2015-0752
Frigatto, E. A., Machado, A. M., Pignatari, A. C., and Gales, A. C. (2005). Is the cefoxitin disk test reliable enough to detect oxacillin resistance in coagulase-negative Staphylococci? J. Clin. Microbiol. 43, 2028–2029. doi: 10.1128/JCM.43.4.2028-2029.2005
Gould, I. M. (2010). VRSA-doomsday superbug or damp squib? Lancet Infect. Dis. 10, 816–818. doi: 10.1016/S1473-3099(10)70259-0
Grundmann, H., Aanensen, D. M., Wijngaard, V. D., Spratt, C. C., Harmsen, B. G., Friedrich, A. W., et al. (2010). Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 7:e1000215. doi: 10.1371/journal.pmed.1000215
Hadžiosmanović, M., Gasparik-Reichardt, J., Smajlović, M., Vesković-Moračanin, S., and Zdolec, N. (2005). Possible use of bacteriocins and starter cultures in upgrading of quality and safety of traditionally fermented sausages. Tehnol. Mesa 46, 194–211.
Han, J. E., Hwang, S. Y., Kim, J. H., Shin, S. P., Jun, J. W., Chai, J. Y., et al. (2013). CPRMethicillin resistant coagulase-negative staphylococci isolated from South Korean ducks exhibiting tremor. Acta Vet. Scand. 55:88. doi: 10.1186/1751-0147-55-88
Hanson, B. M., Dressler, A. E., Harper, A. L., Scheibel, R. P., Wardyn, S. E., Roberts, L. K., et al. (2011). Prevalence of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus (MRSA) on retail meat in Iowa. J. Infect. Public Health 4, 169–174. doi: 10.1016/j.jiph.2011.06.001
Hathwar, S. C., Rai, A. K., Modi, V. K., and Narayan, B. (2012). Characteristics and consumer acceptance of healthier meat and meat product formulations—a review. J. Food Sci. Technol. 49, 653–664. doi: 10.1007/s13197-011-0476-z
He, W., Liu, Y., Qi, J., Chen, H., Zhao, C., Zhang, F., et al. (2013). Food-Animal related Staphylococcus aureus multidrug-resistant ST9 strains with toxin genes. Foodborne Pathog. Dis. 10, 782–788. doi: 10.1089/fpd.2012.1452
Hung, K. H., Yan, J. J., Lu, Y. C., Chen, H. M., and Wu, J. J. (2011). Evaluation of discrepancies between oxacillin and cefoxitin susceptibility in coagulase-negative staphylococci. Eur. J. Clin. Microbiol. Infect. Dis. 30, 785–788. doi: 10.1007/s10096-011-1156-7
Ibrahem, S., Salmenlinna, S., Virolainen, A., Kerttula, A. M., Lyttikäinen, O., Jägerroos, H., et al. (2009). Carriage of methicillin-resistant staphylococci and their SCCmec types in a long-term-care facility. J. Clin. Microbiol. 47, 32–37. doi: 10.1128/JCM.01085-08
Islam, N. N., Akter, M., Farzana, Z., Bin Kader, A. J., Uddin, I., Zonaed Siddiki, A. M. A. M., et al. (2014). Detection of Staphylococcus aureus in frozen chicken rinse through bacteriological and Nuc gene specific PCR methods and their drug resistance patterns in Southern Chittagong, Bangladesh. Res. J. Microbiol. 9, 251–264. doi: 10.3923/jm.2014.251.264
Jean-Baptiste, N., Benjamin, D. K., Wolkowiez, C. M., Fowler, V. G., Laughon, M., Clark, R. H., et al. (2011). Coagulase negative staphylococcal infections in the neonatal intensive care unit. Infect. Control Hosp. Epidemiol. 32, 679–686. doi: 10.1086/660361
Jørgensen, H., Mørk, T., Caugant, D., Kearns, A., and Rørvik, L. (2005). Genetic variation among Staphylococcus aureus strains from Norwegian bulk milk. Appl. Environ. Microbiol. 71, 8352–8361. doi: 10.1128/AEM.71.12.8352-8361.2005
Kadariya, J., Smith, T. C., and Thapaliya, D. (2014). Staphylococcus aureus and Staphylococcal Food-Borne disease: an ongoing challenge in public health. BioMed Res. Int. 2014:827965. doi: 10.1155/2014/827965
Kehrenberg, C., and Schwarz, S. (2006). Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50, 1156–1163. doi: 10.1128/AAC.50.4.1156-1163.2006
Khallaf, M., Benbakhta, B., Nasri, I., Sarhane, B., Senouci, S., and Ennaji, M. M. (2014). Prevalence of Staphylococcus aureus isolated from chicken meat marketed in Rabat, Morocco. Int. J. Innovation Appl. Studies 7, 1665–1670.
Kitai, S., Shimizu, A., Kawano, J., Sato, E., Nakano, C., Uji, T., et al. (2005). Characterization of methicillin-resistant Staphylococcus aureus isolated from retail raw chicken meat in Japan. J. Vet. Med. Sci. 67, 107–110. doi: 10.1292/jvms.67.107
Kluytmans, J., Van Leeuwen, W., Goessens, W., Hollis, R., Messer, S., Herwaldt, L., et al. (1995). Food-initiated outbreak of methicillin-resistant Staphylococcus aureus analyzed by pheno- and genotyping. J. Clin. Microbiol. 33, 1121–1128.
Lee, J. H. (2003). Methicillin (Oxacillin)-Resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl. Environ. Microbiol. 69, 6489–6494. doi: 10.1128/AEM.69.11.6489-6494.2003
Lee, J. H., Jeong, J., Park, Y., Choi, S., Kim, Y., Chae, J., et al. (2004). Evaluation of the methicillin-resistant Staphylococcus aureus (MRSA)- screen latex agglutination test for detection of MRSA of animal origin. J. Clin. Microbiol. 42, 2780–2782. doi: 10.1128/JCM.42.6.2780-2782.2004
Lim, S. K., Nam, H. M., Park, H. J., Lee, H. S., Choi, M. J., Jung, S. C., et al. (2010). Prevalence and characterization of methicillin-resistant Staphylococcus aureus in raw meat in Korea. J. Microbiol. Biotechnol. 20, 775–778. doi: 10.4014/jmb.0912.12022
Lin, J., Yeh, K. S., Liu, H. T., and Lin, J. H. (2009). Staphylococcus aureus isolated from pork and chicken carcasses in Taiwan: prevalence and antimicrobial susceptibility. J. Food Prot. 72, 608–211.
Linde, H. J., Schmidt, M., Fuchs, E., Reischl, U., Niller, H. H., and Lehn, N. (2001). In vitro activities of six Quinolones and mechanisms of resistance in Staphylococcus aureus and coagulase-negative Staphylococci. Antimicrob. Agents Chemother. 45, 1553–1557. doi: 10.1128/AAC.45.5.1553-1557.2001
Livermore, D. M. (2000). Antibiotic resistance in staphylococci. Int. J. Antimicrob. Agents 16, S3–S10. doi: 10.1016/s0924-8579(00)00299-5
Lowy, F. (2013). “Staphylococcal infections,” in Harrison's Principles of Internal Medicine, 18th Edn., eds A. Fauci, E. Braunwald, and D. Casper (New York, NY: McGraw-Hill), 386–399.
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Marra, A. R., Camargo, L. F., Pignatari, A. C., Sukiennik, T., Behar, P. R., Medeiros, E. A., et al. (2011). Nosocomial bloodstream infections in Brazilian hospitals: analysis of 2,563 cases from a prospective nationwide surveillance study. J. Clin. Microbiol. 49, 1866–1871. doi: 10.1128/JCM.00376-11
Marshall, B. M., and Levy, S. B. (2011). Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 24, 718–733. doi: 10.1128/CMR.00002-11
Martins, P. D., de Almeida, T. T., Basso, A. P., de Moura, T. M., Frazzon, J., Tondo, E. C., et al. (2013). Coagulase-Positive staphylococci isolated from chicken meat: pathogenic potential and vancomycin resistance. Foodborne Pathog. Dis. 10, 771–776. doi: 10.1089/fpd.2013.1492
Mathema, B., Mediavilla, J., Chen, L., and Kreiswirth, B. (2009). “Evolution and taxonomy of Staphylococci,” in. Staphylococci in Human Disease, 2nd Edn., eds D. Crossley, K. Jefferson, G. Archer, and V. Fowler (Singapore: Wiley-Blackwell), 31–64.
deKraker, M. E., Davey, P. G., and Grundmann, H. (2011). Mortality and hospital stay associated with resistant Staphylococcus aureus and Escherichia coli bacteremia:estimating the burden of antibiotic resistance in Europe. PLoS Med. 8:e1001104. doi: 10.1371/journal.pmed.1001104
Momtaz, H., Dehkordi, F. S., Rahimi, E., Asgarifar, A., and Momeni, M. (2013). Virulence genes and antimicrobial resistance profiles of Staphylococcus aureus isolated from chicken meat in Isfahan province, Iran. J. Appl. Poult. Res. 22, 913–921. doi: 10.3382/japr.2012-00673
Normanno, G., Corrente, M., Salandra, G., Dambrosio, A., Quaglia, N. C., Parisi, A., et al. (2007). Methicillin-resistant Staphylococcus aureus (MRSA) in foods of animal origin product in Italy. Int. J. Food Microbiol. 117, 219–222. doi: 10.1016/j.ijfoodmicro.2007.04.006
Oliveira, D. C., and de Lencastre, H. (2002). Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 46, 2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002
Omurtag, I., Paulsen, P., Hilbert, F., and Smulders, F. J. M. (2013). The risk of transfer of foodborne bacterial hazards in Turkey through the consumption of meat; risk ranking of muscle foods with the potential to transfer Campylobacter spp. Food Secur. 5, 117–127. doi: 10.1007/s12571-012-0230-z
One Health Commission (2014). Why One Health? Apex, NC. Available online at: https://www.onehealthcommission.org/en/why_one_health/.
Osman, K. M., Abd El-Razik, K. A., Marie, H. S. H., and Arafa, A. (2016a). Coagulase-negative staphylococci collected from bovine milk: species and antimicrobial gene diversity. J. Food Saf. 36, 89–99. doi: 10.1111/jfs.12216
Osman, K. M., Amer, A. M., Badr, J. M., Helmy, N. M., Elhelw, R. A., Orabi, A., et al. (2016b). Antimicrobial resistance, biofilm formation and mecA characterization of methicillin-susceptible S. aureus and Non-S. aureus of Beef Meat Origin in Egypt. Front. Microbiol. 7:222. doi: 10.3389/fmicb.2016.00222
Otto, M. (2012). Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection. Bioessays 35, 4–11. doi: 10.1002/bies.201200112
Owuna, G., Abimiku, R. H., Nkene, I. H., Joseph, G. W., and Ijalana, O. O. (2015). Isolation and Antibiotic Susceptibility of Staphylococcus aureus from Fresh Poultry Meat Sold in Keffi Metropolis, Nigeria. Int. J. Res. Studies Biosci. 3, 1–5.
Pantosti, A. (2012). Methicillin-resistant Staphylococcus aureus associated with animals and its relevance to human health. Front. Microbiol. Antimicrob. Resis. Chemother. 3:127. doi: 10.3389/fmicb.2012.00127
Persoons, D., Van Hoorebeke, S., Hermans, K., Butaye, P., de Kruif, A., Haesebrouck, F., et al. (2009). Methicillin-Resistant Staphylococcus aureus in Poultry. Emerging Infect. Dis. 15, 452–453. doi: 10.3201/eid1503.080696
Pesavento, G., Ducci, B., Comodo, N., and Lo Nostro, A. (2007). Antimicrobial resistance profile of Staphylococcus aureus isolated from raw meat: a research for methicillin resistant Staphylococcus aureus (MRSA). Food Control 18, 196–200. doi: 10.1016/j.foodcont.2005.09.013
Petrelli, D., Repetto, A., D'Ercole, S., Rombini, S., Ripa, S., Prenna, M., et al. (2008). Analysis of meticillin-susceptible and meticillin-resistant biofilm-forming Staphylococcus aureus from catheter infections isolated in a large Italian hospital. J. Med. Microbiol. 57, 364–372. doi: 10.1099/jmm.0.47621-0
Phillips, I., Casewell, M., Cox, T., De Groot, B., Friis, C., Jones, R., et al. (2004). Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. J. Antimicrob. Chemother. 53, 28–52. doi: 10.1093/jac/dkg483
Pinto, J. B., Rossatto, F. C. P., Martins, P. D., and Frazzon, A. P. G. (2015). Genetic relationships and virulence factors in Staphylococcus aureus isolated from raw poultry in South Brazil. Ann. Microbiol. 65, 1933–1940. doi: 10.1007/s13213-014-1031-8
Plata, K., Rosato, A. E., and Wegrzyn, G. (2009). Staphylococcus aureus as an infectious agent: overview of biochemistry and molecular genetics of its pathogenicity. Acta Biochim. Pol. 56, 597–612.
Podkowik, M., Park, J. Y., Seo, K. S., Bystro, J., and Bania, J. (2013). Enterotoxigenic potential of coagulase-negative staphylococci. Int. J. Food Microbiol. 163, 34–40. doi: 10.1016/j.ijfoodmicro.2013.02.005
Quddoumi, S. S., Bdour, S. M., and Mahasneh, A. M. (2006). Isolation and characterization of methicillin-resistant Staphylococcus aureus from livestock and poultry meat. Ann. Microbiol. 56, 155–161. doi: 10.1007/BF03174998
Ruban, S. W., and Fairoze, N. (2011). Effect of proceesing conditions on microbiological quality of market poultry meats in bangalore, india. J. Anim. Vet. Adv. 10, 188–191. doi: 10.3923/javaa.2011.188.191
Schraft, H., Kleinlein, N., and Untermann, F. (1992). Contamination of pig hindquarters with Staphylococcus aureus. Int. J. Food Microbiol. 15, 191–194. doi: 10.1016/0168-1605(92)90148-V
Spanu, T., Sanguinetti, M., D'Inzeo, T., Ciccaglione, D., Romano, L., Leone, F., et al. (2004). Identification of methicillin-resistant isolates of Staphylococcus aureusand coagulase-negative Staphylococci responsible for bloodstreaminfections with the Phoenix™ system. Diag. Microbiol. Infect. Dis. 48, 221–227. doi: 10.1016/j.diagmicrobio.2003.11.004
Šušković, J., Kos, B., Beganović, J., Leboš Pavunc, A., Habjanič, K., and Matošić, S. (2010). Antimicrobial activity - the most important property of probiotic and starter lactic acid bacteria. Food Technol. Biotechnol. 48, 296–307.
Taponen, S., and Pyörälä, S. (2009). Coagulase-negative staphylococci as cause of different from Staphylococcus aureus. Vet. Microbiol. 134, 29–36. doi: 10.1016/j.vetmic.2008.09.011
Teramoto, H., Salaheen, S., and Biswas, D. (2016). Contamination of post-harvest poultry products with multidrug resistant Staphylococcus aureus in Maryland-Washington DC metro area. Food Control 65, 132–135. doi: 10.1016/j.foodcont.2016.01.024
Tong, S. Y. C., Davis, J. S., Eichenberger, E., Holland, T. L., and Fowler, V. G. Jr. (2015). Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661. doi: 10.1128/CMR.00134-14
Vanderhaeghen, W., Hermans, K., Haesebrouck, F., and Butaye, P. (2010). Methicillin-resistant Staphylococcus aureus (MRSA) in food production animals. Epidem. Infect. 138, 606–625. doi: 10.1017/S0950268809991567
van Loo, I., Huijsdens, X., Tiemersma, E., de Neeling, A., van de Sande-Bruinsma Beaujean, D., et al. (2007). Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerging Infect. Dis. 13, 1834–1839. doi: 10.3201/eid1312.070384
Venkatesh, M. P., Placencia, F., and Weisman, L. E. (2006). Coagulase negative staphylococcal infections in the neonate and child: an update. Semin. Pediatr. Infect. Dis. 17, 120–127. doi: 10.1053/j.spid.2006.06.005
Verraes, C., Van Boxstael, S., Van Meervenne, E., Van Coillie, E., Butaye, P., Catry, B., et al. (2013). Antimicrobial resistance in the food chain: A Review. Int. J. Environ. Res. Public Health 10, 2643–2669. doi: 10.3390/ijerph10072643
Vitali, L. A., Petrelli, D., Lamikanra, A., Prenna, M., and Akinkunmi, E. O. (2014). Diversity of antibiotic resistance genes and staphylococcal cassette chromosome mec elements in faecal isolates of coagulase-negative staphylococci from Nigeria. BMC Microbiol. 14:106. doi: 10.1186/1471-2180-14-106
von Wintersdorff, C. J. H., Penders, J., van Niekerk, J. M., Mills, N. D., Majumder, S., van Alphen, L. B., et al. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7:173. doi: 10.3389/fmicb.2016.00173
Vuong, C., and Otto, M. (2002). Staphylococcus epidermidis infections. Microbes Infect. 4, 481–489. doi: 10.1016/S1286-4579(02)01563-0
WHO (2012). World Health Organization The Evolving Threat of Antimicrobial Resistance - Options for Action. Geneva: World Health Organization.
WHO (2013). World Health Organization Integrated Surveillance of Antimicrobial Resistance - Guidance from a WHO Advisory Group. Geneva: World Health Organization.
WHO (2015). 19th WHO Model List of Essential Medicines (April 2015). Geneva: World Health Organization Essential Medicines and Health Products.
Xin, W., Guanghui, L., Xiaodong, X., Baowei, Y., Meili, X., and Jianghong, M. (2014). Antimicrobial susceptibility and molecular typing of methicillin-resistant Staphylococcus aureus in retail foods in shaanxi, China. Foodborne Pathog. Dis. 11, 281–286. doi: 10.1089/fpd.2013.1643
Yurdakul, N. E., Erginkaya, Z., and Ünal, E. (2013). Antibiotic resistance of enterococci, coagulase negative staphylococci and Staphylococcus aureus isolated from chicken meat. Czech J. Food Sci. 31, 14–19.
Zaraket, H., Otsuka, T., Saito, K., Dohmae, S., Takano, T., Higuchi, W., et al. (2007). Molecular characterization of methicillin-resistant Staphylococcus aureus in hospitals in Niigata, Japan: divergence and transmission. Microbiol. Immunol. 51, 171–176. doi: 10.1111/j.1348-0421.2007.tb03898.x
Zarfel, G., Galler, H., Luxner, J., Petternel, C., Reinthaler, F. F., Haas, D., et al. (2014). Multiresistant bacteria isolated from chicken meat in Austria. Int. J. Environ. Res. Public Health 11, 12582–12593. doi: 10.3390/ijerph111212582
Zdolec, N., Dobranić, V., and Filipović, I. (2012a). Occurrence of resistant coagulase-negative staphylococci in meat and dairy products. in Zbornik radova, 5. hrvatski veterinarski kongres, ed I. Harapin (Zagreb: Veterinarski fakultet u Zagrebu, Hrvatska veterinarska komora), 71–76.
Zdolec, N., Dobranić, V., Filipović, I., and Marcincakova, D. (2012b). Antimicrobial resistance of coagulase-negative staphylococci isolated from spontaneously fermented wild boar sausages. Folia Vet. 56(Suppl. 1), 60–62.
Zdolec, N., Dobranić, V., Horvatić, A., and Vučinić, S. (2013a). Selection and application of autochthonous functional starter cultures in traditional Croatian fermented sausages. Int. Food Res. J. 20, 1–6.
Zdolec, N., Račić, I., Vujnović, A., Zdelar-Tuk, M., Matanović, K., Filipović, I., et al. (2013b). Antimicrobial resistance of coagulase-negative staphylococci isolated from spontaneously fermented sausages. Food Technol. Biotechnol. 51, 240–246.
Zhang, K., McClure, J., Elsayed, S., Louie, T., and Conly, J. M. (2008). Novel multiplex PCR assay for simultaneous identification of community-associated methicillin-resistant Staphylococcus aureus strains USA300 and USA400 and detection of mecA and Panton–Valentine leukocidin genes, with discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 46, 1118–1122. doi: 10.1128/JCM.01309-07
Zhang, K., Sparling, J., Chow, B. L., Elsayed, S., Hussain, Z., Church, D. L., et al. (2004). New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 42, 4947–4955. doi: 10.1128/JCM.42.11.4947-4955.2004
Keywords: antimicrobial resistance phenotype, resistance in the quinolone-resistance-determining-regions, biofilm, chicken meat, mecA gene
Citation: Osman K, Badr J, Al-Maary KS, Moussa IMI, Hessain AM, Girah ZMSA, Abo-shama UH, Orabi A and Saad A (2016) Prevalence of the Antibiotic Resistance Genes in Coagulase-Positive-and Negative-Staphylococcus in Chicken Meat Retailed to Consumers. Front. Microbiol. 7:1846. doi: 10.3389/fmicb.2016.01846
Received: 22 August 2016; Accepted: 03 November 2016;
Published: 22 November 2016.
Edited by:
Octavio Luiz Franco, Universidade Católica de Brasília, BrazilReviewed by:
Ildinete Silva-Pereira, University of Brasília, BrazilElizabete De Souza Cândido, Catholic University Dom Bosco, Brazil
Copyright © 2016 Osman, Badr, Al-Maary, Moussa, Hessain, Girah, Abo-shama, Orabi and Saad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kamelia Osman, a2FtZWxpYS1vc21hbkBob3RtYWlsLmNvbQ==
 Kamelia Osman
Kamelia Osman Jihan Badr2
Jihan Badr2 Ihab M. I. Moussa
Ihab M. I. Moussa