- Systems and Synthetic Biology Lab, Department of Cell and Molecular Biology, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, Brazil
Environmental bacteria are endowed with several regulatory systems that have potential applications in biotechnology. In this report, we characterize the arsenic biosensing features of the ars response system from Chromobacterium violaceum in the heterologous host Escherichia coli. We show that the native Pars/arsR system of C. violaceum outperforms the chromosomal ars copy of E. coli when exposed to micromolar concentrations of arsenite. To understand the molecular basis of this phenomenon, we analyzed the interaction between ArsR regulators and their promoter target sites as well as induction of the system at saturating concentrations of the regulators. In vivo titration experiments indicate that ArsR from C. violaceum has stronger binding affinity for its target promoter than the regulator from E. coli does. Additionally, arsenite induction experiments at saturating regulator concentration demonstrates that although the Pars/arsR system from E. coli displays a gradual response to increasing concentration of the inducer, the system from C. violaceum has a steeper response with a stronger promoter induction after a given arsenite threshold. Taken together, these data demonstrate the characterization of a novel arsenic response element from an environmental bacterium with potentially enhanced performance that could be further explored for the construction of an arsenic biosensor.
Introduction
Bacteria that thrive in environments contaminated with toxic compounds are usually endowed with diverse molecular mechanisms related to tolerance to these chemicals (Top and Springael, 2003; Permina et al., 2006). From a physiological point of view, mechanisms for resistance to stressors are usually energy dependent and should be tightly regulated in order to avoid wasting valuable resources when stressors are absent (Nojiri et al., 2004; Diaz et al., 2013). Many bacteria have evolved molecular mechanisms to handle exposure to toxic metals and metalloids. The genes encoding such systems are usually transcriptionally controlled by a plethora of regulatory systems that coordinate gene expression in response to the presence of the cognate chemical.
Several regulatory systems dedicated to the sensing of toxic metals and metalloids have been characterized in bacteria (Busenlehner et al., 2003; Kaur et al., 2006; Permina et al., 2006; Reyes-Caballero et al., 2011). Genomic studies have demonstrated that most of these systems are broadly distributed among different bacterial phyla (Paez-Espino et al., 2009). For instance, transcription factors belonging to the SmtB/ArsR family have been expensively characterized for their role in sensing and controlling gene expression in response to divalent metals (e.g., zinc, nickel, and cadmium) or toxic metalloids (e.g., arsenic and antimonite) (Busenlehner et al., 2003; Eicken et al., 2003; Qin et al., 2007). Members of this family are usually small (~100 aa) regulatory proteins that repress gene expression by blocking access of RNA polymerase to target promoters in the absence of the cognate inducer (VanZile et al., 2000; Kar et al., 2001; Morita et al., 2001; Cavet et al., 2002). These proteins usually act as dimers that bind to target DNA sequences in the apo form. Once in complex with their specific target metal/metalloid, the regulators strongly decrease their affinity for DNA allowing dissociation from the promoter and subsequently, gene expression activation (Kar et al., 1997; Morita et al., 2002, 2003; Eicken et al., 2003; Chauhan et al., 2009).
Understanding the molecular mechanisms behind the transcriptional response to toxic metals and metalloids in bacteria has led to a growing interest in repurposing these systems to construct biosensors for detection of these chemicals in the environment (Stocker et al., 2003; Siddiki et al., 2011; Cortés-Salazar et al., 2013; Chen et al., 2014). Such efforts have been made for the detection of arsenic, a highly abundant and extremely toxic metalloid released to the environment as a result of anthropogenic activity (Matschullat, 2000; Mandal and Suzuki, 2002). In comparison to analytical chemistry methods, biosensors (which encompass a biological sensing component and easily detectable output) would provide reliable, specific, and an inexpensive means for the in situ detection of target compounds in environmental samples (Baeumner, 2003).
Constructed biosensors for environmental purposes have generally coupled well-characterized components (usually from bacteria) that are responsive to the target compound with a reporter gene that gives rise to a colorimetric, luminescent, or fluorescent output. In this sense, developed biosensors proved to be useful tools for arsenite and arsenate detection in groundwater (Siegfried et al., 2012) and river water (Siegfried et al., 2015) as a low cost, suitable and transportable alternative to detect the metalloid to prevent or diminish arsenic exposure. Furthermore, the advent of biological circuit design approaches in the field of synthetic biology has allowed the re-wiring of basic molecular components (regulators, DNA binding sites, and operators), which can be reinserted into the host cell to give rise to sensors with enhanced performance (Stocker et al., 2003; Trang et al., 2005; Fernandez et al., 2016; Merulla and van der Meer, 2016). In fact, the addition of ArsR operator downstream of its target promoter generates reduction in the background noise, which reduces the detection limits to as lower as one microgram per liter (Merulla and van der Meer, 2016).
Although changing the circuit design can improve the efficiency of the biosensor, the utilization of molecular components with intrinsically enhanced transcriptional performance when induced with the target compound could lead to a system with superior behavior. With this reasoning in mind, we aimed to identify a natural system that displayed enhanced transcriptional response to arsenite (AsIII). We focused on the ars system from Chromobacterium violaceum, a gram-negative bacterium with a low arsenic tolerance level but endowed with a fully functional Pars and ArsR regulatory system (Azevedo et al., 2008; Silva-Rocha et al., 2013a). We demonstrate that the ars system from C. violaceum has superior arsenic induction performance when compared to the chromosomal prototype system in Escherichia coli and that these differences can be traced to the binding affinity of ArsR regulators to their DNA targets and to the occurrence of a stronger transcriptional response under inducing conditions in the C. violaceum system. Taken together, the results shown here demonstrate the potential of environmental bacteria as a reservoir of molecular components with enhanced performance for biosensor design, as well as a characterization of a novel arsenic ArsR transcription factor in bacteria.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains, plasmids, and primers used in this study are listed in Table 1. E. coli DH5α cells were used for cloning procedures. E. coli W3110 was used as the wild type strain, whereas E. coli AW3110 (Δars operon) was used as the mutant host for testing the circuits. E. coli strains were grown at 37°C in LB media (Sambrook et al., 1989) or M9 minimal media (6.4 g/L Na2HPO4⋅7H2O, 1.5 g/L KH2PO4, 0.25 g/L NaCl, and 0.5 g/L NH4Cl) supplemented with 2 mM MgSO4, 0.1 mM casamino acid, and 1% glycerol as the sole carbon source. When required, kanamycin (Km, 50 μg/mL) or chloramphenicol (34 μg/mL) was added to the media to ensure plasmid retention. When cells were grown in minimal media, antibiotics were used at half concentrations. For induction experiments, benzoic acid (Sigma–Aldrich, St. Louis, MO, USA) and sodium arsenite (Sigma–Aldrich) were used at different concentrations.
Plasmid and Strain Construction
For analysis of the ars system under negative feedback, DNA fragments containing the Pars/arsR elements from C. violaceum and E. coli were PCR amplified from genomic DNA using primers 5arscvEco/3arscvBam and 5arsecEco/3arsecBam, respectively (Table 1). DNA fragments were amplified using Phusion High-Fidelity DNA polymerase (New England Biolabs, Ipswich, MA, USA) according to the manufacturer’s protocol. The resulting DNA fragments were cloned into the pMR1 reporter vector (Guazzaroni and Silva-Rocha, 2014), which carries the gfplva, a short lived variant of GFP. The resulting recombinant plasmids were named pMR1-Parscvi::ArsRcvi and pMR1-Parseco::ArsReco (Table 1; Supplementary Material). The correct DNA sequence was verified by sequencing using the dideoxy terminal method. Next, the recombinant plasmids were introduced into E. coli W3110 using chemically competent cells (Sambrook et al., 1989). The resulting reporter strains were used for induction experiments. For cloning and analysis of individual promoters, Parscvi and Parseco were PCR amplified using primers 5arscvEco/3ParscvBam and 5arsecEco/3ParsecBam, gel purified, and cloned into pRV2 using EcoRI/BamHI restriction sites. The resulting plasmids, pRV2-Parscvi and pRV2-Parseco, were transformed into E. coli W3110 (wild type) and E. coli AW3110 (mutant) strains. To construct the uncoupled circuit, the arsR genes from each organism were cloned under the control of the XylS/Pm system, which is inducible by benzoate (Blatny et al., 1997). Each regulator was amplified using primers 5arsRcvEco/3arscvBam and 5arsRecEco/3arsecBam (forward primers introduced a strong RBS sequence at the 5′ region of the gene) and resulting fragments were cloned into pSEVA438 (Silva-Rocha et al., 2013b) using EcoRI/BamHI enzymes. The resulting plasmids were named pSEVA438-arsRcvi and pSEVA438-arsReco and co-transformed along with the cognate GFP reporter plasmid (pRV2-Parscvi or pRV2-Parseco) into E. coli AW3110 to generate the reporter strains.
GFP Fluorescence Assay and Data Processing
To measure promoter activity, freshly plated single colonies were grown overnight in LB media, washed, and resuspended in fresh M9 media. Ten microliters of culture was assayed in 96-well microplates in biological triplicate with 170 μL of M9 media supplemented with required antibiotics and different AsIII or benzoate concentrations. Cell growth and GFP fluorescence was quantified using a Victor X3 plate reader (PerkinElmer, Waltham, MA, USA) and calculated as arbitrary units by dividing the fluorescence level by the optical density at 600 nm (reported as GFP/OD600) after background correction. Background signal was evaluated with the same strain harboring the pMR1 (Guazzaroni and Silva-Rocha, 2014) or pRV2 (Silva-Rocha and de Lorenzo, 2011) empty plasmid. Unless otherwise indicated, measurements were taken at 30 min intervals over 8 h. All experiments were performed at least three times. Raw data were processed using ad hoc R script1) and plots were constructed using Microsoft Excel, R, or MeV2.
Results
The Natural ars System from C. violaceum Displays Enhanced Induction by AsIII
In order to characterize the arsenic response system, we initially focused on the response system from C. violaceum, an environmental bacterium with low tolerance to arsenic (Azevedo et al., 2008; Silva-Rocha et al., 2013a). Because this bacterium is sensitive to micromolar doses of AsIII, we reasoned that in the natural environment, this organism would have to trigger a strong transcriptional response to very low concentrations of this metalloid and thus, would be endowed with an intrinsically sensitive ars response system. In order to compare the performance of the ars system from C. violaceum with that of E. coli (the prototype system used for arsenic biosensor construction), we cloned the Pars/arsR elements from both organisms into a GFP reporter vector and introduced the resulting construct into E. coli. This allowed us to faithfully assess the response of the native systems (i.e., retaining the negative feedback loop, Figure 1A) in the same bacterial host.
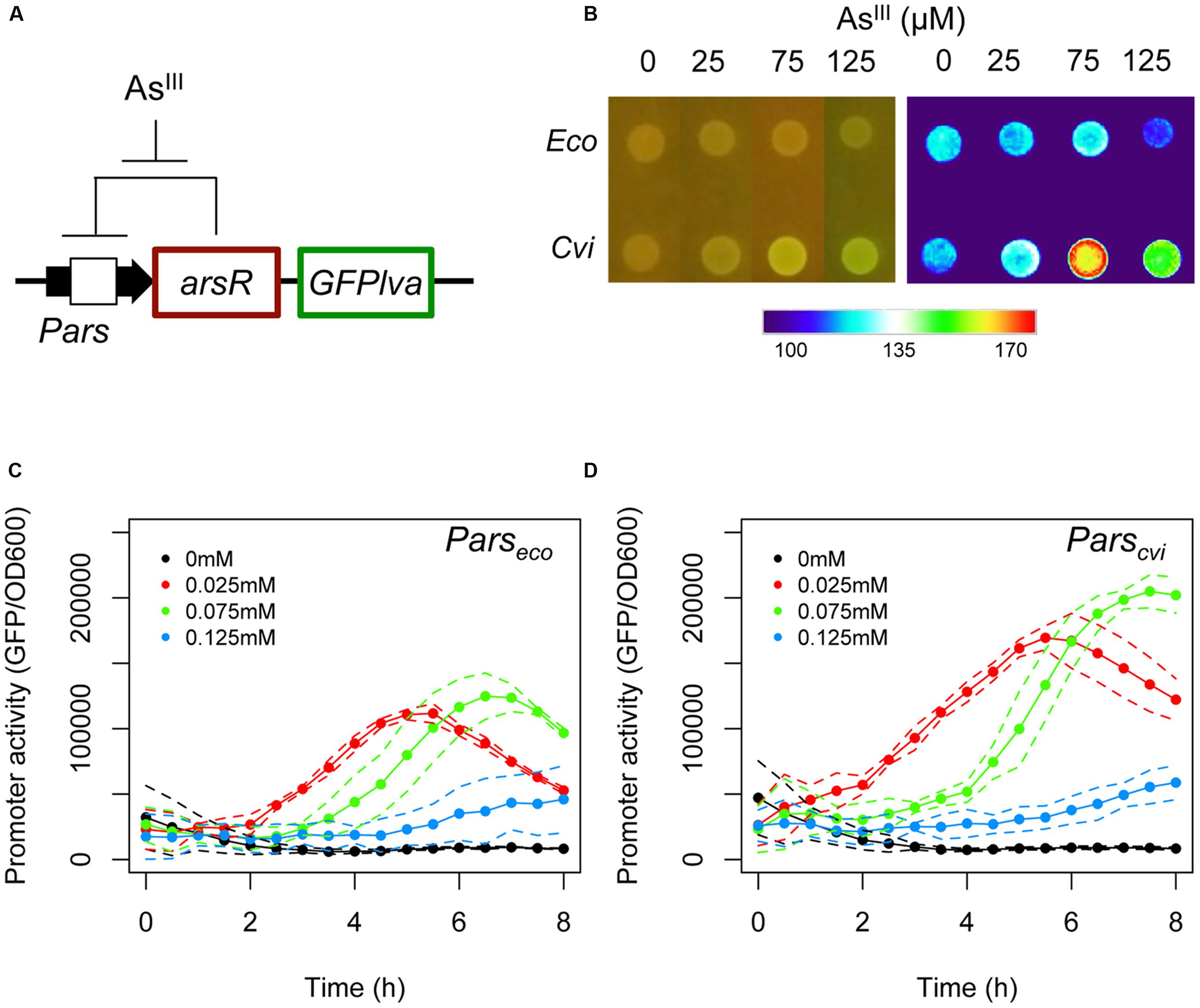
FIGURE 1. Analysis of the natural ars circuits from Escherichia coli and Chromobacterium violaceum. (A) Schematic representation of the feedback circuit controlling GFPlva expression. (B) Induction of the ars system from E. coli and C. violaceum on agar plates using increasing concentrations of AsIII for 6 h. The left panel represents bacterial colonies grown in agar plates and at the right panel the GFP intensities were converted to a false color scale to facilitate the visualization of the differences. (C,D) Induction kinetics of the ars system from E. coli (C) and C. violaceum (D) in wild type (W3110) E. coli in M9 liquid media exposed to increasing concentrations of AsIII. Solid lines represent the average of three independent experiments, whereas dashed lines represents the upper and lower limits of standard deviations.
As seen in Figure 1B, the Pars/arsR system from C. violaceum had a higher GFP output upon exposure to 75 μM of AsIII when assayed on agar plates compared to the E. coli system. In order to analyze the two systems quantitatively, we performed induction experiments in liquid minimal media and quantified GFP production at fixed time intervals. When we compared the performance of the two systems, we observed that the Pars/arsR genes from C. violaceum displayed GFP induction dynamics significantly higher then that from E. coli (Figures 1C,D). It is worth mentioning that the concentration of 125 μM produces a reduced output due to the strong toxicity of arsenite to the strain. This result confirms our hypothesis that the Pars/arsR system from C. violaceum has a more efficient transcriptional response to AsIII. Taking into account the architecture of the ars regulatory elements, the results observed in Figure 1 could be due to differences in three parameters of the two systems. First, the Parscvi promoter could have a stronger intrinsic activity than Parseco. In this scenario, releasing ArsR repression would allow increased promoter activity at Parscvi. Second, ArsRcvi could have stronger binding affinity by its target DNA sequence than ArsReco. If that were the case, the feedback loop would stabilize in lower amounts of ArsR in C. violaceum, which could be easily inactivated by changes in AsIII concentrations, leading to higher promoter output. Finally, ArsRcvi could have higher affinity for AsIII and thus, small changes in concentration of the inducer would lead to increased inactivation of the repressor, resulting in higher promoter activity. In order to investigate these possibilities, we conducted a number of in vivo experiments to quantify the relative promoter strengths, the apparent repressor-promoter binding affinities, and the repressor-effector interactions as described in next sections.
Parscvi and Parseco Have Similar Strengths with Different Kinetics in E. coli
In order to understand the molecular differences leading to the observed behavior of the ars systems from C. violaceum and E. coli, we first compared the relative promoter strengths in the absence of repression. For this experiment, Parscvi and Parseco were cloned upstream of a GFPlva reporter gene and introduced into wild type and ars mutant strains of E. coli. As shown in Figure 2A, both promoters displayed similar maximal activities in wild type and mutant E. coli, indicating that promoter activity alone could not explain the differences observed in Figure 1. When we compared promoter dynamics alongside the growth curve, we observed that Parseco displays high initial activity that tends to stabilize after 2 h of growth, whereas Parscvi activity increases dramatically in the first 4 h and then reaches a similar steady-state level (Figure 2B).
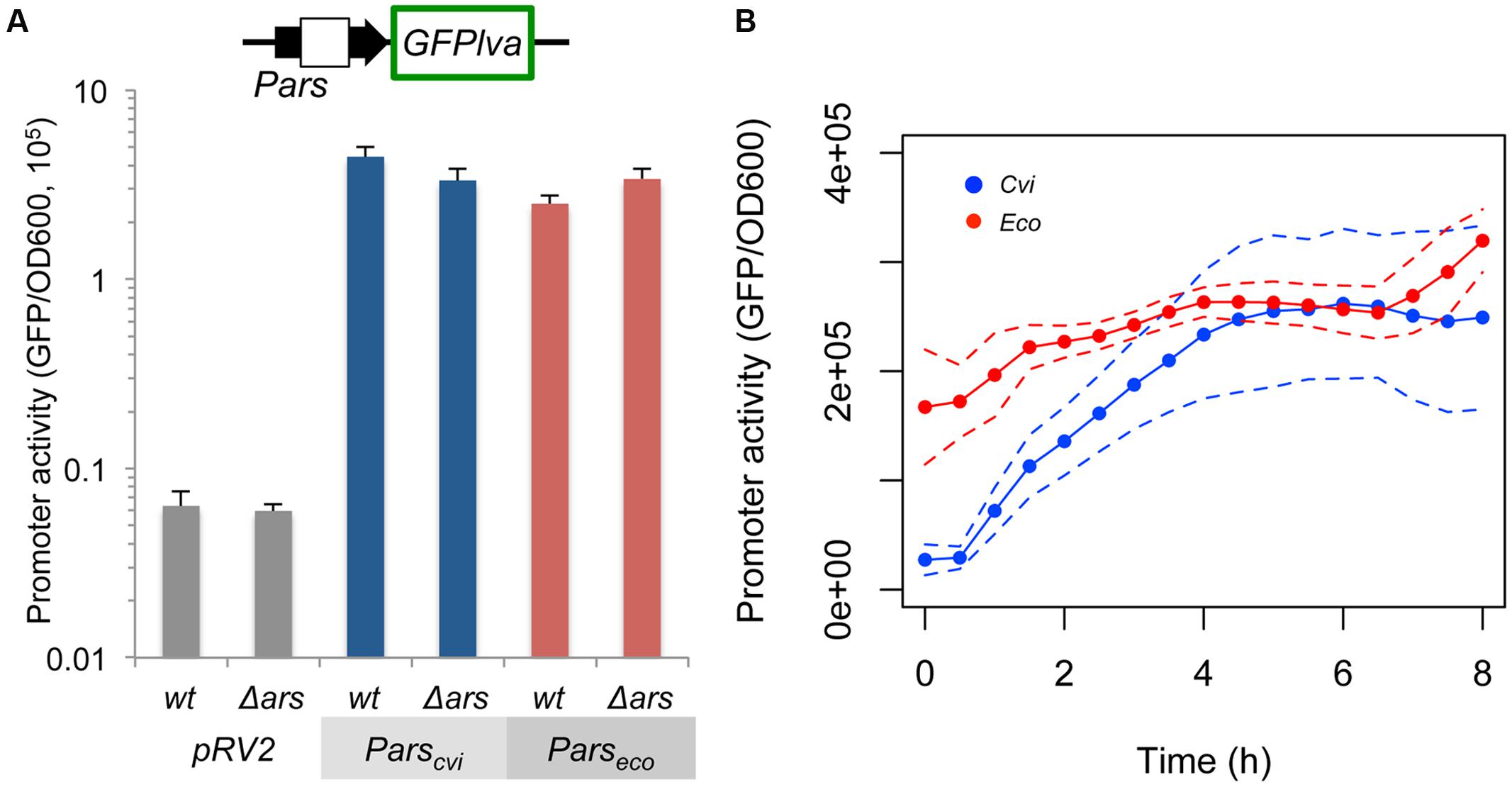
FIGURE 2. Quantification of the intrinsic Parscvi and Parseco activities. Wild type (wt) E. coli or mutant (Δars) strains harboring the reporter plasmids containing the GFPlva gene under the control of the specific promoters were assayed in M9 media in the absence of AsIII. Empty pRV2 plasmid was used as control. (A) Maximal activity of the specific promoter after 6 h of growth. (B) Promoter dynamics during growth of E. coli AW3110 (Δars) harboring pRV2-Parscvi (Cvi, blue) or pRV2-Parseco (Eco, red). Solid lines represent the average of three independent experiments, whereas dashed lines represents the upper and lower limits of standard deviations.
The ArsRcvi Regulator Has a Higher Apparent Affinity for Its Target Promoter
Once we determined the maximal transcriptional activities of both systems, we investigated the effect of increasing concentrations of ArsR on the activity of Parscvi and Parseco promoters. We constructed an uncoupled inducible system (Figure 3A) where ArsR production is under the control of a benzoate inducible system based on the XylS/Pm element (Blatny et al., 1997). The specific Pars promoter is fused to a GFP reporter gene in order to provide a fluorescence output. In this system, the RBS sequences for both regulators are changed to the tir element, thus ensuring similar translation rates for both proteins. Using this setup, we induced the cells with increasing concentrations of benzoate to generate a stepwise decrease in promoter outputs (Figures 3B,C) (Brewster et al., 2014). Assuming that both ArsReco and ArsRcvi are produced from the same transcription and translation signals, the protein level produced should be equivalent and thus, should allow us to indirectly infer the binding affinity of each repressor for its target DNA sequence, similarly to the approach used by Wang and coworkers (Wang et al., 2015). The results are shown in Figures 3D,E, which represents the effect of increasing benzoate concentrations on target promoter activity at 4 and 6 h, respectively. These in vivo titration experiments indicate that Parscvi has a higher decay in activity with increasing benzoate concentration. These results show that ArsRcvi has a higher in vivo affinity for its target DNA. By comparing the benzoate concentration required to reduce promoter activity in each system, we find that approximately five times more inducer is required to reduce Parseco activity than Parscvi activity (62 μM vs. 12 μM).
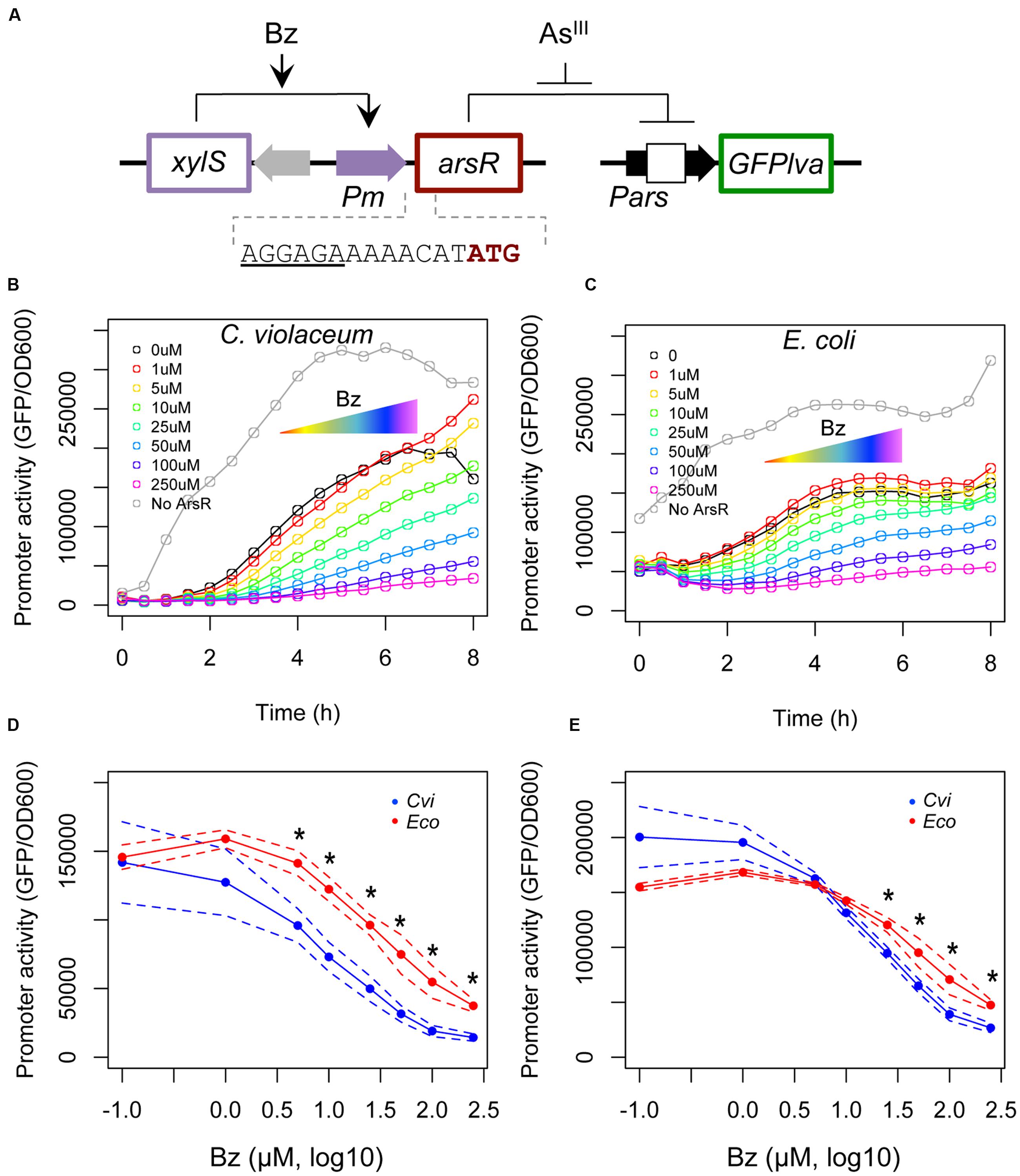
FIGURE 3. In vivo titration experiments using a benzoate inducible system. (A) Schematic representation of the circuit used. In this system, the target regulator is placed under the control of the XylS/Pm expression cassette from pSEVA438 (Silva-Rocha et al., 2013b). To ensure similar levels of ArsR regulators were produced, a strong RBS element was introduced between the Pm promoter and the start codon of the gene. A second plasmid based on pRV2 was used where expression of a GFPlva reporter gene is placed under the control of the target promoter. Promoter activities were assayed during the growth curve analysis using increasing concentrations of benzoate (Bz, from 0 to 250 μM). (B) In vivo titration of ArsR/Pars from C. violaceum. (C) In vivo titration of ArsR/Pars from E. coli. Gray lines represent control strains harboring the empty pSEVA438 vector (i.e., no ArsR production). (D,E) Repression kinetics of Parscvi (blue) and Parseco (red) upon exposure to increasing benzoate concentrations after four (D) or six (E) hours of induction. Solid lines represent the average of three independent experiments, whereas dashed lines represents the upper and lower limits of standard deviations. Statistics differences are highlighted by (∗) as analyzed using Student’s t-test with p-value p < 0.05.
The ars System from C. violaceum Shows Stronger Transcriptional Response to Arsenic
Once we observed that the ArsR repressor from C. violaceum has a higher apparent affinity for its target than the ArsR repressor from E. coli, we analyzed the effect of arsenic binding during allosteric derepression of the system. We used the maximal benzoate concentration (250 μM) to ensure the ArsR concentration was high enough to fully repress Parscvi and Parseco. Using this setup, we exposed the reporter strain harboring the circuits from Figure 3A to increasing concentrations of AsIII. As shown in Figure 4A, under saturating regulator concentrations, the ars system from E. coli is gradually induced with AsIII concentrations above 0.25 μM. However, under similar conditions, the ars system from C. violaceum is insensitive to AsIII concentrations below 1.0 μM (Figure 4B). This differential induction behavior could be observed in the dose-response curve for both systems at 6 h post-induction (Figure 4C) where the ars system from C. violaceum is insensitive to AsIII concentrations below 1.0 μM and displays pronounced induction above this threshold. This remarkable difference between the responses of the systems indicates that the ars from C. violaceum displays steeper induction kinetics, where the system is completely OFF at low concentrations of inducer but displays strong induction upon reaching a certain threshold. In order to test whether this behavior was the result of the high level of ArsRcvi produced (due to the elevated benzoate concentration used), we performed induction experiments in which we varied both benzoate and AsIII concentrations. As shown in Figure 5, the ars system from E. coli displayed a gradual induction by AsIII under varying benzoate concentrations, whereas the induction of the C. violaceum system presented steeper slope even at reduced benzoate concentrations (and thus low ArsRcvi levels). Taken together, these results indicate that the differences in the induction profiles of both systems are the result of intrinsic differences in the way the ArsR regulators interact with AsIII.
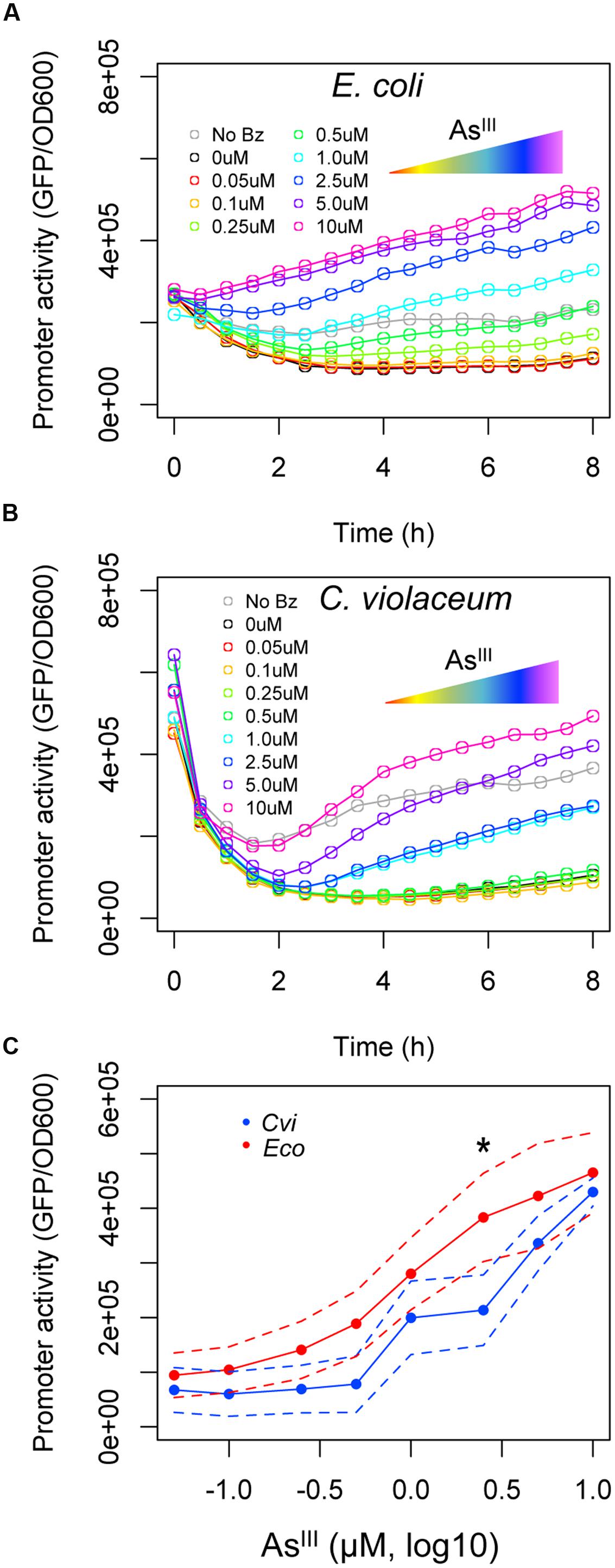
FIGURE 4. Induction of ars systems with increasing concentrations of AsIII. E. coli AW3110 (Δars) harboring the circuits represented in Figure 3A were exposed to a saturating concentration of benzoate (250 μM) and varying concentrations of AsIII (from 0 to 10 μM). (A) The expression profile of the ars system from E. coli induced with increasing concentrations of AsIII. (B) The expression profile of the ars system from C. violaceum induced with increasing concentrations of AsIII. (C) Induction kinetics of Parscvi (blue) and Parseco (red) upon exposure to increasing AsIII concentrations after 6 h of induction. Solid lines represent the average of three independent experiments, whereas dashed lines represents the upper and lower limits of standard deviations. Statistics differences are highlighted by (∗) as analyzed using Student’s t-test with p-value p < 0.05.
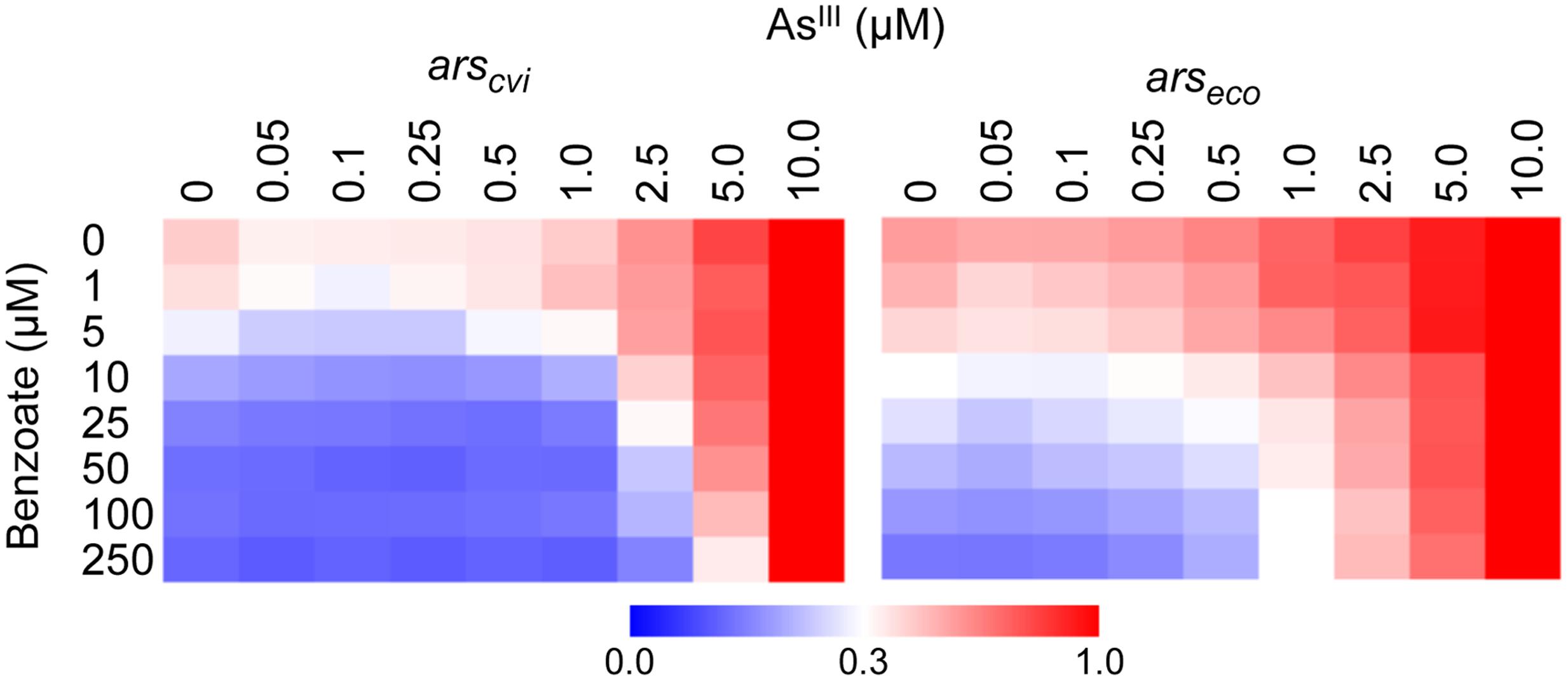
FIGURE 5. Expression dynamics of ars systems from E. coli and C. violaceum under varying concentrations of benzoate and AsIII. E. coli AW3110 (Δars) harboring the circuits represented in Figure 3A were exposed to varying concentrations of benzoate and AsIII and incubated for 6 h to allow induction. Promoter activities (calculated as GFP/OD600) were normalized to the maximal level obtained with 10 μM of AsIII. Data are representative of three independent experiments.
Discussion
Bacteria able to thrive in contaminated environments are endowed with very efficient detoxification mechanisms controlled at the gene expression level. In this context, different metalloregulatory proteins have evolved to control gene expression in response to different metals and metalloids. In vivo functional characterization of the ars system from several bacteria has revealed strong variation in the number of resistance genes, operon organization and, final resistance levels (Carlin et al., 1995; Paez-Espino et al., 2009). However, a conserved feature of the ars system is that the first gene of the operon encodes the ArsR regulator, which repress its own expression in a feedback loop (Wu and Rosen, 1993; Xu and Rosen, 1997). The ArsR from both E. coli and C. violaceum are members of the SmtB/ArsR family. This class of transcription factors include small proteins that share a core secondary structure formed by five alpha helixes and two anti-parallel beta strands in the form ααααββα (Cook et al., 1998; Busenlehner et al., 2003). Although DNA binding occurs through the helix-turn-helix domain formed by helix three and four, members of this family have a more degenerate inducer-binding site that could be located in helix three (Type 1 regulators – ArsReco) or in helix five at the dimerization interface of the protein [Type 2 regulators – ArsRcvi (Qin et al., 2007)]. Considering the metal binding site, ArsR proteins have a conserved metalloid-protein interaction interface, in which cysteine residues are required to coordinate the ligand (Busenlehner et al., 2003). ArsReco has an AsIII binding site formed by Cys residues located at helix three (Wu and Rosen, 1993), whereas ArsRcvi belongs to the class of regulators where binding is formed by Cys residues at helix five (Qin et al., 2007; Azevedo et al., 2008). These differences between the two classes of regulators also imposes differences on the intramolecular allosteric switch upon inducer binding because type 1 regulators have the ligand-binding site close to the DNA binding domain, whereas signal transmission in type 2 regulators must occur from a distance (Qin et al., 2007).
While differences in the localization of metalloid biding site of the regulatory proteins might account for the different expression behaviors observed, other parameters such as protein-DNA interaction might play an important role in the process. For instance, Li et al. (2015) have used in vitro mutagenesis to tune the arsenic response of the ars system from the R773 plasmid of E. coli. By analyzing the variants with enhanced performance, they found several mutations into the ArsR binding site (ABS) at the DNA. This found is particularly relevant since, while members of the SmtB/ArsR family recognize palindromic DNA sequences, the ABS sequence found in the ars promoter of E. coli has an imperfect palindromic sequence (Wu and Rosen, 1993). As the ars systems from E. coli and C. violaceum have no conserved ABS sequence (Carepo et al., 2004; Azevedo et al., 2008), this difference on protein-DNA interface may explain the stronger binding affinity of ArsRcvi and the difference in the dynamics of gene expression. Altogether, these structural differences between the two regulators analyzed here could explain the discrepant behavior (i.e., gradient vs. steeper) during expression of the ars systems.
While the ArsR from E. coli proved to be more sensible to lower arsenic concentrations when assayed in the decoupled system (Figures 4 and 5), the C. violaceum system displayed a better triggering system at higher concentrations, with a steeper slope promoter induction. Although the particular features of the two classes of regulators have not been systematically investigated, different molecular mechanisms for an allosteric switch induced by AsIII binding could be the reason for the different induction profiles observed for ArsR regulators of C. violaceum and E. coli. Uncovering these mechanisms should be a target of future research in order to further understand the evolution of SmtB/ArsR protein family members and for biosensor development.
Microorganisms are a valuable source of molecular components for the construction of biological circuits for biotechnological applications (Cheng and Lu, 2012; Brophy and Voigt, 2014). From a historical perspective, most synthetic circuits constructed in bacteria have been implemented using components from the model organism E. coli (Church et al., 2014). This has indeed been the case for biosensors designed to detect arsenic in environmental samples with several designs having been constructed by shuffling the ars components of E. coli (Siegfried et al., 2012, 2015; Wang et al., 2015; Merulla and van der Meer, 2016). Since the World Health Organization (WHO) recommended acceptable limit for drinking water of 10 μg/L arsenic, any modification of natural arsenic sensing systems for biosensing purposes should aim increasing responsivity of the systems to concentrations close to this limit. The characterization of the ars components of an environmentally relevant bacterium, C. violaceum, shows that orthologous systems dedicated to the same function (in this case, arsenic detoxification from the cell) could have different dynamic properties. In the case of the ars system from C. violaceum, increased ArsR binding affinity by the target DNA sequence together with a stronger induction profile makes these components very attractive for novel designs of arsenic biosensing. Additionally, to the best of our knowledge, this is the first report of a naturally occurring arsenic response system in bacteria with such an atypical induction profile, which provides new venues for the investigation of ars evolution and for the application of this pathway in biosensor design.
Author Contributions
RS-R and LA designed the experimental strategy. LA and LM performed the experiments. LA, LM, and RS-R analyzed and interpreted the data. LA and RS-R wrote the manuscript. All authors have read and approved the final version of the manuscript.
Funding
This work was supported by the Young Investigator Award of Sao Paulo State Foundation (FAPESP, grant number 2012/22921-8 and 2015/12553-0) and by the National Council for Scientific and Technological Development (CNPq, grant number 441833/2014-4).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We thank the lab members and Dr. Maria-Eugenia Guazzaroni for critical discussion of this work.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01851/full#supplementary-material
Footnotes
References
Azevedo, J. S., Silva-Rocha, R., Silva, A., Peixe Carepo, M. S., and Cruz Schneider, M. P. (2008). Gene expression of the arsenic resistance operon in Chromobacterium violaceum ATCC 12472. Can. J. Microbiol. 54, 137–142. doi: 10.1139/w07-123
Bachmann, B. J. (1972). Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev. 36, 525–557.
Baeumner, A. J. (2003). Biosensors for environmental pollutants and food contaminants. Anal. Bioanal. Chem. 377, 434–445. doi: 10.1007/s00216-003-2158-9
Blatny, J. M., Brautaset, T., Winther-Larsen, H. C., Karunakaran, P., and Valla, S. (1997). Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in gram-negative bacteria. Plasmid 38, 35–51. doi: 10.1006/plas.1997.1294
Brewster, R. C., Weinert, F. M., Garcia, H. G., Song, D., Rydenfelt, M., and Phillips, R. (2014). The transcription factor titration effect dictates level of gene expression. Cell 156, 1312–1323. doi: 10.1016/j.cell.2014.02.022
Brophy, J. A., and Voigt, C. A. (2014). Principles of genetic circuit design. Nat. Methods 11, 508–520. doi: 10.1038/nmeth.2926
Busenlehner, L. S., Pennella, M. A., and Giedroc, D. P. (2003). The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27, 131–143. doi: 10.1016/S0168-6445(03)00054-8
Carepo, M. S., Azevedo, J. S., Porto, J. I., Bentes-Sousa, A. R., Batista Jda, S., Silva, A. L., et al. (2004). Identification of Chromobacterium violaceum genes with potential biotechnological application in environmental detoxification. Genet. Mol. Res. 3, 181–194.
Carlin, A., Shi, W., Dey, S., and Rosen, B. P. (1995). The ars operon of Escherichia coli confers arsenical and antimonial resistance. J. Bacteriol. 177, 981–986.
Cavet, J. S., Meng, W., Pennella, M. A., Appelhoff, R. J., Giedroc, D. P., and Robinson, N. J. (2002). A nickel-cobalt-sensing ArsR-SmtB family repressor. Contributions of cytosol and effector binding sites to metal selectivity. J. Biol. Chem. 277, 38441–38448.
Chauhan, S., Kumar, A., Singhal, A., Tyagi, J. S., and Krishna Prasad, H. (2009). CmtR, a cadmium-sensing ArsR-SmtB repressor, cooperatively interacts with multiple operator sites to autorepress its transcription in Mycobacterium tuberculosis. FEBS J. 276, 3428–3439. doi: 10.1111/j.1742-4658.2009.07066.x
Chen, J., Sun, S., Li, C.-Z., Zhu, Y.-G., and Rosen, B. P. (2014). Biosensor for organoarsenical herbicides and growth promoters. Environ. Sci. Technol. 48, 1141–1147. doi: 10.1021/es4038319
Cheng, A. A., and Lu, T. K. (2012). Synthetic biology: an emerging engineering discipline. Annu. Rev. Biomed. Eng. 14, 155–178. doi: 10.1146/annurev-bioeng-071811-150118
Church, G. M., Elowitz, M. B., Smolke, C. D., Voigt, C. A., and Weiss, R. (2014). Realizing the potential of synthetic biology. Nat. Rev. Mol. Cell Biol. 15, 289–294. doi: 10.1038/nrm3767
Cook, W. J., Kar, S. R., Taylor, K. B., and Hall, L. M. (1998). Crystal structure of the cyanobacterial metallothionein repressor SmtB: a model for metalloregulatory proteins. J. Mol. Biol. 275, 337–346. doi: 10.1006/jmbi.1997.1443
Cortés-Salazar, F., Beggah, S., van der Meer, J. R., and Girault, H. H. (2013). Electrochemical As(III) whole-cell based biochip sensor. Biosens. Bioelectron. 47, 237–242. doi: 10.1016/j.bios.2013.03.011
Diaz, E., Jimenez, J. I, and Nogales, J. (2013). Aerobic degradation of aromatic compounds. Curr. Opin. Biotechnol. 24, 431–442. doi: 10.1016/j.copbio.2012.10.010
Eicken, C., Pennella, M. A., Chen, X., Koshlap, K. M., VanZile, M. L., Sacchettini, J. C., et al. (2003). A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins. J. Mol. Biol. 333, 683–695. doi: 10.1016/j.jmb.2003.09.007
Fernandez, M., Morel, B., Ramos, J. L., and Krell, T. (2016). Paralogous regulators ArsR1 and ArsR2 of Pseudomonas putida KT2440 as a basis for arsenic biosensor development. Appl. Environ. Microbiol. 82, 4133–4144. doi: 10.1128/AEM.00606-16
Guazzaroni, M. E., and Silva-Rocha, R. (2014). Expanding the logic of bacterial promoters using engineered overlapping operators for global regulators. ACS Synth. Biol. 3, 666–675. doi: 10.1021/sb500084f
Kar, S. R., Adams, A. C., Lebowitz, J., Taylor, K. B., and Hall, L. M. (1997). The cyanobacterial repressor SmtB is predominantly a dimer and binds two Zn2+ ions per subunit. Biochemistry 36, 15343–15348. doi: 10.1021/bi971690v
Kar, S. R., Lebowitz, J., Blume, S., Taylor, K. B., and Hall, L. M. (2001). SmtB-DNA and protein-protein interactions in the formation of the cyanobacterial metallothionein repression complex: Zn2+ does not dissociate the protein-DNA complex in vitro. Biochemistry 40, 13378–13389. doi: 10.1021/bi0151519
Kaur, A., Pan, M., Meislin, M., Facciotti, M. T., El-Gewely, R., and Baliga, N. S. (2006). A systems view of haloarchaeal strategies to withstand stress from transition metals. Genome Res. 16, 841–854. doi: 10.1101/gr.5189606
Li, L., Liang, J., Hong, W., Zhao, Y., Sun, S., Yang, X., et al. (2015). Evolved bacterial biosensor for arsenite detection in environmental water. Environ. Sci. Technol. 49, 6149–6155. doi: 10.1021/acs.est.5b00832
Mandal, B. K., and Suzuki, K. T. (2002). Arsenic round the world: a review. Talanta 58, 201–235. doi: 10.1016/S0039-9140(02)00268-0
Matschullat, J. (2000). Arsenic in the geosphere–a review. Sci. Total Environ. 249, 297–312. doi: 10.1016/S0048-9697(99)00524-0
Merulla, D., and van der Meer, J. R. (2016). Regulatable and modulable background expression control in prokaryotic synthetic circuits by auxiliary repressor binding sites. ACS Synth. Biol. 5, 36–45. doi: 10.1021/acssynbio.5b00111
Morita, E. H., Wakamatsu, M., and Hayashi, H. (2001). Studies on the molecular mechanism of the interactions between the cyanobacterial transcription factor, SmtB, and its recognition DNA sequences. Nucleic Acids Res. Suppl. 1, 251–252. doi: 10.1093/nass/1.1.251
Morita, E. H., Wakamatsu, M., Kawamoto, S., Nishiyama, Y., and Hayashi, H. (2003). Studies on the protein-DNA complex formation between the cyanobacterial transcription factors, SmtB and its homologues, functioning as zinc-ion sensors and the recognition DNA sequences. Nucleic Acids Res. Suppl. 3, 203–204. doi: 10.1093/nass/3.1.203
Morita, E. H., Wakamatsu, M., Uegaki, K., Yumoto, N., Kyogoku, Y., and Hayashi, H. (2002). Zinc ions inhibit the protein-DNA complex formation between cyanobacterial transcription factor SmtB and its recognition DNA sequences. Plant Cell Physiol. 43, 1254–1258. doi: 10.1093/pcp/pcf140
Nojiri, H., Shintani, M., and Omori, T. (2004). Divergence of mobile genetic elements involved in the distribution of xenobiotic-catabolic capacity. Appl. Microbiol. Biotechnol. 64, 154–174. doi: 10.1007/s00253-003-1509-y
Paez-Espino, D., Tamames, J., de Lorenzo, V., and Canovas, D. (2009). Microbial responses to environmental arsenic. Biometals 22, 117–130. doi: 10.1007/s10534-008-9195-y
Permina, E. A., Kazakov, A. E., Kalinina, O. V., and Gelfand, M. S. (2006). Comparative genomics of regulation of heavy metal resistance in Eubacteria. BMC Microbiol. 6:49. doi: 10.1186/1471-2180-6-49
Qin, J., Fu, H. L., Ye, J., Bencze, K. Z., Stemmler, T. L., Rawlings, D. E., et al. (2007). Convergent evolution of a new arsenic binding site in the ArsR/SmtB family of metalloregulators. J. Biol. Chem. 282, 34346–34355. doi: 10.1074/jbc.M706565200
Reyes-Caballero, H., Campanello, G. C., and Giedroc, D. P. (2011). Metalloregulatory proteins: metal selectivity and allosteric switching. Biophys. Chem. 156, 103–114. doi: 10.1016/j.bpc.2011.03.010
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor.
Siddiki, M. S. R., Kawakami, Y., Ueda, S., and Maeda, I. (2011). Solid phase biosensors for arsenic or cadmium composed of A trans factor and cis element complex. Sensors 11, 10063–10073. doi: 10.3390/s111110063
Siegfried, K., Endes, C., Bhuiyan, A. F., Kuppardt, A., Mattusch, J., van der Meer, J. R., et al. (2012). Field testing of arsenic in groundwater samples of Bangladesh using a test kit based on lyophilized bioreporter bacteria. Environ. Sci. Technol. 46, 3281–3287. doi: 10.1021/es203511k
Siegfried, K., Hahn-Tomer, S., Koelsch, A., Osterwalder, E., Mattusch, J., Staerk, H. J., et al. (2015). Introducing simple detection of bioavailable arsenic at rafaela (Santa Fe Province, Argentina) using the ARSOlux biosensor. Int. J. Environ. Res. Public Health 12, 5465–5482. doi: 10.3390/ijerph120505465
Silva-Rocha, R., de Azevedo, J. S. N., Carepo, M. S. P., Lopes de Souza, R., Silva, A., de Lorenzo, V., et al. (2013a). Vestigialization of arsenic resistance phenotypes/genotypes in Chromobacterium violaceum strains thriving in pristine Brazilian sites. Biocatal. Biotransform. 31, 281–291. doi: 10.3109/10242422.2013.843170
Silva-Rocha, R., and de Lorenzo, V. (2011). Implementing an OR-NOT (ORN) logic gate with components of the SOS regulatory network of Escherichia coli. Mol. Biosyst. 7, 2389–2396. doi: 10.1039/c1mb05094j
Silva-Rocha, R., Martinez-Garcia, E., Calles, B., Chavarria, M., Arce-Rodriguez, A., de Las Heras, A., et al. (2013b). The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 41, D666–D675. doi: 10.1093/nar/gks1119
Stocker, J., Balluch, D., Gsell, M., Harms, H., Feliciano, J., Daunert, S., et al. (2003). Development of a set of simple bacterial biosensors for quantitative and rapid measurements of arsenite and arsenate in potable water. Environ. Sci. Technol. 37, 4743–4750. doi: 10.1021/es034258b
Top, E. M., and Springael, D. (2003). The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr. Opin. Biotechnol. 14, 262–269. doi: 10.1016/S0958-1669(03)00066-1
Trang, P. T. K., Berg, M., Viet, P. H., Mui, N. V., and van der Meer, J. R. (2005). Bacterial bioassay for rapid and accurate analysis of arsenic in highly variable groundwater samples. Environ. Sci. Technol. 39, 7625–7630. doi: 10.1021/es050992e
VanZile, M. L., Cosper, N. J., Scott, R. A., and Giedroc, D. P. (2000). The zinc metalloregulatory protein Synechococcus PCC7942 SmtB binds a single zinc ion per monomer with high affinity in a tetrahedral coordination geometry. Biochemistry 39, 11818–11829. doi: 10.1021/bi001140o
Wang, B., Barahona, M., and Buck, M. (2015). Amplification of small molecule-inducible gene expression via tuning of intracellular receptor densities. Nucleic Acids Res. 43, 1955–1964. doi: 10.1093/nar/gku1388
Wu, J., and Rosen, B. P. (1993). Metalloregulated expression of the ars operon. J. Biol. Chem. 268, 52–58.
Keywords: regulatory network, arsenic response system, cis-regulatory elements, ars operon, ArsR/SmtB family
Citation: Arruda LM, Monteiro LMO and Silva-Rocha R (2016) The Chromobacterium violaceum ArsR Arsenite Repressor Exerts Tighter Control on Its Cognate Promoter Than the Escherichia coli System. Front. Microbiol. 7:1851. doi: 10.3389/fmicb.2016.01851
Received: 29 August 2016; Accepted: 03 November 2016;
Published: 21 November 2016.
Edited by:
Michael Benedik, Texas A&M University, USAReviewed by:
Jan Roelof Van Der Meer, University of Lausanne, SwitzerlandJun-Jie Zhang, Indiana University School of Medicine, USA
Copyright © 2016 Arruda, Monteiro and Silva-Rocha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rafael Silva-Rocha, c2lsdmFyb2NoYXJAZ21haWwuY29t
 Letícia M. Arruda
Letícia M. Arruda Lummy M. O. Monteiro
Lummy M. O. Monteiro Rafael Silva-Rocha
Rafael Silva-Rocha