- 1Department of Biology, The Gandhigram Rural Institute – Deemed University, Gandhigram, India
- 2Biodiversity Research Center, Academia Sinica, Taipei, Taiwan
This study focuses on isolation, screening, and characterization of novel probiotics from gastrointestinal tract of free-range chicken (Gallus gallus domesticus). Fifty seven colonies were isolated and three isolates (FR4, FR9, and FR12) were selected and identified as Lactobacillus gasseri FR4, Bacillus tequilensis FR9, and L. animalis FR12 by 16S rRNA sequencing. Three strains were able to survive in stimulated acidic and bile conditions and inhibit the growth of pathogens. Especially, FR9 exhibited maximum inhibition against Listeria monocytogenes and none of them exhibited hemolytic activity. Native-PAGE revealed the presence of low molecular weight (3.4–5.0 KDa) antimicrobial peptide. The peptide was further purified by Sephadex G-50 column and RP-HPLC using C18 column. N-terminal amino acid sequencing of antimicrobial peptide showed 100% consensus to antilisterial peptide Subtilosin A and SboA gene was amplified from FR9 genome. FR9 showed maximum aggregation activity, exopolysaccharide production (85.46 mg/L) and cholesterol assimilation (63.12 ± 0.05 μg/mL). Strong adhesion property (12.6%) and pathogen invasion protection ability was revealed by B. tequilensis FR9 towards HCT-116 human colon carcinoma cell line. This is the first study to demonstrate antilisterial Subtilosin A production of B. tequilensis. Our results indicate that B. tequilensis FR9 strain furnish the essential characteristics of a potential probiotics and might be incorporated into human and animal food supplements.
Introduction
Probiotic microorganisms are live bacteria, which modulates the host immune system and maintain the intestinal microbial balance (Joint FAO/WHO, 2002). Majority of the probiotic bacteria belongs to the genus Lactobacillus and Bifidobacterium, and considered as “Generally Recognized As Safe” (GRAS) by the United States Food and Drug Administration (FDA; Rubio et al., 2014). However, recent researches demonstrated that several Bacillus species also furnish the essential probiotic characteristic and produce group of antimicrobial peptides with broader inhibition spectrum (Khochamit et al., 2015). Probiotic Lactobacillus and Bacillus strains isolated from diverse sources are used as probiotic candidates and it is unlikely that each species/strain possesses all the preferred characteristics that will make it a felicitous probiotic (Swain et al., 2014). A potential probiotic bacteria must fulfill certain fundamental criteria, such as ability to adhere on intestinal epithelium, to overcome potential obstruction, including low pH of the stomach, presence of bile acids in intestines, compete/antagonize enteric pathogens, susceptibility to commonly used antibiotics, cholesterol lowering effect and colonize the intestinal cell wall (Garriga et al., 1998; Chiang and Pan, 2012; Anandharaj et al., 2015). Probiotic microbes also synthesis various biologically active compounds including enzymes, bacteriocins, exopolysaccharides (EPS), vitamins, and organic acids and these compounds has immense industrial applications (Nel et al., 2001; Walling et al., 2005).
Contrast to the gram-positive lactic acid bacteria (LAB), Bacillus spp. produce diverse antimicrobial compounds (i.e., bacteriocins), exhibiting broader inhibition spectra against various food borne pathogens (Abriouel et al., 2011; Khochamit et al., 2015). Antimicrobial peptides from various Bacillus spp. such as B. subtilis, B. megaterium, B. cereus, B. coagulans, B. thuringiensis have been studied intensively, however, the bacteriocins of B. tequilensis has not yet been studied. In this work, we have demonstrated the Subtilosin A production by B. tequilensis FR9. Subtilosin A is an unique cyclic bacteriocin with three intra molecular bridges produced by B. tequilensis and expressed by sboA-albABCDEFG gene cluster, which is responsible for the antibacterial activity against the common food borne pathogen Listeria monocytogenes (Huang et al., 2009; Fluhe et al., 2012; Khochamit et al., 2015). Moreover, several probiotics have been reported for their EPS production which enhances probiotic colonization by cell-cell interaction in gastrointestinal tract (GIT; Kanmani et al., 2013) and exhibits antimicrobial, immunostimulatory, antioxidant, and antitumor activities (Pan and Mei, 2010; Zhang et al., 2013; Ibarburu et al., 2015). Abundance of EPS around the cells greatly influences bacterial aggregation (Wang et al., 2014; Dertli et al., 2015), cell surface hydrophobicity (Collado et al., 2008), biofilm formation, thus competitively excludes adhesion of pathogens on intestinal cell lines (Collado et al., 2008; Walter et al., 2008). Previously, pathogen invasion protective ability of various LAB strains were demonstrated against Salmonella typhimurium (Golowczyc et al., 2011), Klebsiella pneumonia, and Pseudomonas aeruginosa (Khan and Kang, 2016).
Free-range chickens are mainly fed household food/conventional chicken feeds devoid of antibiotics making them susceptible to a variety of infections which may be controlled by endogenous probiotic microbial communities. The selection and screening processes for strain isolation from broiler chickens have been extensively studied, whereas characterization of probiotic microorganisms isolated from free-range chicken is scarce (Garriga et al., 1998; Musikasang et al., 2009; Taheri et al., 2009). Hence, in this study we intend to isolate the potential probiotic bacteria from GIT of free-range chicken (Gallus gallus domesticus) and investigate their probiotic potential and antimicrobial activity against several food borne pathogens. Further, we have also purified the antimicrobial peptide and demonstrated their pathogen invasion protection ability against L. monocytogenes and Enterococcus faecalis to verify its intestinal barrier function.
Materials and Methods
Ethical Statement
Free-range Chickens used in this study were purchased from local farmers. Since the free-range chickens are wild animals, the ethical review process is not needed for this study. However, the chickens were sacrificed according to the guidelines of Committee for the Purpose of Control and Supervision of Experimentation on Animals (CPCSEA), India.
Bacterial Isolation and Identification
Five healthy, 20-days old indigenous free-range chickens were sacrificed by cervical dislocation. Contents of GIT (i.e., crop, gizzard, small intestine, large intestine, and caeca) were washed with 70% ethanol followed by sterile H2O. The contents were plated on De Man Rogosa and Sharpe (MRS) agar (HiMedia, India), supplemented with 0.02% bromocresol purple and incubated at 37°C for 36 h. Totally, 57 colonies were isolated from the higher dilution plates and 12 isolates displayed clear halo zone around the colonies confirming the lactic acid production. To further scrutinize the selected isolates, we performed acid and bile tolerance assay. Among 12 isolates, FR4, FR9, and FR12 exhibited more resistance to tested pH, bile concentration and was used for further study. FR4 and FR12 isolates were identified as Lactobacillus spp. and FR9 isolate was identified as Bacillus sp.. To identify the species level, 16S rRNA sequencing was performed. Obtained sequences were assembled using BioEdit software (7.00) and identified using nucleotide Basic Local Alignment Search Tool (BLASTn). Phylogenetic relationship was confirmed by neighbor-joining phylogenetic tree.
Evaluation of Probiotic Properties under Stimulated Conditions
Acid Tolerance
Tolerance to acidic conditions was examined by inoculating overnight grown bacterial cells in pH adjusted MRS broth (1.0, 2.0, 3.0, and control 7.0) and incubated for 3 h at 37 ± 2°C. Viable cell count was determined by plating on MRS agar and incubated for 24 h at 37 ± 2°C. The cell count was expressed as log value of colony-forming units per mL (log CFU/mL). Survival percentage was calculated as follows: % survival = final (log CFU/mL)/control (log CFU/mL) × 100.
Bile Tolerance
Bile tolerance ability of bacterial strains was determined by using MRS broth supplemented with (0.3 or 0.5% w/v) bile salts (Oxgall, Merck) and MRS without bile salts served as control. Overnight grown bacterial cells (A600 = 1.5) were harvested, washed with distilled H2O, resuspended in MRS broth and incubated at 37 ± 2°C. One mL sample was withdrawn after 3 and 5 h, subsequently plated onto MRS agar. Bacterial viability was assessed as mentioned in the previous experiment.
Antibiotic Resistance Profile
Antibiotic susceptibility was examined using the method recommended by Clinical and Laboratory Standards Institute (CLSI). Eleven different antibiotic disks (HiMedia, India) such as, Ampicillin (10 μg), Amoxicillin (5 μg), Cefuroxime (30 μg), Chloramphenicol (30 μg), Erythromycin (20 μg), Kanamycin (30 μg), Gentamicin (10 μg), Metronidazole (20 μg), Penicillin (30 μg), Tetracycline (20 μg), and Novobiocin (30 μg) were used. MRS agar plates were seeded with bacterial strains and incubated according to National committee for clinical laboratory standards (NCCLS) guidelines for Kirby Bauer test (NCCLS, 1997). Breakpoints for the interpretation of inhibition zone were expressed in terms of resistance (++), Intermediate susceptible (+), and susceptible (-) as described by CLSI (2011).
Antimicrobial Activity
Antimicrobial activity was performed using agar well-diffusion method (Anandharaj et al., 2015). Cell-free culture supernatants (CFCS) were collected by centrifugation (10,000 × g, 4°C, 20 min) and filtered through 0.22 μm membrane filter (Millipore, USA). To demonstrate the antimicrobial activity, 10 μL test pathogens (107–109 CFU/mL) were spread onto agar plates, 100 μL pH neutralized CFCS (pH 6.5) was added to each well and incubated (Supplementary Table S1). Antimicrobial activity was expressed as Arbitrary unit (AU) using following formula (Abbasiliasi et al., 2014).
Where, Lz = clear zone area (mm2), Ls = well area (mm2), V = volume of sample (mL).
Purification of Bacteriocin
Partial Purification
Partial purification of bacteriocin (PPB) was performed according to Khochamit et al. (2015). Bacteriocin was precipitated with 40% (w/v) ammonium sulfate (AS), collected by centrifugation (10000 × g for 20 min, 4°C) and dissolved in phosphate buffered saline (PBS). Dialysis was performed against same buffer using 1 KDa cutoff membrane (16 h, 4°C). Protein concentration was measured by Bradford method (Protein Assay Kit, BioRad) and antagonistic activity was determined.
Purification
Partial purification of bacteriocin was purified using Sephadex G-50 (Sigma, USA) column connected with AKTA prime plus (GE healthcare, Uppsala, Sweden) protein purification system equilibrated with 10 mM Phosphate buffer (pH 7). Three milliliter PPB was loaded on to pre-equilibrated column and elution was performed at the flow rate of 1 mL/min. The elution was observed at 280 nm using UV detector. Fractions showed peaks were collected (3 mL) and antimicrobial activity was analyzed. Active fractions were pooled together and further purified using Reverse-phase (RP) HPLC coupled with C18 column. Briefly, 20 μL of purified fractions were injected into the column and eluted at the flow rate of 1 mL/min using solvent A (60% methanol) and solvent B (40% water) for 45 min. Protein concentration was monitored at 220 and 284 nm. Fractions were collected manually and antimicrobial activity was assessed.
Characterization of Bacteriocin
SDS-PAGE and Native-PAGE
The molecular weight mass of purified bacteriocin was determined by SDS-PAGE (5% stacking and 12.5% resolving) stained with coomassie brilliant blue R250. Native-PAGE (5% stacking and 12.5% resolving) was performed without heating the protein. Subsequently, the gel was overlaid on nutrient agar plate coated with L. monocytogenes and incubated for 24 h at 37 ± 2°C.
Amino Acid Sequencing
To determine the amino acid sequence of bacteriocin, single band with antimicrobial activity from SDS-PAGE was transferred to polyvinylidene fluoride (PVDF) membrane and excised for N-terminal sequencing using Edman degradation method. The resulted sequence was identified using protein BLAST.
Fourier Transform Infra-Red (FTIR) Analysis
Surface functional groups of purified BLIS were unraveled by Perkin-Elmer infrared spectrophotometer (India). Purified BLIS was mixed with KBr (spectroscopic grade) and pellet was prepared with the size of about 10–13 mm diameter and 1 mm thickness. Sample was scanned in transmission mode with a resolution of 4 cm-1 at 4000–400 cm-1 range and functional groups were compared with previously published literature (Coates, 2000; Ajesh et al., 2013).
Molecular Identification of Bacteriocin Gene
To identify the specific bacteriocin gene, primers were designed (Table 1) for five putative Bacillus sp. antimicrobial genes (cerein, Subtilosin A, thuricin H, lichenicidin, and ericins) and was amplified from FR9 genomic DNA.
Evaluation of Safety and Functional Properties
Hemolytic and β-Glucosidase Activity
Probiotic strains were streaked on nutrient agar plates supplemented with 5% (w/v) sterile defibrinated sheep blood and incubated for 48 h at 37 ± 2°C. Hemolytic activity was observed post partial hydrolysis and formation of green hued zones (α hemolysis), clear zone of hydrolysis (β hemolysis), or no zone around the colonies (γ hemolysis). The β-glucosidase activity was determined using p-nitrophenyl-D-glucose (pNPG) as substrate. Briefly, the culture supernatant was mixed with 50 mM pNPG, 50 mM citrate buffer, 15 mM CaCl2, and incubated at 40°C for 10 min. Absorbance was measured at 410 nm and p-nitrophenol release was calculated using standards.
Bacterial Aggregation Activity
Autoaggregation
Autoaggregation was performed as described in Collado et al. (2008). Briefly, 107 cells/mL of probiotic cells were harvested, washed with PBS (pH 7.2) and resuspended in same buffer. Consecutively, bacterial suspensions were incubated at 37 ± 2°C and monitored at different time intervals (0, 1, 2, 3, 4, and 5 h). The percentage of autoaggregation was expressed as: A% = (A0-At)/A0∗100
Where, A0 represents the absorbance (A600 nm) at 0 h and At represents the absorbance at different time intervals.
Coaggregation
Coaggregation was demonstrated as described in Collado et al. (2008). Probiotic cell suspensions were prepared as illustrated in autoaggregation analysis. Equal volumes (500 μL) of various probiotic and pathogenic (L. monocytogenes, Escherichia coli, and Enterococcus faecalis) cell suspensions were mixed and incubated at room temperature. Absorbance (A600 nm) of above mixtures as well as individual bacterial suspensions was monitored during incubation. Coaggregation was calculated as:
Where, Apat and Aprobio represents absorbance at A600 nm of individual bacterial suspensions in control tubes, Amix represents the absorbance of mixed bacterial suspension at different time tested.
Microbial Adhesion to Hydrocarbons (MATH)
Microbial Adhesion to Hydrocarbons was determined as described in Collado et al. (2008). Probiotic cells were harvested, washed and resuspended in 3 mL of PBS. A600 was adjusted to 0.2–0.3 to obtain 107 CFU/mL of bacteria (A0). One mL hydrocarbons (xylene, n-hexadecane and toluene) were added with cell suspension to form a two-phase system. After pre-incubation (10 min), the two-phase system was vortexed (2 min), incubated (20 min), and A600 of aqueous phase was measured (A1). Percentage of MATH was calculated according to the following equation.
Screening for EPS Production
Exopolysaccharide production was examined by streaking the probiotic strains on MRS agar supplemented with 2 or 4% (w/v) glucose/lactose/sucrose. After incubation (24–48 h at 37 ± 2°C), plates were observed for the appearance of colonies exhibiting a mucoid feature. The production of biopolymer was confirmed by mixing a scrap of mucoid substance in 2 mL of absolute alcohol additionally by observing the mucoid substance in scanning electron microscopy (SEM).
Qualitative Determination of Bile Salt Hydrolase (BSH)
Bile salt deconjugation ability was studied as described by Kumar et al. (2012). Bacterial cells were plated on MRS-Thio agar fortified with 0.5% (w/v) taurodeoxycholate (TDC) and 0.37 g of CaCl2/L preceded by incubation at 30 ± 2°C for 24–48 h. MRS-Thio agar medium plates without TDC served as control.
Cholesterol Assimilation Assay
Water-soluble cholesterol (polyoxyethanyl-cholesteryl sebacate, Sigma, USA) was dissolved in 50% ethanol (5 mg/mL), filter sterilized and added to MRS-Thio broth supplemented with 0.3% ox-bile at a final concentration of 50–200 μg/mL. Medium was inoculated (1% v/v) with probiotic strains and incubated at 37 ± 2°C for 20 h. Subsequently, cells were harvested (10,000 × g at 48°C for 10 min) and remaining cholesterol concentration in broth was determined using the method of Rudel and Morris (1973). Cholesterol assimilated by probiotic strains was determined as follows:
Where, C1 and C2 represent the cholesterol concentration of the uninoculated and inoculated medium, respectively; W1 and W2 represent the weight of culture per milliliter of medium before and after the incubation period.
HCT-116 Cell Line Growth Condition
Adhesion assay and pathogen invasion protection by B. tequilensis FR9 was demonstrated using human colon carcinoma cell line HCT-116. The HCT-116 cells were cultured in Roswell Park Memorial Institute (RPMI)1640 medium supplemented with 25 mM HEPES buffer, 25 mM sodium bicarbonate, 300 mM L-glutamate, and 10% heat inactivated fetal bovine serum at 37°C in 5% CO2 and 95% air. Cells were passaged every 2 days and experiments were conducted after 20–25 passages of cell line (undifferentiated state).
Adhesion Assay on HCT-116 Cell Line
In vitro adhesion of B. tequilensis FR9 on HCT-116 human colon carcinoma epithelial cells was conducted as described by Das and Goyal (2014) with some modifications. To obtain HCT-116 monolayers, each well of the tissue culture plates (six well) were seeded with 4 × 104 cells per cm2 and incubated at 37°C for 24 h. RPMI-1640 medium was aspirated out after reaching 80% confluency, and cells were washed with PBS (pH 7.4) buffer. Simultaneously, overnight grown B. tequilensis FR9 cells were harvested by centrifugation, washed with PBS buffer and resuspended in RPMI-1640 medium (devoid of antibiotic and serum) at 1 × 108 CFU/mL concentration. To perform the adhesion assay, 1 mL bacterial suspension was added to HCT-116 monolayers and incubated in humidified CO2 incubator (5% CO2) for 4 h at 37°C. Non-adhered cells were removed by PBS buffer wash (five times). To determine the percentage of adhesion, cells were lysed by sterile distilled water and plated on nutrient agar. Adhesion rate was calculated using the ratio between number of inoculated cells and number of colonies observed on nutrient agar plates. For the microscopic observation, HCT-116 cells along with adhered probiotic bacteria were fixed with 3 mL methanol and incubated at room temperature for 10 min. Methanol was removed completely and cells were stained with 0.1 % crystal violet (10 min), washed with ethanol to remove excess stain and examined under light microscope using bright field (BF) and Differential interference contrast (DIC) with 100× magnification (oil immersion).
Pathogen Invasion Protection Assay
The pathogen invasion protection ability of FR9 strain was demonstrated using human colon cancer cell line HCT-116 according to Khan and Kang (2016) and Golowczyc et al. (2011). HCT-116 cells were seeded in cell culture plates (4 × 104 cell per cm2) to form monolayer. Then the monolayer of HCT-116 (post-confluence stage) was washed with sterile PBS (pH 7.4) and pre-incubated with 0.5 mL of well grown B. tequilensis FR9 (2 × 108 CFU/mL in PBS) for 1 h at 37 ± 2°C in a 5% CO2 to 95% air atmosphere. After the incubation, monolayer along with probiotic microbes was washed twice with sterile PBS to remove the unbound microbes. Subsequently, 0.5 mL (2 × 108 CFU/mL in PBS) bacterial pathogens (L. monocytogenes MTCC 657 and E. faecalis MTCC 439) as well as 0.5 mL RPMI-1640 media (Gibco, USA) were added to the plate and incubated for 1 h at 37 ± 2°C in a 5% CO2 to 95% air atmosphere. HCT-116 monolayer along with probiotics and test pathogens were washed twice with PBS and lysed by sterile distilled water. Finally, CFU/mL of invaded pathogen was counted on nutrient agar plates.
Scanning Electron Microscopic (SEM) Analysis
Scanning Electron Microscopic was performed to identify the EPS production as well as binding of cholesterol onto the bacterial cell surface. EPS producing cells or cholesterol treated cells were harvested by centrifugation (12,000 × g for 15 min at 4°C), resuspended in PBS buffer (pH 7.0), and air dried at 25°C to remove the moisture. Then the dried bacterial cells were mounted on the gold-coated SEM specimen stub, subsequently observed under the SEM (VEGA 3.0 TE Scan, USA).
Statistical Analysis
All experiments were carried out in triplicates and the mean ± standard deviations (SD) values are represented in tables and figures. Significant differences between the samples were calculated by one way ANOVA with significant level P < 0.05. SPSS 13.0 (SPSS Inc., Chicago, IL, USA) and OriginPro 9.0 (MicroCal Software, Northampton, MA, USA) software’s were used for data analysis and graphical representation, respectively.
Results and Discussion
Screening and Identification of Bacterial Isolates
Totally, 57 isolates (i.e., 11-crop, 8-gizzard, 7-small intestine, 13-large intestine, and 18-caeca) were isolated from the higher dilutions plates. Among them, 12 isolates exhibited clear yellow halo zone around colonies on MRS-bromocresol purple plates. The selected 12 isolates were tested for their basic probiotic characteristics, such as acid and bile tolerance. The isolates FR4, FR9, and FR12 demonstrated higher resistance to the challenged acidic and bile conditions, hence selected for further characterization (Supplementary Table S2). FR4 and FR12 strains were identified as Lactobacillus spp. and FR9 was identified as Bacillus sp. (Hammes and Vogel, 1995). Lactobacillus strains were gram-positive rods, catalase negative and non-spore forming. Bacillus strain was gram-positive, spore forming rod, and catalase positive (Supplementary Figures S1 and S2). Results of morphological, biochemical, and fermentative characterizations are represented in Supplementary Tables S2 and S3.
Bacterial strains were identified by 16S rRNA sequencing and BLASTn analysis was performed. Lactobacillus FR4 revealed 100% identity to Lactobacillus gasseri. Similarly, Lactobacillus FR12 revealed 99% identity to Lactobacillus animalis, while Bacillus FR9 revealed 99% identity to Bacillus tequilensis. These three 16S rRNA gene sequences were deposited at NCBI GenBank under accession numbers KU587452, KU587453, and KU587454, respectively. Phylogenetic relationships are represented in Supplementary Figure S3. This is the first report to isolate and identify B. tequilensis FR9 from GIT of free-range chicken.
Tolerance to Acidic pH
Acid tolerance is generally considered as an essential assessment criterion for probiotic evaluation, since the strains have to survive the acidic conditions of stomach and small intestine (Anandharaj et al., 2015). The viable cell counts were found to be <1.5 log CFU/mL after 3 h of exposure to pH 3, signifying their resistance and exhibiting survival of 82.22% (L. gasseri FR4), 82.86% (B. tequilensis FR9), and 80.91% (L. animalis FR12; Table 2). Among them, B. tequilensis FR9 exhibited more resistance to all tested acidic levels after 3 h. Similar kind of acid tolerance was observed in various Lactobacillus and Bacillus strains (Ahn et al., 2002; Beasley et al., 2004; Messaoudi et al., 2012; Khochamit et al., 2015; Nguyen et al., 2015).
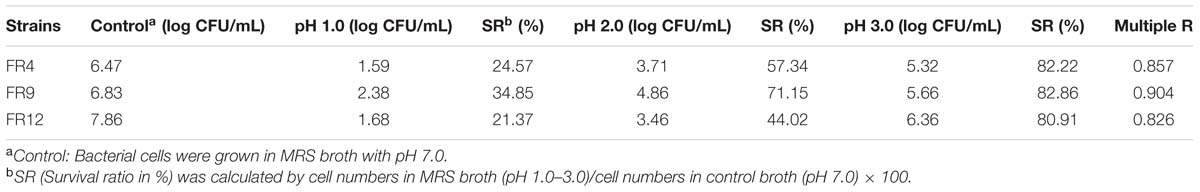
TABLE 2. Effect of pH on the viability of FR4, FR9, and FR12 strains, incubated at various pH range (7, 1, 2, and 3), values are expressed as log CFU/mL, survival percentage and regression coefficient.
Tolerance to Bile salts
Tolerance to bile salts is considered as a vital characteristic for colonization, metabolic activity of the strains in host gut, maintaining the equilibrium of gut microflora as well as lowering the host’s serum cholesterol (Anandharaj et al., 2014). The mean intestinal bile concentration is around 0.3% (w/v; Prasad et al., 1998), hence we challenged our probiotic strains with 0.3 and 0.5% bile concentration. All of the strains showed varying degrees of resistance to bile salts after 5 h exposure (Table 3). L. gasseri FR4, B. tequilensis FR9, and L. animalis FR12 strains were highly tolerant to 0.3% bile and exhibited slight reduction at 0.5% bile. In comparison, FR4 was found to be the least bile tolerant while FR9 was the most resistant strain. Our results are in agreement with the previously published works, in which the Lactobacillus and Bacillus strains retained their viability even after 5 h exposure to 0.3–0.5% bile concentration (Ahn et al., 2002; Messaoudi et al., 2012; Anandharaj et al., 2015).
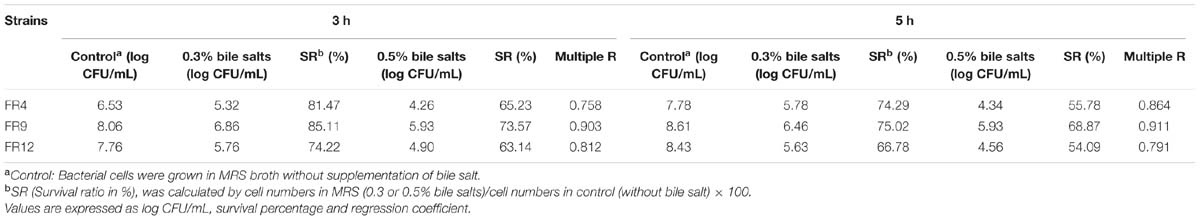
TABLE 3. Effect of bile salt on the viability of FR4, FR9 and FR12 strains, incubated at various bile salt concentrations (0.3% to 0.5%).
Antibiotic Resistance Profile
Bacteria predestined for probiotic use should be screened for antibiotic resistance to prevent any potential transfer of undesirable antibiotic resistance into the intestinal niche (Anandharaj and Sivasankari, 2014). Hence, probiotics strains were subjected to antibiotic susceptibility tests using eleven antibiotics. They were sensitive to 30 μg kanamycin and 30 μg novobiocin. However, FR4 and FR9 showed resistance to 50 μg amoxicillin, 30 μg chloramphenicol, and 10 μg gentamicin. B. tequilensis FR9 strain showed resistance to most of the antibiotics except kanamycin and novobiocin (Supplementary Table S4). Antibiotic resistance of these bacterial strains is considered intrinsic; hence it is non-transmissible (Argyri et al., 2013). Coetzee (2015) reported that B. tequilensis 5A2 was resistant to penicillin, tetracycline, streptomycin, and trimethoprim. Antibiotic resistance of Lactobacillus and Bacillus strains were also reported by other authors (Messaoudi et al., 2012; Anandharaj and Sivasankari, 2014; Nguyen et al., 2015).
Determination of Antimicrobial Spectrum
Antagonist activity of probiotic bacteria facilitates the maintenance of gut microbiota balance and other physiological functions such as reduction of inflammatory bowel disease or colorectal cancer (Heilpern and Szilagyi, 2008). All three probiotic strains were able to inhibit growth of all Gram-positive, negative, and pathogenic yeast with varying extent (Table 4). Inhibition was not observed in E. faecalis by L. animalis FR12 whereas B. tequilensis FR9 strain showed broadest antimicrobial spectrum against all tested pathogens with the maximum inhibitory effect on L. monocytogenes (3250 AU/mL). Similar kind of results has been reported previously by various authors (Ahn et al., 2002; Strompfova and Laukova, 2007; Argyri et al., 2013; Anandharaj and Sivasankari, 2014; Khochamit et al., 2015). This is the first study reporting a broad spectrum antimicrobial activity of B. tequilensis FR9. Antagonism of Lactobacillus and Bacillus sp. is mainly due to the production of antimicrobial proteins and chemical compounds synthesized by secondary metabolic pathways (Anandharaj and Sivasankari, 2014).
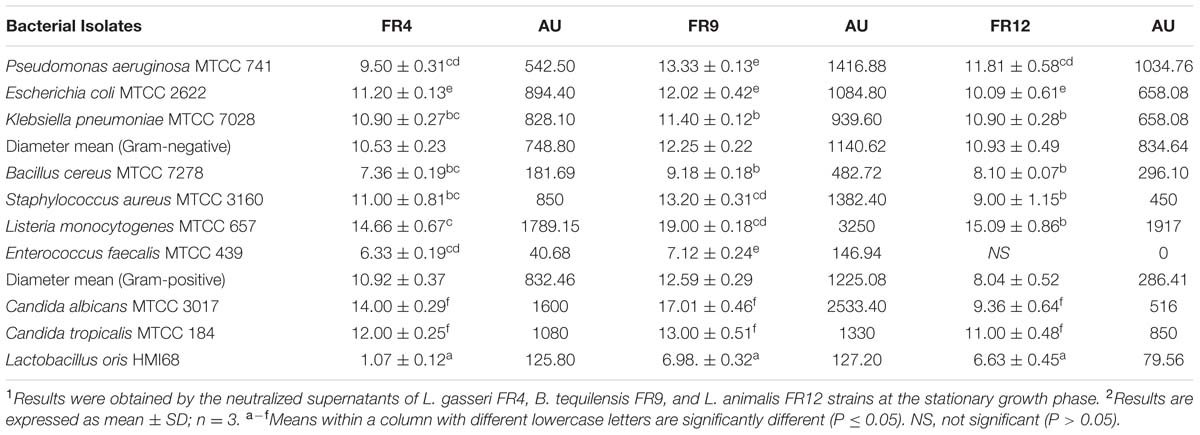
TABLE 4. Antimicrobial activity of cell free culture supernatants (CFCS) of L. gasseri FR4, B. tequilensis FR9, and L. animalis FR12 strains against various pathogens (inhibition zone in mm ± standard deviation).
Antimicrobial Spectrum of FR9-PPB
Due to the hyper activity against L. monocytogenes, further characterization was performed. Antagonistic activity of FR9 PPB increased upto 1.5-fold compared to its CFCS, which indicates that the potency of bacteriocin was enhanced after partial purification (Supplementary Table S5). Percentage of difference in inhibition zone size was maximum (in terms of AU/ml) for L. monocytogenes (51.69%) and minimum for E. faecalis (10.73%).
Purification of B. tequilensis FR9 Bacteriocin
The FR9-PPB was purified using Sephadex G50 column, six fractions were collected (Figure 1A) and their antimicrobial activity was determined. The fractions F2 and F3 showed activity. They were pooled together and further purified using semi-preparative RP-HPLC. A single distinctive peak was observed at retention time 13.81 min (Figure 1B). Molecular weight of purified fraction was determined by SDS and Native-PAGE. SDS-PAGE of PPB showed major band around 4.3 KDa (Lane 1), but a single band was observed at 4.3 KDa in RP-HPLC purified fraction (Lane 2), indicating that the bacteriocin was a low molecular weight protein (Figure 2A). These results were further supported by overlaid native-PAGE, which showed an inhibition of L. monocytogenes growth in the range of around 3.4–5 KDa (Figure 2B). Amino acid sequencing results revealed 100% consensus to Subtilosin A of B. subtilis and B. vallismortis (Figure 3A). This is the first report to identify the Subtilosin A from B. tequilensis. Similarly, Khochamit et al. (2015) also purified the antilisterial peptide from B. subtilis KKU213 and identified as Subtilosin A. Zheng et al. (1999) demonstrated the antilisterial activity of Subtilosin A from B. subtilis. Moreover, bacteriocins of Bacillus spp. have been used as natural preservatives in food, as substitutes to conventional antimicrobials against human and animal diseases (Sass et al., 2008).
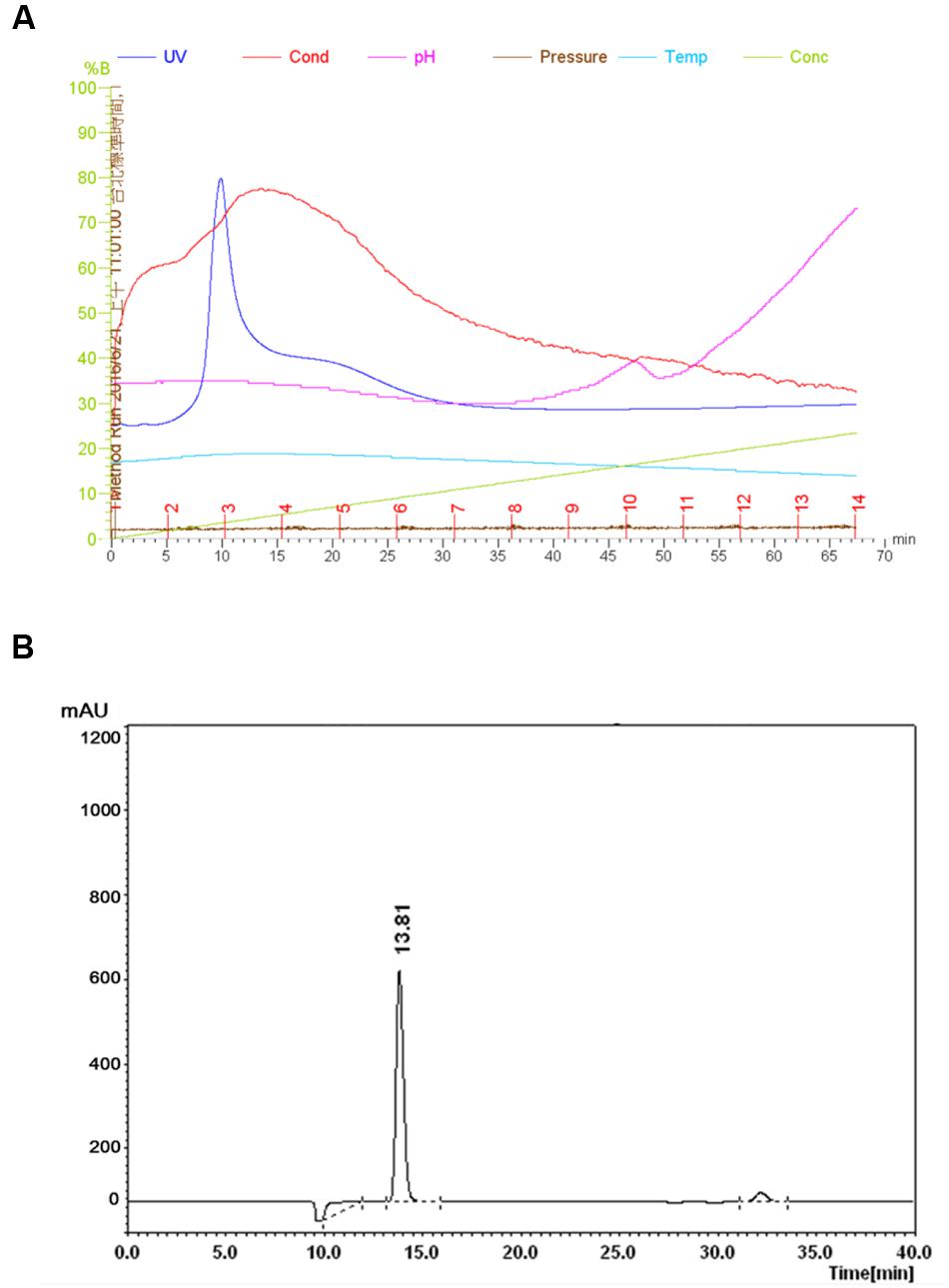
FIGURE 1. Purification of Subtilosin A. Sephadex G-50 chromatogram of Subtilosin A (A). Reverse-phase (RP)-HPLC chromatogram of Subtilosin A (B).
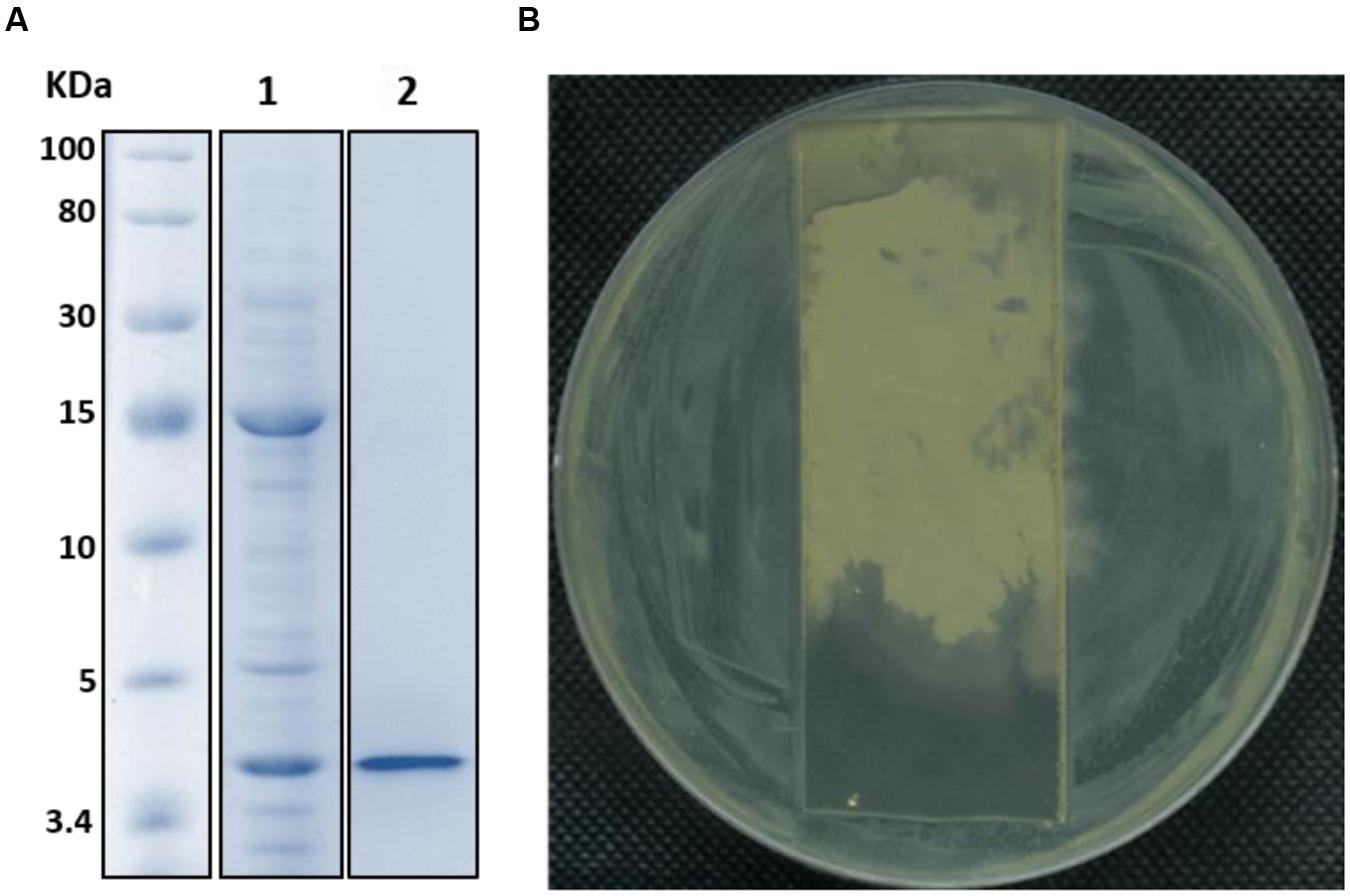
FIGURE 2. SDS-PAGE (A) and Native-PAGE (B) analysis of partially purified (Lane 1) and purified Subtilosin A (Lane 2).
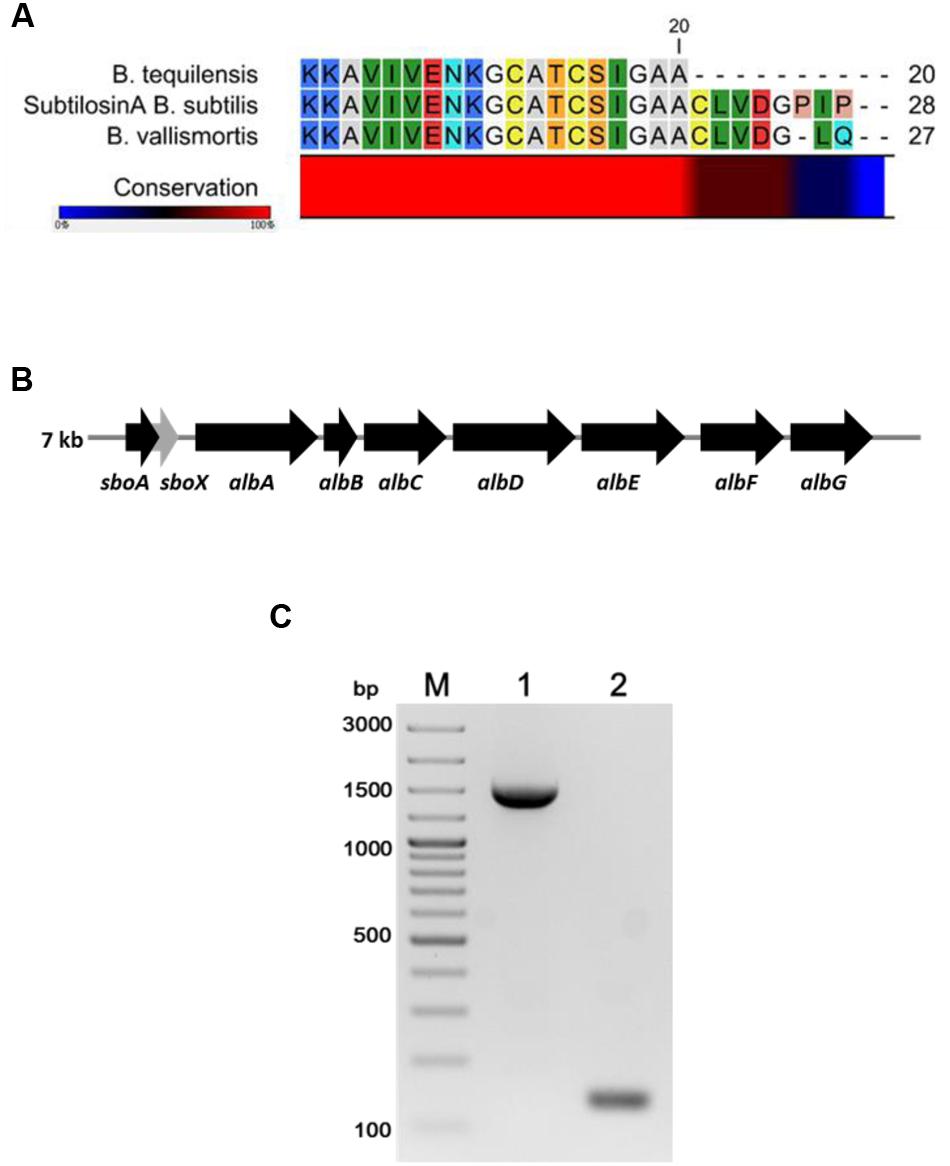
FIGURE 3. Multiple sequence alignment of N-terminal amino acid sequence of B. tequilensis FR9 Subtilosin A with Sequence of B. subtilis and B. vallismortis (A). Organization of structural genes in SboAX-albABCDEFG operon (7.0 kb) (B). PCR amplification of albA (Lane 1) and SboA (Lane 2) from B. tequilensis FR9 genome (C).
Amplification of Subtilosin A Gene
Gene specific primers (Table 1) were designed to amplify the Subtilosin A gene (SboA) and subtilosin maturase gene (albA) from SboAX-albABCDEFG operon to confirm the presence of entire gene cluster in B. tequilensis FR9 genome (Figures 3B,C). The amplified sboA (132 bp) and albA (1347 bp) genes were confirmed by sequencing and multiple sequence alignment showed 100% similarity to Subtilosin A gene cluster of B. subtilis.
FTIR Analysis of Subtilosin A
Fourier Transform Infra-Red is a reliable technique to identify the functional groups and thus unravel the putative mode of action of a bacteriocin (Motta et al., 2008). The FTIR spectra of Subtilosin A showed peaks at 1540, 1644, and 3233 cm-1 which confirms the existence of peptide bonds in the sample (Figure 4). The intense peak at 1644 cm-1 confirms the presence of primary amino group (amide I) with C = O stretch (Ajesh et al., 2013). Similarly, another intense peak at 1181 cm-1 revealed the presence of secondary amino group (amide II) having C-N stretch. Peaks at 2753 and 3534 cm-1 were due to the presence of N-CH3 (methylamino group) and dimeric OH stretch, respectively. Finally, the peak at 3136 cm-1 confirmed aromatic C-H stretch (Coates, 2000).
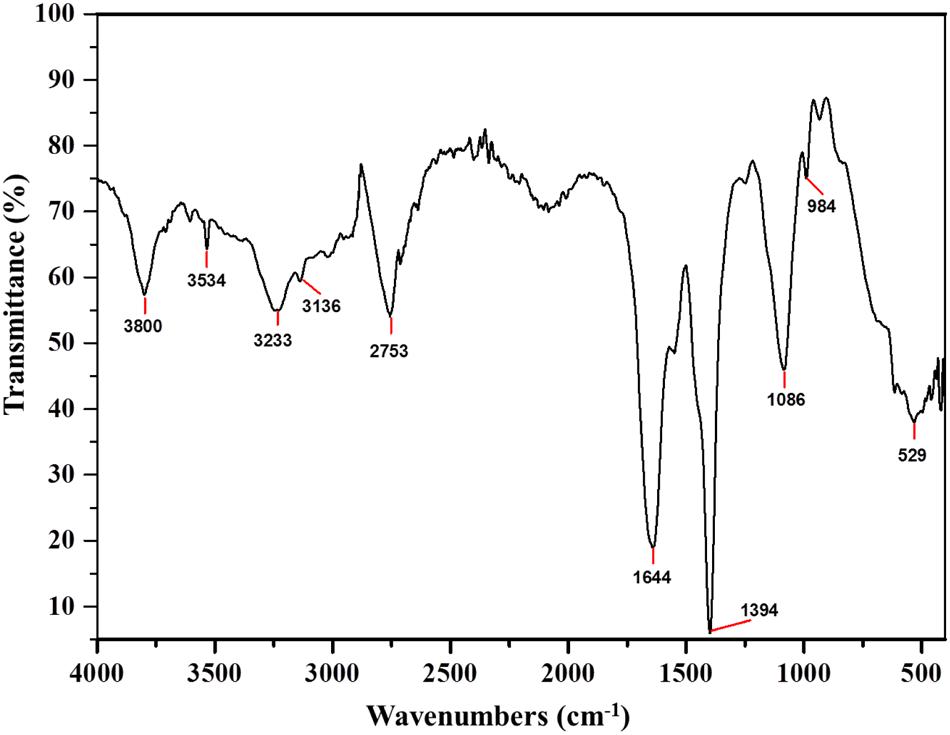
FIGURE 4. Fourier Transform Infra-Red (FTIR) absorption spectra of Subtilosin A produced by B. tequilensis FR9.
Hemolytic Activity Assay
In vitro assessment of hemolytic activity for probiotics is one of the safety necessities used to assess potential probiotic strains (Joint FAO/WHO, 2002). All three strains exhibited γ hemolytic activity (i.e., no hemolysis) which further proves that these isolates are safe and reliable to use based on FDA, USA. Our results are supported by Luis-Villaseñor et al. (2011) in which B. tequilensis YC5-2 exhibited γ hemolytic activity and thus confirmed that, this isolate does not cause any health hazard.
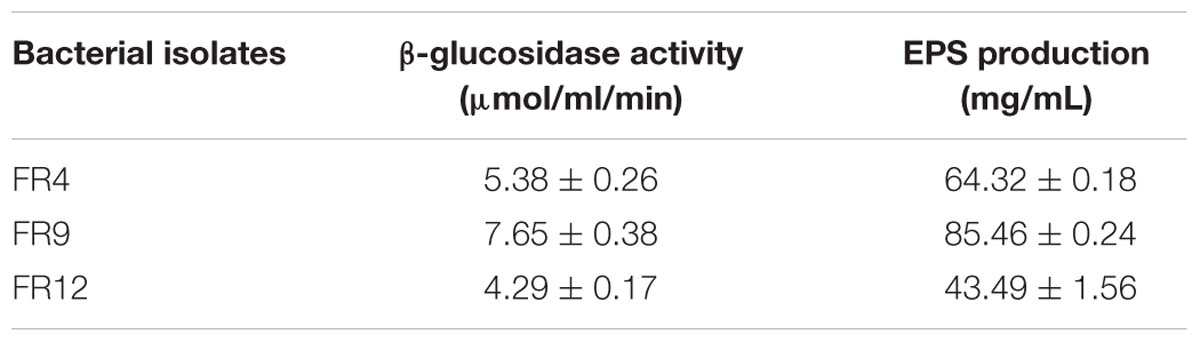
β-Glucosidase Activity
The β-galactosidase production is a common characteristic of several Lactobacillus and Bacillus species. In our study, all three strains produced β-galactosidase and B. tequilensis FR9 exhibited highest activity among others (7.65 ± 0.38 μmol/ml/min; Table 5). Our results are in agreement with the previous report of Palaniswamy and Govindaswamy (2016).
Aggregation Activity
In vitro evaluation of autoaggregation and coaggregation ability with potential enteric pathogens could be used for primary screening and selection of best probiotic strains (Jankovic et al., 2012). Increased autoaggregation plays an important role in the adhesion of bacterial cells on intestinal epithelium and thus maintain the bacterial load in GIT (Del Re et al., 2000). Our results showed that all three strains displayed autoaggregation ability. Among them, B. tequilensis FR9 exhibited a strong autoaggregation (78%), followed by L. gasseri FR4 63% and L. animalis FR12 (45%; Figure 5). Similar effects were previously observed by various authors (Kos et al., 2003; Orłowski and Bielecka, 2006; Anandharaj et al., 2015).
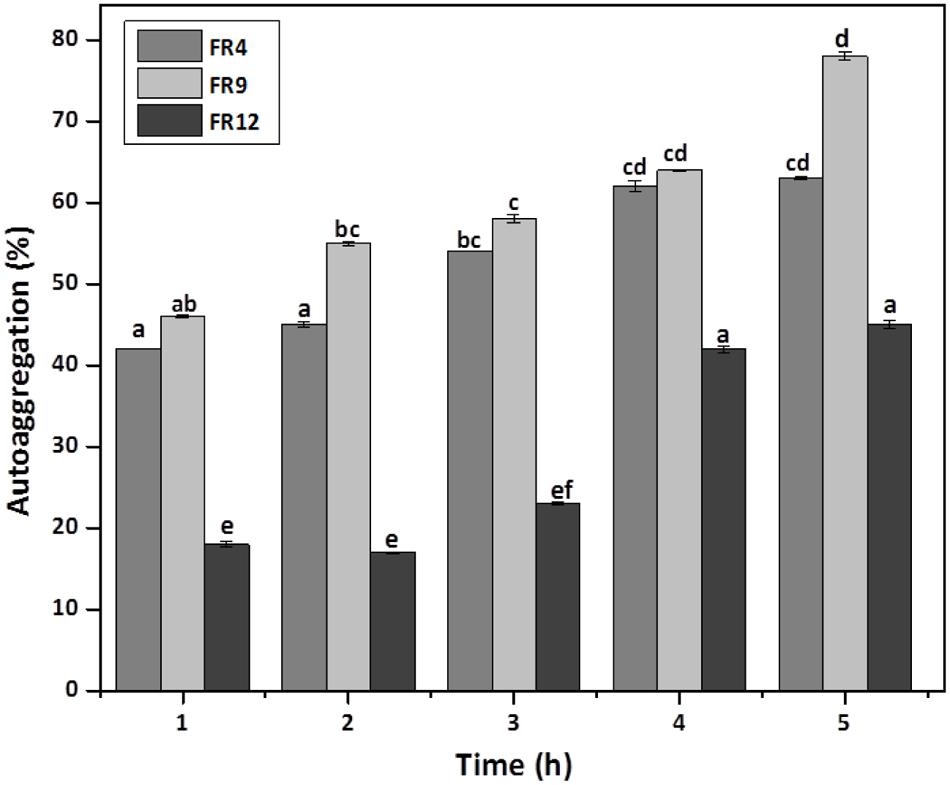
FIGURE 5. Autoaggregation percentage of probiotic strains. Values are expressed as mean ± SD (n = 3). The each bars with no common letters are significantly different (P < 0.05) according to the least significant difference (LSD) mean comparison test.
All three strains demonstrated coaggregation with L. monocytogenes ranging from 26 to 45% (Figure 6). B. tequilensis FR9 exhibited maximum coaggregation with L. monocytogenes (45%) followed by L. animalis FR12 (28%) while minimum coaggregation was recorded in L. gasseri FR4 strain (26%) after 5 h incubation. Bacillus sp. possessing ability to coaggregate with numerous pathogens is of unique interest with regard to its potential applications. Ability of Bacillus and Lactobacillus sp. to coaggregate with pathogens was observed by other authors (Collado et al., 2008; Anandharaj et al., 2015; Palaniswamy and Govindaswamy, 2016).
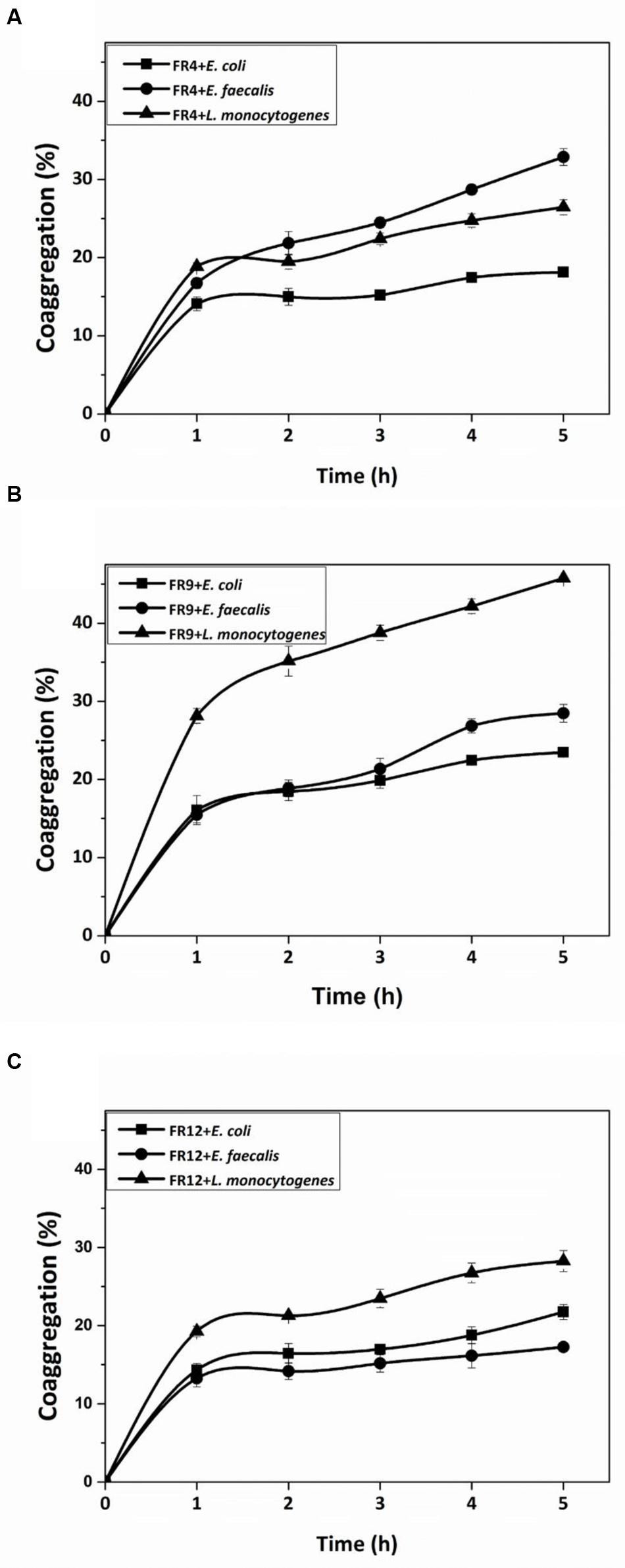
FIGURE 6. Coaggregation percentages of L. gasseri FR4 (A), B. tequilensis FR9 (B), and L. animalis FR12 (C) strains with three different pathogens (E. coli, L. monocytogenes, and E. faecalis). Values are expressed as mean ± SD (n = 3).
Cell Surface Hydrophobicity
The high cell surface hydrophobicity of probiotic strains could indicate the ability to attach on intestinal epithelial cells thereby resist the digestive tract movement (Yu et al., 2007). Significant differences (P < 0.05) in hydrophobicity values were found among the probiotic strains. All strains showed greater hydrophobicity toward hexadecane and lowest toward xylene (Figure 7). Hydrophobicity for hexadecane was observed ranging from 74 to 94%. Similar hydrophobicity was observed by Torshizi et al. (2008) in L. rhamnosus (93.53%). The higher affinity toward hexadecane (apolar solvent) might be due to hydrophobic cell surface of the strains. Studies on physicochemistry of microbial cell surface has previously revealed that presence of glycoproteinaceous substance at the cell surface would lead to higher hydrophobicity (Kos et al., 2003).
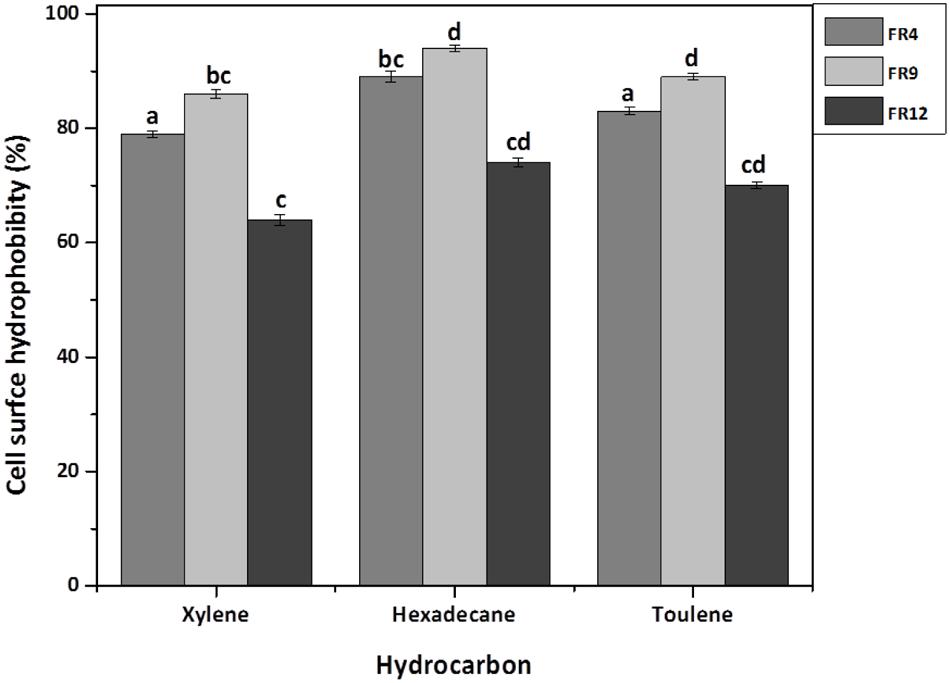
FIGURE 7. Cell surface hydrophobicity [Microbial Adhesion to Hydrocarbons (MATH)] of probiotic using Xylene, Hexadecane, and Toluene. Values are expressed as mean ± SD (n = 3). Each bars with no common letters are significantly different (P < 0.05) according to the LSD mean comparison test.
Screening for EPS Production
Exopolysaccharide is a major component present in cell surface of probiotic microorganisms. It has an extensive impact on their surface characteristics and acts as a protective barrier against harmful conditions (Anandharaj et al., 2015). In this study, B. tequilensis FR9 exhibited mucoid colony pattern in 4% sucrose supplemented MRS medium. Maximum EPS production was observed in B. tequilensis FR9 (85.46 mg/L; Table 5). Figure 8B provides strong evidence for the presence of EPS on FR9 cell surface. Our study reveals a positive correlation with the work done by Dertli et al. (2015), in which the cell surface of L. johnsonii FI9785 was covered by homo-polymeric EPS-1 and hetero-polymeric EPS-2. It has been reported that EPS may contribute to the aggregation properties as well as cell surface hydrophobicity of probiotic microorganism (Deepika et al., 2009).
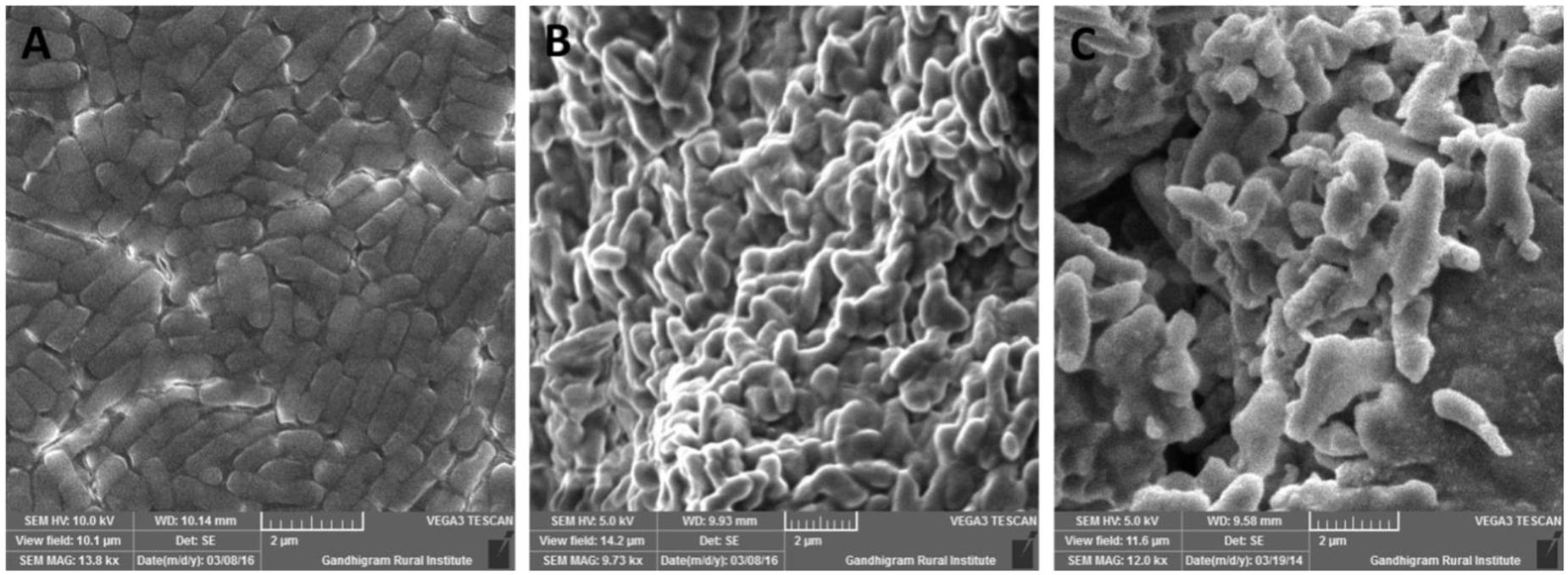
FIGURE 8. Scanning electron microscopy (SEM) micrograph of B. tequilensis FR9 grown in (A) MRS broth and (B) MRS broth with Sucrose for exopolysaccharide (EPS) production (C) MRS broth containing cholesterol fermented at 37°C for 20 h.
BSH Assay
Deconjugation of bile salts could prompt the reduction in serum cholesterol level (Begley et al., 2006). BSH activity was tested for all three selected strains by qualitative direct plate assay and all of them were able to hydrolyse TDC to precipitate deoxycholate on MRS-TDC agar. However, B. tequilensis FR9 was noted to display well defined silvery shine aspect.
Cholesterol Assimilation Assay
All three strains exhibited higher cholesterol reduction ranging from 59.12 to 63.12 μg/mL in MRS broth. Among them, maximal (P ≤ 0.05) reduction was observed in B. tequilensis FR9 (63.12 μg/ml) whilst the minimum in L. animalis FR12 (59.12 μg/mL; Figure 9). Maximum cholesterol assimilation was observed with 50 μg/mL supplementation and thus illustrates that cholesterol concentration in media influences the cholesterol reduction. Through this study, it was observed that B. tequilensis FR9 has the innate ability to assimilate cholesterol which could play a pivotal role in controlling serum cholesterol levels. Furthermore, adherence of cholesterol on the surface of B. tequilensis FR9 was analyzed by SEM, which confirms the cholesterol assimilation mechanism (Figures 8A,C). This is the first study to report cholesterol assimilation of B. tequilensis FR9. Our results are in agreement with the work done by Lin and Chenn (2000) in which cholesterol-reducing abilities of L. acidophilus was investigated and found that hypocholesterolemic ability is due to the assimilation of cholesterol as well as its attachment to the surface. Anandharaj and Sivasankari (2014) reported that cholesterol assimilation by L. oris HMI68 ranged from 43.04 to 61.05 μg/mL after 24 h incubation. Similarly, Ramasamy et al. (2010) reported that L. salivarius I29 isolated from chicken intestine removed cholesterol (50.16%) from growth media (P < 0.005).
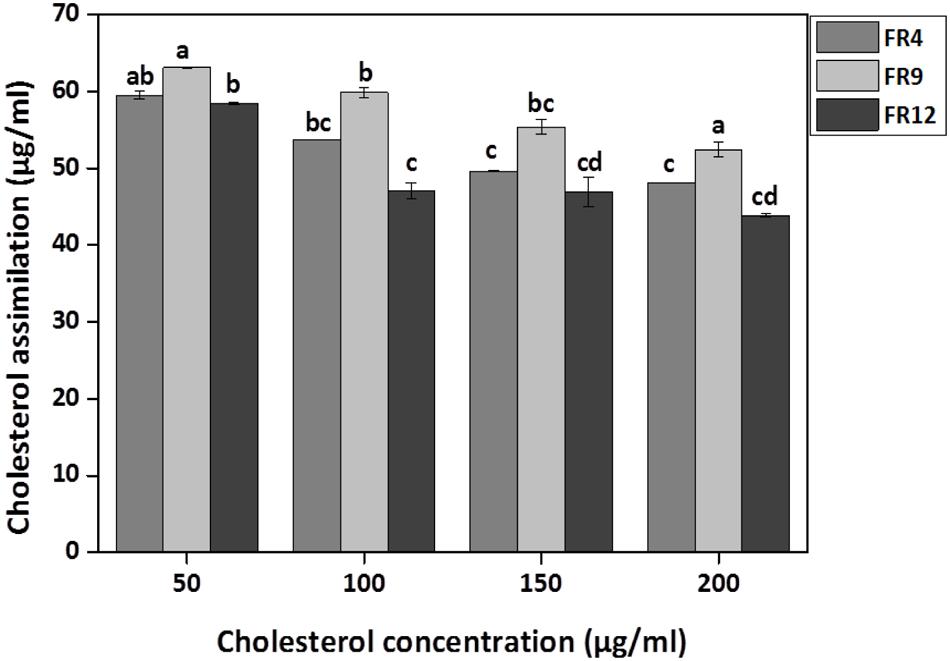
FIGURE 9. In vitro cholesterol assimilation by probiotic strains. Values are expressed as mean ± SD (n = 3). Each bars with no common letters are significantly different (P < 0.05) according to the LSD mean comparison test.
Adhesion Assay
Probiotic persistence and its colonization to digestive tract is the key necessity for bacteria to illustrate its advantageous effects for human wellbeing (Skrzypczak et al., 2015). In our study, HCT-116 colon carcinoma cell monolayer was used to demonstrate the adhesion properties of B. tequilensis FR9 both qualitatively and quantitatively. Qualitative examination was done by Crystal violet staining technique, thus provided visual confirmation of adhesion attributes (Figure 10). In BF and DIC micrographs (Figures 10C,D), bacterial adhesion on the HCT-116 surfaces was indicated with arrow marks. Subsequently, adhesion of B. tequilensis FR9 from initial inoculum was assessed by CFU cell counting. The relative percentage of adhesion was 12.6 ± 0.73% during post incubation. Hong et al. (2008) observed similar adhesion by B. subtilis strains on HT-29-16E, a mucus secreting colon carcinoma cell line. Similarly, Das and Goyal (2014) demonstrated a good adhesion percentage of 8.63 ± 3.03% by L. plantarum DM5 with HT-29 cell line. Adhesive property extensively relies on the origin, dosage, and is strain dependent. In our study, the B. tequilensis FR9 exhibited a good cell surface hydrophobicity of 86–94% and autoaggregation of 78%, which is an essential criterion for adhesion. These results support the adhesive capability of B. tequilensis FR9, and it may adhere on GIT and withstand the peristaltic movement of the human intestine.
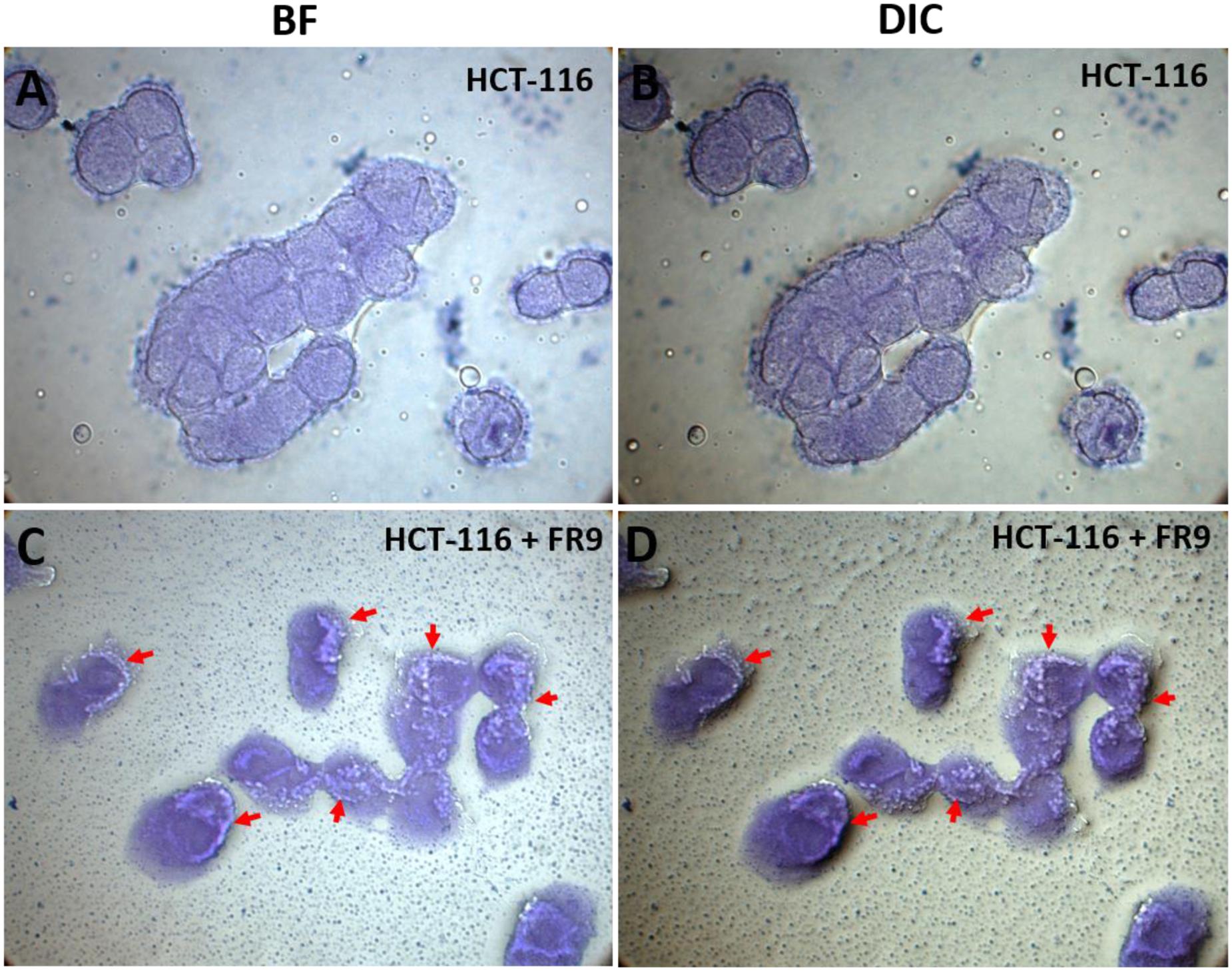
FIGURE 10. In vitro adhesion assay of B. tequilensis FR9 on HCT-116 colon carcinoma cell line observed under light microscope with 100× magnification (oil immersion). The cells were stained with crystal violet. HCT-116 cell line alone observed under bright field (BF) (A) and differential interference contrast (DIC) (B). HCT-116 cell line with B. tequilensis FR9 observed under BF (C) and DIC (D). The arrows indicate the adhesion of FR9 cells on HCT-116 cell line.
Pathogen Invasion Protection Assay
Probiotics should adhere to the intestinal epithelial cells and eliminate the adhesion of intestinal pathogens via competitive binding (Das et al., 2013). Inhibition of pathogen invasion by B. tequilensis FR9 was demonstrated using HCT-116 colon carcinoma cells. Invasion of L. monocytogenes and E. faecalis decreased with the addition of either B. tequilensis FR9 or purified Subtilosin A. B. tequilensis FR9 demonstrated increased invasion protection against L. monocytogenes MTCC 657 (5.83 CFU/mL) compared to E. faecalis MTCC 439 (7.12 CFU/mL; Figure 11). We hypothesize that the increased activity against L. monocytogenes might be due to the production of antilisterial protein (Subtilosin A), hence we have used the purified Subtilosin A instead of whole bacterial cell. Results showed increased protection against L. monocytogenes (4.63 CFU/mL), which confirms that the Subtilosin A plays a major role in pathogen invasion protection. Similar invasion protection by competitive exclusion as well as antimicrobial production has previously been recorded in many Bacillus and Lactobacillus spp. (Golowczyc et al., 2011; Das et al., 2013; Khan and Kang, 2016).
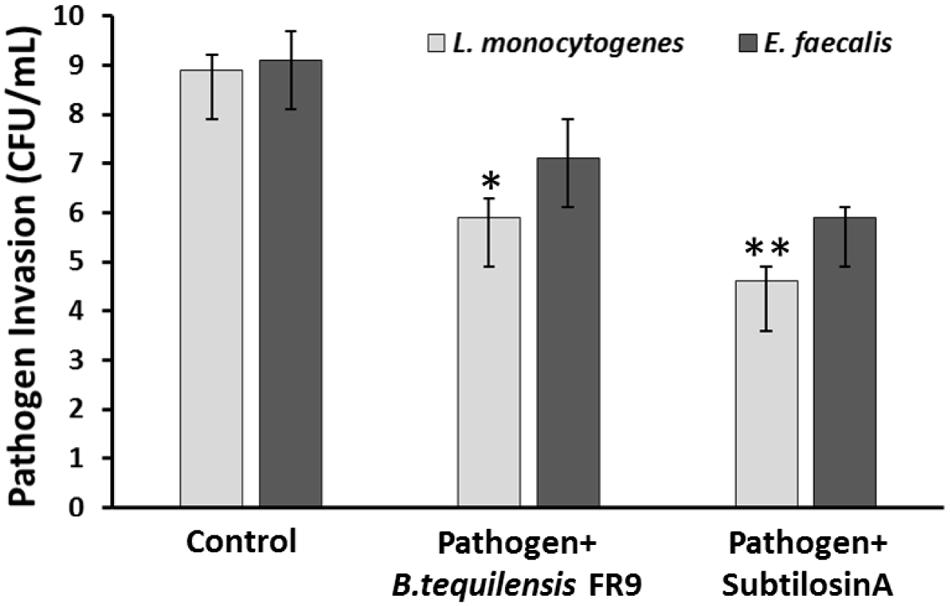
FIGURE 11. Invasion protection analysis of pathogen into HCT 116 colon carcinoma cell line with B. tequilensis FR9 or purified Subtilosin A. Results are expressed as mean ± SD (n = 3). ∗P ≤ 0.05, ∗∗P ≤ 0.01.
Conclusion
In conclusion, the present study has revealed that L. gasseri FR4, B. tequilensis FR9, and L. animalis FR12 strains isolated from healthy free-range chicken’s GIT displayed better performance in all in vitro assays, which is essential to be considered as probiotics. Among them, B. tequilensis FR9 was recognized as a reliable probiotic candidate, since they produce antilisterial Subtilosin A and inhibit the invasion of L. monocytogenes in human colon cells. Greater in vitro adhesion of B. tequilensis FR9 to HCT 116 exemplify its higher retention time and decreased adhesion of pathogen to gut which in turn enhances functional efficacy. The production of bacteriocins together with sporulation capacity supports Bacillus species with fortified supremacy with respect to their survival in diverse niches. For the first time, we have demonstrated the Subtilosin A production by B. tequilensis and have elucidated their probiotic properties. B. tequilensis FR9 could be used as a novel probiotic adjuvant for both human and animal foods.
Author Contributions
ADR, RPR, and MA contributed with the conception and experimental design. RPR, RD, and SH carried out all experiments, MA performed cell line studies and RP-HPLC analysis. MA and RPR analyze and interpret the results. RPR write the manuscript and corrected by ADR and MA. All authors performed a critical revision of the manuscript and approved the final version.
Funding
Author RPR thankful to Department of Science and Technology (DST), Ministry of Science and Technology, India for providing fellowship under DST-INSPIRE (IF140278) fellowship.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are thankful to Department of Biology, The Gandhigram Rural Institute-Deemed University for providing laboratory facility to carry out the entire research. The authors thank GRI-Scanning Electron Microscopy facility for SEM analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01910/full#supplementary-material
References
Abbasiliasi, S., Tan, J. S., Ibrahim, T. A., Kadkhodaei, S., Ng, H. S., Vakhshiteh, F., et al. (2014). Primary recovery of a bacteriocin-like inhibitory substance derived from Pediococcus acidilactici Kp10 by an aqueous two-phase system. Food Chem. 151, 93–100. doi: 10.1016/j.foodchem.2013.11.019
Abriouel, H., Franz, C. M., Ben Omar, N., and Galvez, A. (2011). Diversity and applications of Bacillus bacteriocins. FEMS Microbiol. Rev. 35, 201–232. doi: 10.1111/j.1574-6976.2010.00244.x
Ahn, Y. T., Lim, K. L., Ryu, J. C., Kang, D. K., Ham, J. S., Jang, Y. H., et al. (2002). Characterization of Lactobacillus acidophilus isolated from piglets and chicken. Asian Aust. J. Anim. Sci 15, 1790–1797. doi: 10.5713/ajas.2002.1790
Ajesh, K., Sudarslal, S., Arunan, C., and Sreejith, K. (2013). Kannurin, a novel lipopeptide from Bacillus cereus strain AK1: isolation, structural evaluation and antifungal activities. J. Appl. Microbiol. 115, 1287–1296. doi: 10.1111/jam.12324
Anandharaj, M., and Sivasankari, B. (2014). Isolation of potential probiotic Lactobacillus oris HMI68 from mother’s milk with cholesterol-reducing property. J. Biosci. Bioeng. 118, 153–159. doi: 10.1016/j.jbiosc.2014.01.015
Anandharaj, M., Sivasankari, B., and Parveen Rani, R. (2014). Effects of probiotics, prebiotics, and synbiotics on Hypercholesterolemia: a review. Chin. J. Biol. 2014, 7. doi: 10.1155/2014/572754
Anandharaj, M., Sivasankari, B., Santhanakaruppu, R., Manimaran, M., Parveen Rani, R., and Sivakumar, S. (2015). Determining the probiotic potential of cholesterol-reducing Lactobacillus and Weissella strains isolated from gherkins (fermented cucumber) and south Indian fermented koozh. Res. Microbiol. 166, 428–439. doi: 10.1016/j.resmic.2015.03.002
Argyri, A. A., Zoumpopoulou, G., Karatzas, K. A., Tsakalidou, E., Nychas, G. J., Panagou, E. Z., et al. (2013). Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol. 33, 282–291. doi: 10.1016/j.fm.2012.10.005
Beasley, S. S., Takala, T. M., Reunanen, J., Apajalahti, J., and Saris, P. E. (2004). Characterization and electrotransformation of Lactobacillus crispatus isolated from chicken crop and intestine. Poult. Sci. 83, 45–48. doi: 10.1093/ps/83.1.45
Begley, M., Hill, C., and Gahan, C. G. (2006). Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 72, 1729–1738. doi: 10.1128/AEM.72.3.1729-1738.2006
Chiang, S. S., and Pan, T. M. (2012). Beneficial effects of Lactobacillus paracasei subsp. paracasei NTU 101 and its fermented products. Appl. Microbiol. Biotechnol. 93, 903–916. doi: 10.1007/s00253-011-3753-x
CLSI (2011). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-First Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute.
Coates (2000). “Interpretation of infrared spectra, a practical approach,” in Encyclopedia of Analytical Chemistry, ed. R. Meyers (Chichester: John Wiley & Sons Ltd), 1–23.
Coetzee, I. (2015). Antibiotic Resistance in Triclosan Heterotrophic Plate Count Bacteria from Sewage Water. Ph.D thesis, North-West University, Potchefstroom.
Collado, M. C., Meriluoto, J., and Salminen, S. (2008). Adhesion and aggregation properties of probiotic and pathogen strains. Eur. Food Res. Technol. 226, 1065–1073. doi: 10.1007/s00217-007-0632-x
Das, D., and Goyal, A. (2014). Characterization and biocompatibility of glucan: a safe food additive from probiotic Lactobacillus plantarum DM5. J. Sci. Food Agric. 94, 683–690. doi: 10.1002/jsfa.6305
Das, J. K., Mishra, D., Ray, P., Tripathy, P., Beuria, T. K., Singh, N., et al. (2013). In vitro evaluation of anti-infective activity of a Lactobacillus plantarum strain against Salmonella enterica serovar Enteritidis. Gut. Pathog. 5, 1–11. doi: 10.1186/1757-4749-5-11
Deepika, G., Green, R. J., Frazier, R. A., and Charalampopoulos, D. (2009). Effect of growth time on the surface and adhesion properties of Lactobacillus rhamnosus GG. J. Appl. Microbiol. 107, 1230–1240. doi: 10.1111/j.1365-2672.2009.04306.x
Del Re, B., Sgorbati, B., Miglioli, M., and Palenzona, D. (2000). Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett. Appl. Microbiol. 31, 438–442. doi: 10.1046/j.1365-2672.2000.00845.x
Dertli, E., Mayer, M. J., and Narbad, A. (2015). Impact of the exopolysaccharide layer on biofilms, adhesion and resistance to stress in Lactobacillus johnsonii FI9785. BMC Microbiol. 15:8. doi: 10.1186/s12866-015-0347-342
Fluhe, L., Knappe, T. A., Gattner, M. J., Schafer, A., Burghaus, O., Linne, U., et al. (2012). The radical SAM enzyme AlbA catalyzes thioether bond formation in subtilosin A. Nat. Chem. Biol. 8, 350–357. doi: 10.1038/nchembio.798
Garriga, M., Pascual, M., Monfort, J. M., and Hugas, M. (1998). Selection of lactobacilli for chicken probiotic adjuncts. J. Appl. Microbiol. 84, 125–132. doi: 10.1046/j.1365-2672.1997.00329.x
Golowczyc, M. A., Silva, J., Teixeira, P., De Antoni, G. L., and Abraham, A. G. (2011). Cellular injuries of spray-dried Lactobacillus spp. isolated from kefir and their impact on probiotic properties. Int. J. Food Microbiol. 144, 556–560. doi: 10.1016/j.ijfoodmicro.2010.11.005
Hammes, W. P., and Vogel, R. F. (1995). “The genus Lactobacillus,” in The Genera of Lactic Acid Bacteria, eds B. J. B. Wood and W. H. Holzapfel (Boston, MA: Springer), 19–54.
Heilpern, D., and Szilagyi, A. (2008). Manipulation of intestinal microbial flora for therapeutic benefit in inflammatory bowel diseases: review of clinical trials of probiotics, pre-biotics and synbiotics. Rev. Recent Clin. Trials 3, 167–184. doi: 10.2174/157488708785700302
Hong, H. A., Huang, J. M., Khaneja, R., Hiep, L. V., Urdaci, M. C., and Cutting, S. M. (2008). The safety of Bacillus subtilis and Bacillus indicus as food probiotics. J. Appl. Microbiol. 105, 510–520. doi: 10.1111/j.1365-2672.2008.03773.x
Huang, T., Geng, H., Miyyapuram, V. R., Sit, C. S., Vederas, J. C., and Nakano, M. M. (2009). Isolation of a variant of subtilosin A with hemolytic activity. J. Bacteriol. 191, 5690–5696. doi: 10.1128/JB.00541-09
Ibarburu, I., Puertas, A. I., Berregi, I., Rodríguez-Carvajal, M. A., Prieto, A., and Dueñas, M. T. (2015). Production and partial characterization of exopolysaccharides produced by two Lactobacillus suebicus strains isolated from cider. Int. J. Food Microbiol. 214, 54–62. doi: 10.1016/j.ijfoodmicro.2015.07.012
Jankovic, T., Frece, J., Abram, M., and Gobin, I. (2012). Aggregation ability of potential probiotic Lactobacillus plantarum strains. Int. J. Sanit. Eng. Res. 6, 19–24.
Joint FAO/WHO (2002). Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food. Report of a Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of probiotics in Food, London.
Kanmani, P., Satish Kumar, R., Yuvaraj, N., Paari, K. A., Pattukumar, V., and Arul, V. (2013). Probiotics and its functionally valuable products-a review. Crit. Rev. Food Sci. Nutr. 53, 641–658. doi: 10.1080/10408398.2011.553752
Khan, I., and Kang, S. C. (2016). Probiotic potential of nutritionally improved Lactobacillus plantarum DGK-17 isolated from Kimchi–A traditional Korean fermented food. Food Control 60, 88–94. doi: 10.1016/j.foodcont.2015.07.010
Khochamit, N., Siripornadulsil, S., Sukon, P., and Siripornadulsil, W. (2015). Antibacterial activity and genotypic-phenotypic characteristics of bacteriocin-producing Bacillus subtilis KKU213: potential as a probiotic strain. Microbiol. Res. 170, 36–50. doi: 10.1016/j.micres.2014.09.004
Kos, B., Suskovic, J., Vukovic, S., Simpraga, M., Frece, J., and Matosic, S. (2003). Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 94, 981–987. doi: 10.1046/j.1365-2672.2003.01915.x
Kumar, R., Grover, S., and Batish, V. K. (2012). Bile salt hydrolase (Bsh) activity screening of lactobacilli: in vitro selection of indigenous Lactobacillus strains with potential bile salt hydrolysing and cholesterol-lowering ability. Probiotics Antimicrob. Proteins 4, 162–172. doi: 10.1007/s12602-012-9101-3
Lin, M.-Y., and Chenn, T.-W. (2000). Reduction of cholesterol by Lactobacillus acidophilus in culture broth. J. Food Drug Anal. 8, 97–102.
Luis-Villaseñor, I. E., Macías-Rodríguez, M. E., Gómez-Gil, B., Ascencio-Valle, F., and Campa-Córdova, ÁI. (2011). Beneficial effects of four Bacillus strains on the larval cultivation of Litopenaeus vannamei. Aquaculture 321, 136–144. doi: 10.1016/j.aquaculture.2011.08.036
Messaoudi, S., Madi, A., Prevost, H., Feuilloley, M., Manai, M., Dousset, X., et al. (2012). In vitro evaluation of the probiotic potential of Lactobacillus salivarius SMXD51. Anaerobe 18, 584–589. doi: 10.1016/j.anaerobe.2012.10.004
Motta, A. S., Flores, F. S., Souto, A. A., and Brandelli, A. (2008). Antibacterial activity of a bacteriocin-like substance produced by Bacillus sp. P34 that targets the bacterial cell envelope. Antonie Van Leeuwenhoek 93, 275–284. doi: 10.1007/s10482-007-9202-2
Musikasang, H., Tani, A., H-kittikun, A., and Maneerat, S. (2009). Probiotic potential of lactic acid bacteria isolated from chicken gastrointestinal digestive tract. World J. Microbiol. Biotechnol. 25, 1337–1345. doi: 10.1007/s11274-009-0020-8
NCCLS (1997). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 4th Edn, Vol. 17. Wayne, PA: NCCLS.
Nel, H. A., Bauer, R., Vandamme, E. J., and Dicks, L. M. (2001). Growth optimization of Pediococcus damnosus NCFB 1832 and the influence of pH and nutrients on the production of pediocin PD-1. J. Appl. Microbiol. 91, 1131–1138. doi: 10.1046/j.1365-2672.2001.01486.x
Nguyen, A. T., Nguyen, D. V., Tran, M. T., Nguyen, L. T., Nguyen, A. H., and Phan, T. N. (2015). Isolation and characterization of Bacillus subtilis CH16 strain from chicken gastrointestinal tracts for use as a feed supplement to promote weight gain in broilers. Lett. Appl. Microbiol. 60, 580–588. doi: 10.1111/lam.12411
Orłowski, A., and Bielecka, M. (2006). Preliminary characteristics of Lactobacillus and Bifidobacterium strains as probiotic candidates. Pol. J. Food Nutr. Sci. 15, 269–275.
Palaniswamy, S. K., and Govindaswamy, V. (2016). In-vitro probiotic characteristics assessment of feruloyl esterase and glutamate decarboxylase producing Lactobacillus spp. isolated from traditional fermented millet porridge (kambu koozh). LWT Food Sci. Technol. 68, 208–216. doi: 10.1016/j.lwt.2015.12.024
Pan, D., and Mei, X. (2010). Antioxidant activity of an exopolysaccharide purified from Lactococcus lactis subsp. lactis 12. Carbohydr. Polym. 80, 908–914. doi: 10.1016/j.carbpol.2010.01.005
Prasad, J., Gill, H., Smart, J., and Gopal, P. K. (1998). Selection and characterisation of Lactobacillus and Bifidobacterium strains for use as probiotics. Int. Dairy J. 8, 993–1002. doi: 10.1016/S0958-6946(99)00024-2
Ramasamy, K., Abdullah, N., Wong, M. C., Karuthan, C., and Ho, Y. W. (2010). Bile salt deconjugation and cholesterol removal from media by Lactobacillus strains used as probiotics in chickens. J. Sci. Food Agric. 90, 65–69. doi: 10.1002/jsfa.3780
Rubio, R., Jofré, A., Martín, B., Aymerich, T., and Garriga, M. (2014). Characterization of lactic acid bacteria isolated from infant faeces as potential probiotic starter cultures for fermented sausages. Food Microbiol. 38, 303–311. doi: 10.1016/j.fm.2013.07.015
Rudel, L. L., and Morris, M. D. (1973). Determination of cholesterol using o-phthalaldehyde. J. Lipid Res. 14, 364–366.
Sass, P., Jansen, A., Szekat, C., Sass, V., Sahl, H. G., and Bierbaum, G. (2008). The lantibiotic mersacidin is a strong inducer of the cell wall stress response of Staphylococcus aureus. BMC Microbiol. 8:186. doi: 10.1186/1471-2180-8-186
Skrzypczak, K., Gustaw, W., and Waśko, A. (2015). Health-promoting properties exhibited by Lactobacillus helveticus strains. Acta Biochim. Pol. 62, 713–720. doi: 10.18388/abp.2015_1116
Strompfova, V., and Laukova, A. (2007). In vitro study on bacteriocin production of Enterococci associated with chickens. Anaerobe 13, 228–237. doi: 10.1016/j.anaerobe.2007.07.002
Swain, M. R., Anandharaj, M., Ray, R. C., and Parveen Rani, R. (2014). Fermented fruits and vegetables of Asia: a potential source of probiotics. Biotechnol. Res. Int. 2014:19. doi: 10.1155/2014/250424
Taheri, H. R., Moravej, H., Tabandeh, F., Zaghari, M., and Shivazad, M. (2009). Screening of lactic acid bacteria toward their selection as a source of chicken probiotic. Poult. Sci. 88, 1586–1593. doi: 10.3382/ps.2009-00041
Torshizi, M. A. K., Rahimi, S. H., Mojgani, N., Esmaeilkhanian, S., and Grimes, J. L. (2008). Screening of indigenous strains of lactic acid bacteria for development of a probiotic for poultry. Asian Aust. J. Anim. Sci. 21, 1495–1500. doi: 10.5713/ajas.2008.80081
Walling, E., Dols-Lafargue, M., and Lonvaud-Funel, A. (2005). Glucose fermentation kinetics and exopolysaccharide production by ropy Pediococcus damnosus IOEB8801. Food Microbiol. 22, 71–78. doi: 10.1016/j.fm.2004.04.003
Walter, J., Schwab, C., Loach, D. M., Ganzle, M. G., and Tannock, G. W. (2008). Glucosyltransferase A (GtfA) and inulosucrase (Inu) of Lactobacillus reuteri TMW1.106 contribute to cell aggregation, in vitro biofilm formation, and colonization of the mouse gastrointestinal tract. Microbiology 154(Pt 1), 72–80. doi: 10.1099/mic.0.2007/010637-0
Wang, G., Zhao, Y., Tian, F., Jin, X., Chen, H., Liu, X., et al. (2014). Screening of adhesive lactobacilli with antagonistic activity against Campylobacter jejuni. Food Control 44, 49–57. doi: 10.1016/j.foodcont.2014.03.042
Yu, B., Liu, J. R., Chiou, M. Y., Hsu, Y. R., and Chiou, P. W. S. (2007). The effects of probiotic Lactobacillus reuteri Pg4 strain on intestinal characteristics and performance in broilers. Asian Aust. J. Anim. Sci. 20, 1243–1251. doi: 10.5713/ajas.2007.1243
Zhang, L., Liu, C., Li, D., Zhao, Y., Zhang, X., Zeng, X., et al. (2013). Antioxidant activity of an exopolysaccharide isolated from Lactobacillus plantarum C88. Int. J. Biol. Macromol. 54, 270–275. doi: 10.1016/j.ijbiomac.2012.12.037
Keywords: probiotic, Bacillus tequilensis, bacteriocin, Subtilosin A, adhesion assay, cholesterol reduction
Citation: Parveen Rani R, Anandharaj M, Hema S, Deepika R and David Ravindran A (2016) Purification of Antilisterial Peptide (Subtilosin A) from Novel Bacillus tequilensis FR9 and Demonstrate Their Pathogen Invasion Protection Ability Using Human Carcinoma Cell Line. Front. Microbiol. 7:1910. doi: 10.3389/fmicb.2016.01910
Received: 23 August 2016; Accepted: 15 November 2016;
Published: 01 December 2016.
Edited by:
Joaquin Bautista-Gallego, Rovira i Virgili University, SpainReviewed by:
Cristian Botta, University of Turin, ItalyManas Ranjan Swain, Indian Oil Corporation Ltd., India
Copyright © 2016 Parveen Rani, Anandharaj, Hema, Deepika and David Ravindran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abraham David Ravindran, ZGF2aWRfZ3JpYmlvbG9neUByZWRpZmZtYWlsLmNvbQ==
 Rizwana Parveen Rani
Rizwana Parveen Rani Marimuthu Anandharaj
Marimuthu Anandharaj Subramani Hema1
Subramani Hema1 Abraham David Ravindran
Abraham David Ravindran