- 1Department of Microbial Diseases, UCL Eastman Dental Institute, Faculty of Medical Sciences, University College London, London, UK
- 2Department of Bacteriology, Animal and Plant Health Agency, Addlestone, UK
Antibiotic resistance in human bacterial pathogens and commensals is threatening our ability to treat infections and conduct common medical procedures. As novel antibiotics are discovered and marketed it is important that we understand how resistance to them may arise and know what environments may act as reservoirs for such resistance genes. In this study a tetracycline and tigecycline resistant clone was identified by screening a human saliva metagenomic library in Escherichia coli EPI300 on agar containing 5 μg/ml tetracycline. Sequencing of the DNA insert present within the tetracycline resistant clone revealed it to contain a 7,765 bp fragment harboring novel ABC half transporter genes, tetAB(60). Mutagenesis studies performed on these genes confirmed that they were responsible for the tetracycline and tigecycline resistance phenotypes. Growth studies performed using E. coli EPI300 clones that harbored either the wild type, the mutated, or none of these genes indicated that there was a fitness cost associated with presence of these genes, with the isolate harboring both genes exhibiting a significantly slower growth than control strains. Given the emergence of E. coli strains that are sensitive only to tigecycline and doxycycline it is concerning that such a resistance mechanism has been identified in the human oral cavity.
Introduction
Tetracyclines are a group of broad spectrum antibiotics that have recently faced a reduction in clinical use due to the increase in prevalence of tetracycline resistance (Chopra and Roberts, 2001; Bishburg and Bishburg, 2009). In the UK tetracyclines are the most sold antibiotic for animal use and represent 10% of prescribed antibiotics for human clinical use. This widespread use of tetracyclines exerts a selection pressure on microorganisms to maintain tetracycline resistance genes (Roberts, 2003; Martinez, 2009; Wu et al., 2010). Resistance is mainly attributed to the production of efflux pumps, ribosomal protection proteins (RPPs) that prevent tetracycline binding to the ribosome, and less often, tetracycline degrading enzymes (Aminov et al., 2002; Connell et al., 2003; Yang et al., 2004; Forsberg et al., 2015).
Although resistance to this group of antibiotics is prevalent they are still used in the treatment of some human infections, including Chlamydia infections and some eye infections such as trachoma (Hu et al., 2010; Dukers-Muijrers et al., 2015). Tigecycline is a novel semi-synthetic derivative of tetracycline and the first of the glycylcyclines. It contains a bulky N,N-dimethylglycylamido side group that allows it to overcome RPP and efflux mechanisms of resistance to earlier generation precursors such as tetracycline (Someya et al., 1995; Olson et al., 2006). Tigecycline is used in the treatment of skin and abdominal infections as well as some cases of community acquired pneumonia (Rubinstein and Vaughan, 2005; Shen et al., 2015; Van Berkel et al., 2016). It has been shown that tetracycline resistance genes can obtain mutations that broaden the activity of their products to new tetracycline derivatives (Linkevicius et al., 2015). It is important that we understand the mechanisms of resistance to our current generation of tetracyclines in order for us to identify environments that may harbor genes that could confer resistance to novel tetracyclines, including those that are still in development.
The microbiota of the human oral cavity constitutes a reservoir of tetracycline resistance genes. RPP genes such as tet(M) are the most abundant tetracycline resistant genes in bacteria of the oral cavity (Villedieu et al., 2003; Seville et al., 2009). Tetracycline efflux genes such as major facilitator superfamily (MFS) exporters including tet(L) and the ATP Binding Cassette (ABC) transporter tetAB(46) have also been detected in bacteria in the oral cavity (Lancaster et al., 2005; Seville et al., 2009; Warburton et al., 2013).
ABC transporters are a functionally and structurally diverse family of proteins. They may comprise a single peptide, four peptides or two half transporters. Bacterial half ABC transporters typically contain a transmembrane domain (TMD) and a highly conserved nucleotide binding domain (NBD). Functional dimeric ABC transporters are composed of two half ABC transporter subunits each contributing a TMD spanning the membrane to form a substrate channel (Biemans-Oldehinkel et al., 2006; Dawson and Locher, 2006). The two NBDs interact to form the ABC of the transporter at the cytoplasmic face of the membrane that can bind two ATP molecules (Jones and George, 1999). It is here that ATP binding and hydrolysis triggers conformational changes in the substrate channel (Hollenstein et al., 2007). Cycles of ATP binding and hydrolysis allow the substrate channel to alternate between being open to the cytoplasm for substrate binding and open to the cells external environment or periplasm for substrate efflux (Higgins, 2001; Hellmich et al., 2012). ABC transporters that confer multidrug resistance (MDR) and biocide resistance to bacteria including human pathogens have been described such as YheH/I, LmrCD, PatAB and EfrAB from Bacillus subtilis, Lactococcus lactis, Streptococcus pneumoniae and Enterococcus faecalis, respectively (Lee et al., 2003; Lubelski et al., 2004; Torres et al., 2009; Baylay et al., 2015).
The aim of this study was to identify the gene(s) conferring resistance in a tetracycline and tigecycline resistant clone that was identified from a human saliva metagenomic library in Escherichia coli. Two genes from the clone, tetA(60) and tetB(60) were found to encode for two half transporter proteins which were shown to be responsible for the observed antibiotic resistance phenotype and reduced fitness.
Materials and Methods
Strains and Culture Conditions
The strains used in this study are listed in Table 1. E. coli EPI300 strains were cultured in Luria-Bertani broth (LB; Sigma-Aldrich®) and LB agar (LA; Life TechnologiesTM) at 37°C with shaking at 200 rpm for liquid culture. When antibiotic selection was required the media was supplemented with chloramphenicol (12.5 μg/ml; Sigma-Aldrich®) and tetracycline (5 μg/ml; Sigma-Aldrich®). Mueller Hinton (MH; Sigma-Aldrich®) agar was used in disk diffusion assays.
Sample Collection and Metagenomic DNA Extraction
Saliva samples were collected from 11 healthy individuals who had not taken antibiotics within the previous 3 months. Saliva was expectorated into sterile tubes (approximately 5 ml per individual) and samples were pooled. Metagenomic DNA was extracted in 1.5 ml aliquots using a modified protocol of the Gentra Puregene Yeast/Bact. Kit (Qiagen) as previously described (Seville et al., 2009). Ethical approval to collect human saliva from volunteers was granted by the UCL Research Ethics Committee (Project ID Number 5017/001).
Creation of a Metagenomic Library
Saliva metagenomic DNA was partially digested using HindIII, ligated into pCC1BAC and transformed into E. coli EPI300 as described previously (Seville et al., 2009).
After transformation, cells were recovered in SOC media (New England Biolabs®), cultured on LA containing chloramphenicol, 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG; Promega©) and 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal; Promega©) for 16 h. White clones were cultured in LB with chloramphenicol in individual wells of 96-well plates at 37°C for 16 h. The cultures were then stored at -80°C in 20% glycerol.
Screening of Metagenomic Library and Resistant Clone Isolation
Approximately 27,000 clones of the metagenomic library were screened for tetracycline resistance by plating the library onto LA with chloramphenicol (12.5 μg/ml) and tetracycline (5 μg/ml) and incubating them at 37°C for 16 h. A tetracycline resistant clone, PS9, was selected for further study.
DNA Sequencing, Analysis and Annotation
A list of the primers used in this study is detailed in Supplementary Table S1. Sequencing of the BAC clone in PS9 was accomplished using 454 sequencing as described previously (Card et al., 2014). Sequencing of subclones and mutants was conducted using primer extension Sanger sequencing by Beckman Coulter Genomics Inc. Contigs were assembled using SeqMan Pro (Lasergene software, DNASTAR, Madison, WI, USA) and sequence gaps were closed using PCR and Sanger sequencing (Sanger et al., 1977). Sequences were analyzed using the tools on NCBI. Two open reading frames (ORFs) encoding hypothetical ABC half transporter genes were named tetA(60) and tetB(60) by the Stuart B. Levy lab according to tetracycline resistance gene nomenclature guidelines (Levy et al., 1999). The sequences for tetA(60) and tetB(60) were submitted to GenBank (accession numbers KX000272.1and KX000273.1). The full 7,765 bp insert sequence was also submitted to Genbank (accession number KX887332; Supplementary Figure S1C). The putative amino acid sequences of TetA(60) and TetB(60) were compared to other phenotypically validated tetracycline and multidrug ABC transporter protein sequences from Gram-positive bacteria [TetAB(60), YheH/I, LmrCD, PatAB and EfrAB] by alignment using Clustal Omega at http://www.ebi.ac.uk/Tools/msa/clustalo/.
Subcloning
Primers used for subcloning are detailed in Supplementary Table S1. Regions of PS9 were amplified using primer pairs that introduced flanking HindIII and BamHI sites. The amplified fragments were ligated into pHSG396 and transformed into E. coli EPI300.
Mutagenesis
Primers used for mutagenesis are listed in Supplementary Table S1. In frame deletions of the Walker-A motifs of tetA(60) and tetB(60) were made using the NEB Q5 Site Directed Mutagenesis kit. Two pairs of non-overlapping primers were designed to amplify the pHSG396 vector containing both transporter genes. The first primer pair amplified pHSG396::tetAB(60) without a 69 bp region containing the tetA(60) Walker-A motif, keeping tetB(60) full length. The second primer pair amplified pHSG396::tetAB(60) without a 57 bp region containing the Walker-A motif of tetB(60) keeping tetA(60) intact. The resulting PCR products were circularized and transformed into E. coli EPI300.
Disk Diffusion Assays
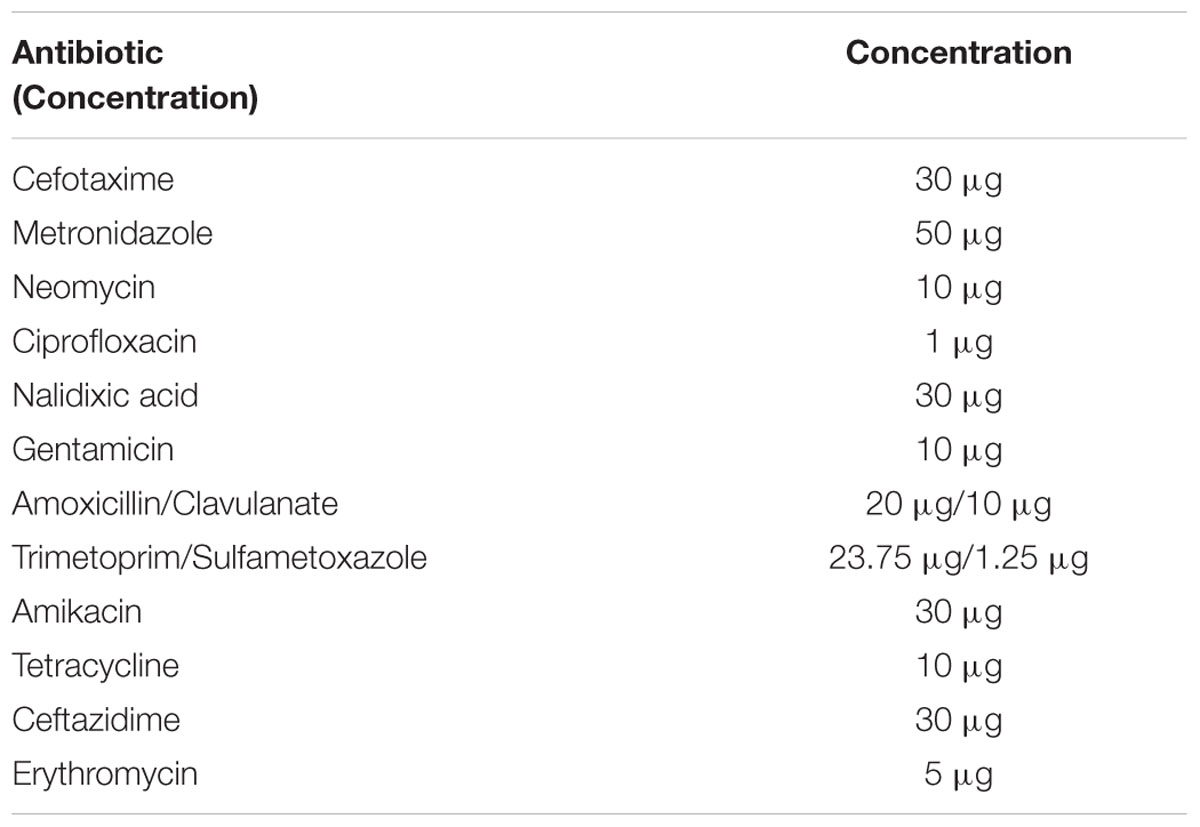
The susceptibilities of E. coli EPI300, E. coli::pHSG396 and E. coli::pHSG396tetAB(60) to various antibiotics (cefotaxime, ceftazidime metronidazole, neomycin, ciprofloxacin, nalidixic acid, gentamicin, amikacin, amoxicillin/clavulanate and trimetoprim/sulfametoxazole and erythromycin) were evaluated using the disk diffusion assay according to BSAC guidelines (Andrews, 2001). The antibiotic disks and concentrations used in this study are listed in Table 2.
Minimum Inhibitory Concentration (MIC) Assays
Minimum Inhibitory Concentrations of tetracycline, minocycline, and tigecycline were determined using the microbroth dilution method according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines (EUCAST, 2015). For MIC determination, overnight cultures grown in LB were adjusted to an OD600 of 0.1; 10 μl of the adjusted overnight cultures were used to inoculate 90 μl fresh LB containing varying concentrations of tetracycline (0.25–32 μg/ml), minocycline (0.25–10 μg/ml) or tigecycline (0.25–10 μg/ml) in a 96 well plate format. These plates were incubated overnight at 37°C with shaking at 200 rpm. Growth was determined by spectrophotometry at OD600 and the MIC was determined as the lowest concentration of antibiotic that inhibited growth.
Growth Curves
Overnight cultures of E. coli::pHSG396, E. coli::pHSG396tetAB(60), E. coli::pHSG396tetB(60)ΔtetA(60) and E. coli::pHSG396tetA(60)ΔtetB(60) grown in LB with chloramphenicol (12.5 μg/ml) and tetracycline (5 μg/ml; when required) were adjusted to an OD600 of 0.05 in LB and chloramphenicol (12.5 μg/ml). Cell suspensions were grown at 37°C with shaking at 200 rpm for 7 h and their cell density was measured every 30 min using spectrophotometry (OD600). E. coli::pHSG396tetAB(60)was also grown in LB and chloramphenicol (12.5 μg/ml) with tetracycline (5 μg/ml) to determine if the presence of this antibiotic affected the clones. Growth rates were measured for each clone as the slope of the line between two time points on the growth curve. The equation Nt = N0∗(1 + r)t, was used to calculate the maximum growth rate between 60 and 240 min. Technical triplicates and biological triplicates were conducted for all growth curves and growth rate calculations.
Statistical Analysis
Standard deviations were calculated for each clone using the data obtained from the growth curve assays, which included nine data points encompassing biological and technical replicates. Standard deviations were used as error bars in Figure 2 for comparison of the mean OD600 for each clone. Two tailed t-tests with 95% confidence intervals were used to determine the significance of differences between clones and the control (E. coli::pHSG396) in terms of OD600 at 420 min and growth kinetics.
Results
A library of 27,000 clones was constructed from the pooled human saliva of 11 individuals. Screening of this metagenomic library for tetracycline resistant clones resulted in the isolation of two of clones capable of growing on tetracycline (5 μg/ml), including PS9.
Analysis of the 7,765 bp PS9 insert revealed it to have nucleotide similarity along its entire length with Streptococcus sp. 263_SSPC (accession: GCA_001071995.1, 98% cover and 90% identity) and Granulicatella adiacens ATCC 49175 (accession: NZ_ACKZ00000000, 94% cover and 92% identity). The alignments also identified an inversion in the PS9 insert between 1,600 bp and 1,789 bp (Supplementary Figures S1A,B). BlastX analysis of the insert predicted it to contain five putative ORFs (Figure 1A). The hypothetical products of the five ORFs had a homolog with >90% amino acid identity from Streptococcus sp. 263_SSPC and G. adiacens ATCC 49175. Of the ORFs identified, three encoded a UDP-galactose mutase, sulfurtransferase and amidohydrolase. The remaining ORFs were predicted to encode two half ABC transporters that were named TetA(60) and TetB(60) as their putative amino acid sequences had less than 80% similarity to any other tetracycline resistance protein amino acid sequence and therefore fulfilled the criteria for a new tetracycline resistance gene. Interestingly, in the Streptococcus sp. 263_SSPC genome sequence two transposase genes were found 634 bp and 6,196 bp upstream of the UDP-galactose mutase. No such genes were identified in the G. adiacens ATCC 49175 genome. Clustal Omega alignments of the putative amino acid sequences of TetA(60) and TetB(60) to characterized antibiotic resistance heterodimeric ABC transporters showed that they were more closely related to TetA(46) and TetB(46) (39.27% and 42.28% identity, respectively) and YheH and YheI (40.93% and 46.61%, respectively). TetAB(60) were less related to the MDR ABC transporters EfrAB, PatAB and LmrCD of E. faecalis, S. pneumoniae and L. lactis, respectively (≤34.46%), Table 3.
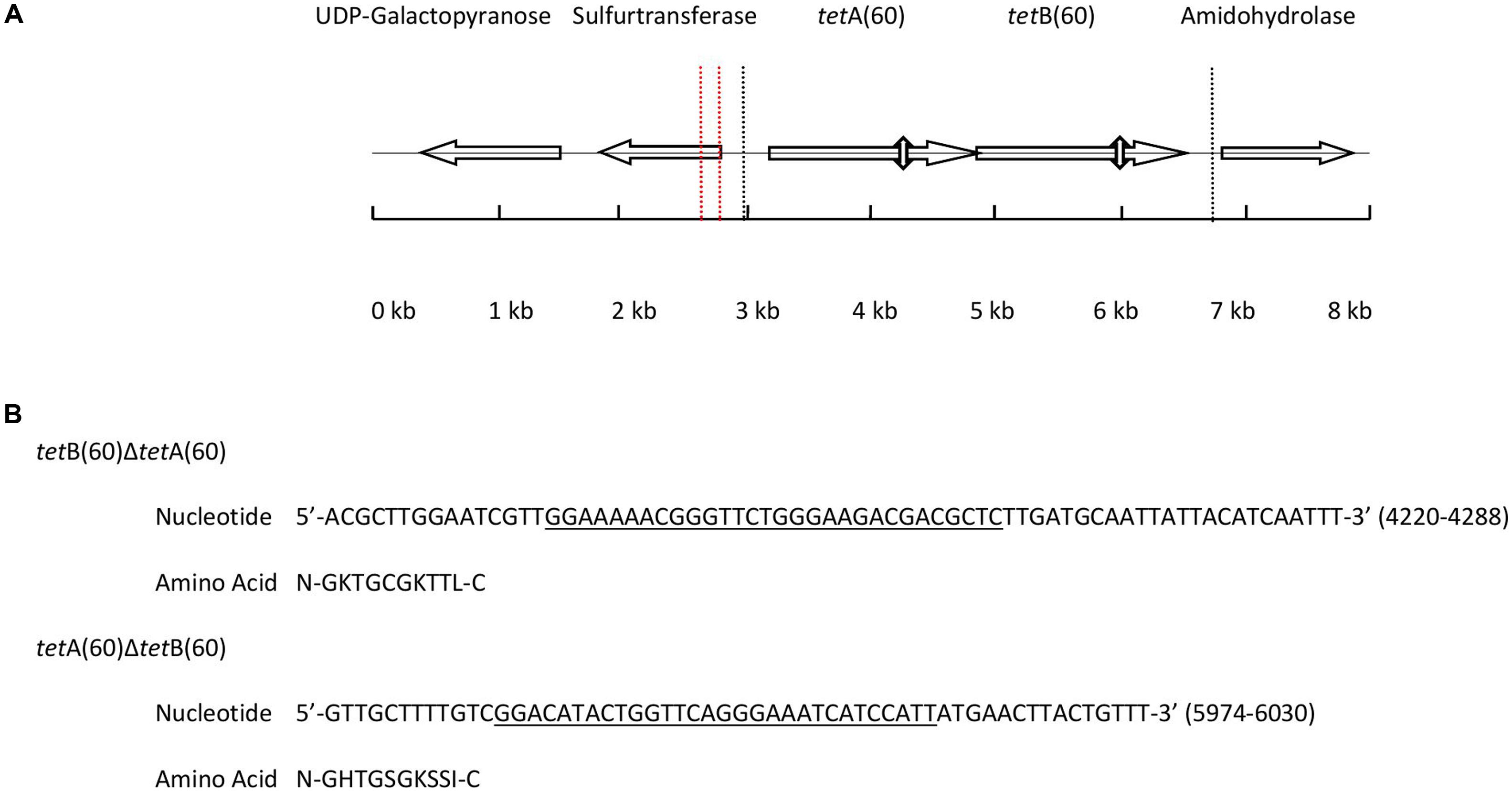
FIGURE 1. (A) Diagram depicting the position and orientation of the ORFs present in the 7,765 base pair insert of PS9 according to BlastX. The 3, 703 base pair region containing tetAB(60) that were subcloned to create pHSG396::tetAB(60) is marked by dashed lines. The positions of the Walker A motifs that were deleted to make pHSG396::tetB(60)ΔtetA(60) and pHSG396::tetA(60)ΔtetB(60) are marked by vertical double headed arrows and the inversion in the sequence is indicated by vertical dashed red lines. (B) The nucleotide sequences of the deleted regions are given above with the Walker A motif of each gene underlined and translated.
Each putative ABC half transporter peptide was predicted to be 579 amino acids and both contained a predicted NBD and a TMD, which are hallmarks of ABC transporters. Additionally, there is a 4 bp overlap of the genes, with the start codon of tetB(60) being contained within tetA(60), although the genes are in different reading frames.
To determine if these two genes were responsible for the observed tetracycline resistance phenotype of PS9, tetA(60) and tetB(60) were individually and jointly subcloned into E. coli EPI300 using the pHSG396 cloning vector. Only E. coli::pHSG396tetAB(60) grew on 5 μg/ml of tetracycline, showing that both tetA(60) and tetB(60) were required for the tetracycline resistance.
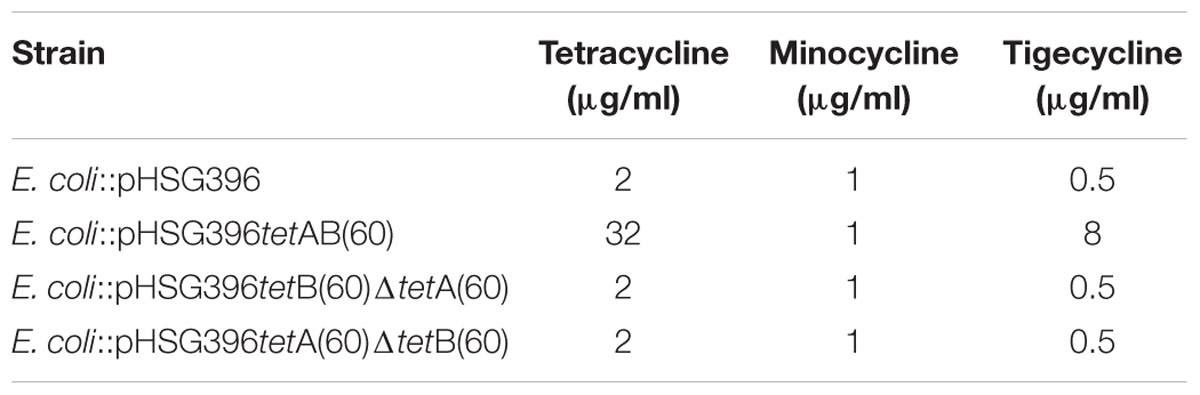
In order to ascertain whether the gene products function as a heterodimeric ABC transporter that confers resistance to tetracycline, a 69 and 57 base pair deletion was made to remove the Walker A motif of the NBD from either tetA(60) or tetB(60), respectively (Figure 1B). Both mutants, E. coli::pHSG396 tetB(60)ΔtetA(60) and E. coli::pHSG396tetA(60)ΔtetB(60) were susceptible to tetracycline. This confirmed that the ABC transporter activity of these gene products is responsible for the tetracycline resistance in PS9 and the E. coli::pHSG396tetAB(60), Table 4.
Using the broth dilution method, the MIC of tetracycline for E. coli EPI300, E. coli::pHSG396, E. coli::pHSG396tetAB(60), E. coli::pHSG396tetB(60)ΔtetA(60) and E. coli::pHSG396 tetA(60)ΔtetB(60) was determined (Table 4). The MIC of tetracycline for E. coli::pHSG396tetAB(60) was found to be 32 μg/ml. The MICs for the mutants and the controls strains were 16-fold lower at 2 μg/ml. To determine if tetAB(60) was able to confer resistance to later generation tetracycline derivatives, MIC assays were conducted using minocycline and tigecycline. The MIC of minocycline for all strains and clones was 1 μg/ml. The MIC of tigecycline for E. coli::pHSG396tetAB(60) was 16-fold higher than the control and mutant strains at 8 μg/ml, which was above the clinical break point for Enterobacteriaceae (0.5 μg/ml).
Disk diffusion assays were used to discern the spectrum of resistance for this transporter. E. coli::pHSG396tetAB(60) was less sensitive to tetracycline but equally sensitive to cefotaxime, ceftazidime, metronidazole, neomycin, ciprofloxacin, nalidixic acid, gentamicin, amikacin, amoxicillin/clavulanate and trimetoprim/sulfametoxazole as E. coli EPI300 and E. coli::pHSG396. E. coli EPI300 was intrinsically resistant to erythromycin which has been described previously and attributed to AcrAB-TolC mediated efflux and membrane impermeability (Vaara, 1993; Chollet et al., 2004).
We observed, when it was first isolated, that PS9 grew slower and formed smaller colonies than E. coli::pCC1BAC even in the absence of tetracycline in the growth media; this phenotype was also observed for the E. coli::pHSG396tetAB(60) subclone. Furthermore, it was noted that the E. coli::pHSG396tetB(60)ΔtetA(60) and E. coli::pHSG396tetA(60)ΔtetB(60) mutants did not have such a large growth defect. Growth curves revealed that although there were significant differences between E. coli::pHSG396 and E. coli::pHSG396tetA(60)ΔtetB(60) maximum growth rates (0.993 ± 0.05 and 0.917 ± 0.05, respectively; p = 0.005) there was no significant difference in their OD600 of cultures at 7 h (1.65 ± 0.08 and 1.565 ± 0.16; p = 0.1807) when grown in the absence of tetracycline (Figure 2).
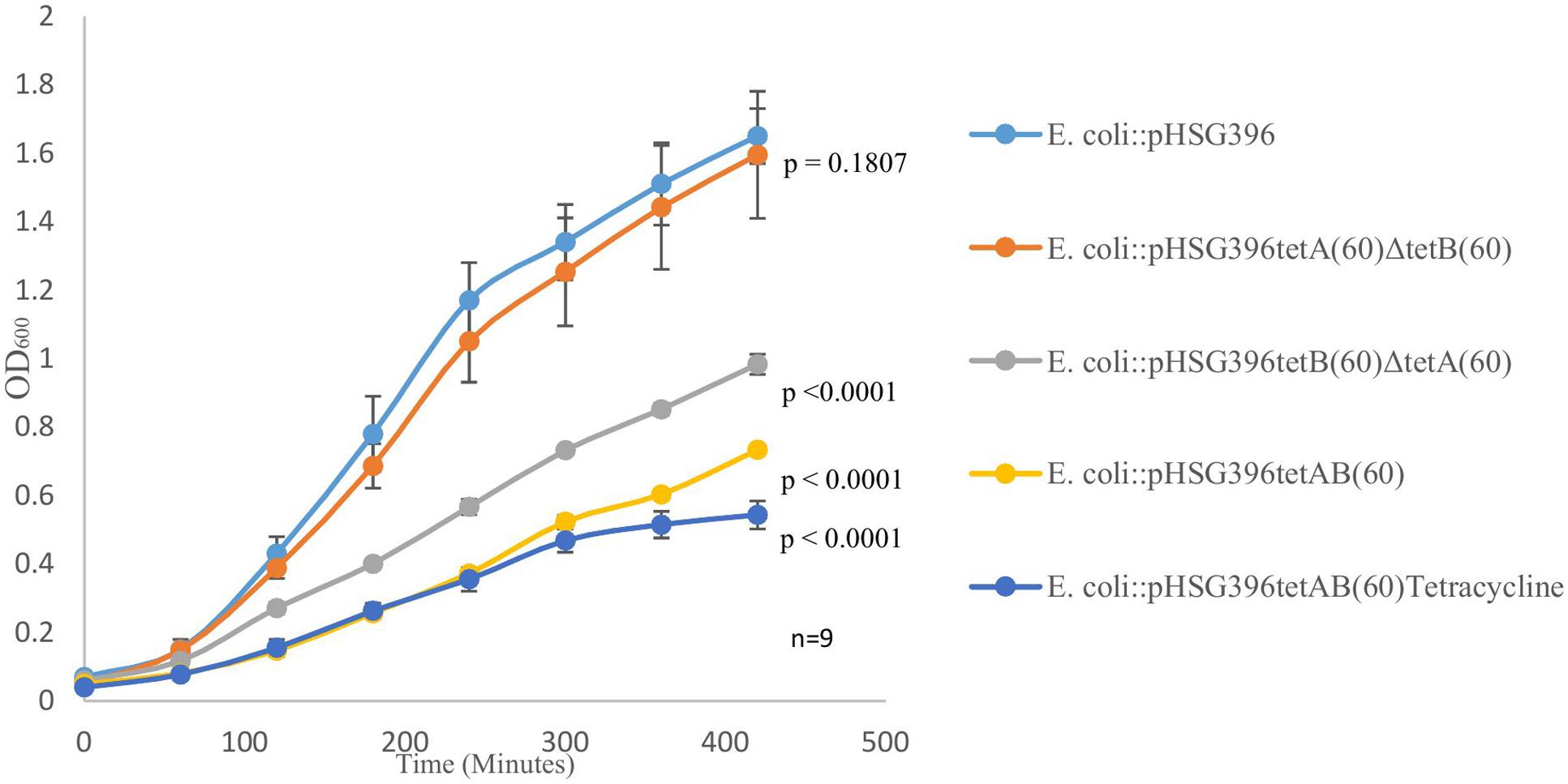
FIGURE 2. The above graph depicts growth curves for E. coli::pHSG396, E. coli::pHSG396tetB(60)ΔtetA(60) and E. coli::pHSG396tetA(60)ΔtetB(60) grown in LB and chloramphenicol for 7 h. It also shows the growth curve for E. coli::pHSG396tetAB(60) grown in LB and chloramphenicol with and without tetracycline to determine how the antibiotic effected the clone’s growth. P-values for OD600 at 7 h were calculated from biological triplicate OD600 measurements at 420 min for each clone compered to E. coli::pHSG396 are indicated beside each growth curve.
Compared to E. coli::pHSG396, E. coli::pHSG396tetB(60)ΔtetA(60) and E. coli::pHSG396tetAB(60) reached lower OD600 at 7 h when they were grown in the absence of tetracycline (0.983 ± 0.03, 0.733 ± 0.01, respectively, p < 0.0001). Additionally, when grown in the presence of tetracycline, E. coli::pHSG396tetAB(60) reached an even lower OD600 at 7 h (0.543 ± 0.04, p < 0.0001). Whilst the maximum growth rates of E. coli::pHSG396tetB(60)ΔtetA(60) grown without tetracycline and E. coli::pHSG396tetAB(60) grown without or with tetracycline were not significantly different from each other (0.702 ± 0.06, 0.68 ± 0.03 and 0.692 ± 0.09, respectively; p = 0.435 to 0.879), they were 1.14-1.46 fold lower than the maximum growth rate of E. coli::pHSG396 grown without tetracycline (p < 0.0001). We therefore suggest that there is a fitness cost due to the activity of TetAB(60) rather than the carriage of the plasmid itself and that TetB(60) contributes more to this fitness cost than TetA(60).
Discussion
There has been a resurgence in the use of tetracyclines in human therapy due to the recent development of a number of semisynthetic derivatives of the antibiotic that are efficacious against antibiotic resistant pathogens (Rubinstein and Vaughan, 2005; Anstead et al., 2014; Shen et al., 2015; Lin et al., 2016; Van Berkel et al., 2016). Tigecycline is the first of this new generation of tetracyclines to enter clinical use, being effective against MDR pathogens including carbapenem and colistin resistant microorganisms and those expressing specific tetracycline efflux systems and RPPs (Fluit et al., 2005; Cai et al., 2011). However, it is worrisome that resistance to tigecycline has already been described and associated with multidrug efflux systems and ribosomal mutations (Villa et al., 2014; Zhong et al., 2014; Lupien et al., 2015).
In this study we characterized a clone, PS9, isolated from a human oral saliva metagenomic library that exhibited high levels of resistance to tetracycline and tigecycline. BlastN alignments of the clone with Streptococcus sp. 263_SSPC and G. adiacens ATCC 49175 revealed the PS9 insert to have 90% and 92% nucleotide similarity to these species, respectively, indicating a probable Gram-positive origin for the insert. Streptococcus spp. are predominant in the oral cavity although to the best of our knowledge Streptococcus sp. 263_SSPC has not been identified (Segata et al., 2012). Granulicatella spp. including G. adiacens are also abundant in the oral cavity, typically inhabiting the mucosa (Aas et al., 2005). Tetracycline resistance mediated by Tn916 encoded tet(M) has been described for oral Streptococcus and Granulicatella spp. (Lancaster et al., 2005). Additionally, MFS and ABC transporter genes conferring resistance to tetracyclines have also been identified in oral Streptococcus spp. including tet(L) and tetAB(46), respectively (Chen et al., 2013; Warburton et al., 2013). Although tetracycline resistance has been observed in Granulicatella spp., minimal characterisation studies have been conducted (Zheng et al., 2004; De Luca et al., 2013). Although Streptococcus and Granulicatella spp. are abundant in the oral cavity, it is not known how prevalent tetAB(60) is in the oral cavity and further work beyond the scope of our characterisation study is required to address this.
Two transposase genes were located up stream of the UDP-galactose mutase gene in Streptococcus sp. 263_SSPC, however, it is unknown if these transposases are found in the host genome of the PS9 sequence or if they are capable of transposition of tetAB(60). The alignments also identified an inverted region in the PS9 insert when compared to the Streptococcus sp. 263_SSPC and G. adiacens ATCC 49175 genomes which may have resulted from a DNA breakage followed by repair or a transposition event.
Analysis of the insert revealed it contained five ORFs, predicted to encode a putative UDP-galactose mutase, a sulfurtransferase, an amidohydrolase and two ABC half transporters. Each predicted protein had amino acid sequences with high similarity (>90% identity) to proteins from Streptococcus sp. 263_SSPC and G. adiacens. Numerous heterodimeric ABC transporters capable of conferring antibiotic resistance have been described, including the MDR transporters EfrAB of E. faecalis (Lee et al., 2003), PatAB from S. pneumoniae (Baylay et al., 2015), LmrCD from L. lactis (Lubelski et al., 2004) and the recently characterized EfrCD of E. faecalis (Hurlimann et al., 2016). These transporters have been shown to confer resistance to fluoroquinolones, tetracyclines and biocides among other antimcrobials. Alignment of the putative amino acid sequences of TetAB(60) to other antibiotic resistance heterodimeric ABC transporters showed that they were most closely related to TetAB(46) and YheH/I and less so to the MDR ABC transporters EfrAB, PatAB and LmrCD. As TetAB(46) has been shown to be most closely related to YheH/I this suggested that TetAB(60) was also tetracycline specific (Warburton et al., 2013).
We showed that both tetA(60) and tetB(60) were required to confer tetracycline resistance in E. coli EPI300, suggesting that the product of these genes formed a heterodimeric ABC transporter with each gene product containing a TMD and NBD as revealed by BlastX (Dawson and Locher, 2006). Previous studies have used E. coli as a host to characterize Gram-positive antimicrobial ABC transporters including EfrAB from E. faecalis and LmrA from L. lactis (Lee et al., 2003; Achard-Joris et al., 2005). As E. coli::pHSG396tetAB(60) did not contain the inverted sequence it is likely that this sequence does not affect expression of tetAB(60).
Walker A motifs are found in many ATP utilizing enzymes including ABC transporters and are required for binding and stabilizing ATP (Ramakrishnan et al., 2002). Deletion of these motifs from ATP transporters has been shown to result in a loss of function (Warburton et al., 2013). Individual in-frame deletions of these motifs from either tetA(60) or tetB(60) led to a loss of the tetracycline and tigecycline resistance phenotype providing further evidence that the products of these genes form a heterodimeric ABC transporter.
Compared to E. coli::pHSG396, E. coli::pHSG396tetAB(60) was 16-fold more resistant to tetracycline (MIC of 32 μg/ml) and tigecycline (MIC of 8 μg/ml). Although E. coli::pHSG396tetAB(60) showed levels of resistance to tetracycline and tigecycline beyond the EUCAST breakpoints, it was as susceptible to minocycline as E. coli::pHSG396, indicating that minocycline is not a substrate for this transporter (Olson et al., 2006; Ramos et al., 2009). Efflux mediated tigecycline resistance has been described previously in Pseudomonas aeruginosa and Klebsiella pneumoniae, being attributed to the activity of an ABC and a resistance nodulation division (RND) transporter, respectively (Dean et al., 2003; He et al., 2015; McDaniel et al., 2016). TetAB(46) was also shown to confer low tigecycline resistance in S. australis (Warburton et al., 2013).
TetAB(60) appeared to be specific for tetracycline and tigecycline as disk diffusion assays demonstrated E. coli::pHSG396 to be as susceptible as E. coli::pHSG396tetAB(60) to cefotaxime, ceftazidime, metronidazole, neomycin, ciprofloxacin, nalidixic acid, gentamicin, amikacin, amoxicillin/clavulanate and trimetoprim/sulfametoxazole and erythromycin, providing further evidence for the tetracycline specificity of the ABC transporter.
The observed fitness cost associated with tetAB(60) was not observed in either mutant as although E. coli::pHSG396tetA(60)ΔtetB(60) had a lower maximum growth rate than the control it had a comparable final OD600 to E. coli::pHSG396 andE. coli::pHSG396tetB(60)ΔtetA(60) exhibited faster growth than E. coli::pHSG396tetAB(60). This indicated that the growth defect was a result of TetAB(60) activity rather than from maintenance of the plasmid. Additionally, as E. coli::pHSG396tetB(60)ΔtetA(60) grew less well than E. coli::pHSG396tetA(60)ΔtetB(60) it suggests that TetB(60) produces a greater cost to the E. coli host than TetA(60).
Conclusion
We have identified two novel genes from the human oral cavity that likely produce a heterodimeric ABC transporter, TetAB(60). TetAB(60) specifically exports tetracycline and tigecycline conferring high levels of resistance to these antibiotics in an E. coli host. A limitation of this work is that we do not know the prevalence of these genes in the human oral cavity. Further work should be undertaken to survey its prevalence in various niches, to determine how common these genes are, and their possible clinical relevance for treating bacterial infections with tetracycline derivatives. This work also shows that the human oral cavity harbors unknown tetracycline resistance determinants in the absence of any obvious selection pressure. There is potential for these genes to be acquired by mobile genetic elements and transferred to bacterial pathogens, which is particularly worrying given the recent identification of a carbapenem and colistin resistant strains of E. coli some of which could only be inhibited by doxycycline and tigecycline (Liu et al., 2016; Mediavilla et al., 2016; Yao et al., 2016). However, the associated fitness cost of tetAB(60) observed in E. coli may limit any possible fixation following dissemination of the genes from their native host to E. coli strains in the absence of a tetracycline or tigecycline selective pressure.
Author Contributions
LR contributed to the design of the experiments as well as to the acquisition, analysis, interpretation of the data included in this manuscript, wrote initial and revised drafts of the manuscript and approves of the final manuscript being submitted and also agrees to be accountable for the work detailed in the submitted manuscript. AR conceived the project, contributed to the design of experiments conducted throughout it, interpreted results and contributed to the drafting and revising of the manuscript being submitted and also approved the final draft of the manuscript and agrees to be accountable for the work detailed in the submitted manuscript. MA conceived the project, contributed to the design and direction of experiments within it, interpreted results and made revisions to the final manuscript, approved the manuscript being submitted and also agrees to be accountable for the work represented in the submitted manuscript.
Funding
LR was jointly funded by the Seedcorn Programme at the Animal and Plant Health Agency and a UCL IMPACT studentship at UCL.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We would like thank Mr. Supathep Tansirichaiya (UCL) for providing us with the human saliva metagenomic DNA used in this study and Dr. Philip J. Warburton (University of Plymouth) for providing us with E. coli EPI300::pCC1BAC.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01923/full#supplementary-material
References
Aas, J. A., Paster, B. J., Stokes, L. N., Olsen, I., and Dewhirst, F. E. (2005). Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43, 5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005
Achard-Joris, M., van den Berg van Saparoea, H. B., Driessen, A. J. M., and Bourdineaud, J.-P. (2005). Heterologously expressed bacterial and human multidrug resistance proteins confer cadmium resistance to Escherichia coli. Biochemistry 44, 5916–5922. doi: 10.1021/bi047700r
Aminov, R. I., Chee-Sanford, J. C., Garrigues, N., Teferedegne, B., Krapac, I. J., White, B. A., et al. (2002). Development, validation, and application of PCR primers for detection of tetracycline efflux genes of gram-negative bacteria. Appl. Environ. Microbiol. 68, 1786–1793. doi: 10.1128/AEM.68.4.1786-1793.2002
Andrews, J. M. (2001). BSAC standardized disc susceptibility testing method. J Antimicrob Chemother. 48(Suppl. 1), 43–57. doi: 10.1093/jac/48.2.322
Anstead, G. M., Cadena, J., and Javeri, H. (2014). Treatment of infections due to resistant Staphylococcus aureus. Methods Mol. Biol. 1085, 259–309. doi: 10.1007/978-1-62703-664-1_16
Baylay, A. J., Ivens, A., and Piddock, L. J. (2015). A novel gene amplification causes upregulation of the PatAB ABC transporter and fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 59, 3098–3108. doi: 10.1128/AAC.04858-14
Biemans-Oldehinkel, E., Doeven, M. K., and Poolman, B. (2006). ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett. 580, 1023–1035. doi: 10.1016/j.febslet.2005.11.079
Bishburg, E., and Bishburg, K. (2009). Minocycline–an old drug for a new century: emphasis on methicillin-resistant Staphylococcus aureus (MRSA) and Acinetobacter baumannii. Int. J. Antimicrob. Agents 34, 395–401. doi: 10.1016/j.ijantimicag.2009.06.021
Cai, Y., Wang, R., Liang, B., Bai, N., and Liu, Y. (2011). Systematic review and meta-analysis of the effectiveness and safety of tigecycline for treatment of infectious disease. Antimicrob. Agents Chemother. 55, 1162–1172. doi: 10.1128/AAC.01402-10
Card, R. M., Warburton, P. J., MacLaren, N., Mullany, P., Allan, E., and Anjum, M. F. (2014). Application of microarray and functional-based screening methods for the detection of antimicrobial resistance genes in the microbiomes of healthy humans. PLoS ONE 9:e86428. doi: 10.1371/journal.pone.0086428
Chen, L., Song, Y., Wei, Z., He, H., Zhang, A., and Jin, M. (2013). Antimicrobial susceptibility, tetracycline and erythromycin resistance genes, and multilocus sequence typing of Streptococcus suis isolates from diseased pigs in China. J. Vet. Med. Sci. 75, 583–587. doi: 10.1292/jvms.12-0279
Chollet, R., Chevalier, J., Bryskier, A., and Pagès, J.-M. (2004). The AcrAB-TolC pump is involved in macrolide resistance but not in telithromycin efflux in Enterobacter aerogenes and Escherichia coli. Antimicrob. Agents Chemother. 48, 3621–3624. doi: 10.1128/AAC.48.9.3621-3624.2004
Chopra, I., and Roberts, M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260. doi: 10.1128/MMBR.65.2.232-260.2001
Connell, S. R., Tracz, D. M., Nierhaus, K. H., and Taylor, D. E. (2003). Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrob. Agents Chemother. 47, 3675–3681. doi: 10.1128/AAC.47.12.3675-3681.2003
Dawson, R. J., and Locher, K. P. (2006). Structure of a bacterial multidrug ABC transporter. Nature 443, 180–185. doi: 10.1038/nature05155
De Luca, M., Amodio, D., Chiurchiu, S., Castelluzzo, M. A., Rinelli, G., Bernaschi, P., et al. (2013). Granulicatella bacteraemia in children: two cases and review of the literature. BMC Pediatr. 13:61. doi: 10.1186/1471-2431-13-61
Dean, C. R., Visalli, M. A., Projan, S. J., Sum, P.-E., and Bradford, P. A. (2003). Efflux-mediated resistance to tigecycline (GAR-936) in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 47, 972–978. doi: 10.1128/AAC.47.3.972-978.2003
Dukers-Muijrers, N. H., van Liere, G. A., Wolffs, P. F., Den Heijer, C., Werner, M. I., and Hoebe, C. J. (2015). Antibiotic use before chlamydia and gonorrhea genital and extragenital screening in the sexually transmitted infection clinical setting. Antimicrob. Agents Chemother. 59, 121–128. doi: 10.1128/aac.03932-14
EUCAST (2015). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 5.0, 2015. Basel: European Committee on Antimicrobial Susceptibility Testing. Available at: http://www.eucast.org
Fluit, A. C., Florijn, A., Verhoef, J., and Milatovic, D. (2005). Presence of tetracycline resistance determinants and susceptibility to tigecycline and minocycline. Antimicrob. Agents Chemother. 49, 1636–1638. doi: 10.1128/aac.49.4.1636-1638.2005
Forsberg, K. J., Patel, S., Wencewicz, T. A., and Dantas, G. (2015). The tetracycline destructases: a novel family of tetracycline-inactivating enzymes. Chem. Biol. 22, 888–897. doi: 10.1016/j.chembiol.2015.05.017
He, F., Fu, Y., Chen, Q., Ruan, Z., Hua, X., Zhou, H., et al. (2015). Tigecycline susceptibility and the role of efflux pumps in tigecycline resistance in KPC-producing Klebsiella pneumoniae. PLoS ONE 10:e119064. doi: 10.1371/journal.pone.0119064
Hellmich, U. A., Lyubenova, S., Kaltenborn, E., Doshi, R., van Veen, H. W., Prisner, T. F., et al. (2012). Probing the ATP hydrolysis cycle of the ABC multidrug transporter LmrA by pulsed EPR spectroscopy. J. Am. Chem. Soc. 134, 5857–5862. doi: 10.1021/ja211007t
Higgins, C. F. (2001). ABC transporters: physiology, structure and mechanism–an overview. Res. Microbiol. 152, 205–210. doi: 10.1016/S0923-2508(01)01193-7
Hollenstein, K., Dawson, R. J., and Locher, K. P. (2007). Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 17, 412–418. doi: 10.1016/j.sbi.2007.07.003
Hu, V. H., Harding-Esch, E. M., Burton, M. J., Bailey, R. L., Kadimpeul, J., and Mabey, D. C. (2010). Epidemiology and control of trachoma: systematic review. Trop. Med. Int. Health 15, 673–691. doi: 10.1111/j.1365-3156.2010.02521.x
Hurlimann, L. M., Corradi, V., Hohl, M., Bloemberg, G. V., Tieleman, D. P., and Seeger, M. A. (2016). The Heterodimeric ABC Transporter EfrCD mediates multidrug efflux in Enterococcus faecalis. Antimicrob. Agents Chemother. 60, 5400–5411. doi: 10.1128/AAC.00661-16
Jones, P. M., and George, A. M. (1999). Subunit interactions in ABC transporters: towards a functional architecture. FEMS Microbiol. Lett. 179, 187–202. doi: 10.1111/j.1574-6968.1999.tb08727.x
Lancaster, H., Bedi, R., Wilson, M., and Mullany, P. (2005). The maintenance in the oral cavity of children of tetracycline-resistant bacteria and the genes encoding such resistance. J. Antimicrob Chemother. 56, 524–531. doi: 10.1093/jac/dki259
Lee, E.-W., Huda, M. N., Kuroda, T., Mizushima, T., and Tsuchiya, T. (2003). EfrAB, an ABC multidrug efflux pump in Enterococcus faecalis. Antimicrob. Agents Chemother. 47, 3733–3738. doi: 10.1128/AAC.47.12.3733-3738.2003
Levy, S. B., McMurry, L. M., Barbosa, T. M., Burdett, V., Courvalin, P., Hillen, W., et al. (1999). Nomenclature for new tetracycline resistance determinants. Antimicrob. Agents Chemother. 43, 1523–1524.
Lin, S. Y., Huang, C. H., Ko, W. C., Chen, Y. H., and Hsueh, P. R. (2016). Recent developments in antibiotic agents for the treatment of complicated intra-abdominal infections. Expert Opin. Pharmacother. 17, 339–354. doi: 10.1517/14656566.2016.1122756
Linkevicius, M., Sandegren, L., and Andersson, D. I. (2015). Potential of tetracycline resistance proteins to evolve tigecycline resistance. Antimicrob Agents Chemother. 60, 789–796. doi: 10.1128/aac.02465-15
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Lubelski, J., Mazurkiewicz, P., van Merkerk, R., Konings, W. N., and Driessen, A. J. M. (2004). ydaG and ydbA of Lactococcus lactis encode a heterodimeric ATP-binding cassette-type multidrug transporter. J. Biol. Chem. 279, 34449–34455. doi: 10.1074/jbc.M404072200
Lupien, A., Gingras, H., Leprohon, P., and Ouellette, M. (2015). Induced tigecycline resistance in Streptococcus pneumoniae mutants reveals mutations in ribosomal proteins and rRNA. J. Antimicrob. Chemother. 70, 2973–2980. doi: 10.1093/jac/dkv211
Martinez, J. L. (2009). Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 157, 2893–2902. doi: 10.1016/j.envpol.2009.05.051
McDaniel, C., Su, S., Panmanee, W., Lau, G. W., Browne, T., Cox, K., et al. (2016). A putative ABC transporter permease is necessary for resistance to acidified nitrite and EDTA in Pseudomonas aeruginosa under aerobic and anaerobic planktonic and biofilm conditions. Front. Microbiol. 7:291. doi: 10.3389/fmicb.2016.00291
Mediavilla, J. R., Patrawalla, A., Chen, L., Chavda, K. D., Mathema, B., Vinnard, C., et al. (2016). Colistin- and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. MBio 7, e01191–e01216 doi: 10.1128/mBio.01191-16
Olson, M. W., Ruzin, A., Feyfant, E., Rush, T. S. III, O’Connell, J., and Bradford, P. A. (2006). Functional, biophysical, and structural bases for antibacterial activity of tigecycline. Antimicrob. Agents Chemother. 50, 2156–2166. doi: 10.1128/AAC.01499-05
Ramakrishnan, C., Dani, V. S., and Ramasarma, T. (2002). A conformational analysis of Walker motif A [GXXXXGKT (S)] in nucleotide-binding and other proteins. Protein Eng. 15, 783–798. doi: 10.1093/protein/15.10.783
Ramos, M. M. B., Gartti-Jardim, E. C., and Gaetti-Jardim Junior, E. (2009). Resistance to tetracycline and 2-lactams and distribution of resistance markers in enteric microorganisms and pseudomonads isolated from the oral cavity. J. Appl. Oral Sci. 17, 13–18. doi: 10.1590/S1678-77572009000700004
Roberts, M. C. (2003). Tetracycline therapy: update. Clin. Infect. Dis. 36, 462–467. doi: 10.1086/367622
Rubinstein, E., and Vaughan, D. (2005). Tigecycline: a novel glycylcycline. Drugs 65, 1317–1336. doi: 10.2165/00003495-200565180-00009
Sanger, F., Nicklen, S., and Coulson, A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 74, 5463–5467. doi: 10.1073/pnas.74.12.5463
Segata, N., Haake, S. K., Mannon, P., Lemon, K. P., Waldron, L., Gevers, D., et al. (2012). Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 13:R42. doi: 10.1186/gb-2012-13-6-r42
Seville, L. A., Patterson, A. J., Scott, K. P., Mullany, P., Quail, M. A., Parkhill, J., et al. (2009). Distribution of tetracycline and erythromycin resistance genes among human oral and fecal metagenomic DNA. Microb. Drug Resist. 15, 159–166. doi: 10.1089/mdr.2009.0916
Shen, F., Han, Q., Xie, D., Fang, M., Zeng, H., and Deng, Y. (2015). Efficacy and safety of tigecycline for the treatment of severe infectious diseases: an updated meta-analysis of RCTs. Int. J. Infect. Dis. 39, 25–33. doi: 10.1016/j.ijid.2015.08.009
Someya, Y., Yamaguchi, A., and Sawai, T. (1995). A novel glycylcycline, 9-(N,N-dimethylglycylamido)-6-demethyl-6-deoxytetracycline, is neither transported nor recognized by the transposon Tn10-encoded metal-tetracycline/H+ antiporter. Antimicrob. Agents Chemother. 39, 247–249. doi: 10.1128/AAC.39.1.247
Torres, C., Galian, C., Freiberg, C., Fantino, J. R., and Jault, J. M. (2009). The YheI/YheH heterodimer from Bacillus subtilis is a multidrug ABC transporter. Biochim. Biophys. Acta 1788, 615–622. doi: 10.1016/j.bbamem.2008.12.012
Vaara, M. (1993). Outer membrane permeability barrier to azithromycin, clarithromycin, and roxithromycin in gram-negative enteric bacteria. Antimicrob. Agents Chemother. 37, 354–356. doi: 10.1128/AAC.37.2.354
Van Berkel, M. A., Twilla, J. D., and England, B. S. (2016). Emergency department management of a myasthenia gravis patient with community-acquired pneumonia: does initial antibiotic choice lead to cure or crisis? J. Emerg. Med. 50, 281–285. doi: 10.1016/j.jemermed.2015.04.019
Villa, L., Feudi, C., Fortini, D., Garcia-Fernandez, A., and Carattoli, A. (2014). Genomics of KPC-producing Klebsiella pneumoniae sequence type 512 clone highlights the role of RamR and ribosomal S10 protein mutations in conferring tigecycline resistance. Antimicrob. Agents Chemother. 58, 1707–1712. doi: 10.1128/aac.01803-13
Villedieu, A., Diaz-Torres, M. L., Hunt, N., McNab, R., Spratt, D. A., Wilson, M., et al. (2003). Prevalence of tetracycline resistance genes in oral bacteria. Antimicrob. Agents Chemother. 47, 878–882. doi: 10.1128/AAC.47.3.878-882.2003
Warburton, P. J., Ciric, L., Lerner, A., Seville, L. A., Roberts, A. P., Mullany, P., et al. (2013). TetAB46, a predicted heterodimeric ABC transporter conferring tetracycline resistance in Streptococcus australis isolated from the oral cavity. J. Antimicrob. Chemother. 68, 17–22. doi: 10.1093/jac/dks351
Wu, N., Qiao, M., Zhang, B., Cheng, W. D., and Zhu, Y. G. (2010). Abundance and diversity of tetracycline resistance genes in soils adjacent to representative swine feedlots in China. Environ. Sci. Technol. 44, 6933–6939. doi: 10.1021/es1007802
Yang, W., Moore, I. F., Koteva, K. P., Bareich, D. C., Hughes, D. W., and Wright, G. D. (2004). TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics. J. Biol. Chem. 279, 52346–52352. doi: 10.1074/jbc.M409573200
Yao, X., Doi, Y., Zeng, L., Lv, L., and Liu, J.-H. (2016). Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect. Dis. 16, 288–289. doi: 10.1016/S1473-3099(16)00057-8
Zheng, X., Freeman, A. F., Villafranca, J., Shortridge, D., Beyer, J., Kabat, W., et al. (2004). Antimicrobial susceptibilities of invasive pediatric Abiotrophia and Granulicatella isolates. J. Clin. Microbiol. 42, 4323–4326. doi: 10.1128/JCM.42.9.4323-4326.2004
Keywords: tetracycline, tigecycline, metagenomics, ABC transporter, fitness, antibiotic resistance
Citation: Reynolds LJ, Roberts AP and Anjum MF (2016) Efflux in the Oral Metagenome: The Discovery of a Novel Tetracycline and Tigecycline ABC Transporter. Front. Microbiol. 7:1923. doi: 10.3389/fmicb.2016.01923
Received: 16 July 2016; Accepted: 16 November 2016;
Published: 06 December 2016.
Edited by:
Yuji Morita, Aichi Gakuin University, JapanReviewed by:
Yaqi You, University of Nevada, Reno, USAMarkus Seeger, University of Zurich, Switzerland
Jennifer Karin Bender, Robert Koch Institute, Germany
Copyright © 2016 Reynolds, Roberts and Anjum. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liam J. Reynolds, TGlhbS5yZXlub2xkcy4xMkB1Y2wuYWMudWs=
 Liam J. Reynolds
Liam J. Reynolds Adam P. Roberts
Adam P. Roberts Muna F. Anjum
Muna F. Anjum