- 1Department of Molecular Genetics, Groningen Biomolecular Sciences and Biotechnology Institute, University of Groningen, Groningen, Netherlands
- 2Department of Bioinformatics and Biotechnology, Government College University, Faisalabad, Pakistan
- 3Department of Microbiology, Tumor and Cell Biology, Karolinska Institutet, Stockholm, Sweden
In this study, we investigated the transcriptomic response of Streptococcus pneumoniae D39 to cysteine. Transcriptome comparison of the D39 wild-type grown at a restricted concentration of cysteine (0.03 mM) to one grown at a high concentration of cysteine (50 mM) in chemically-defined medium (CDM) revealed elevated expression of various genes/operons, i.e., spd-0150, metQ, spd-0431, metEF, gshT, spd-0618, fhs, tcyB, metB-csd, metA, spd-1898, yvdE, and cysK, likely to be involved in the transport and utilization of cysteine and/or methionine. Microarray-based data were further confirmed by quantitative RT-PCR. Promoter lacZ-fusion studies and quantitative RT-PCR data showed that the transcriptional regulator CmbR acts as a transcriptional repressor of spd-0150, metEF, gshT, spd-0618, tcyB, metA, and yvdE, putatively involved in cysteine uptake and utilization. The operator site of CmbR in the promoter regions of CmbR-regulated genes is predicted and confirmed by mutating or deleting CmbR operator sites from the promoter regions of these genes.
Introduction
The major human pathogen Streptococcus pneumoniae colonizes the human nasopharynx and is the causal agent of many diseases, including pneumonia, sepsis, meningitis, and others. Pneumococcal nitrogen metabolism and regulation have been studied extensively as the appropriate acquisition and metabolism of nutrients are important for its lifestyle (Hendriksen et al., 2008). Sulfur is an integral part of many essential components of the cell, such as cysteine, methionine, thiamine, biotin, lipoic acid, coenzyme A, etc. Among these compounds, cysteine plays a key role, as it is the most important sulfur-containing compound-forming metabolite and its de novo synthesis signifies the central pathway of sulfur acquisition in microorganisms and plants (Sperandio et al., 2005). Many important proteins (such as cytochromes and aconitase) also have cysteine as an essential amino acid in their catalytic domains. Moreover, cysteine (and the dimer cysteine) helps in protein folding, assembly and stability, being involved in the formation of disulfide bounds. Cysteine-derived proteins (such as thioredoxin and glutathione) help in countering oxidative stress (Sperandio et al., 2005). Methionine is another sulfur-containing amino acid, regulating the initiation of translation and is vital to several methyl-transferase reactions (Sperandio et al., 2005). Microorganisms can synthesize methionine by converting homoserine to homocysteine through addition of a sulfur group from either cysteine (requiring MetABC), sulfide (requiring MetA and CysD) or by using the SAM (S-adenosylmethionine) recycling pathway (MetK, Pfs, and LuxS) (Kovaleva and Gelfand, 2007). Homocysteine is then methylated by methionine synthase (MetE) in conjunction with a methylenetetrahydrofolate reductase (MetF), with the methyl group supplied by 5-methyltetrahydrofolate, to form methionine (Kovaleva and Gelfand, 2007).
Cysteine and methionine concentrations might be regulating bacterial growth in different conditions, such as pathogenic events or fermentation processes, as these amino acids have essential roles in metabolism (Schell, 1993). In Brucella melitensis (Lestrate et al., 2000), Haemophilus parasuis (Hill et al., 2003) and, Salmonella enterica (Ejim et al., 2004), sulfur-containing amino acid biosynthesis genes have been characterized as virulence factors. The cysDNC operon involved in the sulfate activation pathway forms a stress-induced operon in Mycobacterium tuberculosis (Pinto et al., 2004), whereas several thiol- and cysteine metabolism genes comprise the sigH regulon necessary for optimal existence of the bacterium in macrophages (Manganelli et al., 2002). Furthermore, cysteine metabolism also controls the regulation of toxin formation in Bordetella pertussis (Bogdan et al., 2001). Similarly, cysteine regulates a signaling molecule derivative of sulfur metabolism, autoinducer 2, which is conserved in both Gram-positive and -negative bacteria and is involved in interspecies communication and regulation of virulence factors (Sperandio et al., 1999; Marouni and Sela, 2003).
Our current study elucidates the effect of cysteine on the global gene expression of S. pneumoniae and characterizes the role of the transcriptional regulator CmbR in regulation of spd-0150, metEF, gshT, spd-0618, tcyB, metA, and yvdE. The transcriptional regulator CmbR acts as a transcriptional repressor for a number of genes/operons involved in cysteine uptake and utilization. The putative operator site (5′-GYGATAAAAAWWAYTTATMAC-3′ where Y = T/C, W = A/T and M = A/C) of CmbR in the promoter regions of spd-0150, metEF, gshT, spd-0618, tcyB, metA, and yvdE is predicted and confirmed by promoter mutational/deletion experiments. Moreover, this site is found highly conserved in other pneumococcal strains and streptococci.
Materials and Methods
Bacterial Strains and Growth Conditions
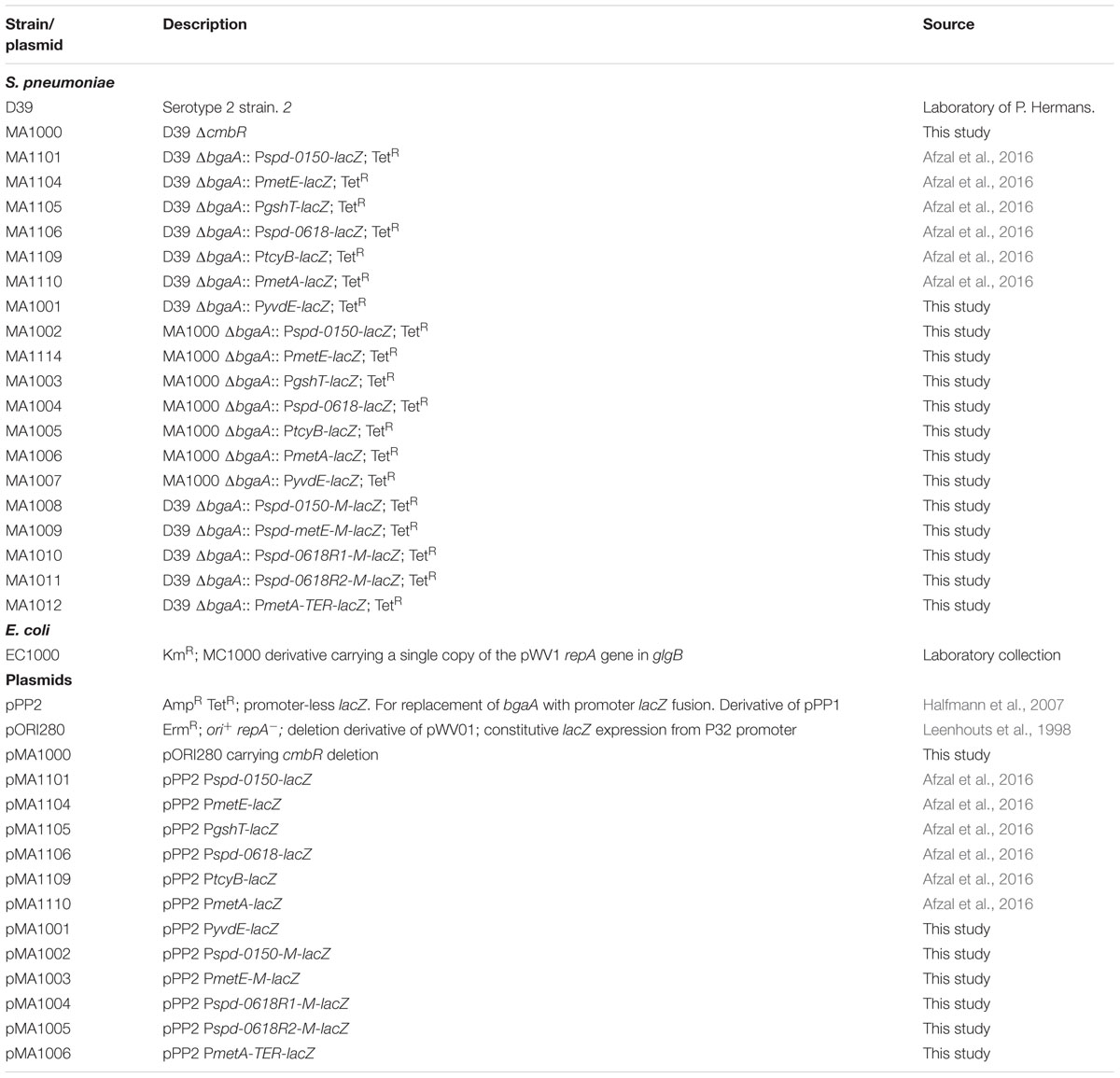
Bacterial strains and plasmids used in this study are listed in Table 1. S. pneumoniae D39 was grown as described previously (Kloosterman et al., 2006a; Afzal et al., 2014). For β-galactosidase assays, derivatives of S. pneumoniae D39 were grown in a chemically defined medium (CDM) (Kloosterman and Kuipers, 2011) supplemented either with 0.03 or 50 mM cysteine. CDM was prepared without cysteine. For selection on antibiotics, the medium was supplemented with the following concentrations of antibiotics: tetracycline: 2.5 μg/ml for S. pneumoniae; ampicillin: 100 μg/ml for Escherichia coli and erythromycin: 0.25 μg/ml for S. pneumoniae and 120 μg/ml for E. coli. All bacterial strains used in this study were stored in 10% (v/v) glycerol at -80°C. For PCR amplification, chromosomal DNA of S. pneumoniae D39 (Lanie et al., 2007) was used as a template. Primers used in this study are based on the sequence of the S. pneumoniae D39 genome and are listed in Table 2.
Construction of a cmbR Mutant
A markerless cmbR mutant (MA1000) was constructed in S. pneumoniae D39 using pORI280, as described before (Kloosterman et al., 2006a). Primer pairs cmbR-1/cmbR-2 and cmbR-3/cmbR-4 were used to generate PCR fragments of the left and right flanking regions of cmbR. The integrity of the cmbR mutant (MA1000) was further confirmed by PCR and DNA sequencing.
Construction of Promoter lacZ-Fusions and β-Galactosidase Assays
Chromosomal transcriptional lacZ-fusions to the spd-0150, metE, gshT, spd-0618, tcyB, metA, and yvdE promoters were constructed in our previous study (Afzal et al., 2016). These constructs were further introduced into the D39 ΔcmbR (MA1000) strain resulting in strains MA1002-06, respectively. Transcriptional lacZ-fusion to the yvdE promoter was constructed in pPP2 (Halfmann et al., 2007) with primer pairs mentioned in Table 2 resulting in pMA1001. This construct was further introduced into the D39 wild-type and the D39 ΔcmbR (MA1000) strains resulting in strains MA1001 and MA1007, respectively. The following sub-clones of Pspd-0150, PmetE, Pspd-0618, and PmetA were made in pPP2 (Halfmann et al., 2007) using the primer pairs mentioned in Table 2: Pspd-0150-M (mutation in the cmbR site), PmetE-M (mutation in the cmbR site), Pspd-0618R1-M (mutation in the cmbR site 1), Pspd-0618R2-M (mutation in the cmbR site 2), and PmetA-TER (termination of the cmbR site), resulting in plasmids pMA1002-06, respectively. These constructs were introduced into the S. pneumoniae D39 wild-type, resulting in strains MA1008-12, respectively. All plasmid constructs were checked for the presence of the insert by PCR and DNA sequencing.
β-galactosidase assays were performed as described before (Israelsen et al., 1995; Halfmann et al., 2007) using cells that were harvested in the mid-exponential growth phase, and grown in CDM supplemented either with 0.03 or 50 mM cysteine.
Microarray Analysis
Microarray analysis was performed as described before (Afzal et al., 2015a; Shafeeq et al., 2015). For DNA microarray analysis of S. pneumoniae in the presence of cysteine, the transcriptomes of S. pneumoniae D39 wild-type, grown in replicates in CDM with 0.03 mM cysteine, was compared to that grown in CDM with 50 mM cysteine and harvested at respective mid-exponential growth phases. For the identification of differentially expressed genes, a Bayesian p-value of <0.001 and a fold-change cut-off >1.5 was applied. RNA isolation was performed as described before (Afzal et al., 2015a). All other procedures regarding the DNA microarray experiments and data analysis were performed as previously described (Shafeeq et al., 2011a,b; Afzal et al., 2015b). Microarray data have been submitted to GEO under the accession number GSE89458.
Reverse Transcription (RT)-PCR and Purification for Quantitative RT-PCR
For quantitative RT-PCR, S. pneumoniae D39 wild-type and D39 ΔcmbR were grown in replicates in CDM supplemented with either 0.03 mM or 50 mM cysteine. RNA isolation was done as described before (Afzal et al., 2015a). First, strand cDNA synthesis was performed on RNA (Shafeeq et al., 2011b). cDNA (2 μl) was amplified in a 20 μl reaction volume that contained 3 pmol of each primer (Table 2) and the reactions were performed in three technical replicates on two biological replicates of RNA (Shafeeq et al., 2011b). The transcription level of specific genes was normalized to gyrA transcription, amplified in parallel with gyrA-F and gyrA-R primers. The results were interpreted using the comparative CT method (Schmittgen and Livak, 2008). Differences in expression of twofold or greater relative to control were considered as significant.
Results
Cysteine-Dependent Gene Regulation in S. pneumoniae D39
Cysteine is one of the most important amino acids for bacteria. It is also present in human blood plasma at concentration of 0.03 mM (Lopez, 2013). To study the impact of cysteine on the transcriptome of S. pneumoniae D39 wild-type, we performed microarray comparison of S. pneumoniae D39 grown in CDM with 0.03–50 mM cysteine. 0.03 mM concentration was chosen, as this is the concentration of cysteine in human blood plasma (Lopez, 2013). 50 mM concentration of cysteine is normally used to prepare CDM. A number of genes/gene clusters were differentially regulated under our tested conditions (Table 3). The expression of spd-0447-49 and spd-1098-99 was altered under our tested conditions. These genes belong to the glutamine regulon and their expression has been reported to be downregulated in the presence of a nitrogen source (Kloosterman et al., 2006b). The expression of important metal-related genes (prtA, psaBC and spd-1402) was downregulated under our tested conditions. These genes belong to the PsaR regulon and repressed by transcriptional regulator PsaR in the presence of manganese (Johnston et al., 2006). These genes have been shown to have role in virulence of pneumococcus (Kloosterman et al., 2008). Therefore, it might be interesting to further explore the role of cysteine in the regulation of these genes.
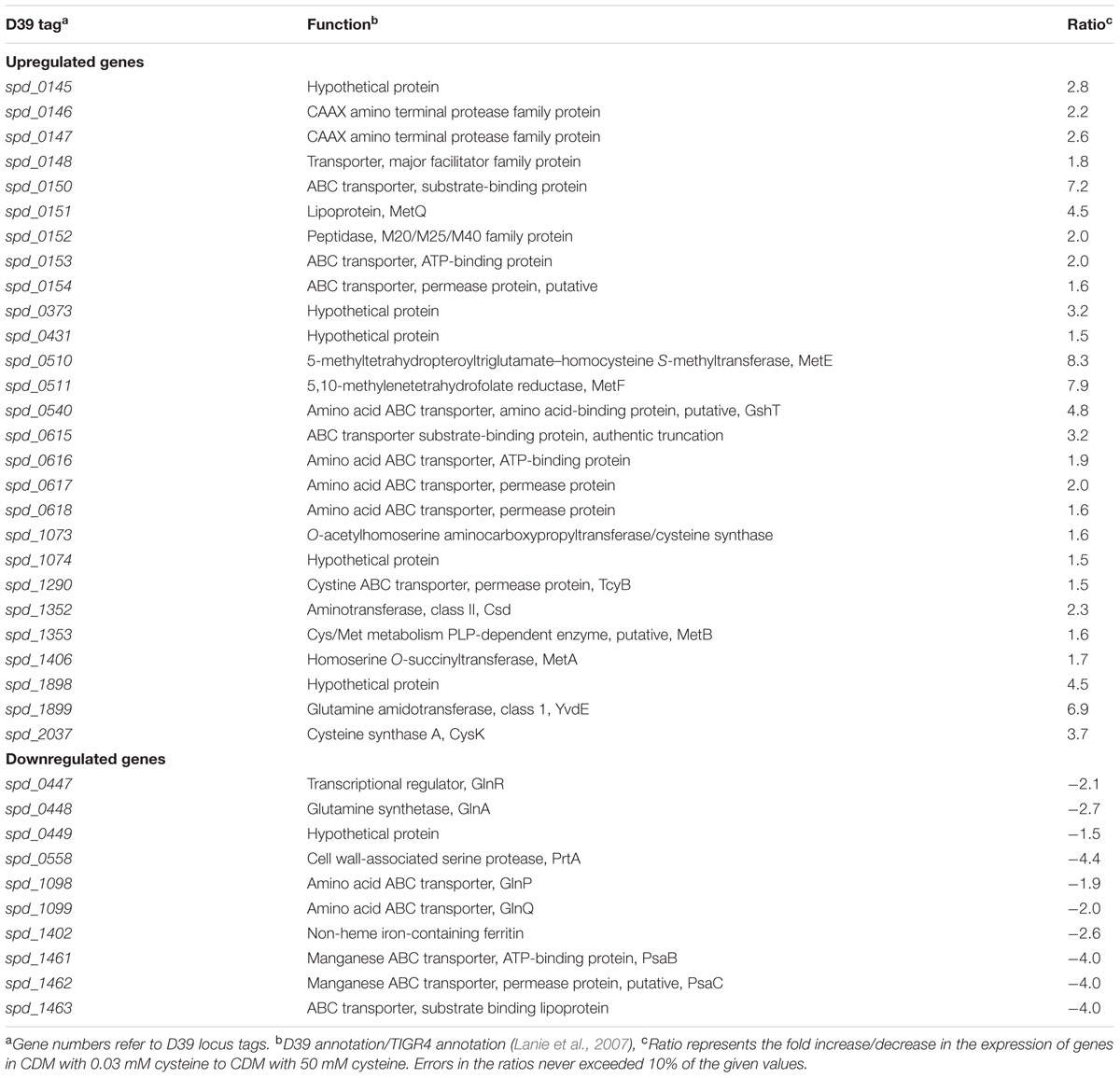
TABLE 3. Summary of transcriptome comparison of S. pneumoniae D39 wild-type grown in CDM with 0.03 mM cysteine to grown in CDM with 50 mM cysteine.
The expression of putative methionine/cysteine transport and biosynthesis pathway genes (spd-0150, metQ, spd-0431, metEF, gshT, spd-0618, fhs, tcyB, metB-csd, metA, spd-1898, yvdE, and cysK) was significantly upregulated in the presence of 0.03 mM cysteine. The role of methionine in regulation of these genes has been demonstrated in our recent study (Afzal et al., 2016). Furthermore, we showed that a transcriptional regulator CmhR acts as a transcriptional activator of fhs, folD, metB, metEF, metQ, and spd-0431. However, regulatory mechanism of spd-0150, gshT, spd-0618, tcyB, metA, spd-1898, yvdE, and cysK is not explored. Therefore, we decided to further explore the role of cysteine in the regulation of these genes.
Confirmation of Cysteine-Dependent Expression of spd-0150, metQ, spd-0431, metEF, gshT, spd-0618, fhs, tcyB, metB-csd, metA, spd-1898, yvdE, and cysK
To confirm our microarray results and to study the expression of spd-0150, metQ, spd-0431, metEF, gshT, spd-0618, fhs, tcyB, metB-csd, metA, spd-1898, yvdE, and cysK under limiting cysteine concentration in CDM, we performed quantitative RT-PCR on these genes. Our quantitative RT-PCR results demonstrated that the expression of these genes was increased significantly in CDM with 0.03 mM cysteine, when compared to 50 mM (Figure 1). These data not only confirms our microarray results mentioned above, but also suggests a direct role of these genes in cysteine transport and biosynthesis.
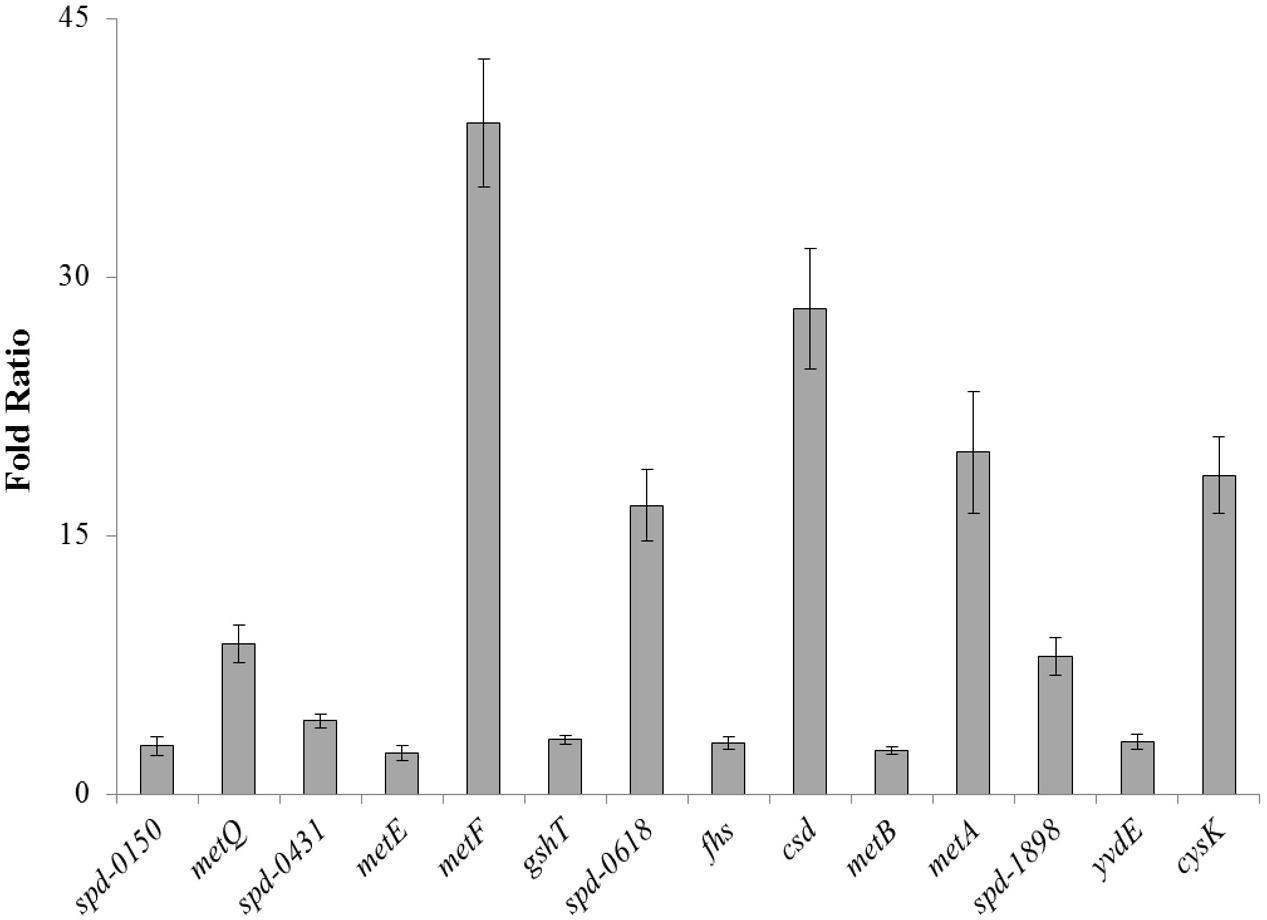
FIGURE 1. The relative increase in the expression of spd-0150, metQ, spd-0431, metEF, gshT, spd-0618, fhs, tcyB, metB-csd, metA, spd-1898, yvdE, and cysK in S. pneumoniae D39 wild-type grown in CDM with 0.03 mM cysteine compared to that grown in CDM with 50 mM cysteine. The expression of these genes was normalized with housekeeping gene gyrA.
There are three LysR-type transcriptional regulators in different bacteria, which have been shown to be involved in the regulation of sulfur amino acids (Sperandio et al., 2005, 2010). S. pneumoniae also has two LysR-type transcriptional regulators (CmhR and CmbR), which are proposed to be involved in the regulation of the sulfur amino acids (Novichkov et al., 2010). Our recent study has revealed the regulatory mechanism of CmhR in S. pneumoniae and demonstrates that CmhR acts as a transcriptional activator of the fhs, folD, metB-csd, metEF, metQ, and spd-0431 in the presence of methionine (Afzal et al., 2016). The presence of CmbR (putative Cysteine Methionine Biosynthesis Regulator) in S. pneumoniae suggests its involvement in the regulation of cysteine-responsive genes. Therefore, we decided to further study the role of transcriptional regulator CmbR in the regulation of cysteine transport and biosynthesis genes.
Prediction of the CmbR Regulatory Site and the Role of CmbR as a Transcriptional Repressor of spd-0150, metEF, gshT, spd-0618, tcyB, metA, and yvdE
The presence of cmbR in the S. pneumoniae genome suggests its involvement in the regulation of cysteine-responsive genes. cmbR codes for the putative transcriptional regulator CmbR, which belongs to the LysR family of proteins. CmbR is a homolog of a LysR-type regulator (also called FhuR) of Lactococcus lactis and Streptococcus mutans (Fernández et al., 2002; Sperandio et al., 2005, 2010). To study the role of CmbR in S. pneumoniae D39, we analysed the promoter regions of cysteine-regulated genes and predicted a 21-bp palindromic-like sequence (that has high homology with the FhuR binding site of L. lactis and S. mutans) in the promoter regions of spd-0150, metEF, gshT, spd-0618, tcyB, metA, and yvdE indicating that CmbR regulon in S. pneumoniae D39 is comprised of these genes. This DNA sequence might serve as the CmbR operator site in S. pneumoniae. The CmbR site present in the promoter regions of spd-0150, metEF, gshT, spd-0618, tcyB, metA, and yvdE is shown in Figure 2A. A weight matrix of these CmbR sites (5′-GYGATAAAAAWWAYTTATMAC-3′) was constructed (Figure 2B). Promoter regions of these genes were also examined in other streptococcal species (Streptococcus mitis, Streptococcus agalactiae, Streptococcus gallolyticus, Streptococcus gordonii, S. mutans, Streptococcus sanguinis, Streptococcus suis, and Streptococcus thermophilus) to check whether the CmbR site is also conserved in those streptococci. The CmbR site is highly conserved in these streptococci as well. Moreover, we constructed a phylogenetic tree of CmbR present in different streptococci, which shows that it is highly conserved in these streptococci (Figure 3).

FIGURE 2. Identification of the CmbR operator site. (A) Weight matrix of the identified CmbR operator site in the promoter region of spd-0150, metEF, gshT, spd-0618, tcyB, metA, and yvdE. (B) Position of the CmbR operator site in the promoter region of spd-0150, metEF, gshT, spd-0618, tcyB, metA, and yvdE. Translational start sites are italic and putative CmbR operator sites are bold and underlined.
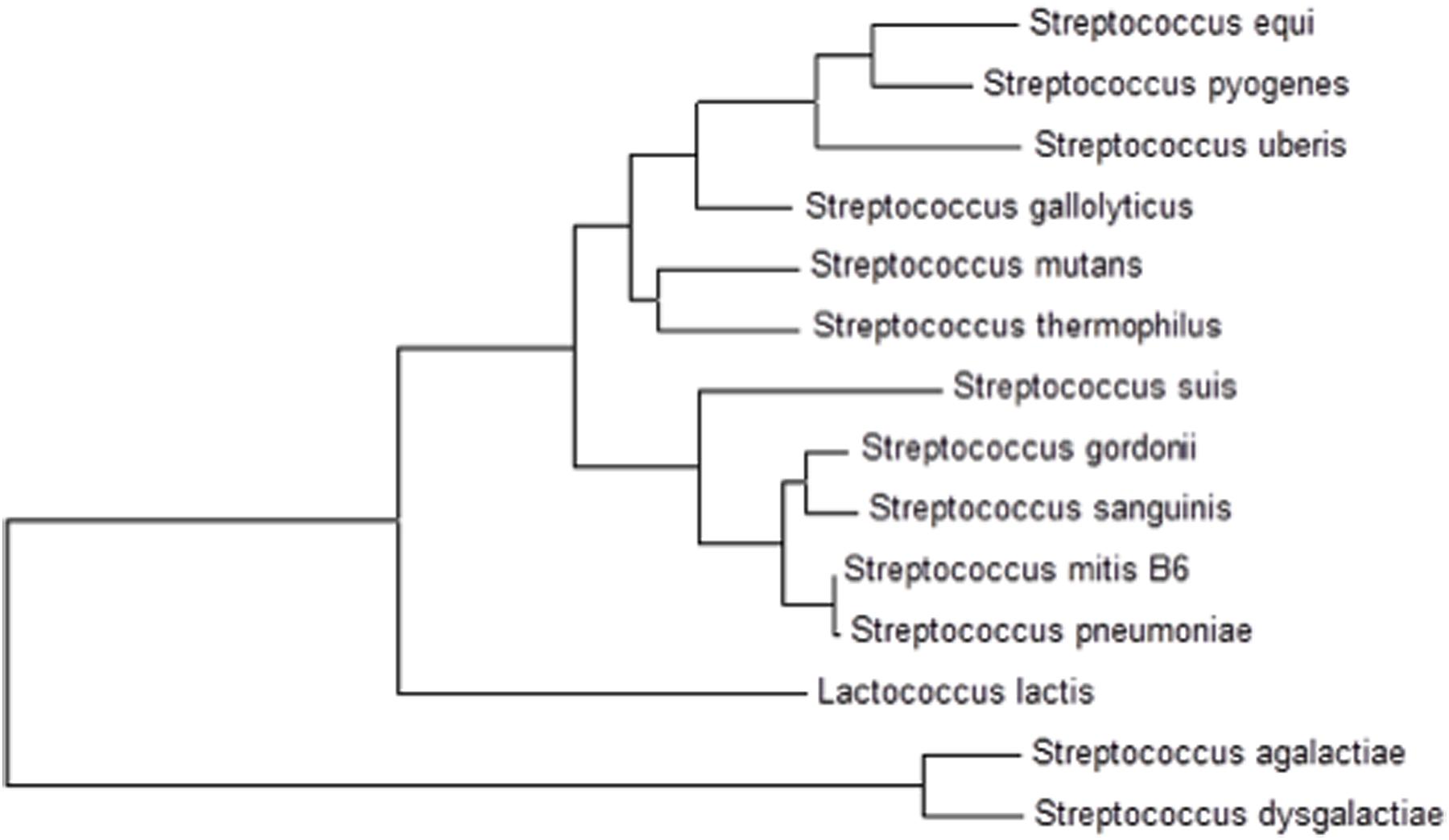
FIGURE 3. A phylogenetic tree of CmbR in different streptococci showing conservation of CmbR in these streptococci.
To investigate the role of CmbR in the regulation of spd-0150, metEF, gshT, spd-0618, tcyB, metA, and yvdE, we performed β-galactosidase assays with the promoter lacZ-fusions that were constructed in the S. pneumoniae D39 wild-type and D39 ΔcmhR in CDM with 50 mM cysteine. We used CDM with 50 mM cysteine, because we assumed the role of CmbR as a transcriptional repressor of these genes in the presence of cysteine. The results of the β-galactosidase assays showed that the activity of Pspd-0150-lacZ, PmetE-lacZ, PgshT-lacZ, Pspd-0618-lacZ, PtcyB-lacZ, PmetA-lacZ, and PyvdE-lacZ increased significantly in D39 ΔcmbR compared to D39 wild-type in CDM with 50 mM cysteine (Figure 4). Increased expression of these promoters in D39 ΔcmbR indicates the role of CmbR as a transcriptional repressor of spd-0150, metEF, gshT, spd-0618, tcyB, metA, and yvdE.
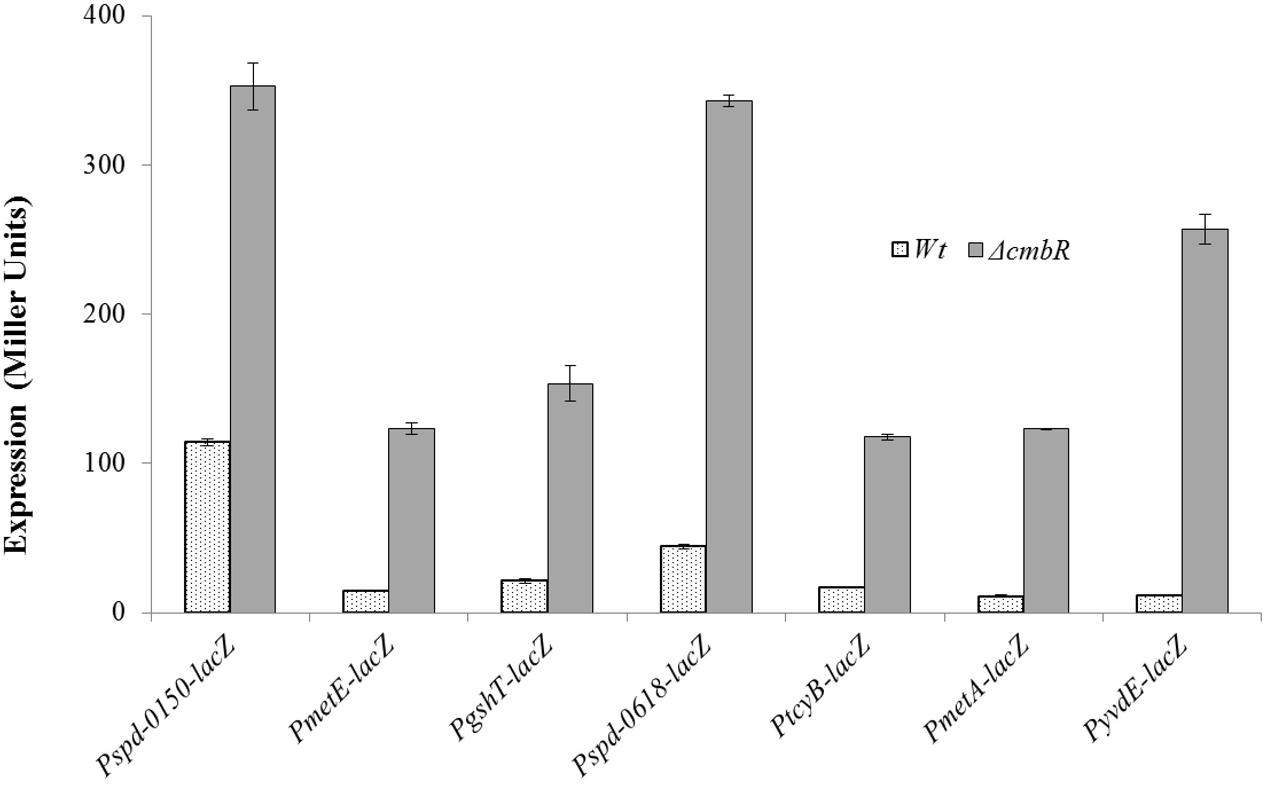
FIGURE 4. Expression levels (in Miller units) of Pspd-0150-lacZ, PmetE-lacZ, PgshT-lacZ, Pspd-0618-lacZ, PtcyB-lacZ, PmetA-lacZ, and PyvdE-lacZ in CDM with 50 mM cysteine in S. pneumoniae D39 wild-type and D39 ΔcmbR.
Confirmation of CmbR-Dependent Expression of spd-0150, metEF, gshT, spd-0618, tcyB, metA, and yvdE
To further confirm the role of CmbR as a transcriptional repressor of spd-0150, metEF, gshT, spd-0618, tcyB, metA, and yvdE, we performed quantitative RT-PCR on these genes in the presence of 50mM cysteine. Our quantitative RT-PCR results demonstrated that the expression of spd-0150, metEF, gshT, spd-0618, tcyB, metA, and yvdE increased significantly in D39 ΔcmbR compared to the D39 wild-type (Figure 5). These data provide further confirmation of our results that CmbR acts as a transcriptional repressor of spd-0150, metEF, gshT, spd-0618, tcyBC, metA, and yvdE.
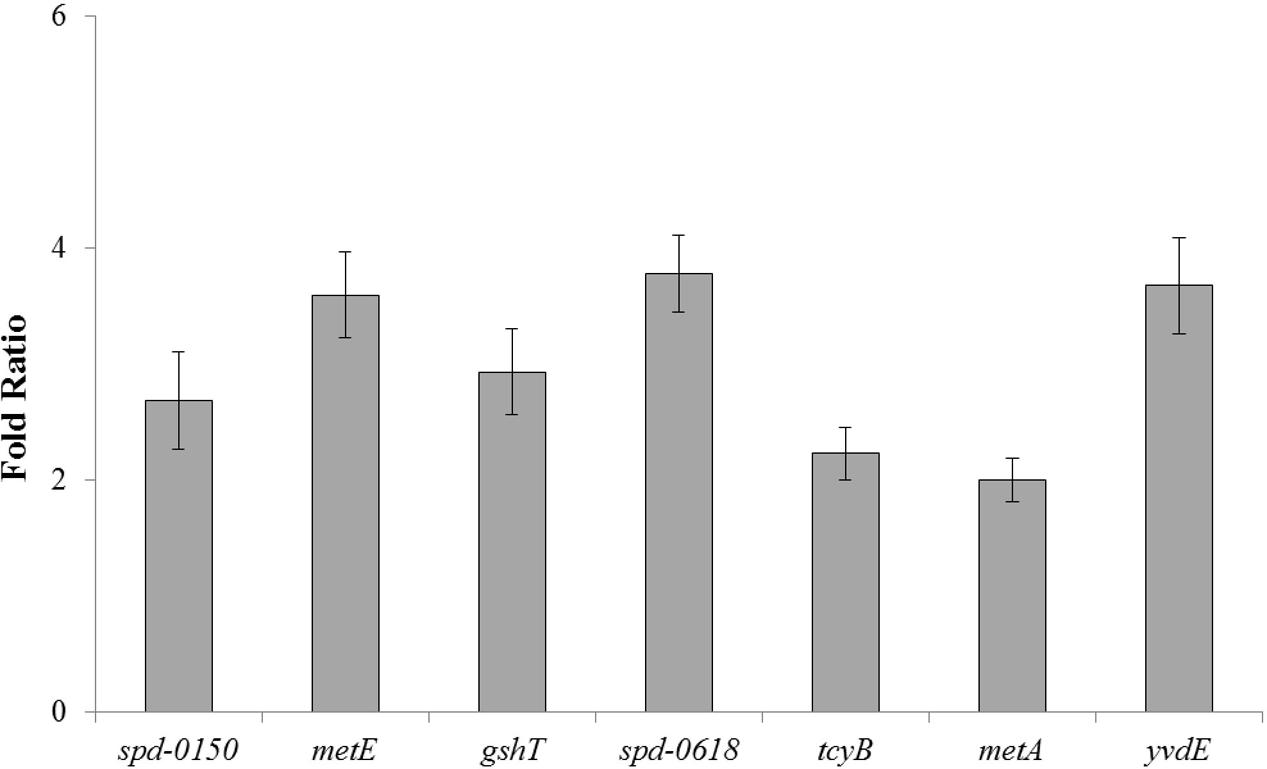
FIGURE 5. The relative increase in the expression of spd-0150, metEF, gshT, spd-0618, tcyB, metA, and yvdE in S. pneumoniae D39 ΔcmbR compared to D39 wild-type grown in CDM with 50 mM cysteine. The expression of these genes was normalized with that of housekeeping gene gyrA.
Confirmation of a CmbR Operator Site in CmbR-Regulated Genes
To verify the functionality of the predicted CmbR operator site present in the promoter regions of the CmbR regulated genes (spd-0150, metEF, gshT, spd-0618, tcyBC, metA, and yvdE), we made transcriptional lacZ-fusions of Pspd-0150, PmetE, Pspd-0618, and PmetA, where conserved bases in the cmbR sites were mutated in Pspd-0150 (5′-CCCATAAAAAAAACTTATCAC-3′ to 5′-CCCGCGAAAAAAACTTATCAC-3′), PmetE (5′-CTTATAAGAATTACTAATAAC-3′ to 5′-CTTGCAAGAATTACTAGCAAC-3′), Pspd-0618 (R1: 5′-ATGGTAAAAAATCCTTATAAC-3′ to 5′-ATGGCCAAAAATCCTGCGAAC-3′ and R2: 5′-GCAATAAAAAATAGATATTAT-3′ to 5′-GCAGCGAAAAATAGAGCGTAT-3′), and terminated in PmetA (CmbR site is deleted), and β-galactosidase assays were performed on cells grown in CDM with 50 mM cysteine. β-galactosidase assays data revealed that mutating the CmbR operator site in Pspd-0150 and PmetE, and deletion of the CmbR operator site in PmetA led to significantly increased expression of these promoter lacZ-fusions in the presence of 50 mM cysteine (Figure 6). These data confirm that the predicted CmbR sites present in the promoter regions of these genes are functional and intact in S. pneumoniae D39. Two putative operator sites of CmbR are present in Pspd-0618 (R1 and R2). We mutated both sites individually. We could only observe derepression (caused by CmbR) of Pspd-0618 when operator site 1 (R1) was mutated and did not witness any change in the activity of Pspd-0618 due to mutations in operator site 2 (R2). This suggests that only operator site 1 (R1) is a functional operator site for CmbR in Pspd-0618.
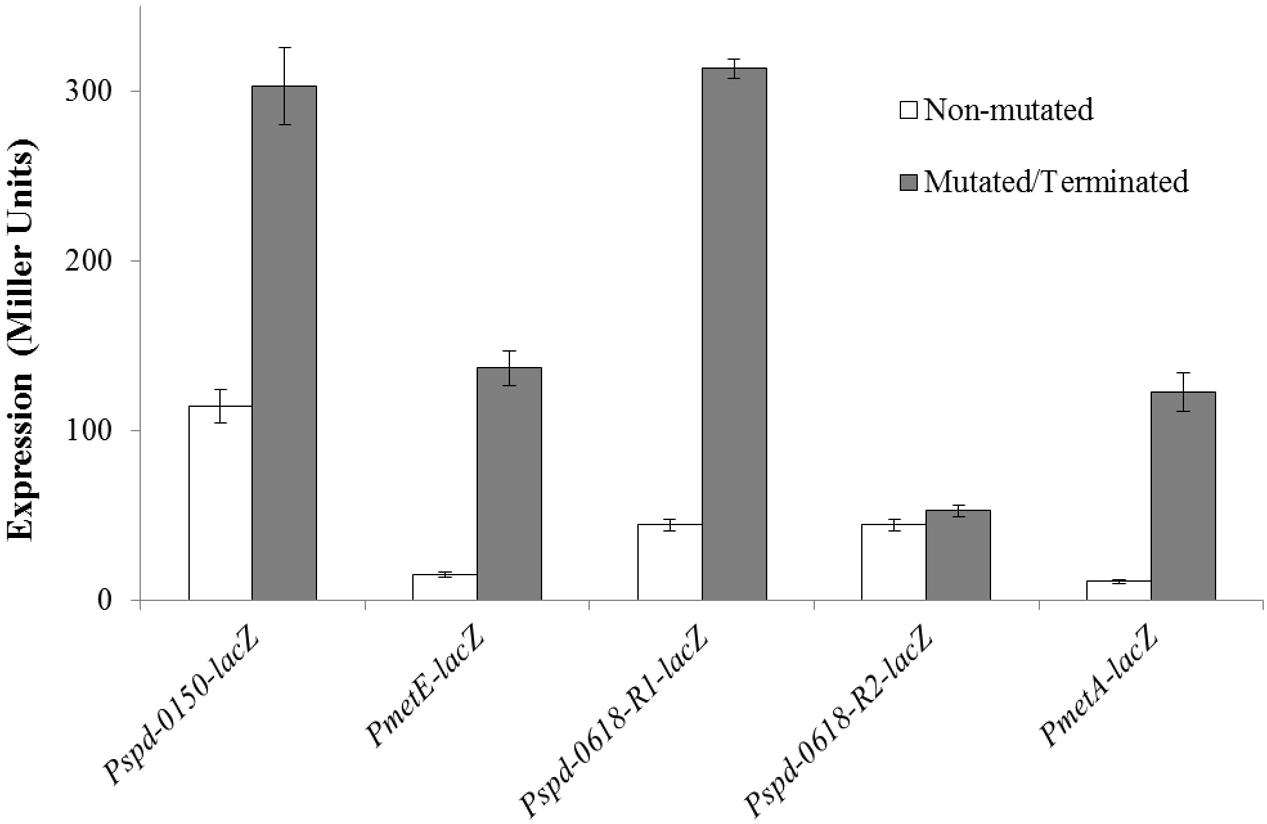
FIGURE 6. Expression levels (in Miller units) of mutated/terminated and non-mutated CmbR sites in Pspd-0150-lacZ, PmetEF-lacZ, Pspd-0618-lacZ, and PmetA-TER-lacZ in S. pneumoniae D39 wild-type grown in CDM with 50 mM cysteine.
Discussion
The sulfur-containing amino acids cysteine and methionine play essential role in various metabolic processes in the cell, especially because of their sulfur group, which plays a vital role in the catalytic sites of many enzymes and participates in ion- and redox metabolism (Ayala-Castro et al., 2008). Involvement of multiple mechanisms in the regulation of these pathways in various groups of bacteria makes the regulatory phenomenon even more interesting, as it seems to evolve faster than that of many other regulatory pathways (Fernández et al., 2002; Sperandio et al., 2005, 2010). CmbR regulates most of the cysteine and methionine genes in L. lactis, where it binds to a 13-bp box centered 46–53 bp upstream of transcriptional start sites, with a second box with a same consensus sequence is located upstream of the first binding box (separated by 8–10 bp) (Fernández et al., 2002; Golic et al., 2005; Sperandio et al., 2005). In other members of the closely related Streptococcaceae family, the existence of a different motif upstream of several potential cysteine genes (Kovaleva and Gelfand, 2007) suggests that the regulation of sulfur amino acid metabolism may be diverse. In this study, we studied the cysteine-dependent gene expression and the role of CmbR in S. pneumoniae D39, and demonstrated that CmbR acts as a transcriptional repressor of spd-0150, metEF, gshT, spd-0618, tcyB, metA, and yvdE.
The number of transcriptional factors regulating cysteine/methionine genes varies among different bacteria. This number is three in S. mutans (MetR, CysR and HomR), where these transcriptional regulators are phylogenetically related (Sperandio et al., 2010). CysR activates the transcription of cysK (codes for the cysteine biosynthesis enzyme), tcyABC, gshT (code for the cysteine and glutathione transporter systems), and homR. HomR is needed for the activation of metBC (code for the methionine biosynthesis enzymes), tcyDEFGH (involved in cysteine transport) and thiosulfate metabolism genes. Control of HomR by CysR emulates a cascade regulation for sulfur amino acid metabolism in S. mutans. Similarly, MtaR has been found to have a role in the regulation of the cysteine/methionine metabolism in S. agalactiae (Shelver et al., 2003). MetJ and MetR, regulate the expression of methionine biosynthetic genes in E. coli and S. enterica serovar Typhimurium (Weissbach and Brot, 1991). The E. coli met genes (except for metH) are negatively regulated by MetJ, a transcriptional repressor, with SAM (S-adenosylmethionine) serving as a co-repressor (Saint-Girons et al., 1988). These genes are also under the positive influence of a LysR-type transcriptional regulator MetR, with homocysteine as a co-effector (Cai et al., 1989; Mares et al., 1992; Cowan et al., 1993). CmbR in L. lactis also activates most genes involved in the methionine and cysteine biosynthesis pathway in the absence of cysteine (Sperandio et al., 2005). The regulatory proteins mentioned above belong to LysR family of transcription factors, which is the most abundant family of regulators in bacteria (Maddocks and Oyston, 2008). These regulators control diverse biological pathways such as central metabolism, cell division, quorum sensing, virulence, motility, nitrogen fixation, oxidative stress responses, toxin production, attachment, and secretion. These transcriptional regulators act as either activators or repressors, and often are transcribed divergently with one of the regulated genes (Schell, 1993). LysR-family transcriptional regulators consist of two characteristic domains, an N-terminal HTH DNA binding domain (PF00126) and a C-terminal substrate binding domain (PF03466). There are two transcriptional regulators in S. pneumoniae that control the expression of the cysteine and methionine genes (CmhR and CmbR) (Novichkov et al., 2010). CmhR in S. pneumoniae belongs to the LysR family of transcriptional factors and has an HTH (helix-turn-helix) domain and a substrate binding domain of LysR-type transcriptional regulators (LTTRs). CmhR acts as a transcriptional activator of the fhs, folD, metB, metEF, metQ, and spd-0431 genes and a 17-bp palindromic sequence (5′-TATAGTTTSAAACTATA-3′) is present in the promoter regions of CmhR-regulated genes in S. pneumoniae D39 (Afzal et al., 2016). There are two methionine transport systems [the methionine ABC uptake transporter (MUT)] (Merlin et al., 2002; Hullo et al., 2004) and a secondary transporter BcaP (den Hengst et al., 2006) in bacteria. metD codes for the MUT system in E. coli and also have the MetQ substrate binding protein (SBP), MetL trans-membrane permease and the MetN cytoplasmic ATP hydrolyzing protein (ATPase) (Merlin et al., 2002). The spd-0150–54 locus codes for a methionine uptake ABC transporter in S. pneumoniae D39 and deletion of the gene encoding the lipoprotein MetQ resulted in a strain that had hampered growth in methionine-restricted medium and no measurable uptake of radioactive methionine (Basavanna et al., 2013). Furthermore, deletion of locus encoding MetEF (which is also part of the CmhR regulon) resulted in an increase in the growth defect of the metQ deletion strain in methionine-restricted medium and in blood plasma, reinforcing a role for the products of these genes in methionine synthesis (Basavanna et al., 2013). CmhR-regulated genes have important roles in the transport and biosynthesis of methionine. Csd and MetE are part of methionine synthesis pathway as Csd coverts cystathionine into homocysteine and MetE converts homocysteine into methionine (Kanehisa et al., 2014). Cystathionine and homocysteine can also be formed from homoserine, where O-acetyl-L-homoserine is converted into cystathionine by MetB. O-acetyl-L-homoserine can also be converted into homocysteine by MetB and SPD-1073-74 (spd-1073-1074 encode an O-acetylhomoserine aminocarboxypropyltransferase/cysteine synthase and a hypothetical protein, respectively) (Kanehisa et al., 2014). Methionine can also be synthesized by other microbes as they may convert homoserine to homocysteine through addition of a sulfur group from either cysteine (involving MetABC), sulfide (involving MetA and CysD) or by using the SAM recycling pathway (MetK, Pfs and LuxS) (Kovaleva and Gelfand, 2007). MetE (methionine synthase) then methylates homocysteine in combination with MetF (methylenetetrahydrofolate reductase) and 5-methyltetrahydrofolate (FolD) provides it with the methyl group to form methionine (Ravanel et al., 1998; Kovaleva and Gelfand, 2007).
CmbR is the second one and acts as transcriptional repressor for spd-0150, metEF, gshT, spd-0618, tcyB, metA, and yvdE. The deletion of CmbR led to a significant increase in the activity of the CmbR-regulated promoters. Thus, it represents a different mode of regulation of the cysteine/methionine genes than in other related streptococci.
A number of metal-related genes are found to be differentially expressed in our tested conditions. These genes include psaBC and prtA, which belong to the PsaR regulon. psaBCA encode a Mn2+-dependent ABC transporter PsaBCA and prtA codes for a serine protease PrtA. The expression of these genes was also altered in our recent transcriptome of S. pneumoniae D39 grown in CDM with 0–10 mM methionine (Afzal et al., 2016). The DtxR-family transcriptional regulator PsaR represses the expression of the PsaR regulon in the presence of Mn2+ (Johnston et al., 2006). Zn2+, Ni2+ and Co2+ have been demonstrated to bind with PsaR and relieve the Mn2+-dependent repression of the PsaR regulon (Kloosterman et al., 2008; Manzoor et al., 2015a,b). The interplay or competition of metal ions plays a significant role in the regulation of metal-responsive genes. Competition of Mn2+ with Zn2+, Co2+, or Ni2+ in the regulation of the PsaR regulon by transcriptional regulator PsaR has already extensively been studied in S. pneumoniae (Kloosterman et al., 2008; Manzoor et al., 2015a,b). The significant changes in the expression of some of these metal-responsive genes in our study indicate the involvement of S-containing amino acids in this interplay as well, in an as yet unknown way.
Author Contributions
MA planned the experiments, performed experiments and wrote the manuscript. SS planned the experiments and wrote the manuscript. IM performed the experiments. OK planned the experiments and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
MA and IM are supported by the Government College University, Faisalabad, Pakistan under the faculty development program of HEC Pakistan.
References
Afzal, M., Kuipers, O. P., and Shafeeq, S. (2016). Methionine-mediated gene expression and characterization of the CmhR regulon in Streptococcus pneumoniae. Microb. Genomics. 2. doi: 10.1099/mgen.0.000091
Afzal, M., Manzoor, I., and Kuipers, O. P. (2015a). A fast and reliable pipeline for bacterial transcriptome analysis case study: serine-dependent gene regulation in Streptococcus pneumoniae. J. Vis. Exp. 98:e52649. doi: 10.3791/52649
Afzal, M., Shafeeq, S., Henriques-Normark, B., and Kuipers, O. P. (2015b). UlaR activates expression of the ula operon in Streptococcus pneumoniae in the presence of ascorbic acid. Microbiol. Read. Engl. 161, 41–49. doi: 10.1099/mic.0.083899-0
Afzal, M., Shafeeq, S., and Kuipers, O. P. (2014). LacR is a repressor of lacABCD and LacT an activator of lacTFEG, constituting the lac-gene cluster in Streptococcus pneumoniae. Appl. Environ. Microbiol. 80, 5349–5358. doi: 10.1128/AEM.01370-14
Ayala-Castro, C., Saini, A., and Outten, F. W. (2008). Fe-S cluster assembly pathways in bacteria. Microbiol. Mol. Biol. Rev. 72, 110–125. doi: 10.1128/MMBR.00034-07
Basavanna, S., Chimalapati, S., Maqbool, A., Rubbo, B., Yuste, J., Wilson, R. J., et al. (2013). The effects of methionine acquisition and synthesis on Streptococcus pneumoniae growth and virulence. PLoS ONE 8:e49638. doi: 10.1371/journal.pone.0049638
Bogdan, J. A., Nazario-Larrieu, J., Sarwar, J., Alexander, P., and Blake, M. S. (2001). Bordetella pertussis autoregulates pertussis toxin production through the metabolism of cysteine. Infect. Immun. 69, 6823–6830. doi: 10.1128/IAI.69.11.6823-6830.2001
Cai, X. Y., Maxon, M. E., Redfield, B., Glass, R., Brot, N., and Weissbach, H. (1989). Methionine synthesis in Escherichia coli: effect of the MetR protein on metE and metH expression. Proc. Natl. Acad. Sci. U.S.A. 86, 4407–4411. doi: 10.1073/pnas.86.12.4407
Cowan, J. M., Urbanowski, M. L., Talmi, M., and Stauffer, G. V. (1993). Regulation of the Salmonella typhimurium metF gene by the MetR protein. J. Bacteriol. 175, 5862–5866. doi: 10.1128/jb.175.18.5862-5866.1993
den Hengst, C. D., Groeneveld, M., Kuipers, O. P., and Kok, J. (2006). Identification and functional characterization of the Lactococcus lactis CodY-regulated branched-chain amino acid permease BcaP (CtrA). J. Bacteriol. 188, 3280–3289. doi: 10.1128/JB.188.9.3280-3289.2006
Ejim, L. J., D’Costa, V. M., Elowe, N. H., Loredo-Osti, J. C., Malo, D., and Wright, G. D. (2004). Cystathionine beta-lyase is important for virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 72, 3310–3314. doi: 10.1128/IAI.72.6.3310-3314.2004
Fernández, M., Kleerebezem, M., Kuipers, O. P., Siezen, R. J., and van Kranenburg, R. (2002). Regulation of the metC-cysK operon, involved in sulfur metabolism in Lactococcus lactis. J. Bacteriol. 184, 82–90. doi: 10.1128/JB.184.1.82-90.2002
Golic, N., Schliekelmann, M., Fernández, M., Kleerebezem, M., and van Kranenburg, R. (2005). Molecular characterization of the CmbR activator-binding site in the metC-cysK promoter region in Lactococcus lactis. Microbiol. Read. Engl. 151, 439–446. doi: 10.1099/mic.0.27411-0
Halfmann, A., Hakenbeck, R., and Bruckner, R. (2007). A new integrative reporter plasmid for Streptococcus pneumoniae. FEMS Microbiol. Lett. 268, 217–224. doi: 10.1111/j.1574-6968.2006.00584.x
Hendriksen, W. T., Bootsma, H. J., Estevao, S., Hoogenboezem, T., de, J. A., de, G. R., et al. (2008). CodY of Streptococcus pneumoniae: link between nutritional gene regulation and colonization. J. Bacteriol. 190, 590–601. doi: 10.1128/JB.00917-07
Hill, C. E., Metcalf, D. S., and MacInnes, J. I. (2003). A search for virulence genes of Haemophilus parasuis using differential display RT-PCR. Vet. Microbiol. 96, 189–202. doi: 10.1016/S0378-1135(03)00212-8
Hullo, M.-F., Auger, S., Dassa, E., Danchin, A., and Martin-Verstraete, I. (2004). The metNPQ operon of Bacillus subtilis encodes an ABC permease transporting methionine sulfoxide, D- and L-methionine. Res. Microbiol. 155, 80–86. doi: 10.1016/j.resmic.2003.11.008
Israelsen, H., Madsen, S. M., Vrang, A., Hansen, E. B., and Johansen, E. (1995). Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl. Environ. Microbiol. 61, 2540–2547.
Johnston, J. W., Briles, D. E., Myers, L. E., and Hollingshead, S. K. (2006). Mn2+-dependent regulation of multiple genes in Streptococcus pneumoniae through PsaR and the resultant impact on virulence. Infect. Immun. 74, 1171–1180. doi: 10.1128/IAI.74.2.1171-1180.2006
Kanehisa, M., Goto, S., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2014). Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205. doi: 10.1093/nar/gkt1076
Kloosterman, T. G., Bijlsma, J. J. E., Kok, J., and Kuipers, O. P. (2006a). To have neighbour’s fare: extending the molecular toolbox for Streptococcus pneumoniae. Microbiol. Read. Engl. 152, 351–359. doi: 10.1099/mic.0.28521-0
Kloosterman, T. G., Hendriksen, W. T., Bijlsma, J. J., Bootsma, H. J., van Hijum, S. A., Kok, J., et al. (2006b). Regulation of glutamine and glutamate metabolism by GlnR and GlnA in Streptococcus pneumoniae. J. Biol. Chem. 281, 25097–25109. doi: 10.1074/jbc.M601661200
Kloosterman, T. G., and Kuipers, O. P. (2011). Regulation of arginine acquisition and virulence gene expression in the human pathogen Streptococcus pneumoniae by transcription regulators ArgR1 and AhrC. J. Biol. Chem. 286, 44594–44605. doi: 10.1074/jbc.M111.295832
Kloosterman, T. G., Witwicki, R. M., van der Kooi-Pol, M. M., Bijlsma, J. J., and Kuipers, O. P. (2008). Opposite effects of Mn2+ and Zn2+ on PsaR-mediated expression of the virulence genes pcpA, prtA, and psaBCA of Streptococcus pneumoniae. J. Bacteriol. 190, 5382–5393. doi: 10.1128/JB.00307-08
Kovaleva, G. Y., and Gelfand, M. S. (2007). Transcriptional regulation of the methionine and cysteine transport and metabolism in streptococci. FEMS Microbiol. Lett. 276, 207–215. doi: 10.1111/j.1574-6968.2007.00934.x
Lanie, J. A., Ng, W. L., Kazmierczak, K. M., Andrzejewski, T. M., Davidsen, T. M., Wayne, K. J., et al. (2007). Genome sequence of Avery’s virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189, 38–51. doi: 10.1128/JB.01148-06
Leenhouts, K., Venema, G., and Kok, J. (1998). A lactococcal pWV01 based integration toolbox for bacteria. Methods Cell Sci. 20, 35–50. doi: 10.1023/A:1009862119114
Lestrate, P., Delrue, R. M., Danese, I., Didembourg, C., Taminiau, B., Mertens, P., et al. (2000). Identification and characterization of in vivo attenuated mutants of Brucella melitensis. Mol. Microbiol. 38, 543–551. doi: 10.1046/j.1365-2958.2000.02150.x
Lopez, J. (2013). Carl A. Burtis Edward R. Ashwood and David E. Bruns (eds): tietz textbook of clinical chemistry and molecular diagnosis (5th edition). Indian J. Clin. Biochem. 28, 104–105. doi: 10.1007/s12291-012-0287-7
Maddocks, S. E., and Oyston, P. C. F. (2008). Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiol. Read. Engl. 154, 3609–3623. doi: 10.1099/mic.0.2008/022772-0
Manganelli, R., Voskuil, M. I., Schoolnik, G. K., Dubnau, E., Gomez, M., and Smith, I. (2002). Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45, 365–374. doi: 10.1046/j.1365-2958.2002.03005.x
Manzoor, I., Shafeeq, S., Kloosterman, T. G., and Kuipers, O. P. (2015a). Co(2+)-dependent gene expression in Streptococcus pneumoniae: opposite effect of Mn(2+) and Co(2+) on the expression of the virulence genes psaBCA, pcpA, and prtA. Front. Microbiol. 6:748. doi: 10.3389/fmicb.2015.00748
Manzoor, I., Shafeeq, S., and Kuipers, O. P. (2015b). Ni2+-dependent and PsaR-mediated regulation of the virulence genes pcpA, psaBCA, and prtA in Streptococcus pneumoniae. PLoS ONE 10:e0142839. doi: 10.1371/journal.pone.0142839
Mares, R., Urbanowski, M. L., and Stauffer, G. V. (1992). Regulation of the Salmonella typhimurium metA gene by the metR protein and homocysteine. J. Bacteriol. 174, 390–397. doi: 10.1128/jb.174.2.390-397.1992
Marouni, M. J., and Sela, S. (2003). The luxS gene of Streptococcus pyogenes regulates expression of genes that affect internalization by epithelial cells. Infect. Immun. 71, 5633–5639. doi: 10.1128/IAI.71.10.5633-5639.2003
Merlin, C., Gardiner, G., Durand, S., and Masters, M. (2002). The Escherichia coli metD locus encodes an ABC transporter which includes Abc (MetN), YaeE (MetI), and YaeC (MetQ). J. Bacteriol. 184, 5513–5517. doi: 10.1128/JB.184.19.5513-5517.2002
Novichkov, P. S., Laikova, O. N., Novichkova, E. S., Gelfand, M. S., Arkin, A. P., Dubchak, I., et al. (2010). RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 38, D111–D118. doi: 10.1093/nar/gkp894
Pinto, R., Tang, Q. X., Britton, W. J., Leyh, T. S., and Triccas, J. A. (2004). The Mycobacterium tuberculosis cysD and cysNC genes form a stress-induced operon that encodes a tri-functional sulfate-activating complex. Microbiol. Read. Engl. 150, 1681–1686. doi: 10.1099/mic.0.26894-0
Ravanel, S., Gakière, B., Job, D., and Douce, R. (1998). The specific features of methionine biosynthesis and metabolism in plants. Proc. Natl. Acad. Sci. U.S.A. 95, 7805–7812. doi: 10.1073/pnas.95.13.7805
Saint-Girons, I., Parsot, C., Zakin, M. M., Bârzu, O., and Cohen, G. N. (1988). Methionine biosynthesis in Enterobacteriaceae: biochemical, regulatory, and evolutionary aspects. CRC Crit. Rev. Biochem. 23(Suppl. 1), S1–S42. doi: 10.3109/10409238809083374
Schell, M. A. (1993). Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47, 597–626. doi: 10.1146/annurev.mi.47.100193.003121
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shafeeq, S., Afzal, M., Henriques-Normark, B., and Kuipers, O. P. (2015). Transcriptional profiling of UlaR-regulated genes in Streptococcus pneumoniae. Genomics Data 4, 57–59. doi: 10.1016/j.gdata.2015.02.004
Shafeeq, S., Kloosterman, T. G., and Kuipers, O. P. (2011a). Transcriptional response of Streptococcus pneumoniae to Zn(2+) limitation and the repressor/activator function of AdcR. Metallomics 3, 609–618. doi: 10.1039/c1mt00030f
Shafeeq, S., Yesilkaya, H., Kloosterman, T. G., Narayanan, G., Wandel, M., Andrew, P. W., et al. (2011b). The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol. Microbiol. 81, 1255–1270. doi: 10.1111/j.1365-2958.2011.07758.x
Shelver, D., Rajagopal, L., Harris, T. O., and Rubens, C. E. (2003). MtaR, a regulator of methionine transport, is critical for survival of group B streptococcus in vivo. J. Bacteriol. 185, 6592–6599. doi: 10.1128/JB.185.22.6592-6599.2003
Sperandio, B., Gautier, C., Pons, N., Ehrlich, D. S., Renault, P., and Guédon, E. (2010). Three paralogous LysR-type transcriptional regulators control sulfur amino acid supply in Streptococcus mutans. J. Bacteriol. 192, 3464–3473. doi: 10.1128/JB.00119-10
Sperandio, B., Polard, P., Ehrlich, D. S., Renault, P., and Guedon, E. (2005). Sulfur amino acid metabolism and its control in Lactococcus lactis IL1403. J. Bacteriol. 187, 3762–3778. doi: 10.1128/JB.187.11.3762-3778.2005
Sperandio, V., Mellies, J. L., Nguyen, W., Shin, S., and Kaper, J. B. (1999). Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 96, 15196–15201. doi: 10.1073/pnas.96.26.15196
Keywords: Cysteine, CmbR, pneumococcus, MetE, MetA
Citation: Afzal M, Manzoor I, Kuipers OP and Shafeeq S (2016) Cysteine-Mediated Gene Expression and Characterization of the CmbR Regulon in Streptococcus pneumoniae. Front. Microbiol. 7:1929. doi: 10.3389/fmicb.2016.01929
Received: 04 August 2016; Accepted: 16 November 2016;
Published: 01 December 2016.
Edited by:
Marc Bramkamp, Ludwig Maximilian University of Munich, GermanyReviewed by:
Patricia Coutinho Dos Santos, Wake Forest University, USAAndrew T. Ulijasz, Loyola University Chicago, USA
Copyright © 2016 Afzal, Manzoor, Kuipers and Shafeeq. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oscar P. Kuipers, by5wLmt1aXBlcnNAcnVnLm5s
 Muhammad Afzal1,2
Muhammad Afzal1,2 Oscar P. Kuipers
Oscar P. Kuipers Sulman Shafeeq
Sulman Shafeeq