- 1Department of Microbiology and Immunology, College of Medicine, Chang Gung University, Tao-yuan, Taiwan
- 2Graduate Institute of Biomedical Sciences, College of Medicine, Chang Gung University, Tao-Yuan, Taiwan
- 3Molecular Infectious Disease Research Center, Chang Gung Memorial Hospital, Tao-yuan, Taiwan
- 4Department of Medical Laboratory Science and Biotechnology, College of Medicine, National Cheng Kung University, Tainan, Taiwan
- 5Department of Biotechnology and Laboratory Science in Medicine, School of Biomedical Science and Engineering, National Yang-Ming University, Taipei, Taiwan
- 6Department of Pediatrics, Chang Gung Children’s Hospital, Tao-yuan, Taiwan
CovR/CovS is an important two-component regulatory system in human pathogen group A Streptococcus (GAS). Epidemiological studies have shown that inactivation of the sensor kinase CovS is correlated with invasive clinical manifestations. The phosphorylation level of response regulator CovR decreases dramatically in the absence of CovS, resulting in the derepression of virulence factor expression and an increase in bacterial invasiveness. Streptococcal pyrogenic exotoxin B (SpeB) is a cysteine protease and is negatively regulated by CovR; however, the expression of SpeB is almost completely repressed in the covS mutant. The present study found that in the emm1-type A20 strain, non-phosphorylated CovR acts as a transcriptional repressor for SpeB-positive regulator Rgg. In addition, the expression of Rgg-negative regulator LacD.1 is upregulated in the covS mutant. These results suggest that inactivation of Rgg in the covS mutant would directly mediate speB repression. The current study showed that overexpression of rgg but not inactivation of lacD.1 in the covS mutant partially restores speB expression, indicating that only rgg repression, but not lacD.1 upregulation, contributes to the speB repression in the covS mutant.
Introduction
Streptococcus pyogenes (group A Streptococcus, GAS) is an important human pathogen causing diseases including pharyngitis, tonsillitis, scarlet fever, cellulitis, necrotizing fasciitis, and toxic shock syndrome. There are more than 500,000 deaths each year due to GAS-related diseases (Carapetis et al., 2005), indicating that invasive GAS infections are still a major clinical problem around the world. Mutations in the covS gene are detected more frequently in clinical isolates from severe invasive infections than in non-invasive isolates (Sumby et al., 2006; Ato et al., 2008; Ikebe et al., 2010; Lin et al., 2014; Friaes et al., 2015). CovS is the sensor kinase that phosphorylates an aspartic residue (D53) of the response regulator CovR (Levin and Wessels, 1998; Horstmann et al., 2014, 2015). Acquisition of null covS alleles causes a repression of streptococcal pyrogenic exotoxin B (SpeB) and upregulation of many virulence factor-encoding genes, resulting in increased GAS virulence during infection (Sumby et al., 2006; Walker et al., 2007; Ikebe et al., 2010; Friaes et al., 2015).
Streptococcal pyrogenic exotoxin B is a secreted cysteine protease of GAS (Chiang-Ni and Wu, 2008; Olsen and Musser, 2010). A DNase I footprint assay showed that recombinant CovR binds to the -53 to +36 region of the speB promoter (+1, transcription start site) (Miller et al., 2001). In addition, phosphorylation of CovR further enhances its binding to the speB DNA probe (Miller et al., 2001). Furthermore, deletion of the covR gene results in an increase in speB expression (Heath et al., 1999; Miller et al., 2001; Graham et al., 2002), indicating that CovR acts as a transcriptional repressor of speB. However, expression of speB is almost completely repressed in the covS mutant (Sumby et al., 2006; Walker et al., 2007; Trevino et al., 2009; Tran-Winkler et al., 2011). Transcriptome analysis showed that expression of the SpeB positive regulator Rgg is decreased accordingly in the covS mutant compared with that in the wild type strain (Sumby et al., 2006), suggesting that the decrease in speB transcription may be caused by the repression of rgg. However, the role of Rgg in the regulation of speB expression in the covS mutant has not been clearly demonstrated.
The location of rgg is adjacent to that of the speB gene in GAS chromosome. Rgg protein binds to the speB promoter specifically and is essential for the transcription of speB (Neely et al., 2003). Expression of both rgg and speB is only detected in late-exponential to stationary phases of growth (Unnikrishnan et al., 1999; Neely et al., 2003). However, expression of rgg from a promoter expressed during the early-log phase of growth cannot trigger speB expression, indicating that the regulatory activity of Rgg is controlled by other regulatory molecules. (Neely et al., 2003). Loughman and Caparon (2006) showed that Rgg activity is regulated by an aldolase-like protein, LacD.1. On NaCl supplementation and at neutral pH conditions, LacD.1 could bind to Rgg protein to restrict its activation of speB transcription (Loughman and Caparon, 2006). In addition, Anbalagan et al. (2012) showed that DNA-binding specificity of Rgg is responsive to environmental changes in a LacD.1-dependent manner. Rgg coordinates virulence factor expression, catabolic activity, and thermal and oxidative stress responses (Chaussee, 2002; Chaussee et al., 2003, 2004; Pulliainen et al., 2008). Therefore, interactions between LacD.1 and Rgg could be important in the pathogen’s virulence and adaptation to environmental changes during infection.
The present study found that expression of lacD.1 was significantly increased in the covS mutant compared with that in the wild type strain. Therefore, our aim was to elucidate the role of the Rgg-LacD.1 system in speB repression in the covS mutant. The results indicate that only rgg repression, but not the upregulation of lacD.1, contributes directly to speB inhibition in the covS mutant.
Materials and Methods
Bacterial Strains and Culture Conditions
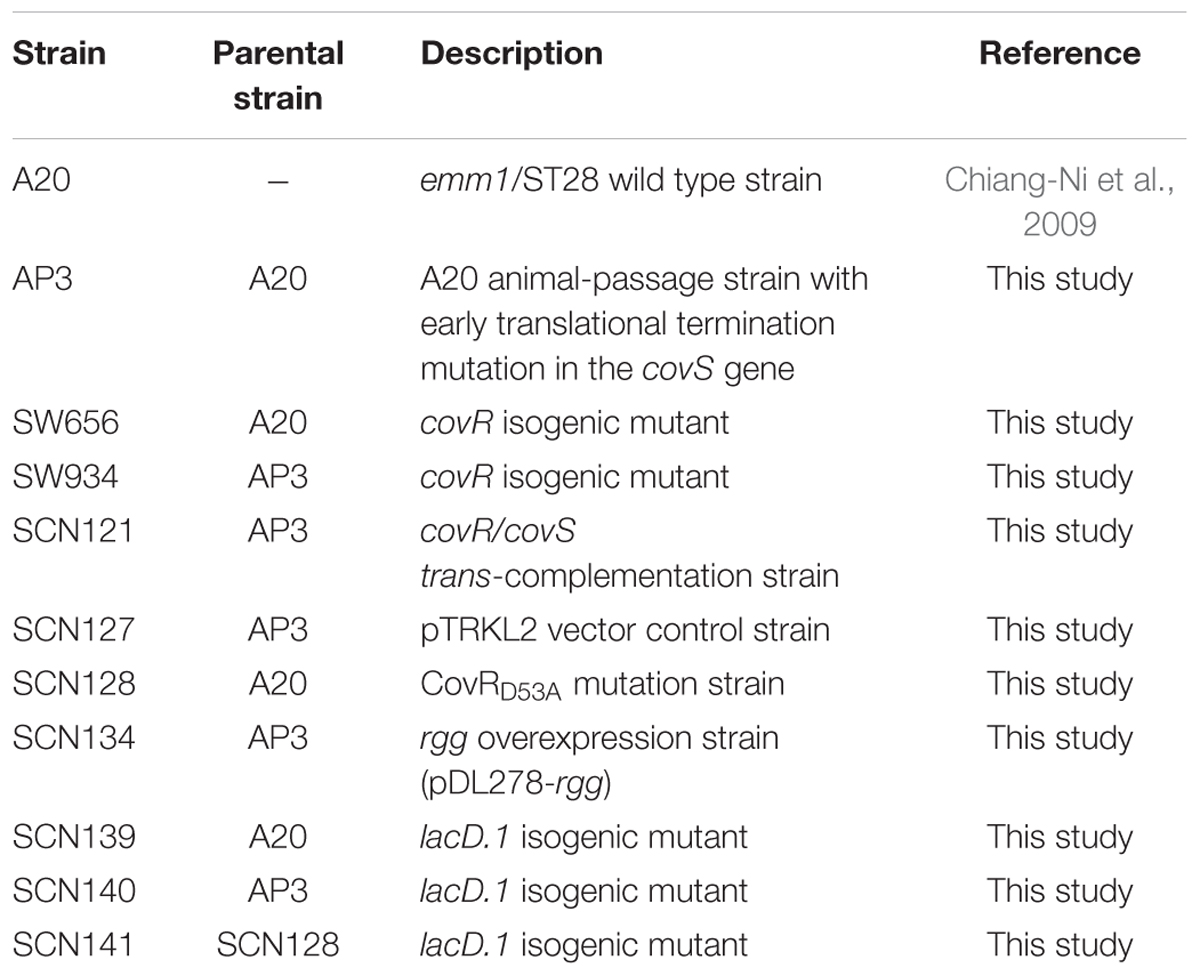
Group A Streptococcus strain A20 (wild type strain) is an emm1-type strain and has been previously described (Chiang-Ni et al., 2009). Strain AP3 was provided by Prof. Jiunn Jong Wu (Department of Medical Laboratory Science and Biotechnology, College of Medicine, National Cheng Kung University, Tainan, Taiwan). This strain was isolated from the spleen of A20-infected BALB/c mouse (subcutaneous infection) after 3 days of infection. In this study, the genome sequence of AP3 was generated using the Miseq sequencer (Illumina) according to the manufacturer’s protocols. Sequencing reads were mapped; and SNPs and Indels were called relative to the A20 genome sequence (NCBI accession number: NC_018936). The 143T deletion in the covS gene was found and confirmed by traditional Sanger sequencing method. In addition, another six SNPs and an Indel were found in the repeat sequence regions of transposases or rRNA genes (data not shown). GAS strains were cultured on the trypticase soy agar with 5% sheep blood or in the tryptic soy broth (Becton, Dickinson and Company, Sparks, MD) supplemented with 0.5% yeast extract (TSBY) and described in Table 1. E. coli DH5α was purchased from Yeastern (Yeastern Biotech Co., LTD, Taipei, Taiwan) and was cultured in Luria-Bertani (LB) broth at 37°C with vigorous aeration. When appropriate, antibiotics chloramphenicol (25 and 3 μg/ml for E. coli and GAS, respectively), erythromycin (125 and 5 μg/ml for E. coli and GAS, respectively) and spectinomycin (100 μg/ml) were used for selection.
To treat GAS strains with neutral and acidic broths, bacterial pellets (collected from cultures in 40 ml of TSBY for 12-16 h at 37°C) were washed and resuspended in 1 ml of fresh TSBY broth. Seventy-five microliter of bacterial suspension was transferred to TSBY broth that was adjusted to pH 7.5 (supplemented with 0.1 M of HEPES) or pH 6.0 (supplemented with 0.1 M of MES) by 1 N HCl or NaOH (Dalton and Scott, 2004). Although culture broths were buffered by salts, pH of bacterial suspensions was decreased significantly after 4-5 h of incubation. Therefore, GAS strains were cultured at different pH conditions for 3 h at 37°C. The O.D.600 of the bacterial suspensions (both pH 7.5 and pH 6.0) after 3 h incubation were around 0.4-0.6.
DNA and RNA Manipulations
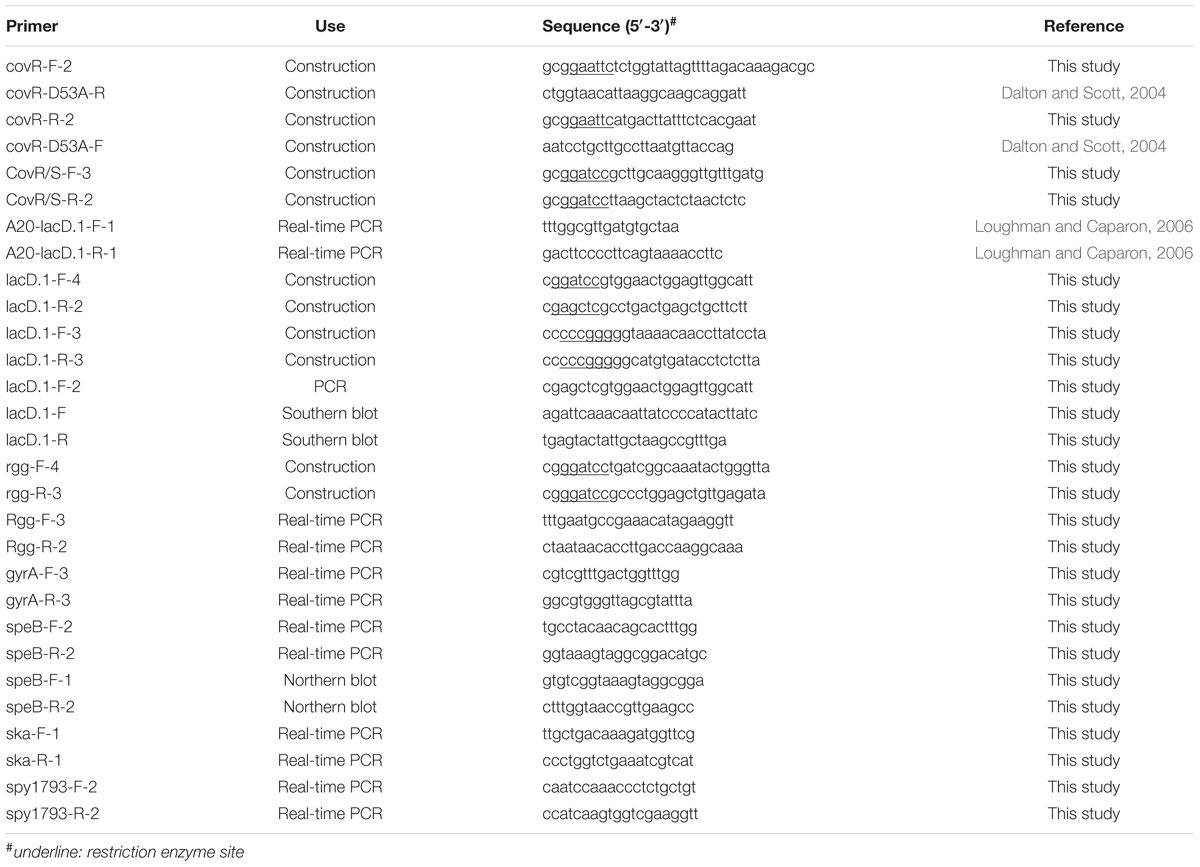
Group A Streptococcus genomic DNA extraction, RNA extraction, and reverse transcription were performed as described previously (Chiang-Ni et al., 2009). Real-time PCR reactions were performed in a 20 μl mixture containing 1 μl of cDNA, 0.8 μl of primers (10 μM), and 10 μl of SensiFASTTM SYBR Lo-ROX pre-mixture (Bioline Ltd, London, UK) according to the manual. The expression level of the target gene was normalized to the gyrA and analyzed using the ΔΔCt method (7500 software v2.0.5, Applied Biosystem®, Thermo Fisher Scientific Inc.). In addition, all values of control and experiment groups were divided by the mean of control samples before statistical analysis (Valcu and Valcu, 2011). Primers used for real-time PCR analysis were designed by Primer31 (v.0.4.0) according to the MGAS5005 sequence (NCBI reference sequence: NC_007297.1) and are described in Table 2. Southern blot and Northern blot analyses were performed as described previously (Chiang-Ni et al., 2012). Ten microgram of genomic DNAs extracted from GAS strains were digested by restriction enzymes for Southern blot analysis. Three microgram of RNAs extracted from GAS strains under different culture conditions were subjected to Northern blot analysis. DNA probes used for detecting the lacD.1 gene and chloramphenicol cassette were amplified by primers described in Table 2 and labeled with alkaline phosphatase (AlkPhos Direct Labeling and Detection System, GE Healthcare, Chicago, IL, USA) according to the manual. After hybridization, the membrane was washed and the signal was detected by the Gel Doc XR+ system (BioRad, Hercules, CA, USA).
Construction of covR Deletion, covR/covS trans-Complemented, and Vector Control Strains
Plasmid pMW506, the covR gene interrupted by a chloramphenicol cassette via the MslI site on E. coli vector pSF152, was provided by Prof. Jiunn Jong Wu. The plasmid pMW506 was electroporated into A20 and AP3 by the method described previously (Chiang-Ni et al., 2008) to interrupt the covR gene by a double-cross homologous recombination (designed SW656 and SW934, respectively).
To construct covR/covS-complemented strain, the covR/S allele (2505 bp, including promoter region) was amplified from wild type A20 strain using primers CovR/S-F-3 and CovR/S-R-2 (Table 2) and ligated into the BamHI site of E. coli-GAS shuttle vector pTRKL2 to create pCN111. The plasmid pTRKL2 was kindly provided by T. R. Klaenhammer (Department of Food Science, Southeast Dairy Foods Research Center, North Carolina State University, Raleigh, USA). The backbone of pTRKL2 is a pAMß1-derived Gram-positive vector pIL252. The copy number of pTRKL2 is corresponded to pIL252, which is 6-9 copies in a streptococcal and lactococcal host and 30-40 copies in E. coli (O’Sullivan and Klaenhammer, 1993). The plasmid pCN111 and vector pTRKL2 were electroporated into AP3 to generate the covR/S-complemented strain (SCN121) and vector control strain (SCN127).
Construction of CovRD53A Mutation, lacD.1 Deletion, and rgg Over-Expression Strains
Phosphorylation site mutation (D53A) in the covR gene was generated by overlap PCR with primers covR-F-2, covR-D53A-R, covR-R-2, and covR-D53A-F listed in Table 2. The PCR product (1.1 kb) with D53A amino acid substitution was confirmed by sequencing and ligated into temperature-sensitive E. coli-Lactococcus shuttle vector pG+HOST9 via the EcoRI site. The constructed plasmid was transformed into wild type A20 strain and cultured at 30°C. Transformants were transferred to 37°C to force plasmid integration via a single homologous recombination. Finally, transformants in which the plasmid excised from chromosome via a second recombination were selected in the antibiotic-free plate at 30°C. The phosphorylation site mutation was further verified by sequencing and the strain was designed as SCN128.
To construct the lacD.1 isogenic mutant, pG+HOST9 (erythromycin resistant) and E. coli vector pSF152 (spectinomycin resistant) were ligated into the EcoRI site. The erythromycin cassette of the constructed vector was further removed by BamHI digestion to generate plasmid pCN143. The lacD.1 gene with upstream (833 bp) and downstream (921 bp) regions was amplified by primers lacD.1-F-4 and lacD.1-R-2 and ligated into pCN143 with the BamHI site. The lacD.1 gene was removed by inverted PCR with primers lacD.1-F-3 and lacD.1-R-3 (Table 2) and replaced by a chloramphenicol cassette from Vector 78 (Tsou et al., 2010) to generate pCN145. The plasmid pCN145 was transformed into A20, AP3, and SCN128 and transformants were selected by chloramphenicol at 30°C. Transformants were transferred to 37°C with chloramphenicol selection to force plasmid integration via a single or double homologous recombination. Transformants with double homologous recombination were screened by antibiotics (spectinomycin sensitive and chloramphenicol resistant) and the deletion of lacD.1 gene was further confirmed by sequencing. Isogenic lacD.1 mutants of A20, AP3, and SCN128 were designed as SCN139, SCN140, and SCN141, respectively.
High-copy-number E. coli-Streptococcus shuttle vector pDL278 (Chiang-Ni et al., 2012) was employed to construct the rgg over-expression strain. The rgg gene with its native promoter was amplified by primers rgg-F-4 and rgg-R-3 (1958 bp) and ligated into pDL278 with the BamHI site (designed as pCN138). pCN138 was transformed into covS mutant AP3 to generate rgg over-expression strain SCN134.
Skim-Milk Agar Assay and Western Blot Analysis for SpeB
Skim-milk agar was prepared by Columbia agar base (Becton, Dickinson and Company, Sparks, MD, USA) supplemented with 3% skim milk. GAS strains were sub-cultured on the Skim-milk agar and incubated at 5% CO2, 37°C incubator for 12-16 h. Casein hydrolysis by GAS strains results in the appearance of a clear zone around bacterial colonies. For Western blot analysis, 30 μl of GAS culture supernatant was subjected to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by transfer to the polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA). The membrane was blocked with 5% skim milk in PBST buffer (PBS containing 0.2% of tween-20) at 37°C for 1 h, and SpeB protein was detected by the anti-SpeB antibody (Toxin Technology, Inc., Sarasota, FL, USA) according to the manual. After hybridization, the membrane was washed with PBST and hybridized with the secondary antibody, peroxidase conjugated goat anti-rabbit IgG (1:10,000 dilution; Cell Signaling Technology, Inc., Danvers, MA, USA) at room temperature for 1 h. The blot was developed using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific Inc., Rockford, IL, USA) and the signal was detected by the Gel Doc XR+ system (BioRad).
Phos-Tag Western Blot Analysis for CovR Protein
Group A Streptococcus strains were cultured in TSBY broth supplemented with or without 20 mM Mg++. Bacteria were grown to the stationary phase of growth, and bacterial pellets were washed and resuspended in the Buffer A. Bacterial cells were disrupted by the bead beater (Mini-Beadbeater, BioSpec Products Inc., Bartlesville, OK, USA) and supernatants were collected for further analysis. Phos-tag SDS-PAGE was prepared according to the manual. Briefly, 10 μg of bacterial total protein was mixed with the 4× protein dye and subjected to 10% SDS-PAGE containing 10 μmol/L of Phos-tag (Wako Pure Chemical Industries Ltd, Richmond, VA, USA) and 0.5 μM of MnCl2 directly without boiling. Phosphorylated proteins were separated on Phos-tag SDS-PAGE for 120-140 min at 100 V at 4°C. The gel was washed with transfer buffer (39 mM of glycine, 48 mM of Tris, 0.037% of SDS, and 20% of methanol) supplemented with 1 mM of EDTA for 40 min. The gel was washed again by EDTA-free transfer buffer for another 20 min and proteins were transferred onto the PVDF membrane. The membrane was blocking by 5% skim milk in PBST buffer at 37°C for 1 h. Mouse anti-CovR serum (kindly provided by Prof. Jiunn Jong Wu) in 1:5,000 dilutions was used as the primary antibody for hybridization. After hybridization, the primary antibody was washed from the membrane with PBST buffer and hybridized with the secondary antibody, peroxidase conjugated goat anti-mouse IgG (1:10,000 dilution; Millipore, Billerica, MA, USA) at room temperature for 1 h. The blot was developed using Pierce ECL Western Blotting Substrate and the signal was detected by the Gel Doc XR+ system (BioRad).
Statistical Analysis
Statistical analysis was performed by using the Prism software, version 4 (GraphPad, San Diego, CA, USA). Significant differences in multiple groups were determined using ANOVA. Post-test for AVOVA was analyzed by Tukey’s Honestly Significant Difference Test. A p-value <0.05 was taken as significant.
Results
CovR Negatively Regulates speB Expression in Both Wild Type and covS Mutant Strains
Although CovR protein can be autophosphorylated in the absence of CovS, inactivation of CovS results in a dramatically decreased level of phosphorylated CovR (Miller et al., 2001; Dalton and Scott, 2004; Horstmann et al., 2015). However, expression of speB is downregulated in covS mutants compared with wild type strains (Sumby et al., 2006; Walker et al., 2007; Trevino et al., 2009; Tran-Winkler et al., 2011). These results suggest that CovR acts as a repressor of speB expression, even in the absence of sensor kinase CovS. To elucidate the role of CovR in the regulation of speB expression in the presence or absence of CovS, isogenic mutants lacking functional covR were constructed from wild type A20 (SW656) and covS mutant AP3 (SW934) strains. The interruption of the covR gene by a chloramphenicol (Cm) cassette in SW656 and SW934 was further confirmed by Southern blot analysis. Results showed that the Cm cassette was only detected in SW656 and SW934 with a size of 3.3 kb, but not in their parental strains (Figure 1A). Expression of speB in the stationary phase of growth was upregulated in SW656 compared with A20 (Figure 1B). In addition, expression of speB was also upregulated in SW934 compared with AP3 (Figure 1B). Furthermore, SW934, but not AP3, hydrolyzed casein in the skim-milk agar plate (Figure 1B, lower panel). These results indicate that CovR acts as a negative regulator of speB expression in both wild type and covS mutant strains.
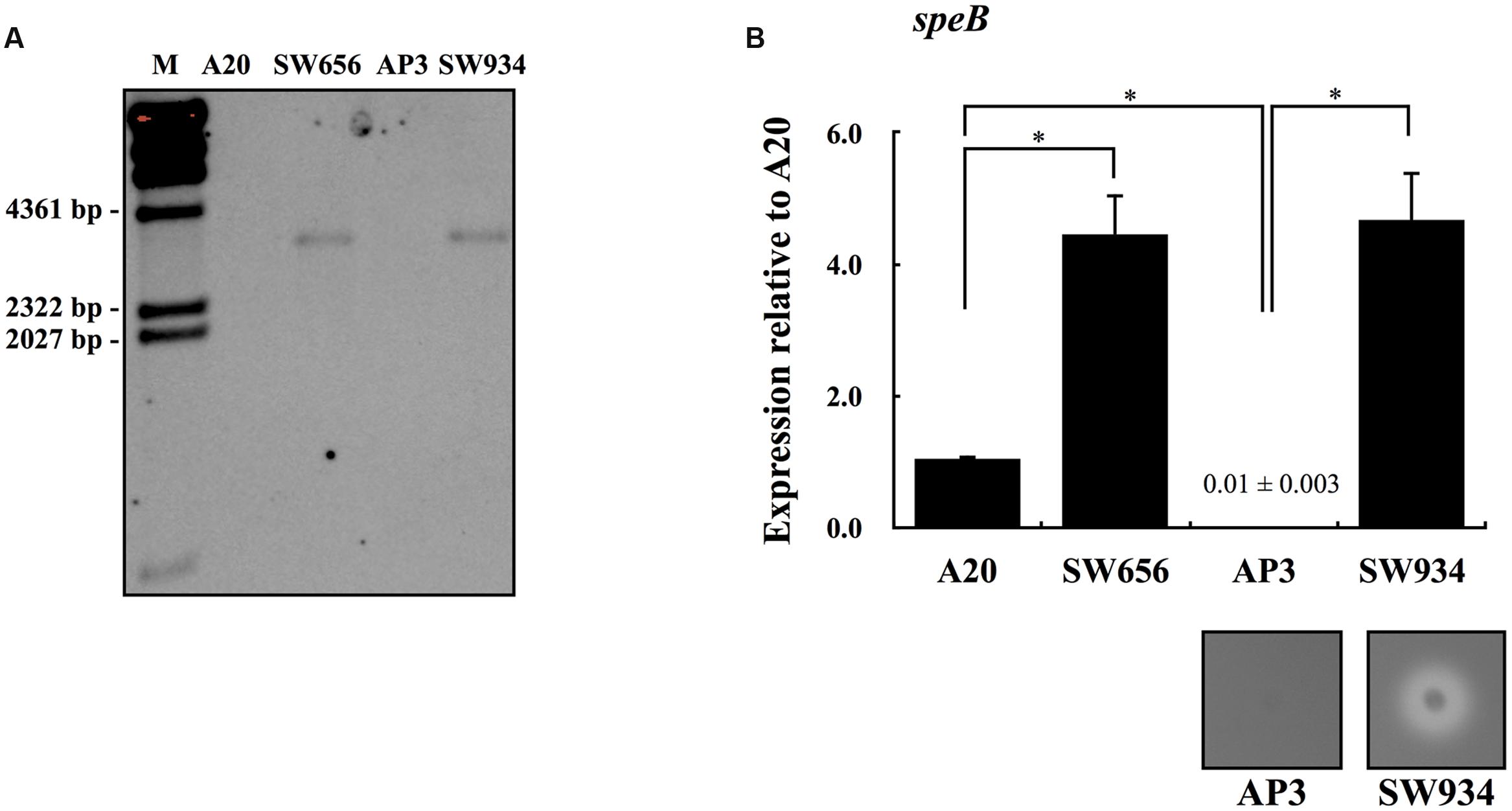
FIGURE 1. The expression of speB in wild type A20 strain, covS mutant AP3, and their covR isogenic mutants. (A) Southern blot analysis for A20, AP3, and their covR isogenic mutants SW656 and SW934. Genomic DNAs were extracted and digested with EcoRV. The interruption of the covR gene by a chloramphenicol (Cm) cassette was detected by the Cm probe. M, λ/HindIII DNA marker. The expression of speB in (B) A20, SW656, AP3, and SW934 in the stationary phase of growth. Group A Streptococcus (GAS) strains were cultured in broths for 7 h and RNAs were extracted for quantitative PCR (qPCR) analysis. Biological replicate experiments were performed using four independent RNA preparations in duplicate. The expression of speB was normalized to the gyrA. ∗p < 0.05. The lower panel in (B) shows the protease activity of AP3 and SW934. Bacteria were cultured in the 3% skim-milk agar for 12-16 h, and the appearance of a clear zone around bacterial colony is considered protease positive.
Overexpression of rgg in the covS Mutant Restores speB Expression
Phosphorylated CovR protein has a greater affinity for the speB promoter DNA than non-phosphorylated CovR (Miller et al., 2001). Therefore, we proposed that CovR does not directly repress the expression of speB in the covS mutant. Sumby et al. (2006) showed that the expression of SpeB-positive regulator Rgg in the covS mutant is significantly decreased compared with that in the wild type strain. In line with the previous observation, we also found that expression of rgg was downregulated in the covS mutant AP3 compared with that in the wild type A20 strain (Figure 2A). In addition, covR/covS trans-complemented strain SCN121, but not vector control strain SCN127, restored rgg expression in the stationary phase of growth of AP3 (Figure 2A). To elucidate the role of rgg repression in speB expression in the covS mutant, rgg was overexpressed by the high-copy number vector pDL278 in AP3 (SCN134). Expression of speB is repressed in the covS mutant; therefore, the value of the fold change in expression cannot be precisely evaluated by a relative quantification PCR method. Therefore, the expression of speB in AP3 and SCN134 was analyzed by Northern blotting. Results showed that speB transcripts were detected in SCN134 but not in AP3 (Figure 2B). Compared to the wild type A20 strain, the expression of speB in the stationary phase of growth in SCN134 was only partially restored (7 h of incubation, Figure 2B). Expression of speB is restricted to the stationary phase of growth (Unnikrishnan et al., 1999; Neely et al., 2003); however, we found that speB transcripts can be detected in SCN134 in the early- and late-exponential phases of growth (3 and 5 h of incubation, respectively, Figure 2B). SpeB protein in the bacterial culture supernatant was further detected by Western blot. SpeB is secreted as an inactive precursor (42 kDa), which is autocatalytically cleaved into an active protease (28 kDa), and produces at least six intermediates during this process (Doran et al., 1999; Collin and Olsen, 2000; Zimmerlein et al., 2005). Our results show that the mature SpeB protein and its intermediates were detected in the culture supernatant of SCN134, but not in AP3 during the stationary phase of growth (Figure 2C). These results indicate that rgg repression contributes directly to speB inhibition in the covS mutant.
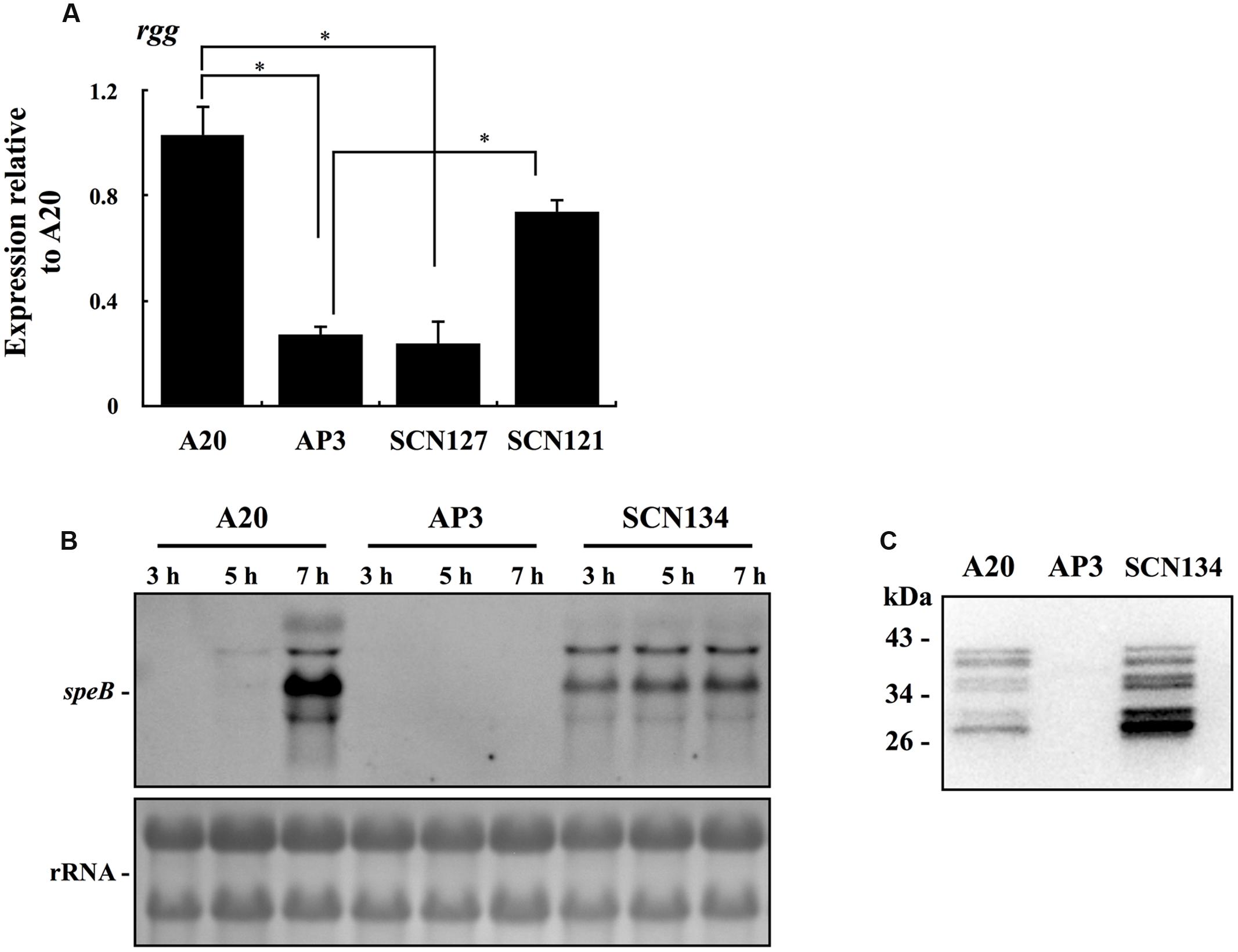
FIGURE 2. The expression of rgg and speB in wild type A20 strain, covS mutant AP3, covR/S-complemented strain SCN121, vector control strain SCN127, and AP3 rgg overexpression strain SCN134. (A) The expression of rgg in A20, AP3, SCN127, and SCN121 in the stationary phase of growth. (B) The expression of speB in A20, AP3, and SCN134 in the early-exponential (3 h), late-exponential (5 h), and stationary phases (7 h) of growth. GAS strains were cultured in TSBY broth to different phases of growth and RNAs were extracted for Northern blot analysis. rRNA (lower panel) is used as the internal loading control. (C) The production of SpeB protein in A20, AP3, and SCN134. GAS strains were cultured in TSBY broth to the stationary phase of growth and 30 μl of bacterial culture supernatants were subjected to Western blot analysis with the anti-SpeB antibody.
Expression of rgg Is Repressed by the Response Regulator CovR in the covS Mutant
Results from Figure 2A show that the expression of rgg was repressed in AP3 compared with that in the wild type A20 strain. To elucidate the role of CovR in regulation of rgg expression in the absence of CovS, the expression of rgg in AP3 and its covR isogenic mutant SW934 was analyzed. The results showed that expression of rgg during the stationary phase of growth increases significantly in SW934 compared with AP3 (Figure 3A), suggesting that CovR negatively regulates rgg expression. AP3 is not the covS isogenic mutant of A20. Although we considered that only a T nucleotide deletion in the covS in AP3 genome is important, the possibility that other unidentified mutations in AP3 contribute to the rgg repression cannot be completely excluded. Regulation by CovS is thought to be solely through CovR (Trevino et al., 2009). Therefore, to study the role of CovS inactivation in rgg expression, a wild type A20 strain was constructed with the mutation in phospho-aspartic acid residue (D53A) of CovR (SCN128) (Dalton and Scott, 2004). Expression of rgg was significantly decreased in SCN128 compared with the wild type A20 strain (Figure 3A). In addition, speB transcripts and protease activity were not detected in SCN128 (Figure 3B and data not shown). These results suggest that the nonphosphorylated CovR protein in the covS mutant acts as a transcriptional repressor of rgg expression.
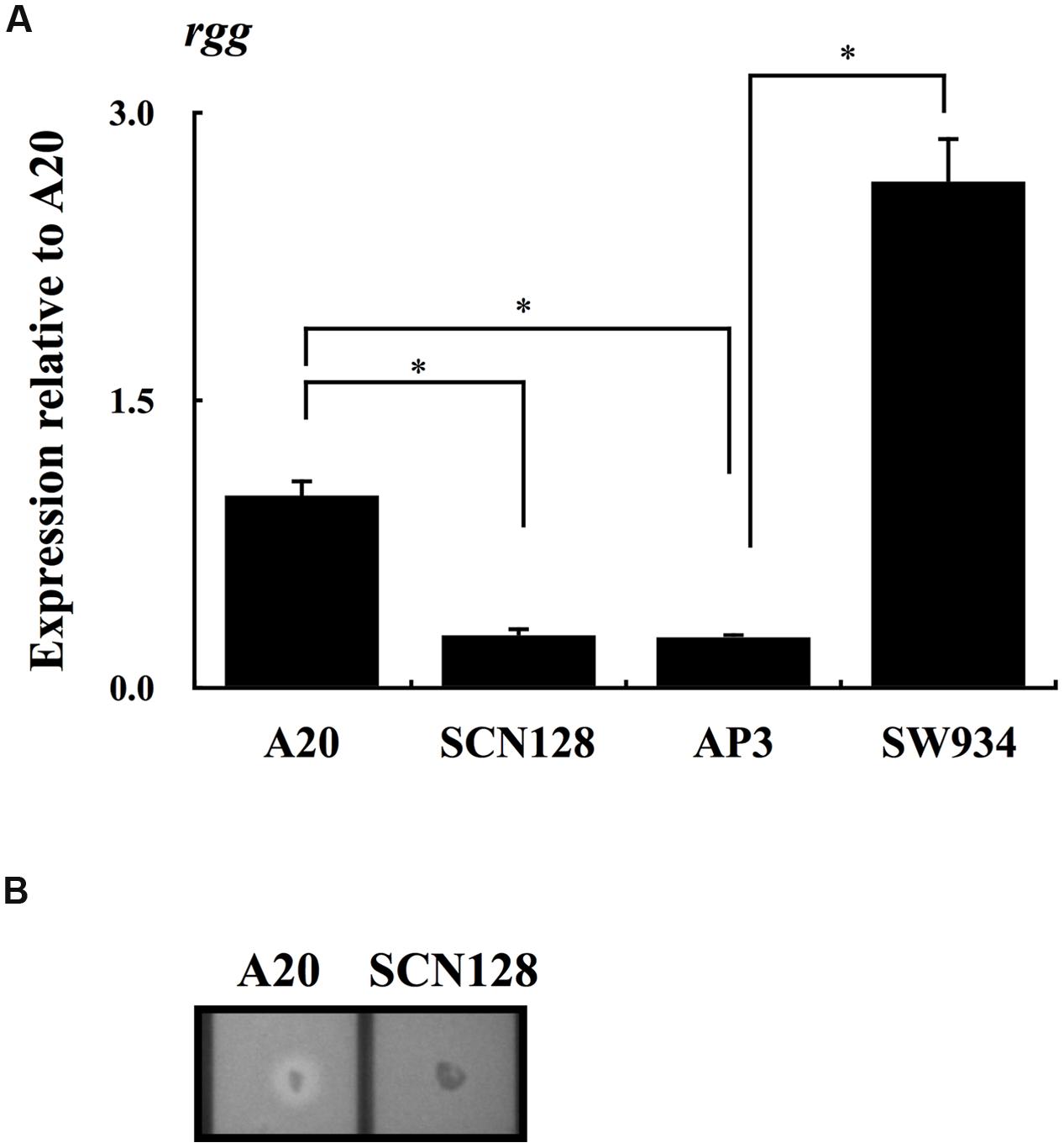
FIGURE 3. The expression of rgg in A20, A20 CovRD53A mutation strain SCN128, covS mutant AP3, AP3 covR mutant SW934. The expression of rgg in (A) A20, SCN128, AP3, and SW934 in the stationary phase of growth. All qPCR analyses were performed using three independent RNA preparations in duplicate. The expression of rgg was normalized to the gyrA. ∗p < 0.05. (B) Shows the protease activity of A20 and SCN128. Bacteria were cultured in the 3% skim-milk agar for 12-16 h, and the appearance of a clear zone around the bacterial colony is considered protease positive.
Expression of rgg Is Negatively Regulated by Both Phosphorylated and Non-phosphorylated CovR
Results showed in Figure 3 suggests that the decrease in the level of phosphorylated CovR results in downregulation of rgg expression. The CovR/CovS system responds to changes in Mg++ concentrations by changing the phosphorylation status of CovR (Tran-Winkler et al., 2011; Horstmann et al., 2015). Therefore, the expression of rgg and the CovR-regulated gene ska in A20 and AP3 under Mg++ stimulation was further analyzed. As expected, the expression of ska was decreased significantly with Mg++ stimulation in A20 but not in AP3 (Figure 4A and data not shown). However, there was no statistical difference in the levels of rgg expression between Mg++ treated and nontreated A20 or AP3 (Figure 4A and data not shown). Horstmann et al. (2015) showed that serotype M1 GAS strains have high levels of phosphorylated CovR in the absence of Mg++ stimulation; therefore, differences in the expression levels of rgg in A20 with or without Mg++ treatment could not be clearly observed. Therefore, CovR phosphorylation levels in A20 and AP3 in the presence with or without Mg++ were further analyzed by Phos-tag western blot (Barbieri and Stock, 2008; Horstmann et al., 2015). Results showed that phosphorylated CovR protein was only clearly observed in A20 but not in AP3 (Figure 4B). In addition, a slight increase in phosphorylated CovR protein in A20 was found under Mg++ stimulation (Figure 4B), indicating that A20 has high levels of phosphorylated CovR in the absence of Mg++ stimulation. Horstmann et al. (2015) showed that levels of phosphorylated CovR in M1 GAS strain decreases significantly in the presence of antimicrobial peptide LL37. To further elucidate the role of phosphorylation level of CovR in the regulation of rgg expression, the wild type A20 strain was treated with LL37 and the expression of ska and rgg was analyzed. A20 strain was grown to the late-exponential phase of growth and then treated by 150 and 300 nM LL37 for another 3 h. After LL37 treatment, the expression of ska increased significantly compared with that in A20 incubated in TSBY broth without supplemented with LL37 (Figure 4C). In addition, the expression of rgg was decreased in the presence of 150 and 300 nM of LL37 (Figure 4C). Furthermore, the role of phosphorylated and non-phosphorylated CovR protein in the regulation of rgg expression was further evaluated by covR isogenic mutants of A20 and AP3. Expression of rgg was significantly increased in SW656 and SW934 compared with the parental strains (Figure 4D), indicating that CovR acts as a transcriptional repressor in both wild type and covS mutant strains. In addition, these results also suggest that non-phosphorylated CovR can more strongly repress rgg expression than phosphorylated CovR.
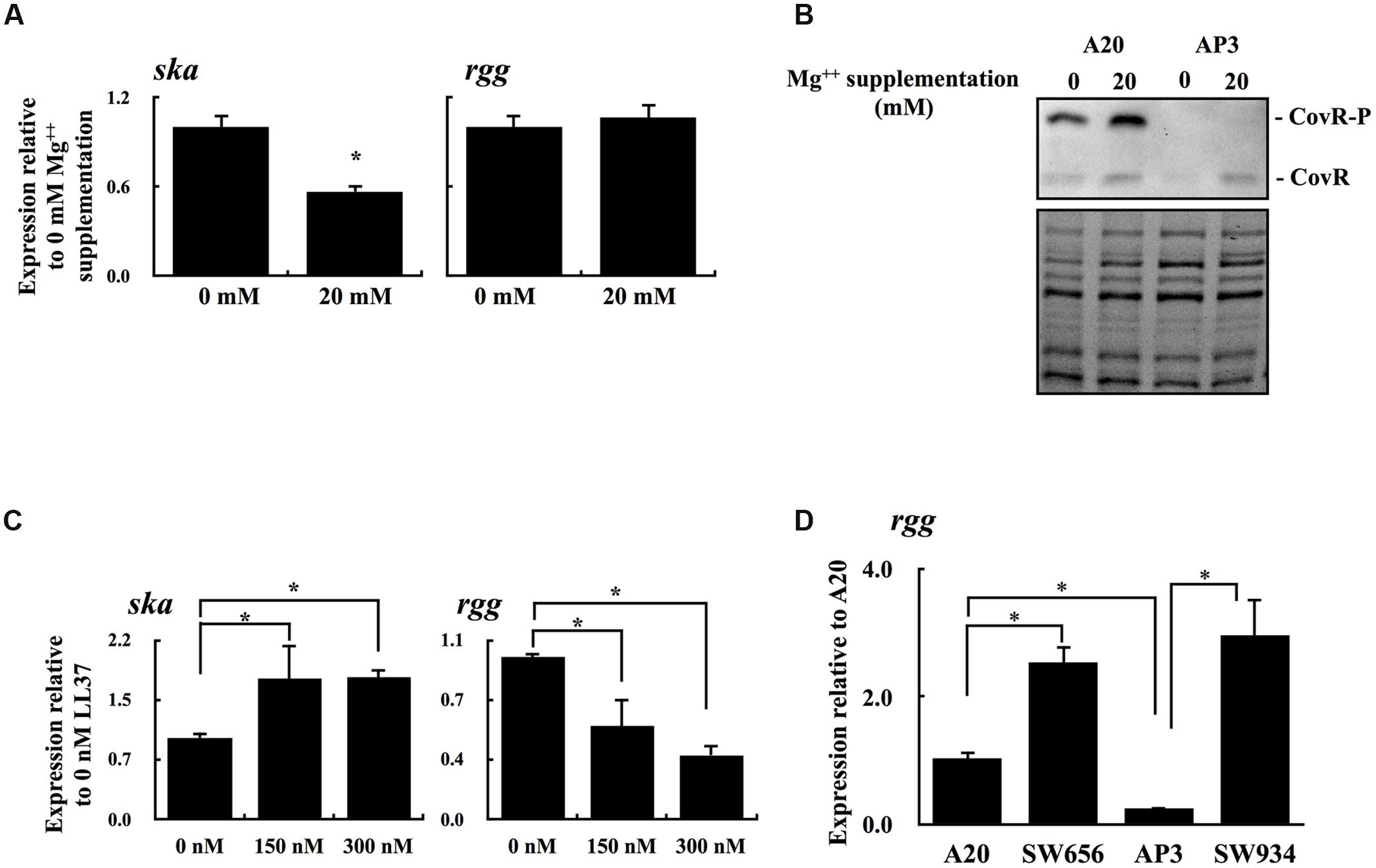
FIGURE 4. The phosphorylation level of CovR protein and the expression of ska and rgg in A20 under Mg++ and LL37 stimulation. (A) The expression of ska and rgg in A20 with or without the high concentration of Mg++ stimulation. (B) Phos-tag Western blot for detecting the phosphorylated and non-phosphorylated CovR in A20 and AP3, with or without a high concentration of Mg++ stimulation. CovR-P indicates the phosphorylated CovR protein; CovR indicates the non-phosphorylated CovR protein. The lower panel shows total protein as a loading control. (C) The expression of ska and rgg in A20 with 0, 150, and 300 nM of LL37 treatments. (D) The expression of rgg in A20, AP3, and their covR isogenic mutants (SW656 and SW934, respectively) in the stationary phase of growth. All qPCR analyses were performed using four independent RNA preparations in duplicate. The expression of ska and rgg were normalized to the gyrA. ∗p < 0.05.
Expression of lacD.1 Is Derepressed in the covS Mutant
Expression of speB in SCN134 was only partially restored (Figure 2B), suggesting that the regulatory activity of Rgg in SCN134 could still be inhibited. Therefore, the expression of lacD.1, a negative regulator of Rgg, was analyzed. The results showed that expression of lacD.1 was significantly increased in the covS mutant AP3 compared with that in the wild type A20 strain (Figure 5). In addition, the AP3 covR/S trans-complemented strain SCN121, but not the vector control strain SCN127, showed similar lacD.1 expression levels to A20 in the stationary phase of growth (Figure 5). The role of CovS inactivation in lacD.1 upregulation was further elucidated by SCN128. Compared to the wild type A20 strain, SCN128 showed a significantly increase in lacD.1 expression. These results indicate that expression of lacD.1 is upregulated in the covS mutant.

FIGURE 5. The expression of lacD.1 in wild type A20 strain, A20 CovRD53A mutation strain SCN128, covS mutant AP3, covR/S-complemented strain SCN121, and vector control strain SCN127. The expression of lacD.1 in A20, SCN128, AP3, SCN127, and SCN121 in the stationary phase of growth. All qPCR analyses were performed using three independent RNA preparations in duplicate. Expression of the lacD.1 was normalized to the gyrA. ∗p < 0.05.
LacD.1 Is Not Involved in speB Repression in the covS Mutant
The increase in lacD.1 expression may lead to Rgg inactivation in the covS mutant AP3 (Loughman and Caparon, 2006). Therefore, the role of LacD.1 in speB inhibition in the covS mutant was further analyzed. Isogenic lacD.1 mutant of A20, AP3, and SCN128 were constructed. The deletion of lacD.1 gene in lacD.1 mutants was confirmed by sequencing analysis (data not shown). In addition, a 2.7 kb region of the lac.1 operon of wild type and lacD.1 mutant strains was amplified by PCR with primers lacD.1-F-2 and lacD.1-R-2 (Table 2), and PCR products were further digested with the restriction enzyme BamHI. The BamHI site is located within the lacD.1 gene; therefore, as expected, only PCR products amplified from the wild type strains were digested by BamHI (Figure 6A). The lacD.1 and lacD.2 genes have 70% sequence identity; therefore, the lacD.1 mutants were further verified by Southern blot analysis. Results showed that no lacD.1 gene was detected in DNA extracted from the lacD.1 isogenic mutants of A20 and AP3 (Figure 6B). In addition, the Cm cassette was only detected in the lacD.1 mutants (Figure 6B). However, deletion of the lacD.1 gene in AP3 (SCN140) and SCN128 (SCN141) did not restore speB expression (Figure 6C). Expression of speB is induced by acidic pH but inhibited by neutral pH (Loughman and Caparon, 2006; Chiang-Ni et al., 2012). LacD.1 has been shown to play critical roles in the inhibition of speB expression under neutral pH culture conditions (Loughman and Caparon, 2006). However, this study found that speB expression was inhibited in both the wild type A20 strain and its lacD.1 isogenic mutant (SCN139) under neutral pH culture conditions (Figure 6D). In addition, the expression level of speB in SCN140 under acidic condition was still lower than that of A20 and SCN139 under pH 7.5 condition (Figure 6D). These results indicate that LacD.1 is not involved in the repression of speB expression in the covS mutant.
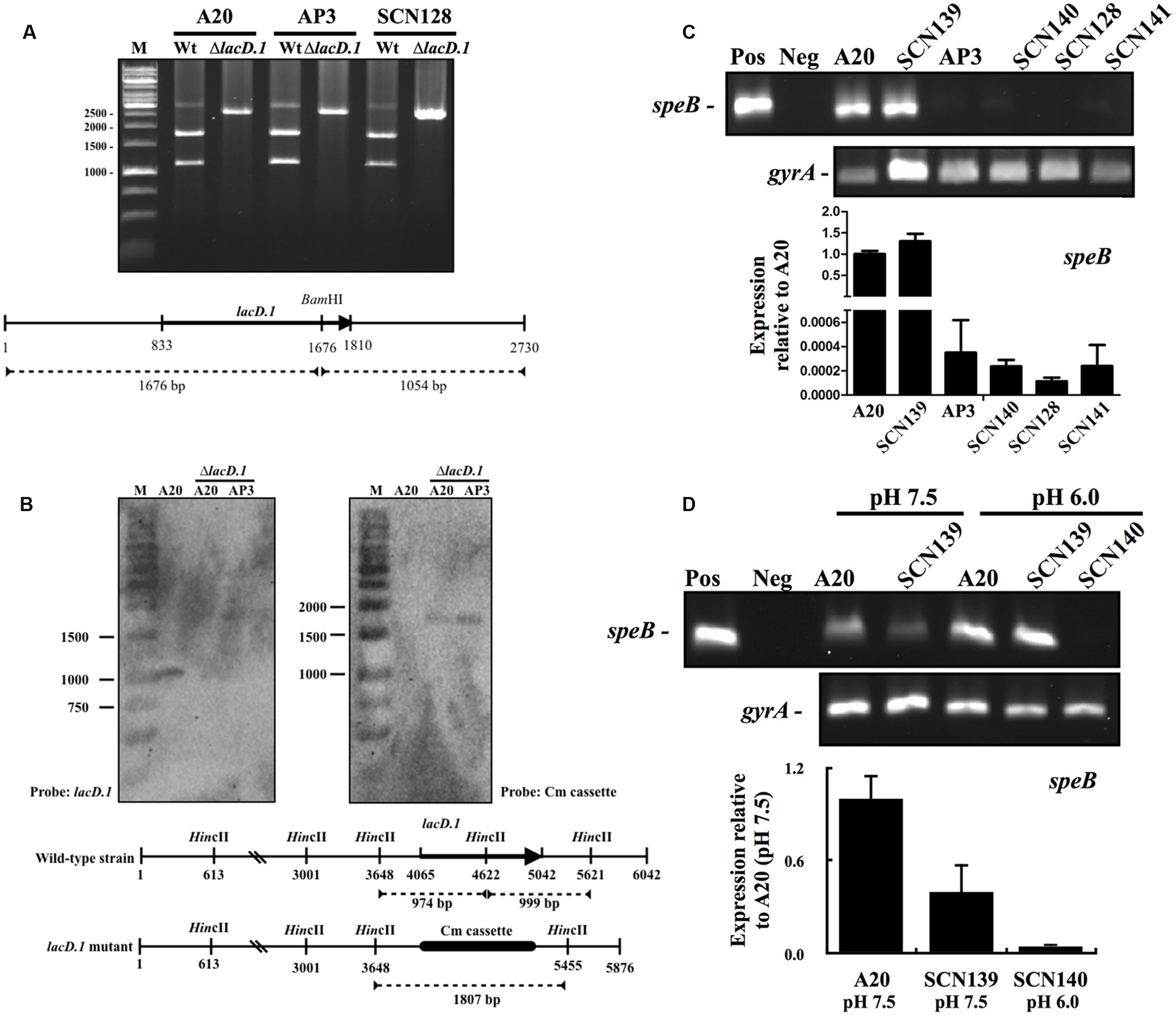
FIGURE 6. The expression of speB in wild type A20 strain, covS mutant AP3, A20 CovRD53A mutation strain SCN128, and their isogenic lacD.1 mutants. Deletion of the lacD.1 gene in lacD.1 mutants was verified by restriction enzyme digestion and Southern blot analysis. (A) The BamHI digestion pattern of the partial lac.1 operon in wild type strains and their isogenic lacD.1 mutants. The lower panel shows the location of the lacD.1 gene, BamHI restriction enzyme sites, and predicted size after BamHI digestion. M, DNA marker. (B) Southern blot analysis for A20, A20 lacD.1 mutant, and AP3 lacD.1 mutant. Genomic DNAs were digested by HincII and separated by 1% agarose gel for Southern blot analysis. The lower panel shows the location of the lacD.1 gene in wild type strains, a Cm cassette in lacD.1 mutants, HincII restriction enzyme sites, and the predicted size for lacD.1 and Cm probe hybridization. M, DNA marker. (C) The expression of speB in A20, AP3, SCN128, and their isogenic lacD.1 mutants (SCN139, SCN140, and SCN141, respectively). GAS strains were cultured in TSBY broth to the stationary phase of growth and RNAs were extracted for RT-PCR and qPCR (lower panel) analyses. (D) The expression of speB in A20, AP3, and their isogenic lacD.1 mutants at pH 7.5 and pH 6.0 conditions. GAS strains were cultured in neutral and acidic pH broths for 3 h and RNAs were extracted for RT-PCR analysis. The expression of gyrA gene is used as the internal control. Pos and Neg: Positive control (genomic DNA as template) and negative control. Lower panel, qPCR analysis for the speB expression in A20 and SCN139 under pH 7.5 condition and SCN140 under pH 6.0 condition. Biological replicate experiments were performed using three independent RNA preparations in duplicate. The expression of speB was normalized to the gyrA.
Discussion
Along with polymorphisms of the covR/S genes, mutations in the rgg gene are detected more frequently in strains isolated from patients with invasive infection (Ikebe et al., 2010). Friaes et al. (2015) suggested that covR/S and rgg genes are under stabilizing selection. However, only the deletion of covS, not rgg, is associated with invasive infections (Friaes et al., 2015). Mutations in covS have been shown to inactivate CovR phosphorylation and derepress target genes expression (Horstmann et al., 2015). However, the expression of CovR-regulated gene, speB, is repressed in the covS mutant. The present study demonstrated that nonphosphorylated CovR in the emm1-type A20 strain represses rgg expression (Figures 2 and 3), and the repression of rgg in covS mutant directly contributes to inhibiting speB expression (Figure 2). Furthermore, these results also suggest that the alteration of Rgg regulon expression in the covS mutant may contribute to higher invasiveness and provide better fitness in specific niches during infection.
CovR acts primarily as a transcriptional repressor to restrict expression of genes that it regulates directly (Churchward, 2007). The present study shows that CovR acts to repress rgg transcription in both wild type and covS mutant strains (Figure 4D). However, the results showed in Figures 3A,4C suggest that nonphosphorylated CovR more strongly inhibits rgg expression than phosphorylated CovR. The AT-rich CovR-binding sequences (ATTARA) have been proposed and identified in the promoter regions of hasA, covR, sagA, ska, and speB (Bernish and van de Rijn, 1999; Miller et al., 2001; Federle and Scott, 2002; Gao et al., 2005; Gusa and Scott, 2005; Gusa et al., 2006; Churchward et al., 2009). A CovR-binding sequence can be found in the coding region of the rgg gene2 (98-113; Virtual Footprint); however, this sequence cannot be identified in the rgg promoter region. In addition, phosphorylation of CovR increases its DNA-binding affinity and specificity for all promoter DNA fragments described above. Based on these results, we suggest that CovR may not bind to the rgg promoter and repress rgg transcription directly in the covS mutant. However, the interactions between phosphorylated and nonphosphorylated CovR proteins and the rgg gene are still uncertain and should be further studied.
The genome of GAS contains two loci (Lac.1 and Lac.2) encoding putative components of the tagatose 6-phosphate pathway (Ferretti et al., 2001; Loughman and Caparon, 2007). A gene encoding tagatose-1,6-bisphosate aldolase is present at both loci (lacD.1 and lacD.2); however, only LacD.2 contributes to utilize carbohydrate (Loughman and Caparon, 2007). LacD.1 has been shown to interact with Rgg to prevent Rgg-mediated gene expression in emm14- and emm49-type strains (Loughman and Caparon, 2006; Anbalagan et al., 2012). In addition, Rgg is a transcriptional regulator of genes associated with stress responses, metabolism, and virulence in emm3- and emm49-type strains (Chaussee, 2002; Chaussee et al., 2004; Pulliainen et al., 2008; Ikebe et al., 2010). However, among different emm-type strains, only the expression of speB and adjacent gene spy2040 are negatively regulated by Rgg (Dmitriev et al., 2008). Therefore, the expression of speB is a reliable marker to evaluate the interaction between LacD.1 and Rgg. The present study shows that the deletion of lacD.1 in AP3 did not restore speB expression (Figure 6C). Overexpression of rgg in AP3 activated speB expression in the early-exponential phase of growth (Figure 2B). In addition, speB expression in the wild type A20 strain and its lacD.1 isogenic mutant was still repressed under neutral culture conditions (Figure 6D). These results suggest two possible mechanisms in this A20 strain: LacD.1 does not interact with Rgg or LacD.1 binds to Rgg but does not inhibit Rgg-mediated speB expression. Both the AP3 strain used in this study and MGAS5005 are emm1-type strains and have an identical T nucleotide deletion in the covS gene. Dmitriev et al. (2008) showed that expression of speB and spy2040 are downregulated and spy1793 is upregulated in an MGAS5005 rgg isogenic mutant, indicating that Rgg negatively regulates spy1793 expression. To further elucidate the role of LacD.1 in the regulation of Rgg activity, the expression of spy1793 was analyzed by qPCR. However, although the expression of spy1793 was upregulated in the covS mutant AP3 compared with that in wild type A20 strain (expression relative to A20: 2.67 ± 0.8), inactivation of rgg in A20 (data not shown) showed to repress spy1793 expression (expression relative to A20: 0.34 ± 0.09), suggesting that spy1793 is regulated by multiple regulators in the covS mutant. These results suggest that spy1793 is not a suitable marker to evaluate the Rgg activity; in addition, more efforts are needed to clarify the interactions between LacD.1 and Rgg among different GAS strains.
Trevino et al. (2009) showed that CovS activates CovR to repress one group of genes, while it simultaneously inhibits the ability of CovR to repress the second group of genes (e.g., speB). Phosphorylated CovR has greater DNA-binding affinity and specificity for target gene promoters than that of nonphosphorylated CovR. Therefore, it is unlikely that CovS phosphorylates CovR leading to phosphorylated CovR failing to bind to the speB promoter. The present study shows that the inactivation of CovS results in a decrease in the level of CovR phosphorylation, and nonphosphorylated CovR mediates rgg repression (Figures 3 and 4). Rgg is essential for triggering speB expression (Neely et al., 2003). Therefore, although non-phosphorylated CovR has a lower binding activity to the speB promoter (Miller et al., 2001), the covS mutant still failed to express speB due to the downregulation of rgg. This study did not reveal the molecular mechanism of the CovR-Rgg interaction; however, these results suggest the importance of Rgg in gene regulation in the invasive covS mutant. Further explorations of the interactions between these regulatory systems in the invasive covS mutant would shed light on the disease pathogenesis of invasive GAS infections.
Author Contributions
The conception or design of the study: CC-N, C-HC, and J-JW. The acquisition, analysis, or interpretation of the data: T-PC and CC-N. Writing of the manuscript: CC-N.
Funding
This work was supported by parts of grants from the Chang Gung Memorial Hospital, LinKou, Taiwan (CMRPD1E0101-3 and BMRPD19) and Ministry of Science and Technology, Taiwan (MOST 103-2320-B-182-025-MY3).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The authors appreciated the constructive comments from Prof. Yi-Ywan Margaret Chen (Department of Microbiology and Immunology, Chang Gung University, Taiwan) for reviewing this work.
Footnotes
References
Anbalagan, S., Dmitriev, A., McShan, W. M., Dunman, P. M., and Chaussee, M. S. (2012). Growth phase-dependent modulation of Rgg binding specificity in Streptococcus pyogenes. J. Bacteriol. 194, 3961–3971. doi: 10.1128/JB.06709-11
Ato, M., Ikebe, T., Kawabata, H., Takemori, T., and Watanabe, H. (2008). Incompetence of neutrophils to invasive group A Streptococcus is attributed to induction of plural virulence factors by dysfunction of a regulator. PLoS ONE 3:e3455. doi: 10.1371/journal.pone.0003455
Barbieri, C. M., and Stock, A. M. (2008). Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tag-based reagents. Anal. Biochem. 376, 73–82. doi: 10.1016/j.ab.2008.02.004
Bernish, B., and van de Rijn, I. (1999). Characterization of a two-component system in Streptococcus pyogenes which is involved in regulation of hyaluronic acid production. J. Biol. Chem. 274, 4786–4793. doi: 10.1074/jbc.274.8.4786
Carapetis, J. R., Steer, A. C., Mulholland, E. K., and Weber, M. (2005). The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5, 685–694. doi: 10.1016/s1473-3099(05)70267-x
Chaussee, M. A., Callegari, E. A., and Chaussee, M. S. (2004). Rgg regulates growth phase-dependent expression of proteins associated with secondary metabolism and stress in Streptococcus pyogenes. J. Bacteriol. 186, 7091–7099. doi: 10.1128/JB.186.21.7091-7099.2004
Chaussee, M. S. (2002). Rgg influences the expression of multiple regulatory loci to coregulate virulence factor expression in Streptococcus pyogenes. Infect. Immun. 70, 762–770. doi: 10.1128/iai.70.2.762-770.2002
Chaussee, M. S., Somerville, G. A., Reitzer, L., and Musser, J. M. (2003). Rgg coordinates virulence factor synthesis and metabolism in Streptococcus pyogenes. J. Bacteriol. 185, 6016–6024. doi: 10.1128/jb.185.20.6016-6024.2003
Chiang-Ni, C., Tsou, C. C., Lin, Y. S., Chuang, W. J., Lin, M. T., Liu, C. C., et al. (2008). The transcriptional terminator sequences downstream of the covR gene terminate covR/S operon transcription to generate covR monocistronic transcripts in Streptococcus pyogenes. Gene 427, 99–103. doi: 10.1016/j.gene.2008.08.025
Chiang-Ni, C., and Wu, J. J. (2008). Effects of streptococcal pyrogenic exotoxin B on pathogenesis of Streptococcus pyogenes. J. Formos. Med. Assoc. 107, 677–685. doi: 10.1016/S0929-6646(08)60112-6
Chiang-Ni, C., Zheng, P. X., Ho, Y. R., Wu, H. M., Chuang, W. J., Lin, Y. S., et al. (2009). emm1/sequence type 28 strains of group A streptococci that express covR at early stationary phase are associated with increased growth and earlier SpeB secretion. J. Clin. Microbiol. 47, 3161–3169. doi: 10.1128/JCM.00202-09
Chiang-Ni, C., Zheng, P. X., Tsai, P. J., Chuang, W. J., Lin, Y. S., Liu, C. C., et al. (2012). Environmental pH changes, but not the LuxS signalling pathway, regulate SpeB expression in M1 group A streptococci. J. Med. Microbiol. 61(Pt 1), 16–22. doi: 10.1099/jmm.0.036012-0
Churchward, G. (2007). The two faces of Janus: virulence gene regulation by CovR/S in group A streptococci. Mol. Microbiol. 64, 34–41. doi: 10.1111/j.1365-2958.2007.05649.x
Churchward, G., Bates, C., Gusa, A. A., Stringer, V., and Scott, J. R. (2009). Regulation of streptokinase expression by CovR/S in Streptococcus pyogenes: CovR acts through a single high-affinity binding site. Microbiology 155(Pt 2), 566–575. doi: 10.1099/mic.0.024620-0
Collin, M., and Olsen, A. (2000). Generation of a mature streptococcal cysteine proteinase is dependent on cell wall-anchored M1 protein. Mol. Microbiol. 36, 1306–1318. doi: 10.1046/j.1365-2958.2000.01942.x
Dalton, T. L., and Scott, J. R. (2004). CovS inactivates CovR and is required for growth under conditions of general stress in Streptococcus pyogenes. J. Bacteriol. 186, 3928–3937. doi: 10.1128/JB.186.12.3928-3937.2004
Dmitriev, A. V., McDowell, E. J., and Chaussee, M. S. (2008). Inter–and intraserotypic variation in the Streptococcus pyogenes Rgg regulon. FEMS Microbiol. Lett. 284, 43–51. doi: 10.1111/j.1574-6968.2008.01171.x
Doran, J. D., Nomizu, M., Takebe, S., Menard, R., Griffith, D., and Ziomek, E. (1999). Autocatalytic processing of the streptococcal cysteine protease zymogen: processing mechanism and characterization of the autoproteolytic cleavage sites. Eur. J. Biochem. 263, 145–151. doi: 10.1046/j.1432-1327.1999.00473.x
Federle, M. J., and Scott, J. R. (2002). Identification of binding sites for the group A streptococcal global regulator CovR. Mol. Microbiol. 43, 1161–1172. doi: 10.1046/j.1365-2958.2002.02810.x
Ferretti, J. J., McShan, W. M., Ajdic, D., Savic, D. J., Savic, G., Lyon, K., et al. (2001). Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U.S.A. 98, 4658–4663. doi: 10.1073/pnas.071559398
Friaes, A., Pato, C., Melo-Cristino, J., and Ramirez, M. (2015). Consequences of the variability of the CovRS and RopB regulators among Streptococcus pyogenes causing human infections. Sci. Rep. 5, 12057. doi: 10.1038/srep12057
Gao, J., Gusa, A. A., Scott, J. R., and Churchward, G. (2005). Binding of the global response regulator protein CovR to the sag promoter of Streptococcus pyogenes reveals a new mode of CovR-DNA interaction. J. Biol. Chem. 280, 38948–38956. doi: 10.1074/jbc.M506121200
Graham, M. R., Smoot, L. M., Migliaccio, C. A., Virtaneva, K., Sturdevant, D. E., Porcella, S. F., et al. (2002). Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc. Natl. Acad. Sci. U.S.A. 99, 13855–13860. doi: 10.1073/pnas.202353699
Gusa, A. A., Gao, J., Stringer, V., Churchward, G., and Scott, J. R. (2006). Phosphorylation of the group A streptococcal CovR response regulator causes dimerization and promoter-specific recruitment by RNA polymerase. J. Bacteriol. 188, 4620–4626. doi: 10.1128/JB.00198-06
Gusa, A. A., and Scott, J. R. (2005). The CovR response regulator of group A Streptococcus (GAS) acts directly to repress its own promoter. Mol. Microbiol. 56, 1195–1207. doi: 10.1111/j.1365-2958.2005.04623.x
Heath, A., DiRita, V. J., Barg, N. L., and Engleberg, N. C. (1999). A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect. Immun. 67, 5298–5305.
Horstmann, N., Sahasrabhojane, P., Saldana, M., Ajami, N. J., Flores, A. R., Sumby, P., et al. (2015). Characterization of the effect of the histidine kinase CovS on response regulator phosphorylation in group A Streptococcus. Infect. Immun. 83, 1068–1077. doi: 10.1128/IAI.02659-14
Horstmann, N., Saldana, M., Sahasrabhojane, P., Yao, H., Su, X., Thompson, E., et al. (2014). Dual-site phosphorylation of the control of virulence regulator impacts group A streptococcal global gene expression and pathogenesis. PLoS Pathog. 10:e1004088. doi: 10.1371/journal.ppat.1004088
Ikebe, T., Ato, M., Matsumura, T., Hasegawa, H., Sata, T., Kobayashi, K., et al. (2010). Highly frequent mutations in negative regulators of multiple virulence genes in group A streptococcal toxic shock syndrome isolates. PLoS Pathog. 6:e1000832. doi: 10.1371/journal.ppat.1000832
Levin, J. C., and Wessels, M. R. (1998). Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol. Microbiol. 30, 209–219. doi: 10.1046/j.1365-2958.1998.01057.x
Lin, J. N., Chang, L. L., Lai, C. H., Lin, H. H., and Chen, Y. H. (2014). Association between polymorphisms in the csrRS two-component regulatory system and invasive group A streptococcal infection. Eur. J. Clin. Microbiol. Infect. Dis. 33, 735–743. doi: 10.1007/s10096-013-2005-7
Loughman, J. A., and Caparon, M. G. (2006). A novel adaptation of aldolase regulates virulence in Streptococcus pyogenes. EMBO J. 25, 5414–5422. doi: 10.1038/sj.emboj.7601393
Loughman, J. A., and Caparon, M. G. (2007). Comparative functional analysis of the lac operons in Streptococcus pyogenes. Mol. Microbiol. 64, 269–280. doi: 10.1111/j.1365-2958.2007.05663.x
Miller, A. A., Engleberg, N. C., and DiRita, V. J. (2001). Repression of virulence genes by phosphorylation-dependent oligomerization of CsrR at target promoters in S. pyogenes. Mol. Microbiol. 40, 976–990. doi: 10.1046/j.1365-2958.2001.02441.x
Neely, M. N., Lyon, W. R., Runft, D. L., and Caparon, M. (2003). Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. J. Bacteriol. 185, 5166–5174. doi: 10.1128/jb.185.17.5166-5174.2003
Olsen, R. J., and Musser, J. M. (2010). Molecular pathogenesis of necrotizing fasciitis. Annu. Rev. Pathol. 5, 1–31. doi: 10.1146/annurev-pathol-121808-102135
O’Sullivan, D. J., and Klaenhammer, T. R. (1993). High- and low-copy-number Lactococcus shuttle cloning vectors with features for clone screening. Gene 137, 227–231. doi: 10.1016/0378-1119(93)90011-Q
Pulliainen, A. T., Hytonen, J., Haataja, S., and Finne, J. (2008). Deficiency of the Rgg regulator promotes H2O2 resistance, AhpCF-mediated H2O2 decomposition, and virulence in Streptococcus pyogenes. J. Bacteriol. 190, 3225–3235. doi: 10.1128/JB.01843-07
Sumby, P., Whitney, A. R., Graviss, E. A., DeLeo, F. R., and Musser, J. M. (2006). Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2:e5. doi: 10.1371/journal.ppat.0020005
Tran-Winkler, H. J., Love, J. F., Gryllos, I., and Wessels, M. R. (2011). Signal transduction through CsrRS confers an invasive phenotype in group A Streptococcus. PLoS Pathog. 7:e1002361. doi: 10.1371/journal.ppat.1002361
Trevino, J., Perez, N., Ramirez-Pena, E., Liu, Z., Shelburne, S. A. III, Musser, J. M., et al. (2009). CovS simultaneously activates and inhibits the CovR-mediated repression of distinct subsets of group A Streptococcus virulence factor-encoding genes. Infect. Immun. 77, 3141–3149. doi: 10.1128/IAI.01560-08
Tsou, C. C., Chiang-Ni, C., Lin, Y. S., Chuang, W. J., Lin, M. T., Liu, C. C., et al. (2010). Oxidative stress and metal ions regulate a ferritin-like gene, dpr, in Streptococcus pyogenes. Int. J. Med. Microbiol. 300, 259–264. doi: 10.1016/j.ijmm.2009.09.002
Unnikrishnan, M., Cohen, J., and Sriskandan, S. (1999). Growth-phase-dependent expression of virulence factors in an M1T1 clinical isolate of Streptococcus pyogenes. Infect. Immun. 67, 5495–5499.
Valcu, M., and Valcu, C. M. (2011). Data transformation practices in biomedical sciences. Nat. Methods 8, 104–105. doi: 10.1038/nmeth0211-104
Walker, M. J., Hollands, A., Sanderson-Smith, M. L., Cole, J. N., Kirk, J. K., and Henningham, A. (2007). DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat. Med. 13, 981–985. doi: 10.1038/nm1612
Keywords: group A Streptococcus, CovR/CovS, Rgg, LacD.1, SpeB
Citation: Chiang-Ni C, Chu T-P, Wu J-J and Chiu C-H (2016) Repression of Rgg But Not Upregulation of LacD.1 in emm1-type covS Mutant Mediates the SpeB Repression in Group A Streptococcus. Front. Microbiol. 7:1935. doi: 10.3389/fmicb.2016.01935
Received: 12 July 2016; Accepted: 17 November 2016;
Published: 29 November 2016.
Edited by:
Awdhesh Kalia, University of Texas MD Anderson Cancer Center, USACopyright © 2016 Chiang-Ni, Chu, Wu and Chiu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuan Chiang-Ni, ZW50Y2h1YW5AZ2FwLmNndS5lZHUudHc=
 Chuan Chiang-Ni
Chuan Chiang-Ni Teng-Ping Chu1
Teng-Ping Chu1 Cheng-Hsun Chiu
Cheng-Hsun Chiu