- School of Biosciences and Biotechnology, Faculty of Science and Technology, Universiti Kebangsaan Malaysia, Bangi, Malaysia
The spread of antibiotic resistance amongst bacterial pathogens has led to an urgent need for new antimicrobial compounds with novel modes of action that minimize the potential for drug resistance. To date, the development of new antimicrobial drugs is still lagging far behind the rising demand, partly owing to the absence of an effective screening platform. Over the last decade, the nematode Caenorhabditis elegans has been incorporated as a whole animal screening platform for antimicrobials. This development is taking advantage of the vast knowledge on worm physiology and how it interacts with bacterial and fungal pathogens. In addition to allowing for in vivo selection of compounds with promising anti-microbial properties, the whole animal C. elegans screening system has also permitted the discovery of novel compounds targeting infection processes that only manifest during the course of pathogen infection of the host. Another advantage of using C. elegans in the search for new antimicrobials is that the worm itself is a source of potential antimicrobial effectors which constitute part of its immune defense response to thwart infections. This has led to the evaluation of effector molecules, particularly antimicrobial proteins and peptides (APPs), as candidates for further development as therapeutic agents. In this review, we provide an overview on use of the C. elegans model for identification of novel anti-infectives. We highlight some highly potential lead compounds obtained from C. elegans-based screens, particularly those that target bacterial virulence or host defense to eradicate infections, a mechanism distinct from the action of conventional antibiotics. We also review the prospect of using C. elegans APPs as an antimicrobial strategy to treat infections.
Introduction
It is undeniable that antibiotics have had an enormous impact on global human health by drastically reducing infection-associated mortality. Nonetheless, the abuse and uncontrolled use of antibiotics has resulted in the emergence and spread of resistant bacteria. Whilst resistance to antibiotics has escalated steadily, the number of new antimicrobial drugs approved, especially those with novel modes of action, continues to decline (Boucher et al., 2013). Among the vast number of Gram-positive and Gram-negative bacteria, the ‘ESKAPE’ group of pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) represent the most common antibiotic-resistance pathogens (Pendleton et al., 2013). The emergence of variant pathogens such as the New Delhi metallo-ß-lactamase 1 (NDM-1)-producing Enterobacteriaceae strains (Kumarasamy et al., 2010) and colistin-resistant Escherichia coli harboring the mcr-1 gene (Liu et al., 2016; McGann et al., 2016) have exacerbated the situation and further underscores the importance of new innovative anti-infective strategies to minimize the spread of drug resistance. A major obstacle in the identification of effective anti-infective therapies is the absence of efficient in vivo screening platforms. Since the first introduction of Caenorhabditis elegans to study developmental biology, the worm has proven to be a versatile host model for elucidating molecular and cellular aspects of various infectious diseases. C. elegans has been adopted as an in vivo infection model to dissect the intricate host–pathogen interaction and the evolutionarily conserved mechanisms employed by pathogens to infect and kill the host (Ermolaeva and Schumacher, 2014; Cohen and Troemel, 2015). The descriptions of an expanding list of bacterial and fungal pathogen infections in C. elegans (Marsh and May, 2012) have led to the use of worms as a whole organism system for antimicrobial and anti-infective drug discovery. This review will assess the use of C. elegans as a platform for the discovery of novel anti-infectives or antimicrobial compounds that target bacterial virulence or host immune responses to attenuate infections. We also highlight the potential of C. elegans as a rich source of antimicrobial proteins and peptides (APPs), key components of innate immunity.
C. elegans Platform for Anti-Infectives Discovery: Why and How?
The increasing popularity of C. elegans as a host model is attributed to its small (∼1 mm) and simple anatomy, short generation time, high fecundity, fully sequenced genome (∼100 Mb) and the relatively easy and inexpensive maintenance. Moreover, working with the worm offers great advantage as many resources of genetic and genomic knowledge and techniques are available to facilitate experimental manipulation (Antoshechkin and Sternberg, 2007). For example, knocking down worm genes can easily be achieved through RNA interference (RNAi) simply by feeding the worms with bacteria harboring a plasmid engineered to express double-stranded RNA (dsRNA) targeting the gene of interest. Two RNAi clone libraries covering almost 94% of the 20 000 genes in C. elegans are available and this allows genome-wide RNAi screens to be performed (Kamath and Ahringer, 2003; Rual et al., 2004). Furthermore, over 3000 phenotypically defined mutant strains are available from the Caenorhabditis Genetics Centre. In addition, the worm is transparent, allowing for in vivo monitoring of cells and the visualization of fluorescently tagged bacterial and host genes and proteins throughout the entire course of an experiment. The nematode defense system is highly homologous to at least three mammalian conserved innate immunity signaling pathways which are crucial for defense against pathogens; the p38 Mitogen-Activated Protein Kinase (p38 MAPK) pathway (Troemel et al., 2006), the Insulin/Growth Factor-1 (IIS) pathway (Garsin et al., 2003) and the Transforming Growth Factor-β (TGF-β) pathway (Mallo et al., 2002). Studies have also indicated that many of the virulence factors involved in the killing of worms were also required for the pathogenesis of mammals (Tan et al., 1999; Sifri et al., 2003) and hence, over the last decade, the utility of the worm has also been extended to facilitate novel antimicrobial drug discovery and development (Ewbank and Zugasti, 2011; Arvanitis et al., 2013). Taken together, these advantages make C. elegans an ideal model organism in various aspects of biology.
The traditional pipeline for antimicrobial drug discovery usually begins with in vitro screening of test compounds followed by subjecting potential hits to in vivo animal testing (Brown and Wright, 2016). However, these potential compounds often end up exhibiting poor pharmacokinetic activities and/or are highly toxic when tested in vivo in animals. Another approach is to directly determine the in vivo effects of the test compounds in conventional mammalian models but this approach presents with serious limitations, including high cost, laborious and time-consuming procedures and ethical constraints (Arvanitis et al., 2013). While mammalian models have been utilized to study potentially new drug leads, screening of a large number of compounds is prohibited with these models. Both these approaches have shortcomings and cause significant delay in antimicrobial discovery. To overcome these limitations, an alternative method is to conduct the initial testing of compounds in the C. elegans whole animal system. Exposing worms to both pathogen and test compounds and subsequently monitoring worm survival throughout the assay can be easily performed as an antimicrobial screen of test compounds in C. elegans. Compounds with potential antimicrobial activity will contribute to prolonged infected worm lifespan when compared to the untreated infected worms. Screening of compounds with the C. elegans whole animal model also allows preliminary assessment of drug toxicity as worms treated with these compounds will show similar or decreased survival when compared to the untreated control (Moy et al., 2009). A further advantage of the C. elegans model is the self-fertilization of hermaphrodite worms which allows for cost effective rapid cultivation of a large population of worms for drug testing without any of the ethical constraints normally encountered when working with rodents and primates. The worm is also amenable to fully automated high-throughput screening whereby 100s of animals can be systematically dispensed into wells of standard 96- and 384- well plates permitting the screen of larger compound libraries, the incorporation of fluorescent dye to ease scoring of dead and alive worms as well as the use of sophisticated hardware and software for image capturing and data analysis (Wahlby et al., 2012; Conery et al., 2014). To increase the efficiency of the high-throughput screen, improvements to the automated worm sorting and transfer technologies, imaging and software for data analysis are constantly being undertaken (O’Reilly et al., 2014).
Another significant advantage of using C. elegans in the search for new antimicrobial compounds is that, as an in vivo model, it allows for the detection of compounds that may not directly target the pathogen viability but instead, may alter pathogen virulence or enhance the host immune response. Since the primary innate immunity signaling pathways identified in C. elegans are highly conserved evolutionarily, the findings from the screen may also be extended to higher organisms, including humans. Nevertheless, although the nematode is a powerful tool to screen for antimicrobials, it cannot completely replace mammalian models and certain limitations must be acknowledged. Firstly, in the laboratory, C. elegans are routinely cultured at 16, 20, or 25°C. The worms cannot grow at 37°C and this restricts the range of pathogens that can be studied. Furthermore, C. elegans also has an efficient detoxification system, which limits the capacity to identify compounds that act via modulation of host defenses (Ewbank and Zugasti, 2011). That being said, the use of C. elegans in drug discovery still holds promise and should be pursued.
Identification of Novel Anti-Infectives
Generally, an anti-infective is an agent capable of preventing or counteracting infection, either by inhibiting the dissemination of an infectious agent or by killing the pathogen directly. Antibiotics are one of the well-known examples of anti-infective agents. Antimicrobial agents have been classified based on their ability to either kill the bacteria (bactericidal) or inhibit bacterial propagation (bacteriostatic) by targeting function essential to bacterial viability. Although these strategies are highly effective in eliminating the bacteria, they result in substantial stress on the target bacterium, leading to rapid selection of resistant sub-populations. Non-antimicrobial approaches to treat infections are therefore needed as alternative therapies to overcome the pitfalls of antibiotic resistance. Anti-infectives that do not affect microbial cell viability, but may instead interfere with virulence-mediated pathways in pathogens, appear as a new therapeutic paradigm (Allen et al., 2014). From the host perspective, selective modulation of the host innate immunity is another emerging concept driven by advances in the current understanding of host defense systems (Nijnik, 2013). Conventional in vitro cell-culture based assays are less likely to probe anti-infectives that target bacterial virulence or host immunity due to the absence of an intact and complex host–pathogen relationship. Following the wide application of C. elegans in infectious diseases modeling, this tiny nematode has emerged as a very useful model for anti-infectives drug discovery and development (Desalermos et al., 2011; Ewbank and Zugasti, 2011). In addition to its ability to identify novel antimicrobials that impair bacterial growth, the simple C. elegans model also presents an advantage, as we are able to select potential anti-virulence and immunomodulatory molecules in the context of a live animal. The possible outcomes of a C. elegans-based anti-infective screen are illustrated in Figure 1.
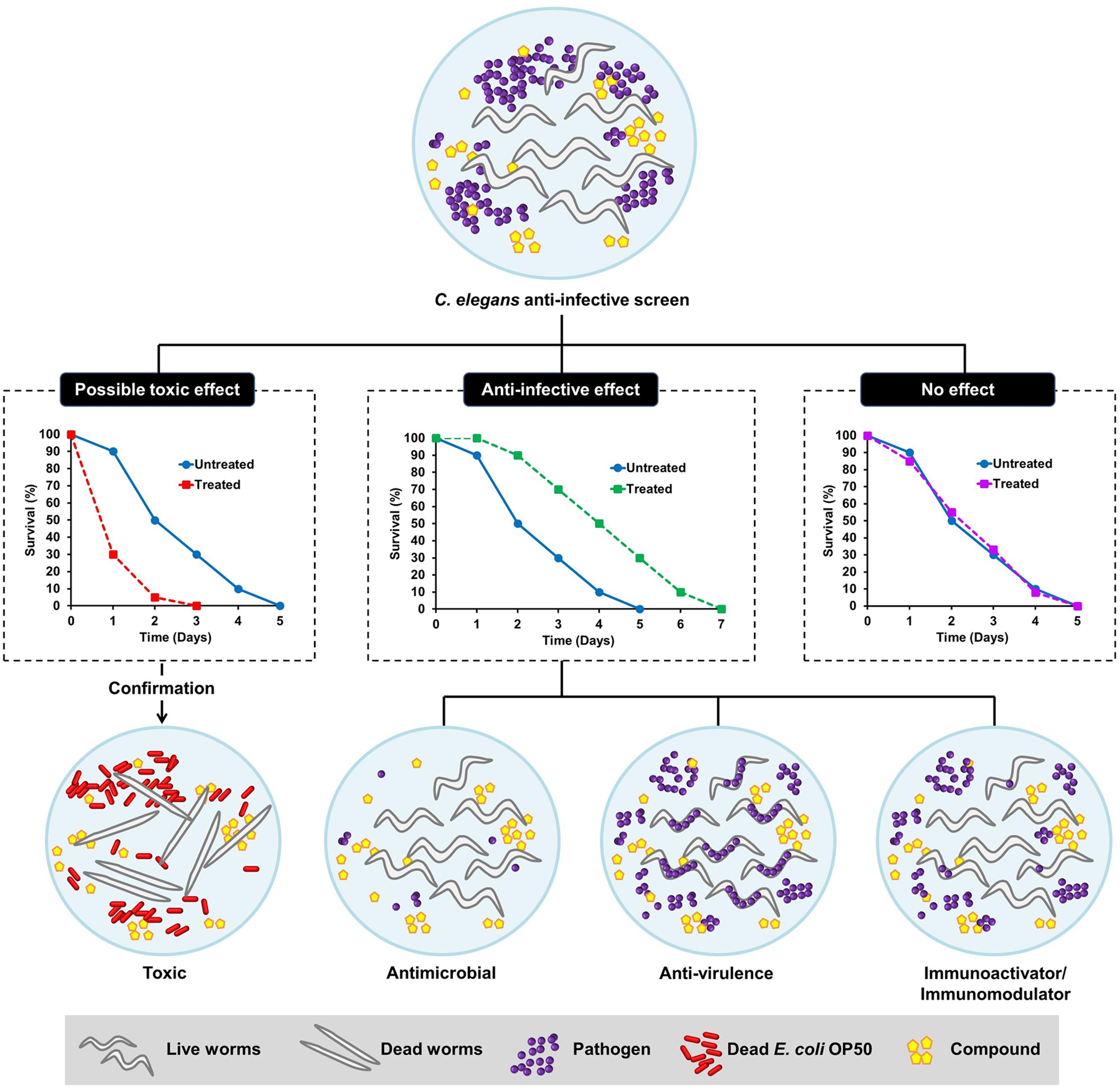
FIGURE 1. Schematic diagram illustrating the possible outcomes of a Caenorhabditis elegans-based anti-infective screen. From a screen, the compounds can be categorized as those with possible toxic effect, potential anti-infective effect or no effect. The hits that promote the survival of C. elegans following an infection can either act as anti-microbial, anti-virulence, or immunoactivator/immunomodulator. The toxic effect of a compound can be further validated by exposing the worms to the particular compound in an uninfected condition where the worms are fed with dead Escherichia coli OP50.
Compounds with In vivo Antimicrobial Activity
In the first antimicrobial screen using C. elegans, the standard agar-based infection assay was modified to form a liquid-based curing assay in a 96-well plate format for high-throughput screening of 6,000 individually synthesized molecules and 1,136 natural extracts, consisting mainly of plant and marine extracts, for their ability to rescue C. elegans following an Enterococcus faecalis persistent infection (Moy et al., 2006). This screen identified 16 synthetic compounds and nine natural extracts that promote survival of E. faecalis infected worms. From the antimicrobial test, 7 of the 16 compounds inhibited the replication of E. faecalis in vitro with minimum inhibitory concentrations (MICs) <31 μg/mL. The screen was refined to a fully automated process which scaled up the throughput of the screen from ∼7000 compounds to ∼37,000 unique molecules and natural product extracts. This allowed the identification of 80 antimicrobial agents that cured C. elegans from a E. faecalis infection, including 62 known antibiotics and 18 candidates with previously reported antibacterial effect or structural analogs of known antibiotics (Moy et al., 2009).
The initial study by Moy et al. (2009) prompted other initiatives to search for novel anti-infectives and these efforts have been fruitful. To identify novel therapeutic molecules toward S. aureus, our group developed a C. elegans–S. aureus screening platform to perform small-scale in vivo screens on extracts from plant and marine resources as well as synthetic molecules (Kong et al., 2014b). In contrast to E. faecalis that persists in the C. elegans intestinal tract, S. aureus infection of worms is transient (Sifri et al., 2003). By constant exposure of C. elegans to S. aureus in liquid medium, 70 candidates were subjected to the C. elegans–S. aureus screen and 28 positive hits (14 extracts and 14 compounds) were obtained. Of the 14 natural extracts, 7 of them demonstrated potential antimicrobial effect against S. aureus when tested in a standard MIC microdilution assay. Among the most effective antimicrobial candidates were the extract from Curcuma longa and its main active component, curcumin (Kong et al., 2014b). An automated high-throughput C. elegans antimicrobial screen was used to screen a compound library comprising of 640 FDA-approved drugs for novel antimicrobial molecules against methicillin-resistant S. aureus (MRSA) (Rajamuthiah et al., 2014). Of note, closantel, the drug used to eradicate helminths, was identified as a promising hit that significantly prolonged the survival of MRSA-infected C. elegans. In vitro MIC microdilution tests showed that closantel exerted antibacterial activity against a wide range of Gram-positive and Gram-negative bacteria at low effective concentrations. Interestingly, closantel was also effective against the vancomycin-resistant S. aureus (VRSA) with an MIC of 0.78 μg/mL. The compound library also contained some clinically relevant antibiotics and almost all of the antibiotics that were previously known to be effective against MRSA were also identified as positive hits in the screen, further validating the robustness of the screen in detecting highly potential anti-staphylococcal molecules for future therapeutics development (Rajamuthiah et al., 2014). Following this, the screen has been expanded to two other compound libraries consisting of 85,000 compounds (Kim W. et al., 2015) and ∼21,000 compounds (Rajamuthiah et al., 2015), respectively. One positive hit from the screen was 1-Hexadecyl-2-methyl-3-(phenylmethyl)-1H-imidazolium iodide (NH125), an antibiotic with well-characterized mode of action (Yamamoto et al., 2000). A further study revealed that NH125 was also able to kill persister cells and eradicate the formation of MRSA biofilm by penetrating the extracellular polymeric substance (EPS) matrix within an established biofilm (Kim W. et al., 2015). Additionally, a novel antibacterial compound, 3-(phenylsulfonyl)-2-pyrazinecarbonitrile (PSPC), rescued MRSA-infected worms. PSPC and its derivatives displayed high antibacterial efficacy toward Gram-positive bacteria, with higher MIC values observed against Gram-negative pathogens (Rajamuthiah et al., 2015). Yang et al. (2013) developed a simple, time-efficient and cost-effective integrated microfluidic-based C. elegans screening system to test the in vivo antimicrobial effect of various natural compounds. The three major active components of the natural product rhubarb (emodin, rhein, and aloe-emodin) enhanced the survival of worms challenged with S. aureus and inhibited S. aureus replication at ≤16 μg/mL but were toxic to the animals at concentrations >60 μg/mL (Yang et al., 2013).
A C. elegans–P. aeruginosa liquid-based slow-killing assay was used to perform an in vivo screen for potential anti-infective substances against multidrug-resistant P. aeruginosa. Screening a collection of 1,300 natural extracts mainly consisting of secondary metabolites from fungal endophytes of medicinal plants, led to the discovery of four extracts from four different fungal species with both in vivo and in vitro antimicrobial activity (Zhou et al., 2011). The in vivo activity and efficacy of naturally occurring antimicrobial peptides isolated from frog skin were also evaluated using a C. elegans infection model (Uccelletti et al., 2010). The peptides, temporin-1Tb and esculentin (1–18), promoted the survival of P. aeruginosa-infected worms. Exposure to the peptides for 24 h significantly reduced the intestinal bacterial load in worms pre-infected with P. aeruginosa. Taking advantage of the transparent nature of C. elegans, the in vivo effect of the peptides on membrane permeability was examined by firstly exposing the P. aeruginosa infected nematodes to Sytox Green followed by treatment with the peptides and subsequently, observation of fluorescence intensity within the intestine of C. elegans. The enhanced fluorescence intensity in treated infected nematodes indicated that the peptide killed P. aeruginosa in vivo by permeating the bacterial membrane. The findings from this study reinforced the fact that C. elegans can be used to investigate the in vivo mode of action of antimicrobial molecules within the context of a living animal.
The high-throughput C. elegans screening platform has been expanded to identify potential antimicrobial therapeutics against other pathogens that cause clinically significant morbidity and mortality such as Burkholderia pseudomallei (Lakshmanan et al., 2014) and A. baumannii (Jayamani et al., 2015). Lakshmanan et al. (2014) described a robust C. elegans based high-throughput screen to search for novel antibiotics to treat melioidosis, a highly lethal infection caused by the Gram-negative B. pseudomallei. As B. pseudomallei is a Tier-1 organism, a closely related species, Burkholderia thailandensis, was substituted as the surrogate pathogen in the screen. One of the hits identified from the pilot screen of ∼30,000 compounds was Clinafloxacin, a fourth generation fluoroquinolone antibiotic, which demonstrated diverse antibacterial activity against B. pseudomallei as well as other Gram-negative pathogens (Lakshmanan et al., 2014). The ability of A. baumannii to infect and kill C. elegans led to the development of a C. elegans–A. baumannii liquid infection assay to screen for potential anti-A. baumannii agents from a collection of synthetic insect-derived antimicrobial peptides. A promising hit was the antimicrobial peptide, cecropin A, isolated from the mosquito Aedes aegypti. Further analysis suggested that this peptide exerted its antimicrobial activity by disrupting the bacterial membrane, leading to cell lysis (Jayamani et al., 2015). The in vivo antimicrobial effect of small molecules identified from an in vitro growth inhibition assay was evaluated using a C. elegans–Burkholderia cenocepacia infection model. Identification of a compound with good in vitro activity without curing B. cenocepacia-infected nematodes from infection highlights the importance of using a live animal model for the discovery of antimicrobial compounds with promising in vivo efficacy (Selin et al., 2015). Other drugs and natural compounds that have been tested for their in vivo antimicrobial properties in the worm model are listed in Table 1.
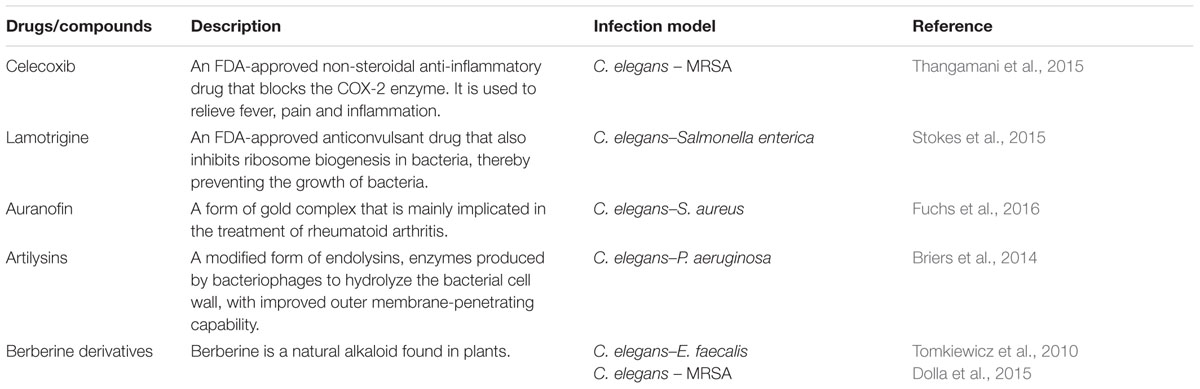
TABLE 1. Additional drugs/compounds evaluated for in vivo antimicrobial effects using a Caenorhabditis elegans model.
Compounds Targeting Bacterial Virulence
In the first antimicrobial screen by Moy et al. (2006) the identification of hits that did not interfere with bacterial growth in vitro or hits with a lower effective concentration in vivo suggests that these compounds and extracts may either act by enhancing host immunity or altering pathogen virulence. The findings from this study also offer proof-of-concept that the C. elegans whole-animal antimicrobial screening platform is able to detect hits that may be missed in a conventional cell-culture based in vitro screen where the live host–pathogen relationship is absent (Moy et al., 2006). In addition to novel antimicrobial compounds, the automated high-throughput C. elegans screen by Moy et al. also identified potential anti-virulence compounds (Moy et al., 2009), including analogs of small molecules that were found to prevent P. aeruginosa biofilm development in another anti-biofilm screen (Junker and Clardy, 2007).
In an earlier study on the effect of compounds on bacterial virulence, the effect of salicylic acid, a type of phenolic compound produced naturally by plants, was tested on P. aeruginosa virulence in the worm model as well as the Arabidopsis thaliana plant model. Feeding the worms with salicylic acid-treated P. aeruginosa prolonged worm survival compared to the worms exposed to untreated bacteria. Microarray and biochemical analyses proposed that this compound reduced the virulence of P. aeruginosa by inhibiting the production of biofilm, pyocyanin, protease, and elastase (Prithiviraj et al., 2005). Following this, the anti-virulence potential of extracts from three medicinal plants (Conocarpus erectus, Callistemon viminalis, and Bucida buceras) was assessed against P. aeruginosa using the C. elegans paralytic killing, fast-killing and slow-killing models. These extracts were known to affect bacterial quorum sensing (Adonizio et al., 2008a). Addition of the individual extracts into the solid agar medium significantly delayed the killing of nematodes by P. aeruginosa to a rate similar to worms fed with the P. aeruginosa lasR quorum sensing mutant, suggesting that these extracts most likely targeted the bacterial quorum sensing pathway (Adonizio et al., 2008b). A similar approach was also used to elucidate the anti-virulence properties of curcumin, the main bioactive component present in turmeric. Curcumin rescued C. elegans from a P. aeruginosa (PAO1) infection via the inhibition of various virulence factors, including pyocyanin, biofilm, acyl homoserine lactone, and bacterial quorum sensing (Rudrappa and Bais, 2008). In addition to its anti-virulence effect on P. aeruginosa, curcumin is also able to rescue worms from a B. pseudomallei-inflicted infection. We showed that curcumin interfered with B. pseudomallei iron acquisition and also inhibited the production of B. pseudomallei lipase, protease as well as biofilm without affecting bacterial viability. This suggests the potential of curcumin as an alternative therapy for the highly lethal melioidosis caused by B. pseudomallei (Eng and Nathan, 2015). In a recent study, curcumin was reported to interfere with bacterial virulence in a mouse model of lung infection, providing an independent validation of the anti-virulence effect identified in a C. elegans model (Wang et al., 2016).
Quorum sensing is a complicated cell-to-cell communication system that regulates the expression of various key virulence factors in Gram-positive and Gram-negative bacteria, making it a target of interest for anti-virulence therapy. The P. aeruginosa quorum sensing system utilizes acyl homoserine lactones as autoinducer molecules to regulate and direct the expression of virulence and biofilm-associated genes (Williams et al., 2007). Other natural inhibitors that have been tested for their anti-virulence effect in vivo using the C. elegans–P. aeruginosa infection model are the methanolic extract of Terminalia chebula fruit (Sarabhai et al., 2013), extract and active compound of Dalbergia trichocarpa bark (Rasamiravaka et al., 2015), tea polyphenols extracted from the leaf of Camellia sinensis L. (Yin et al., 2015), the methanolic extract of Trigonella foenum-graecum L. seed (Husain et al., 2015b), Syzygium aromaticum (clove) oil (Husain et al., 2013), and Mentha piperita (peppermint) oil (Husain et al., 2015a). In addition to targeting quorum sensing circuits, chemical compounds such as phenylacetic acid (Musthafa et al., 2012b), 2,5-piperazinedione (Musthafa et al., 2012a) and meta-bromo-thiolactone (O’Loughlin et al., 2013) as well as the hormonal therapy drug raloxifene (Ho Sui et al., 2012) inhibited P. aeruginosa pyocyanin production and reduced bacterial virulence in the nematode model. Other virulence determinants, including biofilm, proteases and elastase, as well as bacterial motility, were also suppressed by most of these molecules. Compounds targeting the iron-regulation pathway that alleviated P. aeruginosa-mediated killing of C. elegans were identified from a high-throughput liquid-based chemical screen (Kirienko et al., 2013). Recently, an anti-proliferative drug, 5-fluorouracil, was shown to enhance the survival of P. aeruginosa-infected C. elegans via disruption of pyoverdine biosynthesis (Kirienko et al., 2016). Interestingly, in an independent study, 5-fluorocytosine (a 5-fluorouracil precursor) demonstrated the ability to inhibit pyoverdine production in vivo in a mouse model of P. aeruginosa lung infection (Imperi et al., 2013). This compound also diminished P. aeruginosa virulence by targeting biofilm formation and quorum-sensing phenotypes in vitro (Ueda et al., 2009). Moreover, garlic extract also displayed anti-quorum sensing effect against P. aeruginosa in both nematode (Rasmussen et al., 2005) and mice models (Bjarnsholt et al., 2005; Harjai et al., 2010). Together, these findings demonstrate the successfully translation of the anti-virulence effect identified in a C. elegans model in a higher organism and provides a positive correlation between the compound activity identified in a C. elegans model with a more complex mammalian infection model. Through in silico drug design and model simulations, two chemical entities were identified as inhibitors of P. aeruginosa heme oxygenase. One of these compounds prolonged the lifespan of infected nematodes and decreased bacterial colony forming units in the C. elegans intestine, albeit showing poor antimicrobial activity in vitro (Hom et al., 2013). Recently, Zhu et al. (2015) used the C. elegans model to characterize an inhibitor of LasB, a vital virulence factor of P. aeruginosa. Cross-sectional transmission electron microscopy images of the C. elegans intestine displayed less host tissue damage upon treatment with the LasB inhibitor, suggesting that P. aeruginosa was rendered less virulent by the compound (Zhu et al., 2015).
Staphylococcus aureus is continuously acquiring resistance to clinically used antibiotics, underlining the need for new anti-virulence therapeutics against this pathogen. The ability of S. aureus to produce biofilm has complicated treatment by blocking the antibiotics penetration of the cells encased within the exopolysaccharide material. Hence, S. aureus biofilm and the quorum sensing system are attractive targets for anti-virulence therapeutics. Several natural compounds with in vitro anti-biofilm property have been demonstrated to attenuate S. aureus virulence in vivo in a C. elegans model. This includes essential oils from various plants (Lee J.H. et al., 2014; Lee et al., 2014a), red wine (Cho et al., 2015), stilbenes and the natural phenol, resveratrol (Lee J.H. et al., 2014; Lee et al., 2014b). Moreover, these natural products also reduced blood hemolysis caused by S. aureus hemolysins. The colonization of S. aureus in the worm intestine can be determined by staining the bacteria with acridine orange dye. Using this approach, the reduction of in vivo bacterial loads by an extract from a coral actinomycete was reported (Bakkiyaraj and Pandian, 2010). Hamamelitannin, the active component isolated from American witch hazel, has recently been shown to act as an inhibitor of the S. aureus quorum sensing mechanism. This compound and its analogs effectively improved the susceptibility of S. aureus biofilms to multiple antibiotics treatment both in vitro and in vivo (C. elegans and mouse infection models) (Brackman et al., 2016).
The uracil-auxotroph E. coli strain OP50 is routinely used as the laboratory-based food source for C. elegans, however, some pathogenic E. coli strains are lethal on worms. E. coli O157:H7 is a toxin-producing enterohemorrhagic strain with noteworthy medical implications. Broccoli extract and its associated flavonoid compounds were able to impede the virulence of this E. coli strain in the nematode model by inhibiting a number of vital virulence genes as well as the swarming motility of E. coli O157:H7 (Lee et al., 2011). Screening a collection of plant secondary metabolites for anti-biofilm activity against E. coli O157:H7 also identified several coumarin derivatives that were subsequently shown to suppress pathogen virulence in vivo in the C. elegans infection system (Lee J.H. et al., 2014). Lavigne et al. (2008) utilized the C. elegans model to compare virulence of E. coli strains isolated from urine samples of individuals who consumed either cranberry or placebo capsules. E. coli strains from patients who took regular cranberry capsules displayed reduced adherence in vitro and delayed killing of C. elegans (Lavigne et al., 2008). Apart from this, the C. elegans model has also been used to screen for anti-virulence compounds from a collection of ∼250 marine sponge associated bacterial extracts for their ability to rescue the nematodes from a Vibrio alginolyticus infection (Durai et al., 2013). The active ingredients isolated from plants and seawater bacteria inhibited Vibrio cholerae biofilm production and reduced V. cholerae virulence in the killing of nematodes (Kim H.I. et al., 2015; Rajalaxmi et al., 2016). Natural compounds that affected the virulence of Gram-positive Clostridium difficile (Yun et al., 2015) and Listeria monocytogenes (Silva et al., 2015; Sivaranjani et al., 2016) have also been recently described.
Compounds Activating/Modulating Host Immune Responses
Despite lacking an adaptive immune system, C. elegans is still capable of activating protective mechanisms when confronted with pathogenic microorganisms in its natural habitat. The worms mount a complex and non-specific ancestral immune response, involving the activation of multiple signal transduction pathways and the secretion of immune effector molecules (Ermolaeva and Schumacher, 2014). This makes the identification of compounds targeting the host innate immune system possible using the C. elegans-based infection system. Following the initial high-throughput antimicrobial screen by Moy et al. (2009), the compound RPW-24 was shown to extend the lifespan of P. aeruginosa-infected worms without compromising bacterial integrity (Pukkila-Worley et al., 2012). By employing various approaches including microarray analysis using the readily available Affymetrix full-genome C. elegans GeneChip, as well as pathway elucidation studies using transgenic GFP reporter and mutant worms, the effect of the compound on host immunity was further characterized. RPW-24 was found to protect the worms from bacterial infection by inducing the p38 MAP kinase pathway-mediated response and the transcription factor ATF-7, suggesting the utility of the C. elegans model to discover novel strategies effective in treating bacterial infections (Pukkila-Worley et al., 2012). The extract from Swietenia macrophylla seeds promoted the survival of P. aeruginosa-challenged nematodes by boosting the expression of a C. elegans lysozyme encoding gene (lys-7) as observed by increasing fluorescence intensity in lys-7::GFP transgenic worms (Dharmalingam et al., 2012). Similarly, the nematodes subjected to P. aeruginosa infection were rescued by supplementation with dietary selenium. Selenite treatment did not interfere with bacterial quorum sensing and virulence, but elevated the transcript levels of host putative antimicrobial genes such as lys-1, spp-1, and abf-1. It was also demonstrated that the pathogen-resistant phenotype conferred by selenite requires the presence of a functional SKN-1 in C. elegans (Li et al., 2014).
From the C. elegans–S. aureus anti-infective screen discussed in Section “Compounds with In vivo Antimicrobial Activity,” our group identified an extract from a local plant, Orthosiphon stamineus, as a potential immunomodulatory drug that enhanced host tolerance toward a deadly S. aureus infection. This extract was not antimicrobial per se and is non-toxic to the C. elegans model. Expression of the host lys-7 antimicrobial gene in untreated worms was suppressed after 24 and 48 h exposure to S. aureus. This suppression is mediated by the pathogen’s ability to alter the host defense mechanism, particularly the host antimicrobial response. As noted through observations of a fluorescing transgenic strain infected by S. aureus, the down-regulation of lys-7 was restored in the presence of O. stamineus extract. Analysis using a loss-of-function C. elegans mutant established that the protective role of this extract is mediated via the conserved p38 MAPK and daf-2/daf-16 insulin-like signaling pathways. qRT-PCR analysis of the host PMK-1 and DAF-16-regulated antimicrobial genes confirmed a significant positive modulation of these genes in S. aureus-infected worms exposed to O. stamineus extract. Further studies also revealed eupatorin as the major bioactive ingredient that contributed to the immunomodulation effect of O. stamineus (Kong et al., 2014a). In a separate study, O. stamineus extract also modulated the cellular immune response in vitro (Alshawsh et al., 2012). Similarly, the natural polyphenols isolated from Magnolia plant species, honokiol and magnolol, promoted a cellular immune response and slowed down C. elegans killing by S. aureus (Choi et al., 2015).
Host-directed immunomodulators offer a number of potential advantages over antimicrobial drugs. Targeting the host immunity in combating pathogen infection is an appealing approach as the stimulation or modulation of host immune function may promote resistance against a diverse array of pathogens, forming the basis of broad spectrum therapeutics to treat infections (Nijnik, 2013). Additionally, it may lower the risk of selection toward treatment resistance. The water-soluble cranberry extract was reported to up-regulate the expression of C. elegans innate immune genes and confer protection against various major pathogens, namely V. cholerae, E. coli O157:H7, P. aeruginosa, Salmonella typhimurium, S. aureus, and E. faecalis (Dinh et al., 2014). Specifically, the protective effect of this extract against V. cholerae is dependent on the p38 MAPK signaling pathway. The wide spectrum activity of other immunomodulatory compounds has also been seen for the alkaloid compounds harmane and colistin. Harmane promoted survival in nematodes infected by both Gram positive and Gram negative pathogens, including E. coli strain EDL933, S. typhimurium, P. aeruginosa, and E. faecalis (Jakobsen et al., 2013). Cai et al. (2014) developed a C. elegans – based chemical screen for the discovery of immune-inductive drugs using the F35E12.5::gfp transgenic strain and they identified the antibiotic colistin as a potential hit. The nematodes pre-treated with colistin were also resistant against Yersinia pestis and P. aeruginosa infections (Cai et al., 2014).
Concomitantly, some anti-infectives may exhibit both anti-virulence and immunomodulatory effects. The water extract of red seaweed Chondrus crispus exerted dual anti-infective effects by simultaneously disabling the virulence of P. aeruginosa and elevating host immunity to combat pathogen infection. C. crispus down-regulated mRNA levels of quorum sensing and virulence-related genes and retarded the secretion of P. aeruginosa virulence factors such as protease, elastase, pyocyanin, siderophore, hydrogen cyanide, and biofilm (Liu et al., 2013). Furthermore, the protective effect of C. crispus extract was diminished in selected C. elegans mutants (pmk-1, daf-2, daf-16, or skn-1), implying that the effect from the host perspective is mediated via the highly conserved immune pathways. Likewise, concurrent virulence inhibitory and immunostimulatory effects have also been observed in other seaweed-related natural products, including Tasco®, a commercially available animal feed supplement made from the brown seaweed Ascophyllum nodosum (Kandasamy et al., 2012) and another red seaweed species Sarcodiotheca gaudichaudii (Kulshreshtha et al., 2016). Both, S. gaudichaudii and C. crispus extracts disarmed the pathogenicity of Salmonella enteritidis by reducing biofilm formation, swimming and swarming motility and down-regulating expression of virulence and quorum sensing genes in S. enteritidis. These extracts also reduced the intestinal bacterial load in C. elegans by promoting the expression of host immune response genes encoding lectin-family, saponin-like and antibacterial proteins (Kulshreshtha et al., 2016). Whilst no direct evidence of seaweed extract exerting an immunomodulatory effect on animal infection-directed immune responses, is available, Luo et al. (2015) demonstrated that seaweed polysaccharide was immunomodulatory in a cell-culture based assay as well as in a mouse tumor model. The seaweed polysaccharide enhanced the tumor immune response by increasing the number of tumor-infiltrating immune-related cells in the murine lymphoma model. The various experimental procedures used to scrutinize the mode of action of a compound targeting bacterial virulence and/or host immunity are outlined in Figure 2.
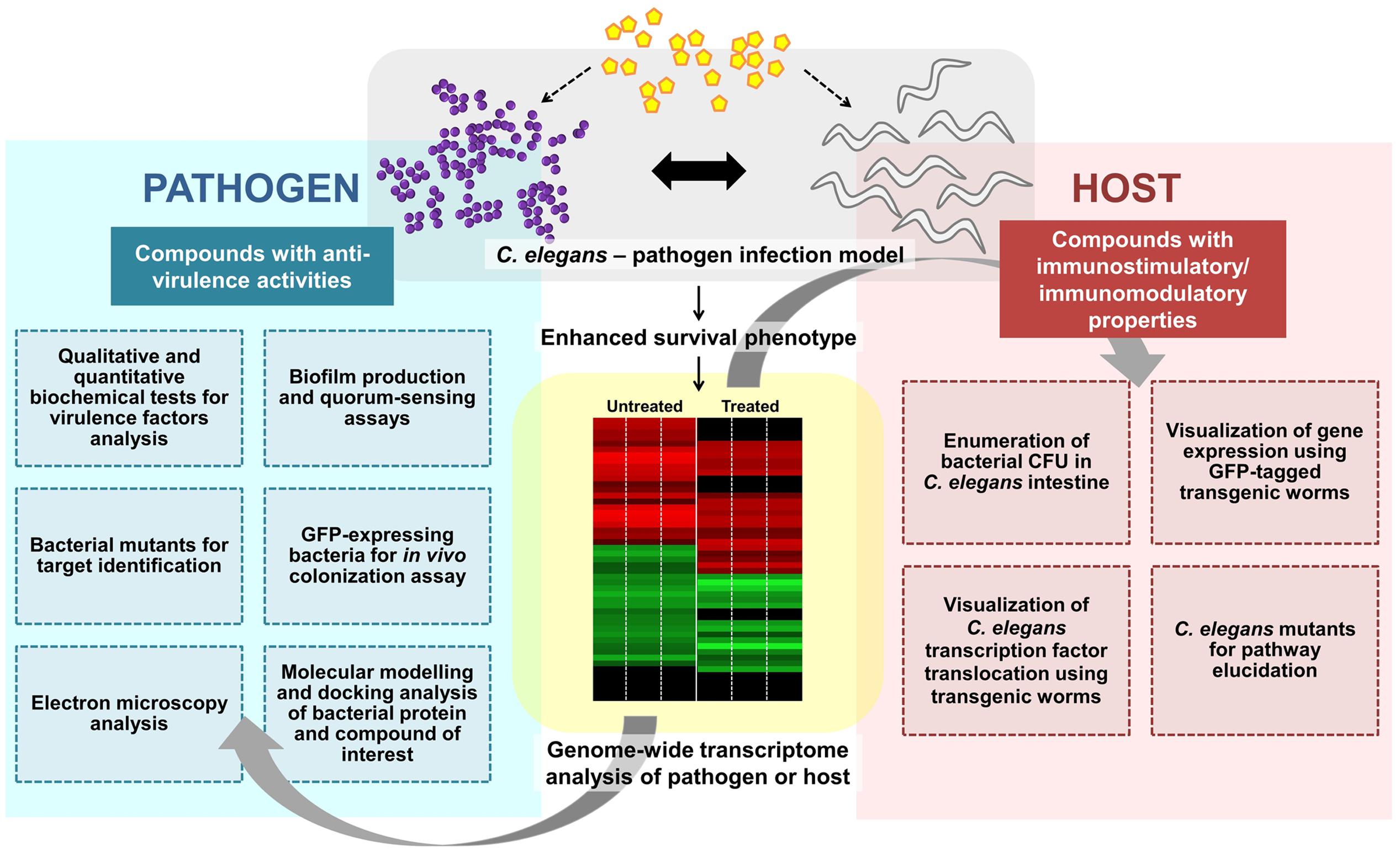
FIGURE 2. Overview of the general experimental approaches used to characterize an anti-infective candidate. A potential anti-infective compound that does not directly target bacterial growth can either impair pathogen virulence and/or enhance/modulate host immune responses. Various strategies from both pathogen and host perspectives can be employed to further characterize the molecular mechanism(s) of the compound of interest. Whole-genome transcriptome profiles of the compound-treated pathogen or host can be generated to study the effect of the compound. To assess the effect on bacterial virulence, qualitative and quantitative biochemical tests as well as biofilm production and quorum sensing assays can be performed. Bacterial mutants are useful for target identification while GFP-expressing bacteria may assist in visualization of in vivo bacterial colonization. Electron microscopy can be used to observe the formation of biofilm or bacterial structures (e.g., flagella). In silico molecular docking provides a clue of the possible binding interference between a bacterial protein and the compound of interest. From the host perspective, the live bacteria can be recovered from the C. elegans intestine and enumerated. The readily available transgenic mutant strains are useful for host target identification and pathway elucidation.
Although, therapeutics that involve the immune system can be broad spectrum against a wide variety of infecting agents, overstimulation or dysregulation can lead to potential harmful side effects such as uncontrolled inflammatory response that may cause tissue damage or even worse, immunosuppression that may result in the shutdown of the entire immune system. For example, the immunomodulatory drug RPW-24 generally induced a C. elegans detoxification pathway and triggered C. elegans aversion behavior under an uninfected condition. Concurrently, the basal lifespan of the uninfected population was also shortened and the development of C. elegans larva was halted upon exposure to this compound (Pukkila-Worley et al., 2012). Therefore, the mechanism of immunostimulatory molecules must be carefully investigated to maximize the beneficial effect and at the same time minimizing the possible toxic effects. Furthermore, the precise mode of drug administration (e.g., prophylactic or during infection) should also be determined to ensure effective treatment.
C. elegans: A Potential Repertoire of Antimicrobial Proteins and Peptides (APPs)
In C. elegans, many of the pathogens are ingested and survive the passage through the worm grinder to establish an infection in the intestinal lumen, including Serratia marcescens (Schulenburg and Ewbank, 2004), S. aureus (Sifri et al., 2003), and P. aeruginosa (Tan et al., 1999). Certain pathogens can stably colonize the intestinal tract and eventually overwhelm the worm, leading to death. Such infections have been demonstrated for S. typhimurium (Aballay et al., 2000) and E. faecalis (Garsin et al., 2001). Similar to the intestine, but to a lesser extent, the epidermis that covers the worm external body surface is also vulnerable to pathogen attack. Some bacteria like Microbacterium nematophilum or Leucobacter spp. Verde adhere to the cuticle, causing swelling of the hypodermal tissue (Hodgkin et al., 2000, 2013) whilst the fungus Drechmeria coniospora adheres to the region of the mouth and vulva, and penetrates through the epidermis (Jansson, 1994). Yang et al. (2005) also demonstrated that the nematophagous fungus Lecanicillium psalliotae secretes serine protease that facilitates mycelia penetration and degradation of worm cuticle (Yang et al., 2005).
With the aid of transcriptomics, it is now clear that infected worms are able to mount an inducible defense system, involving the activation of specific signaling pathways which lead to the release of immune molecules, including APPs (Couillault et al., 2004; O’Rourke et al., 2006; Shapira et al., 2006; Troemel et al., 2006; Zugasti and Ewbank, 2009; Gravato-Nobre et al., 2016). As opposed to conventional antibiotics, many APPs directly act on the membrane of the pathogen and thus, the development of microbial resistance by gene mutation is less likely (Peschel and Sahl, 2006). In some cases, APPs may possess immunomodulatory functions (Lai and Gallo, 2009; Nijnik and Hancock, 2009; Afacan et al., 2012), including (i) reduction in the levels of pro-inflammatory cytokines, (ii) modulation of the expression of chemokines, reactive oxygen species and reactive nitrogen cytokines, (iii) stimulation of angiogenesis, (iv) wound healing, and (v) leukocyte and macrophage differentiation. Up till now, a diverse array of putative APPs has been identified based on their induced expression upon infection or sequence similarities (Kato et al., 2002; Pujol et al., 2008), however, how these different APPs act cooperatively has not yet been characterized. For this review, we only focus on the representative families of APPs that have been documented to exhibit in vitro antimicrobial activity at the peptide/protein level. Figure 3 depicts the tissue-specific induction of APPs in C. elegans.
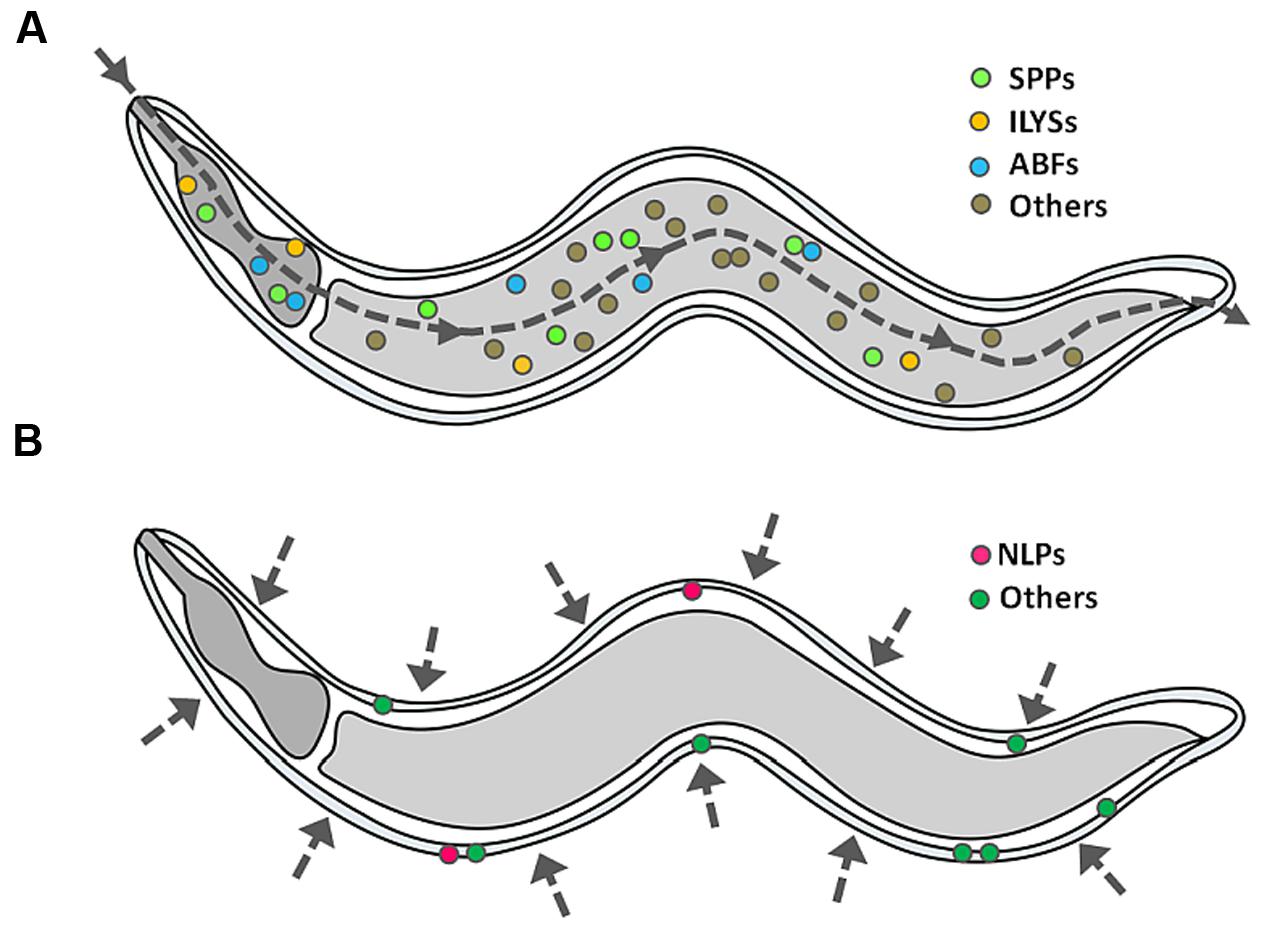
FIGURE 3. Induction of APPs in C. elegans. When C. elegans encounters a single pathogen, it will secrete a mixture of APPs as part of its protective mechanism, specifically (A) caenopores/saposin-like proteins (SPPs), antibacterial factor proteins (ABFs) and invertebrate lysozymes (ILYSs) in the digestive tract, and (B) neuropeptide-like proteins (NLPs) at the epidermis. The dashed line arrow denotes the route of infection.
Caenopores
Examination of the worm’s genome led to the discovery of saposin-like peptides/proteins (SPPs) characterized by an α-helix bundle stabilized by three unique disulphide bonds (Bruhn, 2005). Alternatively, they are named as caenopores owing to their structural and functional resemblance with amoebapores as they consist of a secretory signal peptide followed by saposin-like domain (Roeder et al., 2010). Given that most spp gene expression is regulated by the intestine specific transcription factor ELT-2, it is likely that caenopores are exclusively expressed in the intestine (Hoeckendorf and Leippe, 2012; Hoeckendorf et al., 2012). The direct role for caenopores in the innate immune response of C. elegans was first reported in Banyai and Patthy (1998) when the antibacterial effect of recombinant SPP-1 was observed on E. coli. Like amoebapores, SPP-5 was shown to display pore-forming activity and kill bacteria by permeabilising the cytoplasmic membrane (Roeder et al., 2010). A detailed examination of three-dimensional structure revealed that SPP-5 has five amphiphatic helices, connected by three disulfide bonds arranged like a folded leaf, which is the signature of the SAPLIP family (Mysliwy et al., 2010). It was worth noting that this was the first and only representative structure solved for C. elegans APPs. The molecular mechanism underlying pore formation however remains unclear.
Antibacterial Factor (ABF) Peptides/Proteins
A total of six ASABF (Ascaris suum antibacterial factor) homologs or best known as antibacterial factor (ABF-1 to ABF-6) peptides/proteins have been identified in C. elegans. ASABF is a cysteine-rich microbicidal factor that was first discovered in the body fluid of the nematode A. suum (Kato and Komatsu, 1996). It is possible that ABFs are also part of the C. elegans defense mechanism as exposure to pathogens led to the up-regulation of abf genes (Alegado et al., 2003; Alper et al., 2007; Means et al., 2009). By means of RNAi-mediated gene silencing, the functional significance of ABFs was further characterized. Silencing one of these genes, abf-2, led to increased bacterial load in the intestinal lumen (Alegado et al., 2003). It is well-established that expression of abf genes is localized primarily at the site of contact with bacteria, i.e., in the digestive tract (Alper et al., 2007). To date, only recombinant ABF-2 is known to display broad-spectrum antimicrobial activity, with the greatest effect observed toward Gram-positive bacteria (Kato et al., 2002). The precise role(s) of other members of this family are still unclear and required further analysis.
Invertebrate Lysozymes (ILYS)
Caenorhabditis elegans harbors protist-type (lys-1 to lys-10) and invertebrate type (ilys-1 to ilys-6) lysozyme encoding genes (Schulenburg and Boehnisch, 2008; Tarr, 2012). Representative members of the latter are up-regulated upon exposure to various pathogen insults (Irazoqui et al., 2010), suggesting possible roles involved in host defense. Studies using recombinant ILYS-3 have reported that the protein displays lytic activity against the Gram-positive Micrococcus luteus and M. nematophilum. It is known that invertebrate lysozymes are able to cleave the isopeptide bonds established between D-glutamate and L-Lysine present in peptidoglycans (Tarr, 2012; Van Herreweghe and Michiels, 2012). The importance of invertebrate lysozymes for host defense was recently confirmed when Gravato-Nobre et al. (2016) reported that disruption of ilys-3 renders the mutant worms more susceptible to M. nematophilum. The investigators also elegantly showed that ILYS-3 is primarily produced in the C. elegans digestive tract (Gravato-Nobre et al., 2016). For additional information of the protist-type lysozymes, readers are referred to previous reviews (Schulenburg and Boehnisch, 2008; Ewbank and Zugasti, 2011).
Neuropeptide-Like Peptides/Proteins (NLPs)
In C. elegans, most of the neuropeptide-like peptides/proteins (nlp) genes were named as such because of their limited sequence similarity with YGGXamide neuropeptide genes, sharing the YGGWG and YGGYG motifs (Nathoo et al., 2001). These genes form “the nlp-29 cluster” which comprises nlp-27 to nlp-31 and the adjacent gene nlp-34 on chromosome V (Couillault et al., 2004; Pujol et al., 2008). Upon exposure to the nematode-trapping fungus Monacrosporium haptotylum, several nlp genes were induced in infected worms (Fekete et al., 2008). Overexpression of these genes enhanced worm resistance to D. coniospora infection, further validating their contribution to in vivo host defense (Pujol et al., 2008). Couillault et al. (2004) demonstrated that the 53-amino-acid synthetic NLP-31 has potent antimicrobial activity against fungi and several bacteria. Utilizing transgenic worms expressing GFP under the control of the nlp-31 promoter, these investigators also demonstrated that this gene is exclusively expressed in the hypodermis. Recent work from our group has proposed an immunomodulatory role for NLP-31 as the peptide regulated the expression of inflammatory cytokines in B. pseudomallei-infected macrophage cells. Several lines of evidence suggest that it employs a mechanism that does not involve membrane permeabilisation, but instead interact with cytoplasmic macromolecules to interfere with the viability of B. pseudomallei (Lim et al., 2016).
Concluding Remarks
Active research on the development of anti-infective therapeutics that target bacterial virulence and/or host immune response using a C. elegans host model indicates a paradigm shift in the understanding of the host–pathogen interface. The existence of notable similarities at the molecular and cellular levels between nematode and higher vertebrates opens a path toward the discovery of novel therapeutics for human infections that might exhibit superior function over conventional antibiotics. The findings generated in the simple nematode model provide insights into the modulation of bacterial virulence and host immunity to fight infectious diseases and can readily be translated to higher organisms. Despite numerous efforts to tackle the spread of antimicrobial resistance, bacteria continue to show reduced susceptibility toward antibiotics over time and the rate of new drug discovery is declining rapidly. Developing alternative anti-infectives that do not affect bacterial cell viability hold great potential to compensate for this significant health care challenge. In addition, C. elegans APPs also provide a parallel opportunity to uncover host candidates for drug discovery.
Author Contribution
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
The research utilizing C. elegans as a model for the development of an anti-infective screening system was supported by the Ministry of Science, Technology and Innovation, Malaysia (grant numbers 06-05-16-MB003 and 02-05-20-SF11117) and Universiti Kebangsaan Malaysia (DIP-2015-022). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aballay, A., Yorgey, P., and Ausubel, F. M. (2000). Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr. Biol. 10, 1539–1542. doi: 10.1016/S0960-9822(00)00830-7
Adonizio, A., Kong, K. F., and Mathee, K. (2008a). Inhibition of quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa by South Florida plant extracts. Antimicrob. Agents Chemother. 52, 198–203. doi: 10.1128/AAC.00612-07
Adonizio, A., Leal, S. M. Jr., Ausubel, F. M., and Mathee, K. (2008b). Attenuation of Pseudomonas aeruginosa virulence by medicinal plants in a Caenorhabditis elegans model system. J. Med. Microbiol. 57, 809–813. doi: 10.1099/jmm.0.47802-0
Afacan, N. J., Yeung, A. T., Pena, O. M., and Hancock, R. E. (2012). Therapeutic potential of host defense peptides in antibiotic-resistant infections. Curr. Pharm. Des. 18, 807–819. doi: 10.2174/138161212799277617
Alegado, R. A., Campbell, M. C., Chen, W. C., Slutz, S. S., and Tan, M. W. (2003). Characterization of mediators of microbial virulence and innate immunity using the Caenorhabditis elegans host-pathogen model. Cell. Microbiol. 5, 435–444. doi: 10.1046/j.1462-5822.2003.00287.x
Allen, R. C., Popat, R., Diggle, S. P., and Brown, S. P. (2014). Targeting virulence: can we make evolution-proof drugs? Nat. Rev. Microbiol. 12, 300–308. doi: 10.1038/nrmicro3232
Alper, S., Mcbride, S. J., Lackford, B., Freedman, J. H., and Schwartz, D. A. (2007). Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol. Cell. Biol. 27, 5544–5553. doi: 10.1128/MCB.02070-06
Alshawsh, M. A., Abdulla, M. A., Ismail, S., Amin, Z. A., Qader, S. W., Hadi, H. A., et al. (2012). Free radical scavenging, antimicrobial and immunomodulatory activities of Orthosiphon stamineus. Molecules 17, 5385–5395. doi: 10.3390/molecules17055385
Antoshechkin, I., and Sternberg, P. W. (2007). The versatile worm: genetic and genomic resources for Caenorhabditis elegans research. Nat. Rev. Genet. 8, 518–532. doi: 10.1038/nrg2105
Arvanitis, M., Glavis-Bloom, J., and Mylonakis, E. (2013). C. elegans for anti-infective discovery. Curr. Opin. Pharm. 13, 769–774. doi: 10.1016/j.coph.2013.08.002
Bakkiyaraj, D., and Pandian, S. K. (2010). In vitro and in vivo antibiofilm activity of a coral associated actinomycete against drug resistant Staphylococcus aureus biofilms. Biofouling 26, 711–717. doi: 10.1080/08927014.2010.511200
Banyai, L., and Patthy, L. (1998). Amoebapore homologs of Caenorhabditis elegans. Biochim. Biophys. Acta 1429, 259–264. doi: 10.1016/S0167-4838(98)00237-4
Bjarnsholt, T., Jensen, P. O., Rasmussen, T. B., Christophersen, L., Calum, H., Hentzer, M., et al. (2005). Garlic blocks quorum sensing and promotes rapid clearing of pulmonary Pseudomonas aeruginosa infections. Microbiology 151, 3873–3880. doi: 10.1099/mic.0.27955-0
Boucher, H. W., Talbot, G. H., Benjamin, D. K. Jr., Bradley, J., Guidos, R. J., Jones, R. N., et al. (2013). 10 x ’20 Progress-development of new drugs active against gram-negative bacilli: an update from the Infectious Diseases Society of America. Clin. Infect. Dis. 56, 1685–1694. doi: 10.1093/cid/cit152
Brackman, G., Breyne, K., De Rycke, R., Vermote, A., Van Nieuwerburgh, F., Meyer, E., et al. (2016). The quorum sensing inhibitor hamamelitannin increases antibiotic susceptibility of Staphylococcus aureus biofilms by affecting peptidoglycan biosynthesis and eDNA release. Sci. Rep. 6:20321. doi: 10.1038/srep20321
Briers, Y., Walmagh, M., Van Puyenbroeck, V., Cornelissen, A., Cenens, W., Aertsen, A., et al. (2014). Engineered endolysin-based “Artilysins” to combat multidrug-resistant gram-negative pathogens. MBio 5:e01379-14. doi: 10.1128/mBio.01379-14
Brown, E. D., and Wright, G. D. (2016). Antibacterial drug discovery in the resistance era. Nature 529, 336–343. doi: 10.1038/nature17042
Bruhn, H. (2005). A short guided tour through functional and structural features of saposin-like proteins. Biochem. J. 389, 249–257. doi: 10.1042/BJ20050051
Cai, Y., Cao, X., and Aballay, A. (2014). Whole-animal chemical screen identifies colistin as a new immunomodulator that targets conserved pathways. MBio 5:e01235-14. doi: 10.1128/mBio.01235-14
Cho, H. S., Lee, J. H., Cho, M. H., and Lee, J. (2015). Red wines and flavonoids diminish Staphylococcus aureus virulence with anti-biofilm and anti-hemolytic activities. Biofouling 31, 1–11. doi: 10.1080/08927014.2014.991319
Choi, E. J., Kim, H. I., Kim, J. A., Jun, S. Y., Kang, S. H., Park, D. J., et al. (2015). The herbal-derived honokiol and magnolol enhances immune response to infection with methicillin-sensitive Staphylococcus aureus (MSSA) and methicillin-resistant S. aureus (MRSA). Appl. Microbiol. Biotechnol. 99, 4387–4396. doi: 10.1007/s00253-015-6382-y
Cohen, L. B., and Troemel, E. R. (2015). Microbial pathogenesis and host defense in the nematode C. elegans. Curr. Opin. Microbiol. 23, 94–101. doi: 10.1016/j.mib.2014.11.009
Conery, A. L., Larkins-Ford, J., Ausubel, F. M., and Kirienko, N. V. (2014). High-throughput screening for novel anti-infectives using a C. elegans pathogenesis model. Curr. Protoc. Chem. Biol. 6, 25–37. doi: 10.1002/9780470559277.ch130160
Couillault, C., Pujol, N., Reboul, J., Sabatier, L., Guichou, J. F., Kohara, Y., et al. (2004). TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 5, 488–494. doi: 10.1038/ni1060
Desalermos, A., Muhammed, M., Glavis-Bloom, J., and Mylonakis, E. (2011). Using C. elegans for antimicrobial drug discovery. Exp. Opin. Drug Discov. 6, 645–652. doi: 10.1517/17460441.2011.573781
Dharmalingam, K., Tan, B. K., Mahmud, M. Z., Sedek, S. A., Majid, M. I., Kuah, M. K., et al. (2012). Swietenia macrophylla extract promotes the ability of Caenorhabditis elegans to survive Pseudomonas aeruginosa infection. J. Ethnopharmacol. 139, 657–663. doi: 10.1016/j.jep.2011.12.016
Dinh, J., Angeloni, J. T., Pederson, D. B., Wang, X., Cao, M., and Dong, Y. (2014). Cranberry extract standardized for proanthocyanidins promotes the immune response of Caenorhabditis elegans to Vibrio cholerae through the p38 MAPK pathway and HSF-1. PLoS ONE 9:e103290. doi: 10.1371/journal.pone.0103290
Dolla, N. K., Chen, C., Larkins-Ford, J., Rajamuthiah, R., Jagadeesan, S., Conery, A. L., et al. (2015). On the mechanism of Berberine-INF55 (5-Nitro-2-phenylindole) hybrid qntibacterials. Aust. J. Chem. 67, 1471–1480. doi: 10.1071/CH14426
Durai, S., Vigneshwari, L., and Balamurugan, K. (2013). Caenorhabditis elegans-based in vivo screening of bioactives from marine sponge-associated bacteria against Vibrio alginolyticus. J. Appl. Microbiol. 115, 1329–1342. doi: 10.1111/jam.12335
Eng, S. A., and Nathan, S. (2015). Curcumin rescues Caenorhabditis elegans from a Burkholderia pseudomallei infection. Front. Microbiol. 6:290. doi: 10.3389/fmicb.2015.00290
Ermolaeva, M. A., and Schumacher, B. (2014). Insights from the worm: the C. elegans model for innate immunity. Semin. Immunol. 26, 303–309. doi: 10.1016/j.smim.2014.04.005
Ewbank, J. J., and Zugasti, O. (2011). C. elegans: model host and tool for antimicrobial drug discovery. Dis. Model. Mech. 4, 300–304. doi: 10.1242/dmm.006684
Fekete, C., Tholander, M., Rajashekar, B., Ahren, D., Friman, E., Johansson, T., et al. (2008). Paralysis of nematodes: shifts in the transcriptome of the nematode-trapping fungus Monacrosporium haptotylum during infection of Caenorhabditis elegans. Environ. Microbiol. 10, 364–375. doi: 10.1111/j.1462-2920.2007.01457.x
Fuchs, B. B., Rajamuthiah, R., Souza, A. C., Eatemadpour, S., Rossoni, R. D., Santos, D. A., et al. (2016). Inhibition of bacterial and fungal pathogens by the orphaned drug auranofin. Future Med. Chem. 8, 117–132. doi: 10.4155/fmc.15.182
Garsin, D. A., Sifri, C. D., Mylonakis, E., Qin, X., Singh, K. V., Murray, B. E., et al. (2001). A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. U.S.A. 98, 10892–10897. doi: 10.1073/pnas.191378698
Garsin, D. A., Villanueva, J. M., Begun, J., Kim, D. H., Sifri, C. D., Calderwood, S. B., et al. (2003). Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 300:1921. doi: 10.1126/science.1080147
Gravato-Nobre, M. J., Vaz, F., Filipe, S., Chalmers, R., and Hodgkin, J. (2016). The invertebrate lysozyme effector ILYS-3 is systemically activated in response to danger signals and confers antimicrobial protection in C. elegans. PLoS Pathog. 12:e1005826. doi: 10.1371/journal.ppat.1005826
Harjai, K., Kumar, R., and Singh, S. (2010). Garlic blocks quorum sensing and attenuates the virulence of Pseudomonas aeruginosa. FEMS Immunol. Med. Microbiol. 58, 161–168. doi: 10.1111/j.1574-695X.2009.00614.x
Ho Sui, S. J., Lo, R., Fernandes, A. R., Caulfield, M. D., Lerman, J. A., Xie, L., et al. (2012). Raloxifene attenuates Pseudomonas aeruginosa pyocyanin production and virulence. Int. J. Antimicrob. Agents 40, 246–251. doi: 10.1016/j.ijantimicag.2012.05.009
Hodgkin, J., Felix, M. A., Clark, L. C., Stroud, D., and Gravato-Nobre, M. J. (2013). Two leucobacter strains exert complementary virulence on Caenorhabditis including death by worm-star formation. Curr. Biol. 23, 2157–2161. doi: 10.1016/j.cub.2013.08.060
Hodgkin, J., Kuwabara, P. E., and Corneliussen, B. (2000). A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr. Biol. 10, 1615–1618. doi: 10.1016/S0960-9822(00)00867-8
Hoeckendorf, A., and Leippe, M. (2012). SPP-3, a saposin-like protein of Caenorhabditis elegans, displays antimicrobial and pore-forming activity and is located in the intestine and in one head neuron. Dev. Comp. Immunol. 38, 181–186. doi: 10.1016/j.dci.2012.05.007
Hoeckendorf, A., Stanisak, M., and Leippe, M. (2012). The saposin-like protein SPP-12 is an antimicrobial polypeptide in the pharyngeal neurons of Caenorhabditis elegans and participates in defence against a natural bacterial pathogen. Biochem. J. 445, 205–212. doi: 10.1042/BJ20112102
Hom, K., Heinzl, G. A., Eakanunkul, S., Lopes, P. E., Xue, F., Mackerell, A. D. Jr., et al. (2013). Small molecule antivirulents targeting the iron-regulated heme oxygenase (HemO) of P. aeruginosa. J. Med. Chem. 56, 2097–2109. doi: 10.1021/jm301819k
Husain, F. M., Ahmad, I., Asif, M., and Tahseen, Q. (2013). Influence of clove oil on certain quorum-sensing-regulated functions and biofilm of Pseudomonas aeruginosa and Aeromonas hydrophila. J. Biosci. (Bangalore) 38, 835–844. doi: 10.1007/s12038-013-9385-9
Husain, F. M., Ahmad, I., Khan, M. S., Ahmad, E., Tahseen, Q., and Alshabib, N. A. (2015a). Sub-MICs of Mentha piperita essential oil and menthol inhibits AHL mediated quorum sensing and biofilm of Gram-negative bacteria. Front. Microbiol. 6:420. doi: 10.3389/fmicb.2015.00420
Husain, F. M., Ahmad, I., Khan, M. S., and Al-Shabib, N. A. (2015b). Trigonella foenum-graceum (Seed) extract interferes with quorum sensing regulated traits and biofilm formation in the strains of Pseudomonas aeruginosa and Aeromonas hydrophila. Evid. Based Complement. Alternat. Med. 2015:879540. doi: 10.1155/2015/879540
Imperi, F., Massai, F., Facchini, M., Frangipani, E., Visaggio, D., Leoni, L., et al. (2013). Repurposing the antimycotic drug flucytosine for suppression of Pseudomonas aeruginosa pathogenicity. Proc. Natl. Acad. Sci. U.S.A. 110, 7458–7463. doi: 10.1073/pnas.1222706110
Irazoqui, J. E., Troemel, E. R., Feinbaum, R. L., Luhachack, L. G., Cezairliyan, B. O., and Ausubel, F. M. (2010). Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 6:e1000982. doi: 10.1371/journal.ppat.1000982
Jakobsen, H., Bojer, M. S., Marinus, M. G., Xu, T., Struve, C., Krogfelt, K. A., et al. (2013). The alkaloid compound harmane increases the lifespan of Caenorhabditis elegans during bacterial infection, by modulating the nematode’s innate immune response. PLoS ONE 8:e60519. doi: 10.1371/journal.pone.0060519
Jansson, H. B. (1994). Adhesion of conidia of Drechmeria coniospora to Caenorhabditis elegans wild type and mutants. J. Nematol. 26, 430–435.
Jayamani, E., Rajamuthiah, R., Larkins-Ford, J., Fuchs, B. B., Conery, A. L., Vilcinskas, A., et al. (2015). Insect-derived cecropins display activity against Acinetobacter baumannii in a whole-animal high-throughput Caenorhabditis elegans model. Antimicrob. Agents Chemother. 59, 1728–1737. doi: 10.1128/AAC.04198-14
Junker, L. M., and Clardy, J. (2007). High-throughput screens for small-molecule inhibitors of Pseudomonas aeruginosa biofilm development. Antimicrob. Agents Chemother. 51, 3582–3590. doi: 10.1128/AAC.00506-07
Kamath, R. S., and Ahringer, J. (2003). Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313–321. doi: 10.1016/S1046-2023(03)00050-1
Kandasamy, S., Khan, W., Evans, F., Critchley, A. T., and Prithiviraj, B. (2012). Tasco(R): a product of Ascophyllum nodosum enhances immune response of Caenorhabditis elegans against Pseudomonas aeruginosa infection. Mar. Drugs 10, 84–105. doi: 10.3390/md10010084
Kato, Y., Aizawa, T., Hoshino, H., Kawano, K., Nitta, K., and Zhang, H. (2002). abf-1 and abf-2, ASABF-type antimicrobial peptide genes in Caenorhabditis elegans. Biochem. J. 361, 221–230. doi: 10.1042/bj3610221
Kato, Y., and Komatsu, S. (1996). ASABF, a novel cysteine-rich antibacterial peptide isolated from the nematode Ascaris suum. Purification, primary structure, and molecular cloning of cDNA. J. Biol. Chem. 271, 30493–30498. doi: 10.1074/jbc.271.48.30493
Kim, H. I., Kim, J. A., Choi, E. J., Harris, J. B., Jeong, S. Y., Son, S. J., et al. (2015). In vitro and in vivo antimicrobial efficacy of natural plant-derived compounds against Vibrio cholerae of O1 El Tor Inaba serotype. Biosci. Biotechnol. Biochem. 79, 475–483. doi: 10.1080/09168451.2014.991685
Kim, W., Conery, A. L., Rajamuthiah, R., Fuchs, B. B., Ausubel, F. M., and Mylonakis, E. (2015). Identification of an antimicrobial agent effective against methicillin-resistant Staphylococcus aureus persisters using a fluorescence-based screening strategy. PLoS ONE 10:e0127640. doi: 10.1371/journal.pone.0127640
Kirienko, D. R., Revtovich, A. V., and Kirienko, N. V. (2016). A high-content, phenotypic screen identifies fluorouridine as an inhibitor of pyoverdine biosynthesis and Pseudomonas aeruginosa virulence. mSphere 1, e217-16. doi: 10.1128/mSphere.00217-16
Kirienko, N. V., Kirienko, D. R., Larkins-Ford, J., Wahlby, C., Ruvkun, G., and Ausubel, F. M. (2013). Pseudomonas aeruginosa disrupts Caenorhabditis elegans iron homeostasis, causing a hypoxic response and death. Cell Host Microbe 13, 406–416. doi: 10.1016/j.chom.2013.03.003
Kong, C., Tan, M. W., and Nathan, S. (2014a). Orthosiphon stamineus protects Caenorhabditis elegans against Staphylococcus aureus infection through immunomodulation. Biol Open 3, 644–655. doi: 10.1242/bio.20148334
Kong, C., Yehye, W. A., Abd Rahman, N., Tan, M. W., and Nathan, S. (2014b). Discovery of potential anti-infectives against Staphylococcus aureus using a Caenorhabditis elegans infection model. BMC Complement. Altern. Med. 14:4. doi: 10.1186/1472-6882-14-4
Kulshreshtha, G., Borza, T., Rathgeber, B., Stratton, G. S., Thomas, N. A., Critchley, A., et al. (2016). Red seaweeds Sarcodiotheca gaudichaudii and Chondrus crispus down-regulate virulence factors of Salmonella enteritidis and induce immune responses in Caenorhabditis elegans. Front. Microbiol. 7:421. doi: 10.3389/fmicb.2016.00421
Kumarasamy, K. K., Toleman, M. A., Walsh, T. R., Bagaria, J., Butt, F., Balakrishnan, R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 597–602. doi: 10.1016/S1473-3099(10)70143-2
Lai, Y., and Gallo, R. L. (2009). AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 30, 131–141. doi: 10.1016/j.it.2008.12.003
Lakshmanan, U., Yap, A., Fulwood, J., Yichun, L., Hoon, S. S., Lim, J., et al. (2014). Establishment of a novel whole animal HTS technology platform for melioidosis drug discovery. Comb. Chem. High Throughput Screen 17, 790–803. doi: 10.2174/1386207317666141019195031
Lavigne, J. P., Bourg, G., Combescure, C., Botto, H., and Sotto, A. (2008). In-vitro and in-vivo evidence of dose-dependent decrease of uropathogenic Escherichia coli virulence after consumption of commercial Vaccinium macrocarpon (cranberry) capsules. Clin. Microbiol. Infect. 14, 350–355. doi: 10.1111/j.1469-0691.2007.01917.x
Lee, J. H., Kim, Y. G., Cho, H. S., Ryu, S. Y., Cho, M. H., and Lee, J. (2014). Coumarins reduce biofilm formation and the virulence of Escherichia coli O157:H7. Phytomedicine 21, 1037–1042. doi: 10.1016/j.phymed.2014.04.008
Lee, K., Lee, J. H., Kim, S. I., Cho, M. H., and Lee, J. (2014a). Anti-biofilm, anti-hemolysis, and anti-virulence activities of black pepper, cananga, myrrh oils, and nerolidol against Staphylococcus aureus. Appl. Microbiol. Biotechnol. 98, 9447–9457. doi: 10.1007/s00253-014-5903-4
Lee, K., Lee, J. H., Ryu, S. Y., Cho, M. H., and Lee, J. (2014b). Stilbenes reduce Staphylococcus aureus hemolysis, biofilm formation, and virulence. Foodborne Pathog. Dis. 11, 710–717. doi: 10.1089/fpd.2014.1758
Lee, K. M., Lim, J., Nam, S., Yoon, M. Y., Kwon, Y. K., Jung, B. Y., et al. (2011). Inhibitory effects of broccoli extract on Escherichia coli O157:H7 quorum sensing and in vivo virulence. FEMS Microbiol. Lett. 321, 67–74. doi: 10.1111/j.1574-6968.2011.02311.x
Li, W. H., Chang, C. H., Huang, C. W., Wei, C. C., and Liao, V. H. (2014). Selenite enhances immune response against Pseudomonas aeruginosa PA14 via SKN-1 in Caenorhabditis elegans. PLoS ONE 9:e105810. doi: 10.1371/journal.pone.0105810
Lim, M.-P., Firdaus-Raih, M., and Nathan, S. (2016). Nematode peptides with host-directed anti-inflammatory activity rescue Caenorhabditis elegans from a Burkholderia pseudomallei infection. Front. Microbiol. 7:1436. doi: 10.3389/fmicb.2016.01436
Liu, J., Hafting, J., Critchley, A. T., Banskota, A. H., and Prithiviraj, B. (2013). Components of the cultivated red seaweed Chondrus crispus enhance the immune response of Caenorhabditis elegans to Pseudomonas aeruginosa through the pmk-1, daf-2/daf-16, and skn-1 pathways. Appl. Environ. Microbiol. 79, 7343–7350. doi: 10.1128/AEM.01927-13
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Luo, M., Shao, B., Nie, W., Wei, X. W., Li, Y. L., Wang, B. L., et al. (2015). Antitumor and adjuvant activity of lambda-carrageenan by stimulating immune response in cancer immunotherapy. Sci. Rep. 5:11062. doi: 10.1038/srep11062
Mallo, G. V., Kurz, C. L., Couillault, C., Pujol, N., Granjeaud, S., Kohara, Y., et al. (2002). Inducible antibacterial defense system in C. elegans. Curr. Biol. 12, 1209–1214. doi: 10.1016/S0960-9822(02)00928-4
Marsh, E. K., and May, R. C. (2012). Caenorhabditis elegans, a model organism for investigating immunity. Appl. Environ. Microbiol. 78, 2075–2081. doi: 10.1128/AEM.07486-11
McGann, P., Snesrud, E., Maybank, R., Corey, B., Ong, A. C., Clifford, R., et al. (2016). Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob. Agents Chemother. 60, 4420–4421. doi: 10.1128/AAC.01103-16
Means, T. K., Mylonakis, E., Tampakakis, E., Colvin, R. A., Seung, E., Puckett, L., et al. (2009). Evolutionarily conserved recognition and innate immunity to fungal pathogens by the scavenger receptors SCARF1 and CD36. J. Exp. Med. 206, 637–653. doi: 10.1084/jem.20082109
Moy, T. I., Ball, A. R., Anklesaria, Z., Casadei, G., Lewis, K., and Ausubel, F. M. (2006). Identification of novel antimicrobials using a live-animal infection model. Proc. Natl. Acad. Sci. U.S.A. 103, 10414–10419. doi: 10.1073/pnas.0604055103
Moy, T. I., Conery, A. L., Larkins-Ford, J., Wu, G., Mazitschek, R., Casadei, G., et al. (2009). High-throughput screen for novel antimicrobials using a whole animal infection model. ACS Chem. Biol. 4, 527–533. doi: 10.1021/cb900084v
Musthafa, K. S., Balamurugan, K., Pandian, S. K., and Ravi, A. V. (2012a). 2,5-Piperazinedione inhibits quorum sensing-dependent factor production in Pseudomonas aeruginosa PAO1. J. Basic Microbiol. 52, 679–686. doi: 10.1002/jobm.201100292
Musthafa, K. S., Sivamaruthi, B. S., Pandian, S. K., and Ravi, A. V. (2012b). Quorum sensing inhibition in Pseudomonas aeruginosa PAO1 by antagonistic compound phenylacetic acid. Curr. Microbiol. 65, 475–480. doi: 10.1007/s00284-012-0181-9
Mysliwy, J., Dingley, A. J., Stanisak, M., Jung, S., Lorenzen, I., Roeder, T., et al. (2010). Caenopore-5: the three-dimensional structure of an antimicrobial protein from Caenorhabditis elegans. Dev. Comp. Immunol. 34, 323–330. doi: 10.1016/j.dci.2009.11.003
Nathoo, A. N., Moeller, R. A., Westlund, B. A., and Hart, A. C. (2001). Identification of neuropeptide-like protein gene families in Caenorhabditis elegans and other species. Proc. Natl. Acad. Sci. U.S.A. 98, 14000–14005. doi: 10.1073/pnas.241231298
Nijnik, A. (2013). Immunomodulatory approaches for prevention and treatment of infectious diseases. Curr. Opin. Microbiol. 16, 590–595. doi: 10.1016/j.mib.2013.06.011
Nijnik, A., and Hancock, R. (2009). Host defence peptides: antimicrobial and immunomodulatory activity and potential applications for tackling antibiotic-resistant infections. Emerg. Health Threats J. 2, e1. doi: 10.3134/ehtj.09.001
O’Loughlin, C. T., Miller, L. C., Siryaporn, A., Drescher, K., Semmelhack, M. F., and Bassler, B. L. (2013). A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 110, 17981–17986. doi: 10.1073/pnas.1316981110
O’Reilly, L. P., Luke, C. J., Perlmutter, D. H., Silverman, G. A., and Pak, S. C. (2014). C. elegans in high-throughput drug discovery. Adv. Drug Del. Rev. 6, 247–253. doi: 10.1016/j.addr.2013.12.001
O’Rourke, D., Baban, D., Demidova, M., Mott, R., and Hodgkin, J. (2006). Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 16, 1005–1016. doi: 10.1101/gr.50823006
Pendleton, J. N., Gorman, S. P., and Gilmore, B. F. (2013). Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti Infect. Ther. 11, 297–308. doi: 10.1586/eri.13.12
Peschel, A., and Sahl, H. G. (2006). The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 4, 529–536. doi: 10.1038/nrmicro1441
Prithiviraj, B., Bais, H. P., Weir, T., Suresh, B., Najarro, E. H., Dayakar, B. V., et al. (2005). Down regulation of virulence factors of Pseudomonas aeruginosa by salicylic acid attenuates its virulence on Arabidopsis thaliana and Caenorhabditis elegans. Infect. Immun. 73, 5319–5328. doi: 10.1128/IAI.73.9.5319-5328.2005
Pujol, N., Zugasti, O., Wong, D., Couillault, C., Kurz, C. L., Schulenburg, H., et al. (2008). Anti-fungal innate immunity in C. elegans is enhanced by evolutionary diversification of antimicrobial peptides. PLoS Pathog. 4:e1000105. doi: 10.1371/journal.ppat.1000105
Pukkila-Worley, R., Feinbaum, R., Kirienko, N. V., Larkins-Ford, J., Conery, A. L., and Ausubel, F. M. (2012). Stimulation of host immune defenses by a small molecule protects C. elegans from bacterial infection. PLoS Genet. 8:e1002733. doi: 10.1371/journal.pgen.1002733
Rajalaxmi, M., Beema Shafreen, R., Iyer, P. M., Sahaya Vino, R., Balamurugan, K., and Pandian, S. K. (2016). An in silico, in vitro and in vivo investigation of indole-3-carboxaldehyde identified from the seawater bacterium Marinomonas sp. as an anti-biofilm agent against Vibrio cholerae O1. Biofouling 32, 1–12. doi: 10.1080/08927014.2016.1154545
Rajamuthiah, R., Fuchs, B. B., Jayamani, E., Kim, Y., Larkins-Ford, J., Conery, A., et al. (2014). Whole animal automated platform for drug discovery against multi-drug resistant Staphylococcus aureus. PLoS ONE 9:e89189. doi: 10.1371/journal.pone.0089189
Rajamuthiah, R., Jayamani, E., Majed, H., Conery, A. L., Kim, W., Kwon, B., et al. (2015). Antibacterial properties of 3-(phenylsulfonyl)-2-pyrazinecarbonitrile. Bioorg. Med. Chem. Lett. 25, 5203–5207. doi: 10.1016/j.bmcl.2015.09.066
Rasamiravaka, T., Vandeputte, O. M., Pottier, L., Huet, J., Rabemanantsoa, C., Kiendrebeogo, M., et al. (2015). Pseudomonas aeruginosa biofilm formation and persistence, along with the production of quorum sensing-dependent virulence factors, are disrupted by a triterpenoid coumarate ester isolated from Dalbergia trichocarpa, a tropical legume. PLoS ONE 10:e0132791. doi: 10.1371/journal.pone.0132791
Rasmussen, T. B., Bjarnsholt, T., Skindersoe, M. E., Hentzer, M., Kristoffersen, P., Kote, M., et al. (2005). Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol. 187, 1799–1814. doi: 10.1128/JB.187.5.1799-1814.2005
Roeder, T., Stanisak, M., Gelhaus, C., Bruchhaus, I., Grotzinger, J., and Leippe, M. (2010). Caenopores are antimicrobial peptides in the nematode Caenorhabditis elegans instrumental in nutrition and immunity. Dev. Comp. Immunol. 34, 203–209. doi: 10.1016/j.dci.2009.09.010
Rual, J. F., Ceron, J., Koreth, J., Hao, T., Nicot, A. S., Hirozane-Kishikawa, T., et al. (2004). Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14, 2162–2168. doi: 10.1101/gr.2505604
Rudrappa, T., and Bais, H. P. (2008). Curcumin, a known phenolic from Curcuma longa, attenuates the virulence of Pseudomonas aeruginosa PAO1 in whole plant and animal pathogenicity models. J. Agric. Food Chem. 56, 1955–1962. doi: 10.1021/jf072591j
Sarabhai, S., Sharma, P., and Capalash, N. (2013). Ellagic acid derivatives from Terminalia chebula Retz. downregulate the expression of quorum sensing genes to attenuate Pseudomonas aeruginosa PAO1 virulence. PLoS ONE 8:e53441. doi: 10.1371/journal.pone.0053441
Schulenburg, H., and Boehnisch, C. (2008). Diversification and adaptive sequence evolution of Caenorhabditis lysozymes (Nematoda: Rhabditidae). BMC Evol. Biol. 8:114. doi: 10.1186/1471-2148-8-114
Schulenburg, H., and Ewbank, J. J. (2004). Diversity and specificity in the interaction between Caenorhabditis elegans and the pathogen Serratia marcescens. BMC Evol. Biol. 4:49. doi: 10.1186/1471-2148-4-49
Selin, C., Stietz, M. S., Blanchard, J. E., Gehrke, S. S., Bernard, S., Hall, D. G., et al. (2015). A pipeline for screening small molecules with growth inhibitory activity against Burkholderia cenocepacia. PLoS ONE 10:e0128587. doi: 10.1371/journal.pone.0128587
Shapira, M., Hamlin, B. J., Rong, J., Chen, K., Ronen, M., and Tan, M. W. (2006). A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc. Natl. Acad. Sci. U.S.A. 103, 14086–14091. doi: 10.1073/pnas.0603424103
Sifri, C. D., Begun, J., Ausubel, F. M., and Calderwood, S. B. (2003). Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect. Immun. 71, 2208–2217. doi: 10.1128/IAI.71.4.2208-2217.2003
Silva, A., Genoves, S., Martorell, P., Zanini, S. F., Rodrigo, D., and Martinez, A. (2015). Sublethal injury and virulence changes in Listeria monocytogenes and Listeria innocua treated with antimicrobials carvacrol and citral. Food Microbiol. 50, 5–11. doi: 10.1016/j.fm.2015.02.016
Sivaranjani, M., Gowrishankar, S., Kamaladevi, A., Pandian, S. K., Balamurugan, K., and Ravi, A. V. (2016). Morin inhibits biofilm production and reduces the virulence of Listeria monocytogenes - an in vitro and in vivo approach. Int. J. Food Microbiol. 237, 73–82. doi: 10.1016/j.ijfoodmicro.2016.08.021
Stokes, J. M., Selin, C., Cardona, S. T., and Brown, E. D. (2015). Chemical inhibition of bacterial ribosome biogenesis shows efficacy in a worm infection model. Antimicrob. Agents Chemother. 59, 2918–2920. doi: 10.1128/AAC.04690-14
Tan, M. W., Rahme, L. G., Sternberg, J. A., Tompkins, R. G., and Ausubel, F. M. (1999). Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. U.S.A. 96, 2408–2413. doi: 10.1073/pnas.96.5.2408
Tarr, D. E. (2012). Distribution and characteristics of ABFs, cecropins, nemapores, and lysozymes in nematodes. Dev. Comp. Immunol. 36, 502–520. doi: 10.1016/j.dci.2011.09.007
Thangamani, S., Younis, W., and Seleem, M. N. (2015). Repurposing celecoxib as a topical antimicrobial agent. Front. Microbiol. 6:750. doi: 10.3389/fmicb.2015.00750
Tomkiewicz, D., Casadei, G., Larkins-Ford, J., Moy, T. I., Garner, J., Bremner, J. B., et al. (2010). Berberine-INF55 (5-nitro-2-phenylindole) hybrid antimicrobials: effects of varying the relative orientation of the berberine and INF55 components. Antimicrob. Agents Chemother. 54, 3219–3224. doi: 10.1128/AAC.01715-09
Troemel, E. R., Chu, S. W., Reinke, V., Lee, S. S., Ausubel, F. M., and Kim, D. H. (2006). p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2:e183. doi: 10.1371/journal.pgen.0020183
Uccelletti, D., Zanni, E., Marcellini, L., Palleschi, C., Barra, D., and Mangoni, M. L. (2010). Anti-Pseudomonas activity of frog skin antimicrobial peptides in a Caenorhabditis elegans infection model: a plausible mode of action in vitro and in vivo. Antimicrob. Agents Chemother. 54, 3853–3860. doi: 10.1128/AAC.00154-10
Ueda, A., Attila, C., Whiteley, M., and Wood, T. K. (2009). Uracil influences quorum sensing and biofilm formation in Pseudomonas aeruginosa and fluorouracil is an antagonist. Microb. Biotechnol. 2, 62–74. doi: 10.1111/j.1751-7915.2008.00060.x
Van Herreweghe, J. M., and Michiels, C. W. (2012). Invertebrate lysozymes: diversity and distribution, molecular mechanism and in vivo function. J. Biosci. (Bangalore) 37, 327–348. doi: 10.1007/s12038-012-9201-y
Wahlby, C., Kamentsky, L., Liu, Z. H., Riklin-Raviv, T., Conery, A. L., O’rourke, E. J., et al. (2012). An image analysis toolbox for high-throughput C. elegans assays. Nat. Methods 9, 714–716. doi: 10.1038/nmeth.1984
Wang, J., Zhou, X., Li, W., Deng, X., Deng, Y., and Niu, X. (2016). Curcumin protects mice from Staphylococcus aureus pneumonia by interfering with the self-assembly process of alpha-hemolysin. Sci. Rep. 6:28254. doi: 10.1038/srep28254
Williams, P., Winzer, K., Chan, W. C., and Camara, M. (2007). Look who’s talking: communication and quorum sensing in the bacterial world. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362, 1119–1134. doi: 10.1098/rstb.2007.2039
Yamamoto, K., Kitayama, T., Ishida, N., Watanabe, T., Tanabe, H., Takatani, M., et al. (2000). Identification and characterization of a potent antibacterial agent. NH125 against drug-resistant bacteria. Biosci. Biotechnol., Biochem. 64, 919–923. doi: 10.1271/bbb.64.919
Yang, J., Chen, Z., Ching, P., Shi, Q., and Li, X. (2013). An integrated microfluidic platform for evaluating in vivo antimicrobial activity of natural compounds using a whole-animal infection model. Lab. Chip 13, 3373–3382. doi: 10.1039/c3lc50264c
Yang, J., Huang, X., Tian, B., Wang, M., Niu, Q., and Zhang, K. (2005). Isolation and characterization of a serine protease from the nematophagous fungus, Lecanicillium psalliotae, displaying nematicidal activity. Biotechnol. Lett. 27, 1123–1128. doi: 10.1007/s10529-005-8461-0
Yin, H., Deng, Y., Wang, H., Liu, W., Zhuang, X., and Chu, W. (2015). Tea polyphenols as an antivirulence compound disrupt quorum-sensing regulated pathogenicity of Pseudomonas aeruginosa. Sci. Rep. 5:16158. doi: 10.1038/srep16158
Yun, B., Oh, S., Song, M., Hong, Y. S., Park, S., Park, D. J., et al. (2015). Inhibitory effect of epigallocatechin gallate on the virulence of Clostridium difficile PCR Ribotype 027. J. Food Sci. 80, M2925–M2931. doi: 10.1111/1750-3841.13145
Zhou, Y. M., Shao, L., Li, J. A., Han, L. Z., Cai, W. J., Zhu, C. B., et al. (2011). An efficient and novel screening model for assessing the bioactivity of extracts against multidrug-resistant Pseudomonas aeruginosa using Caenorhabditis elegans. Biosci. Biotechnol. Biochem. 75, 1746–1751. doi: 10.1271/bbb.110290
Zhu, J., Cai, X., Harris, T. L., Gooyit, M., Wood, M., Lardy, M., et al. (2015). Disarming Pseudomonas aeruginosa virulence factor LasB by leveraging a Caenorhabditis elegans infection model. Chem. Biol. 22, 483–491. doi: 10.1016/j.chembiol.2015.03.012
Keywords: Caenorhabditis elegans, antimicrobials, anti-virulence, immunomodulator, antimicrobial peptides
Citation: Kong C, Eng S-A, Lim M-P and Nathan S (2016) Beyond Traditional Antimicrobials: A Caenorhabditis elegans Model for Discovery of Novel Anti-infectives. Front. Microbiol. 7:1956. doi: 10.3389/fmicb.2016.01956
Received: 27 September 2016; Accepted: 22 November 2016;
Published: 02 December 2016.
Edited by:
Luis Cláudio Nascimento Da Silva, CEUMA University, BrazilReviewed by:
Osmar Nascimento Silva, Universidade Católica Dom Bosco, BrazilFrancesco Imperi, Sapienza University of Rome, Italy
Copyright © 2016 Kong, Eng, Lim and Nathan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sheila Nathan, c2hlaWxhQHVrbS5lZHUubXk=
†Present address: Cin Kong, Department of Biomedical Sciences, Faculty of Science, University of Nottingham Malaysia Campus,Semenyih, Malaysia
 Cin Kong
Cin Kong Su-Anne Eng
Su-Anne Eng Mei-Perng Lim
Mei-Perng Lim Sheila Nathan
Sheila Nathan