- 1Institute of Microbiology, College of Life Sciences, Zhejiang University, Hangzhou, China
- 2State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, China
Nicotine is a type of environmental pollutant present in the tobacco waste that is generated during tobacco manufacturing. Sphingomonas melonis TY can utilize nicotine as a sole source of carbon, nitrogen and energy via a variant of the pyridine and pyrrolidine pathway (the VPP pathway). In this study, we report the identification of two novel sets of genes, ndrA1A2A3, and ndrB1B2B3B4, which are crucial for nicotine degradation by strain TY. ndrA1A2A3 and ndrB1B2B3B4 exhibit similarity with both nicotine dehydrogenase ndh from Arthrobacter nicotinovorans and nicotine hydroxylase vppA from Ochrobactrum sp. SJY1. The transcriptional levels of ndrA1A2A3 and ndrB1B2B3B4 in strain TY were significantly upregulated in the presence of nicotine. Furthermore, ndrA1 or ndrB2 knockout resulted in a loss of the ability to degrade nicotine, whereas gene complementation restored the capacity of each mutant to utilize nicotine for growth. Biodegradation assays indicated that the mutant strains retained the ability to degrade the first intermediate in the pathway, 6-hydroxynicotine (6 HN). However, heterologous expression of ndrA1A2A3 and ndrB1B2B3B4 did not confer nicotine dehydrogenase activity to E. coli DH5α, Pseudomonas putida KT2440 or Sphingomonas aquatilis. These results provide information on the VPP pathway of nicotine degradation in S. melonis TY, and we conclude that these two sets of genes have essential functions in the conversion of nicotine to 6 HN in strain TY.
Introduction
Tobacco products are among the most widely popular goods worldwide due to the addictive component in tobacco: nicotine. Nicotine is the most abundant alkaloid in tobacco plants, and a large amount of tobacco waste produced every year is disseminated in the environment through water and soil. Nicotine is very harmful to public health because it can be easily absorbed and can pass the blood-brain barrier (O'neill et al., 2002; Lemay et al., 2004).
As important decomposers in ecosystems, microbes possess powerful, and versatile degradation capacities, as reflected in the type of substrates utilized, the metabolic mechanisms involved, and diversity of catalyzing enzymes. One example is the diversity in metabolic pathways and molecular mechanisms of nicotine degradation by microorganisms. Research on the microbial degradation of nicotine has been studied since the 1950s (Wada and Yamasaki, 1953). To date, various bacteria and fungi are reported to be able to decompose nicotine through at least six pathways, though there are four main degradation pathways. (1) The pyridine pathway, which begins with the hydroxylation of the nicotine pyridine ring to generate 6-hydroxynicotine (6 HN), and then 6-hydroxy-N-methylmyosmine, 6-hydroxypseudooxynicotine (6 HPON), and 2, 6-dihydroxy-pseudooxynicotine, and generates nicotine blue and succinic acid at last, has been described in Arthrobacter (Hochstein and Rittenberg, 1959a,b, 1960; Richardson and Rittenberg, 1961a,b; Gherna et al., 1965; Holmes and Rittenberg, 1972; Holmes et al., 1972; Chiribau et al., 2006), Nocardioides (Ganas et al., 2008) and Rhodococcus (Cobzaru et al., 2011). And the most studied metabolic pathway of the pyridine pathway was encoded by megaplasmid pAO1 of Arthrobacter nicotinovorans (Brandsch, 2006), for which every enzyme involved has been validated. (2) The pyrrolidine pathway, representative of Pseudomonas, begins with oxidation of the pyrrolidine ring to produce N-methylmyosmine, which leads to formation of succinate and 2, 5-dihydroxypyridine (2, 5-DHP); the latter then enters the maleamic acid pathway (Wang et al., 2007; Tang et al., 2012). This pathway has been comprehensively studied in Pseudomonas putida S16, and the final steps are catalyzed sequentially by DHP dioxygenase (Hpo), NFM deformylase (Nfo), maleamate amidase (Ami) and maleate isomerase (Iso) to produce fumaric acid (Tang et al., 2012). (3) The methyl pathway, which is found in some fungi such as Aspergillus and Pellicularia, degrades nicotine via demethylation to nornicotine (Uchida et al., 1983; Meng et al., 2010), while the pathway has not been well-elucidated. (4) A variant of the pyridine and pyrrolidine (VPP) pathway constitutes the fourth main pathway of nicotine degradation. Occurring in several strains, the newly identified VPP pathway first transforms nicotine into 6 HPON through 6-hydroxy-L-nicotine and 6-hydroxy-N-methylmyosmine via the pyridine pathway; the intermediates then enter the pyrrolidine pathway followed by formation of 6-hydroxy-3-succinoylpyridine and 2, 5-DHP. This VPP pathway has been described in Agrobacterium tumefaciens strain S33, Shinella sp. HZN7, and Ochrobactrum sp. strain SJY1 (Wang et al., 2012; Ma et al., 2013; Yu et al., 2014). Surprisingly, a newly isolated strain, Pusillimonas sp. T2, was found to possess both the VPP pathway and a partial pyridine pathway with 2, 6-dihydroxypyridine formation (Ma et al., 2014). Many intermediates, such as 6-hydroxy-3-succinoylpyridine, generated through these diverse nicotine metabolic pathways can potentially be transformed into commercially valuable compounds such as analogs of epibatidine (Wang et al., 2005). In addition, the various nicotine metabolic pathways indicate the diversity of the mechanisms responsible. Although Sphingomonas melonis strain TY, isolated by our research team, can degrade nicotine efficiently, the genes involved are unclear and may differ from the genes described thus far (Wang et al., 2011).
In this work, we studied the nicotine-degrading S. melonis strain TY, a new bacterium able to degrade nicotine via the VPP pathway. The draft genome sequence of S. melonis TY was obtained using the Illumina approach. Sequence analysis revealed two sets of putative nicotine metabolic genes that were found to be essential for nicotine dehydrogenase in strain TY.
Materials and Methods
Chemicals and Reagents
(S)–(-)-Nicotine (>99%) was obtained from Chemsky international Co., Ltd (Shanghai, China). 6 HN was a gift from Jiguo Qiu (Nanjing Agricultural University, China). TransStart® FastPfu DNA Polymerase for fragment amplification was purchased from TransGen Biotech (Beijing, China). Restriction enzymes used for plasmid construction and a premixed protein marker for protein electrophoresis were purchased from Takara Biotechnology Co., Ltd. (Dalian, China). Antibiotics, isopropyl β-D-1-thiogalactopyranoside (IPTG) and other reagents were purchased from Shanghai Sangon Biotech Co., Ltd. (Shanghai, China). A plasmid extraction kit, gel extraction kit and DNA purification kit were obtained from Omega Bio-tek, Inc (Norcross, GA, USA).
PCR Programs Used in this Study
PCR program used for high-fidelity DNA polymerase was according to the manual: pre-denaturing at 95°C for 2 min; denaturing at 95°C for 20 s, annealing at certain temperature (according to the primer pair) for 20 s, extending at 72°C at a speed of 2 kb/min, 32 cycles; extending at 72°C for 5 min; keeping at 4°C. General PCR program used for fragment amplification was pre-denaturing at 94°C for 5 min; denaturing at 94°C for 30 s, annealing at certain temperature (according to the primer pair) for 30 s, extending at 72°C at a speed of 1 kb/min, 32 cycles; extending at 72°C for 5 min, keeping at 4°C. PCR program used for reverse transcription quantitative PCR (RT-qPCR) was: pre-denaturing at 94°C for 30 s; denaturing at 94°C for 5 s, annealing at 60°C for 15 s, extending at 72°C for 12 s, 40 cycles.
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Table 1; the primers used are provided in Table S1. Wild-type S. melonis TY (collection number CGMCC 1.15791) can utilize nicotine as a sole source of carbon, nitrogen and energy for growth (Wang et al., 2011). Wild-type TY and derivatives were cultured aerobically at 30°C in LB medium or inorganic salt medium (ISM) supplemented with nicotine, as described previously (Yang, 2010). Escherichia coli strains were grown in LB broth at 37°C. When necessary, kanamycin and tetracycline were used at final concentrations of 50 and 10 μg/mL, respectively. IPTG was used to induce expression at gradient concentration, and 2, 6-diaminopimelic acid (2, 6-DAP) was used at a final concentration of 0.3 mM for E. coli WM3064.
Draft Genome Sequencing of S. melonis TY and Prediction of Relevant Nicotine Metabolism Genes
The draft genome sequence of S. melonis TY was obtained using Illumina Hiseq2000 paired-end (PE) sequencing (101 bp for each read; 299.8-fold coverage) and then assembled into 68 scaffolds using SOAPdenovo version 2.04 (N50 length, 205,244 bp) (Li et al., 2010). The draft genome sequence of strain TY contains 4,100,783 bp with a GC content of 67.083% and 3748 predicted coding sequences (CDSs). The annotation was performed using Best-placed reference protein set; GeneMarkS+. Previously described nicotine-degrading genes, including ndhLSM, which catalyzes the first step of nicotine transformation in A. nicotinovorans (Dang Dai et al., 1968; Grether-Beck et al., 1994), and vppA and vppB of the upper VPP pathway of Ochrobactrum sp. SJY1 (Yu et al., 2014, 2015), were used for blast searches of the strain TY genome in NCBI.
RT-qPCR Analysis of ndrA1A2A3 and ndrB1B2B3B4
RNAprep Pure Bacteria Kit (Tiangen Biotech, Beijing, China) was used to prepare total RNA from S. melonis TY grown in triplicate in control and nicotine-induction cultures. The RNA was reverse transcribed into cDNA using random hexamer primers and PrimeScript RT Reagent Kit with gDNA Eraser (Perfect Real Time) (Takara, Dalian, China). The respective cDNA fragments were used as templates in PCR with gene-specific primers (Table S1). Real-time quantitative PCR was performed using a Rotor-Gene Q real-time PCR detection system (Qiagen) with TransStart Top Green qPCR SuperMix (TransGen Biotech, Beijing, China). The strain TY genome was used as the positive control, and untranscribed RNA was used as the negative control. Melting curves and agarose gel analyses were applied to confirm the specificity of the PCR products. The threshold cycle (CT) values for each target gene were normalized to the V3 region of the 16S rRNA gene. The 2−ΔΔCT method was used to calculate the relative expression level, where ΔΔCT = (CT, target-CT, 16S)induction—(CT, target-CT, 16S)control; to obtain more reliable results, the theoretical efficiency value 2 was replaced with the estimated PCR efficiency value (Livak and Schmittgen, 2001; Ramakers et al., 2003; Ruijter et al., 2009; Tuomi et al., 2010).
Gene Knockout and Complementation of ndrA1 and ndrB2
In-frame disruption of ndrA1 and ndrB2 in strain S. melonis TY was performed using the suicide plasmid pEX18Tc and a two-step homologous recombination method as described previously (Chen et al., 2014). Gene complementation were conducted the same as described in Chen et al. (2014).
TYΔndrA1 and TYΔndrB2 Growth and Resting Cell Reactions
Biotransformation tests were performed using wild-type TY and derivative strains TYΔndrA1, TYΔndrA1(pRK415- ndrA1), TYΔndrB2, and TYΔndrB2(pRK415-ndrB2) as well as an inactivated wild-type TY strain [heated at 80°C for 5 min to prevent nicotine absorption (Harwood et al., 1994)] and a control consisting only of nicotine. The biotransformation test was performed in a 150 mL flask containing 60 mg dry cell weight of resting cells (approximately 5 OD600nm, with one OD600nm unit equal to 0.40 g/L dry cell weight) and 0.5 g/L nicotine in 30 mL of sterilized ISM medium; the culture was incubated at 30°C and 200 rpm. Three sets of parallel experiments were performed for each strain. Biotransformation tests using 6 HN were conducted in a manner similar to nicotine that described above.
To test whether the two mutant strains retained the ability to metabolize 6 HN, the first product formed in the VPP pathway, strains TYΔndrA1, and TYΔndrB2 were washed three times with sterilized water and streaked onto ISM plates supplemented with 6 HN as the sole source carbon and nitrogen; the plates were placed in an incubator (30°C) for growth; wild-type TY was used as a positive control.
Heterologous Expression of Two Sets of Putative Nicotine-Degrading Genes
Nicotine catalysis activity of ndrA1A2A3 and ndrB1B2B3B4 was tested by heterologous expression. To test for nicotine dehydrogenase activity using whole cell or crude cell extracts, the two gene clusters were cloned (using primers shown in Table S1) into the broad host-range cloning vector pRK415 (Hind III- and EcoR I digested) and mated to P. putida KT2440 and Sphingomonas aquatilis (two non-nicotine-degrading strains) via E. coli WM3064. The culture conditions and enzyme activity detection are those previously reported (Ganas and Brandsch, 2009), while the detection of production 6 HN was replaced by UV-scan, and the host transformed with the control plasmid was used as the negative control, wild-type TY set as positive control.
Genes orf2 (coding for glycosyl transferase) and orf1 (coding for a XshC-Cox1-family protein) just downstream of ndrB4 exhibit similarity with mobA and coxF, respectively, which are essential for nicotine dehydrogenase activity in A. nicotinovorans pAO1 (Ganas and Brandsch, 2009) and may be required for nicotine dehydrogenase cofactor synthesis in strain TY. We cloned orf1 and orf2 together with ndrB1B2B3B4 into vector pRK415 (Hind III- and EcoR I digested) according to a previous report (Ganas and Brandsch, 2009), followed by transformation of the plasmid into E. coli DH5α and mating to S. aquatilis and P. putida KT2440 via E. coli WM3064. Moreover, we also cloned these six genes into vector pET-28a(+) (Hind III- and Nco I-digested) just downstream the T7 promoter and transformed the plasmid into E. coli BL21(DE3) to induce expression with various concentrations of IPTG (at 0, 0.1, 0.2, 0.5 and 1 mM). Activity tests were performed using whole cell and crude cell extracts of the above recombinant strains and corresponding negative controls with the host containing the original plasmid, pRK415 or pET-28a(+). SDS-PAGE was carried out for the E. coli BL21(DE3) derivative strains to detect induction under the test conditions.
Genome Sequence Accession Number
This whole-genome shotgun project has been deposited at DDBJ/ENA/GenBank under accession LQCK00000000. The version described in this paper is version LQCK02000000.
Results
Two Sets of Putative Nicotine Dehydrogenase Genes in Strain TY
Genome sequencing can provide much information for research into the functional mechanisms of microorganisms (Tang et al., 2013). The draft genome sequence of S. melonis TY was obtained by Illumina sequencing, and blast searching of the sequence using representative sequences for nicotine-degrading enzymes from A. nicotinovorans and Ochrobactrum sp. SJY1 was performed. Based on gene similarity and organization, we identified two sets of putative nicotine dehydrogenase genes in TY: ndrA1A2A3 and ndrB1B2B3B4. According to the annotation of genome sequence, ndrA1 encodes an aldehyde dehydrogenase iron-sulfur subunit; ndrA2 encodes a subunit of molybdopterin dehydrogenase; ndrA3 encodes a subunit of xanthine dehydrogenase; ndrB1 encodes an aldehyde dehydrogenase iron-sulfur subunit; ndrB2, ndrB3, and ndrB4 all encode subunit of xanthine dehydrogenase. These two sets of gene clusters, ndrA1A2A3 and ndrB1B2B3B4, show similarity to the three subunits of ndh: nicotine dehydrogenase subunit S (ndhS), nicotine dehydrogenase subunit M (ndhM) and nicotine dehydrogenase subunit L (ndhL). ndrA3, ndrB3, and ndrB4 show similarity to nicotine hydroxylase subunit L (vppAL), and ndrA1 and ndrB1 show similarity to nicotine hydroxylase subunit S (vppAS). All these genes exhibit 15–44% amino acid sequence identity with homologous nicotine-degrading proteins, with similar gene organization. The gene organization and amino acid identity shared with homologous genes ndh and vppA are shown in Figure 1.
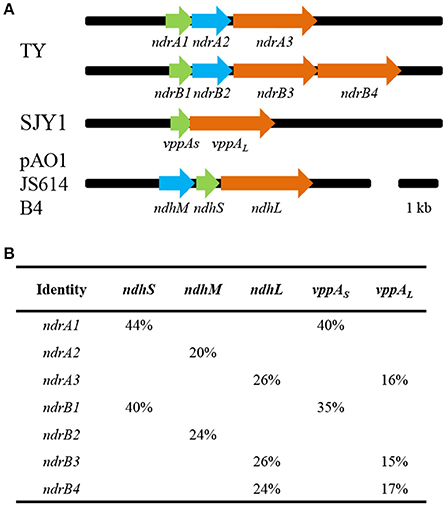
Figure 1. Genetic organization of clusters ndrA1A2A3 and ndrB1B2B3B4 in Sphingomonas melonis TY and amino acid sequence identity with related genes (A). Arrows indicate the size and direction of transcription of each gene. vppAs, nicotine hydroxylase, subunit S; vppAL, nicotine hydroxylase, subunit L; ndhM, nicotine dehydrogenase, subunit M; ndhS, nicotine dehydrogenase, subunit S; ndhL, nicotine dehydrogenase, subunit L. TY, Sphingomonas melonis TY; SJY1, Ochrobactrum sp. SJY1; pAO1, A. nicotinovorans pAO1; JS614, Nocardioides sp. JS614; B4, Rhodococcus opacus B4. (B) Comparison of amino acid sequences; the percentage shows the amino acid identity.
Transcriptional Levels of ndrA1A2A3 and ndrB1B2B3B4 are Upregulated in the Presence of Nicotine
To demonstrate a correlation between nicotine degradation and the two sets of putative nicotine dehydrogenase genes, the levels of mRNA expression of seven putative target genes involved in nicotine degradation in S. melonis TY were estimated using RT-qPCR and the 2ΔΔCT method in the presence or absence of nicotine. The V3 region of the 16S rRNA gene was used as the reference. The results showed that transcription of these genes was significantly upregulated in the presence of nicotine compared to in its absence (Figure 2), suggesting that transcription of ndrA1A2A3 and ndrB1B2B3B4 was induced by nicotine.
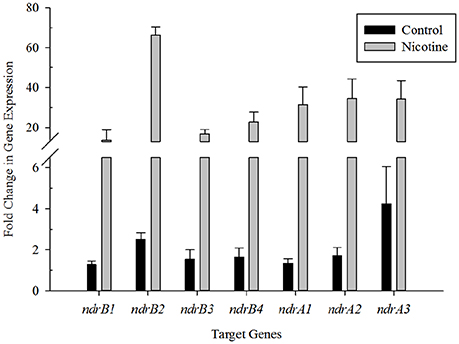
Figure 2. Changes in transcriptional levels of genes putatively involved in nicotine degradation. RT-qPCR analysis of target gene transcripts produced in Sphingomonas melonis TY grown with or without nicotine. The results presented in these histograms are the means of three independent experiments, and error bars indicate the standard error.
The Identification and Characterization of NdrA1A2A3 and NdrB1B2B3B4
To examine whether ndrA1A2A3 or ndrB1B2B3B4 is involved in nicotine degradation in vivo, ndrA1 or ndrB2 was knocked out via kanamycin resistance gene insertion using a gene replacement technique based on homologous recombination. Double crossover was verified by PCR (Figure 3) and sequencing analysis. Although neither mutant strain, TYΔndrA1, or TYΔndrB2, could grow on ISM containing nicotine as the sole carbon and nitrogen source (Figure 4), both mutants were able to grow on ISM supplemented with 1 g/L (NH4)2SO4 and 1 g/L glucose as the nitrogen and carbon sources, respectively. Furthermore, following complementation of the corresponding gene, each mutant displayed restored capacity to grow on nicotine, similar to wild-type TY (Figure 4).
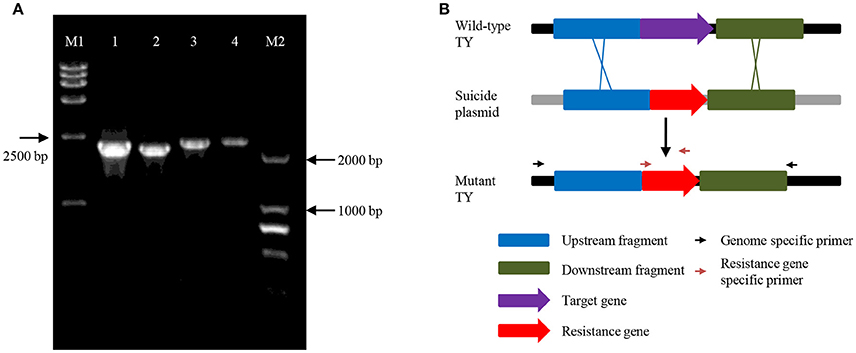
Figure 3. PCR verification of TYΔndrA1 and TYΔndrB2 using specific primers beyond the homologous fragments and primers in the resistance gene. (A) Sample 1, amplified with ndrA1-VF and Kan02, 2260 bp; sample 2, amplified with ndrA1-VR and Kan01, 2172 bp; sample 3, amplified with ndrB2-VF and Kan01, 2308 bp; sample 4, amplified with ndrB2-VR and Kan02, 2330 bp; M1, DL15000 marker; M2, DL2000 marker. Electrophoresis was performed using 0.8% agarose; (B) The in-frame deletion of target genes, and relative site of genomic-specific primers and resistance gene-specific primers.
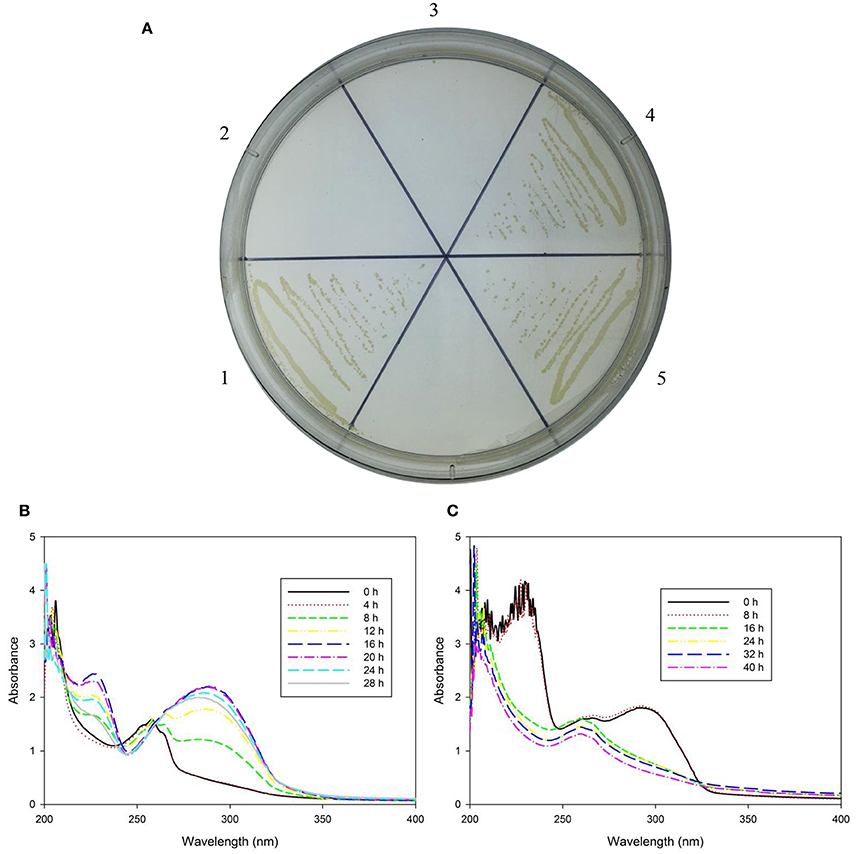
Figure 4. Growth ability of strain TY and its derivatives with nicotine and their biotransformation ability with nicotine and 6-hydroxynicotine. (A) Growth ability of wild-type TY (1), TYΔndrA1 (2), TYΔndrB2 (3), TYΔndrA1(pRK415-ndrA1) (4), and TYΔndrB2(pRK415-ndrB2) (5); (B) Biotransformation of nicotine by wild-type TY and two complementary strains, and example is shown because they are similar spectrometric graphs; (C) Biotransformation of 6-hydroxynicotine by wild-type TY, two mutant strains and two complementary strains, example is shown because they are similar spectrometric graphs.
Based on the results of gene knockout and complementation, it appears that ndrA1 and ndrB2 are involved in the capability of strain TY to utilize nicotine as a sole carbon and nitrogen source, suggesting that one of these two sets of gene clusters encodes a nicotine dehydrogenase. To further confirm the function of ndrA1A2A3 and ndrB1B2B3B4, we performed a nicotine and 6 HN biotransformation test using the wild-type TY strain, the mutant strains and strains complemented with the two genes. Although TYΔndrA1 and TYΔndrB2 lost the ability to degrade and transform nicotine, they retained 6 HN conversion capacity (Figure 4). Complementation of the corresponding gene restored the ability of both TYΔndrA1(pRK415-ndrA1) and TYΔndrB2(pRK415-ndrB2) to degrade and transform nicotine.
Because TYΔndrA1 and TYΔndrB2 can grow on ISM medium supplemented with 6 HN as the sole carbon and nitrogen source (Figure 5), combined with the biotransformation results, it appears that the two mutant strains not only retain the ability to transform 6 HN but can also degrade it completely. This result indicates that disruption of ndrA1 or ndrB2 only affects the first step in nicotine metabolism, supporting our speculation that a nicotine dehydrogenase is encoded by one of these two sets of genes.
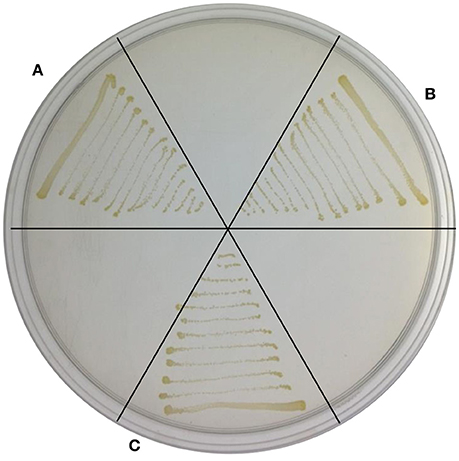
Figure 5. Growth of TYΔndrA1 and TYΔndrB2 in ISM supplemented with 6-hydroxynicotine as a substrate. (A), TYΔndrA1; (B), TYΔndrB2; (C), wild-type TY.
No Catalytic Activity was Detected after Heterologous Expression of NdrA1A2A3 and NdrB1B2B3B4
To shed light on the function of these two gene clusters, we tested the nicotine-degrading activity of ndrA1A2A3 and ndrB1B2B3B4 through heterologous expression in P. putida KT2440 and S. aquatilis; the latter was selected because it is a member of the same genus as strain TY but does not exhibit nicotine degradation ability. However, neither whole cell nor crude cell extracts exhibited nicotine dehydrogenase activity.
The cofactor molybdopterin cytosine dinucleotide (MCD) is needed for Ndh nicotine dehydrogenase function in A. nicotinovorans pAO1 (Sachelaru et al., 2006; Ganas and Brandsch, 2009), we thought MCD or other cofactor is needed by the putative nicotine dehydrogenase in strain TY. Thus, we identified two genes, orf1 and orf2, contiguous on the genome with ndrB1B2B3B4 that show similarity with coxF and mobA (involved in MCD biosynthesis in A. nicotinovorans pAO1), and orf2 shows LAAG amino acid motif as the canonical MocA (Neumann et al., 2011). We cloned these six genes into pRK415 for expression in E. coli DH5α, S. aquatilis and P. putida KT2440 and into pET-28a(+) for expression in E. coli BL21(DE3). However, whole cell and crude cell extracts showed no nicotine dehydrogenase activity. In addition, SDS-PAGE was carried out on extracts from E. coli BL21(DE3)pET28a-ndrB1B2B3B4orf1orf2 and its control strain E. coli BL21(DE3)pET28a, and several bands were appeared in the former compared to the latter (data not shown).
Discussion
S. melonis TY can degrade nicotine efficiently than many other nicotine degraders, and has tolerance to some neonicotinoid insecticides when degrading nicotine, which is suitable for the disposal of tobacco waste and the reduction of nicotine in tobacco leaves and this tolerance was not reported in other nicotine degraders (Wang et al., 2011). In this study, we identified two sets of genes involved in the nicotine metabolism in S. melonis TY. Both mutants were able to grow on glucose and (NH4)2SO4, indicating that disruption of ndrA1 and ndrB2 did not affect basic metabolism. Moreover, following gene complementation, both complemented strains regained the ability to utilize nicotine. The gene knockout and complementation results indicated that ndrA1 and ndrB2 may be involved in converting nicotine to 6 HN, the first step in the VPP pathway.
To elucidate the function of these two gene clusters, we tested the nicotine-degrading activity of ndrA1A2A3 and ndrB1B2B3B4. Many gene products show activity when heterologously expressed in E. coli DH5α and P. putida KT2440, such as the nicotine hydroxylation vppA gene from Ochrobactrum sp. strain SJY1 (Yu et al., 2015), the 6 HN oxidation 6 hlno gene from A. nicotinovorans pAO1 (Ganas and Brandsch, 2009), and the nicotine oxidoreduction nicA gene from P. putida S16. Furthermore, homologous expression is an effective approach for successful protein expression, for example, 6 hlno and ndhMSLcoxFmobA (for nicotine oxidoreduction) (Ganas and Brandsch, 2009) and ndhL and kdhL (for 6 HPON dehydrogenation) (Sachelaru et al., 2006). However, expression of ndrA1A2A3 and ndrB1B2B3B4 in P. putida KT2440 and S. aquatilis did not confer activity on a heterologous host. Based on nicotine-related research of well-established A. nicotinovorans, we speculate that the nicotine dehydrogenase in strain TY might require a cofactor, such as MCD in A. nicotinovorans (Sachelaru et al., 2006). Surprisingly, we found two genes showing similarity to mobA and coxF contiguous with ndrB1B2B3B4 on the strain TY genome. We cloned these two genes, orf1 and orf2, together with ndrB1B2B3B4 into vectors pRK415 and pET-28a(+) and transformed the plasmids into E. coli DH5α, S. aquatilis, P. putida KT2440, and E. coli BL21(DE3). However, no nicotine dehydrogenase activity was detected. Indeed, heterologous expression with a constitutive or inducible promoter showed that ndrA1A2A3 and ndrB1B2B3B4 alone cannot confer nicotine dehydrogenase activity. Considering that TYΔndrA1 and TYΔndrB2 retained the ability to degrade 6 HN and that ndrA1A2A3 and ndrB1B2B3B4 alone could not confer nicotine dehydrogenase activity, we propose that ndrA1A2A3, and ndrB1B2B3B4 are involved and necessary for nicotine dehydrogenase in strain TY but require additional factors. Additionally, disruption of ndrA1 or ndrB2 resulted in no nicotine dehydrogenase activity.
There are possible two speculations for the reasons why the expression of the ndrA1A2A3 and ndrB1B2B3B4 in the tested strains failed. First, the tested strains did not provide a suitable environmental for the transcription, translation or protein folding of the introduced gene sets, such as the different codon usage bias between the gene sets in strain TY and the tested strains and any incorrect step in the formation of functional protein. These errors will lead to the negative results. Second, these two gene sets may take no part in catalyzing the initial degradation of nicotine but perform other essential function in nicotine degradation in strain TY. However, we think that influence of uptake of nicotine by the tested strains can be excluded because both whole cell and crude cell extracts reactions were performed to exhibit activities, and P. putida KT2440 were reported to express functional vppA (Yu et al., 2015).
In studying related genes involved in certain pathway or cellular network, there are several approaches for generating mutants. Nonetheless, some mutants obtained are in genes that do not actually function in the studied pathway due to the influence of gene organization, such as polar effects on downstream genes. However, ndrA1A2A3 and ndrB1B2B3B4 are not contiguous in the strain TY genome, and there are no other nearby putative genes related to nicotine degradation. Knockout of ndrA1 and ndrB2 led to the same superficial phenotypic effect on the undefined nicotine dehydrogenase in strain TY, which was presumably not due to potential downstream effects. Generally, we thought the function of both clusters ndrA1A2A3 and ndrB1B2B3B4 is essential for nicotine degradation in strain TY.
In a word, current experimental data only support that ndrA1A2A3 and ndrB1B2B3B4 are essential in nicotine degradation in strain TY, and the relationship between them is unclear. We have proposed three hypotheses about the relationship between these two gene sets. First, because the details of the reaction mechanism of nicotine conversion to 6 HN are not available and it is classified as different type of reaction according to the source of oxygen in hydroxylation (Hirschberg and Ensign, 1971), and the enzyme is considered as hydroxylase or oxygenase accordingly.. Moreover, reports have speculated some similar mechanisms in nicotinic acid and nicotine degradation (Tang et al., 2012). A previous study showed that 6-hydroxynicotinic acid is formed during nicotinic acid degradation; moreover, the oxygen incorporated into 6-hydroxynicotine acid was from water and not oxygen gas (Hunt et al., 1958; Hirschberg and Ensign, 1971). It was proposed that a dihydro-monohydroxy derivative of nicotinic acid was formed during nicotinic acid degradation, though no such derivative was identified (Hunt et al., 1958). Combining the similarity of the pyridine ring structure and the similar hydroxylation reaction of the pyridine ring in nicotinic acid and nicotine, we speculate that the same molecular mechanism of hydroxylation occurs with nicotine to generate 6 HN, as it does with nicotinic acid. If a type of dihydro-monohydroxy derivative of nicotine is generated, then ndrA1A2A3 and ndrB1B2B3B4 may play essential roles in converting nicotine to its dihydro-monohydroxy derivative and then to 6 HN. Second, one of the two sets of gene is directly related with nicotine degradation and need specific cofactor, and the other one is indirectly related. Third, neither of the two sets of genes is direct related with nicotine degradation, and there is an undefined real nicotine dehydrogenase in strain TY. However, this kind of undirected related function is unknown and need further study.
In summary, this study provides evidence indicating that two novel sets of genes in S. melonis TY are crucial for nicotine degradation. These two sets of genes are first reported here as being involved in the variant of the pyridine and pyrrolidine pathway of nicotine degradation and show similarity with isoenzymes in other nicotine-degrading microbes, an unexpected and surprising result. It will be important to determine whether ndrA1A2A3 and ndrB1B2B3B4 are both involved and/or function together in nicotine dehydrogenase activity as well as what type of reactions are involved in the still-undefined nicotine dehydrogenase in strain TY. Although uncertainty remains regarding the exact function of these two gene clusters, further research is in progress to characterize their function(s). These findings will provide a deeper understanding of the molecular mechanisms of nicotine metabolism in Sphingomonas.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Author Contributions
Revising the work: CX, NZ and ZL. Wrote the paper and final approval of the version to be published: All authors. All authors agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
This work was financially supported by the National Natural Science Foundation of China (No.31170115 and 31422003).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Professor Min Wu from Zhejiang University, for providing several plasmids and strains used in this study. And thank Professor Ping Xu and Hongzhi Tang, associate fellow Ying Xu from Shanghai Jiao Tong University, Jiguo Qiu from Nanjing Agricultural University for experimental suggestion.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.02060/full#supplementary-material
References
Brandsch, R. (2006). Microbiology and biochemistry of nicotine degradation. Appl. Microbiol. Biotechnol. 69, 493–498. doi: 10.1007/s00253-005-0226-0
Chen, Y. F., Chao, H., and Zhou, N. Y. (2014). The catabolism of 2,4-xylenol and p-cresol share the enzymes for the oxidation of para-methyl group in Pseudomonas putida NCIMB 9866. Appl. Microbiol. Biotechnol. 98, 1349–1356. doi: 10.1007/s00253-013-5001-z
Chiribau, C. B., Mihasan, M., Ganas, P., Igloi, G. L., Artenie, V., and Brandsch, R. (2006). Final steps in the catabolism of nicotine. FEBS J. 273, 1528–1536. doi: 10.1111/j.1742-4658.2006.05173.x
Cobzaru, C., Ganas, P., Mihasan, M., Schleberger, P., and Brandsch, R. (2011). Homologous gene clusters of nicotine catabolism, including a new ω-amidase for α-ketoglutaramate, in species of three genera of gram-positive bacteria. Res. Microbiol. 162, 285–291. doi: 10.1016/j.resmic.2011.01.001
Dang D. V., Decker, K., and Sund, H. (1968). Purification and properties of l-6-hydroxynicotine oxidase. Eur. J. Biochem. 4, 95–102. doi: 10.1111/j.1432-1033.1968.tb00177.x
Dehio, C., and Meyer, M. (1997). Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J. Bacteriol. 179, 538–540.
Dennis, J. J., and Zylstra, G. J. (1998). Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64, 2710–2715.
Ganas, P., and Brandsch, R. (2009). Uptake of l-nicotine and of 6-hydroxy-l-nicotine by Arthrobacter nicotinovorans and by Escherichia coli is mediated by facilitated diffusion and not by passive diffusion or active transport. Microbiology 155, 1866–1877. doi: 10.1099/mic.0.028688-0
Ganas, P., Sachelaru, P., Mihasan, M., Igloi, G., and Brandsch, R. (2008). Two closely related pathways of nicotine catabolism in Arthrobacter nicotinovorans and Nocardioides sp. strain JS614. Arch. Microbiol. 189, 511-517. doi: 10.1007/s00203-007-0340-8
Gherna, R. L., Richardson, S. H., and Rittenberg, S. C. (1965). The bacterial oxidation of nicotine: VI. the metabolism of 2,6-dihydroxypseudooxynicotine. J. Biol. Chem. 240, 3669–3674.
Grether-Beck, S., Igloi, G. L., Pust, S., Schilz, E., Decker, K., and Brandsch, R. (1994). Structural analysis and molybdenum-dependent expression of the pAO1-encoded nicotine dehydrogenase genes of Arthrobacter nicotinovorans. Mol. Microbiol. 13, 929–936. doi: 10.1111/j.1365-2958.1994.tb00484.x
Harwood, C. S., Nichols, N. N., Kim, M. K., Ditty, J. L., and Parales, R. E. (1994). Identification of the pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J. Bacteriol. 176, 6479–6488.
Hirschberg, R., and Ensign, J. C. (1971). Oxidation of nicotinic acid by a Bacillus species: source of oxygen atoms for the hydroxylation of nicotinic acid and 6-hydroxynicotinic acid. J. Bacteriol. 108, 757–759.
Hoang, T. T., Karkhoff-Schweizer, R. R., Kutchma, A. J., and Schweizer, H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86. doi: 10.1016/S0378-1119(98)00130-9
Hochstein, L. I., and Rittenberg, S. C. (1959a). The bacterial oxidation of nicotine: I. nicotine oxidation by cell-free preparations. J. Biol. Chem. 234, 151–155.
Hochstein, L. I., and Rittenberg, S. C. (1959b). The bacterial oxidation of nicotine: II. the isolation of the first oxidative product and its identification as (l)-6-hydroxynicotine. J. Biol. Chem. 234, 156–160.
Hochstein, L. I., and Rittenberg, S. C. (1960). The bacterial oxidation of nicotine: III. the isolation and identification of 6-hydroxypseudooxynicotine. J. Biol. Chem. 235, 795–799.
Holmes, P. E., and Rittenberg, S. C. (1972). The bacterial oxidation of nicotine: VII. partial purification and properties of 2,6-dihydroxypyridine oxidase. J. Biol. Chem. 247, 7622–7627.
Holmes, P. E., Rittenberg, S. C., and Knackmuss, H.-J. (1972). The bacterial oxidation of nicotine: VIII. synthesis of 2,3,6-trihydroxypyridine and accumulation and partial characterization of the product of 2,6-dihydroxypyridine oxidation. J. Biol. Chem. 247, 7628–7633.
Hunt, A. L., Hughes, D. E., and Lowenstein, J. M. (1958). The hydroxylation of nicotinic acid by Pseudomonas fluorescens. Biochem. J. 69, 170.
Keen, N. T., Tamaki, S., Kobayashi, D., and Trollinger, D. (1988). Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70, 191–197. doi: 10.1016/0378-1119(88)90117-5
Lee, J. S., Shin, Y. K., Yoon, J. H., Takeuchi, M., Pyun, Y.-R., and Park, Y.-H. (2001). Sphingomonas aquatilis sp. nov., Sphingomonas koreensis sp. nov., and Sphingomonas taejonensis sp. nov., yellow-pigmented bacteria isolated from natural mineral water. Int. J. Syst. Evol. Microbiol. 51, 1491–1498. doi: 10.1099/00207713-51-4-1491
Lemay, S., Chouinard, S., Blanchet, P., Masson, H., Soland, V., Beuter, A., et al. (2004). Lack of efficacy of a nicotine transdermal treatment on motor and cognitive deficits in Parkinson's disease. Prog. Neuro Psychopharmacol. Biol. Psychiatry 28, 31–39. doi: 10.1016/S0278-5846(03)00172-6
Li, R., Zhu, H., Ruan, J., Qian, W., Fang, X., Shi, Z., et al. (2010). De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20, 265–272. doi: 10.1101/gr.097261.109
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, Y., Wei, Y., Qiu, J., Wen, R., Hong, J., and Liu, W. (2013). Isolation, transposon mutagenesis, and characterization of the novel nicotine-degrading strain Shinella sp. HZN7. Appl. Microbiol. Biotechnol. 98, 2625–2636. doi: 10.1007/s00253-013-5207-0
Ma, Y., Wen, R., Qiu, J., Hong, J., Liu, M., and Zhang, D. (2014). Biodegradation of nicotine by a novel strain Pusillimonas. Res. Microbiol. 166, 67–71. doi: 10.1016/j.resmic.2014.12.009
Meng, X. J., Lu, L. L., Gu, G. F., and Xiao, M. (2010). A novel pathway for nicotine degradation by Aspergillus oryzae 112822 isolated from tobacco leaves. Res. Microbiol. 161, 626–633. doi: 10.1016/j.resmic.2010.05.017
Nelson, K., Weinel, C., Paulsen, I. T., Dodson, R. J., Hilbert, H., Martins dos Santos, V., et al. (2002). Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4, 799–808. doi: 10.1046/j.1462-2920.2002.00366.x
Neumann, M., Seduk, F., Iobbi-Nivol, C., and Leimkühler, S. (2011). Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of molybdopterin dinucleotide transferases that determine specificity for binding of guanine or cytosine nucleotides. J. Biol. Chem. 286, 1400–1408. doi: 10.1074/jbc.M110.155671
O'neill, M. J., Murray, T. K., Lakics, V., Visanji, N. P., and Duty, S. (2002). The role of neuronal nicotinic acetylcholine receptors in acute and chronic neurodegeneration. Curr Drug Targets CNS Neurol. Disord. 1, 399–411. doi: 10.2174/1568007023339166
Ramakers, C., Ruijter, J. M., Deprez, R. H., and Moorman, A. F. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66. doi: 10.1016/S0304-3940(02)01423-4
Richardson, S. H., and Rittenberg, S. C. (1961a). The bacterial oxidation of nicotine: IV. the isolation and identification of 2,6-dihydroxy-N-methylmyosmine. J. Biol. Chem. 236, 959–963.
Richardson, S. H., and Rittenberg, S. C. (1961b). The bacterial oxidation of nicotine: V. identification of 2,6-dihydroxypseudooxynicotine as the third oxidative product. J. Biol. Chem. 236, 964–967.
Ruijter, J. M., Ramakers, C., Hoogaars, W. M., Karlen, Y., Bakker, O., van den Hoff, M. J., et al. (2009). Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37:e45. doi: 10.1093/nar/gkp045
Sachelaru, P., Schiltz, E., and Brandsch, R. (2006). A Functional mobA gene for molybdopterin cytosine dinucleotide cofactor biosynthesis is required for activity and holoenzyme assembly of the heterotrimeric nicotine dehydrogenases of Arthrobacter nicotinovorans. Appl. Environ. Microbiol. 72, 5126–5131. doi: 10.1128/aem.00437-06
Saltikov, C. W., and Newman, D. K. (2003). Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. U.S.A. 100, 10983–10988. doi: 10.1073/pnas.1834303100
Tang, H., Wang, L., Wang, W., Yu, H., Zhang, K., Yao, Y., et al. (2013). Systematic unraveling of the unsolved pathway of nicotine degradation in Pseudomonas. PLoS Genet. 9:e1003923. doi: 10.1371/journal.pgen.1003923
Tang, H., Yao, Y., Wang, L., Yu, H., Ren, Y., Wu, G., et al. (2012). Genomic analysis of Pseudomonas putida: genes in a genome island are crucial for nicotine degradation. Sci Rep 2:377. doi: 10.1038/srep00377
Tuomi, J. M., Voorbraak, F., Jones, D. L., and Ruijter, J. M. (2010). Bias in the Cq value observed with hydrolysis probe based quantitative PCR can be corrected with the estimated PCR efficiency value. Methods 50, 313–322. doi: 10.1016/j.ymeth.2010.02.003
Uchida, S., Maeda, S., and Kisaki, T. (1983). Conversion of nicotine into nornicotine and N-methylmyosmine by fungi. Agric. Biol. Chem. 47, 1949–1953. doi: 10.1080/00021369.1983.10865908
Wada, E., and Yamasaki, K. (1953). Mechanism of microbial degradation of nicotine. Science 117, 152–153.
Wang, M., Yang, G., Wang, X., Yao, Y., Min, H., and Lu, Z. (2011). Nicotine degradation by two novel bacterial isolates of Acinetobacter sp. TW and Sphingomonas sp. TY and their responses in the presence of neonicotinoid insecticides. World J. Microbiol. Biotechnol. 27, 1633–1640. doi: 10.1007/s11274-010-0617-y
Wang, S., Huang, H., Xie, K., and Xu, P. (2012). Identification of nicotine biotransformation intermediates by Agrobacterium tumefaciens strain S33 suggests a novel nicotine degradation pathway. Appl. Microbiol. Biotechnol. 95, 1567–1578. doi: 10.1007/s00253-012-4007-2
Wang, S. N., Liu, Z., Tang, H. Z., Meng, J., and Xu, P. (2007). Characterization of environmentally friendly nicotine degradation by Pseudomonas putida biotype A strain S16. Microbiology 153, 1556–1565. doi: 10.1099/mic.0.2006/005223-0
Wang, S. N., Xu, P., Tang, H. Z., Meng, J., Liu, X. L., and Ma, C. Q. (2005). “Green” route to 6-hydroxy-3-succinoyl-pyridine from (S)-nicotine of tobacco waste by whole cells of a PseudomonasâĂĽsp. Environ. Sci. Technol. 39, 6877–6880. doi: 10.1021/es0500759
Woodcock, D. M., Crowther, P. J., Doherty, J., Jefferson, S., DeCruz, E., Noyer-Weidner, M., et al. (1989). Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17, 3469–3478. doi: 10.1093/nar/17.9.3469
Yang, G. (2010). Isolation, Identification and Nicotine Metabolism Pathways Analysis of Two Nicotine-Degrading Bacteria. Master's Thesis, Zhejiang University.
Yu, H., Tang, H., Li, Y., and Xu, P. (2015). Molybdenum-containing nicotine hydroxylase genes in a nicotine degradation pathway that is a variant of the pyridine and pyrrolidine pathways. Appl. Environ. Microbiol. 81, 8330–8338. doi: 10.1128/aem.02253-15
Keywords: nicotine, Sphingomonas melonis TY, ndrA1A2A3, ndrB1B2B3B4 and nicotine dehydrogenase
Citation: Wang H, Xie C, Zhu P, Zhou N-Y and Lu Z (2017) Two Novel Sets of Genes Essential for Nicotine Degradation by Sphingomonas melonis TY. Front. Microbiol. 7:2060. doi: 10.3389/fmicb.2016.02060
Received: 03 August 2016; Accepted: 07 December 2016;
Published: 17 January 2017.
Edited by:
Weiwen Zhang, Tianjin University, ChinaReviewed by:
Qiaoyun Huang, Huazhong Agricultural University, ChinaXu Shi, Texas Commission on Environmental Quality, USA
Copyright © 2017 Wang, Xie, Zhu, Zhou and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenmei Lu, bHpoZW5tZWlAemp1LmVkdS5jbg==
 Haixia Wang1
Haixia Wang1 Zhenmei Lu
Zhenmei Lu