- 1Public Health Ontario Laboratory, Toronto, ON, Canada
- 2Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, ON, Canada
- 3Mount Sinai Hospital and University Health Network, Toronto, ON, Canada
- 4Department of Mathematics and Statistics, University of Laval, Quebec, QC, Canada
- 5The Hospital for Sick Children, Toronto, ON, Canada
Background: Molecular typing is essential for inferring genetic relatedness between bacterial pathogens. In this study, we applied whole genome sequencing (WGS) for rapid prediction of sequence type and antibiotic resistance for invasive pneumococcal isolates.
Methods: 240 isolates from adults (≥50 years old) in Ontario, Canada during 2009 to 2013 were subjected to WGS. Sequence type, antibiotic susceptibility and resistance were predicted directly from short reads. Emerging non-vaccine serotype 22F was further characterized by WGS.
Results: Sequence type was successfully determined for 98.3% of isolates. The overall sensitivity and specificity for antibiotic resistance prediction were 95 and 100% respectively, compared to standard susceptibility testing methods. WGS-based phylogeny divided emerging 22F (ST433) strains into two distinct clades: clade A harboring a 23 kb-prophage and anti-phage PhD/Doc system and clade B with virulence-related proteases. Five isolates in clade A developed macrolide resistance via 5.1 kb mega element recombination (encoding mefE and msrD), while one isolate in clade B displayed quinolone resistance via a gyrA mutation.
Conclusions: WGS is valuable for routine surveillance of pneumococcal clinical isolates and facilitates prediction of genotype and antibiotic resistance. The emergence of 22F in Ontario in the post-vaccine era and evidence of evolution and divergence of the 22F population warrants heightened pneumococcal molecular surveillance.
Introduction
Invasive pneumococcal diseases (IPD) such as septicaemia and meningitis caused by Streptococcus pneumoniae result in a heavy burden of disease among children and adults worldwide (Ausina et al., 1988; O'Brien et al., 2007). After widespread implementation in 2002 of the 7-valent pneumococcal conjugate vaccine (PCV7, covering serotypes 4, 6B, 9V, 14, 18C, 19F, and 23F) in Canadian children, the incidence of IPD due to PCV7 serotypes decreased significantly in all age groups within the Canadian population (Kellner et al., 2009). In 2010 the new PCV13 vaccine (PCV7 serotypes plus 1, 3, 5, 6A, 7F, and 19A) replaced the PCV7 vaccine for routine immunization of children (Desai et al., 2010). Shortly after implementation of PCV13 in Canada, the number of IPD cases due to vaccine-associated serotypes such as 19A and 7F declined (Deng et al., 2015). However, the concurrent increase of IPD due to non-vaccine serotypes 22F, 15A, 33F, and 35B post-introduction of PCV13 has become a concern to clinicians (Deng et al., 2015; Golden et al., 2015; Duvvuri et al., 2016). In addition to use in children, the PCV13 vaccine was also approved by Canadian health authorities in 2012 for use among adults 50 years and older for the prevention of IPD (An Advisory Committee Statement (ACS), 2013). Currently it is also recommended for immunocompromised adults aged ≥ 18 years with high risk of IPD (An Advisory Committee Statement (ACS), 2013; Tomczyk et al., 2014).
Streptococcus pneumoniae is naturally competent, so it can uptake DNA from environment and undergo horizontal gene transfer between strains. Some recombination events such as deletions, insertions and inversions are frequently observed in the pneumococcal genome. In nature, horizontal gene transfer facilitates bacterial evolution, providing S. pneumoniae with easy access to crucial resistance genes and/or virulence factors. A recent analysis of the whole genome sequences of 240 Pneumococcal Molecular Epidemiological Network (PMEN1) pneumococcal strains revealed an exceptionally high recombination/mutation ratio of 7.2, and an average of 72 single nucleotide polymorphisms (SNPs) per recombination site (Croucher et al., 2011).
Molecular typing of Streptococcus pneumoniae is important for delineating genetic structure of bacterial populations and useful for inferring evolutionary relationships between isolates. For years, pulse-field gel electrophoresis (PFGE) that is based on motility variations of restriction fragments was considered the “gold standard” for subtyping of bacterial strains (Tenover et al., 1995). However, this method is technically demanding and time-consuming, and is thus not widely used for epidemiological surveillance. Multi-locus sequence typing (MLST) that measures DNA sequence variations for multiple housekeeping genes gradually replaced PFGE as a popular typing tool in the 21st century (Maiden et al., 1998). However, MLST has its shortcomings, as it is time-consuming, labor intensive and has higher cost than PFGE (Sabat et al., 2013). Recently, next-generation whole genome sequencing (WGS) has become a powerful and attractive tool for rapid typing of bacterial isolates, such as for E. coli (Joensen et al., 2014) and group A streptococcus (Athey et al., 2014). In addition, WGS has been evaluated for rapid prediction of drug susceptibility and resistance in Mycobacterium tuberculosis (Walker et al., 2015) and Staphylococcus aureus isolates (Gordon et al., 2014). With the cost of WGS continuing to decline (to a price close to or lower than MLST), it has the potential of replacing currently used typing methods for clinical isolates in the near future.
To date, MLST remains the primary method for large-scale typing of S. pneumoniae in the clinical laboratory. In this surveillance study, we employed high-throughput whole-genome sequencing for rapid prediction of MLST and antibiotic resistance in silico and further characterization of emerging non-vaccine serotypes of S. pneumoniae following the introduction of PCV13 in Ontario, Canada.
Materials and Methods
Clinical Pneumococcal Isolates and Antimicrobial Susceptibility Testing
Invasive S. pneumoniae isolates (1956 in total, approximately 400 isolates per year) were collected from older adults (≥50 years of age) in Ontario, Canada between 2009 and 2013. Submission of IPD isolates to Public Health Ontario Laboratories (PHOL) for further evaluation is encouraged but not mandatory. Geographic mapping of pneumococcal disease cases in Ontario older adults during 2009–2013 was done with geographic information system software QGIS (Version 2.8, http://www.qgis.org/). First 3 digits of postcode of each patient's residential address were used for the geographic mapping. Clinical isolates were recovered from sterile sites (including blood, cerebrospinal fluid and pleural fluid) and sent to the National Microbiology Laboratory (NML) in Winnipeg, Canada for serotyping using the Quellung reaction. Broth microdilution panels (Thermo Scientific, Waltham, MA, USA) were used for antimicrobial susceptibility testing for penicillin, erythromycin, clindamycin, fluoroquinolones, tetracycline, and cephalosporins. Minimum inhibitory concentrations (MICs) for each antibiotic were determined according to 2015 Clinical and Laboratory Standards Institute (CLSI) guidelines.
Strain Selection for Whole Genome Sequencing
We randomly selected 48 strains per year from 2009 to 2013 (240 isolates in total) from all available isolates in our S. pneumoniae database. The number of isolates selected per serotype was proportional to all isolates in that year, with the exclusion of rare serotypes that had a frequency <1.5%. Illumina multiplex sequencing strategy was used for sequencing the 240 isolates using high-throughput processing. Genomic DNA of clinical isolates was extracted using QIAamp DNA mini kits (Qiagen, Germany). A library of multiplexed samples (96 indexes for Hiseq and 12-24 indexes for Miseq) was prepared according to standard protocols using the Nextera XT kit. Genomic libraries of 192 samples were sequenced using Illumina Hiseq 2500 (100 bp paired end, Illumina, San Diego, USA), 44 samples were sequenced using Illumina Miseq (150 bp paired end), and 4 samples were not sequenced due to poor quality of library.
Prediction of MLST Type and Antibiotic Resistance from Short Reads
The MLST sequence type was determined from short reads for each isolate using SRST2 software (Inouye et al., 2012). Genetic determinants for resistance to antibiotics including chloramphenicol, macrolides, clindamycin and tetracycline were deduced from short reads using SRST2 software by comparison with a database of resistance genes (https://github.com/katholt/srst2). The predicted susceptibilities by whole-genome sequencing were compared with phenotypic drug susceptibility test results. When there was a mismatch between phenotype and genomic prediction, phenotypic drug susceptibility tests were repeated for those isolates. Two types of errors were defined for the susceptibility portion of this study: a very major error (VME) referred to a susceptible genotype with a resistant phenotype, and a major error (ME) was defined as a resistant genotype with a susceptible phenotype.
Genomic Characterization of Emerging Non-Vaccine Serotype 22F
Genomes for 22F isolates were assembled de novo using Velvet 1.2.07 (Zerbino and Birney, 2008) and VelvetOptimiser (mean coverage 208x). The draft genomes had an average of 333 contigs and mean N50 of 61885 bp. The draft genomes were annotated automatically with the RAST pipeline (rast.nmpdr.org). The draft genomes of 22F have been deposited at DDBJ/EMBL/GenBank under accession MLG(H-Z)00000000 and MLH(A-F)00000000. Raw Illumina sequences of 236 isolates have been deposited at SRA/NCBI under accession SRR3211687-729, SRR5011653-846. Core genomes and accessory genomes of 22F were predicted using Pan-Genomes Analysis Pipeline (PGAP) (Zhao et al., 2012) that utilizes BLAST command line tools (NCBI), InParanoid (O'Brien et al., 2005) and MultiParanoid (Alexeyenko et al., 2006) software. The core genes shared by all 22F isolates were retrieved and concatenated to make a “core” genome. MEGA5 (Tamura et al., 2011) was used to align these core genome sequences and to visualize single nucleotide polymorphism (SNP) variants. The genome sequence alignments were imported into BratNextGen (Marttinen et al., 2012) to infer regions of recombination and for clustering analysis (grouping strains into subclusters). Gaps and SNPs due to recombination were removed. The core genome alignment file of 22F isolates was uploaded to CIPRES server (www.phylo.org) for phylogenetic analysis. A phylogenetic tree was constructed with RAxML (Stamatakis, 2014) using a GTR model with a gamma correction for site rate variation. The online tool, Interactive Tree of Life (Letunic and Bork, 2011) was used for manipulation of phylogenetic trees. For comparative genomic analysis, the accessory genomes of different groups (e.g., drug-resistant and susceptible isolates) of 22F were compared in order to identify genetic features unique to any identified groups. Z-test was used to predict significance of difference between the proportions of strains in the two groups possessing a gene of interest. P < 0.01 was considered statistically significant.
Statistical Tests
Chi-square test was used to determine the statistical difference between the proportions of emerging serotypes (i.e., 22F) relative to total isolates in each year between 2009 and 2013 (5 proportions compared using 2 × 5 contingency table). P < 0.05 was considered significant and a simple Z-test was used to compare a pair of proportions (P < 0.05 is considered significant).
Results
Emergence of Non-PCV13 Serotypes Post-Vaccination
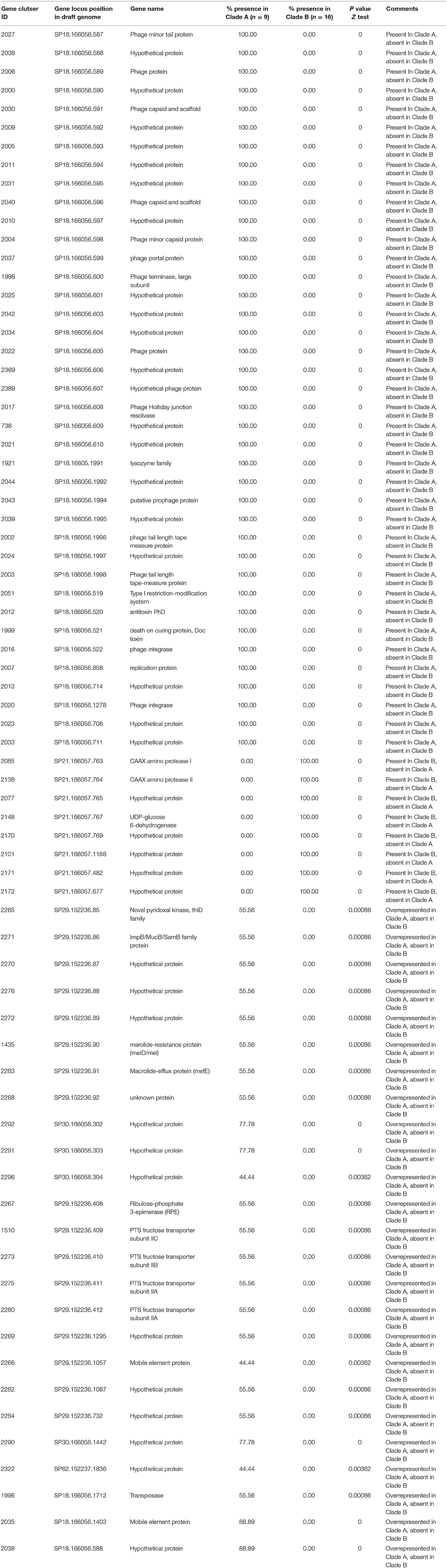
Although, IPD isolates were voluntarily submitted to PHOL from regional health units, distribution map of clinical samples indicated a good representation of geographic regions in the province of Ontario (Figure S1). Overall, during 2009–2013 IPD cases in older adults (≥50 years of age) due to PCV13 serotypes showed the trend of decline (chi-square test p < 0.01 between five ratios), while IPDs due to non-PCV13 serotypes had the trend of increase (chi-square test p < 0.01) (Figure 1). Non-PCV13 serotype 22F increased from 8.5% in 2009 to 14.5% in 2013 (p < 0.01 for chi-square test, and p = 0.0061 bewteen 2009 and 2013) (Figure 2). In 2013, serotype 22F surpassed 19A and 7F, and became the most common S. pneumoniae serotype identified among IPD isolates from Ontario adults.
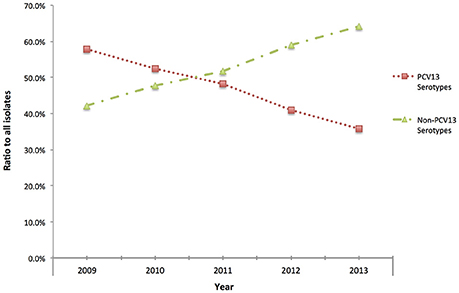
Figure 1. Proportion of Streptococcus pneumoniae isolates causing IPD among older adults (≥50 years of age) expressing PCV13 (1, 3, 4, 5, 6A, 6B, 7F, 9V, 14, 18C, 19A, 19F, and 23F) and non-PCV13 serotypes in Ontario, Canada, during 2009–2013.
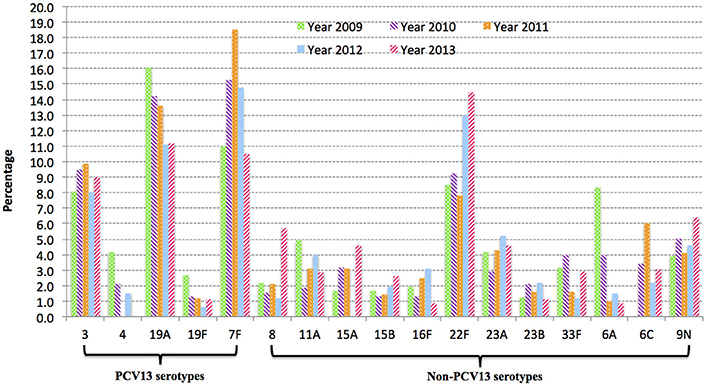
Figure 2. Serotype changes over time in IPD among older adults (≥50 years of age) in Ontario, Canada from 2009 to 2013. Serotype 22F increased significantly after introduction of PCV13 (p < 0.01) between 2009 and 2013, and became most prevalent in 2013.
Sequence Type Prediction In silico and Clonal Lineage Analysis
WGS was performed on a total of 236 IPD isolates (48 isolates per year, with four strains excluded due to extremely low concentrations of DNA following library preparation). MLST sequence type was successfully predicted from Illumina short reads directly for 232 strains, giving a success rate of 98.3% (232/236). The four isolates that failed MLST prediction had low sequence coverage (less than 10, ranging from 2 to 8). A total of 90 different sequence types (ST) were identified and these STs could be further divided into 17 clonal groups and 39 singletons (individual STs) through eBURST analysis. The emerging post-PCV13 serotype 22F was exclusively associated with clonal complex CC433 that include ST433 (25/26, 96%) and ST4533 (1/26, 4%) a single allele variant of ST433. CC63, CC66 and CC100 were the major (>80%) CCs identified among 15A, 9N, and 33F, respectively (Table 1).
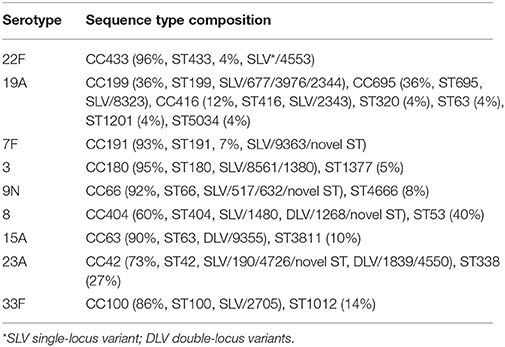
Table 1. Sequence type composition of most common pneumococcal serotypes causing IPD among older patients (≥50 years old) in Ontario, Canada during 2009–2013.
In silico Antibiotic Resistance Prediction and Comparison with Phenotypic Resistance Testing
The presence of acquired genes (Table S1 in supplementary material) including cat, mefA, msrD, ermB, and tetM that are associated with phenotypic resistance was predicted directly from short reads for the IPD isolates sequenced. 226 (95.8%) isolates had complete concordance between genotype and phenotype for the antimicrobial agents tested. The remaining 10 isolates had a total of 11 discrepancies between genotype and phenotype (Table S2 in supplementary material). Repeated susceptibility testing for the isolates with genotype/phenotype mismatches reduced the number of discrepancies from 11 to 7, giving a revised concordance rate of 97%. There were five very major errors (0.55% VME rate) and two major errors (0.22% ME rate) for antibiotic resistance prediction of S. pneumoniae isolates. The overall sensitivity and specificity for predicting phenotypic resistance to the antimicrobials tested using genomic data were 95% (95% CI, 90 to 97%), and 100% (95% CI, 99 to 100%), respectively (Table 2).
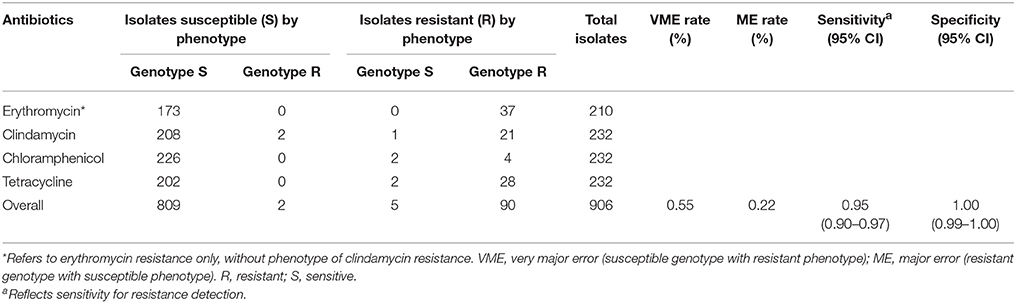
Table 2. Sensitivity and specificity of in silico genomic prediction of antibiotic resistance from short reads for S. pneumoniae isolates.
Phylogenetic Analysis of 22F Reveals Two Divergent Subpopulations
Of 26 serotype 22F isolates, 25 (96%) belonged to the same sequence type, ST433. Core genome alignment (1581 genes) of 22F-ST433 clinical isolates identified a total of 6308 SNPs. After removal of gaps and putative recombination regions predicted by BratNextgen, 3886 high-quality SNPs (2212 parsimony informative sites) were detected from 789,333 sites. A maximum likelihood phylogeny (un-rooted tree) was constructed. All 22F-ST433 isolates can be divided into two sub-clusters: clade A and clade B (Figure 3). In clade A, five isolates that confer resistance to macrolides (both erythromycin and azithromycin) are closely related to each other, forming a sub-cluster clade A1. All isolates in clade B are susceptible to macrolides and other antibiotics, except for isolate SP194, possessing resistance to levofloxacin (MIC 4 μg/mL) and moxifloxacin (MIC 2 μg/mL). Further comparison of quinolone resistance determinant regions (QRDR) between quinolone-resistant and sensitive strains found a non-synonymous mutation on gyrA gene (S81F) only.
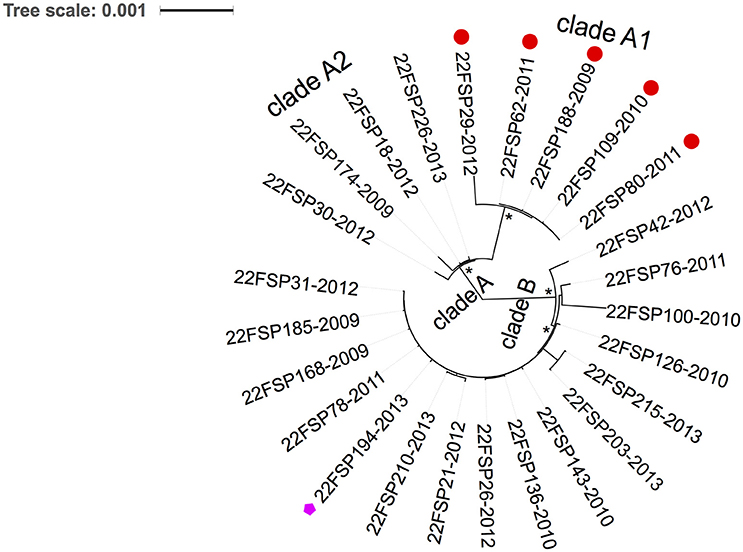
Figure 3. Phylogenetic analysis of emerging 22F-ST433 isolates from Ontario, Canada between 2009 and 2013. An un-rooted maximum-likelihood tree based on the core genome with putative recombination sites excluded. Two clades are revealed by the tree: clade A and clade B. Clade A include two sub-clades: clade A1 (five isolates) and clade A2 (four isolates). All five isolates in clade A1 were resistant to macrolides (shown with red circle at the tip), and one isolate from clade B was resistant to fluoroquinolones (pink pentagon at the tip). * indicates100% bootstrap support. Each isolate at the tip was labeled as: Serotype-isolate ID-year of isolation.
Accessory Genome Comparison Discloses Clade-Specific Genes in 22F
Accessory genomes of clade A and clade B isolates of 22F were compared, and presence or absence of accessory genes in each group was determined. A total of 39 genes were unique to clade A (100% presence in this group, but absent in the other), while clade B had 8 clade-specific genes (Table 3). Thirteen genes (combined length of 22,515 bp) unique to clade A were contiguous to each other (loci SP18.166056.587-610 and SP18.166056.1991-98) in the genome and encoded for a variety of bacteriophage proteins. In addition, all clade A strains possessed Type I restriction system and PhD/Doc toxin-antitoxin (TA) system in their genomes (PhD-Doc locus is surrounded by Type I restriction system and integrase). Five (55.6%) clade A isolates that form clade A1 subclade contained a gene cassette of 5163 bp (loci SP29.152236.85-92) that encode for macrolide resistance determinants mefE and msrD. The 5.1 kb gene cassette shared 96% similarity with mega (macrolide resistance genetic assembly) coding sequences upstream of tetM in the Tn2009 transposon previously described by Del Grosso et al. (2004). All isolates in clade B contained genes encoding for CAAX amino protease, putative UDP-glucose 6-dehydrogenase (loci SP21.166057.763-767) and several hypothetical proteins that clade A lacked.
Discussion
In a previous study we used the Sanger-sequencing based MLST method for the molecular typing of S. pneumoniae isolates from Ontario children (Deng et al., 2015). However, MLST typing is time consuming and very labor intensive. For example, to type 100 pneumococcal isolates using seven gene loci, 700 PCR amplifications plus 1400 Sanger sequencing reactions (forward and reverse direction) are required. Additional repeated sequencing is often needed for ambiguous bases in a sequence due to a variety of reasons such as poor DNA quality and contaminations. Compared to MLST and PFGE, WGS has the highest discriminatory power and has great potential for large-scale typing of bacterial isolates (Joensen et al., 2014). In this surveillance study, we used WGS for rapid determination of sequence type and antibiotic resistance for IPD isolates submitted to a public health laboratory. Illumina high-throughput genome sequencing combined with multiplexing allows rapid sequencing of hundreds of samples in a short period of time (1–5 days in rapid run mode). Our results suggest that MLST type could be accurately predicted from Illumina short reads directly for S. pneumoniae, provided genome sequences have fairly good sequencing depth (at least 10 or better). The overall sensitivity and specificity of genomic prediction of antibiotic resistance for S. pneumoniae were 95% (95% CI, 90 to 97%), and 100% (95% CI, 99 to 100%) respectively, compared to standard susceptibility testing methods. Ninety-seven percent of samples had complete concordance between genotype and phenotype for the antimicrobial agents tested, with low rates of VME (0.55%) and ME (0.22%). These results suggest that WGS prediction of antibiotic resistance mechanisms acquired through horizontal gene transfer events was as sensitive and specific as routine antimicrobial susceptibility assays for pneumococci. However, it is worth noting that only four antibiotics were tested in this study, and our observations in pneumococci may not apply to other bacterial pathogens. In addition, this WGS method is currently unable to reliably predict some resistance mechanisms due to point mutations on gene loci. For example, penicillin resistance among pneumococcal isolates resulting from mutations in penicillin-binding proteins (PBPs) or non-PBP genes cannot be predicted. Although, current phenotypic susceptibility assays are likely to be more reliable for routine testing, WGS methods may be useful for slow growing organisms, or organisms that cannot be cultured.
We have detected some VME and ME errors when comparing WGS methods with standard phenotypic susceptibility tests. Possible explanations for VMEs (genotype-susceptible but phenotype-resistant) may be sequencing error, alternative resistance mechanisms that are not reflected in the resistance gene database and novel resistance mechanisms not previously described. MEs (phenotypically susceptible isolates that contain resistance genes) may be the result of suppressed or delayed gene expression of resistance genes, since expression of a gene is a complex process that involves the interactions between repressors, promoters and other regulatory networks.
In our previous study we reported the increase of 22F prevalence among children (≤ 5 years of age) in Ontario in the post-PCV13 era (Deng et al., 2015). In the present study a significant increase in 22F-associated IPD cases was observed in older adults in Ontario (≥ 50 years old) after the introduction of PCV13. In 2013, 22F was the most common serotype detected in IPD among all ages in Ontario, Canada. The emergence of 22F in the post-vaccine era was also reported in other regions of Canada (Demczuk et al., 2013), and in other countries such as the USA (Kendall et al., 2016), UK (Pichon et al., 2013), Germany (van der et al., 2015), and Norway (Steens et al., 2015). An extensive review of serotype 22F in a MLST database found that it was broadly distributed in over 25 countries and the earliest report of IPD caused by 22F was in 1993 (MLST Databases, 2016). According to the public MLST database, 22F capsular type is mainly associated with the MLST-defined clonal complex CC433 (over 70%), ST1294 (3.3%) and CC698 (3.1%) (MLST Databases, 2016). Although, 24/25 (96%) Ontario 22F isolates mostly shared identical genotype (ST433), WGS-based phylogenetic analysis divided the 22F population into two distinct clusters (clade A and B in Figure 2) that were diverse in their genomic contents. The discovery of clade-specific gene sets by comparative genomics suggests the diversification of 22F-ST433 clone. Interestingly, all strains in clade A shared a 23kb-prophage-associated genomic island that clade B lacked. This chromosomal island comprises 30 genes including lysozyme genes downstream from the phage minor tail protein gene. To combat the bacteriophage invasion, clade A strains possessed anti-phage weaponry of Type-I restriction system that degrades phage DNA (Loenen et al., 2014) and PhD/Doc TA system which promotes cell death and limits phage replication within bacterial populations (Fineran et al., 2009). In addition, clade A1 (5 isolates) acquired macrolide resistance through incorporation of a 5.1 kb mega gene cassette that encodes for mefE and msrD resistance genes possibly via horizontal gene transfer. Antibiotic resistance obtained by 22F might provide selective advantage for them over other drug-susceptible competitors. In the current study 19.2% of 22F isolates carried this macrolide-resistant gene cassette; further dissemination of drug resistance among 22F should be closely monitored. 22F isolates in clade B possessed CAAX proteases that may be virulence-related (Firon et al., 2013) and their exact functions are unknown. One strain in clade B, isolated in 2013, demonstrated fluoroquinolone resistance through a de novo gyrA gene mutation (S81F). These gene differences between distinct clades suggest ongoing evolution and divergence of 22F clone through genetic recombination possibly due to rapid adaptation to environmental stress and selection pressure.
In conclusion, our study suggests that WGS is a valuable tool for routine molecular typing of pneumococcal isolates in the clinical laboratory, including antibiotic resistance prediction. The emergence of 22F in Ontario, Canada and its population divergence warrants heightened surveillance. Before routine use of WGS within clinical laboratories, certain barriers need to be addressed such as sequencing quality assurance, the need for automated WGS data analysis tools, and a shortage of qualified personnel with the necessary skills for data interpretation.
Author Contributions
NM prepared DNA library for genome sequencing. XD performed data analysis and prepared the manuscript. ST and TA performed bioinformatic analysis. MI performed geographic mapping of isolates. TM helped data analysis and manuscript writing. NF and JG designed and supervised this project.
Funding
This study was supported by Pfizer, Canada and Public Health Ontario.
Conflict of Interest Statement
JG has received research grants from GlaxoSmithKline Inc., and Hoffman-La Roche Ltd., to study antiviral resistance in influenza, and from Pfizer Inc., to conduct surveillance of Streptococcus pneumoniae. TM has served on advisory boards for Pfizer Inc., and Merck Inc. All other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dax Toti at the Genome Sequencing Center at University of Toronto and Aimin Li at the Genome Core of Public Health Ontario (PHO) for genome sequencing. We also thank the staff at National Microbiology Laboratory of Canada for serotyping Streptococcus pneumoniae isolates.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.02099/full#supplementary-material
Figure S1. Geographic distribution of invasive pneumonia disease cases among Ontario older adults (≥50 years of age) between 2009 and 2013.
Table S1. List of acquired genes predicted in silico from genome sequences that correlates with phenotype resistance.
Table S2. Details of antibiotic resistance discrepancies between phenotype test and genotype prediction.
References
Alexeyenko, A., Tamas, I., Liu, G., and Sonnhammer, E. L. (2006). Automatic clustering of orthologs and inparalogs shared by multiple proteomes. Bioinformatics 22, e9–e15. doi: 10.1093/bioinformatics/btl213
An Advisory Committee Statement (ACS) (2013). National Advisory Committee on Immunization (NACI). Statement on the Use of Conjugate Pneumococcal Vaccine - 13 Valent in Adults (Pneu-C-13). Can. Commun. Dis. Rep. 39(ACS-5), 1–54.
Athey, T. B., Teatero, S., Li, A., Marchand-Austin, A., Beall, B. W., and Fittipaldi, N. (2014). Deriving group A Streptococcus typing information from short-read whole-genome sequencing data. J. Clin. Microbiol. 52, 1871–1876. doi: 10.1128/JCM.00029-14
Ausina, V., Coll, P., Sambeat, M., Puig, I., Condom, M. J., Luquin, M., et al. (1988). Prospective study on the etiology of community-acquired pneumonia in children and adults in Spain. Eur. J. Clin. Microbiol. Infect. Dis. 7, 342–347. doi: 10.1007/BF01962334
Croucher, N. J., Harris, S. R., Fraser, C., Quail, M. A., and Burton, J. (2011). Rapid pneumococcal evolution in response to clinical interventions. Science 331, 430–435. doi: 10.1126/science.1198545
Del Grosso, M., Scotto d'Abusco, A., Iannelli, F., Pozzi, G., and Pantosti, A. (2004). Tn2009, a Tn916-like element containing mef(E) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 48, 2037–2042. doi: 10.1128/AAC.48.6.2037-2042.2004
Demczuk, W. H., Griffith, A., and Singh, R (2013). National Surveillance of Invasive Streptococcus pneumoniae, Streptococcus pyogenes and Streptococcus agalactiae -Annual summary. Winnipeg: Streptococcus and STI Unit, National Microbiology Laboratory, Public Health Agency of Canada. Available online at: http://www.healthycanadians.gc.ca/publications/drugs-products-medicaments-produits/2013-streptococcus/index-eng.php. (Accessed October 1, 2015)
Deng, X., Arya, G., Memari, N., Mackenzie, R., MacMullin, G., Low, D. E., et al. (2015). Genetic analysis of invasive pneumococcal isolates from children in Ontario, Canada, 2007-2012. Pediatr. Infect. Dis. J. 34, 594–598. doi: 10.1097/INF.0000000000000697
Desai, S., McGeer, A., and Quach, T. C. (2010). Update on the use of conjugate pneumococcal vaccines in childhood. Can. Commun. Dis. Rep. 36(ACS-12), 1–21.
Duvvuri, V. R., Deng, X., Teatero, S., Memari, N., Athey, T., Fittipaldi, N., et al. (2016). Population structure and drug resistance patterns of emerging non-PCV-13 Streptococcus pneumoniae serotypes 22F, 15A, and 8 isolated from adults in Ontario, Canada. Infect. Genet. Evol. 42, 1–8. doi: 10.1016/j.meegid.2016.04.007
Fineran, P. C., Blower, T. R., Foulds, I. J., Humphreys, D. P., Lilley, K. S., and Salmond, G. P. (2009). The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. U.S.A. 106, 894–899. doi: 10.1073/pnas.0808832106
Firon, A., Tazi, A., Da Cunha, V., Brinster, S., Sauvage, E., Dramsi, S., et al. (2013). The Abi-domain protein Abx1 interacts with the CovS histidine kinase to control virulence gene expression in group B Streptococcus. PLoS Pathog. 9:e1003179. doi: 10.1371/journal.ppat.1003179
Golden, A. R., Adam, H. J., Gilmour, M. W., Baxter, M. R., Martin, I., Nichol, K. A., et al. (2015). Assessment of multidrug resistance, clonality and virulence in non-PCV-13 Streptococcus pneumoniae serotypes in Canada, 2011-13. J. Antimicrob. Chemother. 70, 1960–1964. doi: 10.1093/jac/dkv061
Gordon, N. C., Price, J. R., Cole, K., Everitt, R., Morgan, M., Finney, J., et al. (2014). Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J. Clin. Microbiol. 52, 1182–1191. doi: 10.1128/JCM.03117-13
Inouye, M., Conway, T. C., Zobel, J., and Holt, K. E. (2012). Short read sequence typing (SRST): multi-locus sequence types from short reads. BMC Genomics 13:338. doi: 10.1186/1471-2164-13-338
Joensen, K. G., Scheutz, F., Lund, O., Hasman, H., Kaas, R. S., Nielsen, E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13
Kellner, J. D., Vanderkooi, O. G., MacDonald, J., Church, D. L., Tyrrell, G. J., and Scheifele, D. W. (2009). Changing epidemiology of invasive pneumococcal disease in Canada, 1998–2007: update from the Calgary-area Streptococcus pneumoniae research (CASPER) study. Clin. Infect. Dis. 49, 205–212. doi: 10.1086/599827
Kendall, B. A., Dascomb, K. K., Mehta, R. R., Stockmann, C., Mason, E. O., Ampofo, K., et al. (2016). Early Streptococcus pneumoniae serotype changes in Utah adults after the introduction of PCV13 in children. Vaccine 34, 474–478. doi: 10.1016/j.vaccine.2015.12.010
Letunic, I., and Bork, P. (2011). Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478. doi: 10.1093/nar/gkr201
Loenen, W. A., Dryden, D. T., Raleigh, E. A., and Wilson, G. G. (2014). Type I restriction system and their relatives. Nucleic Acids Res. 42, 20–44. doi: 10.1093/nar/gkt847
Maiden, M. C., Bygraves, J. A., Feil, E., Morelli, G., Russell, J. E., Urwin, R., et al. (1998). Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U.S.A. 95, 3140–3145. doi: 10.1073/pnas.95.6.3140
Marttinen, P., Hanage, W. P., Croucher, N. J., Connor, T. R., Harris, S. R., Bentley, S. D., et al. (2012). Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res. 40:e6. doi: 10.1093/nar/gkr928
MLST Databases (2016). Streptococcus pneumoniae, MLST Databases. University of Oxford. Available online at: http://pubmlst.org/spneumoniae (Accessed February 1, 2016).
O'Brien, K. L., Millar, E. V., Zell, E. R., Bronsdon, M., Weatherholtz, R., Reid, R., et al. (2007). Effect of pneumococcal conjugate vaccine on nasopharyngeal colonization among immunized and unimmunized children in a community-randomized trial. J. Infect. Dis. 196, 1211–1220. doi: 10.1086/521833
O'Brien, K. P., Remm, M., and Sonnhammer, E. L. (2005). Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 33, D476–D480. doi: 10.1093/nar/gki107
Pichon, B., Ladhani, S. N., Slack, M. P., Segonds-Pichon, A., Andrews, N. J., Waight, P. A., et al. (2013). Changes in molecular epidemiology of Streptococcus pneumoniae causing meningitis following introduction of pneumococcal conjugate vaccination in England and Wales. J. Clin. Microbiol. 51, 820–827. doi: 10.1128/JCM.01917-12
Sabat, A. J., Budimir, A., Nashev, D., Sá-Leão, R., van Dijl, J., Laurent, F., et al. (2013). Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Euro Surveill. 18:20380.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Steens, A., Caugant, D. A., Aaberge, I. S., and Vestrheim, D. F. (2015). Decreased carriage and genetic shifts in the streptococcus pneumoniae population after changing the seven-valent to the thirteen-valent pneumococcal vaccine in Norway. Pediatr. Infect. Dis. J. 34, 875–883. doi: 10.1097/INF.0000000000000751
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239.
Tomczyk, S., Bennett, N. M., Stoecker, C., Gierke, R., Moore, M. R., Whitney, C. G., et al. (2014). Use of 13-Valent pneumococcal conjugate vaccine and 23-valent pneumococcal polysaccharide vaccine among adults aged ≥65 years: recommendations of the advisory committee on immunization practices (ACIP). MMWR Morb. Mortal Wkly. Rep. 63, 822–826.
van der, L. M., Falkenhorst, G., Perniciaro, S., and Imöhl, M. (2015). Effects of infant pneumococcal conjugate vaccination on serotype distribution in invasive pneumococcal disease among children and adults in germany. PLoS ONE 10:e0131494. doi: 10.1371/journal.pone.0131494
Walker, T. M., Kohl, T. A., Omar, S. V., Hedge, J., Del Ojo Elias, C., Bradley, P., et al. (2015). Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect. Dis. 15, 1193–1202. doi: 10.1016/S1473-3099(15)00062-6
Zerbino, D. R., and Birney, E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829. doi: 10.1101/gr.074492.107
Keywords: invasive pneumococcal disease, whole-genome sequencing, emerging 22F, antibiotic resistance prediction, genomic diversification
Citation: Deng X, Memari N, Teatero S, Athey T, Isabel M, Mazzulli T, Fittipaldi N and Gubbay JB (2016) Whole-genome Sequencing for Surveillance of Invasive Pneumococcal Diseases in Ontario, Canada: Rapid Prediction of Genotype, Antibiotic Resistance and Characterization of Emerging Serotype 22F. Front. Microbiol. 7:2099. doi: 10.3389/fmicb.2016.02099
Received: 17 August 2016; Accepted: 12 December 2016;
Published: 27 December 2016.
Edited by:
Eric Altermann, AgResearch, New ZealandReviewed by:
Suleyman Yildirim, Istanbul Medipol University International School of Medicine, TurkeyTodd Kitten, Virginia Commonwealth University, USA
Copyright © 2016 Deng, Memari, Teatero, Athey, Isabel, Mazzulli, Fittipaldi and Gubbay. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianding Deng, d2F5bmUuZGVuZ0BtYWlsLnV0b3JvbnRvLmNh
†These authors have contributed equally to this work.
 Xianding Deng
Xianding Deng Nader Memari1†
Nader Memari1† Sarah Teatero
Sarah Teatero