- 1Équipe Pathogènes Hydriques Santé Environnements, UMR 5569 HSM, Université de Montpellier, Montpellier, France
- 2Département d'Hygiène Hospitalière, Centre Hospitalier Régional Universitaire (CHRU) de Montpellier, Montpellier, France
- 3Département de Bactériologie, Centre Hospitalier Universitaire (CHU) de Nice, Nice, France
Bacteria of the genus Aeromonas display multicellular behaviors herein referred to as “social life”. Since the 1990s, interest has grown in cell-to-cell communication through quorum sensing signals and biofilm formation. As they are interconnected, these two self-organizing systems deserve to be considered together for a fresh perspective on the natural history and lifestyles of aeromonads. In this review, we focus on the multicellular behaviors of Aeromonas, i.e., its social life. First, we review and discuss the available knowledge at the molecular and cellular levels for biofilm and quorum sensing. We then discuss the complex, subtle, and nested interconnections between the two systems. Finally, we focus on the aeromonad multicellular coordinated behaviors involved in heterotrophy and virulence that represent technological opportunities and applied research challenges.
Introduction
The practices of clinical bacteriology and research in microbiology have long been subjected to the principle of “pure strains”, which limited analysis to unicellular/monoclonal organisms. However, the multicellular/polyclonal lifestyle becomes increasingly important for understanding bacteria, as strengthened by the sociomicrobiology aspects of biofilm formation and quorum sensing (Parsek and Greenberg, 2005; Claessen et al., 2014). These biological mechanisms are particularly studied in the environmental opportunistic pathogen, Pseudomonas aeruginosa and have been confirmed as major virulence factors explaining aspects of P. aeruginosa pathogenesis, mainly in cystic fibrosis and health-care associated infections (Bjarnsholt et al., 2010). Rather than true virulence factors, biofilm formation and quorum sensing are adaptive traits involved in the versatile lifestyle of P. aeruginosa in natural ecosystems, which become patho-adaptive in human and health-care ecosystems.
Aeromonads represent another interesting group of bacteria for such multicellular functioning. The genus Aeromonas belongs to Aeromonadaceae family, Aeromonadales order and Gammaproteobacteria class. These bacteria are gram-negative, facultative anaerobic, oxidase and catalase positive, fermentative, and mostly motile bacilli. Aeromonads are common inhabitants of aquatic environments such as fresh, estuarine, marine waters, and sediments and are found in association with animals. Aeromonas are environmental opportunistic pathogens of animals and human. Aeromonads are responsible for furunculosis and septicemia in fish. In human, they can cause gastroenteritidis, wound infections, bacteraemia, and less frequently respiratory infections, hepatobiliary infections, peritonitis, urinary tract infections, and ocular infections (Janda and Abbott, 2010). Among the 30 species recognized to date in this genus, the most studied are A. dhakensis, A. hydrophila, A. caviae, A. veronii, and A. salmonicida, which correspond to relevant species for human and animal infections. The members of Aeromonas are characterized by a remarkably ability to colonize a wide range of habitats. Typically, many of its colonization aspects rely on biofilm production and cell-cell signaling. Numerous studies have been conducted on these two aspects, and a large amount of data is available but mostly scattered in the literature. These data have never been collected into an integrative perspective of community dynamics.
In this review, we focus on the multicellular behavior of Aeromonas, referred to here as “social life.” First, we review and discuss the available knowledge at the molecular and cellular levels for biofilm and quorum sensing. We then discuss the complex, subtle, and nested interconnections between the two systems and highlight the current gaps in knowledge. Finally, we focus on aeromonad multicellular coordinated behaviors involved in heterotrophy and virulence that represent applied research challenges and technological opportunities.
Biofilm Formation in Aeromonas
A majority of bacteria, including aeromonads, live attached to biofilms on biotic or abiotic surfaces. Natural biofilms are developed and differentiate themselves to build a packed community that is often multi-species and is embedded in a polymeric extracellular matrix of their own production. This matrix contains channels that are included for the circulation of nutrients and water (Donlan and Costerton, 2002). The architecture of bacterial biofilms has largely been documented as confocal scanning laser microscopes have come into use (Donlan and Costerton, 2002). Like P. aeruginosa (Sauer et al., 2002; Klausen et al., 2006), the natural history of biofilm formation in aeromonads includes the classical steps of attachment, microcolony formation, maturation, and dispersion (Figure 1).
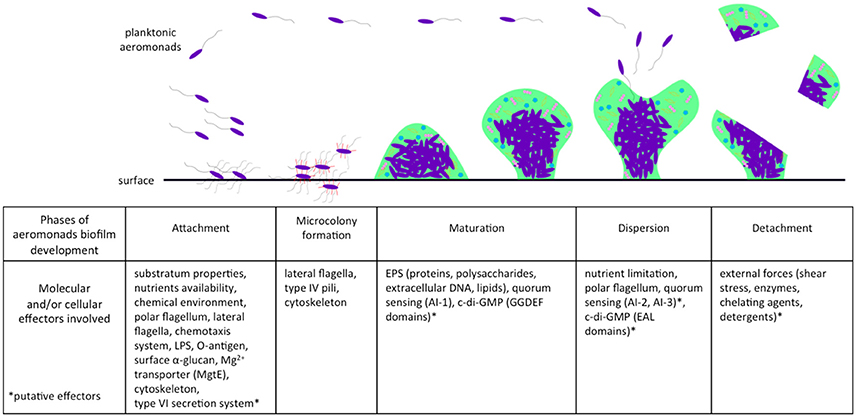
Figure 1. Effectors involved in different phases of biofilm development in aeromonads. Planktonic aeromonads initiate the formation of biofilm on surface under influence of environmental conditions. Several bacterial factors are involved in the attachment step, including flagella and other external structures, chemotaxis system, and cytoskeleton. After division, bacteria that were well-aggregated, attached to the surface to form a microcolony. Biofilm acquires its mechanical stability by the production of an EPS matrix encompassing proteins, polysaccharides, extracellular DNA, and lipids. The AI-1 quorum sensing system enhances the maturation of biofilm, which is likely related to the second messenger c-di-GMP involved in the bacterial transition from planktonic to sessile lifestyle. When the conditions of life in biofilm deteriorate (e.g., nutrient limitation), a dispersion phase occurs and aeromonads escape from biofilm and return to the planktonic lifestyle. In another case, the biofilm can be detached by external stress (e.g., shear forces). AI-1, Autoinducer-1 quorum sensing system; AI-2, Autoinducer-2 quorum sensing system; AI-3, Autoinducer-3 quorum sensing system; EAL, protein domains harboring phosphodiesterase activity involved in the c-di-GMP degradation; EPS, extracellular polymeric substances; GGDEF, protein domains harboring guanylate synthase activity involved in the c-di-GMP synthesis; LPS, lipopolysaccharides.
Attachment and Promoting Factors
This first step, attachment, is pivotal for biofilm formation (Figure 1). Aeromonads are able to colonize both biotic surfaces in plants and animals (Mizan et al., 2015), and abiotic surfaces, notably sediment, steel, glass, and polyvinyl chloride (Zalmum et al., 1998; Béchet and Blondeau, 2003; Bomo et al., 2004; Doğruöz et al., 2009; Balasubramanian et al., 2012). The substratum properties, chemical components, and nutrient availability are critical conditions influencing bacterial attachment. For instance, Jahid et al. (2013, 2015) have shown that low salinity (0.25% wt./vol.) enhances biofilm formation by Aeromonas hydrophila, whereas glucose concentration above 0.05% (wt./vol.) impairs its formation. In addition, Aeromonas spp. harbor several structures and/or mechanisms, including flagella and chemotaxis, lipopolysaccharides (LPS), and other surface polysaccharides (α-glucan), Mg2+ transporters and cytoskeletons that are actively involved in the first steps of biofilm formation (Figure 1).
Motility is decisive for attachment, and any system that promotes motility may stimulate attachment. Among these systems, the constitutive polar flagellum of Aeromonas spp., responsible for swimming in liquid, plays a critical role in biofilm formation and contributes to colonization of surfaces, as demonstrated for Aeromonas caviae strain Sch3 and A. hydrophila {A. piscicola} strain AH-3 (Kirov et al., 2004; Merino et al., 2014). In A. hydrophila, the O-glycosylation of polar flagella seems to be a prerequisite for adhesion and biofilm formation because mutants with reduced flagella glycosylation are unable to form biofilms (Merino et al., 2014; Fulton et al., 2015). In addition to polar flagella, members of Aeromonas spp. display inducible lateral flagella distributed randomly on the cell surface (Kirov et al., 2002). These lateral flagella are responsible for the swarming motility, enabling bacteria to migrate over surfaces by rotative movements and the formation of side-by-side cell groups called rafts (Gavín et al., 2002; Kirov et al., 2002). They also contribute to biofilm formation for Aeromonads (Gavín et al., 2002, 2003). Similarly, swimming, swarming, and twitching motility are known to be pivotal for P. aeruginosa biofilm formation (Barken et al., 2008), but Aeromonas strains do not develop any detectable twitching motility (Kirov et al., 1999). Chemotaxis systems mediated by the histidine kinase CheA allow bacterial cells to navigate in chemical gradients by regulating bacterial flagellar motility and are necessary for swimming, swarming, and for biofilm formation (Porter et al., 2011). Consistent with this, a chemotactic mutant strain ΔcheA of A. caviae is unable to swim or swarm in agar assays, and its biofilm formation ability is decreased by more than 80% (Kirov et al., 2004). Ten clusters of chemotaxis genes have been described in the genome of A. hydrophila ATCC 7966T, including two gene-system homologs to the gene clusters I and V of P. aeruginosa PAO1 (Wuichet and Zhulin, 2010) shown to be essential for chemotactic motility (Masduki et al., 1995). The VgrG proteins corresponding to both components and effectors of the type VI secretion system (T6SS) of the A. hydrophila {A. dhakensis} strain SSU promote biofilm formation. Their function is not fully characterized but may occur at the attachment step because VgrG3 also enhances swimming motility (Sha et al., 2013).
The O-antigen is the most surface-exposed moiety of LPS, which acts as an attachment factor and enhances the formation of biofilm in A. hydrophila strains (Merino et al., 2014; Fulton et al., 2015). The cell surface hydrophobicity and charge conferred by LPS were involved in P. aeruginosa biofilm formation (Ruhal et al., 2015), but their exact roles in the Aeromonas biofilm development are not fully understood. The A. hydrophila surface α-glucan independent of the LPS also improves biofilm formation (Merino et al., 2012). In addition, Mg2+ is suspected to contribute to the integrity and stability of the outer membrane by preventing electrostatic repulsion between LPS molecules (Hancock, 1984). This divalent cation and its transporter MgtE are directly involved in adherence to epithelial cells, in swarming and in biofilm formation of A. hydrophila {A. piscicola} AH-3 (Merino et al., 2001). Genomic data confirm the links between MgtE and cell motility because the gene mgtE is adjacent to the polar flagellar operon flg in the strain A. hydrophila ATCC 7966T (Seshadri et al., 2006). However, the level of evidence is still low for understanding how MgtE enhances biofilm formation. Finally, the protein MinD, a cytoskeletal ATPase involved in the septal placement of bacteria and plastid division sites in many bacteria (Shih and Rothfield, 2006), is also markedly involved in adherence, bacterial motility, and formation of biofilm in the A. hydrophila strain W (Huang et al., 2015).
Microcolony Formation and Maturation
Once attachment is completed, cell division processes maintain bacterium to bacterium bonds and lead to microcolony formation (Figure 1; Lynch et al., 2002). Type IV pili and fimbriae participate in the formation of microcolonies and biofilm for strains of Aeromonas (Béchet and Blondeau, 2003; Kozlova et al., 2008). Three families of type IV pili structures (Bfp, Flp, and Tap) have been involved in microcolony and biofilm formation for several species, e.g., Aggregibacter actinomycetemcomitans and Vibrio vulnificus (Paranjpye and Strom, 2005; Perez-Cheeks et al., 2012). Bacteria of the genus Aeromonas also harbor the three families of type IV pili structures (Kirov et al., 1999; Boyd et al., 2008). Bfp has been shown to be critical for biofilm formation in Aeromonas veronii (Hadi et al., 2012), but the involvement of Flp and Tap has not yet been demonstrated in aeromonads. Further evaluation deserves to be conducted to more precisely specify the role of each type of pili in early steps of Aeromonas biofilm formation.
From microcolonies, colonies grow and the biofilm matures. After 48 h in a stainless steel flow-through model, Lynch et al. (2002) observed mushroom-like “large microcolonies” of A. hydrophila, characteristic of mature biofilms (Figure 1). Aeromonads live embedded in a self-produced matrix of extracellular polymeric substances (EPS), mainly composed of polysaccharides, proteins, nucleic acids, and lipids (Andersson et al., 2011). The resulting structure provides the mechanical stability of biofilms (Peterson et al., 2015). When forming biofilms, A. hydrophila produces more capsular and colloidal EPS compared to planktonic cells (Castro et al., 2014). As in other bacteria including P. aeruginosa (Rasamiravaka et al., 2015), the persistence of a mature biofilm in Aeromonas is largely controlled by signaling systems triggered by high bacterial density (Lynch et al., 2002; Khajanchi et al., 2009). These aspects are developed in the section “How are biofilm and quorum sensing interconnected?”
Detachment and Dispersion
Detachment corresponds to passive escape from a biofilm, occurring under the influence of external factors such as shear stress or degradation of extracellular polymeric matrix by enzymes, chelating agents, or detergents. In contrast, dispersion is an active mechanism of biofilm escape depending on biofilm growth, cell density, and related factors (Figure 1; Petrova and Sauer, 2016). Very few data on Aeromonas biofilm detachment and dispersion are available, and most knowledge relies on data from P. aeruginosa biofilm models. Dispersion is triggered by exogenous factors such as nutrient availability and toxic compounds, and by internal regulatory systems including quorum sensing systems (Kim and Lee, 2016). To enhance its dispersion, it is suspected that aeromonads degrade some compounds of their own extracellular polymeric matrix or other bacteria. Consistent with this, Bansal et al. (2015) showed that the depolymerase produced by an Aeromonas punctata strain is able to degrade the capsular polysaccharides of Klebsiella pneumoniae within a biofilm.
Functions of Biofilms
Biofilm Acts as Niche and Reservoir
Biofilm formation is an emblematic example of niche construction, a process by which an organism alters its own environment in order to increase its chances of survival (Odling-Smee et al., 2003). Indeed, biofilms enhance stability and protect bacteria against external factors (Costerton et al., 1987; Peterson et al., 2015). First, bacterial sessile life is associated with an increased persistence and resistance to stressful conditions including salinity, antimicrobial substances, or oxidative stress, compared to the planktonic lifestyle (Van Acker et al., 2014). Second, biofilms provide cell nutrients in higher concentrations than the surrounding environment via the nutrient-rich solute retained in the interstitial region of the extracellular polymeric matrix (Tsuchiya et al., 2009, 2016).
The formation of biofilm is highly beneficial even if Aeromonas spp. are able to grow and live freely in water. Biofilms act as reservoirs in which some aeromonads are able to persist for several years and emerge later in favorable conditions (Kühn et al., 1997). Only certain clones seem to be able to persist; Villari et al. (2003) found that only two clones of A. hydrophila and A. caviae persisted in natural mineral freshwater over a 3-year study, while molecular heterogeneity was much higher in samples from stream waters running near the spring.
Aeromonads within a biofilm are more resistant to disinfectants than planktonic cells, as shown for A. hydrophila strains (Jahid and Ha, 2014). Aeromonads have thus been recovered from biofilm in drinking-water distribution systems (Chauret et al., 2001; September et al., 2007), even when water supply is chlorinated (Fernández et al., 2000).
Biofilm Promotes Gene Exchange and Antibiotic Resistance
Biofilm structure provides a close cell-to-cell proximity that enhances genetic transfers, mainly conjugation and natural transformation (Hausner and Wuertz, 1999; Hendrickx et al., 2003; Madsen et al., 2012). The two types of biofilm-associated horizontal genetic transfers (HGT) have been demonstrated in the genus Aeromonas (Rhodes et al., 2000; Huddleston et al., 2013), but the transfer by phage transduction has not yet been observed within aeromonads biofilm.
In the genus Aeromonas, conjugation has even been demonstrated in experiments with aeromonads as donor cells and E. coli as recipient cells (Rhodes et al., 2000; Schmidt et al., 2001; Casas et al., 2005). Aeromonas can harbor the machinery for the type IV secretion system (T4SS), enabling the genetic conjugative transfer of mobile genetic elements between bacteria (Rangrez et al., 2006). Within biofilms, high cell density may facilitate conjugation between two aeromonads or between aeromonads and other bacteria.
Additionally, Huddleston et al. (2013) have demonstrated, using direct experimental assays, that most members of aeromonads are naturally competent for transformation (73% of 37 tested strains). The type IV pili of Aeromonas may enable the recipient cell to incorporate extracellular DNA. The analysis of “transformability” and “donatability” between Aeromonas strains showed that transformation of groups was constrained to phylogroups (Huddleston et al., 2013), consistent with some population studies that have highlighted HGT between close relatives in the genus Aeromonas (Silver et al., 2011; Roger et al., 2012b). The high concentration of extracellular DNA within a biofilm may facilitate the transformation of Aeromonas, as in other bacteria capable of natural transformation (Merod and Wuertz, 2014).
Evidence is available for HGT of 16S rDNA and housekeeping genes over the evolutionary history of the Aeromonas genus (Roger et al., 2012a,b). Moreover, mobile genetic elements are frequently recovered from aeromonad genomes, e.g., plasmids, transposons, insertion sequences, and integron-associated gene cassettes, as recently reviewed (Piotrowska and Popowska, 2015). These HGT are important in the evolution and fitness of this genus and may be enhanced in biofilm, as in other bacteria (Madsen et al., 2012).
Mobile genetic elements recovered from Aeromonas strains carry genes involved in virulence (T4SS, T6SS, T3SS compounds, and effectors), stress response (HipAB toxin/antitoxin system) and resistance to heavy metals (mercury) and toxic compounds (quaternary ammoniums), but the most frequently reported elements are antibiotic resistance genes (Piotrowska and Popowska, 2015).
In Aeromonas, acquired resistance increases the level of antibiotic resistance in both environmental and clinical strains (Esteve et al., 2015). The genetic support of these acquired resistances is transferable by chromosomal transposons/integrons or plasmids that carry genes associated with resistance to beta-lactamines, quinolones, macrolides, tetracycline, sulfonamides, and chloramphenicol (Janda and Abbott, 2010; Piotrowska and Popowska, 2015). In vivo transfer of TEM-24 plasmid-borne extended-spectrum β-lactamase, likely from human microbiota, was reported from enterobacteria to Aeromonas (Marchandin et al., 2003). Such antibiotic-resistance gene transfer has not yet been demonstrated within aeromonad biofilms, but it was reported for K. pneumoniae (Hennequin et al., 2012). The co-localization of resistance genes in mobile genetic elements can lead to cross-resistance to multiple families of antibiotics in aeromonads (Maravić et al., 2013).
Consequently, there are some concerns about the spread of resistance genes by transfer events within microbial communities, including aeromonads in surface biofilms, both in natural environments and care units.
Mixed Biofilms
Under environmental conditions, biofilms often mix several bacterial species including Aeromonas sp., as reviewed in Table 1. Aeromonads have been co-isolated with one or more representatives of other genera from natural freshwater and seawater biofilms (Stine et al., 2003; September et al., 2007; Balasubramanian et al., 2012; Farkas et al., 2012; Marti et al., 2014). Aeromonas spp. are also observed in mixed-species biofilms inside dental care plastic lines (Tall et al., 1995) or the surfaces of food plants (Gunduz and Tuncel, 2006).
Bacterial biofilms harbor some inherent degree of structural heterogeneity in response to chemical gradients and adaptation to local microenvironments. Heterogeneity also concerns the mixture of bacterial species in a biofilm; juxtaposition of bacteria occurs such that mutualistic interactions are facilitated (Stewart and Franklin, 2008). Experimentally, the A. hydrophila strain AH-1 and Flavobacterium sp. form a mixed biofilm on chitin-containing particles. The A. hydrophila strain AH-1 is able to degrade chitin due to extracellular chitinases, and Flavobacterium sp. acts as a cheater that uses chitin degradation products as nutrients (Jagmann et al., 2012). The biofilm formation of Aeromonas was enhanced when co-cultured with a Bacillus cereus strain that improves aggregation between bacteria (Cheng et al., 2014). In addition to mutualism, competitive behavior was shown between aquatic bacteria organized in biofilms. For instance, A. hydrophila exhibits antagonism against L. pneumophila through the production of bacteriocine-like substances. This interfering effect enhanced the detachment of Legionella from biofilms, contributing to its dissemination (Guerrieri et al., 2008).
The medicinal leech Hirudo verbana is the natural host of the two symbiotic bacterial species A. veronii and Mucinovorans hirudinis (Graf, 1999; Worthen et al., 2006; Nelson et al., 2015). Synergy occurs between the two bacterial endosymbionts because A. veronii associated with M. hirudinis forms larger mixed microcolonies than each species alone (Kikuchi and Graf, 2007). The mechanisms involved in this synergy are not yet known, and it is unknown whether it could be controlled by QS signaling and whether bacterial cooperation could initiate or influence leech gut colonization in the two symbionts.
Quorum Sensing Systems in Aeromonas
Since its early description, intercellular communication via quorum signaling has gained a central place in bacterial sociobiology (Fuqua et al., 1994). Quorum sensing (QS) systems regulate a wide range of functions including bioluminescence, motility, extracellular virulence factors, and biofilm productions. Three different QS systems have been described in gram-negative bacteria, each composed of a sensor–autoinducer pair: type 1, type 2, and type 3 autoinducer systems.
Type 1 Autoinducer (AI-1) System
The AI-1 system was discovered within the LuxRI bioluminescent system in Vibrio fisheri (Engebrecht and Silverman, 1984) and is widespread among gram-negative bacteria including P. aeruginosa (Parsek et al., 1999). AI-1 signals are small molecules with a chemical structure based on N-acyl homoserine lactone (AHL), which is derived from common components of the bacterial metabolism, i.e., S-adenosyl methionine and acyl-acyl carrier proteins derived from fatty acid biosynthesis (Parsek et al., 1999). Depending on the species, the acyl chain length of AHLs varies from C4 to C18 and can be modified by unsaturation, methyl branches, and oxo- or hydroxyl substituents (Churchill and Chen, 2011).
AI-1 Signaling in Aeromonas
A schematic model of the Aeromonas AI-1 quorum sensing system is presented in Figure 2. The LuxI-type enzyme, known as AhyI in A. hydrophila and AsaI in A. salmonicida, synthetizes AHL molecules (Figure 2A; Swift et al., 1997). AHLs interact with a cytoplasmic homolog of protein LuxR in A. hydrophila called AhyR (Figure 2), which is a transcriptional regulator of target genes, including the gene that encodes AhyI (Figure 2; Swift et al., 1997, 1999). During the exponential phase, AhyR and AhyI interplay in an activation loop of the AI-1 system. This leads to an auto-amplification effect called “autoinduction” (Figure 2A). AHLs freely diffuse across cellular membranes in and out of the Aeromonas cell and accumulate in the bacterial cell environment (Figure 2A). At the stationary phase over an exogenous AHL concentration threshold, the autoinduction phenomenon is suppressed while intercellular activation (i.e., “intercellular communication”) occurs between two bacterial cells and is the only active phenomenon (Figure 2B), as shown in A. hydrophila (Garde et al., 2010). The expression of AhyI is growth phase-dependent. Indeed, Kirke et al. (2004) showed that AhyI is produced during the exponential phase, but not during the post-exponential phase, when it is instead degraded. In the exponential phase, AhyR up-regulates the expression of AhyI and enhances AHL production (Figure 2A). In contrast, AhyR inhibits the expression of AhyI during the post-exponential phase (Figure 2B; Kirke et al., 2004). In addition, the transcription of ahyRI is enhanced by the expression of AI-2 synthase LuxS and the AI-3 response regulator QseB (Figure 2A; Kozlova et al., 2012).
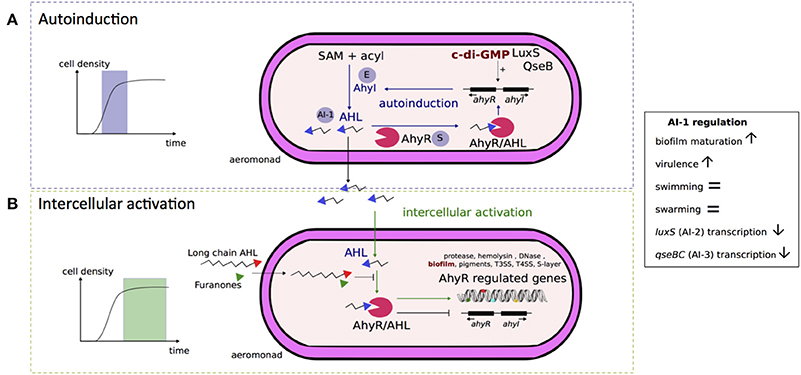
Figure 2. Schematic representation of AI-1 quorum sensing system in Aeromonas. From an in vitro model of autoinducer 1 (AI-1) quorum sensing system of A. hydrophila, Garde et al. (2010) have distinguished two phases since the complex AI-1/receptor (AhyR) activates the quorum sensing loop of the initial AI-1 producer bacterial cell during exponential growth (A) or of other bacterial cells during the stationary phase (B) due to slow decay of the complex AI-1/receptor (AhyR). (A) Autoinduction occurs during exponential growth phase (Garde et al., 2010). In this phase, the enzyme (E) AhyI synthetizes AI-1 signal molecules of acyl-homoserine lactones (AHL) from S-adenosyl-methionine (SAM) and acyl–acyl carrier proteins (acyl) (Swift et al., 1997; Parsek et al., 1999). The protein AhyR is the sensor (S) of the AI-1 system and is activated by AHL molecules (Swift et al., 1997). Once activated, AhyR is a transcriptional regulator for the ahyRI locus encompassing AhyI and AhyR encoding genes, and participates in the auto-amplification loop (Kirke et al., 2004; Garde et al., 2010). The transcription of ahyRI locus is also likely enhanced (discontinuous traits) by the second messenger c-di-GMP and by AI-2 synthase LuxS or by AI-3 transcriptional regulatory protein QseB (Kozlova et al., 2012). The AHL molecules are freely diffusible across bacterial membranes and accumulate in the extracellular environment (Garde et al., 2010). (B) Intercellular activation occurs over an AHL concentration threshold corresponding to high cell density occurring at the stationary phase (Garde et al., 2010). Once activated by AHL molecules, AhyR is a transcriptional regulator for several genes associated to virulence and biofilm formation. In contrast to its action during the autoinduction phase, activated-AhyR negatively regulates the transcription of the ahyRI locus (Kirke et al., 2004). This AI-1 system is inhibited by exogenous long chain AHL or furanones (Swift et al., 1999; Ponnusamy et al., 2010) that may act as competitive inhibitors of AHL for AhyR binding. The AI-1 quorum sensing system negatively regulates the transcription of luxS and qseBC loci, encoding AI-2 synthase and AI-3 two components system, respectively (Kozlova et al., 2011, 2012).
AI-1 QS system may differ among bacterial genera, and may involve several kinds of LuxRI homologs. For example, P. aeruginosa harbors two distinct LuxRI homologs, LasRI and RhlRI (Lee and Zhang, 2015). In Aeromonas, only one AI-1 system has been described and is virtually present in every Aeromonas strain because LuxRI homologs were detected in all 73 tested strains, covering the known diversity in the genus Aeromonas (Jangid et al., 2007). The genes ahyI and ahyR from the locus ahyRI encode this system in A. hydrophila and are transcribed divergently, together with an intergenic region of 62 bp (Swift et al., 1997; Kirke et al., 2004). Homologs for these genes were also identified in other Aeromonas species (e.g., A. salmonicida, Aeromonas molluscorum, A. veronii, A. media, and Aeromonas diversa). A putative binding site for AhyR was identified in the intergenic region 10 bp upstream of the ahyI promoter (Kirke et al., 2004).
As shown in other gram-negative bacteria (e.g., Agrobacterium, Erwinia), the activation of AI-1 systems modulates fitness and virulence in Aeromonas, as reviewed in Table 2. Overall, AI-1 system activation in Aeromonas is associated with enhancement of biofilm maturation (without effects on swimming and swarming; Lynch et al., 2002; Khajanchi et al., 2009) and virulence. Indeed, the AhyRI system enhances the expression of numerous virulence factors in Aeromonas, including production of exoenzymes such as metalloproteases, serine proteases, hemolysin, amylase, DNAse, S-layer production, and pigment production (Table 2; Swift et al., 1999; Bi et al., 2007; Khajanchi et al., 2009; Schwenteit et al., 2011). Concerning the type III secretion system, the effect of AI-1 is unclear because of conflicting data. Some works have observed that T3SS is up-regulated by the AhyRI system, but other works observed no effect on the production and translocation of the T3SS effector AexU (Table 2; Khajanchi et al., 2009; Vilches et al., 2009). Type 1 QS also regulates the switch between two metabolic pathways. For instance, to avoid lethal acidification of the medium in the late growth phase, Aeromonas are able to switch from mixed acid fermentation to butanediol fermentation that produces fewer acid compounds (Table 2; Van Houdt et al., 2007).
Type of AHL Produced
The different types of AHL produced by Aeromonas spp. strains, the methods of AHL identification and the origin of isolates are presented in Table 3. Culture supernatants of A. hydrophila AH-1 and A. salmonicida type strain NCIMB 1102T, as studied by bioassays and high-performance liquid chromatography (HPLC), showed that N-butanoyl homoserine lactone (C4-HSL), and N-hexanoyl homoserine lactone (C6-HSL) are the major autoinducers produced by aeromonads, and the ratio C4:C6 is 70:1 (Swift et al., 1997). In another study, all clinical tested strains of A. hydrophila (n = 20) and Aeromonas sobria (n = 2) produced either C4-HSL and C6-HSL or both, including 10 strains that produced an additional AHL, a putative N-pentanoyl homoserine lactone (C5-HSL; Chan et al., 2011). Other studies showed similar results, e.g., A. caviae YL12 (Table 3), although additional AHLs were not systematically characterized (Bruhn et al., 2005; Morgan-Sagastume et al., 2005; Medina-Martínez et al., 2006; Schwenteit et al., 2011; Chong et al., 2012; Huang et al., 2012; Chu et al., 2013; Ochiai et al., 2013; Lim et al., 2014; Zeng et al., 2014). The HPLC data confirmed results obtained with bioassays (Table 3). However, there were significant discrepancies in the AHL profiles between the HPLC and gas chromatography methods for A. salmonicida type strain (NCIMB1102T = ATCC 33658T) because AHL with longer chains were detected only by gas chromatography (Swift et al., 1997; Cataldi et al., 2007). Compared to Aeromonas, P. aeruginosa produces two types of HSL, N-(3-oxododecanoyl)-HSL, and C4-HSL (Lee and Zhang, 2015).
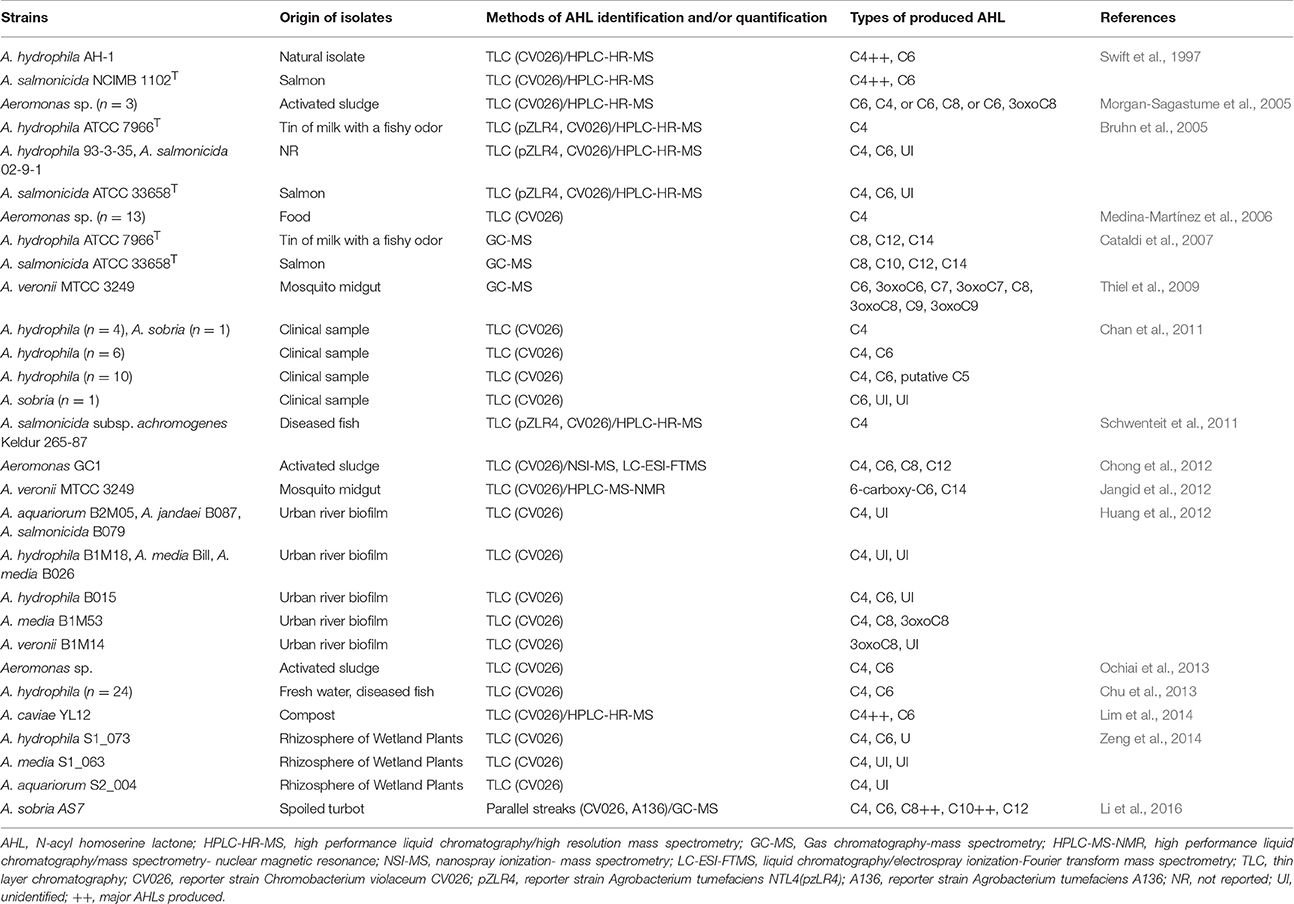
Table 3. Types of N-acyl homoserine lactone (AHL) produced by Aeromonas species from different isolation origins regarding the methods used for AHL analysis.
AI-1 Inhibitors
The Aeromonas AI-1 system is inhibited in vitro by exogenous AHLs harboring long chains, i.e., 10–14 carbons HSL, leading to decreased production of exoproteases (Swift et al., 1999). Consistently, the 3-oxo-C10-HSL produced by Vibrio anguillarum inhibits protease activities from A. salmonicida and A. hydrophila (Rasch et al., 2007). Similarly, the synthetic 2(5H)-furanone derived from the competitive inhibitor of AHL produced by the marine algae Delisea pulchra, exhibited QS inhibition activity against C4-HSL and C6-HSL, molecules usually produced by aeromonads (Ponnusamy et al., 2010).
Type 2 Autoinducer (AI-2) System
The AI-2 QS system was initially described in Vibrio harveyi to control the expression of bioluminescence in response to fluctuation of bacterial population density (Bassler et al., 1994; Surette et al., 1999). A putative schematic model of Aeromonas AI-2 quorum sensing system is presented in Figure 3. AI-2 molecules are produced and detected by many gram-positive and gram-negative bacteria, including Aeromonas spp. and are considered a “universal signal autoinducer” with functions in interspecies cell-to-cell communication (Figure 3; Fong et al., 2001; Miller et al., 2004; Federle, 2009). AI-2 molecules are by-products of S-adenosyl-methionine (as AI-1) and correspond to a furanosyl borate diester in V. harveyi (Chen et al., 2002) or a variant lacking borate in Salmonella enterica (Figure 3; Miller et al., 2004). These molecules are synthesized by the enzyme LuxS (Xavier and Bassler, 2003) and freely diffuse across bacterial membranes (Figure 3). Under conditions of high cell density in V. cholerae, AI-2 molecules bind to a periplasmic receptor and lead indirectly to the derepression of the transcriptional regulator HapR (Figure 3; Henke and Bassler, 2004; Xavier and Bassler, 2005).
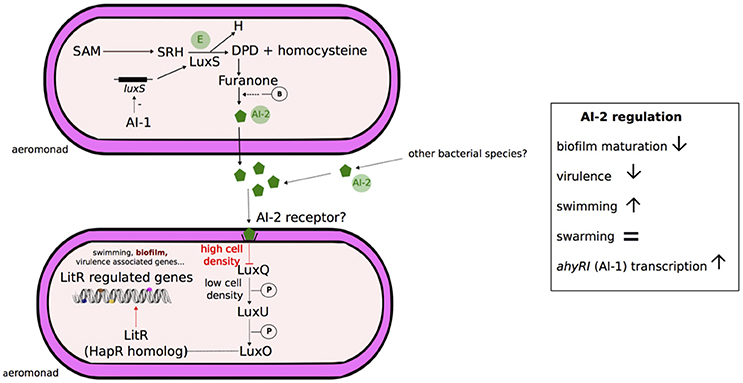
Figure 3. Schematic representation of AI-2 quorum sensing system in Aeromonas. Aeromonads are able to produce the AI-2 synthase enzyme (E) LuxS, and AI-2 (autoinducer 2) quorum sensing system has been detected in the genus (Kozlova et al., 2008). In the bacterial AI-2 quorum sensing systems, LuxS catalyzes the cleavage of S-ribosyl-homocysteine (SRH) derived from S-adenosyl-methionine (SAM) in homocysteine and 4,5-dihydroxy-2,3-pentanedione (DPD) (Xavier and Bassler, 2003). DPD spontaneously cyclizes to form a furanone, which can possibly react with borate (-B) depending on the bacterial species (discontinuous traits), and leading to AI-2 molecule formation (Chen et al., 2002; Miller et al., 2004). Based on studies in Vibrio, it has been shown that in absence of AI-2, LuxQ generates a phosphorylation cascade (-P) via LuxU and ultimately LuxO. LuxO is the response regulator that represses the master regulatory protein HapR (V. cholerae). At high cell density, AI-2 freely diffusible molecules reach a threshold and bind the LuxP periplasmic receptors. The autoinducer signal is transduced by the LuxP/AI-2 complex, inactivating the transmembrane sensor kinase LuxQ and subsequently leading to LuxO inactivation, which lifts repression of HapR and influences gene expression (Bassler et al., 1994; Henke and Bassler, 2004). However, the AI-2-internalization step of aeromonads is not yet known (discontinuous traits) and no luxP homolog were detected into their genomes (Kozlova et al., 2008). Signal transduction may involve the proteins LuxQ, LuxU, LuxO and subsequently the transcriptional regulator LitR (homolog of HapR), but the level of proof is so far only genetic (Kozlova et al., 2011). Overall, the AI-2 activation system in Aeromonas is associated with inhibition of biofilm maturation, enhancement of swimming and a decrease in virulence (Kozlova et al., 2008). The transcription of luxS locus is likely inhibited (discontinuous traits) by AI-1 quorum sensing system (Kozlova et al., 2011).
The genomes of A. hydrophila ATCC 7966T and A. hydrophila {A. dhakensis} SSU contain homologs for AI-2 synthase LuxS and enzymes involved in signal transformation (AI-2 sensor kinase/phosphatase LuxQ, phosphorelay protein LuxU, regulatory protein LuxO) and a LitR encoding-gene, a homolog for the transcriptional regulator HapR of V. cholerae, and Lit-R-regulated genes (Figure 3; Kozlova et al., 2011). The transcription of luxS is negatively regulated by the expression of the locus ahyRI (Kozlova et al., 2011). The functions of AI-2 in the A. hydrophila {A. dhakensis} strain SSU have been studied by constructing ΔluxS deletion mutants. AI-2 is involved in the up-regulation of swimming motility and the down-regulation of biofilm formation and bacterial virulence in a murine model (Kozlova et al., 2008).
Type 3 Autoinducer (AI-3) System
The type 3 autoinducer (AI-3) system is a hormone-like signal transduced by the two-component QseBC system in which QseC is the sensor kinase and QseB the response regulator (Figure 4). A putative schematic model of the Aeromonas AI-3 quorum sensing system is presented in Figure 4. AI-3 is suspected to behave similar to eukaryotic hormones because QseC is also a bacterial adrenergic receptor for the eukaryotic host hormones epinephrine and norepinephrine and is thus involved in interkingdom cross-signaling (Figure 4; Sperandio et al., 2003). AI-3 molecules are usually produced by gastrointestinal microbiota, e.g., in the context of symbiotic relationships between microbiota and host (Clarke et al., 2006). The periplasmic sensing domain of QseC is conserved among several gram-negative bacterial species (e.g., E. coli, S. enterica; Clarke et al., 2006).
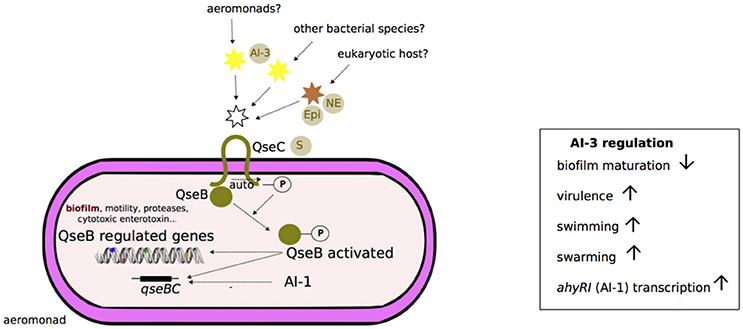
Figure 4. Schematic representation of AI-3 quorum sensing system in Aeromonas. Although, the two-component system QseB/QseC was characterized in Aeromonas (Khajanchi et al., 2012), the synthesis of autoinducer 3 (AI-3) signals is not yet known in this genus (discontinuous traits). According to the Escherichia coli model, the transmembrane protein QseC is a sensor (S) that can bind at its periplasmic domain: (i) signal molecules of AI-3 from other cells of a bacterial clone or from other bacterial species, or (ii) catecholamines (epinephrine, Epi, or norepinephrine, NE) from a eukaryotic host (Sperandio et al., 2002; Clarke et al., 2006). QseC then undergoes autophosphorylation (-P) at its cytoplasmic domain. The signal is then transmitted by phosphorylation (-P) to the transcriptional regulatory protein QseB. Subsequently, activated QseB-P binds to the transcription regulator domains of virulence-associated genes (flagella, shiga-like toxin, type III secretion system components, and effectors) and autoregulates its own operon qseBC (Sperandio et al., 2002; Clarke et al., 2006). Overall, the AI-3 system activation in Aeromonas is associated with inhibition of biofilm maturation, enhancement of swimming and swarming and an increase in virulence (Khajanchi et al., 2012). The transcription of qseBC locus is likely inhibited (discontinuous traits) by AI-1 quorum sensing system (Kozlova et al., 2012).
Khajanchi et al. (2012) have identified and characterized the QseBC QS system in A. dhakensis that encodes a functional homolog for E. coli QseBC. In Aeromonas, AI-3 enhances swarming and swimming motility and virulence (e.g., hemolytic activity, protease and cytotoxic enterotoxin production, toxicity in murine models) and negative regulation of biofilm formation, as shown in the ΔqseB deleted mutant of the A. hydrophila {A. dhakensis} strain SSU (Khajanchi et al., 2012). The transcription of qseB and qseC genes is negatively regulated by the AI-1 QS system (Kozlova et al., 2012; Figure 4).
How Are Biofilm and Quorum Sensing Interconnected in Aeromonas?
Bacteria commonly form dense communities in biofilms (Nadell et al., 2009). Consequently, biofilm lifestyle has emerged as a suitable model to study cell-to-cell signaling mechanisms. The three QS systems described in Aeromonas spp. are able to coordinate and influence biofilm formation, maintenance, and possibly dispersion. In addition, the major intracellular second messenger c-di-GMP has an impact on biofilm formation of Aeromonas spp. through QS systems, which is similar to the role observed in P. aeruginosa (Harmsen et al., 2010).
AI-1 Signaling Positively Regulates Aeromonas Biofilm Maturation
After the initial attachment phase, the AHL synthase AhyI and AHLs (C4-HSL and C6-HSL) are produced within the biofilm during microcolony formation and biofilm maturation (Lynch et al., 2002), as observed in other gram-negative bacteria and in particuliar P. aeruginosa (Davies et al., 1998; Huber et al., 2001; Labbate et al., 2004). Evidence of AI-1 effects on biofilms in aeromonads is provided by mutagenesis experiments and the effect of specific inhibitors.
Mutagenesis experiments clearly demonstrated that AI-1 influence depends on the biofilm development stage. The early attachment phase was not altered in ΔahyI (AI-1 synthase) or ΔahyR (AI-1 sensor/regulator) mutants of the A. hydrophila strain AH-1N (Lynch et al., 2002). Consistent with this, both swimming and swarming motilities were able to influence biofilm formation and were conserved in a double ΔahyRI mutant of A. hydrophila {A. dhakensis} strain SSU (Khajanchi et al., 2009). However, the later phases of biofilm maturation were affected by AI-1; the AH-1N ΔahyI (AI-1 synthase) mutant developed a biofilm less differentiated than that of the wild type (WT; Lynch et al., 2002). In addition, the number of viable cells was reduced in ΔahyI mutant biofilms compared to WT, but there was no difference in the viability of planktonic cells between ΔahyI and WT (Lynch et al., 2002). These defects in biofilm formation were partially restored by addition of exogenous AI-1, C4-HSL (Lynch et al., 2002). Confirming these observations, an A. hydrophila {A. dhakensis} SSU ΔahyRI mutant was defective and unable to form a well-structured biofilm, with alterations in filamentation, strain aggregation, and the (Khajanchi et al., 2009). In contrast to the ΔahyI (AI-1 synthase) mutant, the ΔahyR (AI-1 sensor/regulator) mutant of A. hydrophila strain AH-1N retains the ability to form mature biofilm, suggesting that the role of AHLs in the formation of large mushroom-like microcolonies does not depend on AhyR (Lynch et al., 2002) but presumably depends on transcriptional regulators other than AhyR. For instance, Kozlova et al. (2011) reported the presence of two loci that encode AhyR homologs in the strain A. hydrophila {A. dhakensis} SSU. In summary, these mutagenesis experiments showed that AI-1 molecules and AI-1 synthase enhanced biofilm formation.
Evidence for the role of AI-1 on biofilm formation is reinforced by studies focusing on AI-1 inhibitors, i.e., compounds that bear AI-1 quorum-quenching properties. For instance, chestnut honey decreases the level of C4-HSL molecules produced by the A. hydrophila strain CECT 839T and leads to significant inhibition of biofilm formation (Truchado et al., 2009). Synthetic 2(5H)-furanone, derived from natural furanones produced by the marine algae D. pulchra, inhibits in vitro AHLs, including those usually produced by aeromonads (C4-HSL and C6-HSL), and results in biofilm inhibition in a strain of A. hydrophila (Ponnusamy et al., 2010). Vanillin exhibited quorum-quenching activity against aeromonad AHLs and reduced A. hydrophila biofilm by altering its architecture and decreasing surface cell-density and protein content (Ponnusamy et al., 2013). Mentha piperata essential oil and menthol have a quorum-quenching effect against C6-HSL. Sub-minimal inhibitory concentrations of mint essential oil and menthol decrease extracellular polymeric substance production and biofilm formation in the A. hydrophila strain WAF-28 (Husain et al., 2015). Examples of inhibition on biofilm formation displayed by several AI-1 inhibitors confirm dependence of biofilm formation to AI-1 QS.
AI-2 and AI-3 Operate in Negative Feedback Regulation on Aeromonas Biofilm Formation
Overall, the planktonic form is promoted by AI-2 and AI-3 because these two QS systems operate in negative feedback regulation in the biofilm formation of Aeromonas (Kozlova et al., 2008, 2012; Khajanchi et al., 2012).
A ΔluxS mutant of A. hydrophila {A. dhakensis} strain SSU defective in AI-2 showed enhanced amount of biofilm (Kozlova et al., 2008). Meanwhile, swimming motility, was reduced when luxS was deleted (Kozlova et al., 2008), suggesting that LuxS up-regulates swimming motility. This result should not be viewed as conflicting because it favors a switch to a planktonic lifestyle and is compatible with down-regulation of biofilm formation exhibited by the AI-2 QS system. Indeed, polar flagella are involved in biofilm formation at the initial stage, but this appendage is still fully expressed in the planktonic life of aeromonads (Canals et al., 2006). AI-3 signaling seems to exert similar effects to AI-2 on the down-regulation of biofilm formation and up-regulation of swimming motility as observed in the strain A. hydrophila {A. dhakensis} SSU and also up-regulates swarming motility (Khajanchi et al., 2012; Kozlova et al., 2012).
Role of c-di-GMP on Aeromonas Biofilm Maturation
Cyclic di-guanosine monophosphate (c-di-GMP) is a secondary messenger that is mainly involved in the transition from a planktonic to sessile lifestyle in the domain Bacteria and was well-studied in P. aeruginosa or V. cholerae (Simm et al., 2004; Tischler and Camilli, 2005; Kuchma et al., 2007). For a comprehensive review, see Römling et al. (2013). Synthesis and degradation of c-di-GMP are under control of peptide domains widespread and universally conserved in bacteria: GGDEF (di-guanylate synthase activity) and EAL (phosphodiesterase activity), respectively. The A. hydrophila ATCC 7966T genome contains 32 proteins with GGDEF, 9 proteins with EAL and 13 proteins with both (Rahman et al., 2007). In Aeromonas, the overexpression of the GGDEF domain enhances biofilm formation and inhibits swimming (polar flagellum dependent) and swarming motility (lateral flagella dependent) whereas, overexpression of the EAL domain leads to opposite effects. Thus, it is thought that c-di-GMP positively regulates biofilm maturation (sessile life) and negatively affects bacterial motility for aeromonads (planktonic life; Rahman et al., 2007; Kozlova et al., 2008, 2011, 2012; Khajanchi et al., 2012).
AI-2 and AI-3 QS are involved in the control of biofilm by c-di-GMP. For instance, one gene coding for a GGDEF domain protein, namely AHA0701, is located immediately downstream of the LuxS encoding-gene (AI-2 synthase gene) in the genomes of A. hydrophila ATCC 7966T and A. hydrophila {A. dhakensis} SSU strains (Seshadri et al., 2006; Kozlova et al., 2011). Overexpression of AHA0701 increased the number of surface-attached cells and enhanced biofilm formation (Kozlova et al., 2011) and putatively led to extensive EPS production (Kozlova et al., 2012) by down-regulation of AI-2 synthase transcription. Corroborating the involvement of AI-2 and AI-3 QS in the c-di-GMP dependent regulation of sessile lifestyle in Aeromonas, the phenotype of strains that overproduced GGDEF domains varied in the same direction as ΔluxS (AI-2) and ΔqseB (AI-3) mutants vs. WT, i.e., increasing the maturation of biofilm and decreasing bacterial motility (Kozlova et al., 2008, 2011, 2012; Khajanchi et al., 2012). The regulation of sessile life through c-di-GMP also involves the AI-1 system in Aeromonas, as proved by the increase in C4-HSL production and enhancement of ahyI and ahyR transcription when c-di-GMP increases. In conditions of c-di-GMP overproduction, the transcript levels of luxS, qseB, and qseC genes also increased, but this influence is dependent on the AI-1 system (Kozlova et al., 2011). Finally, the ahyRI locus is required to promote c-di-GMP-mediated biofilm maturation (Rahman et al., 2007; Kozlova et al., 2011). Moreover, several homologs for genes encoding effectors of c-di-GMP signaling involved in biofilm formation of V. cholerae or P. aeruginosa have been found in Aeromonas spp. genomes. Among them, two transcriptional regulators containing the DNA-binding domains VpsR/FleQ and VpsT/CsgAB, and one master regulator for biofilm formation FleN were identified (Kozlova et al., 2011). In A. dhakensis, the transcription of effectors VpsR/FleQ, VpsT/CsgAB, and FleN is enhanced by c-di-GMP and is also modulated by AI-1, AI-2, and AI-3 systems, with opposite effects (Kozlova et al., 2011, 2012).
In summary, there is cross-talk between the three QS systems AI-1, AI-2, and AI-3 in the c-di-GMP regulation of the sessile life in Aeromonas (Kozlova et al., 2011, 2012).
Further Research
A large amount of knowledge is available for understanding biofilm formation, QS systems, and interconnections between biofilms and QS in aeromonads. However, some knowledge is still missing for a comprehensive understanding of aeromonad social life. The type 1 autoinducer (AI-1) system is well-described, but further investigations are needed to specify the AHL profiles actually produced according to species, growth phase and origin of isolates. The AI-2 and AI-3 QS systems are poorly described and deserve to be more deeply investigated. Although, the AI-2 system has proved to be involved in Aeromonas biology thanks to mutagenesis evidence, the system could be better characterized. More specifically, homologs for the AI-2 receptor (LuxP in V. harveyi) have not been detected in the genome of A. hydrophila {A. dhakensis} strain SSU or A. hydrophila ATCC 7966T (Kozlova et al., 2011), and the chemical nature of AI-2 produced from aeromonads should be clarified. Further studies are also needed to explore the AI-3 system and characterize the AI-3 signal(s) at the level of bacterial clone and within host interacting microbiota. Further studies designed to better understand this regulation system in Aeromonas spp. in high-density cell and biofilm situations are needed. Above all, QS and biofilm are the beginning of the understanding of complex and fine systems involved in aeromonad social life. Once the Pandora's box opened, additional exciting, unstudied questions will arise. Mixed biofilm structures and lifestyles likely result from a balance between cooperation and competition that we will have to unravel. Changes in gene expression during different stages of biofilms are another field to explore (Nadell et al., 2009). In addition, packed communities secrete into the local environment and share beneficial metabolites that are costly to produce, with benefits for individuals (Dumas and Kümmerli, 2012). These “public goods” may be exploited by non-producing mutants, i.e., cheaters (Heilmann et al., 2015). How biofilm structure, population dynamics within biofilm is impacted by cheaters is unknown. Understanding these aspects will help us to better understand the social life of aeromonads.
Fields Impacted by the Multi-Cellular Behavior of Aeromonas
Flocs, Biofilms, and Aeromonads in Activated Sludge
Activated sludge is a typical process for treating sewage and industrial wastewater taking advantage of the sociobiology of aeromonads, i.e., biofilm formation, QS and synergistic behavior. Members of the genus Aeromonas have been estimated to account for ~2% of the total biomass of activated sludge (Kampfer et al., 1996). Aeromonads organized in biofilm and aggregates have the biotechnological interest in the removal of organic pollutants (Chong et al., 2012), nitrogen (Chen et al., 2014), and phosphorus (Andersson et al., 2011) and are resistant to extreme conditions of pH, salinity, heavy metals, and temperature (Chen et al., 2014; Mohd Yasin et al., 2014).
Flocs in activated sludge are microbial aggregates enchased in a matrix of EPS composed of carbohydrates, proteins, lipids, and nucleic acids. These polymers come from bacterial cell metabolism, autolysis, and exogenous wastewater particles (Frølund et al., 1996). An analogy between biofilms and flocs is generally drawn because these bioaggregates correspond to a microbial surface producing EPS that improve the cohesion of the supracellular structure. The ability to flocculate was shown in vitro with a strain of Aeromonas sp. isolated from activated sludge (Chong et al., 2012).
To reduce hydraulic retention times, biofilm techniques are increasingly used to increase the surface of biodegradation. In a model of mixed biofilms composed of strains regularly present in activated sludge, Andersson et al. (2011) reported that mixed species biofilms that included A. hydrophila were associated with synergistic effects on denitrification and phosphorus removal. A shift in the composition of the produced EPS was also highlighted, with a significant rise in the carbohydrate fraction in the Brachymonas denitrificans model mixed with A. hydrophila, compared with single species biofilms. New polysaccharides were also detected but only from a mixed culture of A. hydrophila and Comamonas denitrificans or Acinetobacter calcoaceticus (Andersson et al., 2011). Another multi-species biofilm containing Comamonas testosteroni and A. hydrophila strains display synergistic biofilm formation, enabling better resistance of the biofilm to the repetitive physical and chemical shocks occurring in wastewater (Li et al., 2008). These interactive adaptive behaviors within the biofilm have great potential to improve wastewater treatment efficiency.
From typical activated sludge flocs containing Alpha-, Beta-, and Gammaproteobacteria, as well as members of Bacteroidetes and Actinobacteria, Li and Zhu (2014) showed by a quorum quenching method that AI-1 QS is necessary for the aerobic granulation of flocs and qualitatively and quantitatively regulates the content of exopolymeric substances. Aeromonas sp. in activated sludge are reported to produce AHLs (Chong et al., 2012; Ochiai et al., 2013). Interestingly, AHLs were localized in flocs community but absent from the aqueous phase (Chong et al., 2012). Moreover, the exogenous addition of AHLs led to increase the chitinase activity of the aeromonads isolated from activated sludge and of the whole activated sludge (Chong et al., 2012). Furthermore, QS signaling within activated sludge can also be modulated by bacteria degrading AHLs, such as Acinetobacter (Ochiai et al., 2013). All these data support that Aeromonas in flocs display QS-dependent social behavior improving water treatment.
Role in Virulence
Biofilm formation is involved in the pathogenesis of numerous human colonizations and/or infections (e.g., healthcare-associated infections) and persistent infections (e.g., chronic infection during cystic fibrosis, endocarditis, biliary tract infections, periodontitis, otitis media, wounds; Lebeaux et al., 2013). Aeromonads are occasionally involved in biofilm-associated colonization of medical devices, leading to infection such as lenses-associated keratitis (Willcox et al., 2001; Pinna et al., 2004) and central venous catheter-associated bloodstream infection (Andreoli-Pinto and Graziano, 1999; Tang et al., 2014). These reports suggest that biofilm acts as a virulence or patho-adaptive factor in Aeromonas infections. Consequently, biofilms are constantly evaluated in virulence studies. However, the correlation between biofilm producer and aeromonad pathogenesis is still weak. Presumably, the ability to produce biofilm in vitro may explain long-term digestive epithelium colonization by some strains whether they are associated with an infection (Janda and Abbott, 2010). Bfp type IV pili and polar/lateral flagella were suspected to be factors of intestinal colonization (Kirov et al., 1999, 2004). However, the production of lateral flagella is unlikely to be compulsory in digestive colonization because it is inconstantly expressed in Aeromonas spp. clinical isolates recovered from diarrheal feces (Kirov et al., 2002). The level of evidence for the exact contribution of biofilm to Aeromonas pathogenicity needs to be improved, and further studies are welcomed.
Because it was shown that several virulence determinants (e.g., alpha-hemolysin, cholesterol acetyltransferase, lipase, and serine protease) were overproduced at high cell density in late exponential/stationary phase, cell density could be a prerequisite for aeromonad pathogenic behavior and virulence expression (MacIntyre and Buckley, 1978; Whitby et al., 1992; Anguita et al., 1993). This suggests a pivotal role for QS in the course of infection. Both AI-1 and AI-3 signaling enhance the expression of some virulence determinants of aeromonads (Table 2; Figures 2, 4) and increase lethality in a murine model, as proved by the virulence-reduced phenotypes exhibited by ΔahyRI and ΔqseB mutants vs. WT (Khajanchi et al., 2009, 2012). In contrast, AI-2 signaling is associated with a down-regulation of bacterial virulence in a murine model (Kozlova et al., 2008), although molecular targets have yet to be identified. The pathogenesis of Aeromonas infections remain to be better understood, but QS-based cross-talk is indubitably involved in the expression of virulence from a threshold of cell density.
As described in P. aeruginosa, AHL molecules (e.g., 3-oxo-C12-HSL) are not only involved in bacterial virulence regulation but also interact with several eukaryotic cells and play a role in the immuno-modulation of the host response (Liu et al., 2015). Khajanchi et al. (2011) have reported a similar immune-modulatory effect with C4-HSL, C6-HSL, and N-3-oxo-C12-HSL in septicemic mice infected with A. hydrophila {A. dhakensis}. Indeed, AHL-based pretreatment was associated with reduced levels of cytokines/chemokines in tissues, increased neutrophil recruitment from blood (C6-HSL), enhanced bacterial clearance, limitation of clinical symptoms, and increase in survivability for infected mice. In addition, the AHL-based pretreatment increased in vitro bacterial phagocytosis by murine macrophages. Thus, it seems that exogenous AHLs act as a signal for the activation of host response to infection (Khajanchi et al., 2011). Both promising QS inhibitors that interfere with aeromonad QS, e.g., menthol (Husain et al., 2015), and some purified QS molecules (Khajanchi et al., 2011), represent new therapeutic perspectives to fight against aeromonosis threatening multi-drug antibiotic resistance.
There is increasing evidence that suggests the influence of multicellular microbial interaction during the course of infection, in the case of mixed infections driven by cooperation and/or competition (Trejo-Hernández et al., 2014; Armbruster et al., 2016). Microbial interactions may be particularly critical in the Aeromonas genus. Indeed, infections caused by Aeromonas are frequently polymicrobial, i.e., caused by Aeromonas and other bacterial genera (30–80% of the cases), (Lamy et al., 2009; Lay et al., 2010; Figueras and Beaz-Hidalgo, 2015); aeromonads are mainly associated with enterobacteria, Staphylococcus aureus and anaerobes. In 5–10% of human aeromonosis, infections are caused by heterogeneous populations associating several clones of Aeromonas spp. (Lamy et al., 2009). For some pairs of aeromonads co-isolated from clinical samples, the virulence during infection in Caenorhabditis elegans was higher for pairs than for each individual strain (Mosser et al., 2015). The exact mechanism of pathogenicity and aeromonads interactions during mixed infections remains unsolved, but intercellular communication likely plays a critical role in cooperation and/or competition behaviors. Ponnusamy et al. (2016) have also reported complex cross-talk in which two different clones of A. hydrophila affect each other, leading to a changing course of infection in a murine model. Indeed, an antagonistic effect through direct and host-mediated elimination was shown. In parallel, authors have observed a synergistic effect on the dissemination via local tissue barrier damage (Ponnusamy et al., 2016).
Conclusion
Aeromonads display diverse multicellular behaviors that represent many strategies to grow and persist in natural and anthropized environments. These adaptive behaviors potentially influence the virulence of Aeromonas. Living in biofilm provides aeromonads with a high cell-density that enhances interaction between bacteria through QS systems. The three QS systems play an important regulatory role in the expression of a wide range of functions for aeromonads including biofilm formation, motility, and virulence, and these systems harbor differences in their regulative influences. The AI-1 system enhances sessile life by promoting biofilm while AI-2 and AI-3 enhance planktonic life by inhibiting biofilm maturation and increasing bacterial motility. These effects on planktonic and sessile life are also influenced by the second messenger c-di-GMP. Considerable interconnections exist between the three QS systems and the biofilm formation in the genus Aeromonas, as in other bacterial species. Efforts should be made to develop models in order to obtain accurate and robust data to better understand multicellular aspects of Aeromonas biology. In human infections, the role of Aeromonas biofilm in human gastro-intestinal colonization and the influence of bacterial clones in aeromonad-containing mixed infections deserve to be studied to better decipher aeromonosis patho-physiology.
Author Contributions
Conceived and designed the work: ET, EJ, BL; Performed survey and drafted the paper: ET; Critically revised the manuscript: EJ, BL. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Association des Biologistes de l'Ouest, and by the Association pour la recherche et le développement en microbiologie et pharmacie (ADEREMPHA).
References
Andersson, S., Dalhammar, G., and Kuttuva Rajarao, G. (2011). Influence of microbial interactions and EPS/polysaccharide composition on nutrient removal activity in biofilms formed by strains found in wastewater treatment systems. Microbiol. Res. 166, 449–457. doi: 10.1016/j.micres.2010.08.005
Andreoli-Pinto, T. J., and Graziano, K. U. (1999). Important aspects of the colonization of central venous catheter. Boll. Chim. Farm. 138, 19–23.
Anguita, J., Rodríguez Aparicio, L. B., and Naharro, G. (1993). Purification, gene cloning, amino acid sequence analysis, and expression of an extracellular lipase from an Aeromonas hydrophila human isolate. Appl. Environ. Microbiol. 59, 2411–2417.
Armbruster, C. R., Wolter, D. J., Mishra, M., Hayden, H. S., Radey, M. C., Merrihew, G., et al. (2016). Staphylococcus aureus protein A mediates interspecies interactions at the cell surface of Pseudomonas aeruginosa. mBio 7:e00538-16. doi: 10.1128/mBio.00538-16
Balasubramanian, V., Palanichamy, S., Subramanian, G., and Rajaram, R. (2012). Development of polyvinyl chloride biofilms for succession of selected marine bacterial populations. J. Environ. Biol. Acad. Environ. Biol. India 33, 57–60.
Bansal, S., Harjai, K., and Chhibber, S. (2015). Aeromonas punctata derived depolymerase improves susceptibility of Klebsiella pneumoniae biofilm to gentamicin. BMC Microbiol. 15:119. doi: 10.1186/s12866-015-0455-z
Barken, K. B., Pamp, S. J., Yang, L., Gjermansen, M., Bertrand, J. J., Klausen, M., et al. (2008). Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 10, 2331–2343. doi: 10.1111/j.1462-2920.2008.01658.x
Bassler, B. L., Wright, M., and Silverman, M. R. (1994). Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13, 273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x
Béchet, M., and Blondeau, R. (2003). Factors associated with the adherence and biofilm formation by Aeromonas caviae on glass surfaces. J. Appl. Microbiol. 94, 1072–1078. doi: 10.1046/j.1365-2672.2003.01931.x
Bi, Z. X., Liu, Y. J., and Lu, C. P. (2007). Contribution of AhyR to virulence of Aeromonas hydrophila J-1. Res. Vet. Sci. 83, 150–156. doi: 10.1016/j.rvsc.2007.01.003
Bjarnsholt, T., Jensen, P. Ø., Jakobsen, T. H., Phipps, R., Nielsen, A. K., Rybtke, M. T., et al. (2010). Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS ONE 5:e10115. doi: 10.1371/journal.pone.0010115
Bomo, A. M., Storey, M. V., and Ashbolt, N. J. (2004). Detection, integration and persistence of aeromonads in water distribution pipe biofilms. J. Water Health 2, 83–96.
Boyd, J. M., Dacanay, A., Knickle, L. C., Touhami, A., Brown, L. L., Jericho, M. H., et al. (2008). Contribution of type IV pili to the virulence of Aeromonas salmonicida subsp. salmonicida in Atlantic salmon (Salmo salar L.). Infect. Immun. 76, 1445–1455. doi: 10.1128/IAI.01019-07
Bruhn, J. B., Dalsgaard, I., Nielsen, K. F., Buchholtz, C., Larsen, J. L., and Gram, L. (2005). Quorum sensing signal molecules (acylated homoserine lactones) in gram-negative fish pathogenic bacteria. Dis. Aquat. Organ. 65, 43–52. doi: 10.3354/dao065043
Canals, R., Ramirez, S., Vilches, S., Horsburgh, G., Shaw, J. G., Tomás, J. M., et al. (2006). Polar flagellum biogenesis in Aeromonas hydrophila. J. Bacteriol. 188, 542–555. doi: 10.1128/JB.188.2.542-555.2006
Casas, C., Anderson, E. C., Ojo, K. K., Keith, I., Whelan, D., Rainnie, D., et al. (2005). Characterization of pRAS1-like plasmids from atypical North American psychrophilic Aeromonas salmonicida. FEMS Microbiol. Lett. 242, 59–63. doi: 10.1016/j.femsle.2004.10.039
Castro, L., Zhang, R., Muñoz, J. A., González, F., Blázquez, M. L., Sand, W., et al. (2014). Characterization of exopolymeric substances (EPS) produced by Aeromonas hydrophila under reducing conditions. Biofouling 30, 501–511. doi: 10.1080/08927014.2014.892586
Cataldi, T. R., Bianco, G., Palazzo, L., and Quaranta, V. (2007). Occurrence of N-acyl-L-homoserine lactones in extracts of some Gram-negative bacteria evaluated by gas chromatography-mass spectrometry. Anal. Biochem. 361, 226–235. doi: 10.1016/j.ab.2006.11.037
Chan, K. G., Puthucheary, S. D., Chan, X. Y., Yin, W. F., Wong, C. S., Too, W. S. S., et al. (2011). Quorum sensing in Aeromonas species isolated from patients in Malaysia. Curr. Microbiol. 62, 167–172. doi: 10.1007/s00284-010-9689-z
Chauret, C., Volk, C., Creason, R., Jarosh, J., Robinson, J., and Warnes, C. (2001). Detection of Aeromonas hydrophila in a drinking-water distribution system: a field and pilot study. Can. J. Microbiol. 47, 782–786. doi: 10.1139/w01-070
Chen, M., Wang, W., Feng, Y., Zhu, X., Zhou, H., Tan, Z., et al. (2014). Impact resistance of different factors on ammonia removal by heterotrophic nitrification-aerobic denitrification bacterium Aeromonas sp. HN-02. Bioresour. Technol. 167, 456–461. doi: 10.1016/j.biortech.2014.06.001
Chen, X., Schauder, S., Potier, N., Van Dorsselaer, A., Pelczer, I., Bassler, B. L., et al. (2002). Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415, 545–549. doi: 10.1038/415545a
Cheng, Z., Meng, X., Wang, H., Chen, M., and Li, M. (2014). Isolation and characterization of broad spectrum coaggregating bacteria from different water systems for potential use in bioaugmentation. PLoS ONE 9:e94220. doi: 10.1371/journal.pone.0094220
Chong, G., Kimyon, O., Rice, S. A., Kjelleberg, S., and Manefield, M. (2012). The presence and role of bacterial quorum sensing in activated sludge. Microb. Biotechnol. 5, 621–633. doi: 10.1111/j.1751-7915.2012.00348.x
Chu, W., Liu, Y., Jiang, Y., Zhu, W., and Zhuang, X. (2013). Production of N-acyl homoserine lactones and virulence factors of waterborne Aeromonas hydrophila. Indian J. Microbiol. 53, 264–268. doi: 10.1007/s12088-013-0381-4
Churchill, M. E., and Chen, L. (2011). Structural basis of acyl-homoserine lactone-dependent signaling. Chem. Rev. 111, 68–85. doi: 10.1021/cr1000817
Claessen, D., Rozen, D. E., Kuipers, O. P., Søgaard-Andersen, L., and van Wezel, G. P. (2014). Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat. Rev. Microbiol. 12, 115–124. doi: 10.1038/nrmicro3178
Clarke, M. B., Hughes, D. T., Zhu, C., Boedeker, E. C., and Sperandio, V. (2006). The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 10420–10425. doi: 10.1073/pnas.0604343103
Costerton, J. W., Cheng, K. J., Geesey, G. G., Ladd, T. I., Nickel, J. C., Dasgupta, M., et al. (1987). Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41, 435–464. doi: 10.1146/annurev.mi.41.100187.002251
Davies, D. G., Parsek, M. R., Pearson, J. P., Iglewski, B. H., Costerton, J. W., and Greenberg, E. P. (1998). The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280, 295–298. doi: 10.1126/science.280.5361.295
Doğruöz, N., Göksay, D., Ilhan-Sungur, E., and Cotuk, A. (2009). Pioneer colonizer microorganisms in biofilm formation on galvanized steel in a simulated recirculating cooling-water system. J. Basic Microbiol. 49(Suppl. 1), S5–S12. doi: 10.1002/jobm.200800250
Donlan, R. M., and Costerton, J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193. doi: 10.1128/CMR.15.2.167-193.2002
Dumas, Z., and Kümmerli, R. (2012). Cost of cooperation rules selection for cheats in bacterial metapopulations. J. Evol. Biol. 25, 473–484. doi: 10.1111/j.1420-9101.2011.02437.x
Engebrecht, J., and Silverman, M. (1984). Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. U.S.A. 81, 4154–4158. doi: 10.1073/pnas.81.13.4154
Esteve, C., Alcaide, E., and Giménez, M. J. (2015). Multidrug-resistant (MDR) Aeromonas recovered from the metropolitan area of Valencia (Spain): diseases spectrum and prevalence in the environment. Eur. J. Clin. Microbiol. Infect. Dis. 34, 137–145. doi: 10.1007/s10096-014-2210-z
Farkas, A., Drăgan-Bularda, M., Ciatarâs, D., Bocoş, B., and Tigan, S. (2012). Opportunistic pathogens and faecal indicators in drinking water associated biofilms in Cluj, Romania. J. Water Health 10, 471–483. doi: 10.2166/wh.2012.148
Federle, M. J. (2009). Autoinducer-2-based chemical communication in bacteria: complexities of interspecies signaling. Contrib. Microbiol. 16, 18–32. doi: 10.1159/000219371
Fernández, M. C., Giampaolo, B. N., Ibañez, S. B., Guagliardo, M. V., Esnaola, M. M., Conca, L., et al. (2000). Aeromonas hydrophila and its relation with drinking water indicators of microbiological quality in Argentine. Genetica 108, 35–40. doi: 10.1023/A:1004025907858
Figueras, M. J., and Beaz-Hidalgo, R. (2015). “Aeromonas infections in humans,” in Aeromonas, ed J. Graf (Norfolky: Caister Academic Press), 65–68.
Fong, K. P., Chung, W. O., Lamont, R. J., and Demuth, D. R. (2001). Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect. Immun. 69, 7625–7634. doi: 10.1128/IAI.69.12.7625-7634.2001
Frølund, B., Palmgren, R., Keiding, K., and Nielsen, P. H. (1996). Extraction of extracellular polymers from activated sludge using a cation exchange resin. Water Res. 30, 1749–1758. doi: 10.1016/0043-1354(95)00323-1
Fulton, K. M., Mendoza-Barberá, E., Twine, S. M., Tomás, J. M., and Merino, S. (2015). Polar glycosylated and lateral non-glycosylated flagella from Aeromonas hydrophila strain AH-1 (serotype O11). Int. J. Mol. Sci. 16, 28255–28269. doi: 10.3390/ijms161226097
Fuqua, W. C., Winans, S. C., and Greenberg, E. P. (1994). Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176, 269–275. doi: 10.1128/jb.176.2.269-275.1994
Garde, C., Bjarnsholt, T., Givskov, M., Jakobsen, T. H., Hentzer, M., Claussen, A., et al. (2010). Quorum sensing regulation in Aeromonas hydrophila. J. Mol. Biol. 396, 849–857. doi: 10.1016/j.jmb.2010.01.002
Gavín, R., Merino, S., Altarriba, M., Canals, R., Shaw, J. G., and Tomás, J. M. (2003). Lateral flagella are required for increased cell adherence, invasion and biofilm formation by Aeromonas spp. FEMS Microbiol. Lett. 224, 77–83. doi: 10.1016/S0378-1097(03)00418-X
Gavín, R., Rabaan, A. A., Merino, S., Tomás, J. M., Gryllos, I., and Shaw, J. G. (2002). Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Mol. Microbiol. 43, 383–397. doi: 10.1046/j.1365-2958.2002.02750.x
Graf, J. (1999). Symbiosis of Aeromonas veronii Biovar sobria and hirudo medicinalis, the medicinal leech: a novel model for digestive tract associations. Infect. Immun. 67, 1–7.
Guerrieri, E., Bondi, M., Sabia, C., de Niederhäusern, S., Borella, P., and Messi, P. (2008). Effect of bacterial interference on biofilm development by Legionella pneumophila. Curr. Microbiol. 57, 532–536. doi: 10.1007/s00284-008-9237-2
Gunduz, G. T., and Tuncel, G. (2006). Biofilm formation in an ice cream plant. Antonie Van Leeuwenhoek 89, 329–336. doi: 10.1007/s10482-005-9035-9
Hadi, N., Yang, Q., Barnett, T. C., Tabei, S. M., Kirov, S. M., and Shaw, J. G. (2012). Bundle-forming pilus locus of Aeromonas veronii bv. Sobria. Infect. Immun. 80, 1351–1360. doi: 10.1128/IAI.06304-11
Hancock, R. E. (1984). Alterations in outer membrane permeability. Annu. Rev. Microbiol. 38, 237–264. doi: 10.1146/annurev.mi.38.100184.001321
Harmsen, M., Yang, L., Pamp, S. J., and Tolker-Nielsen, T. (2010). An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol. Med. Microbiol. 59, 253–268. doi: 10.1111/j.1574-695X.2010.00690.x
Hausner, M., and Wuertz, S. (1999). High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 65, 3710–3713.
Heilmann, S., Krishna, S., and Kerr, B. (2015). Why do bacteria regulate public goods by quorum sensing?-How the shapes of cost and benefit functions determine the form of optimal regulation. Front. Microbiol. 6:767. doi: 10.3389/fmicb.2015.00767
Hendrickx, L., Hausner, M., and Wuertz, S. (2003). Natural genetic transformation in monoculture Acinetobacter sp. strain BD413 biofilms. Appl. Environ. Microbiol. 69, 1721–1727. doi: 10.1128/AEM.69.3.1721-1727.2003
Henke, J. M., and Bassler, B. L. (2004). Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J. Bacteriol. 186, 6902–6914. doi: 10.1128/JB.186.20.6902-6914.2004
Hennequin, C., Aumeran, C., Robin, F., Traore, O., and Forestier, C. (2012). Antibiotic resistance and plasmid transfer capacity in biofilm formed with a CTX-M-15-producing Klebsiella pneumoniae isolate. J. Antimicrob. Chemother. 67, 2123–2130. doi: 10.1093/jac/dks169
Huang, L., Qin, Y., Yan, Q., Lin, G., Huang, L., Huang, B., et al. (2015). MinD plays an important role in Aeromonas hydrophila adherence to Anguilla japonica mucus. Gene 565, 275–281. doi: 10.1016/j.gene.2015.04.031
Huang, Y., Zhang, J., Yu, Z., Zeng, Y., and Chen, Y. (2012). Isolation and characterization of acyl homoserine lactone-producing bacteria during an urban river biofilm formation. Arch. Microbiol. 194, 1043–1048. doi: 10.1007/s00203-012-0849-3
Huber, B., Riedel, K., Hentzer, M., Heydorn, A., Gotschlich, A., Givskov, M., et al. (2001). The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiol. Read. Engl. 147, 2517–2528. doi: 10.1099/00221287-147-9-2517
Huddleston, J. R., Brokaw, J. M., Zak, J. C., and Jeter, R. M. (2013). Natural transformation as a mechanism of horizontal gene transfer among environmental Aeromonas species. Syst. Appl. Microbiol. 36, 224–234. doi: 10.1016/j.syapm.2013.01.004
Husain, F. M., Ahmad, I., Khan, M. S., Ahmad, E., Tahseen, Q., Khan, M. S., et al. (2015). Sub-MICs of Mentha piperita essential oil and menthol inhibits AHL mediated quorum sensing and biofilm of Gram-negative bacteria. Front. Microbiol. 6:420. doi: 10.3389/fmicb.2015.00420
Jagmann, N., von Rekowski, K. S., and Philipp, B. (2012). Interactions of bacteria with different mechanisms for chitin degradation result in the formation of a mixed-species biofilm. FEMS Microbiol. Lett. 326, 69–75. doi: 10.1111/j.1574-6968.2011.02435.x
Jahid, I. K., and Ha, S. D. (2014). Inactivation kinetics of various chemical disinfectants on Aeromonas hydrophila planktonic cells and biofilms. Foodborne Pathog. Dis. 11, 346–353. doi: 10.1089/fpd.2013.1682
Jahid, I. K., Lee, N. Y., Kim, A., and Ha, S. D. (2013). Influence of glucose concentrations on biofilm formation, motility, exoprotease production, and quorum sensing in Aeromonas hydrophila. J. Food Prot. 76, 239–247. doi: 10.4315/0362-028X.JFP-12-321
Jahid, I. K., Mizan, M. F., Ha, A. J., and Ha, S. D. (2015). Effect of salinity and incubation time of planktonic cells on biofilm formation, motility, exoprotease production, and quorum sensing of Aeromonas hydrophila. Food Microbiol. 49, 142–151. doi: 10.1016/j.fm.2015.01.016
Janda, J. M., and Abbott, S. L. (2010). The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23, 35–73. doi: 10.1128/CMR.00039-09
Jangid, K., Kong, R., Patole, M. S., and Shouche, Y. S. (2007). luxRI homologs are universally present in the genus Aeromonas. BMC Microbiol. 7:93. doi: 10.1186/1471-2180-7-93
Jangid, K., Parameswaran, P. S., and Shouche, Y. S. (2012). A variant quorum sensing system in Aeromonas veronii MTCC 3249. Sensors 12, 3814–3830. doi: 10.3390/s120403814
Kampfer, P., Erhart, R., Beimfohr, C., Bohringer, J., Wagner, M., and Amann, R. (1996). Characterization of bacterial communities from activated sludge: culture-dependent numerical identification versus in situ identification using group- and genus-specific rRNA-targeted oligonucleotide probes. Microb. Ecol. 32, 101–121. doi: 10.1007/BF00185883
Khajanchi, B. K., Kirtley, M. L., Brackman, S. M., and Chopra, A. K. (2011). Immunomodulatory and protective roles of quorum-sensing signaling molecules N-acyl homoserine lactones during infection of mice with Aeromonas hydrophila. Infect. Immun. 79, 2646–2657. doi: 10.1128/IAI.00096-11
Khajanchi, B. K., Kozlova, E. V., Sha, J., Popov, V. L., and Chopra, A. K. (2012). The two-component QseBC signalling system regulates in vitro and in vivo virulence of Aeromonas hydrophila. Microbiol. Read. Engl. 158, 259–271. doi: 10.1099/mic.0.051805-0
Khajanchi, B. K., Sha, J., Kozlova, E. V., Erova, T. E., Suarez, G., Sierra, J. C., et al. (2009). N-acylhomoserine lactones involved in quorum sensing control the type VI secretion system, biofilm formation, protease production, and in vivo virulence in a clinical isolate of Aeromonas hydrophila. Microbiol. Read. Engl. 155, 3518–3531. doi: 10.1099/mic.0.031575-0
Kikuchi, Y., and Graf, J. (2007). Spatial and temporal population dynamics of a naturally occurring two-species microbial community inside the digestive tract of the medicinal leech. Appl. Environ. Microbiol. 73, 1984–1991. doi: 10.1128/AEM.01833-06
Kim, S. K., and Lee, J. H. (2016). Biofilm dispersion in Pseudomonas aeruginosa. J. Microbiol. Seoul Korea 54, 71–85. doi: 10.1007/s12275-016-5528-7
Kirke, D. F., Swift, S., Lynch, M. J., and Williams, P. (2004). The Aeromonas hydrophila LuxR homologue AhyR regulates the N-acyl homoserine lactone synthase, AhyI positively and negatively in a growth phase-dependent manner. FEMS Microbiol. Lett. 241, 109–117. doi: 10.1016/j.femsle.2004.10.011
Kirov, S. M., Castrisios, M., and Shaw, J. G. (2004). Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect. Immun. 72, 1939–1945. doi: 10.1128/IAI.72.4.1939-1945.2004
Kirov, S. M., O'Donovan, L. A., and Sanderson, K. (1999). Functional characterization of type IV pili expressed on diarrhea-associated isolates of Aeromonas species. Infect. Immun. 67, 5447–5454.
Kirov, S. M., Tassell, B. C., Semmler, A. B., O'Donovan, L. A., Rabaan, A. A., and Shaw, J. G. (2002). Lateral flagella and swarming motility in Aeromonas species. J. Bacteriol. 184, 547–555. doi: 10.1128/JB.184.2.547-555.2002
Klausen, M., Gjermansen, M., Kreft, J. U., and Tolker-Nielsen, T. (2006). Dynamics of development and dispersal in sessile microbial communities: examples from Pseudomonas aeruginosa and Pseudomonas putida model biofilms. FEMS Microbiol. Lett. 261, 1–11. doi: 10.1111/j.1574-6968.2006.00280.x
Kozlova, E. V., Khajanchi, B. K., Popov, V. L., Wen, J., and Chopra, A. K. (2012). Impact of QseBC system in c-di-GMP-dependent quorum sensing regulatory network in a clinical isolate SSU of Aeromonas hydrophila. Microb. Pathog. 53, 115–124. doi: 10.1016/j.micpath.2012.05.008
Kozlova, E. V., Khajanchi, B. K., Sha, J., and Chopra, A. K. (2011). Quorum sensing and c-di-GMP-dependent alterations in gene transcripts and virulence-associated phenotypes in a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 50, 213–223. doi: 10.1016/j.micpath.2011.01.007
Kozlova, E. V., Popov, V. L., Sha, J., Foltz, S. M., Erova, T. E., Agar, S. L., et al. (2008). Mutation in the S-ribosylhomocysteinase (luxS) gene involved in quorum sensing affects biofilm formation and virulence in a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 45, 343–354. doi: 10.1016/j.micpath.2008.08.007
Kuchma, S. L., Brothers, K. M., Merritt, J. H., Liberati, N. T., Ausubel, F. M., and O'Toole, G. A. (2007). BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189, 8165–8178. doi: 10.1128/JB.00586-07
Kühn, I., Allestam, G., Huys, G., Janssen, P., Kersters, K., Krovacek, K., et al. (1997). Diversity, persistence, and virulence of Aeromonas strains isolated from drinking water distribution systems in Sweden. Appl. Environ. Microbiol. 63, 2708–2715.
Labbate, M., Queck, S. Y., Koh, K. S., Rice, S. A., Givskov, M., and Kjelleberg, S. (2004). Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J. Bacteriol. 186, 692–698. doi: 10.1128/JB.186.3.692-698.2004
Lamy, B., Kodjo, A., and Laurent, F. (2009). Prospective nationwide study of Aeromonas infections in France. J. Clin. Microbiol. 47, 1234–1237. doi: 10.1128/JCM.00155-09
Lay, C. J., Zhuang, H. J., Ho, Y. H., Tsai, Y. S., Wang, L. S., and Tsai, C. C. (2010). Different clinical characteristics between polymicrobial and monomicrobial Aeromonas bacteremia–a study of 216 cases. Intern. Med. Tokyo Jpn. 49, 2415–2421. doi: 10.2169/internalmedicine.49.4117
Lebeaux, D., Chauhan, A., Rendueles, O., and Beloin, C. (2013). From in vitro to in vivo models of bacterial biofilm-related infections. Pathog. Basel Switz. 2, 288–356. doi: 10.3390/pathogens2020288
Lee, J., and Zhang, L. (2015). The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6, 26–41. doi: 10.1007/s13238-014-0100-x
Li, M., Peng, L., Ji, Z., Xu, J., and Li, S. (2008). Establishment and characterization of dual-species biofilms formed from a 3,5-dinitrobenzoic-degrading strain and bacteria with high biofilm-forming capabilities. FEMS Microbiol. Lett. 278, 15–21. doi: 10.1111/j.1574-6968.2007.00913.x
Li, T., Cui, F., Bai, F., Zhao, G., and Li, J. (2016). Involvement of acylated homoserine lactones (AHLs) of Aeromonas sobria in spoilage of refrigerated turbot (Scophthalmus maximus L.). Sensors 16:1083. doi: 10.3390/s16071083
Li, Y. C., and Zhu, J. R. (2014). Role of N-acyl homoserine lactone (AHL)-based quorum sensing (QS) in aerobic sludge granulation. Appl. Microbiol. Biotechnol. 98, 7623–7632. doi: 10.1007/s00253-014-5815-3
Lim, Y. L., Ee, R., Yin, W. F., and Chan, K. G. (2014). Quorum sensing activity of Aeromonas caviae strain YL12, a bacterium isolated from compost. Sensors 14, 7026–7040. doi: 10.3390/s140407026
Liu, Y.-C., Chan, K.-G., and Chang, C.-Y. (2015). Modulation of Host biology by Pseudomonas aeruginosa quorum sensing signal molecules: messengers or traitors. Microb. Immunol. 6:1226. doi: 10.3389/fmicb.2015.01226
Lynch, M. J., Swift, S., Kirke, D. F., Keevil, C. W., Dodd, C. E., and Williams, P. (2002). The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ. Microbiol. 4, 18–28. doi: 10.1046/j.1462-2920.2002.00264.x
MacIntyre, S., and Buckley, J. T. (1978). Presence of glycerophospholipid: cholesterol acyltransferase and phospholipase in culture supernatant of Aeromonas hydrophila. J. Bacteriol. 135, 402–407.
Madsen, J. S., Burmølle, M., Hansen, L. H., and Sørensen, S. J. (2012). The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 65, 183–195. doi: 10.1111/j.1574-695X.2012.00960.x
Maravić, A., Skočibušić, M., Šamanić, I., Fredotović, Ž., Cvjetan, S., Jutronić, M., et al. (2013). Aeromonas spp. simultaneously harbouring blaCTX-M-15, blaSHV-12, blaPER-1 and blaFOX-2, in wild-growing Mediterranean mussel (Mytilus galloprovincialis) from Adriatic Sea, Croatia. Int. J. Food Microbiol. 166, 301–308. doi: 10.1016/j.ijfoodmicro.2013.07.010
Marchandin, H., Godreuil, S., Darbas, H., Jean-Pierre, H., Jumas-Bilak, E., Chanal, C., et al. (2003). Extended-spectrum β-lactamase TEM-24 in an Aeromonas clinical strain: acquisition from the prevalent enterobacter aerogenes clone in France. Antimicrob. Agents Chemother. 47, 3994–3995. doi: 10.1128/AAC.47.12.3994-3995.2003
Marti, E., Huerta, B., Rodríguez-Mozaz, S., Barceló, D., Jofre, J., and Balcázar, J. L. (2014). Characterization of ciprofloxacin-resistant isolates from a wastewater treatment plant and its receiving river. Water Res. 61, 67–76. doi: 10.1016/j.watres.2014.05.006
Masduki, A., Nakamura, J., Ohga, T., Umezaki, R., Kato, J., and Ohtake, H. (1995). Isolation and characterization of chemotaxis mutants and genes of Pseudomonas aeruginosa. J. Bacteriol. 177, 948–952. doi: 10.1128/jb.177.4.948-952.1995
Medina-Martínez, M. S., Uyttendaele, M., Demolder, V., and Debevere, J. (2006). Influence of food system conditions on N-acyl-L-homoserine lactones production by Aeromonas spp. Int. J. Food Microbiol. 112, 244–252. doi: 10.1016/j.ijfoodmicro.2006.04.025
Merino, S., Bouamama, L., Knirel, Y. A., Senchenkova, S. N., Regué, M., and Tomás, J. M. (2012). Aeromonas surface glucan attached through the O-antigen ligase represents a new way to obtain UDP-glucose. PLOS ONE 7:e35707. doi: 10.1371/journal.pone.0035707
Merino, S., Gavín, R., Altarriba, M., Izquierdo, L., Maguire, M. E., and Tomás, J. M. (2001). The MgtE Mg2+ transport protein is involved in Aeromonas hydrophila adherence. FEMS Microbiol. Lett. 198, 189–195. doi: 10.1111/j.1574-6968.2001.tb10641.x
Merino, S., Wilhelms, M., and Tomás, J. M. (2014). Role of Aeromonas hydrophila flagella glycosylation in adhesion to Hep-2 cells, biofilm formation and immune stimulation. Int. J. Mol. Sci. 15, 21935–21946. doi: 10.3390/ijms151221935
Merod, R. T., and Wuertz, S. (2014). Extracellular polymeric substance architecture influences natural genetic transformation of Acinetobacter baylyi in biofilms. Appl. Environ. Microbiol. 80, 7752–7757. doi: 10.1128/AEM.01984-14
Miller, S. T., Xavier, K. B., Campagna, S. R., Taga, M. E., Semmelhack, M. F., Bassler, B. L., et al. (2004). Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 15, 677–687. doi: 10.1016/j.molcel.2004.07.020
Mizan, M. F., Jahid, I. K., and Ha, S. D. (2015). Microbial biofilms in seafood: a food-hygiene challenge. Food Microbiol. 49, 41–55. doi: 10.1016/j.fm.2015.01.009
Mohd Yasin, N. H., Sanchez-Torres, V., and Maeda, T. (2014). Enhanced reduction of waste activated sludge at a low temperature by locally isolated strains Pseudomonas sp. VNT and Aeromonas sp. VNT. Bioresour. Technol. 174, 134–141. doi: 10.1016/j.biortech.2014.10.005
Morgan-Sagastume, F., Boon, N., Dobbelaere, S., Defoirdt, T., and Verstraete, W. (2005). Production of acylated homoserine lactones by Aeromonas and Pseudomonas strains isolated from municipal activated sludge. Can. J. Microbiol. 51, 924–933. doi: 10.1139/w05-077
Mosser, T., Talagrand-Reboul, E., Colston, S. M., Graf, J., Figueras, M. J., Jumas-Bilak, E., et al. (2015). Exposure to pairs of Aeromonas strains enhances virulence in the Caenorhabditis elegans infection model. Front. Microbiol. 6:1218. doi: 10.3389/fmicb.2015.01218
Nadell, C. D., Xavier, J. B., and Foster, K. R. (2009). The sociobiology of biofilms. FEMS Microbiol. Rev. 33, 206–224. doi: 10.1111/j.1574-6976.2008.00150.x
Nelson, M. C., Bomar, L., Maltz, M., and Graf, J. (2015). Mucinivorans hirudinis gen. nov., sp. nov., an anaerobic, mucin-degrading bacterium isolated from the digestive tract of the medicinal leech Hirudo verbana. Int. J. Syst. Evol. Microbiol. 65, 990–995. doi: 10.1099/ijs.0.000052
Ochiai, S., Morohoshi, T., Kurabeishi, A., Shinozaki, M., Fujita, H., Sawada, I., et al. (2013). Production and degradation of N-acylhomoserine lactone quorum sensing signal molecules in bacteria isolated from activated sludge. Biosci. Biotechnol. Biochem. 77, 2436–2440. doi: 10.1271/bbb.130553
Odling-Smee, F. J., Laland, K. N., and Feldman, M. W. (2003). Niche Construction: The Neglected Process in Evolution. Princeton, NJ: Princeton University Press.
Paranjpye, R. N., and Strom, M. S. (2005). A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infect. Immun. 73, 1411–1422. doi: 10.1128/IAI.73.3.1411-1422.2005
Parsek, M. R., and Greenberg, E. P. (2005). Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 13, 27–33. doi: 10.1016/j.tim.2004.11.007
Parsek, M. R., Val, D. L., Hanzelka, B. L., Cronan, J. E., and Greenberg, E. P. (1999). Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. U.S.A. 96, 4360–4365. doi: 10.1073/pnas.96.8.4360
Perez-Cheeks, B. A., Planet, P. J., Sarkar, I. N., Clock, S. A., Xu, Q., and Figurski, D. H. (2012). The product of tadZ, a new member of the parA/minD superfamily, localizes to a pole in Aggregatibacter actinomycetemcomitans. Mol. Microbiol. 83, 694–711. doi: 10.1111/j.1365-2958.2011.07955.x
Peterson, B. W., He, Y., Ren, Y., Zerdoum, A., Libera, M. R., Sharma, P. K., et al. (2015). Viscoelasticity of biofilms and their recalcitrance to mechanical and chemical challenges. FEMS Microbiol. Rev. 39, 234–245. doi: 10.1093/femsre/fuu008
Petrova, O. E., and Sauer, K. (2016). Escaping the biofilm in more than one way: desorption, detachment or dispersion. Curr. Opin. Microbiol. 30, 67–78. doi: 10.1016/j.mib.2016.01.004
Pinna, A., Sechi, L. A., Zanetti, S., Usai, D., and Carta, F. (2004). Aeromonas caviae keratitis associated with contact lens wear. Ophthalmology 111, 348–351. doi: 10.1016/j.ophtha.2003.05.012
Piotrowska, M., and Popowska, M. (2015). Insight into the mobilome of Aeromonas strains. Front. Microbiol. 6:494. doi: 10.3389/fmicb.2015.00494
Ponnusamy, D., Kozlova, E. V., Sha, J., Erova, T. E., Azar, S. R., Fitts, E. C., et al. (2016). Cross-talk among flesh-eating Aeromonas hydrophila strains in mixed infection leading to necrotizing fasciitis. Proc. Natl. Acad. Sci. U.S.A. 113, 722–727. doi: 10.1073/pnas.1523817113
Ponnusamy, K., Kappachery, S., Thekeettle, M., Song, J. H., and Kweon, J. H. (2013). Anti-biofouling property of vanillin on Aeromonas hydrophila initial biofilm on various membrane surfaces. World J. Microbiol. Biotechnol. 29, 1695–1703. doi: 10.1007/s11274-013-1332-2
Ponnusamy, K., Paul, D., Sam Kim, Y., and Kweon, J. H. (2010). 2(5H)-Furanone: a prospective strategy for biofouling-control in membrane biofilm bacteria by quorum sensing inhibition. Braz. J. Microbiol. 41, 227–234. doi: 10.1590/S1517-83822010000100032
Porter, S. L., Wadhams, G. H., and Armitage, J. P. (2011). Signal processing in complex chemotaxis pathways. Nat. Rev. Microbiol. 9, 153–165. doi: 10.1038/nrmicro2505
Rahman, M., Simm, R., Kader, A., Basseres, E., Römling, U., and Möllby, R. (2007). The role of c-di-GMP signaling in an Aeromonas veronii biovar sobria strain. FEMS Microbiol. Lett. 273, 172–179. doi: 10.1111/j.1574-6968.2007.00803.x
Rangrez, A. Y., Dayananda, K. M., Atanur, S., Joshi, R., Patole, M. S., and Shouche, Y. S. (2006). Detection of conjugation related type four secretion machinery in Aeromonas culicicola. PLoS ONE 1:e115. doi: 10.1371/journal.pone.0000115
Rasamiravaka, T., Vandeputte, O. M., Pottier, L., Huet, J., Rabemanantsoa, C., Kiendrebeogo, M., et al. (2015). Pseudomonas aeruginosa biofilm formation and persistence, along with the production of quorum sensing-dependent virulence factors, are disrupted by a triterpenoid coumarate ester isolated from Dalbergia trichocarpa, a tropical legume. PLoS ONE 10:e0132791. doi: 10.1371/journal.pone.0132791
Rasch, M., Kastbjerg, V. G., Bruhn, J. B., Dalsgaard, I., Givskov, M., and Gram, L. (2007). Quorum sensing signals are produced by Aeromonas salmonicida and quorum sensing inhibitors can reduce production of a potential virulence factor. Dis. Aquat. Organ. 78, 105–113. doi: 10.3354/dao01865
Rhodes, G., Huys, G., Swings, J., McGann, P., Hiney, M., Smith, P., et al. (2000). Distribution of oxytetracycline resistance plasmids between Aeromonads in hospital and aquaculture environments: implication of tn1721 in dissemination of the tetracycline resistance determinant tet A. Appl. Environ. Microbiol. 66, 3883–3890. doi: 10.1128/AEM.66.9.3883-3890.2000
Roger, F., Lamy, B., Jumas-Bilak, E., Kodjo, A., and Marchandin, H. (2012a). Ribosomal multi-operon diversity: an original perspective on the genus Aeromonas. PLoS ONE 7:e46268. doi: 10.1371/journal.pone.0046268
Roger, F., Marchandin, H., Jumas-Bilak, E., Kodjo, A., and Lamy, B. (2012b). Multilocus genetics to reconstruct aeromonad evolution. BMC Microbiol. 12:62. doi: 10.1186/1471-2180-12-62
Römling, U., Galperin, M. Y., and Gomelsky, M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52. doi: 10.1128/MMBR.00043-12
Ruhal, R., Antti, H., Rzhepishevska, O., Boulanger, N., Barbero, D. R., Wai, S. N., et al. (2015). A multivariate approach to correlate bacterial surface properties to biofilm formation by lipopolysaccharide mutants of Pseudomonas aeruginosa. Colloids Surf. B Biointerfaces 127, 182–191. doi: 10.1016/j.colsurfb.2015.01.030
Sauer, K., Camper, A. K., Ehrlich, G. D., Costerton, J. W., and Davies, D. G. (2002). Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184, 1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002
Schmidt, A. S., Bruun, M. S., Dalsgaard, I., and Larsen, J. L. (2001). Incidence, distribution, and spread of tetracycline resistance determinants and integron-associated antibiotic resistance genes among motile Aeromonads from a fish farming environment. Appl. Environ. Microbiol. 67, 5675–5682. doi: 10.1128/AEM.67.12.5675-5682.2001
Schwenteit, J., Gram, L., Nielsen, K. F., Fridjonsson, O. H., Bornscheuer, U. T., Givskov, M., et al. (2011). Quorum sensing in Aeromonas salmonicida subsp. achromogenes and the effect of the autoinducer synthase AsaI on bacterial virulence. Vet. Microbiol. 147, 389–397. doi: 10.1016/j.vetmic.2010.07.020
September, S. M., Els, F. A., Venter, S. N., and Brözel, V. S. (2007). Prevalence of bacterial pathogens in biofilms of drinking water distribution systems. J. Water Health 5, 219–227. doi: 10.2166/wh.2007.004
Seshadri, R., Joseph, S. W., Chopra, A. K., Sha, J., Shaw, J., Graf, J., et al. (2006). Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J. Bacteriol. 188, 8272–8282. doi: 10.1128/JB.00621-06
Sha, J., Rosenzweig, J. A., Kozlova, E. V., Wang, S., Erova, T. E., Kirtley, M. L., et al. (2013). Evaluation of the roles played by Hcp and VgrG type 6 secretion system effectors in Aeromonas hydrophila SSU pathogenesis. Microbiol. Read. Engl. 159, 1120–1135. doi: 10.1099/mic.0.063495-0
Sharma, D. K., Saini, H. S., Singh, M., Chimni, S. S., and Chadha, B. S. (2004). Biodegradation of acid blue-15, a textile dye, by an up-flow immobilized cell bioreactor. J. Ind. Microbiol. Biotechnol. 31, 109–114. doi: 10.1007/s10295-004-0121-1
Shih, Y.-L., and Rothfield, L. (2006). The bacterial cytoskeleton. Microbiol. Mol. Biol. Rev. 70, 729–754. doi: 10.1128/MMBR.00017-06
Silver, A. C., Williams, D., Faucher, J., Horneman, A. J., Gogarten, J. P., and Graf, J. (2011). Complex evolutionary history of the Aeromonas veronii group revealed by host interaction and DNA sequence data. PLoS ONE 6:e16751. doi: 10.1371/journal.pone.0016751
Simm, R., Morr, M., Kader, A., Nimtz, M., and Römling, U. (2004). GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53, 1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x
Sperandio, V., Torres, A. G., Jarvis, B., Nataro, J. P., and Kaper, J. B. (2003). Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. U.S.A. 100, 8951–8956. doi: 10.1073/pnas.1537100100
Sperandio, V., Torres, A. G., and Kaper, J. B. (2002). Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43, 809–821. doi: 10.1046/j.1365-2958.2002.02803.x
Stewart, P. S., and Franklin, M. J. (2008). Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6, 199–210. doi: 10.1038/nrmicro1838
Stine, O. C., Carnahan, A., Singh, R., Powell, J., Furuno, J. P., Dorsey, A., et al. (2003). Characterization of microbial communities from coastal waters using microarrays. Environ. Monit. Assess. 81, 327–336. doi: 10.1023/A:1021353814020
Surette, M. G., Miller, M. B., and Bassler, B. L. (1999). Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. U.S.A. 96, 1639–1644. doi: 10.1073/pnas.96.4.1639
Swift, S., Karlyshev, A. V., Fish, L., Durant, E. L., Winson, M. K., Chhabra, S. R., et al. (1997). Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J. Bacteriol. 179, 5271–5281. doi: 10.1128/jb.179.17.5271-5281.1997
Swift, S., Lynch, M. J., Fish, L., Kirke, D. F., Tomás, J. M., Stewart, G. S., et al. (1999). Quorum sensing-dependent regulation and blockade of exoprotease production in Aeromonas hydrophila. Infect. Immun. 67, 5192–5199.
Tall, B. D., Williams, H. N., George, K. S., Gray, R. T., and Walch, M. (1995). Bacterial succession within a biofilm in water supply lines of dental air-water syringes. Can. J. Microbiol. 41, 647–654. doi: 10.1139/m95-088
Tang, H. J., Lai, C. C., Lin, H. L., and Chao, C. M. (2014). Clinical manifestations of bacteremia caused by Aeromonas species in southern Taiwan. PLoS ONE 9:e91642. doi: 10.1371/journal.pone.0091642
Thiel, V., Kunze, B., Verma, P., Wagner-Döbler, I., and Schulz, S. (2009). New structural variants of homoserine lactones in bacteria. Chembiochem 10, 1861–1868. doi: 10.1002/cbic.200900126
Tischler, A. D., and Camilli, A. (2005). Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect. Immun. 73, 5873–5882. doi: 10.1128/IAI.73.9.5873-5882.2005
Trejo-Hernández, A., Andrade-Domínguez, A., Hernández, M., and Encarnación, S. (2014). Interspecies competition triggers virulence and mutability in Candida albicans-Pseudomonas aeruginosa mixed biofilms. ISME J. 8, 1974–1988. doi: 10.1038/ismej.2014.53
Truchado, P., Gil-Izquierdo, A., Tomás-Barberán, F., and Allende, A. (2009). Inhibition by chestnut honey of N-Acyl-L-homoserine lactones and biofilm formation in Erwinia carotovora, Yersinia enterocolitica, and Aeromonas hydrophila. J. Agric. Food Chem. 57, 11186–11193. doi: 10.1021/jf9029139
Tsuchiya, Y., Eda, S., Kiriyama, C., Asada, T., and Morisaki, H. (2016). Analysis of dissolved organic nutrients in the interstitial water of natural biofilms. Microb. Ecol. 72, 85–95. doi: 10.1007/s00248-016-0749-1
Tsuchiya, Y., Ikenaga, M., Kurniawan, A., Hiraki, A., Arakawa, T., Kusakabe, R., et al. (2009). Nutrient-rich microhabitats within biofilms are synchronized with the external environment. Microbes Environ. 24, 43–51. doi: 10.1264/jsme2.ME08547
Van Acker, H., Van Dijck, P., and Coenye, T. (2014). Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol. 22, 326–333. doi: 10.1016/j.tim.2014.02.001
Van Houdt, R., Aertsen, A., and Michiels, C. W. (2007). Quorum-sensing-dependent switch to butanediol fermentation prevents lethal medium acidification in Aeromonas hydrophila AH-1N. Res. Microbiol. 158, 379–385. doi: 10.1016/j.resmic.2006.11.015
Vilches, S., Jimenez, N., Tomás, J. M., and Merino, S. (2009). Aeromonas hydrophila AH-3 type III secretion system expression and regulatory network. Appl. Environ. Microbiol. 75, 6382–6392. doi: 10.1128/AEM.00222-09
Villari, P., Crispino, M., Montuori, P., and Boccia, S. (2003). Molecular typing of Aeromonas isolates in natural mineral waters. Appl. Environ. Microbiol. 69, 697–701. doi: 10.1128/AEM.69.1.697-701.2003
Whitby, P. W., Landon, M., and Coleman, G. (1992). The cloning and nucleotide sequence of the serine protease gene (aspA) of Aeromonas salmonicida ssp. salmonicida. FEMS Microbiol. Lett. 78, 65–71. doi: 10.1111/j.1574-6968.1992.tb05543.x
Willcox, M. D., Harmis, N., Cowell, Williams, T., and Holden. (2001). Bacterial interactions with contact lenses; effects of lens material, lens wear and microbial physiology. Biomaterials 22, 3235–3247. doi: 10.1016/S0142-9612(01)00161-2
Worthen, P. L., Gode, C. J., and Graf, J. (2006). Culture-independent characterization of the digestive-tract microbiota of the medicinal leech reveals a tripartite symbiosis. Appl. Environ. Microbiol. 72, 4775–4781. doi: 10.1128/AEM.00356-06
Wuichet, K., and Zhulin, I. B. (2010). Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 3, ra50–ra50. doi: 10.1126/scisignal.2000724
Xavier, K. B., and Bassler, B. L. (2003). LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6, 191–197. doi: 10.1016/S1369-5274(03)00028-6
Xavier, K. B., and Bassler, B. L. (2005). Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187, 238–248. doi: 10.1128/JB.187.1.238-248.2005
Zalmum, A. A., Marialegite, K., and Ghenghesh, K. S. (1998). Bacterial composition of the biofilm on the surface of course sediment of the Danube: with special reference to the clinically important bacteria. Arch. Inst. Pasteur Tunis 75, 205–209.
Keywords: biofilm, quorum sensing, multicellularity, coordination, cooperation, social life, bacterial communities, virulence
Citation: Talagrand-Reboul E, Jumas-Bilak E and Lamy B (2017) The Social Life of Aeromonas through Biofilm and Quorum Sensing Systems. Front. Microbiol. 8:37. doi: 10.3389/fmicb.2017.00037
Received: 11 October 2016; Accepted: 06 January 2017;
Published: 20 January 2017.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Giovanna Batoni, University of Pisa, ItalyJuan M Tomas, University of Barcelona, Spain
Copyright © 2017 Talagrand-Reboul, Jumas-Bilak and Lamy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brigitte Lamy, YnJpZ2l0dGVfbGFteUB5YWhvby5mcg==
 Emilie Talagrand-Reboul
Emilie Talagrand-Reboul Estelle Jumas-Bilak
Estelle Jumas-Bilak Brigitte Lamy
Brigitte Lamy