- 1Department of Life Sciences, Taishan Medical University, Tai'an, China
- 2Department of Basic Medicine, Taishan Medical University, Tai'an, China
- 3Department of Pediatrics, Maternal and Child Health hospital of Laiwu, Laiwu, China
- 4Disease Controlling Center, Veterinary Bureau of Daiyue, Tai'an, China
This study aimed to investigate antimicrobial resistance and molecular epidemiology of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli (E. coli) isolated from outpatients in town hospitals of Shandong province, China. Antimicrobial susceptibility of ESBL-producing E. coli was tested using the disk diffusion and resistance genes encoding for β-lactamases (blaTEM, blaCTXM, and blaSHV) were detected by polymerase chain reaction (PCR). Multilocus sequence typing (ST) of ESBL-producing E. coli was analyzed in this study. Our results showed that of 320 E. coli isolates, 201 carried ESBL genes (201/320, 62.8%), and these isolates all carried blaCTX-M genes, the most common being blaCTX-M-14 (116/201, 57.7%), followed by blaCTX-M-55 (47/201, 23.4%) and blaCTX-M-15 (31/201, 15.4%). ESBL-producing E. coli exhibited highly resistant to penicillin derivatives, fluoroquinolones, folate pathway inhibitors, and third-generation cephalosporins, but no carbapenem-resistant isolates were found in this study. Forty-two STs were found among the 201 ESBL-producing E. coli, and the most common ST was ST131 (27/201, 13.4%), followed by ST405 (19/201, 9.5%) and ST69 (15/201, 7.5%). Taken together, a high isolation rate of ESBL-producing E. coli (62.8%) was found among outpatients in town hospitals. blaCTX-M gene was most dominant and was composed of a variety of subtypes. No dominant ST was detected among ESBL-producing E. coli, indicating that these ESBL-producing E. coli isolates derive from different clones.
Introduction
Beta-lactam antimicrobials are first line anti-bacterial infection drugs for humans due to their high potency, broad anti-bacterial spectrum, and minimal side effects. They are widely used in the treatment of various infections, such as those of the lungs, urinary tract, and the bloodstream. However, widespread use of antibiotics has intensified the problem of antibiotic resistance in bacteria (Paterson and Bonomo, 2005; Biedenbach et al., 2014; D'Angelo et al., 2016). The production of extended-spectrum beta-lactamases (ESBLs) is an important mechanism of antimicrobial resistance in Enterobacteriaceae, especially Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pneumoniae), and the enzyme can hydrolyze penicillin, cephalosporin, and monocyclic amide antibiotics, but its activity is usually inhibited by beta-lactamase inhibitors, such as sulbactam, clavulanic acid, and tazobactam (Bush et al., 1995; Bradford, 2001).
Currently, over hundreds of ESBLs have been identified; the most prevalent genotypes are blaTEM, blaSHV, and blaCTX−M. Within the past decade, the genotype blaCTX−M has rapidly increased and is now widely found in clinically isolated E. coli across the world (Paterson and Bonomo, 2005; Livermore et al., 2007). In practice, blaCTX−M genes have already become the major ESBL genotype in American, European, and Asian countries (Pitout et al., 2005; Livermore et al., 2007; Ben-Ami et al., 2009; Zhang et al., 2014). Emergence of community-associated infections caused by ESBL-producing E. coli has been reported in Europe and the United States (Ben-Ami et al., 2009). Moreover, relevant studies from Oceania, Asia, and South America have also reported that ESBL-positive E. coli are the key pathogens in community-onset infections (Baas and Ahmad, 2001; Bell et al., 2002; Munday et al., 2004; da Silva Dias et al., 2008; Baurin et al., 2009; Rawat et al., 2013).
Numerous studies in China have already demonstrated that ESBL-producing E. coli in tertiary and county hospitals is becoming an epidemic (Xiao et al., 2011, 2012, 2013; Zhang et al., 2014; Liu et al., 2015). Previous studies that monitored infections in tertiary hospitals of China indicated that the prevalence of ESBL-producing E. coli was rapidly on the rise, increasing from an ESBL-positive rate of <20% in 2000 to 72.2% in 2011 (Xiao et al., 2011, 2012, 2013). A similar study that examined infections in county hospitals across China also reported an ESBL-positive rate of up to 46.5% in E. coli (Zhang et al., 2014). However, these studies were focused on city hospitals, and there are very few reports that have examined ESBL-producing E. coli in town hospitals of rural areas in China. Therefore, this study was undertaken to investigate drug-resistance and molecular epidemiology of ESBL-producing E. coli isolated from outpatients in town hospitals of Shandong province, in order to provide comprehensive and reliable epidemiological information for preventing dissemination of resistance genes.
Materials and Methods
Ethics Statement
This study was in compliance with the various requirements of the Research Ethics Committee of Taishan Medical University (Permit No.: TSMC20141012). All participants signed an informed consent.
Sample Collection
Sputum and urine samples of outpatients were collected from 15 town hospitals across three regions of the Shandong province (five hospitals per region from October 2014 to September 2015), for E. coli isolation (Figure 1). The outpatients were selected according to the following three conditions: (1) they had not stayed at the hospital within the past 3 months, (2) they had no long-term intubation, and (3) they had not taken antimicrobial medication for over 72 h before treatment.
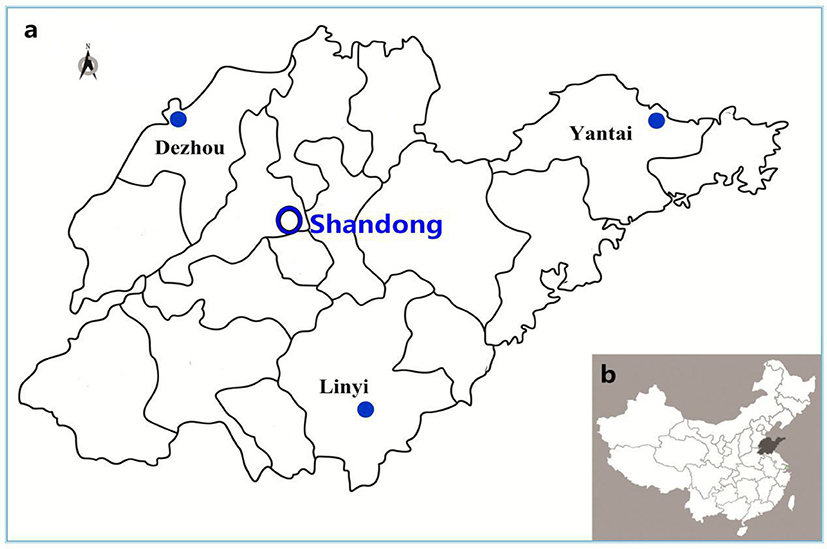
Figure 1. Sampling sites in this study. (A): The enlarged map of Shandong province, in which sampling sites in three administrative districts was marked. (B): The location of Shandong province was highlighted in China.
Isolation and Identification of E. coli
Samples were transported back to the lab on ice within 6–10 h of collection, for E. coli isolation and identification. Samples were inoculated onto MacConkey agar plates using sterile cotton swabs and were incubated overnight at 37°C in aerobic conditions. Five single red colonies from each patient sample were selected for further colony purification, and the colonies were subsequently identified using conventional biochemical methods and API20 assays (bioMérieux, Durham, NC, USA). All positively identified E. coli strains (one strain per patient) were stored at −80°C in Luria-Bertani (LB) broth containing 30% glycerol.
Antimicrobial Susceptibility and ESBL Phenotypic Confirmatory Tests
E. coli susceptibility to 17 antibiotics, including ampicillin, piperacillin, ampicillin-sulbactam, piperacillin-tazobactam, cefotaxime, cefriaxone, cefuroxime, cefepime, ceftazidime, aztreonam, imipenem, meropenem, amikacin, gentamicin, ciprofloxacin, levofloxacin, and trimethoprim-sulfamethoxazole, was tested using disk diffusion. All drug susceptibility testing were performed in accordance with the CLSI 2014 criteria (Clinical Laboratory Standards Institute, 2014). E. coli ATCC25922 and K. pneumoniae ATCC700603 were used as quality control strains.
ESBL phenotypic confirmatory test was performed on E. coli using the double-disc synergy procedure with paper disks that contained ceftazidime and cefotaxime alone, or in combination with clavulanic acid (30 μg ceftazidime, 30/10 μg ceftazidime/clavulanic acid, 30 μg cefotaxime, 30/10 μg cefotaxime/clavulanic acid) (Oxoid Limited, UK; Clinical Laboratory Standards Institute, 2014).
Bacterial DNA Extraction
Single colonies of ESBL-producing E. coli were inoculated into LB media and cultured overnight at 37°C with 220 rpm shaking. Bacterial culture (1 mL) was transferred to an Eppendorf tube, centrifuged at 12,000 rpm for 5 min, before the pellet was resuspended in 60 μl of sterile ultrapure water. The solution was then placed in boiling water for 10 min, immediately transferred to an ice bath for 5 min, and centrifuged at 12,000 rpm for 5 min to obtain the extracted bacterial DNA in the supernatant.
Detection of Beta-Lactamase Gene by PCR
Polymerase chain reaction (PCR) amplification for the beta-lactamase genes (TEM, SHV, and CTX-M) were carried out as previously described (Yu et al., 2007; Dallenne et al., 2010; Sun et al., 2010; Zhang et al., 2011, 2014). The PCR products were sequenced following purification, and ESBL genotype was determined by comparing to GenBank sequences (http://www.ncbi.nlm.nih.gov/BLAST).
MLST
Multilocus sequence typing of the ESBL-producing E. coli was performed according to the experimental procedures on the Environmental Research Institute, University College Cork website (http://mlst.ucc.ie/mlst/dbs/Ecoli; Lau et al., 2008a). The E. coli strains were grouped according to the eBURST algorithm based on their allelic properties, where strains with the same six out of seven alleles were assigned to the same group (Feil et al., 2004).
Statistical Analysis
Statistical analysis was performed using SAS 8.2 (SAS Institute, Cary, NC, USA). Continuous variables and categorical variables were compared using the Student's t-test and chi-squared test or Fisher's exact test, respectively. A two-tailed P < 0.05 was considered to be statistically significant.
Results
Outpatient Demographics
A total of 320 outpatients were recruited in this study, including 110 from Yantai (YT), 90 from Dezhou (DZ), and 120 from Linyi (LY). Among 320 outpatients aged 10–85 years, there were 32 between 10 and 18 years of age (10.0%), 161 between 19 and 45 years (50.3%), 107 between 46 and 65 years (33.4%), and 20 over 65 years (6.3%). There were more male (185/320, 57.8%) than female (135/320, 42.2%) outpatients.
Isolation and Identification of E. coli
A total of 320 E. coli isolates were recovered, comprising 231 isolated from urine (72.2%) and 89 (27.8%) from sputum. Among these 320 E. coli, 201 carried ESBL genes (201/320, 62.8%), including 67 (67/110, 60.9%), 79 (79/120, 65.8%), and 55(55/90, 61.1%) isolates from YT, LY, and DZ, respectively. The isolation rates of ESBL-producing E. coli among three regions did not differ significantly (P > 0.05), but the isolation rate of ESBL-producing E. coli in urine (170/231, 73.6%) was significantly higher than that in sputum (31/89, 34.8%; P < 0.05).
Antimicrobial Resistance Characteristics
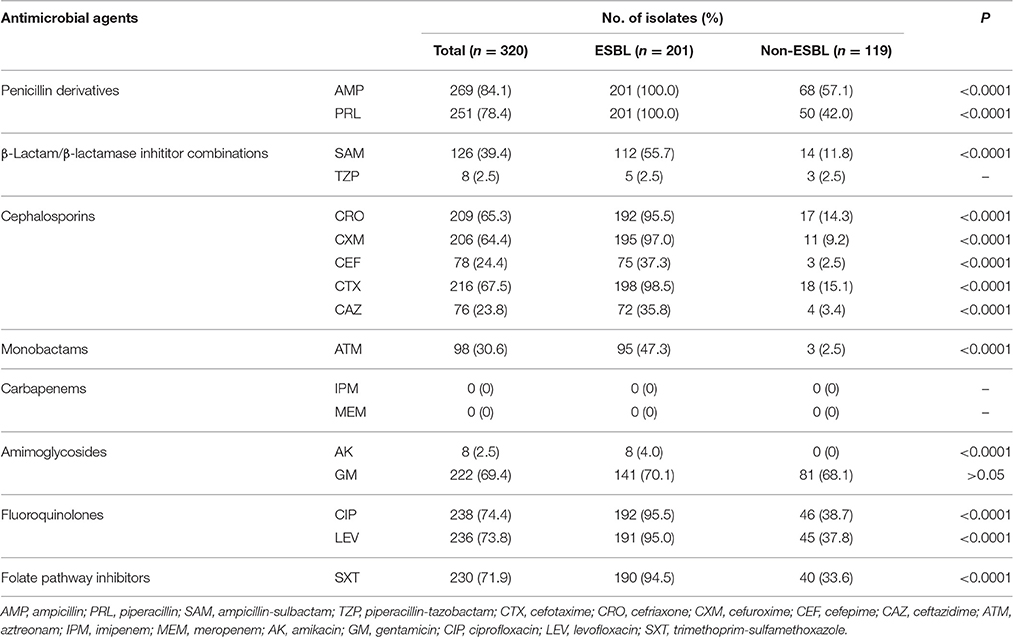
All of the 320 E. coli were susceptible to imipenem and meropenem, and showed high resistance rates to ampicillin (269/320, 84.1%), piperacillin (251/320, 78.4%), ciprofloxacin (238/320, 74.4%), levofloxacin (236/320, 73.8%), trimethoprim-sulfamethoxazole (230/320, 71.9%), gentamicin (222/320, 69.4%), cefotaxime (216/320, 67.5%), ceftriaxone (209/320, 65.3%), and cefuroxime (206/320, 64.4%). By contrast, these isolates exhibited low resistance rates to piperacillin/tazobactam (8/320, 2.5%), amikacin (8/320, 2.5%), ceftazidime (76/320, 23.8%), and cefepime (78/320, 24.4%). In addition, ESBL-producing E. coli showed significantly higher resistance rates to most antibiotics compared with those of non-ESBL-producing E. coli (P < 0.0001). However, the low resistance rate against piperacillin-tazobactam and high resistance rate to gentamicin in the two populations did not differ significantly (P > 0.05; Table 1).
ESBL Gene Characteristics
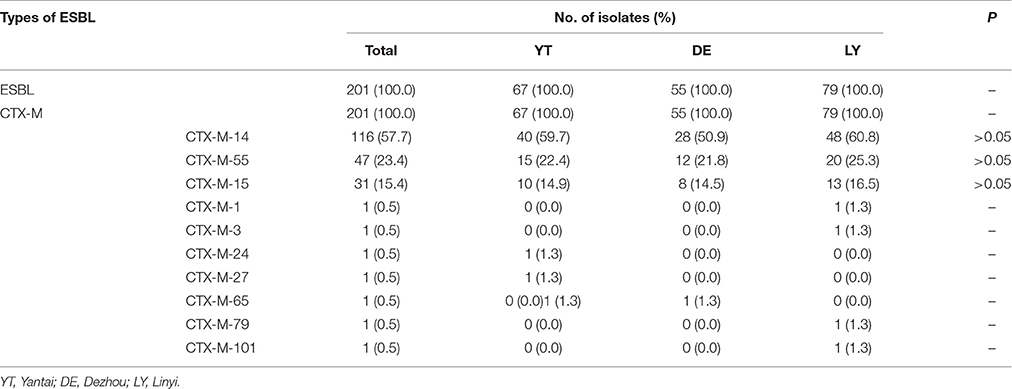
All of the 201 ESBL-producing E. coli strains carried blaCTX-M genes, with the most common being blaCTX-M-14 (116/201, 57.7%), followed by blaCTX-M-55 (47/201, 23.4%) and blaCTX-M-15 (31/201, 15.4%). Additionally, 122 of the 201 ESBL-producing E. coli strains simultaneously carried blaTEM-1 genes. blaSHV genes were not detected in this study. There was no significant difference in the prevalence of blaCTX-M genes among the ESBL-producing E. coli isolated from the three regions (P > 0.05; Table 2).
Multilocus Sequence Typing of ESBL-Producing E. coli
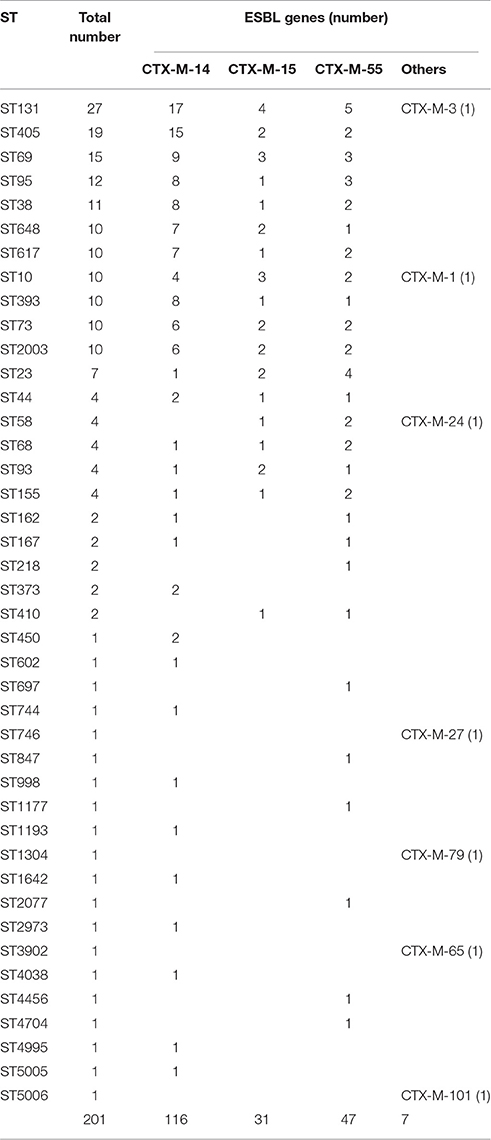
A total of 42 different STs were found, which were grouped in 5 non-overlapping groups, 1 clonal complex, and 23 singletons (Figure 2). The most common ST were ST131 (27/201, 13.4%), followed by ST405 (19/201, 9.5%) and ST69 (15/201, 7.5%). There were 27 ST131 detected from town hospitals across the three regions, including 17 strains that were blaCTX-M-14-positive, 4 strains that were blaCTX-M-15-positive, 5 strains that were blaCTX-M-55-positive, and 1 strain that carried blaCTX-M-3 gene (Table 3).
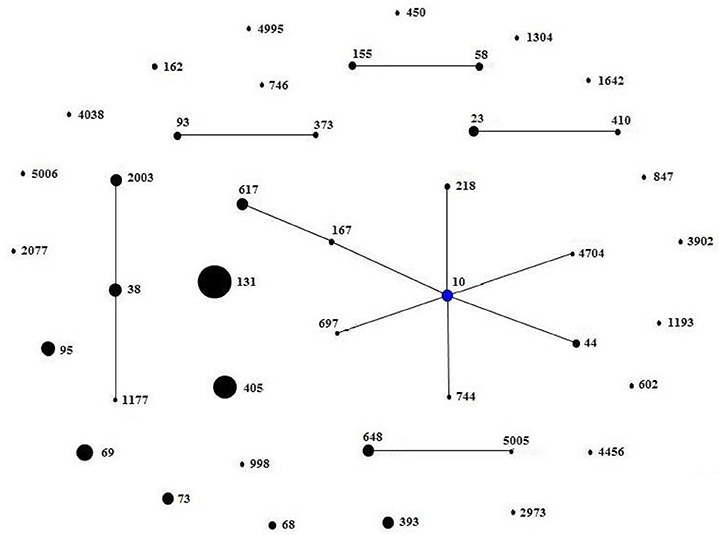
Figure 2. Minimum spanning tree constructed based on the MLST profiles of ESBL-producing E. coli. There were 23 singletons, 5 groups (group1: ST58, ST155; group2: ST93, ST373; group3: ST23, ST410; group4: ST2003, ST38, ST1177; group5: ST648, ST5005), and 1 clonal complex (ST617, ST167, ST10, ST44, ST218, ST744, ST697, ST4704), which was radial. The blue dot in it indicated putative founder. The area of each black circle corresponded to the prevalence of the ST in the MLST data of this study.
Discussion
To our best knowledge, this study was the first time to investigate drug resistance and molecular characteristics of ESBL-producing E. coli from outpatients in town hospitals of Shandong province, China. The isolation rate of ESBL-producing E. coli in our study was 62.8%, which was similar with Zhao's (62.5%) and Wang's (67.8%) results conducted in Shanghai, China (Zhao et al., 2015; Wang et al., 2016), but higher than those reported in Argentina (18.1%), Mexico (48.4%), Chile (23.8%), and Brazil (12.8%; Gales et al., 2012). All ESBL-producing E. coli from these three regions carried blaCTX-M genes, which was composed of 10 genotypes including blaCTX-M-1, -3, -14, -15, -24, -27, -55, -65, -79, and-101. This indicates that ESBL-producing E. coli from Shandong province have diverse CTX-M genotypes, and similar results were also found in the tertiary and county hospitals of China (Zhang et al., 2014; Zhao et al., 2015; Wang et al., 2016).
E. coli isolated in this study were found to be highly resistant to penicillin derivatives, fluoroquinolone, folate pathway inhibitors, and third generation cephalosporins, but were 100% susceptible to imipenem and meropenem. In addition, these isolates displayed low resistance to amikacin, piperacillin/tazobactam, ceftazidime, and cefepime. The antibiotics to which the E. coli was found to be highly resistant are common medications used in Shandong town hospitals, and therefore our findings should caution clinicians for the rational use of antibiotics.
We found that blaCTX-M-14 was the most prevalent genotype of ESBL-producing E. coli in Shandong town hospitals, followed by blaCTX-M-55 and blaCTX-M-15, which is consistent with findings reported in Chinese county hospitals between 2010 and 2011 (Zhang et al., 2014), as well as in 3 Shanghai hospital studies between 2011 and 2013 (Zhao et al., 2015). CTX-M-55 genotype, which only has 1 amino acid site mutation (Ala-77-Val) compared to CTX-M-15 genotype, was first discovered in clinically isolated E. coli and K. pneumoniae from Thailand in 2007 (Kiratisin et al., 2007), and was subsequently detected in Salmonella in China, US, Korea, and Switzerland (Shi et al., 2009; Sjölund-Karlsson et al., 2011). At present, CTX-M-55 genotype is frequently detected in ESBL-producing E. coli that originates from animals (Dinubile et al., 2005; Ma et al., 2012; Zheng et al., 2012; Zurfluh et al., 2013; Li et al., 2016). In China, two nationwide breeding farm studies have shown that CTX-M-55 was, respectively, the second (26.1%, 29/111) and third (18.5%, 10/54) most frequently detected ESBL gene (Li et al., 2010; Zheng et al., 2012). These findings demonstrated that blaCTX-M-55 gene may have already been passed from animals to humans through the food chain. The subjects of this study were outpatients from rural town hospitals. Since residents from these regions have greater exposure to food animals and breeding farms, compared to those living in the cities, the chance of transmission of drug-resistant bacteria from animals to humans is therefore increased. However, the transmission mechanism of drug-resistant bacteria from animals to humans is currently unclear, and further studies are required to elucidate this process. Additionally, it is an interesting finding that the resistance to cefotaxime and ceftriaxone is not 100% while only CTX-M-type ESBLs were found in this study, which is needed to be further studied.
Although ST131 was the most common ST among the 201 ESBL-producing E. coli strains, it only accounted for 13.4% of the total ST. Similarly, some recent nationwide studies in tertiary and county hospitals have also shown that ST131 was found in 9.6% and 12.7% of ESBL-producing E. coli, respectively, indicating that no predominant ESBL-producing E. coli ST epidemic was found in China (Cao et al., 2011; Zhang et al., 2014). In contrast, the percentage of ST131 ESBL-producing E. coli in many European and American countries is far greater than that in China. For example, a community infection study in U.S showed that 53% of ESBL gene CTX-M-carrying E. coli were ST131 (Pitout et al., 2005). Another community infection study in the U.K also reported that ST131 comprised 64% of the cephalosporin-resistant E. coli (Lau et al., 2008b). Furthermore, a similar community infection study in Belgium between 2006 and 2007 showed that 64% of CTX-M-15-carrying E. coli was also ST131 (Smet et al., 2010). Of note, some sequence types found in this work belong to known international clonal complexes, such as ST131, ST393, and ST405 (Wirth et al., 2006; Hrabák et al., 2009). These international clonal complexes have been described as E. coli clones disseminating on a global scale (Coque et al., 2008; Nicolas-Chanoine et al., 2008; Literacka et al., 2009; Lee et al., 2010).
Conclusions
Taken together, These findings demonstrated the high isolation rate of ESBL-producing E. coli (62.8%) detected in outpatients in town hospitals, China, and the blaCTX-M gene was most dominant and was composed of a variety of subtypes. More importantly, this study spotlights the necessity to carry out long-term surveillance of ESBL-producing E. coli in hospital environments, especially in underdeveloped areas.
Author Contributions
ZM and SL conceived and designed the experiment. LW, YZ, and SL collected these isolates. SL, LW, YZ, and WS performed the experiments. ZM and SL analyzed the data and wrote the paper.
Funding
This study was supported by the National Natural Science Foundations of China (81501357) and Science and Technology Development Project of Shandong Province (2014GSF118044).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Baas, P. W., and Ahmad, F. J. (2001). Force generation by cytoskeletal motor proteins as a regulator of axonal elongation and retraction. Trends Cell Biol. 11, 244–249. doi: 10.1016/S0962-8924(01)02005-0
Baurin, S., Vercheval, L., Bouillenne, F., Falzone, C., Brans, A., Jacquamet, L., et al. (2009). Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases. Biochemistry 48, 11252–11263. doi: 10.1021/bi901548c
Bell, J. M., Turnidge, J. D., Gales, A. C., Pfaller, M. A., Jones, R. N., and Sentry, A. S. G. (2002). Prevalence of extended spectrum beta-lactamase (ESBL)-producing clinical isolates in the Asia-Pacific region and South Africa: regional results from SENTRY Antimicrobial Surveillance Program (1998-99). Diagn. Microbiol. Infect. Dis. 42, 193–198. doi: 10.1016/S0732-8893(01)00353-4
Ben-Ami, R., Rodríguez-Ba-o, J., Arslan, H., Pitout, J. D., Quentin, C., Calbo, E. S., et al. (2009). A multinational survey of risk factors for infection with extended-spectrum beta-lactamase-producing enterobacteriaceae in nonhospitalized patients. Clin. Infect. Dis. 49, 682–690. doi: 10.1086/604713
Biedenbach, D. J., Bouchillon, S. K., Hoban, D. J., Hackel, M., Phuong, D. M., Nga, T. T., et al. (2014). Antimicrobial susceptibility and extended-spectrum beta-lactamase rates in aerobic gram-negative bacteria causing intra-abdominal infections in Vietnam: report from the Study for Monitoring Antimicrobial Resistance Trends (SMART-2011). Diagn. Microbiol. Infect. Dis. 79, 463–467. doi: 10.1016/j.diagmicrobio.2014.05.009
Bradford, P. A. (2001). Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14, 933–951. doi: 10.1128/CMR.14.4.933-951.2001
Bush, K., Jacoby, G. A., and Medeiros, A. A. (1995). A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211. doi: 10.1128/AAC.39.6.1211
Cao, X., Cavaco, L. M., Lv, Y., Li, Y., Zheng, B., Wang, P., et al. (2011). Molecular characterization and antimicrobial susceptibility testing of Escherichia coli isolates from patients with urinary tract infections in 20 Chinese hospitals. J. Clin. Microbiol. 49, 2496–2501. doi: 10.1128/JCM.02503-10
Clinical Laboratory Standards Institute (2014). Performance Standards for Antimicrobial Susceptibility Testing; 24th Informational Supplement, M100-S24. Wayne, PA.
Coque, T. M., Novais, A., Carattoli, A., Poirel, L., Pitout, J., Peixe, L., et al. (2008). Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg. Infect. Dis. 14, 195–200. doi: 10.3201/eid1402.070350
Dallenne, C., Da Costa, A., Decre, D., Favier, C., and Arlet, G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65, 490–495. doi: 10.1093/jac/dkp498
D'Angelo, R. G., Johnson, J. K., Bork, J. T., and Heil, E. L. (2016). Treatment options for extended-spectrum beta-lactamase (ESBL) and AmpC-producing bacteria. Expert Opin. Pharmacother. 17, 953–967. doi: 10.1517/14656566.2016.1154538
da Silva Dias, R. C., Borges-Neto, A. A., D'Almeida Ferraiuoli, G. I., de-Oliveira, M. P., Riley, L. W., and Moreira, B. M. (2008). Prevalence of AmpC and other beta-lactamases in enterobacteria at a large urban university hospital in Brazil. Diagn. Microbiol. Infect. Dis. 60, 79–87. doi: 10.1016/j.diagmicrobio.2007.07.018
Dinubile, M. J., Friedland, I., Chan, C. Y., Motyl, M. R., Giezek, H., Shivaprakash, M., et al. (2005). Bowel colonization with resistant gram-negativebacilli after antimicrobial therapy of intra-abdominal infections: observations from two randomized comparative clinical trials of ertapenem therapy. Eur. J. Clin. Microbiol. Infect. Dis. 24, 443–449. doi: 10.1007/s10096-005-1356-0
Feil, E. J., Li, B. C., Aanensen, D. M., Hanage, W. P., and Spratt, B. G. (2004). eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186, 1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004
Gales, A. C., Castanheira, M., Jones, R. N., and Sader, H. S. (2012). Antimicrobial resistance among Gram-negative bacilli isolated from Latin America: results from SENTRY Antimicrobial Surveillance Program (Latin America, 2008–2010). Diagn. Microbiol. Infect. Dis. 73, 354–360. doi: 10.1016/j.diagmicrobio.2012.04.007
Hrabák, J., Empel, J., Bergerová, T., Fajfrlík, K., Urbásková, P., Kern-Zdanowicz, I., et al. (2009). International clones of Klebsiella pneumoniae and Escherichia coli with extended-spectrum beta-lactamases in a Czech hospital. J. Clin. Microbiol. 47, 3353–3357. doi: 10.1128/JCM.00901-09
Kiratisin, P., Apisarnthanarak, A., Saifon, P., Laesripa, C., Kitphati, R., and Mundy, L. M. (2007). The emergence of a novel ceftazidime-resistant CTX-M extended spectrum β-lactamase, CTX-M-55, in both community-onset and hospital-acquired infections in Thailand. Diagn. Microbiol. Infect. Dis. 58, 349–355. doi: 10.1016/j.diagmicrobio.2007.02.005
Lau, S. H., Kaufmann, M. E., Livermore, D. M., Woodford, N., Willshaw, G. A., Cheasty, T., et al. (2008b). UK epidemic Escherichia coli strains A–E, with CTX-M-15 β -lactamase, all belong to the international O25: H4-ST131 clone. J. Antimicrob. Chemother. 62, 1241–1244. doi: 10.1093/jac/dkn380
Lau, S. H., Reddy, S., Cheesbrough, J., Bolton, F. J., Willshaw, G., Cheasty, T., et al. (2008a). Major uropathogenic Escherichia coli strain isolated in the northwest of England by multilocus sequence typing. J. Clin. Microbiol. 46, 1076–1080. doi: 10.1128/JCM.02065-07
Lee, M. Y., Choi, H. J., Choi, J. Y., Song, M., Song, Y., Kim, S. W., et al. (2010). Dissemination of ST131 and ST393 community-onset, ciprofloxacin-resistant Escherichia coli clones causing urinary tract infections in Korea. J. Infect. 60, 146–153. doi: 10.1016/j.jinf.2009.11.004
Li, J., Ma, Y., Hu, C., Jin, S., Zhang, Q., Ding, H., et al. (2010). Dissemination of cefotaxime-M-producing Escherichia coli isolates in poultry farms, but not swine farms, in China. Foodborne Pathog. Dis. 7, 1387–1392. doi: 10.1089/fpd.2010.0581
Li, S., Zhao, M., Liu, J., Zhou, Y., and Miao, Z. (2016). Prevalence and Antibiotic Resistance profiles of extended-spectrum β-lactamase-producing Escherichia coli isolated from healthy broilers in shandong province, China. J. Food Prot. 79, 1169–73. doi: 10.4315/0362-028X.JFP-16-025
Literacka, E., Bedenic, B., Baraniak, A., Fiett, J., Tonkic, M., Jajic-Bencic, I., et al. (2009). blaCTX-M genes in Escherichia coli strains from croatian hospitals are located in new (blaCTX-M-3a) and widely spread (blaCTX-M-3a and blaCTX-M-15) genetic structures. Antimicrob. Agents Chemother. 53, 1630–1635. doi: 10.1128/AAC.01431-08
Liu, H., Wang, Y., Wang, G., Xing, Q., Shao, L., Dong, X., et al. (2015). The prevalence of Escherichia coli strains with extended spectrum beta-lactamases isolated in China. Front. Microbiol. 6:335. doi: 10.3389/fmicb.2015.00335
Livermore, D. M., Canton, R., Gniadkowski, M., Nordmann, P., Rossolini, G. M., Arlet, G., et al. (2007). CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59, 165–174. doi: 10.1093/jac/dkl483
Ma, J., Liu, J. H., Lv, L., Zong, Z., Sun, Y., Zheng, H., et al. (2012). Characterization of extended-spectrum beta-lactamase genes found among Escherichia coli isolates from duck and environmental samples obtained on a duck farm. Appl. Environ. Microbiol. 78, 3668–3673. doi: 10.1128/AEM.07507-11
Munday, C. J., Xiong, J., Li, C., Shen, D., and Hawkey, P. (2004). Dissemination of CTX-M type β-lactamases in Enterobacteriaceae isolates in the people's republic of China. Int. J. Antimicrob. Agents. 23, 175–180. doi: 10.1016/j.ijantimicag.2003.07.004
Nicolas-Chanoine, M. H., Blanco, J., Leflon-Guibout, V., Demarty, R., Alonso, M. P., Caniça, M. M., et al. (2008). Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61, 273–281. doi: 10.1093/jac/dkm464
Paterson, D. L., and Bonomo, R. A. (2005). Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev. 18, 657–686. doi: 10.1128/CMR.18.4.657-686.2005
Pitout, J. D., Gregson, D. B., Church, D. L., Elsayed, S., and Laupland, K. B. (2005). Community-wide outbreaks of clonally related CTX-M-14 beta-lactamase-producing Escherichia coli strains in the Calgary health region. J. Clin. Microbiol. 43, 2844–2849. doi: 10.1128/JCM.43.6.2844-2849.2005
Rawat, V., Singhai, M., and Verma, P. K. (2013). Detection of different beta-Lactamases and their co-existence by using various discs combination methods in clinical isolates of enterobacteriaceae and pseudomonas spp. J. Lab. Phys. 5, 21–25. doi: 10.4103/0974-2727.115918
Shi, W.-F., Zhou, J., and Qin, J.-P. (2009). Transconjugation and genotyping of the plasmid-mediated AmpC beta-lactamase and extended-spectrum beta-lactamase genes in Klebsiella pneumoniae. Chin. Med. J. (Engl.). 122, 1092–1096. doi: 10.3760/cma.j.issn.0366-6999.2009.09.015
Sjölund-Karlsson, M., Howie, R., Krueger, A., Rickert, R., Pecic, G., Lupoli, K., et al. (2011). CTX-M-producing non-Typhi Salmonella spp. Isolated from humans, United States. Emerg. Infect. Dis. 17, 97–99. doi: 10.3201/eid1701.100511
Smet, A., Martel, A., Persoons, D., Dewulf, J., Heyndrickx, M., Claeys, G., et al. (2010). Characterization of extended-spectrum β -lactamases produced by Escherichia coli isolated from hospitalized and nonhospitalized patients: emergence of CTX-M-15-producing strains causing urinary tract infections. Microb. Drug Resist. 16, 129–134. doi: 10.1089/mdr.2009.0132
Sun, Y., Zeng, Z., Chen, S., Ma, J., He, L., Liu, Y., et al. (2010). High prevalence of bla(CTX-M) extended-spectrum beta-lactamase genes in Escherichia coli isolates from pets and emergence of CTX-M-64 in China. Clin. Microbiol. Infect. 16, 1475–1481. doi: 10.1111/j.1469-0691.2010.03127.x
Wang, S., Zhao, S. Y., Xiao, S. Z., Gu, F. F., Liu, Q. Z., Tang, J., et al. (2016). Antimicrobial resistance and molecular epidemiology of Escherichia coli causing bloodstream infections in three hospitals in Shanghai, China. PLoS ONE 11:e0147740. doi: 10.1371/journal.pone.0147740
Wirth, T., Falush, D., Lan, R., Colles, F., Mensa, P., Wieler, L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 60, 1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x
Xiao, Y. H., Giske, C. G., Wei, Z. Q., Shen, P., Heddini, A., and Li, L. J. (2011). Epidemiology and characteristics of antimicrobial resistance in China. Drug Resist. Updat. 14, 236–250. doi: 10.1016/j.drup.2011.07.001
Xiao, Y., Shen, P., Wei, Z., Chen, Y., Kong, H., Yang, Q., et al. (2012). National surveillance of antimicrobial resistance of Mohnarin. Chin. J. Nosocomiol. 22, 4946–4952.
Xiao, Y., Zhang, J., Zheng, B., Zhao, L., Li, S., and Li, L. (2013). Changes in Chinese policies to promote the rational use of antibiotics. PLoS Med. 10:e1001556. doi: 10.1371/journal.pmed.1001556
Yu, Y., Ji, S., Chen, Y., Zhou, W., Wei, Z., Li, L., et al. (2007). Resistance of strains producing extended-spectrum beta-lactamases and genotype distribution in China. J. Infect. 54, 53–57. doi: 10.1016/j.jinf.2006.01.014
Zhang, J., Zheng, B., Zhao, L., Wei, Z., Ji, J., Li, L., et al. (2014). Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect. Dis. 14:659. doi: 10.1186/s12879-014-0659-0
Zhang, W., Luo, Y., Li, J., Lin, L., Ma, Y., Hu, C., et al. (2011). Wide dissemination of multidrug-resistant Shigella isolates in China. J. Antimicrob. Chemother. 66, 2527–2535. doi: 10.1093/jac/dkr341
Zhao, S. Y., Wang, Y. C., Xiao, S. Z., Jiang, X. F., Guo, X. K., Ni, Y. X., et al. (2015). Drug susceptibility and molecular epidemiology of Escherichia coli in bloodstream infections in Shanghai, China, 2011–2013. Infect. Dis. (Lond). 47, 310–318. doi: 10.3109/00365548.2014.990509
Zheng, H., Zeng, Z., Chen, S., Liu, Y., Yao, Q., Deng, Y., et al. (2012). Prevalence and characterisation of CTX-M beta-lactamases amongst Escherichia coli isolates from healthy food animals in China. Int. J. Antimicrob. Agents 39, 305–310. doi: 10.1016/j.ijantimicag.2011.12.001
Keywords: outpatient, antimicrobial resistance, ESBL, ST, town hospital
Citation: Miao Z, Li S, Wang L, Song W and Zhou Y (2017) Antimicrobial Resistance and Molecular Epidemiology of ESBL-Producing Escherichia coli Isolated from Outpatients in Town Hospitals of Shandong Province, China. Front. Microbiol. 8:63. doi: 10.3389/fmicb.2017.00063
Received: 21 November 2016; Accepted: 10 January 2017;
Published: 24 January 2017.
Edited by:
Fatah Kashanchi, George Mason University, USAReviewed by:
Ákos Tóth, National Center for Epidemiology, HungaryJozsef Soki, University of Szeged, Hungary
Copyright © 2017 Miao, Li, Wang, Song and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zengmin Miao, emVuZ21pbm1pYW9AMTI2LmNvbQ==
†These authors have contributed equally to this work.
 Zengmin Miao
Zengmin Miao Song Li2†
Song Li2†