- 1Departamento de Microbiología y Parasitología, CIBUS-Facultad de Biología, Universidade de Santiago de Compostela, Santiago de Compostela, Spain
- 2Department of Bioinformatics and Genomics, The University of North Carolina at Charlotte, Charlotte, NC, USA
Vibrio toranzoniae is a Gram-negative bacterium of the Splendidus clade within the Vibrio genus. V. toranzoniae was first isolated from healthy clams in Galicia (Spain) but recently was also identified associated to disease outbreaks of red conger eel in Chile. Experimental challenges showed that the Chilean isolates were able to produce fish mortalities but not the strains isolated from clams. The aim of the present study was to determine the differences at the genomic level between the type strain of the species (CECT 7225T) and the strain R17, isolated from red conger eel in Chile, which could explain their different virulent capacity. The genome-based comparison showed high homology between both strains but differences were observed in certain gene clusters that include some virulence factors. Among these, we found that iron acquisition systems and capsule synthesis genes were the main differential features between both genomes that could explain the differences in the pathogenicity of the strains. Besides, the studied genomes presented genomic islands and toxins, and the R17 strain presented CRISPR sequences that are absent on the type strain. Taken together, this analysis provided important insights into virulence factors of V. toranzoniae that will lead to a better understanding of the pathogenic process.
Introduction
Members of Vibrionaceae family are a predominant group of Gram-negative bacteria found in aquatic environment (Thompson et al., 2004). This bacterial group live as planktonic forms in the water column or associated in biofilms or with host organisms as pathogenic, commensal or mutualistic bacteria. Vibrio is the largest genera within this family with at least 130 recognized species (http://www.bacterio.net/vibrio.html). Many Vibrio species are recognized as pathogens of different organisms, including humans and aquatic animals. For instance, several species of the Splendidus clade, that comprises 17 species (Romalde et al., 2014; Pérez-Cataluña et al., 2016), are causing significant losses in the aquaculture industry worldwide.
Vibrio splendidus has been described as affecting fish species such as the turbot Scophtalmus maximus (Gatesoupe et al., 1999) but also, associated with mortalities of different molluscan species as scallops, Pecten maximus (Nicolas et al., 1996), Pactinopecten yessonensis (Liu et al., 2013) and Argopecten purpurartus (Rojas et al., 2015), oysters, Crassostrea gigas (Le Roux et al., 2002; Domeneghetti et al., 2014) and Crassostrea virginica (Elston and Leibovitz, 1980), and mussels (Mytilus galloprovincialis; Domeneghetti et al., 2014). Even though the virulence mechanisms involved in the pathogenesis of the V. splendidus-related species are not well-characterized, some virulence factors have been identified such as the invasion porine OmpU (Duperthuy et al., 2010), the metalloprotease Vsm (Le Roux et al., 2007; Binesse et al., 2008) and the invasive vesicular serine protease Vsp (Vanhove et al., 2014).
The virulence of pathogenic Vibrio strains isolated from molluscs has been shown to be related to their ability to produce extracellular products (Elston and Leibovitz, 1980; Labreuche et al., 2006; Hasegawa et al., 2008). For instance, Vibrio lentus has been recognized as a potential pathogen that caused skins lesions, colonize the internal organs and induce mortality in octopus (Farto et al., 2003). On subsequent studies (Farto et al., 2006), the presence of a lethal extracellular protease was detected in the ECP (extracellular products) of V. splendidus-V. lentus related group.
V. kanalaoe is also considered as a potential pathogen for aquatic animals including fish and crustaceans (Austin et al., 2005). Experimental infections of this species in C. gigas produced extensive lesions in the adductor muscle (Gay et al., 2004). Unfortunately, no clear discrimination between V. kanaloae and V. pomeroyi was possible, even using a polyphasic approach to identify the strains. Another member of the Splendidus clade, Vibrio celticus, showed potential pathogenic activity for adult clams (R. phillipinarum and R. decussatus) in virulence assays (Beaz-Hidalgo et al., 2010).
V. toranzoniae was first isolated from healthy clams (R. phillipinarum and R. decussatus) in Galicia (Spain) and designated as a new member of the Splendidus clade (Lasa et al., 2013). Further studies (Gulla et al., 2015; Kwan and Bolch, 2015) indicated that the geographical and host distribution of this species could be wider than expected. In fact, a group of strains, identified as V. toranzoniae using a polyphasic approach, were isolated from cultured red conger eel (Genypterus chilensis) in Chile during an episode of mortalities at one aquaculture rearing system (Lasa et al., 2015). Inoculation experiments in turbot of the Chilean isolates and the type strain of the species (CECT 7225T) demonstrated the pathogenic potential for fish of the strains isolated from red conger eel but not for the original strains isolated from clams.
The aim of the present study was to determine the differences, at the genome level, between the clam and Chilean isolates. In order to elucidate the genomic base of the pathogenic potential of V. toranzoniae species, the type strain CECT 7225T and a representative strain of the Chilean isolates, R17, were selected for the genome comparison and the identification of virulence factors. The identification of putative pathogenicity islands common in the Vibrio genus was also evaluated.
Materials and Methods
Genomic DNA Extraction and Sequencing
DNA extraction and sequencing project of strain R17 was performed following the protocol described previously for the type strain of the species (Lasa et al., 2016). Briefly, CECT 7225T and R17 strains were routinely cultured in marine agar (MA; Difco) 24 h at 25°C. Genomic DNA was extracted using the QIAamp DNA minikit (Qiagen), according to the manufacturer's protocol. The purified DNA was used to prepare a library, following the Illumina TruSeq DNA sample prep protocol (Illumina, San Diego, CA). The genomes of V. toranzoniae strains were sequenced at David H. Murdock Research Institute (DHMRI) of the University of North Carolina (Kannapolis, North Carolina), using a HiSeq 2500 sequencing technology (Illumina) with 2 × 100-bp paired-end reads.
Genome Assembly, Annotation, and Analysis of the Vibrio toranzoniae Genomes
The Illumina reads were analyzed for quality control using FASTQC (Brabaham Bioinformatics). Reads were trimmed and filtered to remove adapters and low quality bases, using Trimmomatic 0.32 (Bolger et al., 2014) program. The remaining reads were used for the genome assembly, performed with the SPAdes 3.6.1 the novo assembler tool (Nurk et al., 2013), and QUAST (Gurevich et al., 2013) software was used to evaluate the assembly.
The draft genome of the type strain was annotated using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP; Angiuoli et al., 2008). Additionally, the genomes were analyzed on the Rapid Annotations using Subsystems Technology (RAST) server (Overbeek et al., 2014). CRISPRfinder tool (Grissa et al., 2007) was used to assess the presence of CRISPR repeats in both genomes.
A sequenced-based comparison analysis was performed using the RAST annotation server and Blast Ring Image Generator (BRIG; Alikhan et al., 2011) was used to obtain a genomic map showing similarity percentages. The upper and lower identity thresholds were set at 90 and 70%, respectively.
The presence of recognized virulence factors in other fish pathogens was determined. Identification of Genomic Islands (GIs) in the genomes of V. toranzoniae strains was performed by comparative analysis against some of the main fish pathogenic Vibrio species including V. splendidus, V. vulnificus, and V. anguillarum with IslandViewer (Waack et al., 2006; Langille et al., 2008; Dhillon et al., 2015) which uses the IslandPick, SIGI-HMM and IslandPath-DIMOB tools for the prediction.
Genome Deposit in Public Databases
The draft genome sequencing project of Vibrio toranzoniae strain CECT 7225T numbers LMXU00000000. Additionally, annotated contigs of the type strain are available under the accession numbers LMXU01000001-LMXU01000165 (Lasa et al., 2016). The Whole Genome Shotgun project of R17 has been deposited at DDBJ/ENA/GenBank under the accession MRTB00000000. The version described in this paper is version MRTB01000000.
Results
Genomic Features
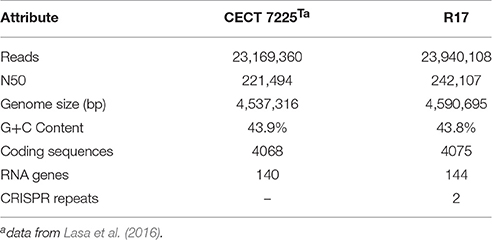
The sequencing experiments of CECT 7225T and R17 strains yielded 23,169,360 and 23,940,108 reads and the final draft assemblies contained 165 and 156 contigs, respectively, for each strain. The first draft genome summarizes 4,537,316 bp and the second 4,590,695 bp (Table 1).
The G+C content of V. toranzoniae CECT 7225T was 43.9 mol%. A total of 4058 protein-coding sequences were predicted using RAST annotation server and 140 were RNA genes. Eighty one percent of coding sequences were assigned to putative functions, while 19% remained as hypothetical proteins. On the other hand, the G+C content of V. toranzoniae R17 strain was 43.8 mol%. The annotation performed on the RAST annotation server revealed 4075 protein-coding sequences and 144 RNA genes. Of the total, 80.8% of the coding sequences were assigned to putative functions and 19.2% remained as hypothetical proteins. The search of CRISPR repeats showed the presence of two sequences in the R17 strain, while the type strain lacked these sequences (Table 1).
The genomic map revealed a high homology between the type strain and R17 strain (Figure 1). Regions of major differences between both strains contained mobile elements, phage genes and virulence factors.
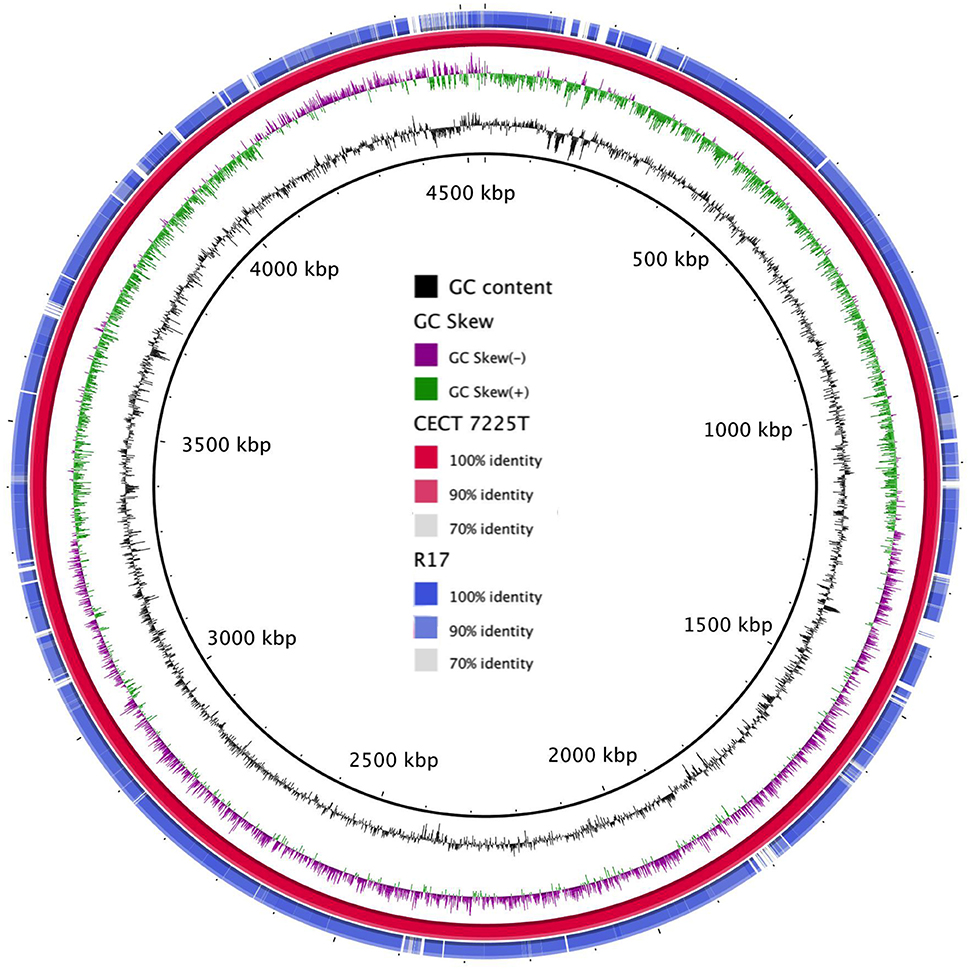
Figure 1. Graphical map of the BLASTN comparison of the two V. toranzoniae draft genomes. From center to outside: GC Content (ring 1), GC skew (ring 2), V. toranzoniae CECT 7225T (ring 3), and V. toranzoniae R17 (ring 4).
Genomic and Pathogenicity Islands of the V. toranzoniae Genomes
In the Vibrio species analyzed, GIs could be identified by the SIGI-HMM and IslandPath-DIMOB methods, but not using the IslandPick method. In both genomes, the highest number of GIs were found using SIGI-HMM software. The highest number of proteins within identified GIs were found when comparing against V. anguillarum (443) for the two studied genomes (Table 2). Interestingly, among the identified proteins different virulence factors were found, namely hemolysins, toxins, or iron acquisition system.
Pathogenic and Virulence Related Features
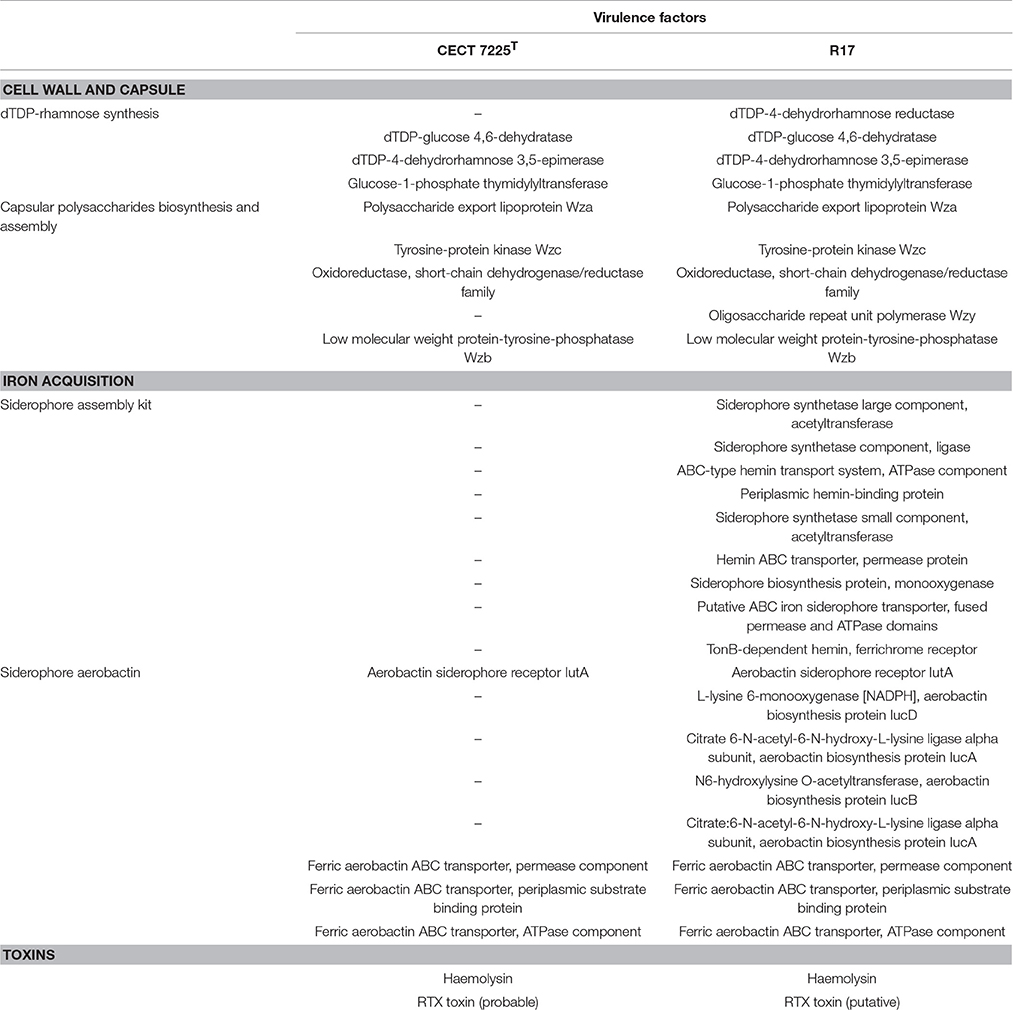
Based on established studies, we scanned both V. toranzoniae genomes for virulence-related features that could explain the different ability of these strains to produce disease in fish (summarized in Table 3).
Related to these virulence factors, we found differences in the gene content of the systems involved in the synthesis of certain capsular polysaccharides. Thus, in the strain CECT 7225T the gene of the Wzy polymerase is absent and also a reductase gene of the dTDP-rhamnose pathway.
Additionally, we found significant differences in the iron metabolism between both genomes. In this sense, R17 strain posseses an entire cluster of genes for siderophore assembly kit that encodes ligase, acetyltransferases, monooxygenase, an ABC-type transport system and a ferrichrome receptor, which is absent in the CECT 7225T strain. Besides, we found the presence of other iron uptake proteins in both genomes coding an aerobactin system, however in the CECT 7225T genome the genes that encode the aerobactin are absent and only the transporter and receptor genes are present. The region of the aerobactin iron transport system observed in both genomes showed the absence of the genes IucABCD genes in the type strain (Figure 2). The predicted IucABCD proteins showed 70–82% identity to the corresponding proteins of other Vibrio species.
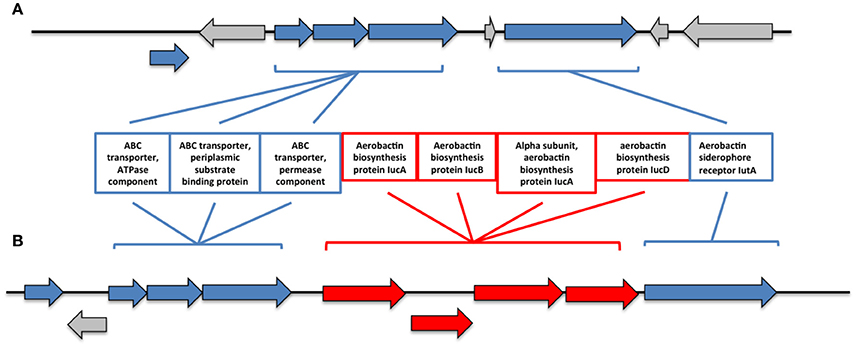
Figure 2. Genomic region of the aerobactin system in the genomes of CECT 7225T (A) and R17 (B) strains. In red are indicated the genes that are absent on the type strain.
The presence of proteins with cytotoxic activity was assessed and in the genome of R17 strain a toxin was found and annotated as putative RTX toxin. Also in the CECT 7225T genome this toxin was present but the protein was annotated as a probable toxin. The difference between these two proteins seems to be related to the length of the gene, which is shorter in the CECT 7225T genome. Another virulence factor with cytotoxic activity that we found and is present in both strains was a haemolysin with high homology to other haemolysins present in the Splendidus clade species.
Discussion
Our analysis depicted the first step to understand the pathogenic potential of V. toranzoniae for fish. The results obtained may explain the specificity of the species to produce disease in fish but not in clams, since the main virulence factors found here are related to iron uptake. However, it cannot be discard the pathogenic potential for molluscs and further experimental assays will be necessary.
The capsule is a recognized virulence factor that contributes to virulence in other Vibrio pathogens (Williams et al., 2014; Oliver, 2015). The proposed function of this factor is to protect the organism from phagocytosis, thus initiating the infection. Capsulated strains grow on solid media forming opaque colonies, while non-capsulated strains develop translucent colonies. Therefore, we have observed that R17 strain form opaque colonies and the type strain CECT 7225T develop into translucent colonies when grown on solid media. The formation of translucent colonies of the CECT 7225T strain can be explained by the loss of important proteins that are involved in the synthesis of capsular polysaccharides. The Wzy polymerase is required for the extension of polysaccharides of the capsule and also of the O-antigen (Islam and Lam, 2014). Additionally, the rhamnose synthesis pathway, which seems to be interrupted on CECT 7225T strain, is involved in the synthesis of these capsular polysaccharides. Other virulence factor that would be crucial for the initial infection and disease development are the CRISPR arrays. The presence of CRISPR sequences in the R17 strain would allow this strain not only to resist the infection of virus but also to avoid the host immune system (Louwen et al., 2014) promoting the infection and the dissemination within the fish.
Genomic islands are large genomic regions that contain multiple genes, probably with horizontal transfer origin. GIs are classified based on the different functions they encode, including metabolic islands, degradation islands, symbiosis islands, and pathogenicity islands (Murphy and Boyd, 2008). Pathogenicity islands are unstable regions that exhibit virulence-related genes that are involved in virulence, antibiotic resistance and other adaptations to the infective process (Langille et al., 2010). Most of the annotated proteins within the GIs of the V. toranzoniae genomes were identified as hypothetical proteins, transposases, or ribosomal proteins and are present in all Vibrio (Lux et al., 2014). Important virulence factors were found within these predicted GIs, including haemolysin, RTX toxin or iron uptake systems.
Iron uptake systems and siderophores production allows fish pathogens to avoid the natural defense mechanisms of the fishes using iron-binding proteins (such as trasnferrin, lactoferrin, or ferritin). Interestingly, the siderophore assembly kit was present only in the R17 strain and the presence of this system could provide important advantages in regards to infection, pathogenicity, and resistance. In addition, the CECT 7225T strain may have lost these advantages by losing the group of genes that encode the aerobactin, despite this strain are able to produce the transporter system and the receptor.
Cytotoxic activity may be possible through the codification of toxins. Homologs to putative RTX toxins have been identified in V. vulnificus and may play a role in cell adhesion and/or pathogenesis, and in V. anguillarum are major component of virulences. These toxins exhibit a cytotoxic pore-forming activity and the secretion occurs via type I secretion system. We found that the coding gene for this toxin is present in both strains, but the open reading frame of the CECT 7225T strain appear to be disrupted. Other virulence factor that contributes to the cytotoxic activity is the haemolysin and both strains have these coding genes. In fact, these strains produce β haemolysis when grew on blood agar plates.
Other potential virulence factors, such as Vsm metalloproteases, Vsp proteases, or extracellular proteases, were not found within the annotated proteins or the GIs. However, cannot be discard the presence of this virulence related proteins among the genes that were not identified and they remained as hypothetical proteins.
Conclusions
The genome-based comparison presented here reveal common basic features between the two V. toranzoniae strains. However the distribution of virulence factors was different between the isolates, especially those systems related to iron acquisition systems and capsule production. In general, these results are in congruence with the previous observations on the pathogenic ability of the isolates, although further in vivo analysis, including different aquatic animal models and the construction of mutants for the different virulence-related genes, are required to confirm the role of this factors in the pathogenic process.
Author Contributions
The experiments, data analysis, and manuscript writing were performed by AL and JR, while CG provided vital guidance and technical support.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported in part by grants AGL2013-42628R and AGL2016-77539R from Ministerio de Economia y Competitividad (Spain). We are grateful to Dr. Mike Wang from David H. Murdock Research Institute's (DHMRI), University of North Carolina (UNC) for sequencing assistance.
References
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Angiuoli, S. V., Gussman, A., Klimke, W., Cochrane, G., Field, D., Garrity, G. M., et al. (2008). Toward an online repository of standard operating procedures (SOPs) for (Meta)genomic annotation. OMICS 12, 137–141. doi: 10.1089/omi.2008.0017
Austin, B., Austin, D., Sutherland, R., Thompson, F., and Swings, J. (2005). Pathogenicity of vibrios to rainbow trout (Oncorhynchus mykiss, Walbaum) and Artemia nauplii. Environ. Microbiol. 7, 1488–1495. doi: 10.1111/j.1462-2920.2005.00847.x
Beaz-Hidalgo, R., Diéguez, A. L., Cleenwerck, I., Balboa, S., Doce, A., de Vos, P., et al. (2010). Vibrio celticus sp. nov., a new Vibrio species belonging to the Splendidus clade with pathogenic potential for clams. Syst. Appl. Microbiol. 33, 311–315. doi: 10.1016/j.syapm.2010.06.007
Binesse, J., Delsert, C., Saulnier, D., Champomier-Vergès, M.-C., Zagorec, M., Munier-Lehmann, H., et al. (2008). Metalloprotease vsm is the major determinant of toxicity for extracellular products of Vibrio splendidus. Appl. Environ. Microbiol. 74, 7108–7117. doi: 10.1128/AEM.01261-08
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:btu170. doi: 10.1093/bioinformatics/btu170
Dhillon, B. K., Laird, M. R., Shay, J. A., Winsor, G. L., Lo, R., Nizam, F., et al. (2015). IslandViewer 3: more flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res. 43, W104–W108. doi: 10.1093/nar/gkv401
Domeneghetti, S., Varotto, L., Civettini, M., Rosani, U., Stauder, M., Pretto, T., et al. (2014). Mortality occurrence and pathogen detection in Crassostrea gigas and Mytilus galloprovincialis close-growing in shallow waters (Goro lagoon, Italy). Fish Shellfish Immunol. 41, 37–44. doi: 10.1016/j.fsi.2014.05.023
Duperthuy, M., Binesse, J., Le Roux, F., Romestand, B., Caro, A., Got, P., et al. (2010). The major outer membrane protein OmpU of Vibrio splendidus contributes to host antimicrobial peptide resistance and is required for virulence in the oyster Crassostrea gigas. Environ. Microbiol. 12, 951–963. doi: 10.1111/j.1462-2920.2009.02138.x
Elston, R., and Leibovitz, L. (1980). Pathogenesis of experimental vibriosis in larval American oysters, Crassostrea virginica. Can. J. Fish. Aquat. Sci. 37, 964–978. doi: 10.1139/f80-126
Farto, R., Armada, S. P., Montes, M., Guisande, J. A., Pérez, M. J., and Nieto, T. P. (2003). Vibrio lentus associated with diseased wild octopus (Octopus vulgaris). J. Invertebr. Pathol. 83, 149–156. doi: 10.1016/S0022-2011(03)00067-3
Farto, R., Armada, S. P., Montes, M., Perez, M. J., and Nieto, T. P. (2006). Presence of a lethal protease in the extracellular products of Vibrio splendidus-Vibrio lentus related strains. J. Fish Dis. 29, 701–707. doi: 10.1111/j.1365-2761.2006.00746.x
Gatesoupe, F. J., Lambert, C., and Nicolas, J. L. (1999). Pathogenicity of Vibrio splendidus strains associated with turbot larvae, Scophthalmus maximus. J. Appl. Microbiol. 87, 757–763. doi: 10.1046/j.1365-2672.1999.00922.x
Gay, M., Berthe, F. C. J., and Le Roux, F. (2004). Screening of Vibrio isolates to develop an experimental infection model in the Pacific oyster Crassostrea gigas. Dis. Aquat. Org. 59, 49–56. doi: 10.3354/dao059049
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52–W57. doi: 10.1093/nar/gkm360
Gulla, S., Sørum, H., Vågnes, Ø., and Colquhoun, D. J. (2015). Phylogenetic analysis and serotyping of Vibrio splendidus-related bacteria isolated from salmon farm cleaner fish. Dis. Aquat. Organ. 117, 121–131. doi: 10.3354/dao02938.
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hasegawa, H., Lind, E., Boin, M., and Häse, C. (2008). The extracellular metalloprotease of Vibrio tubiashii is a major virulence factor for Pacific oyster (Crassostrea gigas) larvae. Appl. Environ. Microbiol. 74, 4101–4110. doi: 10.1128/AEM.00061-08
Islam, S. T., and Lam, J. S. (2014). Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can. J. Microbiol. 60, 697–716. doi: 10.1139/cjm-2014-0595
Kwan, T. N., and Bolch, C. J. (2015). Genetic diversity of culturable Vibrio in an Australian blue mussel Mytilus galloprovincialis hatchery. Dis. Aquat. Organ. 116, 37–46. doi: 10.3354/dao02905
Labreuche, Y., Soudant, P., Gonçalves, M., Lambert, C., and Nicolas, J. L. (2006). Effects of extracellular product from the pathogenic Vibrio aestuarianus strain 01/32 on lethality and cellular immune response of the oyster Crassostrea gigas. Dev. Comp. Immunol. 30, 367–379. doi: 10.1016/j.dci.2005.05.003
Langille, M. G. I., Hsiao, W. W. L., and Brinkman, F. S. L. (2008). Evaluation of genomic island predictors using a comparative genomics approach. BMC Bioinformatics 9:329. doi: 10.1186/1471-2105-9-329
Langille, M. G., Hsiao, W. W., and Brinkman, F. S. (2010). Detecting genomic islands using bioinformatics approaches. Nat. Rev. Microbiol. 8, 373–782. doi: 10.1038/nrmicro2350
Lasa, A., Avendño-Herrera, R., Estrada, J. M., and Romalde, J. L. (2015). Isolation and identification of Vibrio toranzoniae associated with diseased red conger eel (Genypterus chilensis) farmed in Chile. Vet. Microbiol. 179, 327–331. doi: 10.1016/j.vetmic.2015.06.003
Lasa, A., Diéguez, A. L., and Romalde, J. L. (2013). Vibrio toranzoniae sp. nov., a new member of the Splendidus clade in the genus Vibrio. Syst. Appl. Microbiol. 36, 96–100. doi: 10.1016/j.syapm.2012.11.005
Lasa, A., Gibas, C. J., and Romalde, J. L. (2016). Draft genome sequence of Vibrio toranzoniae strain CECT 7225T. Genome Announc. 4:e00212-16. doi: 10.1128/genomeA.00212-16
Le Roux, F., Binesse, J., Saulnier, D., and Mazel, D. (2007). Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl. Environ. Microbiol. 73, 777–784. doi: 10.1128/AEM.02147-06
Le Roux, F., Gay, M., Lambert, C., Waechter, M., Poubalanne, S., Chollet, B., et al. (2002). Comparative analysis of Vibrio splendidus related strains isolated during Crassostrea gigas mortality events. Aquat. Living Ressour. 15, 251–258. doi: 10.1016/S0990-7440(02)01176-2
Liu, R., Qiu, L., Yu, Z., Zi, J., Yue, F., Wang, L., et al. (2013). Identification and characterisation of pathogenic Vibrio splendidus from Yesso scallop (Patinopecten yessoensis) cultured in a low temperature environment. J. Invertebr. Pathol. 114, 144–150. doi: 10.1016/j.jip.2013.07.005
Louwen, R., Staals, R. H. J., Endtz, H. P., van Baarlen, P., and van der Oost, J. (2014). The role of CRISPR-Cas systems in virulence of pathogenic bacteria. Microbiol. Mol. Biol. Rev. 78, 74–88. doi: 10.1128/MMBR.00039-13
Lux, T. M., Lee, R., and Love, J. (2014). Genome-wide phylogenetic analysis of the pathogenic potential of Vibrio furnissii. Front. Microbiol. 5:435. doi: 10.3389/fmicb.2014.00435.
Murphy, R. A., and Boyd, E. F. (2008). Three pathogenicity islands of Vibrio cholerae can excise from the chromosome and form circular intermediates. J. Bacteriol. 190, 636–647. doi: 10.1128/JB.00562-07
Nicolas, J. L., Corre, S., Gauthier, G., Robert, R., and Ansquer, D. (1996). Bacterial problems associated with scallop Pecten maximus larval culture. Dis. Aquat. Org. 27, 67–76. doi: 10.3354/dao027067
Nurk, S., Bankevich, A., Antipov, D., Gurevich, A. A., Korobeynikov, A., Lapidus, A., et al. (2013). Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 20, 714–737. doi: 10.1089/cmb.2013.0084
Oliver, J. D. (2015). The biology of Vibrio vulnificus. Microbiol. Spectr. 3:VE-0001-2014. doi: 10.1128/microbiolspec.VE-0001-2014
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Pérez-Cataluña, A., Lucena, T., Tarazona, E., Arahal, D. R., Macián, M. C., and Pujalte, M. J. (2016). An MLSA approach for the taxonomic update of the Splendidus clade, a lineage containing several fish and shellfish pathogenic Vibrio spp. Syst. Appl. Microbiol. 39, 361–369. doi: 10.1016/j.syapm.2016.03.010
Rojas, R., Miranda, C. D., Opazo, R., and Romero, J. (2015). Characterization and pathogenicity of Vibrio splendidus strains associated with massive mortalities of commercial hatchery-reared larvae of scallop Argopecten purpuratus (Lamarck, 1819). J. Invertebr. Pathol. 124, 61–69. doi: 10.1016/j.jip.2014.10.009
Romalde, J. L., Dieguez, A. L., Lasa, A., and Balboa, S. (2014). New Vibrio species associated to molluscan microbiota: a review. Front Microbiol. 4:413. doi: 10.3389/fmicb.2013.00413
Thompson, F. L., Iida, T., and Swings, J. (2004). Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 68, 403–431. doi: 10.1128/MMBR.68.3.403-431.2004
Vanhove, A. S., Duperthuy, M., Charrière, G. M., Le Roux, F., Goudenège, D., Gourbal, B., et al. (2014). Outer membrane vesicles are vehicles for the delivery of Vibrio tasmaniensis virulence factors to oyster immune cells. Environ Microbiol. 17, 1152–1165. doi: 10.1111/1462-2920.12535
Waack, S., Keller, O., Asper, R., Brodag, T., Damm, C., Fricke, W. F., et al. (2006). Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinformatics 7:142. doi: 10.1186/1471-2105-7-142
Keywords: Vibrio toranzoniae, fish pathogen, genomic comparison, pathogenicity islands, virulence factors
Citation: Lasa A, Gibas CJ and Romalde JL (2017) Comparative Genomic Analysis of Two Vibrio toranzoniae Strains with Different Virulence Capacity Reveals Clues on Its Pathogenicity for Fish. Front. Microbiol. 8:86. doi: 10.3389/fmicb.2017.00086
Received: 09 November 2016; Accepted: 12 January 2017;
Published: 30 January 2017.
Edited by:
Martha E. Trujillo, University of Salamanca, SpainReviewed by:
Brian Austin, University of Stirling, UKJose Gustavo Ramirez-Paredes, University of Stirling, UK
Copyright © 2017 Lasa, Gibas and Romalde. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jesús L. Romalde, amVzdXMucm9tYWxkZUB1c2MuZXM=
 Aide Lasa
Aide Lasa Cynthia J. Gibas2
Cynthia J. Gibas2 Jesús L. Romalde
Jesús L. Romalde