- State Key Laboratory of Microbial Technology, School of Life Science, Shandong University, Jinan, China
dnaE is an alpha subunit of the tripartite protein complex of DNA polymerase III that is responsible for the replication of bacterial genome. The dnaE gene is often duplicated in many bacteria, and the duplicated dnaE gene was reported dispensable for cell survivals and error-prone in DNA replication in a mystery. In this study, we found that all sequenced myxobacterial genomes possessed two dnaE genes. The duplicate dnaE genes were both highly conserved but evolved divergently, suggesting their importance in myxobacteria. Using Myxococcus xanthus DK1622 as a model, we confirmed that dnaE1 (MXAN_5844) was essential for cell survival, while dnaE2 (MXAN_3982) was dispensable and encoded an error-prone enzyme for replication. The deletion of dnaE2 had small effects on cellular growth and social motility, but significantly decreased the development and sporulation abilities, which could be recovered by the complementation of dnaE2. The expression of dnaE1 was always greatly higher than that of dnaE2 in either the growth or developmental stage. However, overexpression of dnaE2 could not make dnaE1 deletable, probably due to their protein structural and functional divergences. The dnaE2 overexpression not only improved the growth, development and sporulation abilities, but also raised the genome mutation rate of M. xanthus. We argued that the low-expressed error-prone DnaE2 played as a balancer for the genome mutation rates, ensuring low mutation rates for cell adaptation in new environments but avoiding damages from high mutation rates to cells.
Introduction
DNA polymerase III is an enzyme complex responsible for prokaryotic genome replication (Kelman and O'donnell, 1995). The holoenzyme consists of a DNA polymerase (polymerase III α-subunit, PolIIIα), a processivity factor β-clamp and a clamp loader protein (McHenry, 2011a,b; Robinson et al., 2012). There are two types of PolIIIα: dnaE and PolC, both of which belong to the C-family of DNA polymerase (Ito and Braithwaite, 1991). While PolC exists in low-GC Gram-positive bacteria such as Bacillus subtilis, dnaE is universally distributed in different kinds of bacterial cells (Evans et al., 2008). The dnaE gene is essential in bacteria, and its functions have been well elucidated in some bacteria, such as Escherichia coli (Lamers et al., 2006). In addition to the essential dnaE gene, many bacterial species have evolved a second dnaE copy, usually named as dnaE2. For example, duplicated dnaE genes are often existed in those bacteria possessing large-sized genomes with high G+C contents (Zhao et al., 2007). dnaE2 normally exists in a three-gene operon in Proteobacteria, accompanying with imuA and imuB, and the operon is regulated by LexA2, a possible transcriptional regulator associated with DNA damage response (Abella et al., 2004). Previous genetic studies indicated that dnaE2 was non-essential for the chromosomal DNA replication in bacterial cells (Boshoff et al., 2003; Abella et al., 2004; Warner et al., 2010; Tsai et al., 2012). In Streptomyces, DnaE2 was reported to be an error-prone enzyme, associating with the DNA damage-inducible translesion DNA synthesis (Tsai et al., 2012). In Mycobacterium tuberculosis, the error-prone DnaE2 was proved in vitro to act as a primary mediator for cell survival through inducing mutagenesis and thus contributing directly to the emergence of drug resistance (Boshoff et al., 2003). However, this function is not in prevalence. For example, researches in Pseudomonas and Streptomyces indicated that dnaE2 was not required for replication, end patching, or ultraviolet resistance and mutagenesis (Koorits et al., 2007; Tsai et al., 2012). The role of the replicated DnaE2 remains mostly unclear yet.
Myxobacteria are phylogenetically located in the delta division of the Proteobacteria (Shimkets et al., 2006). The bacteria are widely distributed in various environmental conditions, playing as micropredators by feeding on other microbial cells or macromolecules (Reichenbach, 1999; Jiang et al., 2010; Brinkhoff et al., 2012; Li et al., 2012; Zhou et al., 2014). Myxobacteria are characterized among the Prokaryotes by their complex multicellular social behaviors: cells locomote on solid surfaces in swarms to collaboratively prey on other microbial cells and, when food is scarce, aggregate to develop multicellular fruiting bodies, inside which differentiate metabolically quiescent myxospores (Shimkets, 1990; Kaiser and Losick, 1993; Dworkin, 1996; Shimkets et al., 2006). In this study, we bioinformatically analyzed those sequenced myxobacteria, and found that each of the myxobacterial genomes possessed two dnaE genes. The duplicate myxobacterial dnaE genes were both highly conserved but evolved divergently. Using Myxococcus xanthus DK1622 as a model, we confirmed that one dnaE gene was essential for cell survival, while the other was dispensable and error-prone. We evaluated functions of the dnaE genes and assayed their expressions in vegetative growth stage and developmental stage. It is known that there are two processes involving the genome replication in M. xanthus, one in the growth stage and the other in the early development stage (Tzeng and Singer, 2005). We found that the non-essential dnaE gene played functions in both of the two replication processes in M. xanthus DK1622. We suggested that the error-prone DnaE2 played as a balancer for the genome mutation rates via expression regulation, thus ensuring low mutation rates for the adaptation in new environments and avoiding high mutation rates to damage cells.
Results
dnaE1 Is Essential While dnaE2 Is Dispensable and Encodes An Error-prone DNA Polymerase in M. xanthus
Myxobacterial genomes are normally larger than nine Mbp in size, but with some exceptions of approximate five-Mbp-genomes in Anaeromyxobacter strains. The G+C contents of the sequenced myxobacterial genomes ranged from 67.4% in Enhygromyxa salina DSM 15201 to 74.9% in Anaeromyxobacter dehalogenans 2CP-C. All the sequenced myxobacterial genomes, no matter in large size or small size, contained two dnaE genes (Table S1). Phylogenetic analysis indicated that the two protein sequences encoded by the duplicate myxobacterial dnaE genes were clustered separately with determined DnaE1 and DnaE2 of other bacteria, forming the DnaE1 and DnaE2 groups (Figure S1). The single DnaE gene of E. coli was in the DnaE1 group, clearly distant from the DnaE2 group; and the two DnaE-group trees had highly similar topologies. Consistent with the previous results (Timinskas et al., 2014; Wu et al., 2014), the emergence of myxobacterial dnaE2 was derived from an early duplication event of the primordial dnaE. High conservation but great divergence suggested that the duplicate DnaE polymerases were both important for myxobacterial cells, but functioned divergently.
M. xanthus DK1622 is the model strain of myxobacteria. In the genome of DK1622, two genes, i.e., MXAN_3982 and MXAN_5844, were predicted to be dnaE, encoding for the alpha subunit of DNA polymerase III (Goldman et al., 2006). The proteins encoded by the two dnaE genes contain 1185 and 1013 amino acids (MXAN_5844 and MXAN_3982), respectively. These two proteins were phylogenetically distant, having 48% similarity and 30% identity of their amino acid sequences. According to their phylogenetic locations (Figure S1), the MXAN_3982 gene was designated as dnaE2, and the MXAN_5844 gene was dnaE1.
To evaluate their essentiality, we made deletion mutations of the dnaE1 and dnaE2 gene in M. xanthus DK1622, respectively. While dnaE2 was deletable, producing the mutant of YL1601, the dnaE1 gene could not be deleted, which was further confirmed by failed attempts to delete the dnaE1 gene in a dnaE2-overexpressing mutant (see below). Compared with that of the wild-type strain DK1622, the OD values of the dnaE2 deletion mutant YL1601 were smaller than the wild type at different time points during the exponential growth stage in CTT growth medium (two-way ANOVA, P < 0.01; Figure 1A). To confirm the function of dnaE2 in growth, we further constructed a mutant by inserting the dnaE2 gene following its own promoter at the attB site in the YL1601 genome, forming the YL1606 mutant. As expected, the complementary strain recovered the delayed growth in YL1601, showing almost the same growth curve as the wild type strain DK1622 (Figure 1A).
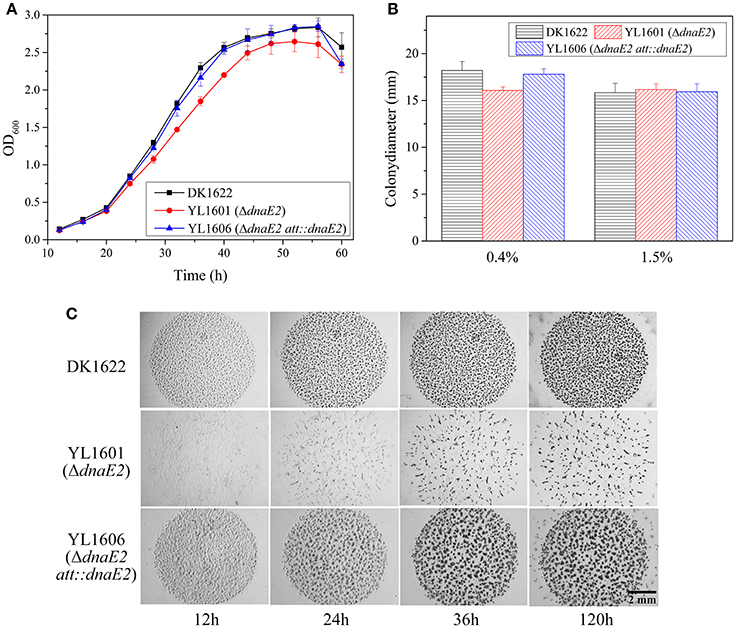
Figure 1. Comparison of phenotypic characteristics of the dnaE2 mutant (YL1601), the dnaE2-complementary mutant (YL1606) and the wild type strain DK1622. (A) Growth curves of YL1601, YL1606, and DK1622 in CTT liquid medium. OD600 values were measured every 4 h until 60 h of incubation. The data were analyzed using two-way ANOVA. (B) Cellular swarming abilities on 0.4 and 1.5% agar CTT plate. (C) Aggregation and formation abilities of fruiting bodies on TPM development plate. The error bars represent the standard deviation of three independent experiments.
To confirm whether the DnaE2 protein was an error-prone enzyme, we constructed two more mutants by deleting two DNA-repair genes of mutL (MXAN_4026) and mutS (MXAN_3897) from DK1622, respectively, producing the mutants of YL1602 and YL1603. The wild type strain DK1622 and the three mutants of YL1601, YL1602 and YL1603 were cultivated on CTT growth medium supplemented with nalidixic acid (40 μg/ml) to evaluate their production abilities of resistant mutations. It is known that the inhibition of nalidixic acid on DNA repair can be reversed by the MutL and MutS enzymes of the DNA repairing system in E. coli cells, thus the deletion mutant of mutL or mutS will produce more genome mutations than the wild type strain during the chromosome replication process (Sniegowski et al., 1997). In M. xanthus, the mutation rate of DK1622 was approximately 3.69 × 10−8 per nucleotide in the presence of nalidixic acid; whereas the YL1602 and YL1603 mutants produced mutations of 5.41 × 10−7 and 1.77 × 10−6, respectively (Table 1). The mutation rates of the mutL and mutS mutants were both significantly higher than that of DK1622 (t-test, P < 0.01). This result was consistent with that reported in E. coli (Sniegowski et al., 1997), suggesting the method was also reliable in M. xanthus for the evaluation of mutation rates. As expected, the deletion of dnaE2 made the mutant, possessing only dnaE1, to have nearly 10 times lower of mutation rate than that of the wild type strain DK1622, which harbors both dnaE1 and dnaE2 (Table 1). This result confirmed the error-prone characteristics of the DnaE2 protein in M. xanthus cells.
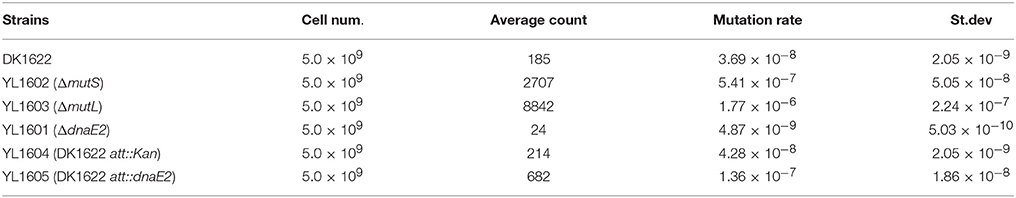
Table 1. Mutation rates of M. xanthus DK1622 and mutants grown on CTT plates supplemented with nalidixic acid.
Effects of the Deletion of dnaE2 on M. xanthus Social Behaviors
The above results indicated that dnaE1 was essential for the survival of M. xanthus cells, while the error-prone DnaE2 gene was dispensable for cell survival. dnaE2 also involved in cellular growth, but in weak effects and low fidelity. We further assayed effects of the deletion of dnaE2 in the swarming and developmental processes in M. xanthus. Similar to the growth change, the swarming ability of the YL1601 mutant was weakly decreased on 0.4% CTT plate, comparing with that of DK1622 (t-test, P = 0.051), while the motility on 1.5% CTT plate showed no difference between DK1622 and YL1601 (Figure 1B).
When cultivated on the TPM developmental medium, cells of the dnaE2 deletion mutant aggregated irregularly (Figure 1C). From the phenotypes of the mutant on TPM, the dnaE2 gene probably played functions in the early stage of the aggregation in the development process. The sporulation ability of the dnaE2 mutant was decreased to approximately 70% of that of DK1622 after 5 days of incubation on TPM medium (1.49 × 106 ± 9.61 × 104 vs. 2.14 × 106 ± 1.70 × 105; Table 2). If the dnaE2 gene was complemented into the dnaE2 deletion mutant (YL1606), the irregular aggregation phenotype on TPM developmental medium was completely recovered (Figure 1C). This result suggested that the DnaE2 protein also played functions in the early development stage of M. xanthus DK1622 cells. Seemly, the function of the dnaE2 gene was more obvious in the development stage than its function in the growth stage.
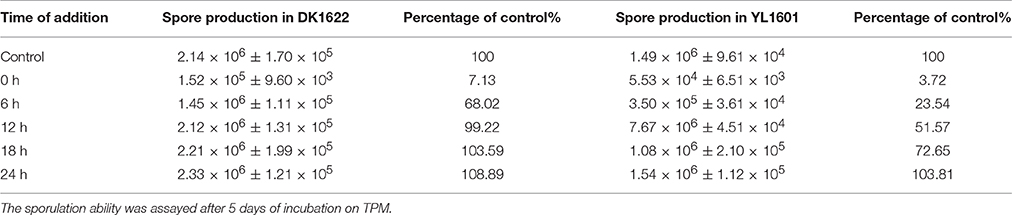
Table 2. Sporulation abilities of M. xanthus DK1622 and YL1601 (ΔdnaE2) cells grown on TPM plates treated with nalidixic acid for1 h at different time points.
Functions of dnaE2 in the Chromosome Replication Progress in Development
Tzeng and Singer found that there is a DNA replication process during the development of fruiting bodies and sporulation in M. xanthus DK1622 (Errington and Wake, 1991; Tzeng and Singer, 2005). Accordingly, there are two processes involving the chromosome replication during the lifecycle of M. xanthus cells: one in the growth stage and the other in the development stage. To determine DnaE2 function in development, we inoculated concentrated cells of DK1622 and YL1601 on the TPM plates, and exposed culture plates with nalidixic acid solution at the final concentration of 20 μg/ml for 1 h at different cultivation time points. After 120 h of incubation, the sporulation rates of the cultures were calculated and compared with that of their respective controls without the exposure of nalidixic acid. Consistent with the previous report (Tzeng et al., 2006), the developmental progress of DK1622 was arrested by nalidixic acid in the early stage of development. After 12 h of incubation, the nalidixic acid exposure had nearly no effect on the sporulation of M. xanthus DK1622 cells (Table 2). In contrast, the sporulation ability of the dnaE2 deletion mutant YL1601 was lower than that of DK1622 after the nalidixic acid exposure at each time point. The developmental arrest in YL1601 by the exposure of nalidixic acid was released after 24 h, which was nearly two-times long of that in DK1622. This result indicated that the deletion of dnaE2 markedly prolonged the process of chromosome replication in development. Thus, together with the dnaE1 gene, the dnaE2 gene involved in the two processes of chromosome replication during the lifecycle of M. xanthus cells; its function in the development progress seemed to be more significant.
dnaE1 Expression Is Greatly Higher than That of dnaE2
There are two possible reasons for the differentiated functions of the dnaE1 and dnaE2 genes in the growth and developmental stages: structure differences and/or expression levels. We further assayed expression levels of the dnaE1 and dnaE2 genes under either the growth or the developmental conditions. Quantitative real-time PCR amplification showed that the expression levels of dnaE1 were greatly higher than that of dnaE2 at different time points under the CTT nutritional growth conditions (Figure 2A). For example, after 12 h of incubation in CTT liquid medium, the expression of dnaE1 was more than 10 times of that of dnaE2. With the increase of incubation time, the expressed products of dnaE1 were gradually decreased and reached to less than one-tenth (8.7%) of the 12-h-level at 48 h of incubation. Similarly, the expressions of dnaE2 were also gradually decreased and were always significantly lower than that of dnaE1 at each time point (t-test, P < 0.01).
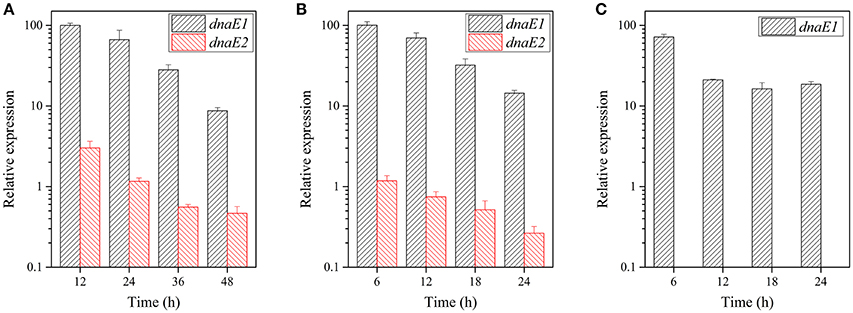
Figure 2. Expression levels of the dnaE1 and dnaE2 genes at different time points under the nutritional and developmental conditions. (A) M. xanthus DK1622 cells grown in the CTT growth medium. (B) The DK1622 strain grown on the TPM development plate. (C). The YL1601 mutant cells grown on TPM plate. The expression of the dnaE1 gene in CTT at 12 h of incubation was set as 100, and the others were the relative expressions. The error bars represent the standard deviation of three independent experiments.
The expressions of the two dnaE-genes were in similar patterns on the TPM development plate as they were in the growth conditions (Figure 2B). In contrast, the expressions of dnaE2 in the developmental conditions were even lower than that in the growth conditions. After the deletion of dnaE2, no expression of dnaE2 was detectable in the mutant, and the expression of the dnaE1 gene was lower than that of the wild type strain at 6 h of incubation on the TPM plate (71.5%; Figure 2C). The results suggested that the deficiencies in the fruiting body formation and sporulation in the YL1601 mutant were probably resulted not only from the loss of dnaE2, but also from the decreased expression level of dnaE1.
Overexpression of dnaE2 Improves Growth, Development, and Mutation Rate
To further investigate effects of the dnaE2 expression, we introduced a second copy of the gene into the attB site of DK1622 chromosome by the Mx8 integrase in pSWU19 (Wu and Kaiser, 1995), producing the YL1605 mutant (containing a local dnaE2 and an introduced dnaE2). The introduced dnaE2 gene was designed to follow the pilA promoter, a high efficient promoter in M. xanthus cells. Meanwhile, a mutant containing the empty pSWU19 plasmid was also constructed into the attB site of DK1622 chromosome as control (the mutant YL1604). The total expression levels of the two dnaE2 genes were approximately 10 times higher than that of dnaE1 in the YL1605 mutant under either the nutritional (Figure 3A; at 24 h of incubation in CTT; t-test, P < 0.01) or the developmental (Figure 3B; at 6 h of incubation on TPM; t-test, P < 0.01) conditions. In contrast, the expressions of dnaE2 in YL1605 were approximately 500 times and 1500 times higher than that in YL1604 under the nutritional and the developmental conditions, respectively. These results suggested that the overexpression of dnaE2 was mostly resulted from the introduced dnaE2 gene. When cultivated in CTT liquid medium, the OD values of the dnaE2 overexpression mutant YL1605 were higher than the control mutant YL1604 during the early exponential growth stage (Figure 3C). The growth differences were statistically significant (two-way ANOVA, P < 0.01). This result indicated that while the deletion of dnaE2 led to weakly delayed growth, overexpression of the gene could weakly increase cellular growth.
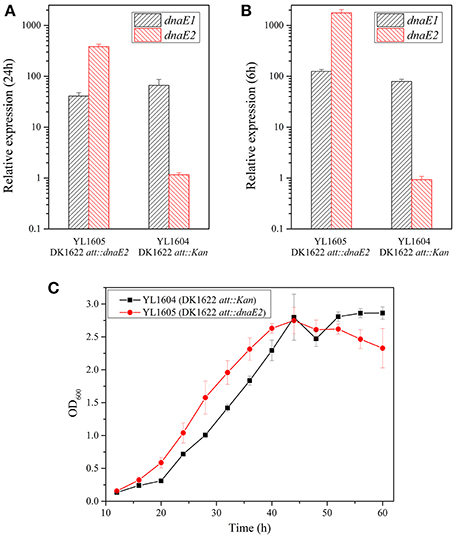
Figure 3. Overexpression of the dnaE2 gene in M. xanthus cells and cellular growth abilities. (A) Expressions of dnaE1 and dnaE2 in YL1605 (DK1622 att::dnaE2) and YL1604 (DK1622 att::Kan) in CTT growth medium; (B) Expressions of dnaE1 and dnaE2 in the two mutants on TPM plate; (C) Growth curves of YL1605 and YL1604 in CTT liquid medium supplemented with kanamycin (40 μg/ml). OD600 values were measured every 4 h until 60 h of incubation. The data were analyzed using two-way ANOVA. The error bars represent the standard deviation of three independent experiments. The expressions of dnaE1 and dnaE2 in (A) and (B) of this Figure were the relative expressions, compared with the expression of the dnaE1 gene in CTT at 12 h of nutritional condition of Figure 2A.
Interestingly, the overexpression of dnaE2 made the YL1605 mutant to have nearly three times higher of mutation rate than that of YL1604 cultivated on CTT plates supplemented with high concentrations of nalidixic acid (1.36 × 10−7 vs. 4.28 × 10−8, referred to Table 1). The difference was statistically significant (t-test, P < 0.01). In order to verify whether the overexpression changed the functions of dnaE2 in the development progress, we exposed the TPM cultures of YL1605 and YL1604 mutants with nalidixic acid solution for 1 h at the same time points described above (Table 3). The results showed that the dnaE2-overexpressing mutant also had higher sporulation ability than the control when cells were treated with nalidixic acid in the early stage of development (before 12 h of incubation). Thus, overexpression of the dnaE2 gene was not only able to increase the genome mutation rate, but also speed up the development progress. However, compared with effects of the dnaE2 deletion, overexpression of dnaE2 produced rather weak effects. The dnaE1 gene still played major roles in the chromosome replication.
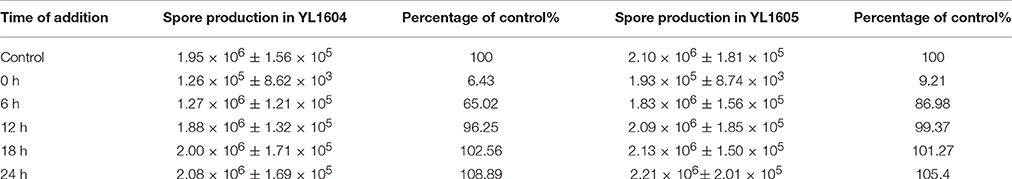
Table 3. Sporulation abilities of M. xanthus YL1604 (DK1622 att::Kan) and YL1605 (DK1622 att:: dnaE2) cells grown on TPM plates treated with nalidixic acid for 1 h at different time points.
Overexpression of dnaE2 Does Not Take Place of dnaE1
Imbalanced effects of the deletion and overexpression of dnaE2 suggested that the structural differences of the two DnaE proteins played more important roles for function than their expressions. Structural differences between DnaE2 and DnaE1 have been thoroughly investigated (Warner et al., 2010; McHenry, 2011a,b; Timinskas et al., 2014). To determine whether the essentiality of dnaE1 was due to high expressions in DK1622, we retried to knock out the dnaE1 gene in the dnaE2-overexpressing mutant (YL1605) using the pBJ113Cm-5844 plasmid but failed. This result indicated that dnaE2 could not replace dnaE1 even if the dnaE2 gene was overexpressed, which confirmed that the non-fungibility of DnaE1 was due to the structural differences between DnaE1 and DnaE2. We modeled three-dimensional structures of DK1622 DnaE1 and DnaE2 (Figure 4A), using the threading approach (Yang et al., 2015). The estimated TM-scores are 0.84 ± 0.08 and 0.87 ± 0.07 for the modeled structures of DnaE1 and DnaE2, respectively. In general, the modeled protein structure with a TM-score > 0.5 is acceptable with the correct topology (Yang et al., 2015). The DnaE1 and DnaE2 proteins of M. xanthus DK1622 had highly similar three-dimensional structures, even though their sequences were highly different. The active sites and some of the DNA binding surfaces were also highly conserved in the DnaE1 and DnaE2 polymerases. In addition, the two DnaE proteins were also considerably conserved of their surface amino acid positions (Figure 4A, right panels). However, the DnaE1 polymerase contained five domains, including PHP, Pol3, HhH, OB, and CTD domains, while DnaE2 had four domains, lacking the CTD domain (Figure 4A, left panels), which is important for binding to Pol III τ-subunit (Liu et al., 2013).
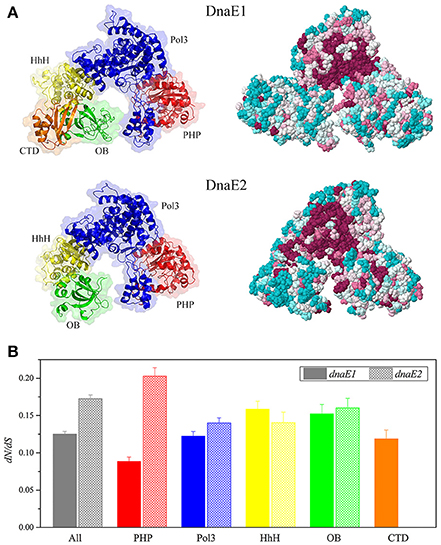
Figure 4. Structure modeling of DK1622 DnaE1 and DnaE2 (A) and comparison of the domain conservation of myxobacterial dnaE1 and dnaE2 (B). (A) Three-dimensional structures showing different domains in different colors (left panels) and evolutionary conservation of surface amino acids (right panels); (B) The dN/dS values of dnaE1 and dnaE2 and the different domains of those sequenced myxobacteria.
In order to get insight into the domain conservation, we estimated the dN/dS values, an indicator of the selective pressure acting on a protein-coding gene, for the duplicated dnaE genes of those sequenced myxobacteria. The dN/dS values of the complete dnaE1 and dnaE2 sequences were 0.125 (95% confidence interval: 0.121–0.129) and 0.172 (95% confidence interval: 0.167–0.178), respectively, suggesting both polymerases were subjected to a strong negative selection pressure for their strict functional constraint. The dN/dS values were close between the Pol3, HhH, and OB domain sequences of the dnaE1 and dnaE2 genes, but had a great difference in the PHP sequence (t-test, P < 0.01, Figure 4B), which has a binding site of the proofreading ε-subunit (Wieczorek and McHenry, 2006; Ozawa et al., 2013). The dN/dS value of the dnaE2 PHP domain was more than twice than that of dnaE1. Accordingly, dnaE1 is more evolutionarily conservative than dnaE2 in myxobacteria. Stano et al. reported that the PHP domain of DnaE-type polymerases had 3′-5′ exonuclease activity (Stano et al., 2006). Similarly, the PHP domain of Mtb DnaE1 retains all amino acids associated with exonuclease activity in M. tuberculosis, but the DnaE2 PHP lost some key residues (Wieczorek and McHenry, 2006; Baños et al., 2008). It was speculated that the loss of conserved residues in the DnaE2 PHP domain affected relative fidelity. The DNA polymerase III complexes constructed with the two DnaE proteins thus probably had divergent functions.
Discussion
DNA polymerase III holoenzyme is the main polymerase for bacterial DNA replication. While most bacterial cells contain single copies of dnaE, encoding for the alpha subunit of the polymerase, some bacteria contain duplicate dnaE genes (Timinskas et al., 2014). There are several reports dealt with the functions of duplicated dnaE genes and have revealed some key characteristics of dnaE2, for example, the second dnaE gene encodes an error-prone enzyme and is dispensable. However, the function of the duplicate dnaE is still an enigma. For example, what is the role of the second copy of dnaE in bacteria, why do bacterial cells need an error-prone DNA polymerase enzyme, and how does the error-prone enzyme play functions in bacterial cells?
Myxobacteria have complex multicellular social behaviors, and all sequenced myxobacterial genomes contain duplicated dnaE genes. Given the high conservation in myxobacteria (Figure S1), the DnaE2 subunit should be important for myxobacterial cells. The expression of dnaE1 was greatly higher than that of the dnaE2 gene throughout the lifecycle of M. xanthus cells. Obviously, the dnaE1 gene plays the major and essential role in chromosome replication in M. xanthus cells, not only in the growth stage but also in the development stage, thus ensuring the population genetically stable. In contrast, the dnaE2 gene expressed approximately ten times lower than the dnaE1 gene did in either the growth or the developmental stage, which is consistent with their weak cellular effects. Although weakly affected the growth and social motility, phenotypic characteristics of dnaE2 mutants supported that dnaE2 strongly affected the development progress. However, because the expression of dnaE1 was also decreased, the deficiencies of development and sporulation in the dnaE2 deletion mutant were the results of the synergetic effects of the two dnaE genes.
In nutrition-limited conditions, M. xanthus populations aggregate and develop fruiting bodies, which contain stress resistant myxospores (Shimkets, 1990; Shimkets et al., 2006). Unlike the formation progress of endospores in Bacillus subtilis (Errington and Wake, 1991), M. xanthus cells carry out a round of chromosome replication in the early stage of development (Tzeng and Singer, 2005). We demonstrated that the deletion of dnaE2 caused obvious phenotypic deficiencies in development and sporulation. The development program of M. xanthus is a multicellular process requiring the coordinated expression of numerous regulation pathways to produce extracellular A- to E-signals and intracellular signals (Kaiser, 2004). Stringent response to the starvation of amino acids, the (p)ppGpp synthase enzyme (RelA) produces the chemical signal (p)ppGpp to initiate the development process (Singer and Kaiser, 1995; Harris et al., 1998). Rosario and Singer showed that inhibition of DNA replication within the first 6 h of development results in a block in developmental program, and the dnaA gene that encodes the initiator protein for DNA replication is not essential for development but allow for the proper timing and maximum efficiency of the sporulation process (Rosario and Singer, 2010). Our data improve the understanding of chromosome replication in the early stage of development of M. xanthus. We determined that, together with the essential DnaE1 polymerase, the error-prone DnaE2 enzyme participated in the chromosome replication of development process in M. xanthus DK1622, but in low fidelity for the genome replication due to its error-prone characteristic. It is known that suitable mutation rates can increase the adaptation abilities, whereas high mutation rates damage bacterial cells (Oliver et al., 2000; Giraud et al., 2001; Wielgoss et al., 2013). In new environments, the mutations probably increase for rapid adaptation, while in stable conditions, the mutations may decrease to reduce damages to cells. Thus, bacterial cells have to balance the mutation rates. The previous studies suggested that the bacterial land colonization is dominated by the emergence of dnaE2, followed by a series of niche-specific genomic adaptations, including GC content increase, intensive horizontal gene transfer and constant genome expansion (Wu et al., 2014). We suggested that the DnaE2 is rather a balancer to control the genome mutations to a suitable rate, based on its error-prone characteristic and its low expressions in either the growth or development stage. Low concentrations make the replication carried out by the DnaE2 polymerases occupy small fractions. We found that artificial overexpression of the dnaE2 gene significantly increased the genome mutation rate, which suggested to be damaged for cell survivals. In addition, because the deletion of dnaE2 caused deficiency of development and sporulation, the presence of the gene had some more effects on cellular behaviors. Using the master strategy, M. xanthus populations are able to live through the changing environments.
Experimental Procedures
Strains and Cultivation
The bacterial strains used in this study are listed in Table S2. The M. xanthus strains were cultivated in Casitone-based CTT medium (Hodgkin and Kaiser, 1977) for growth assays and on TPM agar plate (Kearns et al., 2000) for developmental assays. E. coli strains were routinely grown on Luria-Bertani (LB) agar or in LB liquid broth. M. xanthus strains were incubated at 30°C, and E. coli strains were at 37°C. When required, a final concentration of 40 μg/ml of kanamycin (Km) was added to the solid or liquid media.
Construction of M. xanthus Mutants
We performed the in-frame deletions of the dnaE1, dnaE2, mutS, and mutL genes in M. xanthus strains using pBJ113 plasmid, respectively. The plasmid contains a kanamycin resistant cassette for the first round of screening and a galK gene for the negative screening (Ueki et al., 1996). Briefly, homologous arms of dnaE1, dnaE2, mutS, or mutL were cloned with primers (listed in Table S3) and the products were inserted into the EcoRI/XbaI site of pBJ113. The resulting plasmid were introduced into M. xanthus DK1622 strains via electroporation (1.25 kV, 400 W, 25 mF, 0.1-cm cuvette gap), respectively. The second round of screening was then performed on CTT plates containing 1% galactose (Sigma). The deletion mutants that grew on galactose but were sensitive to kanamycin were identified and verified by PCR amplification and sequencing. Restriction enzymes, DNA ligase, and other DNA enzymes were used according to the manufacturers' recommendations. All fragments were validated by Sanger 3730 sequencing.
Complementation and Overexpression of dnaE2 Gene
The dnaE2 gene and upstream 500 bp sequence that contains the whole promoter of dnaE2 gene was cloned using the primer pair of MXAN_3982_NativeF and MXAN_3982_NativeR. The fragment and pSWU19 plasmid (replacing the tet resistant cassette of pSWU30 with a kanamycin resistant cassette) were digested with the XbaI/EcoRI digestion solution. The two fragments were ligated with T4 DNA ligase, producing the recombinant plasmid pSW3982. The promoter of the pilA gene was cloned using the primer pair of MXAN_3982P_UF and MXAN_3982P_UR, and fused with the dnaE2 gene. The fusion product was inserted into XbaI/EcoRI sites of pSWU19, producing the plasmid pSWp3982.
The dnaE2 fusion construct was transferred by electroporation into M. xanthus DK1622. Individual kanamycin-resistant clones were selected, and the mutants of the dnaE2 transcription fusion were verified by PCR amplification (primers are listed in Table S3) and sequencing.
Mutation Rate Assay
The mutation rate assay was conducted by screening the nalidixic acid resistant strains according to previous report (Tzeng et al., 2006). Briefly, approximately 5 × 109 cells of M. xanthus strains were placed on CTT agar containing 40 μg/ml nalidixic acid. Nalidixic acid- resistant candidates were counted and mutation rate was calculated using the following formula, r = Nm/N0, where r is the mutation rate, Nm represents the count of nalidixic acid resistant mutant, and N0 is the count of cells placed on the screening plate.
Development and Sporulation Assays
Developmental experiments were performed on TPM agar plates (10 mM Tris [pH 7.6], 8 mM MgSO4, and 1 mM KH2PO4 containing 1.5% agar), as described below. Cells were allowed to develop in a humidity chamber at 30°C. When indicated, nalidixic acid solution of the 20 μg/ml concentration was added onto different plates at the time of 0, 6, 12, 18, or 24 h for 1 h, and then the extra solution was poured out. After 120 h cultivation at 30°C. Sporulation was conducted as follows: five colonies were collected and resuspended in liquid CTT media. Sonication was conducted at 200 W for 4 s twice to release myxospores from fruiting bodies. The myxospore suspensions were incubated at 55°C for 2 h to kill the vegetative cells. Then the suspensions were serially diluted and aliquots of 50 μL were used to plate. The sporulation rate was counted after 5 days of cultivation on the CTT plate.
RNA Extraction and Quantitative Real-Time PCR Assay
The M. xanthus DK1622 cells were concentrated to approximately 1.75 × 1010 cells per milliliter. The cell suspension was washed three times using TPM buffer. RNA was extracted immediately with a BIOZOL total RNA extraction reagent (BioFlux) following the manufacturer's instructions. Genomic DNA contamination was removed by using DNA eraser supplied in the PrimeScript RT Reagent kit with gDNA Eraser (TaKaRa). The purified RNA extracts were transcribed reversely to cDNA and stored in aliquots at −80°C. Quantitative real-time PCR was performed in a total reaction volume of 25 μl, containing 250 nM primers, 12.5 μl of SYBR Premix Ex Taq GC mix (TaKaRa), 10.5 μl of RNase-free water (TaKaRa), and 1 μl of a 10-fold-diluted cDNA template. PCR was conducted in a Roche LightCycler 480 sequence detection system, following the program: 3 min at 95°C, followed by 40 cycles of 30 s at 95°C, 30 s at 55°C, and 15 s at 72°C. The gapA gene, encoding for glyceraldehyde-3-phosphate dehydrogenase, was used as the normalization signal. Calibration curves of gapA, MXAN_3982, and MXAN_5844 were generated from a 10-fold dilution of M. xanthus DK1622 genomic DNA. The primer pairs used for each gene are listed in Table S3.
Bioinformatics Analyses
The dnaE1 and dnaE2 genes were extracted from the genome of M. xanthus DK1622 strain. All the dnaE genes used for phylogenetic analysis were extracted according to the Genomic Encyclopedia of Bacteria and Archaea (Wu et al., 2009). These genes were translated into amino acid sequences according to the bacterial standard codon. A multiple sequence alignment of the DnaE1 and DnaE2 proteins was established using the MAFFT program (Katoh and Standley, 2013). The phylogenetic tree was constructed using the PhyML program with the LG substitution model and the four substitution rate categories (Guindon et al., 2010). LG model was regarded as the best substitution model by automatic model selection. Branch support was calculated using the approximate likelihood ratio tests (aLRT SH-like) (Anisimova and Gascuel, 2006). The phylogenetic tree was visualized by iTOL (Letunic and Bork, 2016).
The structures of the DnaE1 and DnaE2 were modeled using the I-TASSER program based on a threading approach (Yang et al., 2015). The evolutionary conservation of amino acid positions in the dnaE1 and dnaE2 sequences was estimated by using the ConSurf algorithm (Ashkenazy et al., 2010). The JTT substitution matrix was used and the computation was based on the empirical Bayesian paradigm. The conservation scale was defined from the most variable amino acid positions (grade 1), which were considered to be evolved rapidly, to the most conservative positions (grade 9), which were considered to be evolved slowly. The sequences and modeled structures of the DnaE1 and DnaE2 were shown in nine-color conservation grades.
The number ratio of nonsynonymous substitutions per nonsynonymous site (dN) to synonymous substitutions per synonymous site (dS), dN/dS, is an indicator of the selective pressure acting on a protein-coding gene. The dN/dS calculation of the dnaE1 and dnaE2 locus was set at the 95% confidence intervals, using the Datamonkey web server (Delport et al., 2010). For the calculation, a multiple sequence alignment of the nucleic acid sequences of myxobacterial dnaE1 and dnaE2 genes, based on codon, was established with TranslatorX (Abascal et al., 2010). Domains of DnaE1 and DnaE2 proteins were extracted by blasting against the PFAM database (Finn et al., 2014).
The statistical analysis was conducted using IBM SPSS Statistics.
Author Contributions
Conceived and designed the experiments: YL and RP. Performed the experiments and analyzed the data: RP, JC, WF, ZZ, JY, and ZL. Wrote the paper: YL and RP.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. K. Han for invaluable help with the manuscript. This work was financially supported by the National Natural Science Foundation of China (NSFC) (No. 31471183) and the NSFC Key Program (No. 31130004) awarded to YL.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00122/full#supplementary-material
Figure S1. Phylogenetic analyses of myxobacterial DnaE protein sequences. Some determined DnaE1 and DnaE2 proteins from other bacteria species were also included in the tree.
Table S1. Information of dnaE genes in sequenced myxobacterial genomes.
Table S2. Strains and plasmids used in this study.
Table S3. Primers used in this study.
References
Abascal, F., Zardoya, R., and Telford, M. J. (2010). TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res 38, W7–W13. doi: 10.1093/nar/gkq291
Abella, M., Erill, I., Jara, M., Mazón, G., Campoy, S., and Barbé, J. (2004). Widespread distribution of a lexA-regulated DNA damage-inducible multiple gene cassette in the Proteobacteria phylum. Mol. Microbiol. 54, 212–222. doi: 10.1111/j.1365-2958.2004.04260.x
Anisimova, M., and Gascuel, O. (2006). Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55, 539–552. doi: 10.1080/10635150600755453
Ashkenazy, H., Erez, E., Martz, E., Pupko, T., and Ben-Tal, N. (2010). ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533. doi: 10.1093/nar/gkq399
Baños, B., Lázaro, J. M., Villar, L., Salas, M., and De Vega, M. (2008). Editing of misaligned 3'-termini by an intrinsic 3'-5' exonuclease activity residing in the PHP domain of a family X DNA polymerase. Nucleic Acids Res. 36, 5736–5749. doi: 10.1093/nar/gkn526
Boshoff, H. I., Reed, M. B., Barry, C. E.III, and Mizrahi, V. (2003). DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell 113, 183–193. doi: 10.1016/S0092-8674(03)00270-8
Brinkhoff, T., Fischer, D., Vollmers, J., Voget, S., Beardsley, C., Thole, S., et al. (2012). Biogeography and phylogenetic diversity of a cluster of exclusively marine myxobacteria. ISME J. 6, 1260–1272. doi: 10.1038/ismej.2011.190
Delport, W., Poon, A. F., Frost, S. D., and Kosakovsky Pond, S. L. (2010). Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26, 2455–2457. doi: 10.1093/bioinformatics/btq429
Dworkin, M. (1996). Recent advances in the social and developmental biology of the myxobacteria. Microbiol. Rev. 60, 70–102.
Errington, J., and Wake, R. G. (1991). Chromosome strand segregation during sporulation in Bacillus subtilis. Mol. Microbiol. 5, 1145–1149. doi: 10.1111/j.1365-2958.1991.tb01887.x
Evans, R. J., Davies, D. R., Bullard, J. M., Christensen, J., Green, L. S., Guiles, J. W., et al. (2008). Structure of PolC reveals unique DNA binding and fidelity determinants. Proc. Natl. Acad. Sci. U.S.A. 105, 20695–20700. doi: 10.1073/pnas.0809989106
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Giraud, A., Matic, I., Tenaillon, O., Clara, A., Radman, M., Fons, M., et al. (2001). Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291, 2606–2608. doi: 10.1126/science.1056421
Goldman, B. S., Nierman, W. C., Kaiser, D., Slater, S. C., Durkin, A. S., Eisen, J. A., et al. (2006). Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U.S.A. 103, 15200–15205. doi: 10.1073/pnas.0607335103
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Harris, B. Z., Kaiser, D., and Singer, M. (1998). The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 12, 1022–1035. doi: 10.1101/gad.12.7.1022
Hodgkin, J., and Kaiser, D. (1977). Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. U.S.A. 74, 2938–2942. doi: 10.1073/pnas.74.7.2938
Ito, J., and Braithwaite, D. K. (1991). Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 19, 4045–4057. doi: 10.1093/nar/19.15.4045
Jiang, D. M., Kato, C., Zhou, X. W., Wu, Z. H., Sato, T., and Li, Y. Z. (2010). Phylogeographic separation of marine and soil myxobacteria at high levels of classification. ISME J. 4, 1520–1530. doi: 10.1038/ismej.2010.84
Kaiser, D. (2004). Signaling in myxobacteria. Annu. Rev. Microbiol. 58, 75–98. doi: 10.1146/annurev.micro.58.030603.123620
Kaiser, D., and Losick, R. (1993). How and why bacteria talk to each other. Cell 73, 873–885. doi: 10.1016/0092-8674(93)90268-U
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kearns, D. B., Campbell, B. D., and Shimkets, L. J. (2000). Myxococcus xanthus fibril appendages are essential for excitation by a phospholipid attractant. Proc. Natl. Acad. Sci. U.S.A. 97, 11505–11510. doi: 10.1073/pnas.210448597
Kelman, Z., and O'donnell, M. (1995). DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu. Rev. Biochem. 64, 171–200. doi: 10.1146/annurev.bi.64.070195.001131
Koorits, L., Tegova, R., Tark, M., Tarassova, K., Tover, A., and Kivisaar, M. (2007). Study of involvement of ImuB and DnaE2 in stationary-phase mutagenesis in Pseudomonas putida. DNA Repair (Amst). 6, 863–868. doi: 10.1016/j.dnarep.2007.01.010
Lamers, M. H., Georgescu, R. E., Lee, S. G., O'donnell, M., and Kuriyan, J. (2006). Crystal structure of the catalytic α subunit of E. coli replicative DNA polymerase III. Cell 126, 881–892. doi: 10.1016/j.cell.2006.07.028
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Li, S. G., Zhou, X. W., Li, P. F., Han, K., Li, W., Li, Z. F., et al. (2012). The existence and diversity of myxobacteria in lake mud - a previously unexplored myxobacteria habitat. Environ. Microbiol. Rep. 4, 587–595. doi: 10.1111/j.1758-2229.2012.00373.x
Liu, B., Lin, J., and Steitz, T. A. (2013). Structure of the PolIIIα-tauc-DNA complex suggests an atomic model of the replisome. Structure 21, 658–664. doi: 10.1016/j.str.2013.02.002
McHenry, C. S. (2011a). Bacterial replicases and related polymerases. Curr. Opin. Chem. Biol. 15, 587–594. doi: 10.1016/j.cbpa.2011.07.018
McHenry, C. S. (2011b). Breaking the rules: bacteria that use several DNA polymerase IIIs. EMBO Rep. 12, 408–414. doi: 10.1038/embor.2011.51
Oliver, A., Cantón, R., Campo, P., Baquero, F., and Blázquez, J. (2000). High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288, 1251–1254. doi: 10.1126/science.288.5469.1251
Ozawa, K., Horan, N. P., Robinson, A., Yagi, H., Hill, F. R., Jergic, S., et al. (2013). Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits α, ε, θ, and β reveals a highly flexible arrangement of the proofreading domain. Nucleic Acids Res. 41, 5354–5367. doi: 10.1093/nar/gkt162
Reichenbach, H. (1999). The ecology of the myxobacteria. Environ. Microbiol. 1, 15–21. doi: 10.1046/j.1462-2920.1999.00016.x
Robinson, A., Causer, R. J., and Dixon, N. E. (2012). Architecture and conservation of the bacterial DNA replication machinery, an underexploited drug target. Curr. Drug Targets 13, 352–372. doi: 10.2174/138945012799424598
Rosario, C. J., and Singer, M. (2010). Developmental expression of dnaA is required for sporulation and timing of fruiting body formation in Myxococcus xanthus. Mol. Microbiol. 76, 1322–1333. doi: 10.1111/j.1365-2958.2010.07178.x
Shimkets, L. J. (1990). Social and developmental biology of the myxobacteria. Microbiol. Rev. 54, 473–501.
Shimkets, L. J., Dworkin, M., and Reichenbach, H. (2006). “The myxobacteria,” in The prokaryotes, eds M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (New York, NY: Springer), 31–115.
Singer, M., and Kaiser, D. (1995). Ectopic production of guanosine penta- and tetraphosphate can initiate early developmental gene expression in Myxococcus xanthus. Genes Dev. 9, 1633–1644. doi: 10.1101/gad.9.13.1633
Sniegowski, P. D., Gerrish, P. J., and Lenski, R. E. (1997). Evolution of high mutation rates in experimental populations of E. coli. Nature 387, 703–705. doi: 10.1038/42701
Stano, N. M., Chen, J., and McHenry, C. S. (2006). A coproofreading Zn 2+-dependent exonuclease within a bacterial replicase. Nat. Struct. Mol. Biol. 13, 458–459. doi: 10.1038/nsmb1078
Timinskas, K., Balvociute, M., Timinskas, A., and Venclovas, C. (2014). Comprehensive analysis of DNA polymerase III α subunits and their homologs in bacterial genomes. Nucleic Acids Res. 42, 1393–1413. doi: 10.1093/nar/gkt900
Tsai, H. H., Shu, H. W., Yang, C. C., and Chen, C. W. (2012). Translesion-synthesis DNA polymerases participate in replication of the telomeres in Streptomyces. Nucleic Acids Res. 40, 1118–1130. doi: 10.1093/nar/gkr856
Tzeng, L., Ellis, T. N., and Singer, M. (2006). DNA replication during aggregation phase is essential for Myxococcus xanthus development. J. Bacteriol. 188, 2774–2779. doi: 10.1128/JB.188.8.2774-2779.2006
Tzeng, L., and Singer, M. (2005). DNA replication during sporulation in Myxococcus xanthus fruiting bodies. Proc. Natl. Acad. Sci. U.S.A. 102, 14428–14433. doi: 10.1073/pnas.0506969102
Ueki, T., Inouye, S., and Inouye, M. (1996). Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene 183, 153–157. doi: 10.1016/S0378-1119(96)00546-X
Warner, D. F., Ndwandwe, D. E., Abrahams, G. L., Kana, B. D., Machowski, E. E., Venclovas, C., et al. (2010). Essential roles for imuA'- and imuB-encoded accessory factors in DnaE2-dependent mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 107, 13093–13098. doi: 10.1073/pnas.1002614107
Wieczorek, A., and McHenry, C. S. (2006). The NH2-terminal php domain of the α subunit of the Escherichia coli replicase binds the epsilon proofreading subunit. J. Biol. Chem. 281, 12561–12567. doi: 10.1074/jbc.M513844200
Wielgoss, S., Barrick, J. E., Tenaillon, O., Wiser, M. J., Dittmar, W. J., Cruveiller, S., et al. (2013). Mutation rate dynamics in a bacterial population reflect tension between adaptation and genetic load. Proc. Natl. Acad. Sci. U.S.A. 110, 222–227. doi: 10.1073/pnas.1219574110
Wu, D., Hugenholtz, P., Mavromatis, K., Pukall, R., Dalin, E., Ivanova, N. N., et al. (2009). A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 462, 1056–1060. doi: 10.1038/nature08656
Wu, H., Fang, Y., Yu, J., and Zhang, Z. (2014). The quest for a unified view of bacterial land colonization. ISME J. 8, 1358–1369. doi: 10.1038/ismej.2013.247
Wu, S. S., and Kaiser, D. (1995). Genetic and functional evidence that Type IV pili are required for social gliding motility in Myxococcus xanthus. Mol. Microbiol. 18, 547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x
Yang, J., Yan, R., Roy, A., Xu, D., Poisson, J., and Zhang, Y. (2015). The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8. doi: 10.1038/nmeth.3213
Zhao, X., Zhang, Z., Yan, J., and Yu, J. (2007). GC content variability of eubacteria is governed by the pol III α subunit. Biochem. Biophys. Res. Commun. 356, 20–25. doi: 10.1016/j.bbrc.2007.02.109
Keywords: dnaE2 gene, dispensable, error-prone, chromosome replication, growth, development and sporulation, Myxococcus xanthus
Citation: Peng R, Chen J-h, Feng W-w, Zhang Z, Yin J, Li Z-s and Li Y-z (2017) Error-prone DnaE2 Balances the Genome Mutation Rates in Myxococcus xanthus DK1622. Front. Microbiol. 8:122. doi: 10.3389/fmicb.2017.00122
Received: 05 November 2016; Accepted: 17 January 2017;
Published: 01 February 2017.
Edited by:
Feng Gao, Tianjin University, ChinaReviewed by:
Camilo E. Khatchikian, University of Texas at El Paso, USAHao Wu, University of Gothenburg, Sweden
Copyright © 2017 Peng, Chen, Feng, Zhang, Yin, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yue-Zhong Li, bGlsYWJAc2R1LmVkdS5jbg==
 Ran Peng
Ran Peng Jiang-he Chen
Jiang-he Chen Zheng Zhang
Zheng Zhang Yue-zhong Li
Yue-zhong Li