- 1Department of Clinical Microbiology, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, China
- 2Department of Burns and Plastic Surgery, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, China
- 3Department of Medical Microbiology and Parasitology, Institutes of Medical Sciences, Shanghai Jiaotong University School of Medicine, Shanghai, China
Objectives: To describe the genetic environment, transferability, and antibiotic susceptibility of one clinical Klebsiella pneumoniae isolate harboring both blaOXA-48 and blaNDM-1 on different plasmids from a Chinese hospital.
Methods: The isolate was subjected to antimicrobial susceptibility testing and multilocus sequence typing using Etest and PCR. The plasmids harboring blaOXA-48 and blaNDM-1 were analyzed through conjugation experiments, S1-nuclease pulsed-field gel electrophoresis, and hybridization with specific probes. Plasmid DNA was sequenced using Pacbio RS II and annotated using RAST.
Results: K. pneumoniae RJ119, carrying both blaOXA-48 and blaNDM-1, was resistant to almost all carbapenems, cephalosporins, fluoroquinolone, and aminoglycosides and belonged to ST307. blaOXA-48 was located on a 61,748-bp IncL/M conjugative plasmid, which displayed overall nucleotide identity (99%) to pKPN-E1-Nr.7. blaNDM-1 was located on a 335,317-bp conjugative plasmid, which was a fusion of a blaNDM-1-harboring InA/C plasmid pNDM-US (140,825 bp, 99% identity) and an IncFIB plasmid pKPN-c22 (178,563 bp, 99% identity). The transconjugant RJ119-1 harboring blaNDM-1 was susceptible to carbapenem, and there was an insertion of IS10 into the blaNDM-1 gene.
Conclusion: This is the first report of the coexistence of blaOXA-48 and blaNDM-1 in one K. pneumoniae clinical isolate in China. OXA-48 in RJ119 contributed to the majority to its high resistance to carbapenems, whereas NDM-1 remained unexpressed, most likely due to the insertion of IS10. Our results provide new insight for the relationship between genetic diagnosis and clinical treatment. They also indicate that increased surveillance of blaOXA-48 is urgently needed in China.
Introduction
The emergence of carbapenem-resistant Enterobacteriaceae (CRE) has become a challenge to clinical therapy because of the rapid worldwide dissemination of multidrug resistance. The most important carbapenemase genes in Enterobacteriaceae are those encoding for KPC, VIM, IMP, NDM, and OXA-48 (Queenan and Bush, 2007). NDM-1 was first reported in 2008 as being produced by a Klebsiella pneumoniae isolate from a Swedish patient who had returned from India (Yong et al., 2009). Alarmingly, it was often found on large conjugative plasmids along with additional antibiotic resistance determinants (Hudson et al., 2014). NDM-1-producing K. pneumoniae isolates are considered as endemic in many countries including India, Pakistan, Bangladesh (Nordmann and Poirel, 2014) and show sporadic spread in the USA (Rasheed et al., 2013), Canada (Lowe et al., 2013), Colombia (Ocampo et al., 2016), Greece (Spyropoulou et al., 2016), Singapore (Ling et al., 2015), and China (Qu et al., 2015). OXA-48 was first identified in a K. pneumoniae isolate from Turkey in 2001 (Poirel et al., 2003). Since then, endemic spread of OXA-48-producing K. pneumoniae isolates has been reported in countries such as Turkey, Morocco, Libya, Egypt, Tunisia, and India (Nordmann and Poirel, 2014). The coexistence of genes for at least two classes of carbapenemases in K. pneumoniae also has been reported worldwide, including KPC-3 and VIM-2 in Italy, KPC-2 and VIM-24 in Colombia, NDM-1 and KPC-2 in Brazil, and NDM-1 and OXA-181 in Singapore (Lee et al., 2016).
We studied the genetic environment, transferability, and antibiotic susceptibility of a clinical isolate of K. pneumoniae harboring the blaOXA-48 and blaNDM-1 genes on different plasmids, which was isolated from a Chinese hospital.
Materials and Methods
Bacterial Isolates and Antimicrobial Susceptibility Testing
Klebsiella pneumoniae RJ119, carrying both the blaOXA-48 and blaNDM-1 genes, was isolated from the wound of a female burn patient at Ruijin Hospital, Shanghai, in September 2015. The previous travel history of the patient was not recorded. The genes were detected by the Cepheid Xpert Carba-R assay (Cepheid, Sunnyvale, CA, USA), a qualitative diagnostic test that was designed for rapid detection and differentiation of the blaKPC, blaNDM, blaV IM, blaOXA-48, and blaIMP-1 genes. The minimum inhibitory concentrations (MICs) of ceftazidime, cefotaxime, ceftriaxone, cefepime, aztreonam, piperacillin/tazobactam, ertapenem, meropenem, imipenem, trimethoprim/sulfamethoxazole, ciprofloxacin, and amikacin in the different strains isolated from the patient were determined by using the Etest (bioMérieux, France), and the results were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute [CLSI] (2014). The CLSI does not have interpretative criteria for tigecycline; therefore, European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints were used1. Escherichia coli ATCC 25922 was used as a quality control reference strain.
This study was approved by Ruijin Hospital Ethics Committee (Shanghai Jiao Tong University School of Medicine), and the Review Board exempted the requirement for informed consent because this retrospective study only focused on bacteria and did not affect the patients.
Multilocus Sequence Typing (MLST)
Multilocus sequence typing (MLST) was performed on seven housekeeping genes according to the guidelines given in the K. pneumoniae MLST website2.
Conjugation Experiments, S1-Nuclease Pulsed-Field Gel Electrophoresis (S1-PFGE), and Southern Hybridization
The transferability of blaOXA-48 and blaNDM-1 was assessed in broth culture using E. coli J53 Azr (sodium azide-resistant) as the recipient. Transconjugants were selected on MacConkey agar containing sodium azide (100 mg/L) and meropenem (0.5 mg/L) or ceftazidime (1 mg/L) and were confirmed to have blaOXA-48 and blaNDM-1 by PCR analysis. Plasmid DNA of the parental and transconjugant isolates was visualized by S1-PFGE (Barton et al., 1995) and then transferred to positively charged nylon membranes (Roche Applied Science, Penzberg, Germany). The membranes were hybridized with digoxigenin-labeled blaOXA-48- and blaNDM-1-specific probes, respectively.
DNA Preparation and Sequencing
Plasmid DNA was extracted from the RJ119 strain using the QIAGEN Midi Kit (Qiagen, Hilden, Germany). DNA was sequenced using Pacbio RS II (Pacific Biosciences, Menlo Park, CA, USA). The reads were de novo assembled using HGAP 3.0 of SMRTTM Pipe (Supplementary Images 1). Protein-coding genes were initially identified and annotated using RAST (Aziz et al., 2008). Insertion elements (IS) and antimicrobial resistance genes were identified using IS Finder3 and ResFinder4, respectively. PlasmidFinder5 was used to detect and type the plasmid. BLAST6 searches were used to identify related plasmids carrying blaOXA-48 and blaNDM-1 to guide PCR-based gap closure and Sanger sequencing to assemble contigs into complete plasmids.
Verification of the Fusion Region
To confirm the fusion region, four overlapping PCR were employed with in-house-designed primers P1 (5′-AAATGCGGCTAAGGTTGTGA-3′), P2 (5′-CCCTTCAGGCGTGATTCATA-3′), P3 (5′-GGCAAAGGCGACAAGAAGGA-3′), P4 (5′-GACAGCAGGCTCAGAAGACG-3′), P5 (5′-AGTAGCGGAGCAGGAAGGAC-3′) and P6 (5′-GTTCGGAGATGGAGGGTCAA-3′), respectively (Figure 3).
Nucleotide Sequence Accession Numbers
The complete sequence of plasmids pRJ119-NDM1 and pRJ119-OXA48 in K. pneumoniae RJ119 has been deposited in GenBank under the accession numbers KX636095 and KX636096, respectively (Supplementary Datasheet File 1).
Results
Antimicrobial Susceptibility Testing and MLST
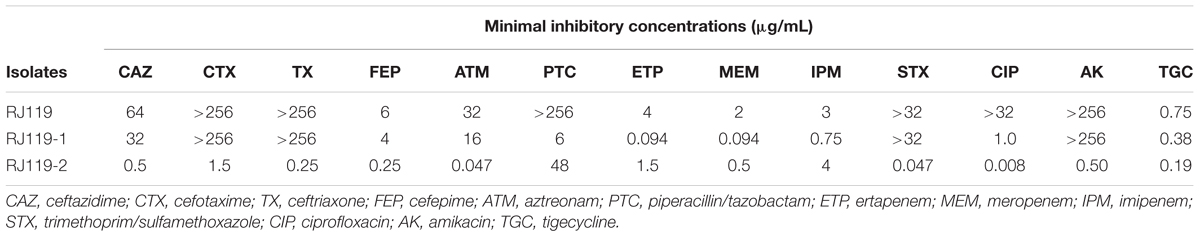
Klebsiella pneumoniae RJ119 belonged to ST307, exhibiting resistance or intermediate resistance to cephalosporins, β-lactam/β-lactamase inhibitor combinations, carbapenems, fluoroquinolones, aminoglycosides, trimethoprim/sulfamet-hoxazole, and tigecycline, as shown in Table 1.
Conjugation Experiments, S1-PFGE, and Southern Hybridization
In the conjugation experiments, two phenotypically different transconjugants (RJ119-1 and RJ119-2), each of which carried one plasmid, were obtained. Phenotypic testing of the transconjugants showed that RJ119-1 was resistant to cephalosporins but susceptible to carbapenems, whereas RJ119-2 was susceptible to cephalosporins but resistant to carbapenems (Table 1). S1-PFGE revealed that RJ119 harbored two plasmids, and the transconjugants RJ119-1 and RJ119-2 each contained a single plasmid. Southern hybridization analysis revealed blaNDM-1 located on a 330-kb (pRJ119-NDM1) plasmid and blaOXA-48 on a 60-kb (pRJ119-OXA48) plasmid (Figure 1).
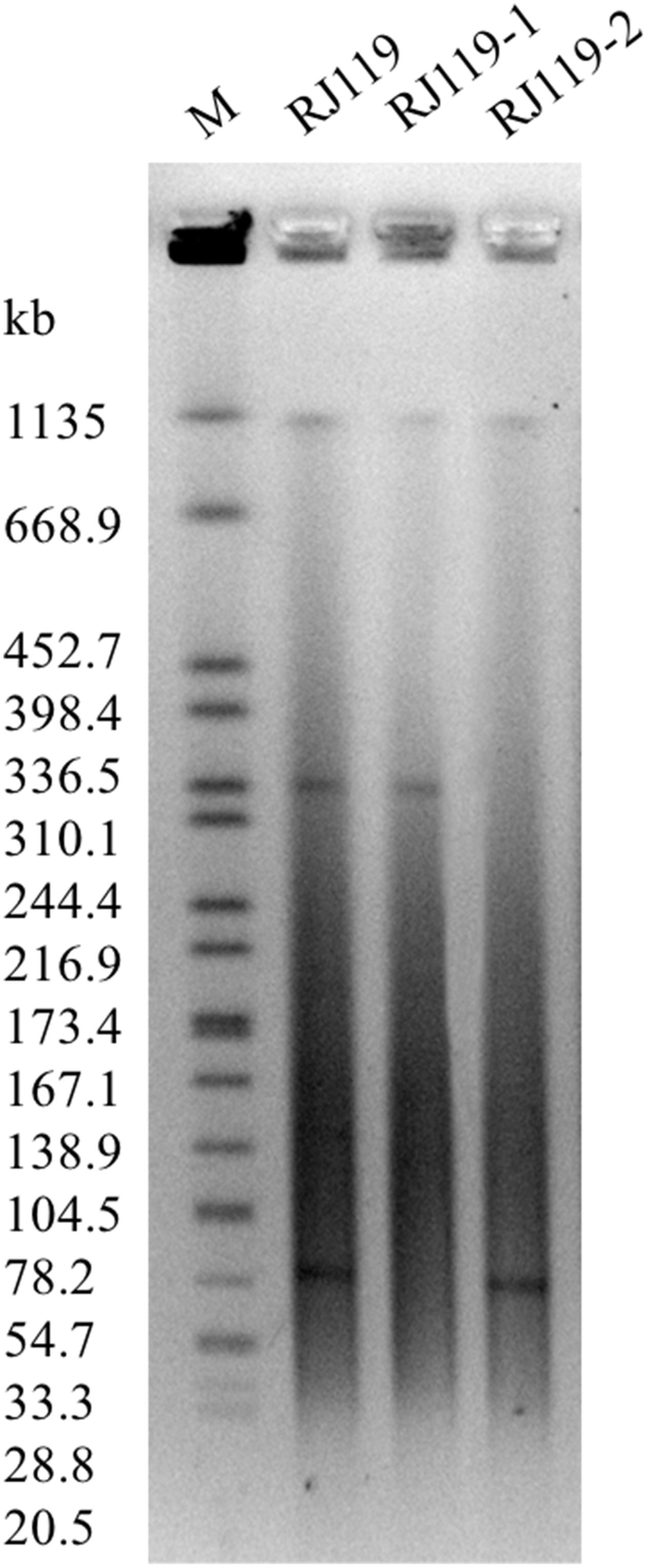
FIGURE 1. S1-nuclease pulsed-field gel electrophoresis (S1-PFGE) profiles of Klebsiella pneumoniae RJ119 and its transconjugants RJ119-1, RJ119-2. M, Salmonella enterica serotype Braenderup H9812 was digested with XbaI and used as a molecular size marker.
Complete Nucleotide Sequence of Plasmids and Verification of the Fusion Region
pRJ119-OXA48 is a 61,748-bp plasmid that belongs to the IncL/M incompatibility group (Figure 2). A BLAST search against all complete sequences indicated that pRJ119-OXA48 displayed overall nucleotide identity (99%) to pKPN-E1-Nr.7, a blaOXA-48-harboring IncL/M plasmid from a K. pneumoniae isolate from Switzerland (Carattoli et al., 2015). The main difference between the plasmids pRJ119-OXA48 and pKPN-E1-Nr.7 was that the Tn1999.2 element (IS1999-LysR- blaOXA-48-IS1-IS1999) harboring blaOXA-48 in pRJ119-OXA48 was in the opposite orientation in pKPN-E1-Nr.7, and there was a deletion of korC (transcriptional repressor) in pR119-OXA48 (Figure 2), which was confirmed by PCR and Sanger sequencing. No resistance genes other than blaOXA-48 were identified in pR119-OXA48.
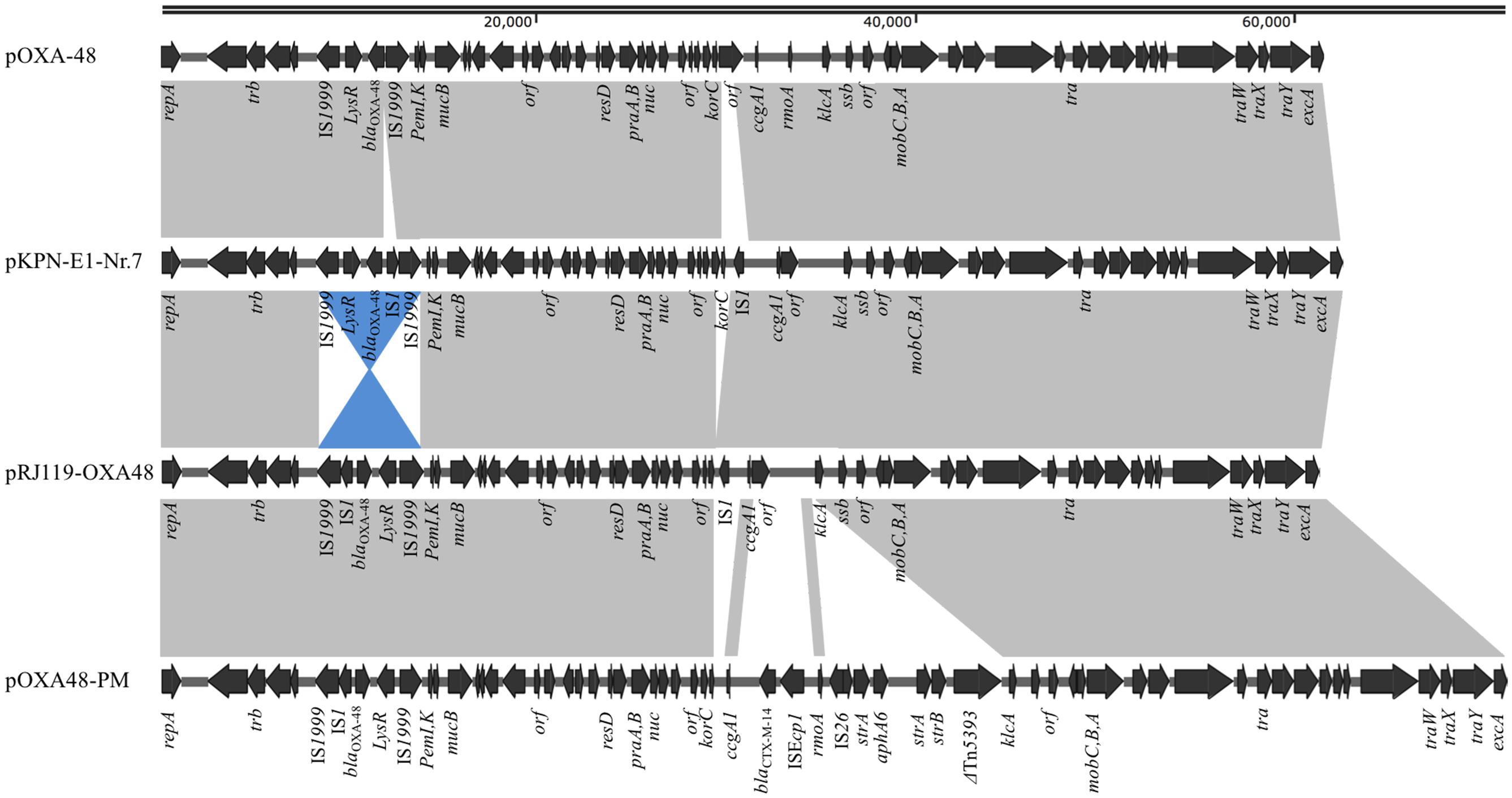
FIGURE 2. Plasmid structures of blaOXA-48-harboring IncL/M plasmids pOXA-48 (JN626286), pKPN-E1-Nr.7 (KM406491), pRJ119-OXA48 (KX636096, this study), and pOXA48-PM (KP025948). Gray shading indicates shared regions of homology, whereas blue shading indicates inversely displayed regions of homology. Open reading frames (ORFs) are indicated by arrows.
pRJ119-NDM1 had a length of 335,317 bp and displayed query coverage (44%) and overall nucleotide identity (99%) to pNDM-US, as well as query coverage (55%) and overall nucleotide identity (99%) to pKPN-c22, according to the BLAST search (Conlan et al., 2014; Hudson et al., 2014) (Figure 3). These two different plasmids had two identical IS1-IS2-ΔIS1 structures (2,381-bp). The first half of pRJ119-NDM1 was a close variant of the previously characterized pNDM-US, without the insertion sequence IS3000, and it harbored a variety of antimicrobial resistance genes including aac(6’)Ib-cr, blaNDM, and blaCMY -6, sul1. Among them, the blaNDM-1 gene cluster was arranged sequentially as ISAba14, ISAba125, blaNDM-1, bleMBL, trpF, and tat. However, it is noteworthy that the blaNDM-1 region found in pRJ119-NDM1 was different from that of pNDM-US; there was an insertion (IS10) in the blaNDM-1 gene. The second part of the pRJ119-NDM1 plasmid showed 99% identity to pKPN-c22 and harbored aac(6’)Ib-cr, aac(3)-IIa, strA, strB, blaOXA-1,blaCTX-M-15, blaTEM-1B, QnrB66, catB3, sul2, tet(A), and dfrA14.
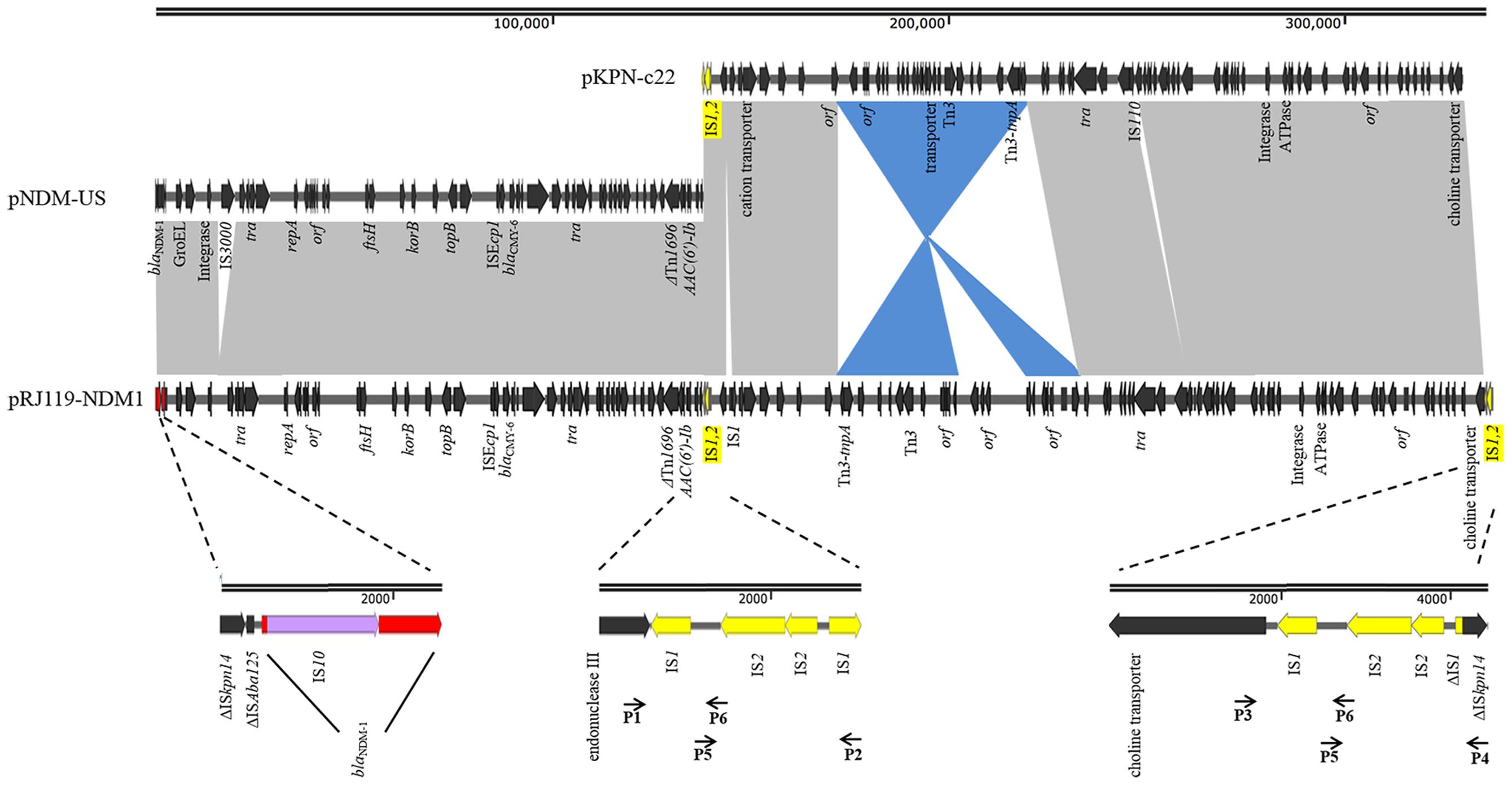
FIGURE 3. Comparisons of pRJ119-NDM1 (KX636095, this study), pNDM-US (CP006661), and pKPN-c22 (CP009879). Gray shading indicates shared regions of homology, whereas blue shading indicates inversely displayed regions of homology. ORFs are indicated by arrows. The IS1-IS2-ΔIS1 structure is indicated by yellow arrows, and four overlapping PCR amplicons are shown by lines with primer names. blaNDM-1 is indicated by red arrows, and IS10 is indicated by purple arrows in the amplification of the blaNDM-1 region.
Discussion
Carbapenemase genes often spread worldwide through clonal expansion in several pathogenic strains (Lee et al., 2016). Over 50% of NDM-producing K. pneumoniae isolates from India belong to either ST11 or ST147 (Lascols et al., 2013), and ST11 has also been associated with OXA-48-like enzymes from isolates found in Argentina, Turkey, and Spain (Oteo et al., 2013). The K. pneumoniae RJ119 isolate in our study belonged to ST307, which is not the part of the major clonal complex (Supplementary Images 2) and most prevalent in Pakistan and Korea (Habeeb et al., 2013; Park et al., 2015).
Thus far, OXA-48, in contrast to KPC-2 or NDM-1, has been rarely reported in China. The molecular epidemiology of OXA-48 in European and North Africa showed that in 92.5% of isolates, the blaOXA-48 was located on a self-conjugative IncL/M type plasmid (Potron et al., 2013). In our study, pRJ119-OXA48 showed high identity with the scaffold of other previously sequenced plasmids carrying blaOXA-48. However, we found a few differences, each discernible as a distinct DNA mobility event, between pRJ119-OXA48 and other blaOXA-48-harboring plasmids, including pOXA-48 (Poirel et al., 2012), pKPN-E1-Nr.7 (Carattoli et al., 2015), and pOXA48-PM (Chen et al., 2015) (Figure 2). Three interesting observations were made: (i) there were two IS1s in pKPN-E1-Nr.7, which were not present in pOXA-48; (ii) Tn1999.2 in pKPN-E1-Nr.7 was only the inversion of the sequence in pRJ119-OXA48, and the korC gene was not present in pRJ119-OXA48; and (iii) the sequence of Tn1999.2 was homologous to that in pOXA48-PM. Therefore, we speculated that pRJ119-OXA48 was an existence form in the evolution of blaOXA-48. Recent studies have reported the emergence of OXA-48-producing K. pneumoniae isolates and nosocomial outbreak of infections in Taiwan and Beijing, respectively (Ma et al., 2015; Guo et al., 2016). blaOXA-48 found in Taiwan was located on a 160-kb IncA/C plasmid, which was identical to the pKPOXA-48N1 plasmid found in France (Berger et al., 2013). blaOXA-48 found in Beijing was located on the IncL/M plasmid. These data from epidemiological investigations, coupled with those of our current report, show that the blaOXA-48 genes found in mainland China and Taiwan are from two different genetic backgrounds, and emphasize the need for increased surveillance of blaOXA-48 in China.
In China, blaNDM-1 was first identified in Acinetobacter baumannii. Subsequently, more blaNDM-1 genes were discovered in Enterobacteriaceae, especially in K. pneumoniae, and the gene was found carried on plasmids of various sizes (Qin et al., 2014; Qu et al., 2015; Zhang et al., 2015). In our study, blaNDM-1 was located on a 335-kb large plasmid. Based on the BLAST results, we suspected that it might be the fusion of a blaNDM-1-harboring InA/C plasmid, pNDM-US, and an IncFIB plasmid, pKPN-c22 (Figure 3). Previous studies have suggested that IncX3-type plasmids contributed to the dissemination of blaNDM-1 in China (Ho et al., 2012; Qu et al., 2015; Yang et al., 2015). However, in our study, the first half of pRJ119-NDM1 (contain blaNDM-1) was found belonging to the IncA/C group and identical to pNDM-US, the second part of pRJ119-NDM1 showed 99% identity to pKPN-c22, a plasmid detected in the USA. We speculated that the pRJ119-NDM1 plasmid might have been introduced from America after the fusion. Moreover, IS1-IS2-ΔIS1, the repeat sequence joint, might play an important role in the fusion of the two plasmids. The fusion of these two plasmids, each harboring an extraordinary number of resistance genes, might enhance antibiotic resistance. However, notably, the transconjugant RJ119-1, harboring truncated blaNDM-1, was susceptible to carbapenem, which might have been due to the insertion of IS10 into the gene. Molecular-based diagnostic methods such as PCR, microarrays, and sequence-based diagnostics are now used in clinical applications due to their celerity and accuracy and are beginning to permeate clinical diagnostic laboratories in many countries (Okeke et al., 2011). However, in the case of our present study (unexpressed blaNDM-1 due to the insertion of IS10), because the PCR results did not match with the actual clinical situation, they may mislead clinical treatment.
Conclusion
This is the first report on the coexistence of blaOXA-48 and blaNDM-1 in one K. pneumoniae clinical isolate in China. The production of OXA-48 in RJ119 contributed to the majority to its high resistance to carbapenems, whereas NDM-1 remained unexpressed, most likely due to the insertion of IS10. This study provides insight on the relationship between genetic diagnosis and clinical treatment in cases similar to the one in this study. Our findings also indicate the possibility of further spread of blaOXA-48 in China and emphasize the need for intensive surveillance and precautions.
Author Contributions
JS and YD designed experiments; LX and KZ carried out experiments; JS, LH, and XG analyzed experimental results; YC and YD analyzed sequencing data; LX and JS wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by National Natural Science Foundation of China (No. 81472010) and the Medical-engineering Cross Project of Shanghai Jiaotong University (No. YG2015MS59).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00133/full#supplementary-material
Footnotes
- ^http://www.eucast.org/clinical_breakpoints
- ^http://www.pasteur.fr/-recherche/genopole/PF8/mlst/Kpneumoniae.html
- ^https://www-is.biotoul.fr/
- ^https://cge.cbs.dtu.dk/services/ResFinder/
- ^https://cge.cbs.dtu.dk/services/PlasmidFinder/
- ^http://blast.ncbi.nlm.nih.gov/Blast.cgi
References
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Barton, B. M., Harding, G. P., and Zuccarelli, A. J. (1995). A general method for detecting and sizing large plasmids. Anal. Biochem. 226, 235–240. doi: 10.1006/abio.1995.1220
Berger, S., Alauzet, C., Aissa, N., Henard, S., Rabaud, C., Bonnet, R., et al. (2013). Characterization of a new blaOXA-48-carrying plasmid in Enterobacteriaceae. Antimicrob. Agents Chemother. 57, 4064–4067. doi: 10.1128/AAC.02550-12
Carattoli, A., Seiffert, S. N., Schwendener, S., Perreten, V., and Endimiani, A. (2015). Differentiation of IncL and IncM plasmids associated with the spread of clinically relevant antimicrobial resistance. PLoS ONE 10:e0123063. doi: 10.1371/journal.pone.0123063
Chen, L., Al Laham, N., Chavda, K. D., Mediavilla, J. R., Jacobs, M. R., Bonomo, R. A., et al. (2015). First report of an OXA-48-producing multidrug-resistant Proteus mirabilis strain from Gaza. Palestine. Antimicrob. Agents Chemother. 59, 4305–4307. doi: 10.1128/AAC.00565-15
Clinical and Laboratory Standards Institute [CLSI] (2014). Performance Standards for Antimicrobial Susceptibility Testing; 24th Informational Supplement, M100-S24. Wayne,PA: Clinical and Laboratory Standards Institute (CLSI).
Conlan, S., Thomas, P. J., Deming, C., Park, M., Lau, A. F., Dekker, J. P., et al. (2014). Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci. Transl. Med. 6:254ra126. doi: 10.1126/scitranslmed.3009845
Guo, L., An, J., Ma, Y., Ye, L., Luo, Y., Tao, C., et al. (2016). Nosocomial outbreak of OXA-48-producing Klebsiella pneumoniae in a Chinese hospital: clonal transmission of ST147 and ST383. PLoS ONE 11:e0160754. doi: 10.1371/journal.pone.0160754
Habeeb, M. A., Haque, A., Nematzadeh, S., Iversen, A., and Giske, C. G. (2013). High prevalence of 16S rRNA methylase RmtB among CTX-M extended-spectrum beta-lactamase-producing Klebsiella pneumoniae from Islamabad. Pak. Int. J. Antimicrob. Agents 41, 524–526. doi: 10.1016/j.ijantimicag.2013.02.017
Ho, P. L., Li, Z., Lo, W. U., Cheung, Y. Y., Lin, C. H., Sham, P. C., et al. (2012). Identification and characterization of a novel incompatibility group X3 plasmid carrying bla NDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg. Microbes Infect. 1:e39. doi: 10.1038/emi.2012.37
Hudson, C. M., Bent, Z. W., Meagher, R. J., and Williams, K. P. (2014). Resistance determinants and mobile genetic elements of an NDM-1-encoding Klebsiella pneumoniae strain. PLoS ONE 9:e99209. doi: 10.1371/journal.pone.0099209
Lascols, C., Peirano, G., Hackel, M., Laupland, K. B., and Pitout, J. D. (2013). Surveillance and molecular epidemiology of Klebsiella pneumoniae isolates that produce carbapenemases: first report of OXA-48-like enzymes in North America. Antimicrob. Agents Chemother. 57, 130–136. doi: 10.1128/AAC.01686-12
Lee, C. R., Lee, J. H., Park, K. S., Kim, Y. B., Jeong, B. C., and Lee, S. H. (2016). Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front. Microbiol. 7:895. doi: 10.3389/fmicb.2016.00895
Ling, M. L., Tee, Y. M., Tan, S. G., Amin, I. M., How, K. B., Tan, K. Y., et al. (2015). Risk factors for acquisition of carbapenem resistant Enterobacteriaceae in an acute tertiary care hospital in Singapore. Antimicrob. Resist. Infect. Control 4:26. doi: 10.1186/s13756-015-0066-3
Lowe, C. F., Kus, J. V., Salt, N., Callery, S., Louie, L., Khan, M. A., et al. (2013). Nosocomial transmission of New Delhi metallo-beta-lactamase-1-producing Klebsiella pneumoniae in Toronto. Can. Infect. Control Hosp. Epidemiol. 34, 49–55. doi: 10.1086/668778
Ma, L., Wang, J. T., Wu, T. L., Siu, L. K., Chuang, Y. C., Lin, J. C., et al. (2015). Emergence of OXA-48-Producing Klebsiella pneumoniae in Taiwan. PLoS ONE 10:e0139152. doi: 10.1371/journal.pone.0139152
Nordmann, P., and Poirel, L. (2014). The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin. Microbiol. Infect. 20, 821–830. doi: 10.1111/1469-0691.12719
Ocampo, A. M., Chen, L., Cienfuegos, A. V., Roncancio, G., Chavda, K. D., Kreiswirth, B. N., et al. (2016). A two-year surveillance in five colombian tertiary care hospitals reveals high frequency of Non-CG258 Clones of carbapenem-resistant Klebsiella pneumoniae with distinct clinical characteristics. Antimicrob. Agents Chemother. 60, 332–342. doi: 10.1128/AAC.01775-15
Okeke, I. N., Peeling, R. W., Goossens, H., Auckenthaler, R., Olmsted, S. S., de Lavison, J. F., et al. (2011). Diagnostics as essential tools for containing antibacterial resistance. Drug Resist. Updat. 14, 95–106. doi: 10.1016/j.drup.2011.02.002
Oteo, J., Saez, D., Bautista, V., Fernandez-Romero, S., Hernandez-Molina, J. M., Perez-Vazquez, M., et al. (2013). Carbapenemase-producing enterobacteriaceae in Spain in 2012. Antimicrob. Agents Chemother. 57, 6344–6347. doi: 10.1128/AAC.01513-13
Park, D. J., Yu, J. K., Park, K. G., and Park, Y. J. (2015). Genotypes of ciprofloxacin-resistant Klebsiella pneumoniae in Korea and their characteristics according to the genetic lineages. Microb. Drug Resist. 21, 622–630. doi: 10.1089/mdr.2015.0001
Poirel, L., Bonnin, R. A., and Nordmann, P. (2012). Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob. Agents Chemother. 56, 559–562. doi: 10.1128/AAC.05289-11
Poirel, L., Heritier, C., Tolun, V., and Nordmann, P. (2003). Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48, 15–22. doi: 10.1128/aac.48.1.15-22.2004
Potron, A., Poirel, L., Rondinaud, E., and Nordmann, P. (2013). Intercontinental spread of OXA-48 beta-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro. Surveill. 18:20549. doi: 10.2807/1560-7917.ES2013.18.31.20549
Qin, S., Fu, Y., Zhang, Q., Qi, H., Wen, J. G., Xu, H., et al. (2014). High incidence and endemic spread of NDM-1-positive Enterobacteriaceae in Henan Province. China. Antimicrob. Agents Chemother. 58, 4275–4282. doi: 10.1128/AAC.02813-13
Qu, H., Wang, X., Ni, Y., Liu, J., Tan, R., Huang, J., et al. (2015). NDM-1-producing Enterobacteriaceae in a teaching hospital in Shanghai, China: incX3-type plasmids may contribute to the dissemination of blaNDM-1. Int. J. Infect. Dis. 34, 8–13. doi: 10.1016/j.ijid.2015.02.020
Queenan, A. M., and Bush, K. (2007). Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20, 440–458. doi: 10.1128/CMR.00001-07
Rasheed, J. K., Kitchel, B., Zhu, W., Anderson, K. F., Clark, N. C., Ferraro, M. J., et al. (2013). New Delhi metallo-beta-lactamase-producing Enterobacteriaceae. U. S. Emerg. Infect. Dis. 19, 870–878. doi: 10.3201/eid1906.121515
Spyropoulou, A., Bartzavali, C., Vamvakopoulou, S., Marangos, M., Anastassiou, E. D., Spiliopoulou, I., et al. (2016). The first NDM metallo-beta-lactamase producing Klebsiella pneumoniae isolate in a University Hospital of Southwestern Greece. J. Chemother. 28, 350–351. doi: 10.1179/1973947815Y.0000000003
Yang, Q., Fang, L., Fu, Y., Du, X., Shen, Y., and Yu, Y. (2015). Dissemination of NDM-1-producing Enterobacteriaceae mediated by the IncX3-type plasmid. PLoS ONE 10:e0129454. doi: 10.1371/journal.pone.0129454
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Keywords: carbapenemase, OXA-48, NDM-1, coexistence, plasmid
Citation: Xie L, Dou Y, Zhou K, Chen Y, Han L, Guo X and Sun J (2017) Coexistence of blaOXA-48 and Truncated blaNDM-1 on Different Plasmids in a Klebsiella pneumoniae Isolate in China. Front. Microbiol. 8:133. doi: 10.3389/fmicb.2017.00133
Received: 04 November 2016; Accepted: 18 January 2017;
Published: 02 February 2017.
Edited by:
Daniela Ceccarelli, Wageningen Bioveterinary Research, NetherlandsReviewed by:
Benjamin Andrew Evans, University of East Anglia, UKThandavarayan Ramamurthy, Translational Health Science and Technology Institute, India
Copyright © 2017 Xie, Dou, Zhou, Chen, Han, Guo and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingyong Sun, MTM2NzE1Nzg4OTlAMTI2LmNvbQ==
†Joint first authors: these authors have contributed equally to this work.
 Lianyan Xie1†
Lianyan Xie1† Jingyong Sun
Jingyong Sun