- 1Department of Biology, Hamilton College, Clinton, NY, USA
- 2Department of Biology, Indiana University, Bloomington, IN, USA
- 3National Center for Genome Analysis Support, Indiana University, Bloomington, IN, USA
- 4Department of Aquatic Biosciences, National Chyai University, Chyai City, Taiwan
- 5Departamento de Aquicultura, Centro de Ciências Agrárias, Universidade Federal de Santa Catarina, Florianópolis, Brazil
- 6Department of Microbiology and Immunology, College of Veterinary Medicine, Cornell University, Ithaca, NY, USA
- 7Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan
Although the presence of endosymbiotic rickettsial bacteria, specifically Candidatus Megaira, has been reported in diverse habitats and a wide range of eukaryotic hosts, it remains unclear how broadly Ca. Megaira are distributed in a single host species. In this study we seek to address whether Ca. Megaira are present in most, if not all isolates, of the parasitic ciliate Ichthyophthirius multifiliis. Conserved regions of bacterial 16S rRNA genes were either PCR amplified, or assembled from deep sequencing data, from 18 isolates/populations of I. multifiliis sampled worldwide (Brazil, Taiwan, and USA). We found that rickettsial rRNA sequences belonging to three out of four Ca. Megaira subclades could be consistently detected in all I. multifiliis samples. I. multifiliis collected from local fish farms tend to be inhabited by the same subclade of Ca. Megaira, whereas those derived from pet fish are often inhabited by more than one subclade of Ca. Megaira. Distributions of Ca. Megaira in I. multifiliis thus better reflect the travel history, but not the phylogeny, of I. multifiliis. In summary, our results suggest that I. multifiliis may be dependent on this endosymbiotic relationship, and the association between Ca. Megaira and I. multifiliis is more diverse than previously thought.
Introduction
Rickettsial bacteria (Order Rickettsiales), members of alphaproteobacteria, are well-known as the causative agents for insect-borne human diseases such as typhus, scrub typhus, and Rocky Mountain spotted fever (Walker and Ismail, 2008). These bacteria are gram-negative, obligate intracellular organisms, and their presence was once thought to be limited to animals, particularly insects and vertebrates (Raoult and Roux, 1997). Recently, surveys of environmental samples revealed that in addition to the pathogenic rickettsia, rickettsia-like bacteria could be found as endosymbionts in a variety of species and from different habitats. However, the functions of these rickettsial endosymbionts in their hosts remain unclear.
Results derived from phylogenetic analyses using 16S rRNA sequences show that rickettsia-like endosymbiotic bacteria can be classified into two monophyletic groups. The first group, the recently described Candidatus Midichloriaceae (Vannini et al., 2005; Epis et al., 2008; Gillespie et al., 2012; Mariconti et al., 2012; Williams-Newkirk et al., 2012; Driscoll et al., 2013; Montagna et al., 2013), is placed as a sister clade to Anaplasmataceae, and comprises endosymbionts found in insects (Epis et al., 2008; Hornok et al., 2008; Erickson et al., 2009; Richard et al., 2009; Matsuura et al., 2012), amoebas (Fritsche et al., 1999), ciliates (Vannini et al., 2010; Boscaro et al., 2013a,b), placozoa (Driscoll et al., 2013), and cnidarians (Fraune and Bosch, 2007; Sunagawa et al., 2009). Furthermore, members of Midichloriaceae have also been detected in fish suffering from strawberry disease (Lloyd et al., 2008, 2011) and red mark syndrome (Metselaar et al., 2010; Cafiso et al., 2016), and in humans and other mammals after tick bites (Mediannikov et al., 2004; Mariconti et al., 2012; Matsuura et al., 2012; Bazzocchi et al., 2013). However, there has been no direct evidence suggesting that these Midichloriaceae are etiological agents of disease.
The other group of rickettsia-like endosymbiotic bacteria, Candidatus Megaira, forms a sister clade to the genus Rickettsia (family Rickettsiaceae) (Schrallhammer et al., 2013). Based on SSU rRNA sequences, Schrallhammer et al. (2013) further classified Ca. Megaira into three subclades. Members of the subclade Ca. Megaira polyxenophila were identified in both marine and freshwater ciliates (Vannini et al., 2005), in green algae (Kawafune et al., 2012), in lake water from the US (Percent et al., 2008) and China, in subsurface water from South Africa, and in aquaria in Greece (Vlahos et al., 2013). The other two subclades, Ca. Megaira B and C, contain species found in diverse hosts and habitats including: ciliate Ichthyophthirius multifiliis (Sun et al., 2009), cnidarians (Fraune and Bosch, 2007; Sunagawa et al., 2009), siphonous green algae (Hollants et al., 2013), lake water from the US (Percent et al., 2008), water from a lagoon in North Pacific (Galand et al., 2012), and a wastewater treatment plant in France (Chouari et al., 2010). There have been no reports that these bacteria are pathogenic, and the growth and reproduction of ciliate Diophrys were not affected when inhabited by Ca. Megaira (Vannini et al., 2003).
While it seems that Ca. Megaira are widely spread, it is not clear how ubiquitous they are. Furthermore, how universal these bacteria are in isolates/populations of particular host species is less well-studied. Research carried out by Kawafune et al. (2012) showed that Ca. Megaira were present only in 1 of 12 isolates of four unicellular green algal species (Cateria), and in one of nine isolates of multicellular green algae Volvox carteri (Kawafune et al., 2014), suggesting that Ca. Megaira might not be ubiquitously found in all isolates of one species. However, despite the works on non-phagotrophic green alga, to our knowledge there have been no other research systematically examining the distribution of Ca. Megaira in one single species, particularly in phagotrophic ones.
The parasitic ciliate I. multifiliis is the etiological agent for the ‘white spot disease’ in freshwater fish (Matthews, 2005; Dickerson, 2011). I. multifiliis contains an oral apparatus (Dickerson, 2006), and are apparently phagotrophic (Lobo-da-Dunha and Azevedo, 1993). Moreover, endosymbiotic Sphingobacteria and rickettsial alphaproteobacteria were detected in two I. multifiliis isolates isolated from the state of Georgia, USA (Sun et al., 2009; Coyne et al., 2011). The rickettsial alphaproteobacteria were later identified as members of the Ca. Megaira subclade C (Schrallhammer et al., 2013). We are therefore intrigued to determine if Ca. Megaira can be detected in most, if not all, isolates of the phagotrophic I. multifiliis. Furthermore, the phylogenetic relationships among different isolates of I. multifiliis can now be well-resolved by using mitochondrial sequences (MacColl et al., 2015). The phylogenies of Ca. Megaira, if they are present in most isolates of I. multifiliis, can then be compared to that of I. multifiliis to help deduce transmission routes of Ca. Megaira.
In this study we show that Ca. Megaira can be detected in 18 isolates of I. multifiliis, collected from Brazil, Taiwan, and the US. Ca. Megaira can now be classified into four subclades based on their rRNA sequences, and at least three of the subclades are capable of inhabiting I. multifiliis. The significance of the ubiquitous distribution of Ca. Megaira in I. multifiliis, and the transmission routes of Ca. Megaira, are discussed.
Materials and Methods
I. multifiliis and DNA Isolation
Ichthyophthirius multifiliis was collected from infected fish in the US, Taiwan, and Brazil, and each isolate likely derived from a distinct population. This collection represents more than 20 years of effort—on many researchers’ part—in the collection and storage of samples from fish farms and pet stores across the world (Table 1). Isolates were named with a letter(s) denoting the state or the country of its origin and a sequential number in the order they were discovered (Table 1). Among the 18 I. multifiliis isolates 9 have been previously reported (Lin et al., 1996; MacColl et al., 2015), but only endosymbionts in the G5 isolate have been studied (Sun et al., 2009). Isolates collected in the US were at one point in time cultivated in the lab following previously established protocols (Noe and Dickerson, 1995), and except for G15 and NY3, all other US isolates were clonal lines. I. multifiliis trophont cells were collected from infected fish either by gently rubbing the skin of fish (Cassidy-Hanley et al., 2011), or by using saline shock (Schmahl et al., 1989). DNA was extracted either following protocols described elsewhere (Cassidy-Hanley et al., 2011; MacColl et al., 2015), or using the Qiagen DNeasy Blood & Tissue Kit (Redwood City, CA, USA) following manufacturer’s protocols. The protocol of using fish (to pass I. multifiliis) was approved by the Institutional Animal Care and Use Committee of Cornell University (protocol number 1996-0083).
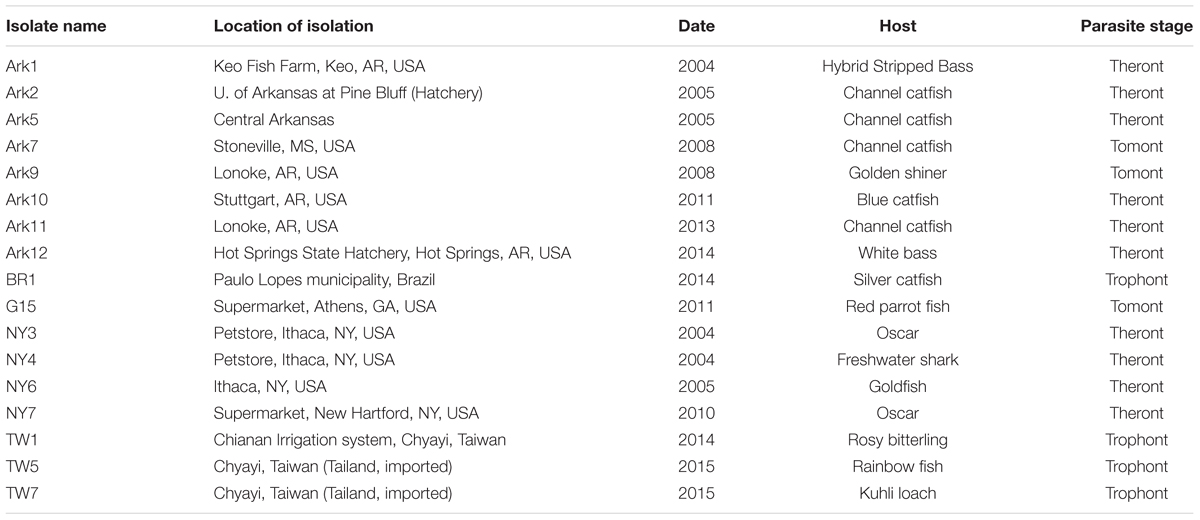
TABLE 1. Histories and characteristics of the 17 Ichthyophthirius multifiliis isolates used in this study.
Amplification, Cloning, and Sequencing
Endosymbiotic bacterial 16S rRNA sequences were either PCR amplified, or derived from whole genome assemblies. PCR mixtures contained 1X GoTaq Green Master Mix (Promega, Madison, WI, USA), each primer at 0.2 μM, and DNA (5–50 ng) in a final volume of 50 μL. A reagent negative control was always included in every PCR experiment. PCR primers were either the bacterial SSU-specific set described elsewhere (Weisburg et al., 1991) (Escherichia coli rRNA positions 8–1,509, GenBank: J01859.1; Forward 5′ AGA GTT TGA TYM TGG CTC AG 3′, Reverse 5′ GGH TAC CTT GTT ACG ACT 3′), or an in-house set more specific against Rickettsia 16S rRNA (approximate E. coli rRNA positions 45–1,345; Forward 5′ TGC TTA ACA CAT GCA AGT CGA ACG A 3′, Reverse 5′ TAG TGA TTC CGA CTT CAT GCT CT 3′). The following cycling conditions were followed: initial denaturation at 94°C for 2 min, denaturation at 94°C for 30 s, annealing at 46°C for 30 s, extension at 72°C for 1.5 min (30 cycles), with a final extension of 68°C for 5 min. Amplified PCR products were cloned into pGEM-T Easy Vector (Promega), and sequences were determined by using Sanger’s sequencing method (Genewiz, South Plainfield, NJ, USA). For each isolate at least 10 positive clones were screened. rRNA sequences are deposited in NCBI GenBank (accession KT851755-851878).
We followed protocols described elsewhere to amplify and determine I. multifiliis mitochondrial cox-1 and nad1_b sequences (MacColl et al., 2015). Briefly, 0.2 μM of each primer (cox-1 Forward: 5′ TATCAGGTGCTGCATTAGCTACT 3′, Reverse: 5′ TAAACCTAAAGTAGATGAAGTGTGAAG 3′; nad1_b Forward: 5′ CTATGACCATAAATCGGAGAAAGTT 3′, Reverse: 5′ GAGTTTATATCATGGAAGCTAACAG 3′), and 2–20 ng of I. multifiliis DNA were added to a PCR mixture containing 1X GoTaq in a final volume of 50 μL. Cycling conditions were: 95°C 2 min followed by 35 cycles of 95°C 30 s, 50°C 1 min, 72°C 1.5 min, with a final extension of 72°C for 5 min. Cox-1 and nad1_b sequences are also deposited in NCBI GenBank (KT783590–KT783607).
Whole genomes of isolates G15, Ark11, and Ark12 were sequenced using Illumina technologies, which generated paired reads with >200X coverages of I. multifiliis genome (MacColl et al., 2015). Raw reads were first corrected using SOAPec v2.01 (Luo et al., 2012), and corrected reads specific to bacterial 16S rRNA were baited using MIRA v4.9.3 (Chevreux et al., 2004) against the bacterial 16S rRNA database v119 downloaded from The SILVA ribosomal RNA database (Quast et al., 2013; Yilmaz et al., 2014) before being assembled by MIRA into contigs.
Phylogenetic Analyses
DNA sequences were first aligned using T-Coffee (Notredame et al., 2000), and alignments were further manually corrected in Jalview (Waterhouse et al., 2009) and/or BioEdit (Hall, 1999). For phylogenetic tree reconstructions protocols described elsewhere were followed (MacColl et al., 2015). Briefly, maximum likelihood (ML) trees were constructed with models predetermined by jModeltest (rRNA sequences: GTR+G; concatenated cox-1 and nad1_b: TIM1+I) (Darriba et al., 2012) and 1,000 bootstrapping replicates (Guindon and Gascuel, 2003). For Bayesian analyses (MB), MrBayes was used with the setting of: GTR+G model, two independent runs (each with three heated chains and one cold chain), 2,500,000 MCMC steps, and a sampling frequency of 1,000 (Ronquist et al., 2012). By the end of MCMC the standard deviation of split frequencies reached 0.0068. A burn-in of 25%, or 625, was used to generate both parameters and the consensus tree.
Results
Phylogeny of Ca. Megaira
We identified more than 50 unique rickettsia-like 16S rRNA sequences from 17 isolates of I. multifiliis, and at least one sequence was detected in each of the isolates. The sequence of the rickettsial endosymbiotic bacterium found in the 18th isolate, G5, was published in an earlier report (Sun et al., 2009). If there were no deviations between two sequences beyond three or more positions, or 0.23% difference among 1,302 positions including gaps, only one was chosen for subsequent analyses. After such filtration 42 sequences were retained.
Phylogenetic analyses were conducted to assess the relationships among the newly obtained sequences and Ca. Megaira 16S rRNA sequences reported in other studies, including those derived from endosymbiotic bacteria found in ciliates (Vannini et al., 2005; Schrallhammer et al., 2013), hydra (Fraune and Bosch, 2007), corals (Sunagawa et al., 2009), siphonous green algae (Hollants et al., 2013), and from environmental samples (lakes (Percent et al., 2008), stratified lagoon (Galand et al., 2012), basins of a wastewater treatment plant (Chouari et al., 2010), aquariums (Vlahos et al., 2013). In addition, sequences derived from representative species in the families of Rickettsiaceae, Midichloriacea, and Anaplasmataceae, from two non-Rickettsiales alphaproteobacteria were included as internal reference points. Candidatus Nebulobacter yamunensis, an endosymbiotic gammaproteobacteria found in the ciliate Euplotes aediculatus (Boscaro et al., 2012), was used as the outgroup (Figure 1).
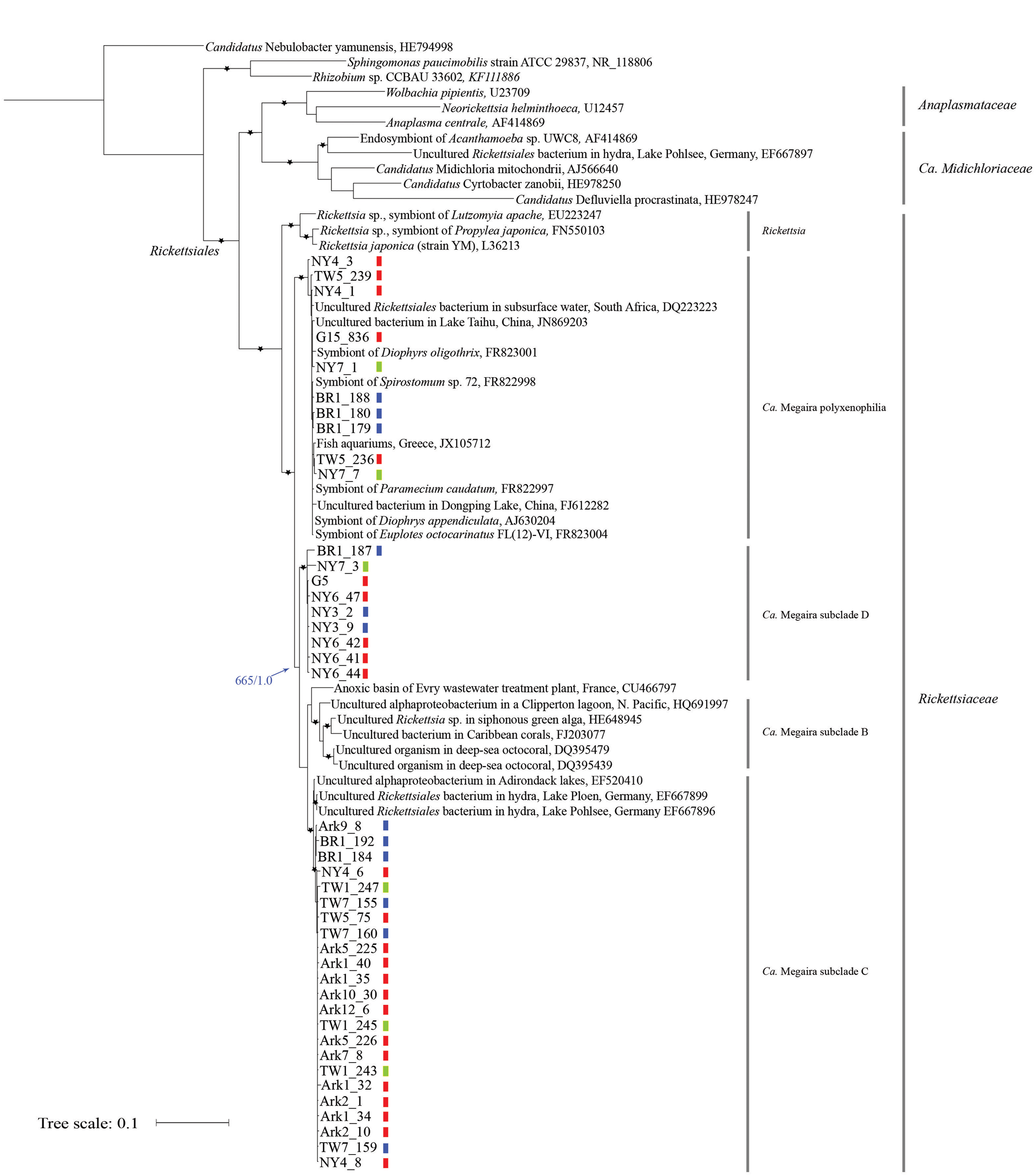
FIGURE 1. Maximum likelihood (ML) tree derived from 16S rRNA sequences of Ca. Megaira and representative Alphaproteobacteria. Asterisks (∗) denote branches with >750 bootstrap values and >0.75 posterior probability in Bayesian (MB) analyses. The blue arrow and numbers indicate bootstrap support from ML and posterior probability from MB analyses on the branch separating Ca. Megaira subclades B, C, and D from subclade polyxenophila. The genus and family names in the order Rickettisales are provided on the right. Short vertical color bars indicate the three phylogroups of Ichthyophthirius multifiliis (see Figure 2). Ark11 share identical Ca. Megaira 16S rRNA to that of Ark12_6 and is not shown. Scale bar represents 0.1 nucleotide substitutions per site.
Both Bayesian (MB) and ML trees place all newly reported rickettsia-like 16S rRNA sequences and Ca. Megaira sequences in a monophyletic group next to the genus Rickettsia, a result consistent with a previous observation (Schrallhammer et al., 2013) (Figure 1). From this point we will follow the nomenclature system established by Schrallhammer et al. (2013) and collectively regard all endosymbiotic bacteria in this monophyletic group as Ca. Megaira.
Based on our phylogenetic analyses, Ca. Megaira can be further divided into four well-supported subclades, and at least three of the subclades are capable of inhabiting I. multifiliis (Figure 1). The early diverging position and the grouping of the subclade Ca. Megaira polyxenophilia are consistent with previous findings (Schrallhammer et al., 2013). However, Ca. Megaira polyxenophilia was not known to inhabit I. multifiliis and here we show ample examples (Figure 1).
The remaining three subclades might have descended from a common ancestor after the split from Ca. Megaira polyxenophilia (blue arrow in Figure 1), but the bootstrapping support of the branching point from ML analyses is only moderate. Among these three subclades, the grouping of the subclade Ca. Megaira B is consistent with findings reported in other studies (Schrallhammer et al., 2013), and this subclade has been the only one that has not been detected in I. multifiliis. Ca. Megaira subclade C includes endosymbionts found in hydra, in a US lake sample, and in I. multifiliis (Figure 1). It should be noted, however, that while the rickettsial endosymbionts in the G5 isolate of I. multifiliis were first classified as members in Ca. Megaira subclade C (Schrallhammer et al., 2013), our results show that these and a few other rickettsial endosymbionts detected in other I. multifiliis isolates should be classified as a new Ca. Megaira subclade D (Figure 1). Moreover, to date rickettsial endosymbionts in Ca. Megaira subclade D have only been detected in I. multifiliis. A nucleotide blast search using the Ca. Megaira 16S rRNA sequence found in G5 against the NCBI nr/nt database failed to identify sequences—including those derived from environmental samples—with higher similarities than those found in Ca. Megaira subclade C (data not shown).
Phylogeny of I. multifiliis
We then determined the phylogeny of the 18 host isolates of I. multifiliis using concatenated mitochondrial cox-1 and nad1_b sequences. In a previous study based on nine isolates I. multifiliis could be classified into two distinct groups with the possibility of a third group (MacColl et al., 2015). With more isolates included in this study it is clear that the 18 isolates of I. multifiliis belong to three distinct groups (Figure 2).
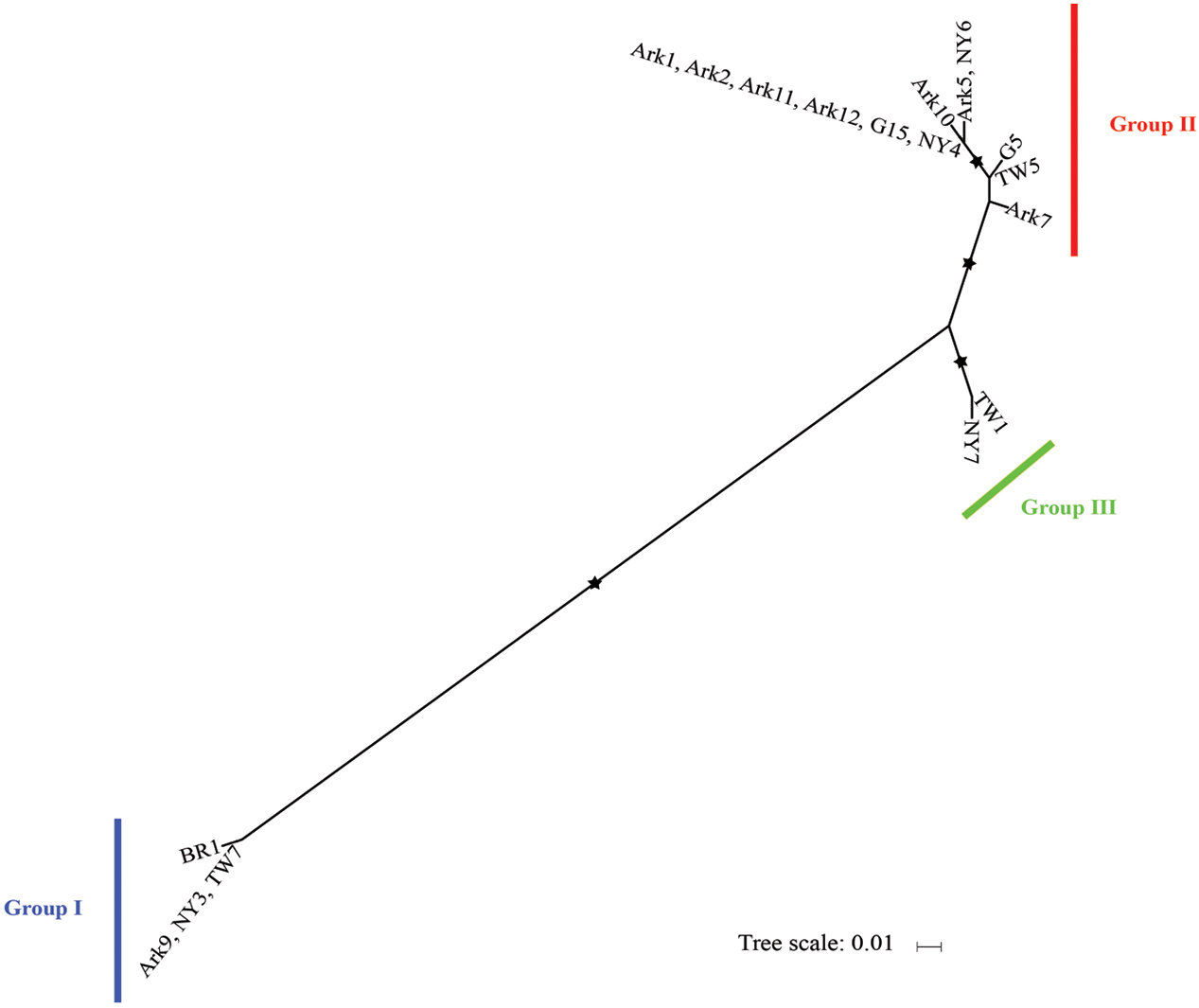
FIGURE 2. Maximum likelihood tree derived from concatenated mitochondrial cox-I and nad1_b sequences of 18 I. mutifiliis isolates. Asterisks (∗) denote branches with >750 bootstrap values. Color bars indicate the three distinct phylogroups. Scale bar represents 0.01 nucleotide substitutions per site.
Group II contains seven out of eight isolates collected from Arkansas, demonstrating a pattern of local, repetitive infection of fish stocks. This pattern matches the isolate histories: most of these isolates were collected from local hatchery farms (Table 1). In contrast, for isolates obtained from pet stores (Table 1) where the sources of I. multifiliis are expected to be variable, such a pattern does not exist. For instance, among the four isolates obtained in New York State, NY3, NY4, and NY7 were obtained from three different pet stores and belong to Groups I, II, and III, respectively (Figure 2). A similar pattern holds for isolates TW5 (Group II) and TW7 (Group I), which were imported from vendors in Thailand to aquarium shops in Taiwan (Figure 2).
Distributions of Ca. Megaira in I. multifiliis
We next mapped groupings of I. multifiliis to the phylogenetic tree of Ca. Megaira (Figure 1, short vertical color bars). While in most I. multifiliis isolates we could only detect Ca. Megaira endosymbionts from one subclade, a few isolates, particularly those derived from pet store aquaria—NY4, NY7, and TW5—harbored endosymbionts from two subclades. BR1 was inhabited with endosymbionts from three Ca. Megaira subclades. The detailed history of this isolate is, however, not clear.
Transmissions of Ca. Megaira
If Ca. Megaira are transmitted solely vertically, i.e., to sister cells through asexual division, we should expect congruent evolution between Ca. Megaira and I. multifiliis. The distributions of Ca. Megaira in I. multifiliis isolates show that a congruent pattern does not exist (Figure 1). The three Ca. Megaira subclades that are found capable of inhabiting I. multifiliis can be detected in all three groups of I. multifiliis. This observation would argue strongly against solely vertical transmission.
Furthermore, we found that all isolated samples from Arkansas, which fell into two distinct groups, were inhabited with Ca. Megaira subclade C, suggesting a strong influence of the local environment on the acquisition of endosymbiont strains.
The Presence of Sphingobacteria in I. multifiliis
Since Sphingobacteria were also detected in two isolates of I. multifiliis collected from Georgia, USA (Sun et al., 2009), we set out to determine whether Sphingobacteria, like Ca. Megaira, were present in all 18 isolates of I. multifiliis. In three isolates, G15, Ark11, and Ark12, where genomic sequences were determined using next generation sequencing technologies with high coverages, assembled contigs with high similarity (>90%) to Sphingobacteria 16S rRNA sequence (GQ870456.1) were not detected (data not shown). We also failed to detect Sphingobacteria or Sphingobacteria-like 16S rRNA sequences in cloned PCR products. These results suggest that Sphingobacteria is not present in all I. multifiliis isolates.
Discussion
In this study we surveyed the presence and distribution of rickettsial endosymbionts Ca. Megaira in 18 isolates of the parasitic ciliate I. multifiliis. In contrast to findings reported in non-phagotrophic green alga, where only a few isolates harbored this endosymbiont (Kawafune et al., 2012, 2014), Ca. Megaira could be detected in all 18 isolates of I. multifiliis, collected from North and South America, and Southeast Asia. Like many other ciliates, I. multifiliis has an oral apparatus and is presumed to be phagocytic when feeding on fish (Ewing et al., 1985; Dickerson, 2006). It is therefore possible that I. multifiliis acquires Ca. Megaira through phagocytosis and Ca. Megaira subsequently escapes from phagolysosomes using a mechanism similar to that used by pathogenic rickettsiae (Teysseire et al., 1995; Whitworth et al., 2005). On the other hand, ciliates are also equipped with a clathrin-mediated endocytosis pathway (Ramoino et al., 2001; Elde et al., 2005). Because rickettsiae invade mammalian cells in a clathrin-dependent manner (Chan et al., 2009), it is possible that Ca. Megaira enters I. multifiliis through clathrin-mediated endocytosis. Further experiments are needed to elucidate which mechanism(s) Ca. Megaira use to enter their hosts.
The ubiquitous presence of Ca. Megaira in I. multifiliis prompts us to consider whether the bacteria and the ciliate host have formed a dependent relationship, which may well be an example of hyperparasitism between bacteria (hyperparasite) and protozoan (pathogen) (Parratt and Laine, 2016). I. multifiliis shows a significant reduction of its somatic genome size (∼50 Mb) compared to two other free-living ciliate species in the same class (Oligohymenophorea) – Paramecium tetraurelia (72 Mb) (Aury et al., 2006) and Tetrahymena thermophila (104 Mb) (Eisen et al., 2006). Although parasitic protozoans tend to have smaller genomes (Ersfeld, 2003; Hertz-Fowler et al., 2005), I. multifiliis might have undergone further genome reduction due to the formation of a mutualistic symbiotic relationships with Ca. Megaira.
Prokaryotic endosymbionts have been identified in more than 200 ciliate species (Fokin, 2004; Schweikert et al., 2013), and these endosymbionts have been shown to provide hosts with nutritional support (Vannini et al., 2003), defense (Beale et al., 1969; Preer et al., 1972; Quackenbush and Burbach, 1983), and/or access to better environments (Finlay and Fenchel, 1989; Fenchel and Finlay, 1991a,b). It is unclear what roles Ca. Megaira may play in I. multifiliis biology. Lobo-da-Cunha and Azevedo showed that endosymbiotic bacteria in I. multifiliis, likely Ca. Megaira, were surrounded by glycogen in the cytoplasm (Lobo-da-Cunha and Azevedo, 1988). While this raises the interesting possibility that Ca. Megaira utilizes glycogen/glucoses derived from I. multifiliis, it does not immediately suggest that Ca. Megaira provides anything to its I. multifiliis host. Further details on metabolic dependencies between Ca. Megaira and I. multifiliis may be revealed when genomic sequences of Ca. Megaira become available.
With the addition of Ca. Megaira 16S rRNA sequences derived from I. multifiliis it is now clear that Ca. Megaira can be further divided into at least four subclades. Three of these subclades (polyxenophila, C and D) are capable of inhabiting I. multifiliis, while subclade B, which is found primarily in seawater samples, is not (Figure 1). The geographic isolation and/or changes in host tropism may contribute to this phenomenon (I. multifiliis only infect freshwater fish). Along the same lines, the fact that Ca. Megaira subclade D appears to be present only in I. multifiliis suggests a specific tropism for these bacteria, although under-sampling may explain this as well.
By comparing the phylogenies of I. multifiliis and Ca. Megaira, we concluded that transmission of Ca. Megaira is not solely vertical. Isolates of I. multifiliis in the same phylogroup may be inhabited with different subclades of Ca. Megaira. Moreover, local Ca. Megaira strains seem to play a more important role in determining which bacteria are present in I. multifiliis. The fact that all Arkansas isolates of I. multifiliis, regardless their phylogroups, were inhabited with Ca. Megaira subclade C, and not with other subclades, supports this idea. In this case, local bacteria may transmit horizontally to I. multifiliis and outcompete Ca. Megaira already inhabiting I. multifiliis. The presence of two subclades of Ca. Megaira in some clonally derived parasite lines (for example, NY4 and NY7), may reflect complex travel histories (exposures to different Ca. Megaira in different locations) and balanced competition between endosymbiont groups. The host/bacterial nature histories may always be more complicated than simple scenario we propose. Multiple gains/losses, in combination with horizontal/vertical transmissions, could result in what we observe today. More molecular sequences from Ca. Megaira will allow us to make better phylogenetic inferences not only between Ca. Megaira subclades, but also within subclades, with which we will be better able to determine the transmission routes of Ca. Megaira.
Finally, host range and tropisms of Ca. Megaira remain to be further investigated. Ca. Megaira polyxenophilia is capable of inhabiting at least six ciliate species encompassing three distinct classes: Hypotrichea (Diophrys oligothrix. Diophrys appendiculata, and Euplotes octocarinatus), Heterotrichea (Spirostomum sp.), and Oligohymenophorea (Paramecium caudatum and I. multifiliis). Do these observations imply that Ca. Megaira (polyxenophilia) may be able to inhabit most, if not all, species in these three classes? Moreover, could Ca. Megaira subclade C and subclade D inhabit ciliate species other than I. multifiliis? In pathogenic rickettsiae it has been shown that several surface proteins, e.g., rOmpB (Sca5), Sca1, and Sca2, play significant roles in host cell adherence and invasion, and may be involved in determining host tropisms (Cardwell and Martinez, 2009; Chan et al., 2009; Riley et al., 2010; Uchiyama, 2012). An extensive survey of variations of these protein sequences from different Ca. Megaira subclades may help us gain insight on the host specificities of Ca. Megaira.
Ethics Statement
The protocol of using fish (to pass I. multifiliis) was approved by the Institutional Animal Care and Use Committee of Cornell University (protocol number 1996-0083).
Author Contributions
KZ, TD, HE, C-HT, M-CY, DC-H, TC, and W-JC conceived and designed the study. KZ, HE, C-HT, MM, and DK conducted experiments. KZ, TD, C-HT, TC, and W-JC analyzed the resulting data. KZ, TD, and W-JC wrote the manuscript. KZ, TD, M-CY, DC-H, TC, and W-JC revised the manuscript.
Funding
This work was supported by the Wertimer Endowed Chair Fund to W-JC; Casstevens Family Fund to KZ; National Science Foundation (MRI-0959297); and Research Corporation Cottrell College Award (20976).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Hamilton students Elisabeth MacColl for coordinating sample collection, and Patricia Taik for assistance on a few cloning experiments; Librarian Glynis Asu for research and instructional support; Ms. Anlee Hsiao for testing experimental procedures; Dr. Harry Dickerson from University of Georgia for insightful comments.
References
Aury, J. M., Jaillon, O., Duret, L., Noel, B., Jubin, C., Porcel, B. M., et al. (2006). Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444, 171–178. doi: 10.1038/nature05230
Bazzocchi, C., Mariconti, M., Sassera, D., Rinaldi, L., Martin, E., Cringoli, G., et al. (2013). Molecular and serological evidence for the circulation of the tick symbiont Midichloria (Rickettsiales: Midichloriaceae) in different mammalian species. Parasit. Vectors 6, 350. doi: 10.1186/1756-3305-6-350
Beale, G. N., Jurand, A., and Preer, J. R. (1969). The classes of endosymbiont of Paramecium aurelia. J. Cell. Sci. 5, 65–91.
Boscaro, V., Petroni, G., Ristori, A., Verni, F., and Vannini, C. (2013a). “Candidatus Defluviella procrastinata” and “Candidatus Cyrtobacter zanobii”, two novel ciliate endosymbionts belonging to the “Midichloria clade”. Microb. Ecol. 65, 302–310. doi: 10.1007/s00248-012-0170-3
Boscaro, V., Schrallhammer, M., Benken, K. A., Krenek, S., Szokoli, F., Berendonk, T. U., et al. (2013b). Rediscovering the genus Lyticum, multiflagellated symbionts of the order Rickettsiales. Sci. Rep. 3, 3305. doi: 10.1038/srep03305
Boscaro, V., Vannini, C., Fokin, S. I., Verni, F., and Petroni, G. (2012). Characterization of “Candidatus nebulobacter yamunensis” from the cytoplasm of euplotes aediculatus (Ciliophora, Spirotrichea) and emended description of the family Francisellaceae. Syst. Appl. Microbiol. 35, 432–440. doi: 10.1016/j.syapm.2012.07.003
Cafiso, A., Sassera, D., Serra, V., Bandi, C., McCarthy, U., and Bazzocchi, C. (2016). Molecular evidence for a bacterium of the family Midichloriaceae (order Rickettsiales) in skin and organs of the rainbow trout Oncorhynchus mykiss (Walbaum) affected by red mark syndrome. J. Fish Dis. 39, 497–501. doi: 10.1111/jfd.12371
Cardwell, M. M., and Martinez, J. J. (2009). The Sca2 autotransporter protein from Rickettsia conorii is sufficient to mediate adherence to and invasion of cultured mammalian cells. Infect. Immun. 77, 5272–5280. doi: 10.1128/IAI.00201-09
Cassidy-Hanley, D., Cordonnier-Pratt, M., Pratt, L. H., Devine, C., Mozammal Hossain, M., Dickerson, H. W., et al. (2011). Transcriptional profiling of stage specific gene expression in the parasitic ciliate Ichthyophthirius multifiliis. Mol. Biochem. Parasitol. 178, 29–39. doi: 10.1016/j.molbiopara.2011.04.004
Chan, Y. G., Cardwell, M. M., Hermanas, T. M., Uchiyama, T., and Martinez, J. J. (2009). Rickettsial outer-membrane protein B (rOmpB) mediates bacterial invasion through Ku70 in an actin, c-Cbl, clathrin and caveolin 2-dependent manner. Cell. Microbiol. 11, 629–644. doi: 10.1111/j.1462-5822.2008.01279.x
Chevreux, B., Pfisterer, T., Drescher, B., Driesel, A. J., Muller, W. E., Wetter, T., et al. (2004). Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14, 1147–1159. doi: 10.1101/gr.1917404
Chouari, R., Le Paslier, D., Daegelen, P., Dauga, C., Weissenbach, J., and Sghir, A. (2010). Molecular analyses of the microbial community composition of an anoxic basin of a municipal wastewater treatment plant reveal a novel lineage of proteobacteria. Microb. Ecol. 60, 272–281. doi: 10.1007/s00248-009-9632-7
Coyne, R. S., Hannick, L., Shanmugam, D., Hostetler, J. B., Brami, D., Joardar, V. S., et al. (2011). Comparative genomics of the pathogenic ciliate Ichthyophthirius multifiliis, its free-living relatives and a host species provide insights into adoption of a parasitic lifestyle and prospects for disease control. Genome Biol. 12, R100. doi: 10.1186/gb-2011-12-10-r100
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772. doi: 10.1038/nmeth.2109
Dickerson, H. W. (2006). “Ichthyophthirius multifiliis and Cryptocaryon irritans (phylum Ciliophora),” in Fish Diseases and Disorders, Vol. Volume 1, ed. P. T. K. Woo (Wallingford: CABI), 116–153.
Dickerson, H. W. (2011). “Ichthyophthirius multifiliis,” in Fish Parasites: Pathobiology and Protection, ed. P. T. K. Woo (Wallingford: CABI), 55–72.
Driscoll, T., Gillespie, J. J., Nordberg, E. K., Azad, A. F., and Sobral, B. W. (2013). Bacterial DNA sifted from the Trichoplax adhaerens (Animalia: Placozoa) genome project reveals a putative rickettsial endosymbiont. Genome Biol. Evol. 5, 621–645. doi: 10.1093/gbe/evt036
Eisen, J. A., Coyne, R. S., Wu, M., Wu, D., Thiagarajan, M., Wortman, J. R., et al. (2006). Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 4:e286. doi: 10.1371/journal.pbio.0040286
Elde, N. C., Morgan, G., Winey, M., Sperling, L., and Turkewitz, A. P. (2005). Elucidation of clathrin-mediated endocytosis in tetrahymena reveals an evolutionarily convergent recruitment of dynamin. PLoS Genet. 1:e52. doi: 10.1371/journal.pgen.0010052.eor
Epis, S., Sassera, D., Beninati, T., Lo, N., Beati, L., Piesman, J., et al. (2008). Midichloria mitochondrii is widespread in hard ticks (Ixodidae) and resides in the mitochondria of phylogenetically diverse species. Parasitology 135, 485–494. doi: 10.1017/S0031182007004052
Erickson, D. L., Anderson, N. E., Cromar, L. M., and Jolley, A. (2009). Bacterial communities associated with flea vectors of plague. J. Med. Entomol 46, 1532–1536. doi: 10.1603/033.046.0642
Ersfeld, K. (2003). Genomes and genome projects of protozoan parasites. Curr. Issues Mol. Biol. 5, 61–74.
Ewing, M. S., Kocan, K. M., and Ewing, S. A. (1985). Ichthyophthirius multifiliis (Ciliophora) Invasion of Gill Epithelium1. J. Protozool. 32, 305–310. doi: 10.1111/j.1550-7408.1985.tb03055.x
Fenchel, T., and Finlay, B. J. (1991a). The biology of free-living anaerobic ciliates. Eur. J. Protistol. 26, 201–215. doi: 10.1016/S0932-4739(11)80143-4
Fenchel, T., and Finlay, B. J. (1991b). Endosymbiotic methanogenic bacteria in anaerobic ciliates: significance for the growth efficiency of the host. J. Protozool 38, 18–22. doi: 10.1016/S0932-4739(11)80143-4
Finlay, B. J., and Fenchel, T. (1989). Hydrogenosomes in some anaerobic protozoa resemble mitochondria. FEMS Microbiol. Lett. 65, 311–314. doi: 10.1111/j.1574-6968.1989.tb03679.x
Fokin, S. I. (2004). Bacterial endocytobionts of ciliophora and their interactions with the host cell. Int. Rev. Cytol. 236, 181–249. doi: 10.1016/S0074-7696(04)36005-5
Fraune, S., and Bosch, T. C. (2007). Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc. Natl. Acad. Sci. U.S.A. 104, 13146–13151. doi: 10.1073/pnas.0703375104
Fritsche, T. R., Horn, M., Seyedirashti, S., Gautom, R. K., Schleifer, K. H., and Wagner, M. (1999). In situ detection of novel bacterial endosymbionts of Acanthamoeba spp. phylogenetically related to members of the order Rickettsiales. Appl. Environ. Microbiol. 65, 206–212.
Galand, P. E., Bourrain, M., De Maistre, E., Catala, P., Desdevises, Y., Elifantz, H., et al. (2012). Phylogenetic and functional diversity of Bacteria and Archaea in a unique stratified lagoon, the Clipperton atoll (N Pacific). FEMS Microbiol. Ecol. 79, 203–217. doi: 10.1111/j.1574-6941.2011.01209.x
Gillespie, J. J., Nordberg, E. K., Sobral, B. W. S., and Azad, A. F. (2012). “Phylogeny and comparative genomics: the shifting landscape in the genomics era,” in Intracellular Pathogens II: Rickettsiales, eds G. Palmer and A. F. Azad (Washington, DC: ASM Press), 84–141.
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hertz-Fowler, C., Berriman, M., and Pain, A. (2005). A feast of protozoan genomes. Nat. Rev. Microbiol. 3, 670–671. doi: 10.1038/nrmicro1237
Hollants, J., Leliaert, F., Verbruggen, H., Willems, A., and De Clerck, O. (2013). Permanent residents or temporary lodgers: characterizing intracellular bacterial communities in the siphonous green alga Bryopsis. Proc. Biol. Sci. 280, 20122659. doi: 10.1098/rspb.2012.2659
Hornok, S., Foldvari, G., Elek, V., Naranjo, V., Farkas, R., and de la Fuente, J. (2008). Molecular identification of Anaplasma marginale and rickettsial endosymbionts in blood-sucking flies (Diptera: Tabanidae, Muscidae) and hard ticks (Acari: Ixodidae). Vet. Parasitol. 154, 354–359. doi: 10.1016/j.vetpar.2008.03.019
Kawafune, K., Hongoh, Y., Hamaji, T., and Nozaki, H. (2012). Molecular identification of rickettsial endosymbionts in the non-phagotrophic volvocalean green algae. PLoS ONE 7:e31749. doi: 10.1371/journal.pone.0031749
Kawafune, K., Hongoh, Y., and Nozaki, H. (2014). A rickettsial endosymbiont inhabiting the cytoplasm of Volvox carteri (Volvocales, Chlorophyceae). Phycologia 53, 95–99. doi: 10.2216/13-193.1
Lin, T. L., Clark, T. G., and Dickerson, H. (1996). Passive immunization of channel catfish (Ictalurus punctatus) against the ciliated protozoan parasite Ichthyophthirius multifiliis by use of murine monoclonal antibodies. Infect. Immun. 64, 4085–4090.
Lloyd, S. J., LaPatra, S. E., Snekvik, K. R., Cain, K. D., and Call, D. R. (2011). Quantitative PCR demonstrates a positive correlation between a Rickettsia-like organism and severity of strawberry disease lesions in rainbow trout, Oncorhynchus mykiss (Walbaum). J. Fish Dis. 34, 701–709. doi: 10.1111/j.1365-2761.2011.01285.x
Lloyd, S. J., LaPatra, S. E., Snekvik, K. R., St-Hilaire, S., Cain, K. D., and Call, D. R. (2008). Strawberry disease lesions in rainbow trout from southern Idaho are associated with DNA from a Rickettsia-like organism. Dis. Aquat. Org. 82, 111–118. doi: 10.3354/dao01969
Lobo-da-Cunha, A., and Azevedo, C. (1988). Association between xenosomes and glycogen in the cytoplasm of the ciliate Ichthyophthirius multifiliis. Endocyt. Cell Res. 5, 225–231.
Lobo-da-Dunha, A., and Azevedo, C. (1993). Processing of food vacuoles in the parasitic ciliate Ichthyophthirius multifiliis after exist from the host. Parasitol. Res. 79, 272–278. doi: 10.1007/BF00932181
Luo, R., Liu, B., Xie, Y., Li, Z., Huang, W., Yuan, J., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1, 18–23. doi: 10.1186/2047-217X-1-18
MacColl, E., Therkelsen, M. D., Sherpa, T., Ellerbrock, H., Johnston, L. A., Jariwala, R. H., et al. (2015). Molecular genetic diversity and characterization of conjugation genes in the fish parasite Ichthyophthirius multifiliis. Mol. Phylogenet. Evol. 86, 1–7. doi: 10.1016/j.ympev.2015.02.017
Mariconti, M., Epis, S., Gaibani, P., Dalla Valle, C., Sassera, D., Tomao, P., et al. (2012). Humans parasitized by the hard tick Ixodes ricinus are seropositive to Midichloria mitochondrii: is Midichloria a novel pathogen, or just a marker of tick bite? Pathog. Glob. Health 106, 391–396. doi: 10.1179/2047773212Y.0000000050
Matsuura, Y., Kikuchi, Y., Meng, X. Y., Koga, R., and Fukatsu, T. (2012). Novel clade of alphaproteobacterial endosymbionts associated with stinkbugs and other arthropods. Appl. Environ. Microbiol. 78, 4149–4156. doi: 10.1128/AEM.00673-12
Matthews, R. A. (2005). Ichthyophthirius multifiliis fouquet and ichthyophthiriosis in freshwater teleosts. Adv. Parasitol. 59, 159–241. doi: 10.1016/S0065-308X(05)59003-1
Mediannikov, O. I., Ivanov, L. I., Nishikawa, M., Saito, R., Sidel’nikov, I., Zdanovskaia, N. I., et al. (2004). Microorganism “Montezuma” of the order Rickettsiales: the potential causative agent of tick-borne disease in the Far East of Russia. Zh. Mikrobiol. Epidemiol. Immunobiol. 1, 7–13.
Metselaar, M., Thompson, K. D., Gratacap, R. M. L., Kik, M. J. L., LaPatra, S. E., Lloyd, S. J., et al. (2010). Association of red-mark syndrome with a Rickettsia-like organism and its connection with strawberry disease in the USA. J. Fish Dis. 33, 849–858. doi: 10.1111/j.1365-2761.2010.01187.x
Montagna, M., Sassera, D., Epis, S., Bazzocchi, C., Vannini, C., Lo, N., et al. (2013). “Candidatus Midichloriaceae” fam. nov. (Rickettsiales), an ecologically widespread clade of intracellular alphaproteobacteria. Appl. Environ. Microbiol. 79, 3241–3248. doi: 10.1128/AEM.03971-12
Noe, J. G., and Dickerson, H. W. (1995). Sustained growth of Ichthyophthirius multifiliis at low temperature in the laboratory. J. Parasitol. 81, 1022–1024. doi: 10.2307/3284065
Notredame, C., Higgins, D. G., and Heringa, J. (2000). T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217. doi: 10.1006/jmbi.2000.4042
Parratt, S. R., and Laine, A. L. (2016). The role of hyperparasitism in microbial pathogen ecology and evolution. ISME J. 10, 1815–1822. doi: 10.1038/ismej.2015.247
Percent, S. F., Frischer, M. E., Vescio, P. A., Duffy, E. B., Milano, V., McLellan, M., et al. (2008). Bacterial community structure of acid-impacted lakes: what controls diversity? Appl. Environ. Microbiol. 74, 1856–1868. doi: 10.1128/AEM.01719-07
Preer, L. B., Jurand, A., Preer, J. R. Jr., and Rudman, B. M. (1972). The classes of kappa in Paramecium aurelia. J. Cell. Sci. 11, 581–600.
Quackenbush, R. L., and Burbach, J. A. (1983). Cloning and expression of DNA sequences associated with the killer trait of Paramecium tetraurelia stock 47. Proc. Natl. Acad. Sci. U.S.A. 80, 250–254. doi: 10.1073/pnas.80.1.250
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ramoino, P., Fronte, P., Fato, M., Beltrame, F., Robello, M., and Diaspro, A. (2001). Fluid phase and receptor-mediated endocytosis in Paramecium primaurelia by fluorescence confocal laser scanning microscopy. Eur. Biophys. J. 30, 305–312. doi: 10.1007/s002490100166
Raoult, D., and Roux, V. (1997). Rickettsioses as paradigms of new or emerging infectious diseases. Clin. Microbiol. Rev. 10, 694–719.
Richard, S., Seng, P., Parola, P., Raoult, D., Davoust, B., and Brouqui, P. (2009). Detection of a new bacterium related to ‘Candidatus Midichloria mitochondrii’ in bed bugs. Clin. Microbiol. Infect. 15(Suppl. 2), 84–85. doi: 10.1111/j.1469-0691.2008.02244.x
Riley, S. P., Goh, K. C., Hermanas, T. M., Cardwell, M. M., Chan, Y. G., and Martinez, J. J. (2010). The Rickettsia conorii autotransporter protein Sca1 promotes adherence to nonphagocytic mammalian cells. Infect. Immun. 78, 1895–1904. doi: 10.1128/IAI.01165-09
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Schmahl, G., Taraschewski, H., and Mehlhorn, H. (1989). Chemotherapy of fish parasites. Parasitol. Res. 75, 503–511. doi: 10.1007/BF00931157
Schrallhammer, M., Ferrantini, F., Vannini, C., Galati, S., Schweikert, M., Gortz, H. D., et al. (2013). ‘Candidatus Megaira polyxenophila’ gen. nov., sp. nov.: considerations on evolutionary history, host range and shift of early divergent rickettsiae. PLoS One 8:e72581. doi: 10.1371/journal.pone.0072581
Schweikert, M., Fujishima, M., and Görtz, H. (2013). “Symbiotic associations between ciliates and prokaryotes,” in The Prokaryotes: Prokaryotic Biology and Symbiotic Associations, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer), 427–463.
Sun, H. Y., Noe, J., Barber, J., Coyne, R. S., Cassidy-Hanley, D., Clark, T. G., et al. (2009). Endosymbiotic bacteria in the parasitic ciliate Ichthyophthirius multifiliis. Appl. Environ. Microbiol. 75, 7445–7452. doi: 10.1128/AEM.00850-09
Sunagawa, S., DeSantis, T. Z., Piceno, Y. M., Brodie, E. L., DeSalvo, M. K., Voolstra, C. R., et al. (2009). Bacterial diversity and white plague disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J. 3, 512–521. doi: 10.1038/ismej.2008.131
Teysseire, N., Boudier, J. A., and Raoult, D. (1995). Rickettsia conorii entry into Vero cells. Infect. Immun. 63, 366–374.
Uchiyama, T. (2012). Tropism and pathogenicity of rickettsiae. Front. Microbiol. 3:230. doi: 10.3389/fmicb.2012.00230
Vannini, C., Ferrantini, F., Schleifer, K. H., Ludwig, W., Verni, F., and Petroni, G. (2010). “Candidatus anadelfobacter veles” and “Candidatus cyrtobacter comes,” two new rickettsiales species hosted by the protist ciliate Euplotes harpa (Ciliophora, Spirotrichea). Appl. Environ. Microbiol. 76, 4047–4054. doi: 10.1128/AEM.03105-09
Vannini, C., Petroni, G., Schena, A., Verni, F., and Rosati, G. (2003). Well-established mutualistic associations between ciliates and prokaryotes might be more widespread and diversified than so far supposed. Eur. J. Protistol. 39, 481–485. doi: 10.1078/0932-4739-00024
Vannini, C., Petroni, G., Verni, F., and Rosati, G. (2005). A bacterium belonging to the Rickettsiaceae family inhabits the cytoplasm of the marine ciliate Diophrys appendiculata (Ciliophora, Hypotrichia). Microb. Ecol. 49, 434–442. doi: 10.1007/s00248-004-0055-1
Vlahos, N., Kormas, K. A., Pachiadaki, M. G., Meziti, A., Hotos, G. N., and Mente, E. (2013). Changes of bacterioplankton apparent species richness in two ornamental fish aquaria. Springerplus 2, 66. doi: 10.1186/2193-1801-2-66
Walker, D. H., and Ismail, N. (2008). Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat. Rev. Microbiol. 6, 375–386. doi: 10.1038/nrmicro1866
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M., and Barton, G. J. (2009). Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Whitworth, T., Popov, V. L., Yu, X. J., Walker, D. H., and Bouyer, D. H. (2005). Expression of the Rickettsia prowazekii pld or tlyC gene in Salmonella enterica serovar Typhimurium mediates phagosomal escape. Infect. Immun. 73, 6668–6673. doi: 10.1128/IAI.73.10.6668-6673.2005
Williams-Newkirk, A. J., Rowe, L. A., Mixson-Hayden, T. R., and Dasch, G. A. (2012). Presence, genetic variability, and potential significance of “Candidatus Midichloria mitochondrii” in the lone star tick Amblyomma americanum. Exp. Appl. Acarol. 58, 291–300. doi: 10.1007/s10493-012-9582-5
Keywords: Ciliophora, alphaproteobacteria, Sphingobacteria, hyperparasitism, phagocytosis, symbiosis
Citation: Zaila KE, Doak TG, Ellerbrock H, Tung C -H, Martins ML, Kolbin D, Yao M -C, Cassidy-Hanley DM, Clark TG and Chang W -J (2017) Diversity and Universality of Endosymbiotic Rickettsia in the Fish Parasite Ichthyophthirius multifiliis. Front. Microbiol. 8:189. doi: 10.3389/fmicb.2017.00189
Received: 23 August 2016; Accepted: 25 January 2017;
Published: 09 February 2017.
Edited by:
Dennis A. Bazylinski, University of Nevada, Las Vegas, USAReviewed by:
Kathleen Scott, University of South Florida, USARodrigo Costa, Universidade de Lisboa, Portugal
Copyright © 2017 Zaila, Doak, Ellerbrock, Tung, Martins, Kolbin, Yao, Cassidy-Hanley, Clark and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Theodore G. Clark, dGdjM0Bjb3JuZWxsLmVkdQ== Wei-Jen Chang, d2NoYW5nQGhhbWlsdG9uLmVkdQ==
 Kassandra E. Zaila
Kassandra E. Zaila Thomas G. Doak2,3
Thomas G. Doak2,3 Wei-Jen Chang
Wei-Jen Chang