- 1Department of Clinical Laboratory, Peking Union Medical College Hospital, Peking Union Medical College, Chinese Academy of Medical Sciences, Beijing, China
- 2Department of Clinical Laboratory, The First Affiliated Hospital of Xiamen University, Xiamen, China
- 3Department of Clinical Laboratory, Bazhou People’s Hospital, Xinjiang, China
- 4Department of Clinical Laboratory, Beijing Youan Hospital, Capital Medical University/Hospital for Infectious Diseases of Baoding, Hebei, China
- 5Charles Sturt University, Orange Campus, Orange, NSW, Australia
- 6Centre for Infectious Diseases and Microbiology Laboratory Services, ICPMR – Pathology West, Westmead Hospital, University of Sydney, Westmead, NSW, Australia
To gain some insights into the molecular evolution of Moraxella catarrhalis macrolide resistance, PCR and sequencing analysis of the 23S rRNA gene, copB typing and multilocus sequence typing (MLST) were performed on 181 M. catarrhalis isolates. The isolates were obtained from children (n = 47) and adults (n = 134) presenting with respiratory disease in the years 2010–2014. Macrolide resistance was highly age-related, and nucleotide position alterations at A2330T could be detected in all macrolide-resistant isolates. copB 0 and copB NT (non-typable) were only found in macrolide-susceptible isolates from adults. Furthermore, copB I/III was the main type in adult or macrolide-susceptible isolates, while copB II was the most common type in children or macrolide-resistant isolates. Twenty-two different MLST clusters (sharing 7 of the 8 identical loci) were detected and only four likely primary founders (ST224, ST363, STN08, and STN10) which belong to clonal complex (CC) 224, CC363, CCN08, and CCN10, were detected, respectively. Macrolide-resistant M. catarrhalis isolates were highly concentrated in two CCs (CCN10 and CC363), which indicates some potential evolutionary advantage or co-evolution to some extent. However, further studies are needed to fully elucidate the evolution of CCN10 and CC363 in macrolide resistance.
Introduction
Moraxella catarrhalis is a prominent pathogen that causes acute otitis media in children and lower respiratory tract infections in adults (such as exacerbations of chronic obstructive pulmonary disease) (Hol et al., 1996; Ahmad, 1998), resulting in significant socio-economic burden on healthcare systems globally.
Previous studies, including our own (Wang et al., 2011; Flamm et al., 2012; Liu et al., 2012, 2015; Tang et al., 2016), have reported on the increased prevalence of macrolide-resistant M. catarrhalis in Mainland China, ranging from 40 to 80% in children, and 5–10% in adults. A better understanding of the evolutionary path of the organism from macrolide-susceptible to macrolide-resistant M. catarrhalis, is crucial for controlling the spread of macrolide resistance. However, based on our previous studies, showing 40–60% macrolide resistance amongst M. catarrhalis isolates from healthy toddlers (12–18 months) (Liu et al., 2012), and that most macrolide resistant isolates (5/7) were obtained from patients who had not received macrolide antibiotics within the previous 30 days (Liu et al., 2015), we think that macrolide resistance is acquired.
Multilocus sequence typing (MLST) has been widely employed in epidemiological investigations of various scales (Cox et al., 1995; Enright and Spratt, 1998; Maiden et al., 1998; Enright et al., 2001; Qin et al., 2009), including population, pathogenicity, and evolution studies, of several bacteria. To gain insights into the molecular evolution of macrolide resistance in M. catarrhalis, PCR and sequence analysis of the 23S rRNA gene, copB typing, MLST, and macrolide susceptibility testing, were performed on 181 M. catarrhalis isolates.
Materials and Methods
Statement
All the authors confirm that all experiments were performed in accordance with relevant guidelines and regulations. Only clinical bacterial isolates, and no human subjects (including the use of tissue samples), were used in the present study. Thus informed consent was not obtained from the patients. The study was approved by the Human Research Ethics Committee of Peking Union Medical College Hospital (No. S-424).
Bacterial Isolates
One hundred and twenty non-duplicate M. catarrhalis isolates were collected from the sputum or broncho-alveolar lavage of adult patients (>18 years of age) in the Peking Union Medical College hospital (PUMCH) (59 wards) between 2010 and 2013. Furthermore, 14 M. catarrhalis isolates from the sputum or ear purulent secretion of adult patients (10 wards), and 47 from the sputum or ear purulent secretion of children (7 wards), were obtained from the First Affiliated Hospital of Xiamen University (FAHXU) in 2014. The isolates were identified by mass spectrometry (MALDI-TOF MS) Bruker Biotyper (Bruker Daltonics, Bremen, Germany). Isolates were stored at -70°C until testing, with a minimum number of passages.
Macrolide Susceptibility Testing
The isolates were tested for susceptibility to erythromycin and azithromycin using the Kirby-Bauer disk diffusion (Thermo Fisher, Oxoid) method in accordance with the CLSI M45-A3 guideline (Clinical and Laboratory Standards Institute [CLSI], 2015). In addition, the E-test method was used to confirm the antimicrobial susceptibility results of macrolide-resistant M. catarrhalis. Staphylococcus aureus ATCC 25923 and ATCC 29213 were used for quality control.
copB Polymerase Chain Reaction-Restriction Fragment Length Polymorphisms
The copB gene was tested for in all the isolates (n = 181), as described by Verhaegh et al. (2008) and Liu et al. (2012). A touchdown thermocycling program was used for all PCRs. The touchdown protocol used an initial annealing temperature of 70°C, which was reduced by 1°C per cycle over 15 cycles of PCR. The final 20 amplification cycles used an annealing temperature of 55°C. The copB PCR products were digested with RsaI (New England Biolabs, Ipswich, MA, USA). Five units of RsaI were used per reaction mix, and the mixture was incubated at 37°C. Agarose gels were prepared at a 1% concentration in 0.5 × TBE buffer (45 mM Tris base, 45 mM boric acid, and 1 mM EDTA). DNA fragments were separated using an electric current of 120 V/cm RT for 30 min. Gels were stained for 15–20 min in ethidium bromide (1 mg/ml) and decolorized in distilled water for 2–5 min. Fluorescent bands were visualized using UV transillumination, and gel images were captured using Bio-Rad GelDocEQ (Bio-Rad). A 50 bp Ladder (NEB) was used as a molecular size standard (TransGen Biotech, Beijing, China). Four types of copB were included: type 0, consisting of 374 and 157-bp; copB types I/III, consisting of 342 and 157-bp; copB type II, with bands of 332 and 187-bp; and copB type IV, consisting of a single band of 519-bp.
PCR and Sequencing Analysis of the 23S rRNA Gene
The 23S rRNA gene was amplified and sequenced using primer pairs described before (MUT-F, 5′-2685CAGGCTGCTGCAACTGTTTA2704-3′, and MUT-R, 5′-3618CAACCGAAACA CCAGAGGTT3599-3′, from M. catarrhalis KCCM: 40056; positions determined according to the bases on GenBank submission of FJ410380) (Liu et al., 2012, 2016).
Multilocus Sequence Typing
Multilocus sequence typing was performed on all the 181 isolates by amplification of internal fragments of the housekeeping genes glyRS (glycyl-tRNA synthetase beta subunit), ppa (Pyrophosphate phospho-hydrolase), efp (elongation factor P), fumC (fumarate hydratase), trpE (anthranilate synthase component I), mutY (adenine glycosylase), adk (Adenylate kinase), and abcZ (ATP-binding protein), in separate PCRs. Based on the sequences, alleles were determined by querying the central database at http://mlst.warwick.ac.uk/mlst/dbs/Mcatarrhalis/ Finally, a graphical representation of the relatedness of isolates was performed using the program enhanced based upon BioNumerics (version 7.6). Through the availability of an MLST plugin, the BioNumerics program automatically analyses batches of sequence trace files, connects to online MLST databases, retrieves corresponding allele numbers, sequence types, as well as available clonal complex (CC) information.
Statistical Analysis
The prevalence of macrolide-resistant M. catarrhalis in adults and children, the frequency of sequence types (STs) or the copB types in adults and children, or the frequency of STs or the copB types in macrolide-susceptible and resistant isolates, was statistically compared using Pearson’s χ2-test (or the Fisher exact test, when appropriate) for categorical data.
Results
M. catarrhalis Genetic Population Study
Among the 181 M. catarrhalis isolates studied, 20 were resistant to macrolide (MIC > 256 μg/ml). Furthermore, a nucleotide position alteration at A2330T was detected in each of the 20 macrolide-resistant isolates. The prevalence of macrolide-resistant M. catarrhalis in adults was much lower (6.0%, 8/134) than in children (25.5%, 12/47; p = 0.006). Using the copB PCR-RFLP typing method, copB II was the most common type amongst the resistant isolates (11/20, 55%), whilst only 33.5% copB II (54/161) was detected in macrolide-susceptible isolates (p = 0.082) (Figure 1). copB I/III (56.0%, 75/134) was the main type in isolates from adults while only 40.4% copB I/III (19/47) was detected in isolates from children (p = 0.089). The copB 0 and non-typeable (NT) isolates were only detected in adults (p = 0.114).
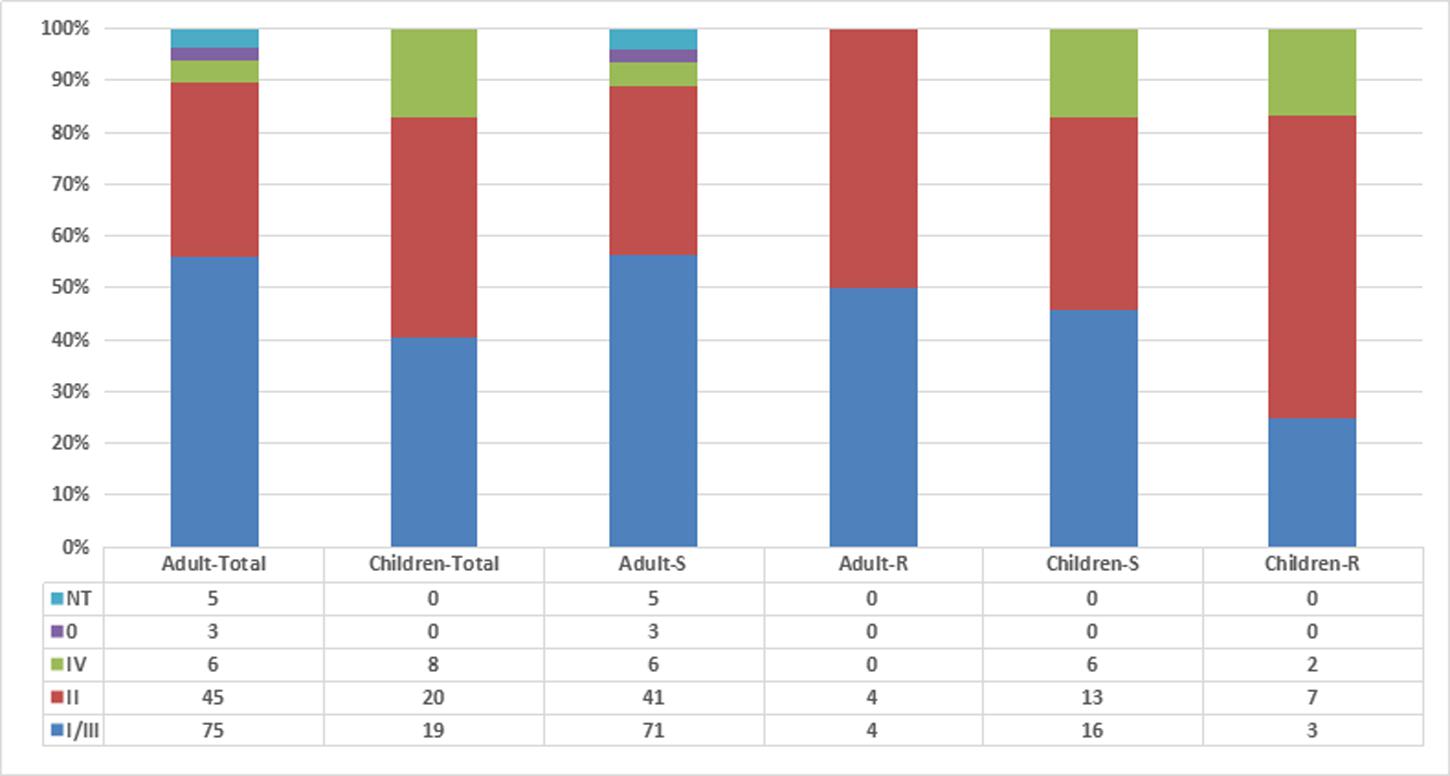
FIGURE 1. Relationship among Moraxella catarrhalis isolates by age, antibiotic resistance, and copB typing. Adult-S: macrolide-susceptible M. catarrhalis isolates from adults, Adult-R: macrolide-resistant M. catarrhalis isolates from adults, Children-S: macrolide-susceptible M. catarrhalis isolates from children, Children-R: macrolide-resistant M. catarrhalis isolates from children.
Relationship between Age, Macrolide Resistance, and MLST Results
Using MLST, the 181 isolates were discriminated into 119 different ST, including 103 novel STs (ST293-ST327, ST329-369, STN01-STN31) (STN01-STN31 were defined in the present study). In adults, ST224, ST105, and ST180 (4.5%, 6/134 each), were the most common STs, followed by STN05 and STN26 (3.7%, 5/134, for each), STN29 and ST308 (3.0%, 4/134, for each), ST64, STN14, ST62, ST156, and ST176 (2.2%, 3/134, for each). Just like in adults, ST224 (8.5%, 4/47) was the most common ST in children, albeit almost doubled in prevalence, followed by ST64 and ST342 (4.3%, 2/47, for each). In PUMCH, only isolates from adults were included in the study whilst in FAHXU, children were the main population. However, even when the isolates from adults and children in FAHXU were divided into two parts, the frequency of STs was almost the same as mentioned above.
A population snapshot of the studied M. catarrhalis isolates based on allelic profiles of MLST is shown in Figure 2. STs that shared alleles at ≥5 of the 8 MLST loci were obtained from the M. catarrhalis MLST website, and a diagram was constructed using Bionumerics. There are 22 different MLST clusters (Definition: Sharing 7 of the 8 identical loci, or single locus variant with one other member of the group) marked in Figure 2 using gray halo surrounding the MLST STs. But only four likely primary founders (Definition: A ST which is positioned centrally in the cluster and with at least three links to other STs) (ST224, ST363, STN08, and STN10) were identified and are marked by yellow rings. The four likely primary founders (ST224, ST363, STN08, and STN10) were defined to belong to CC (Definition: A cluster containing primary founder) CC224, CC363, CCN08, and CCN10. In the 22 different MLST clusters, there were only five clusters that contained macrolide-resistant M. catarrhalis isolates in which only CCN10 and CC363 could be likely defined for diverse STs and few links. The CCN10 contained six STs (STN10, STN11, STN16, ST332, ST347, and ST348) and all the STs consisted of macrolide-resistant isolates. The CC363 contained six STs (ST363, ST318, ST339, ST340, ST361, and ST362) and the likely primary founder ST363 consisted of both macrolide-resistant and susceptible isolates.
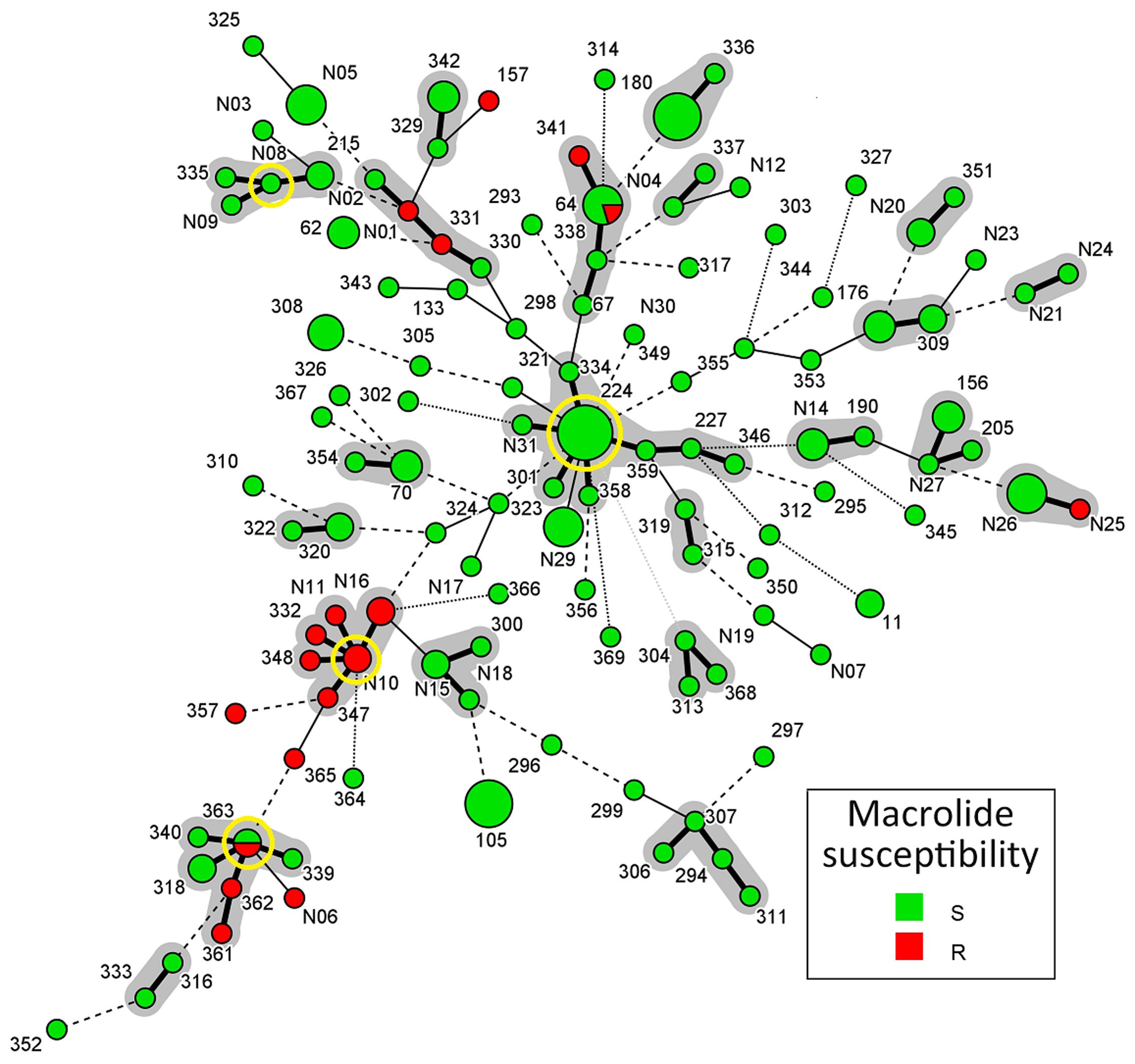
FIGURE 2. Population snapshot of M. catarrhalis based on allelic profiles of MLST. Sequence types (STs) that shared alleles at ≥5 of the eight MLST loci were obtained from the M. catarrhalis MLST website, and a diagram was constructed by using Bionumerics. Each circle in Figure corresponds to a MLST ST, and different circle colors represent different susceptibilities to macrolide. The lines between circles indicate the similarity between profiles: bold line, seven of eight MLST alleles/MLVA loci in common; normal line, six alleles/loci in common; dashed line, five alleles/loci in common; dotted line, ≤4 alleles/loci. The gray halo surrounding the STs in Figure denotes STs belonging to different MLST clusters. The likely primary founders with at least three links to other STs are positioned centrally in the cluster and identified by yellow rings. Cluster 1: ST363/ST340/ST339/ST362/ST361/ST318, Cluster 2: STN10/STN11/STN16/ST332/ST348/ST347, Cluster 3: STN08/ST335/ST215/STN09, Cluster 4: ST224/ST227/ST323/ST334/ST346/ST358/ ST359/STN31, Cluster 5: STN25/STN26, Cluster 6: ST330/ST331/STN01/STN02, Cluster 7: ST64/ST67/ST338/ST341, Cluster 8: ST329/ST342, Cluster 9: ST180/ST336, Cluster 10: ST337/STN04, Cluster 11: STN20/ST351, Cluster 12: STN21/STN24, Cluster 13: ST176/ ST309, Cluster 14: ST156/ST205/STN27, Cluster 15: ST190/STN14, Cluster 16: ST315/ST319, Cluster 17: ST304/ST313/ST368, Cluster 18: ST294/ST306/ST307/ST311, Cluster 19: ST337/STN04, Cluster 20: ST300/STN15/STN18, Cluster 21: ST320/ST322, Cluster 22: ST70/ST354.
Discussion
CopB is one of the several M. catarrhalis outer membrane proteins that elicit a systemic humoral immune response in humans, resulting in the production of serum antibody specific for CopB, which underscores the potential of CopB as a vaccine candidate (Meier et al., 2003; Murphy et al., 2005; Liu et al., 2006). The copB gene is relatively conserved in M. catarrhalis, and has been detected in all isolates examined so far (Liu et al., 2012, 2015). Digestion of the copB products with RsaI restriction enzyme divides the organism into four copB types, namely copB type 0 (374 and 157 bp), type I/III (342 and157 bp), type II (332 and 187 bp) and type IV (519 bp) (Verhaegh et al., 2008), and has already been used in several typing studies (Verhaegh et al., 2008; Liu et al., 2012, 2015).
In our previous studies, we found that there was a trend toward an increase in copB types I/III, and a decrease in copB type II, in adult associated M. catarrhalis isolates, when compared with isolates from children (Liu et al., 2012, 2015). The present findings are somewhat in agreement to these previous results although the difference was not statistically significant, which suggests that the copB type may not be age-related.
When the allelic profiles of MLST were analyzed using Bionumerics software, population diversity was clearly demonstrated (Figure 2). However, four likely primary founders (ST224, ST363, STN08, and STN10) were demonstrated in CCs CC224, CC363, CCN08, CCN10. The predicted primary founder ST224, with the largest number and complicated links to other STs, is likely to be the ancestor of all macrolide- susceptible isolates. And all the five clusters encompassing macrolide-resistant M. catarrhalis isolates, located at the edge of the population snapshot (Figure 2) with less links to CC224, seem to have evolved in several ways from macrolide-susceptible isolates, but not exactly only from CC224.
One important finding of this study is the description of two macrolide-resistant CCs, CCN10 and CC363, and the likely primary founders STN10 and ST363. As illustrated in Figure 2, the two valuable macrolide-resistant CCs, CCN10 and CC363, are significantly separated from other clusters including CC224, and from the year collected. STN10 and ST224 were first isolated in 2010 and 2012, respectively, from which we surmise that CCN10 is less likely to have evolved from CC224. The small number of isolates analyzed over a short duration (2010–2014) makes it difficult to come up with firm conclusions. Some valuable insights might be gained from studying a large number of isolates collected over a 10 or 12 years period.
A limitation of this study is that the isolates originated from two different hospitals; the prevalence and distribution of macrolide-resistance or STs might be totally different in different areas. Actually, antimicrobial susceptibility testing and MLST typing were first performed on isolates from PUMCH adults, and seven macrolide-resistant isolates and some important STs such as ST224, STN10, and STN16, were detected. This encouraged us to examine whether the macrolide-resistant population was concentrated in selected STs. PUMCH is located in the northern part of China and the patients are mainly adults, while FAHXU is situated in the southern part, and is well known for pediatric infectious diseases. So the two hospitals were ideal locations for our study purpose, including comparing the distribution of different STs and macrolide-resistant isolates in the two hospitals (Figure 2; Supplementary Figure S1). From the results we can see that even though the distribution of different STs is definitely related to regions, some important STs such as ST224, STN10, and STN16, were detected in both hospitals. Moreover, most STs of macrolide-resistant isolates (except for STN10 and STN16) detected in PUMCH or FAHXU belonged to the same clusters (such as both STN01 and ST331 belonged to cluster 6) (Figure 2; Supplementary Figure S1), which suggests that these STs might be very critical clones in the process of evolution, especially on macrolide resistance. And we can also see that macrolide resistance is mainly age-related without obvious connection with different regions (Figure 2; Supplementary Figures S1 and S2), albeit limited data from the two hospitals. However, even when the results from adults and children in the same hospital were analyzed (Supplementary Figure S3), most of the derived conclusions are somehow confirmed.
Further study limitations include the relatively small number of macrolide-resistant M. catarrhalis isolates in adults compared with macrolide- susceptible isolates, which may influence the reliability of results to some extent. In addition, STN01–STN31 have not been submitted to http://mlst.warwick.ac.uk/mlst/dbs/Mcatarrhalis/, so these ST names were defined in the present study. Moreover, we compared isolates collected over different time periods in the two hospitals; strains from PUMCH were collected from 2010 to 2013 while the ones from FAHXU were collected in 2014. In our further study, the genomic sequences of these strains would be determined according to the requirements of the Enterobase1 instead of the MLST website, and then the accurate STs would be given.
Author Contributions
Y-LL, MX, TK, and FK wrote the manuscript. J-WC, Z-PX, and H-PX collaborated in molecular investigations of the strains. SY and W-JZ summarized the patient’s medical records. Y-CX designed and supervised the study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported by the Research Special Fund for Public Welfare Industry of Health (Grant No. 201402001).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00201/full#supplementary-material
FIGURE S1 | Population snapshot of M. catarrhalis based on allelic profiles of multilocus sequence typing (MLST) in two hospitals. Sequence types (STs) that shared alleles at ≥5 of the eight MLST loci were obtained from the M. catarrhalis MLST website, and a diagram was constructed by using Bionumerics. Each circle corresponds to a MLST ST, and different circle colors represent different hospitals. The lines between circles indicate the similarity between profiles: bold line, seven of eight MLST alleles/MLVA loci in common; normal line, six alleles/loci in common; dashed line, five alleles/loci in common; dotted line, ≤4 alleles/loci. The gray halo surrounding the STs denotes STs belonging to different MLST clusters. Cluster 1: ST363/ST340/ST339/ST362/ST361/ST318, Cluster 2: STN10/STN11/STN16/ST332/ST348/ST347, Cluster 3: STN08/ST335/ST215/STN09, Cluster 4: ST224/ST227/ST323/ST334/ST346/ST358/ ST359/STN31, Cluster 5: STN25/STN26, Cluster 6: ST330/ST331/STN01/STN02, Cluster 7: ST64/ST67/ST338/ST341, Cluster 8: ST329/ST342, Cluster 9: ST180/ST336, Cluster 10: ST337/STN04, Cluster 11: STN20/ST351, Cluster 12: STN21/STN24, Cluster 13: ST176/ ST309, Cluster 14: ST156/ST205/STN27, Cluster 15: ST190/STN14, Cluster 16: ST315/ST319, Cluster 17: ST304/ST313/ST368, Cluster 18: ST294/ST306/ST307/ST311, Cluster 19: ST337/STN04, Cluster 20: ST300/STN15/STN18, Cluster 21: ST320/ST322, Cluster 22: ST70/ST354.
FIGURE S2 | Population snapshot of M. catarrhalis based on allelic profiles of MLST in adults and children. Sequence types (STs) that shared alleles at ≥5 of the eight MLST loci were obtained from the M. catarrhalis MLST website, and a diagram was constructed by using Bionumerics. Each circle corresponds to a MLST ST, and different circle colors represent different population (A: Adult/C: children).
The lines between circles indicate the similarity between profiles: bold line, seven of eight MLST alleles/MLVA loci in common; normal line, six alleles/loci in common; dashed line, five alleles/loci in common; dotted line, ≤4 alleles/loci. The gray halo surrounding the STs denotes STs belonging to different MLST clusters. Cluster 1: ST363/ST340/ST339/ST362/ST361/ST318, Cluster 2: STN10/STN11/STN16/ST332/ST348/ST347, Cluster 3: STN08/ST335/ST215/STN09, Cluster 4: ST224/ST227/ST323/ST334/ST346/ST358/ ST359/STN31, Cluster 5: STN25/STN26, Cluster 6: ST330/ST331/STN01/STN02, Cluster 7: ST64/ST67/ST338/ST341, Cluster 8: ST329/ST342, Cluster 9: ST180/ST336, Cluster 10: ST337/STN04, Cluster 11: STN20/ST351, Cluster 12: STN21/STN24, Cluster 13: ST176/ ST309, Cluster 14: ST156/ST205/STN27, Cluster 15: ST190/STN14, Cluster 16: ST315/ST319, Cluster 17: ST304/ST313/ST368, Cluster 18: ST294/ST306/ST307/ST311, Cluster 19: ST337/STN04, Cluster 20: ST300/STN15/STN18, Cluster 21: ST320/ST322, Cluster 22: ST70/ST354.
FIGURE S3 | Population snapshot of M. catarrhalis based on allelic profiles of MLST in adults and children of FAHXU. Sequence types (STs) that shared alleles at ≥5 of the eight MLST loci were obtained from the M. catarrhalis MLST website, and a diagram was constructed by using Bionumerics. Each circle corresponds to a MLST ST, and different circle colors represent different population (A: Adult/C: children). The lines between circles indicate the similarity between profiles: bold line, seven of eight MLST alleles/MLVA loci in common; normal line, six alleles/loci in common; dashed line, five alleles/loci in common; dotted line, ≤4 alleles/loci. The gray halo surrounding the STs denotes STs belonging to different MLST clusters.
Footnotes
References
Ahmad, S. (1998). Bronchopulmonary infection due to Moraxella (Branhamella) catarrhalis at a specialist hospital in Saudi Arabia. J. Commun. Dis. 30, 233–236.
Clinical and Laboratory Standards Institute [CLSI] (2015). Antimicrobial Susceptibility Testing of Fastidious or Infrequently Isolated Organisms; Approved Guideline. CLSI document M45-A3. Wayne, PA: CLSI.
Cox, R. A., Conquest, C., Mallaghan, C., and Marples, R. R. (1995). A major outbreak of methicillin-resistant Staphylococcus aureus caused by a new phage-type (EMRSA-16). J. Hosp. Infect. 29, 87–106. doi: 10.1016/0195-6701(95)90191-4
Enright, M. C., and Spratt, B. G. (1998). A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144, 3049–3060. doi: 10.1099/00221287-144-11-3049
Enright, M. C., Spratt, B. G., Kalia, A., Cross, J. H., and Bessen, D. E. (2001). Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69, 2416–2427. doi: 10.1128/IAI.69.4.2416-2427.2001
Flamm, R. K., Sader, H. S., Farrell, D. J., and Jones, R. N. (2012). Macrolide and tetracycline resistance among Moraxella catarrhalis isolates from 2009 to 2011. Diagn. Microbiol. Infect. Dis. 74, 198–200. doi: 10.1016/j.diagmicrobio.2012.06.007
Hol, C., Schalen, C., Verduin, C. M., Van Dijke, E. E., Verhoef, J., Fleer, A., et al. (1996). Moraxella catarrhalis in acute laryngitis: infection or colonization? J. Infect. Dis. 174, 636–638. doi: 10.1093/infdis/174.3.636
Liu, D. F., Xie, X., Mastri, M. G., Fortuna-Nevin, M., Colocillo, C., Fletcher, L., et al. (2006). Polymorphism of the major surface epitope of the CopB outer membrane protein of Moraxella catarrhalis. FEMS Immunol. Med. Microbiol. 47, 343–350. doi: 10.1111/j.1574-695X.2006.00093.x
Liu, Y., Xu, H. P., Xu, Z. P., Kudinha, T., Fan, X., Xiao, M., et al. (2015). High-level macrolide-resistant Moraxella catarrhalis and development of an allele-specific PCR assay for detection of 23S rRNA gene A2330T Mutation: a three-year study at a chinese tertiary hospital. Microb. Drug Resist. 21, 507–511. doi: 10.1089/mdr.2014.0217
Liu, Y., Zhao, C., Zhang, F., Chen, H., Chen, M., and Wang, H. (2012). High prevalence and molecular analysis of macrolide-nonsusceptible Moraxella catarrhalis isolated from nasopharynx of healthy children in China. Microb. Drug Resist. 18, 417–426. doi: 10.1089/mdr.2011.0175
Liu, Y. L., Li, D. F., Xu, H. P., Xiao, M., Cheng, J. W., Zhang, L., et al. (2016). Use of next generation sequence to investigate potential novel macrolide resistance mechanisms in a population of Moraxella catarrhalis isolates. Sci. Rep. 2016:35711. doi: 10.1038/srep35711
Maiden, M. C., Bygraves, J. A., Feil, E., Morelli, G., Russell, J. E., Urwin, R., et al. (1998). Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U.S.A. 95, 3140–3145. doi: 10.1073/pnas.95.6.3140
Meier, P. S., Freiburghaus, S., Martin, A., Heiniger, N., Troller, R., and Aebi, C. (2003). Mucosal immune response to specific outer membrane proteins of Moraxella catarrhalis in young children. Pediatr. Infect. Dis. J. 22, 256–262. doi: 10.1097/01.inf.0000054827.86683.bd
Murphy, T. F., Brauer, A. L., Aebi, C., and Sethi, S. (2005). Antigenic specificity of the mucosal antibody response to Moraxella catarrhalis in chronic obstructive pulmonary disease. Infect. Immun. 73, 8161–8166. doi: 10.1128/IAI.73.12.8161-8166.2005
Qin, L., Masaki, H., Gotoh, K., Furumoto, A., Terada, M., Watanabe, K., et al. (2009). Molecular epidemiological study of Moraxella catarrhalis isolated from nosocomial respiratory infection patients in a community hospital in Japan. Intern. Med. 48, 797–803. doi: 10.2169/internalmedicine.48.2036
Tang, P., Shi, W., Zeng, H. L., Ding, W., Wang, C., Yao, K. H., et al. (2016). Prevalence of Moraxella catarrhalis in the nasopharyngeal specimen from 1082 hospitalized children with respiratory infection and the drug resistance of the isolates. Zhongguo Dang Dai Er Ke Za Zhi 18, 707–712.
Verhaegh, S. J., Streefland, A., Dewnarain, J. K., Farrell, D. J., van Belkum, A., and Hays, J. P. (2008). Age-related genotypic and phenotypic differences in Moraxella catarrhalis isolates from children and adultspresenting with respiratory disease in 2001-2002. Microbiology 154, 1178–1184. doi: 10.1099/mic.0.2007/015057-0
Wang, H., Chen, M. J., Xu, Y. C., Sun, H. L., Yang, Q., Hu, Y. J., et al. (2011). Antimicrobial susceptibility of bacterial pathogens associated with community-acquired respiratory tract infections in Asia: report from the community-acquired respiratory tract infection pathogen surveillance (CARTIPS) study, 2009-2010. Int. J. Antimicrob. Agents 38, 376–383. doi: 10.1016/j.ijantimicag.2011.06.015
Keywords: Moraxella catarrhalis, copB, mlst, clonal complexes, macrolide-resistance
Citation: Liu Y-L, Xiao M, Cheng J-W, Xu H-P, Xu Z-P, Ye S, Zhang W-J, Kudinha T, Kong F and Xu Y-C (2017) Moraxella catarrhalis Macrolide-Resistant Isolates Are Highly Concentrated in Two MLST Clonal Complexes -CCN10 and CC363. Front. Microbiol. 8:201. doi: 10.3389/fmicb.2017.00201
Received: 10 November 2016; Accepted: 27 January 2017;
Published: 10 February 2017.
Edited by:
Yuji Morita, Aichi Gakuin University, JapanReviewed by:
Liang Li, Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center, USAGiovanni Gherardi, Università Campus Bio-Medico, Italy
Haijian Zhou, Chinese Center for Disease Control and Prevention, China
Copyright © 2017 Liu, Xiao, Cheng, Xu, Xu, Ye, Zhang, Kudinha, Kong and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying-Chun Xu, eHljcHVtY2hAMTM5LmNvbQ==
†These authors have contributed equally to this work.
 Ya-Li Liu1†
Ya-Li Liu1† Meng Xiao
Meng Xiao Jing-Wei Cheng
Jing-Wei Cheng He-Ping Xu
He-Ping Xu Timothy Kudinha
Timothy Kudinha Fanrong Kong
Fanrong Kong