- 1Beijing Key Laboratory of Detection Technology for Animal-Derived Food Safety, College of Veterinary Medicine, China Agricultural University, Beijing, China
- 2Guangdong Provincial Key Laboratory of Protein Function and Regulation in Agricultural Organisms, College of Life Sciences, South China Agricultural University, Guangzhou, China
- 3Institute of Microbiology and Epizootics, Centre for Infection Medicine, Department of Veterinary Medicine, Freie Universität Berlin, Berlin, Germany
Increasing emergence of staphylococci resistant to pleuromutilins, lincosamides, and streptogramin A (PLSA) and isolated from humans and pets is a growing public health concern worldwide. Currently, there was only one published study regarding one of the PLSA genes, vga(A) detected in staphylococci isolated from cat. In this study, eleven pleuromutilin-resistant staphylococci from pets and two from their owners were isolated and further characterized for their antimicrobial susceptibilities, plasmid profiles, genotypes, and genetic context of the PLSA resistance genes. The gene sal(A) identified in 11 staphylococcal isolates was found for the first time in Staphylococcus haemolyticus, Staphylococcus epidermidis, and Staphylococcus xylosus. Moreover, these 11 isolates shared the identical regions flanking the sal(A) gene located in the chromosomal DNA. Two S. haemolyticus isolates from a cat and its owner carried similar vga(A)LC plasmids and displayed indistinguishable PFGE patterns. A novel chromosomal multidrug resistance genomic island (MDRGI) containing 13 resistance genes, including lsa(E), was firstly identified in S. epidermidis. In addition, vga(A)LC, sal(A), and lsa(E) were for the first time identified in staphylococcal isolates originating from pet animals. The plasmids, chromosomal DNA region, and MDRGI associated with the PLSA resistance genes vga(A), vga(A)LC, sal(A), and lsa(E) are present in staphylococci isolated from pets and humans and present significant challenges for the clinical management of infections by limiting therapeutic options.
Introduction
Transferable resistance to three chemically distinct classes of antimicrobial agents (pleuromutilins, lincosamides, and streptogramin A; PLSA) in staphylococci has been attributed to ABC transporters of the Vga, Lsa, or Sal families. All of the corresponding resistance genes, including vga(A), vga(A)V, vga(A)LC, vga(B), vga(C), vga(E), vga(E)V, lsa(E), and sal(A), were mainly identified in staphylococci from food-producing isolates (Allignet and El Solh, 1997; Haroche et al., 2000; Kadlec and Schwarz, 2009; Jung et al., 2010; Lozano et al., 2012; Hauschild et al., 2012; Wendlandt et al., 2013; Li et al., 2014; Hot et al., 2014). However, the vga(A) gene was also identified in Staphylococcus epidermidis isolates originating from one cat, clinic environment and an employee (Weiß et al., 2013).
To date, the antimicrobial resistance mechanisms of staphylococci in pets have received less attention than those in food-producing animals. Given the close relationship between pets and their owners, resistant bacteria and their mobile resistance determinants may be exchanged in either direction between pets and humans (Schwarz et al., 2016). The increasingly frequent isolation of methicillin- and multidrug-resistant staphylococci from pets, particularly from dogs with pyoderma and otitis (Cain, 2013), and the occurrence of the same resistance genes in pets and in humans (Butaye et al., 2001; Simjee et al., 2002), may underline the potential role of pets in the transmission of antimicrobial-resistant staphylococci to humans. Currently, there is one published study regarding one of the PLSA genes detected in staphylococci isolated from a cat (Weiß et al., 2013). In the present study, we investigated the distribution of PLSA resistance genes in staphylococci of pet origin and analyzed the locations and genetic environments of these genes.
Materials and Methods
Ethics Statement
This research was carried out according to the principles of the Declaration of Helsinki. The involved pet animals were treated with the best practice veterinary care and the informed consent was obtained from pets’ owners. The study protocol was approved by the Ethics Committee of China Agricultural University.
Bacterial Strains and Antimicrobial Susceptibility Testing
A total of 300 anal swabs or nasal samples were used in this study which were collected from dogs (n = 269), cats (n = 10), and some of their owners (n = 21) in the Veterinary Teaching Hospital of China Agricultural University, Beijing in 2013. The pleuromutilin-resistant isolates were selected by incubation on mannitol salt agar (Luqiao, Beijing, China) supplemented with 2 μg/ml of valnemulin at 37°C for 16–24 h.
The MIC (Minimal Inhibitory Concentration) determinations were done by a standard broth microdilution test following the recommendations of the Clinical and Laboratory Standards Institute [CLSI] (2015). Staphylococcus aureus ATCC 29213 was served as the quality control strain.
Molecular Methods
The genomic DNAs of the valnemulin-resistant isolates were obtained using a Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, USA), and were screened for the vga, lsa, and sal gene types by PCR assays (Novotna and Janata, 2006; Hot et al., 2014; Li et al., 2014; Wendlandt et al., 2015). Furthermore, the species assignment of the resistant isolates was done by 16S rDNA sequencing and confirmed by MALDI-TOF MS (Bruker Daltonik, Bremen, Germany).
The clonality of the valnemulin-resistant isolates was analyzed by pulsed-field gel electrophoresis (PFGE) as described previously (Wang et al., 2011), and the PFGE results were analyzed using InfoQuestFP software (version 4.5).
The S1 nuclease-PFGE (S1-PFGE) and subsequent Southern blot hybridization were performed to locate the valnemulin resistance genes as described before (Barton et al., 1995). The Low Range PFGE Marker (New England BioLabs, Beverly, MA, USA) served as the size marker.
Characterization and Sequence Analysis of the PLSA Genes
Genomic DNAs of S. sciuri isolate 100N carrying sal(A) and S. epidermidis isolate 138N carrying sal(A) and lsa(E) were submitted to high-throughput whole-genome sequencing (WGS), and preceded by library construction on a HiSeq 2500, which produced 150 bp paired-end reads (Berry Genomics Company, Beijing, China).
Draft assembly of the genomic DNA sequences was analyzed by CLC Genomics Workbench 5 (CLC Bio, Aarhus, Denmark). All contigs with the average coverage of >100-fold were searched for the PLSA genes using BLAST analysis. The regions flanking the PLSA genes were identified using de novo assembly as earlier described (Zerbino and Birney, 2008) and the random primer walking strategy (Zhang et al., 2009). Sequence analysis was conducted using the ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and BLAST functions (http://blast.ncbi.nlm.nih.gov/Blast.cgi). To obtain more information about the genetic environments of the PLSA genes in valnemulin-resistant isolates, the random primer walking strategy and inverse PCR were performed.
Nucleotide Sequence Accession Numbers
The PLSA-carrying segments of various isolates in this study have been deposited in GenBank, and their accession numbers are KX712120 (Staphylococcus haemolyticus plasmid p131A carrying vga(A)LC), KX712121 (S. epidermidis plasmid p132R carrying vga(A)), KX712119 (S. epidermidis 138N carrying sal(A)), and KX712118 (S. epidermidis 138N carrying lsa(E)).
Results
Characterization of Valnemulin-Resistant Staphylococcal Isolates and Antimicrobial Resistance Profiles
Amongst the 300 anal swabs and nasal samples, 13 (4.3%) valnemulin-resistant isolates with valnemulin MICs of ≥8 μg/ml were detected, which were identified to species level as Staphylococcus sciuri (n = 8), Staphylococcus haemolyticus (n = 2), S. epidermidis (n = 2), and Staphylococcus xylosus (n = 1) (Table 1).
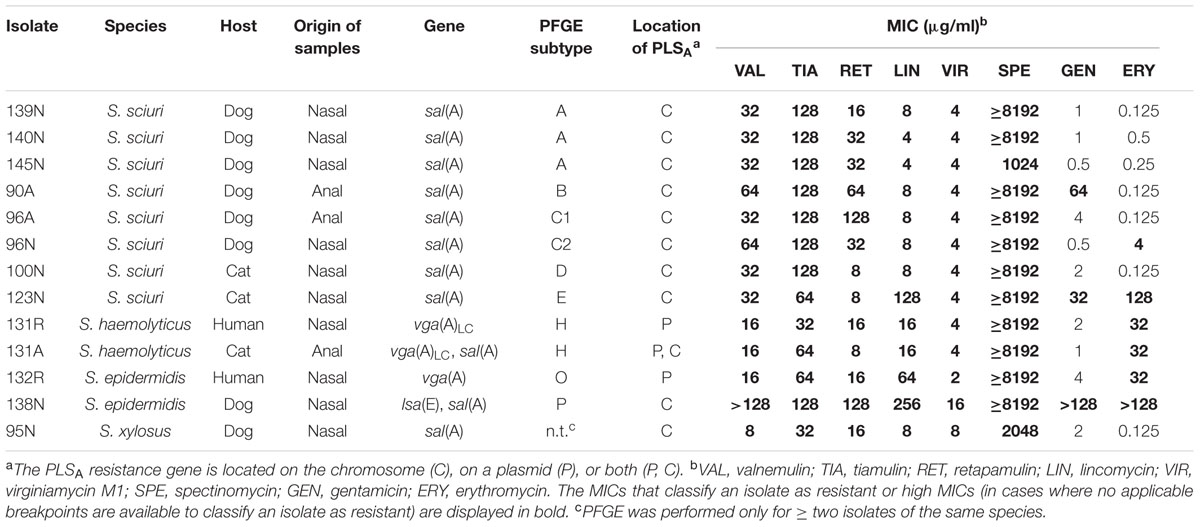
TABLE 1. Characteristics of the 13 valnemulin-resistant staphylococci isolates identified in this study.
Nine (eight S. sciuri and one S. xylosus) of the 13 isolates carried only sal(A), one S. haemolyticus and one S. epidermidis isolate carried vga(A)LC and vga(A), respectively, one S. haemolyticus isolate carried both vga(A)LC and sal(A), and one S. epidermidis isolate carried both lsa(E) and sal(A) (Table 1). Notably, the lsa(E) gene was only detected in one S. epidermidis isolate of dog origin, and the vga genes were detected in staphylococcal isolates originating from both humans and cats. In contrast, sal(A) was widespread among the valnemulin-resistant staphylococcal isolates from pets. As the sal(A) gene has been previously found exclusively in S. sciuri isolates (Hot et al., 2014; Wendlandt et al., 2015), this finding describes for the first time the occurrence of sal(A) in S. haemolyticus, S. epidermidis, and S. xylosus.
All 13 valnemulin-resistant isolates displayed resistance or high MICs to pleuromutilins (valnemulin, tiamulin, and retapamulin), lincomycin, virginiamycin M1 and spectinomycin (Table 1).
Genotyping by PFGE
Using a cut-off of 90% similarity, the eight S. sciuri isolates, which carried highly conserved (>99% nucleotide identity) sal(A) genes, clustered into five PFGE types (A–E), with type A accounting for the majority (3/8) of the isolates (Table 1; Figure 1). Remarkably, the feline S. haemolyticus isolate 131A and the human S. haemolyticus isolate 131R from the corresponding pet owner, which carried identical copies of vga(A)LC, showed the same PFGE type H, indicating that they were closely related. This finding supports the idea that these isolates might have been exchanged between the cat and its owner.
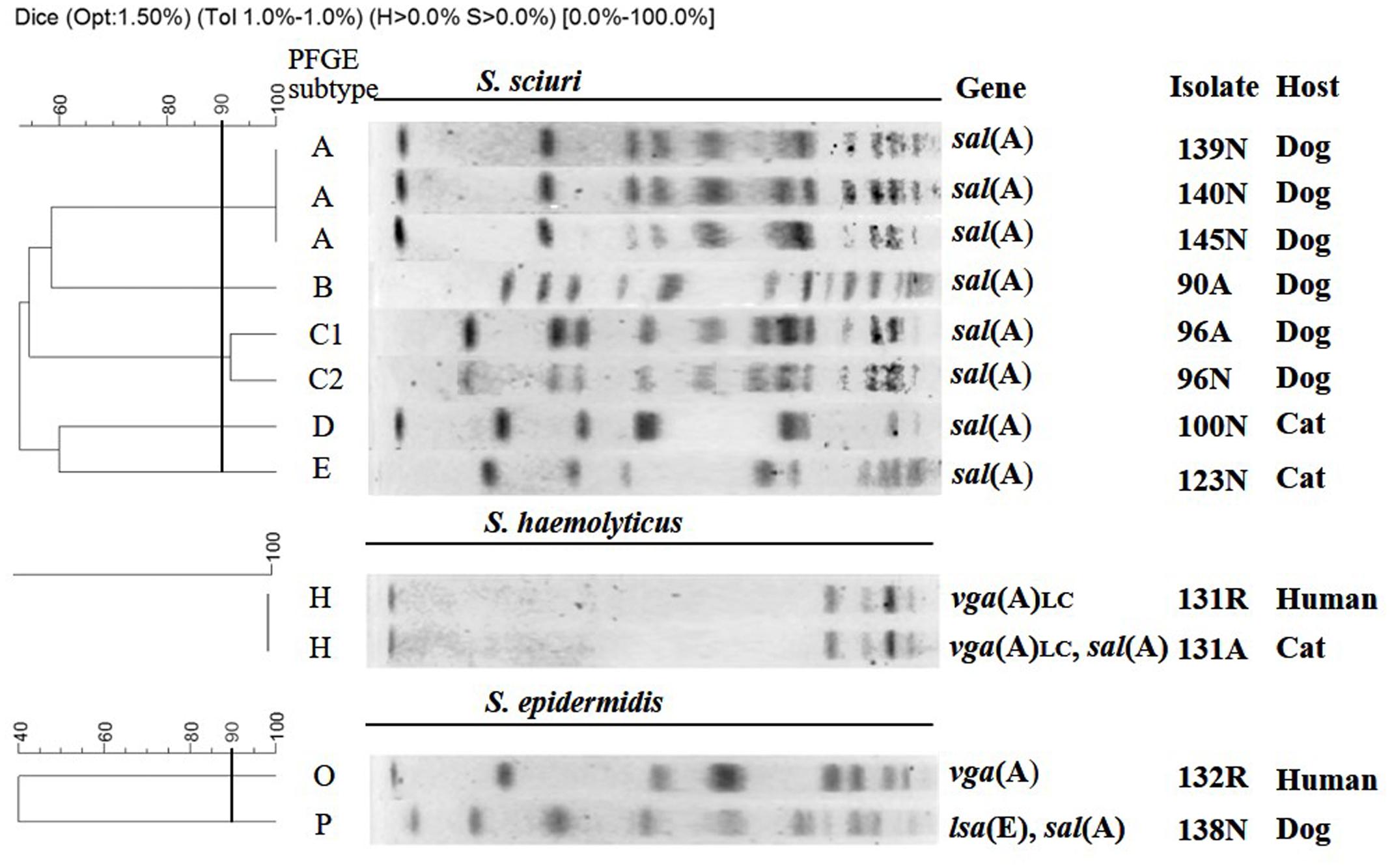
FIGURE 1. SmaI-PFGE typing of 12 valnemulin-resistant staphylococcal isolates for which more than a single isolate per species was available.
Genetic Environment of PLSA Genes
S1-PFGE and Southern blot hybridization indicated that all vga-type genes in the three isolates were located on plasmids, while all lsa(E) and sal(A) genes in the respective staphylococci were located in chromosomal DNA (Table 1; Figure 2).
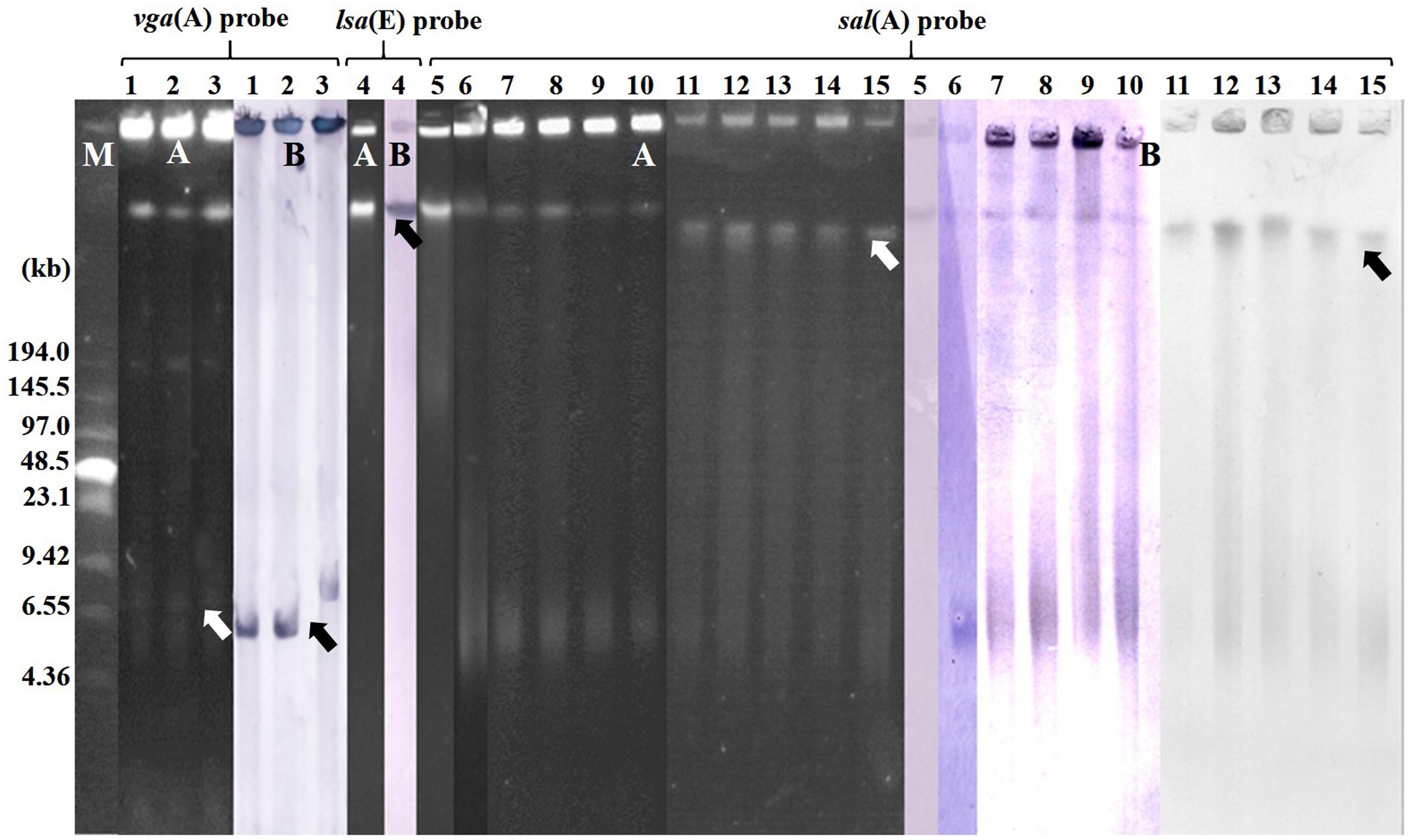
FIGURE 2. Localization of pleuromutilins, lincosamides, and streptogramin A (PLSA) in the valnemulin-resistant staphylococcal isolates by S1 nuclease-PFGE (A) and Southern blot hybridization (B). M indicates the Low Range PFGE Marker. The black arrows indicate the location of PLSA gene in Southern blot hybridization and white arrows indicate the position of PLSA gene in genome according to the S1-PFGE and marker. 131A and 131R (lanes 1–2, respectively) were vga(A)LC-positive S. haemolyticus isolates. 132R (lane 3) was a vga(A)-positive S. epidermidis isolate. 138N (lane 4) was a lsa(E)-positive S. epidermidis isolate. 90A, 95N, 96A, 96N, 100N, 123N, 131A, 138N, 139N, 140N, and 145N (lanes 5–15, respectively) were sal(A)-positive staphylococcal isolates.
To determine the regions flanking the vga-type genes in isolates 131R, 131A, and 132R, inverse PCRs were performed using the primers vga-F (5′-CAAGCTGAAAAGCCAACAAGG-3′) and vga-R (5′-CCTCGTCAATTTCCCATATAGT-3′), which are located inside the conserved regions of vga(A) and vga(A)LC genes. Three amplicons of 6056-bp (plasmids p131A and p131R) and 7209-bp (p132R) were obtained (Figure 3A). Plasmids p131A and p131R from the feline and human S. haemolyticus isolates differed in only nine base pairs, and showed 99% nucleotide sequence identity to plasmid pUR2355 from human S. aureus (accession no. JQ312422). Plasmid p132R shared 99% nucleotide identity with the S. epidermidis plasmid pUR3036 of cat origin (accession no. JQ312423). The most notable difference between p132R and p131A/p131R was the presence of 13 amino acid substitutions in the Vga protein. Surprisingly, the lincomycin MICs of isolates 131A, 131R, and 132R were 16, 16, and 64 μg/ml, respectively, which is in contrast to the finding that vga(A)LC confers higher MICs to lincosamides than does vga(A) (Novotna and Janata, 2006).
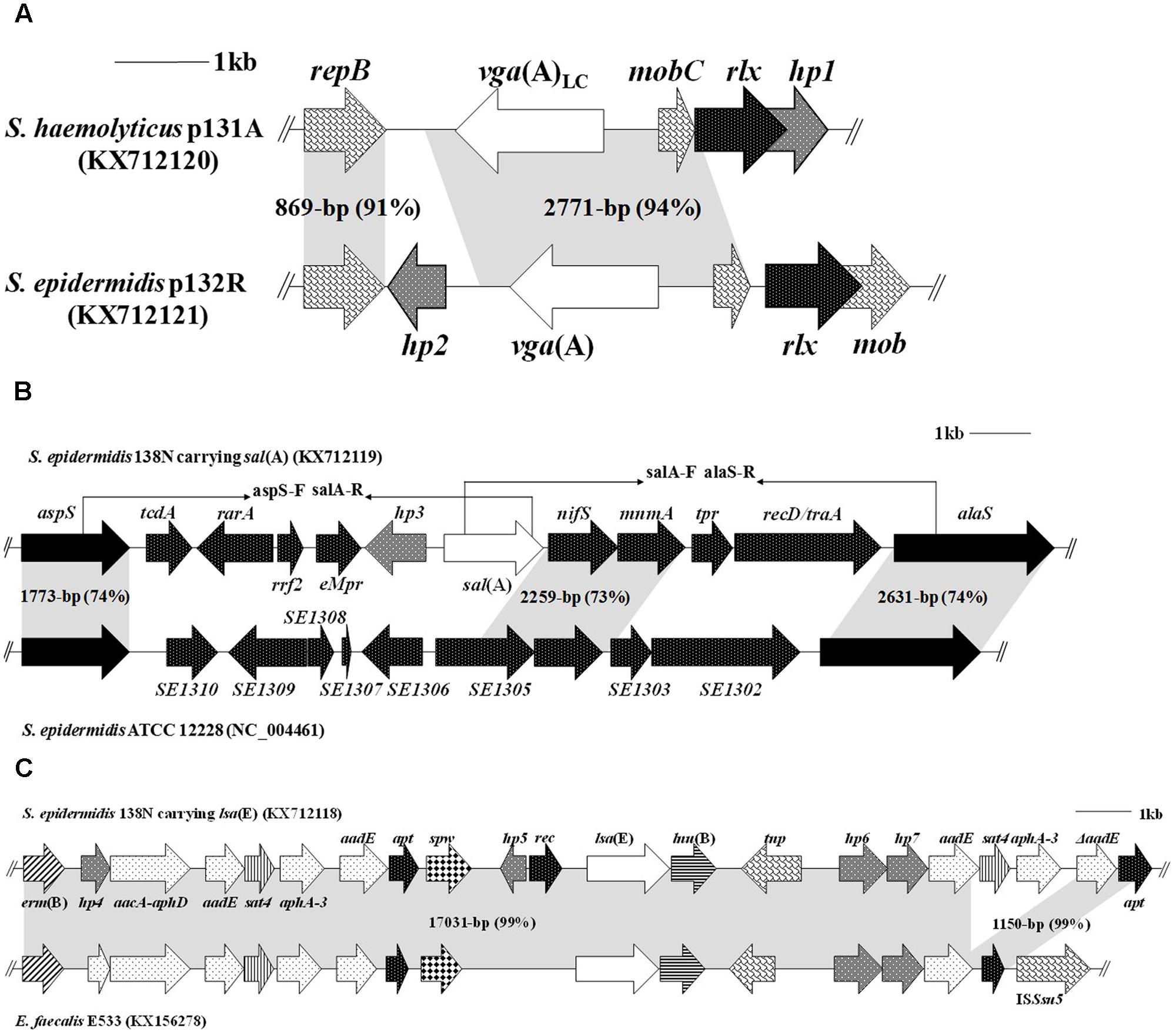
FIGURE 3. Comparison of genetic variations of the PLSA genes (vga(A)LC, vga(A), sal(A), and lsa(E)) of staphylococcal isolates. Four accession numbers (KX712120, KX712121, KX71211, and KX712118) were newly submitted in this study. The arrows indicate the positions and orientations of the genes. The similarities between the different structures are marked by gray shading. (A) Genetic environments of vga(A)LC in plasmid p131A from a S. haemolyticus isolate of cat origin and of vga(A) in p132R from a S. epidermidis isolate of human origin. (B) Structure of the novel genetic environment of sal(A) in 11 staphylococcal isolates (138N was the representative). The locations of primers used to detect and verify the genetic environment of sal(A) in other isolates are indicated by black arrowheads. (C) Genetic environment of lsa(E) in S. epidermidis 138N and structural comparison with the corresponding regions in E. faecalis E533.
Genomic DNA of S. sciuri isolate 100N carrying sal(A) and S. epidermidis isolate 138N carrying sal(A) and lsa(E) were sequenced by WGS. Comparative analysis of the draft genomes of isolates 100N and 138N with the corresponding sequenced genomes of staphylococci from NCBI revealed the presence of a very similar 12.5-kb DNA segment (97% nucleotide identity) containing the sal(A) gene inserted between the housekeeping genes aspS and alaS of isolates 100N and 138N (Figure 3B). This DNA segment contains 10 ORFs, of which only sal(A) is an antimicrobial resistance gene. Additionally, two pairs of primers, aspS-F (5′-CGTTGTTGAAGATGGTGCGTCT-3′), salA-R (5′-GGACCGAACCTTGAAATGATTG-3′), and salA-F (5′-GATGGATACCTTATAGAAGGTG-3′), alaS-R (5′-GTCTGTATCATAGTTCGTTGG-3′), which are located in the conserved regions of the aspS, sal(A), and alaS genes (Figure 3B), respectively, were designed, and long-range PCRs were preformed to investigate the genetic environments of sal(A) in the other nine sal(A)-positive staphylococcal isolates (Table 1). Two amplicons of 7.4 and 7.7 kb were obtained, and identical sequences were found in all nine isolates.
A 20,154-bp lsa(E)-carrying fragment containing 21 ORFs was obtained in the canine S. epidermidis isolate 138N (Figure 3C) after the conjoint analysis of the results of WGS and modified random primer walking strategy. Similar to the corresponding regions in porcine E. faecalis E533 (accession no. KX156278), this lsa(E)-carrying fragment also contained the macrolide-lincosamide-streptogramin B resistance gene erm(B), the aminoglycoside resistance genes aacA-aphD, aadE, and aphA-3, the spectinomycin resistance gene spw, the streptothricin resistance gene sat4, and the lincosamide resistance gene lnu(B). Further downstream of the lnu(B) gene, a second copy of the genes aadE-sat4-aphA-3 was detected (Figure 3C). This multidrug resistance gene region confers resistance to seven classes of antimicrobial agents and explained why isolate 138N exhibited resistance not only to PLSA antibiotics, but also to spectinomycin, gentamicin and erythromycin (Table 1). In addition, the hp4 gene encoded a predicted protein related to an acetyltransferase enzyme GNAT (GCN5-related N-acetyltransferase, with no confirmed function) from Enterococcus.
Discussion
Increasing emergence of staphylococci that are resistant to PLSA and are isolated from humans and pets is a growing public health concern worldwide. Each of these three classes of antimicrobial agents contains important drugs for human and veterinary medicine. The pleuromutilin antibiotics valnemulin and tiamulin are frequently used in veterinary medicine. Retapamulin was the first pleuromutilin approved for topical use in skin infections of humans caused by staphylococci (Jacobs, 2007). Lincosamides, such as clindamycin and lincomycin, have generally been indicated for the treatment of osteomyelitis but also skin and soft tissue infections caused by staphylococci in humans and pets (Xue et al., 1996; Zetner et al., 2003). Virginiamycin is a two-component streptogramin with a broad spectrum of activity against Gram-positive bacteria. Some antimicrobial agents approved for use in human medicine are also applied to nonfood-producing animals under specific regulations, such as the Animal Medicinal Drug Use Clarification Act (AMDUCA) in the USA. In this study, all 13 valnemulin-resistant isolates displayed resistance or high MICs to pleuromutilins (valnemulin, tiamulin, and retapamulin), lincomycin, virginiamycin M1, indicating that all PLSA genes including vga(A), vga(A)LC, sal(A), and lsa(E) of these isolates were functional. Interestingly, genes sal(A) and vga(A)LC coexisted in one feline S. haemolyticus isolate 131A, and genes sal(A) and lsa(E) were co-located in the chromosomal DNA of the canine S. epidermidis isolate 138N. Isolate 138N exhibited significant differences in the MICs of valnemulin, lincomycin, and virginiamycin M1 compared with other isolates that carried only one type of PLSA genes (Table 1). There have been no studies regarding to sal, vga, and lsa coexisting in the same isolate, while both vga(A) and lsa(E) were identified in four bovine staphylococcal isolates (Wendlandt et al., 2015). Notably, vga(A)LC, sal(A), and lsa(E) were for the first time identified in staphylococcal isolates originating from pet animals.
The presence of PLSA resistance genes in pet- and human-associated staphylococci indicates the adaptation of these bacteria to antibiotic pressure, given that PLSA resistance genes confer resistance to antimicrobial agents used in both pets and humans. Additionally, S. epidermidis 138N carried at least 13 resistance genes conferring resistance to seven classes of antimicrobial compounds (pleuromutilins, lincosamides, streptogramins, macrolides, aminoglycosides, aminocyclitols, and streptothricins), indicating that the use of any of the abovementioned antimicrobial agents may lead to co-selection of the MDR genomic island (MDRGI) in isolate 138N. The plasmids, chromosomal DNA region, and MDRGI associated with the multiresistance genes vga(A), vga(A)LC, sal(A), and lsa(E) exist in staphylococci isolated from pets and humans, which enhances the dissemination of PLSA-resistant staphylococci among pets and humans and presents significant challenges for the clinical management of infections by limited therapeutic options.
The three canine sal(A)-positive S. sciuri isolates 139N, 140N, and 145N not only shared the identical regions flanking the sal(A) gene, but also had the uniform PFGE pattern (subtype A), suggesting that these isolates have originated from a single clone. Remarkably, plasmids p131A and p131R, the two almost identical vga(A)LC-carrying plasmids present in isolates with the same PFGE subtype (subtype H), were derived from a cat and its owner. This fact suggests that these two isolates might have been exchanged between the cat and its owner due to their extensive contact. In this regard, it is important to consider the current role of dogs and cats as actual family members in many households and, consequently, the close contact to their owners and other family members (Schwarz et al., 2016).
Conclusion
This is the first description of the PLSA genes in staphylococci of pet origin and also the description of a novel lsa(E)-carrying MDRGI in S. epidermidis. Moreover, we identified the sal(A) gene for the first time in S. haemolyticus, S. epidermidis, and S. xylosus. Pets are likely reservoirs of antimicrobial-resistant bacteria, warranting the prudent use of all antimicrobials in pet animal medicine.
Author Contributions
YW and SS designed research; FD, HW, and YL performed research; FD, JL, AF, and GM analyzed data; FD, SS, and YW wrote the paper. All authors listed have approved research for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was funded by grants from National Natural Science Foundation of China (31422055 and 31370046), and German Federal Ministry of Education and Research (MedVet-Staph 2, grant no. 01KI1301D) provided through the German Aerospace Center.
References
Allignet, J., and El Solh, N. (1997). Characterization of a new staphylococcal gene, vga(B), encoding a putative ABC transporter conferring resistance to streptogramin A and related compounds. Gene 202, 133–138. doi: 10.1016/S0378-1119(97)00464-2
Barton, B. M., Harding, G. P., and Zuccarelli, A. J. (1995). A general method for detecting and sizing large plasmids. Anal. Biochem. 226, 235–240. doi: 10.1006/abio.1995.1220
Butaye, P., Devriese, L. A., and Haesebrouck, F. (2001). Differences in antibiotic resistance patterns of Enterococcus faecalis and Enterococcus faecium strains isolated from farm and pet animal. Antimicrob. Agents Chemother. 45, 1374–1378. doi: 10.1128/AAC.45.5.1374-1378.2001
Cain, C. L. (2013). Antimicrobial resistance in staphylococci in small animals. Vet. Clin. North Am. Small Anim. Pract. 43, 19–40. doi: 10.1016/j.cvsm.2012.09.003
Clinical and Laboratory Standards Institute [CLSI] (2015). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement (M100-S25). Wayne, PA: Clinical and Laboratory Standards Institute.
Haroche, J., Allignet, J., Buchrieser, C., and El Solh, N. (2000). Characterization of a variant of vga(A) conferring resistance to streptogramin A and related compounds. Antimicrob. Agents Chemother. 44, 2271–2275. doi: 10.1128/AAC.44.9.2271-2275.2000
Hauschild, T., Feßler, A. T., Kadlec, K., Billerbeck, C., and Schwarz, S. (2012). Detection of the novel vga(E) gene in methicillin-resistant Staphylococcus aureus CC398 isolates from cattle and poultry. J. Antimicrob. Chemother. 67, 503–504. doi: 10.1093/jac/dkr446
Hot, C., Berthet, N., and Chesneau, O. (2014). Characterization of sal(A), a novel gene responsible for lincosamide and streptogramin A resistance in Staphylococcus sciuri. Antimicrob. Agents Chemother. 58, 3335–3341. doi: 10.1128/AAC.02797-13
Jacobs, M. R. (2007). Retapamulin: a semisynthetic pleuromutilin compound for topical treatment of skin infections in adults and children. Future Microbiol. 2, 591–600. doi: 10.2217/17460913.2.6.591
Jung, Y. H., Shin, E. S., Kim, O., Yoo, J. S., Lee, K. M., Yoo, J. L., et al. (2010). Characterization of two newly identified genes, vga(D) and vat(G), conferring resistance to streptogramin A in Enterococcus faecium. Antimicrob. Agents Chemother. 54, 4744–4749. doi: 10.1128/AAC.00798-09
Kadlec, K., and Schwarz, S. (2009). Novel ABC transporter gene, vga(C), located on a multiresistance plasmid from a porcine methicillin-resistant Staphylococcus aureus ST398 strain. Antimicrob. Agents Chemother. 53, 3589–3591. doi: 10.1128/AAC.00570-09
Li, J., Li, B., Wendlandt, S., Schwarz, S., Wang, Y., Wu, C., et al. (2014). Identification of a novel vga(E) gene variant that confers resistance to pleuromutilins, lincosamides and streptogramin A antibiotics in staphylococci of porcine origin. J. Antimicrob. Chemother. 69, 919–923. doi: 10.1093/jac/dkt482
Lozano, C., Aspiroz, C., Rezusta, A., Gómez-Sanz, E., Simon, C., Gómez, P., et al. (2012). Identification of novel vga(A)-carrying plasmids and a Tn5406-like transposon in methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis of human and animal origin. Int. J. Antimicrob. Agents 40, 306–312. doi: 10.1016/j.ijantimicag.2012.06.009
Novotna, G., and Janata, J. (2006). A new evolutionary variant of the streptogramin A resistance protein, Vga(A)LC, from Staphylococcus haemolyticus with shifted substrate specificity towards lincosamides. Antimicrob. Agents Chemother. 50, 4070–4076. doi: 10.1128/AAC.00799-06
Schwarz, S., Loeffler, A., and Kadlec, K. (2016). Bacterial resistance to antimicrobial agents and its impact on veterinary and human medicine. Vet. Dermatol. 28:82-e19. doi: 10.1111/vde.12362
Simjee, S., White, D. G., McDermott, P. F., Wagner, D. D., Zervos, M. J., Donabedian, S. M., et al. (2002). Characterization of Tn1546 in vancomycin-resistant Enterococcus faecium isolated from canine urinary tract infections: evidence of gene exchange between human and animal enterococci. J. Clin. Microbiol. 40, 4659–4665. doi: 10.1128/JCM.40.12.4659-4665.2002
Weiß, S., Kadlec, K., Feßler, A. T., and Schwarz, S. (2013). Identification and characterization of methicillin-resistant Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus haemolyticus and Staphylococcus pettenkoferi from a small animal clinic. Vet. Microbiol. 167, 680–685. doi: 10.1016/j.vetmic.2013.07.036
Wendlandt, S., Kadlec, K., Feßler, A. T., and Schwarz, S. (2015). Identification of ABC transporter genes conferring combined pleuromutilin-lincosamide-streptogramin A resistance in bovine methicillin-resistant Staphylococcus aureus and coagulase-negative staphylococci. Vet. Microbiol. 177, 353–358. doi: 10.1016/j.vetmic.2015.03.027
Wendlandt, S., Lozano, C., Kadlec, K., Gómez-Sanz, E., Zarazaga, M., Torres, C., et al. (2013). The enterococcal ABC transporter gene lsa(E) confers combined resistance to lincosamides, pleuromutilins and streptogramin A antibiotics in methicillin-susceptible and methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 68, 473–475. doi: 10.1093/jac/dks398
Wang, Y., Zhang, W., Wang, J., Wu, C., Shen, Z., Fu, X., et al. (2011). Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob. Agents Chemother. 56, 1485–1490. doi: 10.1128/AAC.05827-11
Xue, I. B., Davey, P. G., and Phillips, G. (1996). Variation in postantibiotic effect of clindamycin against clinical isolates of Staphylococcus aureus and implications for dosing of patients with osteomyelitis. Antimicrob. Agents Chemother. 40, 1403–1407
Zerbino, D. R., and Birney, E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829
Zetner, K., Schmidt, H., and Pfeiffer, S. (2003). Concentrations of clindamycin in the mandibular bone of companion animals. Vet. Ther. 4, 166–171
Zhang, K., McClure, J. A., Elsayed, S., and Conly, J. M. (2009). Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53, 531–540. doi: 10.1128/AAC.01118-08
Keywords: PLSA genes, staphylococci, pet, human, multidrug resistance
Citation: Deng F, Wang H, Liao Y, Li J, Feßler AT, Michael GB, Schwarz S and Wang Y (2017) Detection and Genetic Environment of Pleuromutilin-Lincosamide-Streptogramin A Resistance Genes in Staphylococci Isolated from Pets. Front. Microbiol. 8:234. doi: 10.3389/fmicb.2017.00234
Received: 22 December 2016; Accepted: 02 February 2017;
Published: 14 February 2017.
Edited by:
Axel Cloeckaert, French National Institute for Agricultural Research, FranceCopyright © 2017 Deng, Wang, Liao, Li, Feßler, Michael, Schwarz and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Wang, d2FuZ3lhbmdAY2F1LmVkdS5jbg== Stefan Schwarz, c3RlZmFuLnNjaHdhcnpAZnUtYmVybGluLmRl
†These authors have contributed equally to this work.
 Fengru Deng1,2†
Fengru Deng1,2† Huiwen Wang
Huiwen Wang Yifei Liao
Yifei Liao Geovana B. Michael
Geovana B. Michael Stefan Schwarz
Stefan Schwarz Yang Wang
Yang Wang